Abstract
Abnormal gene regulation as a consequence of flawed epigenetic mechanisms may be central to the initiation and persistence of many human diseases. However, the association of epigenetic dysfunction with disease and the development of therapeutic agents for treatment are slow. Developing new methodologies used to visualize chromatin modifying enzymes and their function in the human brain would be valuable for diagnosis of brain disorders and drug discovery. We provide an overview of current invasive and noninvasive techniques for measuring expression and functions of chromatin modifying enzymes in the brain, emphasizing tools applicable to histone deacetylase (HDAC) enzymes as a leading example. The majority of current techniques are invasive and difficult to translate to what is happening within a human brain in vivo. However, recent progress in molecular imaging provides new, noninvasive ways to visualize epigenetics in human brain. Neuroimaging tool development presents a unique set of challenges in order to identify and validate CNS radiotracers for HDACs and other histone modifying enzymes. We summarize advances in the effort to image HDACs and HDAC inhibitory effects in the brain using PET and highlight generalizable techniques that can be adapted to investigate other specific components of epigenetic machinery. Translational tools like neuroimaging by PET and MRI provide the best way to link our current understanding of epigenetic changes with in vivo function in normal and diseased brain. These tools will be a critical addition to ex vivo methods to evaluate - and intervene - in CNS dysfunction.
Keywords: epigenetics, chromatin, brain, imaging, probes, translation
Introduction
An overarching goal in basic neuroscience research is to develop knowledge and techniques that can be extended from model systems to the living human brain. Translational tools that can visualize brain function and biochemical events are particularly useful because they can provide insight into the brain as it responds to external cues (e.g. environment, drug, and behavior). Visualizing brain function is a major challenge given that the human brain is inaccessible and difficult to assay directly. Currently, noninvasive imaging tools provide the best way to visualize changes in vivo. Importantly, the same in vivo techniques can be used to visualize changes in animal models and in humans, a major translational advantage. This holds promise for great returns in integrating existing knowledge with observations from an intact, living brain.
In the past decade, epigenetics research has provided new insight into almost all aspects of biology – cellular differentiation, growth, development, and aging (Fass et al., 2012). Changes in DNA methylation and post-translational modification of histone proteins modulate gene expression. These gene expression changes alter diverse signaling pathways in the brain, and impact brain activity from neurotransmission to functional output at the level of behavioral response. Investigation of epigenetic changes in the brain has provided fresh perspective into the mediators of diverse CNS disorders as well as potential targets in developing improved treatments (Hasan et al., 2013). In this review, we highlight ways to visualize epigenetic changes in the brain and emphasize development of in vivo neuroimaging tools using the histone deacetylase (HDAC) enzymes as an example. While this review focuses on HDACs due to their progressed stage in in vivo tool development, the themes presented herein are no less applicable to other targets and processes. Even with the progress made in HDAC imaging, there is much ground left to cover before we can truly link epigenetics and function in the human brain.
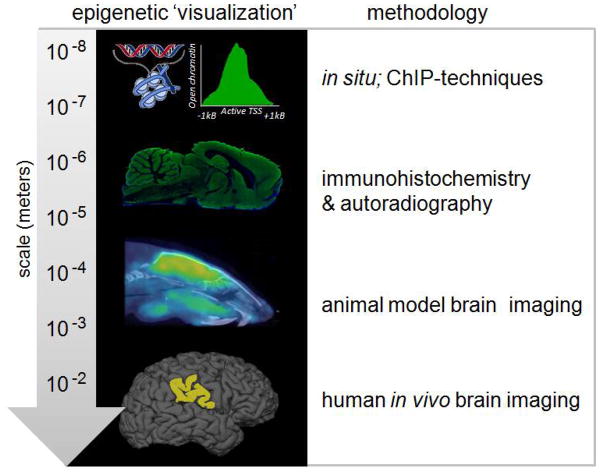
There are two main ways to think about imaging an epigenetic target in the brain: direct observation and functional/indirect observation and we have divided our review into these themes (Figure 1). To this end, the imaging target in the brain could be an epigenetic ‘machine’ - one of the ‘readers,’ ‘writers’ or ‘erasers’ of epigenetic change (Fass et al., 2012). Alternatively, the target could also be an epigenetic ‘mark’ - a modification to a protein or nucleic acid resulting from epigenetic enzyme action. Direct observation has the advantage of providing detailed information on a protein target independent of its activity. This is useful as an enzyme may have a structural as well as functional role regulating brain function and this protein presence can be measured by visualizing a specific, tight binding ligand. One obvious drawback to direct observation is that the assumption is made that enzyme density is related to activity and that the inferred activated changes neural processing (in a phenotypic way). However, using techniques in functional observation, the impact of a protein or enzyme on brain function can be visualized. Catalytic action on a labeled enzyme substrate or differential binding of established neuroimaging probes can provide a surrogate measure of changes in brain activity with robust spatial and temporal resolution. These methods reveal where an enzyme is working as well as the regions of the brain that integrate downstream signaling changes.
Figure 1.
Epigenetic imaging techniques can be used to visualize the presence of chromatin modifying enzymes as well as their function in modulating transcription and brain activity. In the first portion of this review, we discuss a range of ex vivo methods requiring brain removal and homogenization with can resolve details of chromatin modification on the order of nucleic acid enrichment and resolution of subcellular expression (~10−8 – 10−6 meters in resolution). We further review efforts that can evaluate the expression and activity of epigenetic modifiers in intact tissue from histology to autoradiography. In the second half of the review, we highlight radioactive chemicals that have been adapted for neuroimaging in rodent and the potential to translate this work to human (10−3 – 10−2 meters in resolution). Further, we provide details on optimizing probe development design, which has progressed for the histone deacetylaseenzyme family, but is generalizable for creating epigenetic radioligands for any chromatin modifying enzyme.
Evidence from human postmortem brain and animal models has indicated that dysregulation of chromatin modifying enzymes may play a key role in the transcriptional changes thought to underlie diseases including neurodegenerative disorders, schizophrenia, depression, mood-dysregulation and addiction. This includes enzymes that control DNA methylation, as well as acetylation, methylation and phosphorylation of histone proteins.
In this review, we provide an overview of invasive methodologies that have been used to visualize and understand the biological role of chromatin modifying enzymes. A major gap exists between these methods and evaluating how the same enzymes are expressed and function in living human brain. Noninvasive neuroimaging has revolutionized our understanding of brain function. Modalities including positron emission tomography (PET), and magnetic resonance imaging (MRI) provide a window into the physical and biochemical changes evident in vivo. Depending on the nature of the PET radiotracer or MRI pulse sequence, these techniques can be used to probe structure, function, neurochemistry, or drug pharmacokinetics. Combining this information with what has already been determined about chromatin changes and their role in biology can provide new insight into the role of epigenetics in the brain. To this end, we highlight recent advances in neuroimaging epigenetic regulation and provide discussion on the challenge to develop imaging tools to visualize chromatin modifying enzyme expression and function in the brain.
Expression: visualizing evidence of protein presence
In order understand how epigenetic enzymes may alter function in the CNS, an important first step is to investigate the distribution of each enzyme subtype throughout the brain. This can be done using invasive or noninvasive methods. The methods we highlight are well established in basic neuroscience research and can be used to investigate conceivably any epigenetic enzyme. As an example, we focus on the HDAC family of enzymes. This family formally comprises subtypes from class I (HDAC 1,2,3 and 8); class IIa (HDAC 4,5,7,9); class IIb (HDAC 6,10); class III (the sirtuins, SIRT1–7); and class IV (HDAC11). Each class is defined in part on homology to yeast enzymes and cellular localization. Additionally, class I, II and IV HDACs are separated from class III ‘sirtuins’ as the sirtuins require NAD+ as an energy source and do not respond to the prototypical HDAC inhibitor, trichostatin A (TSA). As convention, these subgroups are thus referred to as ‘sirtuins’ and ‘HDACs’.
Invasive imaging: visualizing epigenetic targets in the ex vivo brain (Table 1)
Table 1.
Methods for visualizing evidence of epigenetic change ex vivo
| Method | Pros | Cons |
|---|---|---|
| In situ hybridization |
|
|
| Reductive immunostain (western, ELISA) |
|
|
| Immunohistochemistry |
|
|
| Autoradiography |
|
|
In situ hybridization and transcript measurement
The central dogma of biochemistry - RNA to DNA to protein - provides a fundamental base from which to evaluate messenger RNA levels as a surrogate for direct measurement of translated proteins. In situ hybridization measures the binding of radioisotope- or fluorescent-labeled probes to a histological brain section. Investigating regional transcript expression differences by in situ hybridization allows novel targets to be evaluated without the need for specific antibodies, which can be time consuming and expensive to generate. Major advantages of in situ include the ability to create highly-specific oligomeric probes quickly at low cost, as well as the benefit of visualizing probe binding to tissue sections without destroying cellular/neuroanatomical organization although each probe must be carefully validated and optimized to ensure binding stringency. (Broide et al., 2007, Janssen et al., 2010)
Comprehensive investigation of HDAC expression in the brain has only been addressed by few select reports. For HDAC expression in CNS, beyond early additions to the Allen Brain Atlas, (Available from: http://www.brain-map.org. © 2012 Allen Institute for Brain Science.), the first resource for this information was an in situ hybridization study of class I, II and IV subtype expression (Broide et al., 2007). Broide and colleagues measured HDACs throughout more than 50 regions of the rat brain (Broide et al., 2007). This painstaking effort revealed that the expression of HDAC subtypes were both overlapping and distinct and supported the idea that HDAC subtypes likely had discernible roles in regulating brain activity.
Differences in HDAC subtype transcript levels were investigated in postmortem brain samples from a small cohort of ALS patients and age-matched controls (Janssen et al., 2010) Similar to the work in rat brain (Broide et al., 2007), the study led by Janssen is perhaps the only one of its kind to have examined the expression of each HDAC subtype comprising class I, II and IV. In this case, quantitative PCR was used which provides a highly accurate measure of the amount of transcript present, however with no anatomical resolution as in in situ. Nevertheless, Janssen and colleagues identified robust expression of HDAC 2 and HDAC 11 in human brain. As the authors discussed, a notable difference between the human and rodent datasets underscores high HDAC2 expression in human brain with high HDAC3 expression in rat (Broide et al., 2007, Janssen et al., 2010). This indicates that HDAC2 may play a more predominant role in modulating brain activity in humans.
Although relatively limited differences were found in the expression of HDACs in the brain between rodents and human (Broide et al., 2007, Janssen et al., 2010), there are obviously major differences between the species. Indeed, there may be homology in regional protein expression between rodent and man, however the context in which functional changes modulate complex behaviors will likely be impossible to replicate in any animal model. This supports that tools to visualize epigenetic changes in humans, including those changes related to nuanced behaviors encoded by healthy and diseased brain signaling, will require neuroimaging in people.
Adapting in situ for noninvasive use in humans is not likely. There are significant technological hurdles for an in vivo nucleic acid probe including crossing the blood brain barrier and surviving degradation in the bloodstream. Given the readily available alternatives and the limitations in measuring transcript levels as a surrogate for protein expression, in situ will likely remain a useful tool for use in ex vivo systems.
Measurement of protein and mRNA levels ex vivo can also be done in humans and capture expression information at the molecular level. However, post-mortem brain tissue work has limitations including i) limited tissue resources, ii) confounds of post-mortem interval, iii) disease state- and iv) drug treatments at time of death. These factors can be accounted for using carefully matched control samples. However, post-mortem studies can nevertheless only provide data to be used to help understand disease and design improved treatments for future patients.
Transcript levels encoding for a protein can be sensitively measured using quantitative PCR, however this method typically begins with tissue homogenization. Laser capture microdissection or cell sorting can be used to resolve transcript levels at a single-cell or population level. However coupling these techniques is not practical from the standpoint of investigating a family of epigenetic enzymes, such as HDACs, throughout the brain.
Immunoreactive, reductive methods: Immunoblotting, ELISA and immunoprecipitation
Perhaps the most common methods to investigate protein presence are each based in part on antibody affinity and begin with sample homogenization. Indeed, established techniques including western blotting, enzyme linked immunosorbent assays, and immunoprecipitation allow a visualization of a protein. A shared drawback, however, is that samples are homogenized at the outset, thus, the neuroanatomical detail maintained is limited to the accuracy of brain tissue dissection. To this point, subpopulations of cells may differentially express an epigenetic target within the same brain structure. However homogenizing whole tissue regions dilutes immunoreactive signal and prevents identification of localized expression differences.
Investigating protein expression using western blotting rather than a related nucleic acid sequence circumvents the potential issue that transcript levels may not faithfully represent protein levels. Western blotting can provide a relatively fast way to visualize the presence of a protein in a homogenized sample, accurately resolving molecular weight. This method is frequently applied to HDAC proteins in homogenized brain tissue (Zhou et al., 2001, Mielcarek et al., 2011), developmental HDAC11 (Liu et al., 2008). A disadvantage here is the dependence on an antibody raised against the protein of interest to visualize immunoreactivity. Further, immunoreactive signal captured from a western blot is at best semi-quantitative. Enzyme-linked immunosorbent assays (ELISA) utilize specific antibodies bound to a substrate to capture proteins from a sample preparation. Comparing signal generated from an enzymatic reaction to a within-experiment standard curve is accepted to provide quantitiative results on the amount of protein in the sample. Quantitiative ELISA methods have been adapted into commercially available HDAC kits, however their application to brain tissue homogenates is likely bypassed for alternative methods, or assessment of HDAC function.
We mention here the rich data from immunoprecipitation (I.P.) studies examining the role of HDAC proteins. This methodology utilizes the same antibodies as in western and ELISA, and likewise begins with tissue homogenization, thus neuroanatomy is lost. Nevertheless, this method uniquely provides information on the physical interaction of the proteins and nucleic acids co-precipitated with an antibody against a target protein which can be resolved with subsequent immunoblotting, qPCR or intense sequencing and genomic alignment. This was exemplified by the work of Wang in 2009 (Wang et al., 2009) demonstrating the relationship between the localization of HDAC subtypes and transcriptional regulatory protein across the genome in a cellular system.
Immunohistochemistry
Visualizing epigenetic enzyme expression using histochemical techniques can provide a high-resolution view of protein presence in the brain while maintaining neuroanatomy. Further, visualizing the regional expression patterns of protein targets co-labeled with colorimetric or fluorescent markers using high magnification microscopy can discriminate protein localization to r neural cell types or sub-cellular cellular compartments.
In the example of HDAC enzymes, Broide and colleagues, as an additional component to their in situ work, used immunolabeling to demonstrate that HDAC subtypes had preferential expression in either neurons or glia. (Broide et al., 2007) MacDonald and Roskam extended this in 2008 showing that HDAC subtypes 1 and 2 in mouse brain had differential expression both in neural cell types and in development. (MacDonald and Roskams, 2008). This group more recently showed that HDACs are differentially expressed in neural stem cell subtypes in mouse and that neural migration and differentiation are impacted by HDAC inhibitor treatment (Foti et al., 2013). Takase also investigated HDAC expression in mouse brain using co-labeling of a number of neuronal cell types to resolve a new level of specificity in the expression of class I, II and IV HDAC subtype expression (Takase et al., 2013). HDAC expression was recently examined in sections from non-human primate brain (Yeh et al., 2013) as part of a non-invasive imaging study on a labeled HDAC substrate.
Histological techniques are a necessary component in evaluating pathological differences in clinical samples. To this end, interpreting the immunoreactive signal depends heavily on the specificity of antibodies to visualize a protein of interest. As the tissue remains intact, it is important to validate at a minimum that the immunoreactivity of an antibody corresponds to the expected molecular weight of a protein target. Immunohistochemistry, like western blotting, is a semiquantitative approach. As in the cited examples for HDAC expression, careful scoring can be applied to evaluate ‘high’ or ‘low’ expression, but are not a measure of protein concentration.
As with other invasive techniques mentioned above, immunohistochemistry has a highly limited potential for in vivo translation. Indeed, while new methods are being developed to enhance blood brain barrier (BBB) penetration of antibodies (Bacskai et al., 2001, Frenkel and Solomon, 2002, Atwal et al., 2011) the increased uptake is still orders of magnitude below what would be required for probe development.
Autoradiography
Autoradiography is a technique closely related to immunohistochemistry (IHC). Compared to IHC, a major advantage of autoradiography is the incorporation of a radiolabel to visualize the binding of a ligand to the protein target which provides high-resolution, quantifiable results of protein density and pharmacokinetic properties of the ligand. The benefit autoradiography offers in a quantifiable result bears emphasis. The radioactivity associated with a bound probe can be decay corrected for time and used to determine the number of molecules of probe bound. Expressed relative to tissue volume, this readily provides an absolute measure of protein in any brain region. The most common isotope used for autoradiography experiments is tritium (3H) which has a half-life of ~12.5 years. This long half-life is an important feature and allows radioactive signal to be captured slowly over the course of weeks to provide fine anatomical detail from histological sections. Custom compound labeling with tritium can be applied to almost any molecule, but is cost intense based in large part on the cost of tritium itself. Tritium labeled HDAC inhibitors, particularly those with selectivity for a subset of class I, II and IV subtypes would be a highly useful tool for ex vivo determination of the HDAC binding capacity in brain, as recently described for HDACs1–3 using the [3H]- benzamide, CI-994 (Wang et al., 2013b). Indeed, tritiated acetyl groups have already been utilized in a biochemical assay to measure HDAC activity (further described in section two) but so far only one no studies have yet examined this in samples from brain (Qiu et al., 2006, Lee et al., 2011). As a disadvantage, using radiolabeled compounds requires special training and poses greater safety and workflow challenges than nonradioactive probes used in immunohistochemistry. Further, the issue of non-specific probe binding requires careful control experiments to provide confidence that radioactive signal accurately represents target protein expression.
Animal models: visualizing genetic disruption
At the whole-organism level, genetic mutation or disruption of protein expression can provide a means to visualize the importance of a protein in basic biological function. Classical genetics, cre-transgenic and small RNA-mediated knockdown and viral overexpression have each been used to begin to clarify the role of HDACs in modulating brain function. (Guan et al., 2009, Kennedy et al., 2013, Morris and Monteggia, 2013) Traditional genetic methods can be time consuming and expensive and require multiple filial generations to ensure observed effects are not artefacts of mixed (heterogeneous) genetic backgrounds or by physical disruption of brain tissue in focal injection of viral particles. To this end, optogenetic techniques - gene expression modulated via stimulation with a specific wavelength of light - hold immense promise in providing a way to view gene expression changes in the brain with unprecedented spatial and temporal control as reviewed elsewhere in this edition. Likewise, the cutting-edge methods using CRISPR/Cas systems for rapid genome engineering will no doubt revolutionize the way in which chromatin modifying enzymes are investigated in rodent models, although their application to HDAC enzymes has not been examined (Cong et al., 2013, Li et al., 2013, Wang et al., 2013a).
In any case, genetic models are a critical tool in visualizing the influence of protein expression on brain function and remain an important part of investigating epigenetics in the CNS. A drawback to these techniques is that altered expression of chromatin modifying enzymes changes both the function and expression of the target. Thus, it is difficult to interpret whether effects in a genetic model are the result of structural or functional change in epigenetic enzymes. Subsequent characterization of specific brain regions can provide evidence of epigenetic mechanisms regulating behavioral response – examples relevant to mood disorders are highlighted throughout another review in this special issue of Neuroscience (Fass et al., 2013). Although in any case, this requires that the brain be assayed ex vivo, thus limiting the extent to which analogous work can be done in humans.
Noninvasive techniques for measuring epigenetic expression in the brain
Numerous efforts have been made to develop noninvasive tools for imaging epigenetic modulators which permit detection and quantification of expression in vivo is critical to assess the efficacy of therapies targeting epigenetic mechanisms and to clarify the understanding of the mechanism enzyme dysfunction in disease (Table 2). PET is an excellent tool for the in vivo quantification of HDAC biological processes and is well-suited to evaluate the pattern of HDAC distribution in animals and human. A major advantage of PET as a technique is its extraordinarily high sensitivity (10−9 to 10−12 M), much more than MRI (10−4 M) or magnetic resonance spectroscopy (MRS) (10−3 to 10−5 M). PET is also able to quantify the distribution of radiotracers in the brain providing in vivo correlates of in vitro measurement outcomes with invasive techniques such as autoradiography, immunohistochemistry and western immunoblotting. Importantly, the specificity of PET probes to detect a particular protein or enzyme of interest cannot be readily matched by MRI modalities. Therefore, we consider PET to provide the greatest advantage as an imaging tool in characterizing protein expression of the highly related HDAC family members as well as other chromatin modifying enzymes.
Table 2.
In vivo imaging methods to measure epigenetic ‘function’
| Method | Applications (Examples) | Refs |
|---|---|---|
| Direct observation | • HDAC inhibitor pharmacokinetics [11C]MS-275 | Hooker et al., 2010 |
| • Radiotracer for direct binding [18F]SAHA | Hendricks, et al., 2011 | |
| • Radiotracer for HDAC activity [18F]FAHA | Reid et al., 2009; Yeh et al., 2013; | |
| • Activity of HDAC via MR spectroscopy | Sankaranarayanapillai et al., 2006; 2008. | |
| Indirect observation | • Assess brain glucose metabolism [18F]FDG-PET | Mayberg, et al., 2000; Mosconi et al., 2008; |
| • Measure the effects of drug treatment fMRI, phMRI, BOLD MRI | Febo et al., 2009; Jang et al., 2011; Wang et al., 2012. |
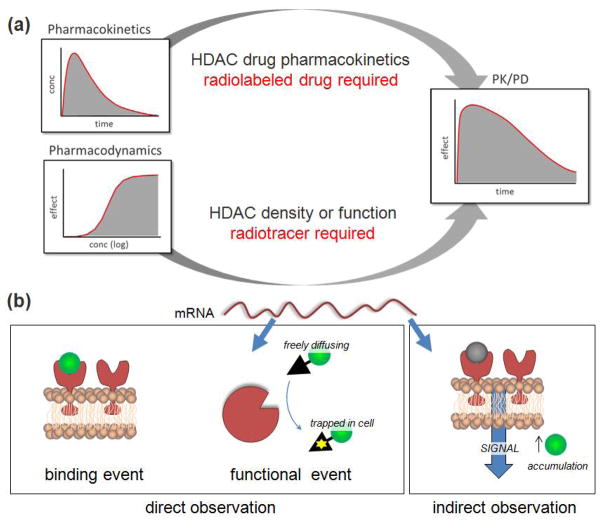
A PET radiotracer is a chemical in which one atom has been replaced by positron emission isotopes (e.g., carbon-11, half-life of 20.38 min; fluorine-18, half-life of 109.8 min), allowing in vivo characterization and measurement of biological processes at the cellular and molecular levels on a timescale of minutes to hours. The development of a PET radiotracer to visualize epigenetic enzymes and their activity would allow characterization of normal epigenetic status in vivo as a function of normal processes. This could be used as a powerful tool for early diagnosis of human diseases with abnormal epigenetic status (e.g. cancer, cognitive and psychiatric disorders, and heart and inflammatory diseases). A PET radiotracer can also be used to measure the effects of therapeutic drug treatment. Radiotracer imaging is very different from PET imaging with radiolabeled drugs (or drug candidates). Radiolabeled drugs can be used to trace the biological pathway of any compound in living humans and allows assessment of pharmacokinetic parameters such as drug absorption, biodistribution, metabolism, excretion in preclinical studies (Figure 2). However, to date, there has been limited progress for the observation of any epigenetic processes. In the following sections, we will continue using HDAC as an example to point out how PET tracers are developed and differentiated from radiolabeled drugs and how each are used in PET brain imaging.
Figure 2.
Information from in vivo imaging (a) Radiolabeled compounds can provide two broad classes of information: I. The pharmacokinetics of a labeled drug (uptake, distribution, clearance, metabolism); II. Using Pharmacodynamics of the target (density/receptor occupancy, enzyme action) using a PET radiotracer. (b) Radiotracer studies can be further divided into three main classes: binding of a radiotracer to its brain target, revealing density and distribution of the target; (middle) accumulation of an activity-dependent probe that is activated or trapped within a cell reveals enzmatic function directly; (right) indirect imaging of the consequence of HDAC modulation. Established neural signaling tracers like [18F]flurodeoxygolucose (FDG) and [11C] raclopride can be used in such indirect observation experiments to provide evidence of a drug- or brain function-stimulated signaling cascade, accumulating in regions with increased activity.
Criteria for development of HDAC radiotracers for PET
Development of novel CNS-penetrant and specific PET radiotracers for HDAC and any epigenetic processes is challenging. There are a number of major factors that determine the success of a CNS-penetrant radiotracer candidate which we describe here:
(a) BBB penetration
Small molecular weight (<400 Daltons) and a high degree of lipophilicity are required for a tracer to pass the BBB which is composed of a lipid bilayer. Generally, increasing lipophilicity increases the permeability of the compound, but it also tends to increase plasma protein binding (PPB), thereby decreasing the concentration of free drug available to cross the BBB. Once a drug is in the brain, specific and nonspecific binding must be taken into account. As such, high BBB penetration may be achieved by increasing lipophilicity, however this may result in increased nonspecific binding in the brain. Thus, BBB penetrance, lipophilicity and binding specificity are inextricably linked properties. In general, for brain radiotracers, a partition coefficient (log P) value between 1.5 and 3 is optimal to enable the labeled molecule to cross the BBB.
(b) Specific and selective binding
Radiolabeled compounds show different types of binding, depending on their physicochemical and pharmacological properties. There is specific binding to the target receptor, and it is saturable, typically reversible and can be inhibited by the unlabeled form of the molecule. Nonspecific binding, due to adsorption to tissue, is linearly proportional with the labeled ligand concentration and non-saturable. By experimentally varying the concentration of radiolabeled compound, unlabeled compound and incubation time, ex vivo experiments and curve fitting can provide a detailed understanding of the level of specific and nonspecific binding of a tracer candidate. A radiotracer should have high specific binding (and low nonspecific binding), which is challenging to predict although some headway is being made. (Li et al., 2009, Poulin and Haddad, 2011) Binding specificity is different from binding selectivity. Selective binding means the radiolabeled compounds only bind to very limited types of receptors, whereas non-selective binding refers radiolabeled compounds that bind to several types of receptors. This is a particularly important concept when considering a family of related targets that may each have a distinct function, such the HDACs. Given a putative HDAC radiotracer, specific binding to HDAC targets is expected. Experimentally, this would be visualized as binding of an unlabeled form of the radiotracer would saturate the HDACs in the brain resulting in decreased binding of radiotracer to its target. Likewise, unlabeled compound binding would also be expected to alter the distribution and pharmacokinetics the radioligand in the brain. Regarding selectivity, it is worth to point out that class- or isoform-selective HDAC radiotracers would greatly facilitate efforts to investigate the distribution and function of each class or subtype HDACs in the brain. As a first step, subtype selective HDAC radiotracers may be constructed from recently reported isoform-specific HDAC inhibitors. (Malvaez et al., 2013, Wagner et al., 2013)
(c) Binding affinity
The binding affinity of a radiotracer for the target must be high enough to produce sufficient signal for detection, but it must dissociate from the target quickly enough to allow binding equilibrium to be reached within the timeframe of the scan (typically 1–2 h). After the administration of carbon-11 or fluorine-18-labeled radiotracers in the subject, the time–activity curve of brain is characterized by uptake and clearance of activity. The rate of tissue clearance is in part determined by the affinity of the tracer. Ligands with higher affinity will remain bound to their target longer, although as binding affinity affects both specific and nonspecific binding, it should not be so high that some kinetic data (e.g. washout data) are unavailable to calculate receptor levels in the brain due to the slow clearance from the brain.
The affinity of the tracer must balance the opposing goals of tight binding and fast washout from the brain. In general, the higher the affinity of a probe for its target, the higher the signal-to-background ratio of the PET radiotracer (with the assumption that non-specific binding is minimal). Additionally, it is preferred if the radiotracers have fast on, slow off binding kinetics to visualize in a relevant time setting. As expected, a slow-on compound would likely not be appropriate for in vivo imaging. For example, the benzamide-based HDAC inhibitors have been shown to exhibit slow-on kinetics in binding to HDACs. (Malvaez et al., 2013) Given radioisotope labeling with carbon-11 for PET imaging, the short half-life of the radioisotope is largely decayed before sufficient binding has occurred; longer targeting time and thus longer-lived isotopes would therefore be required to obtain sufficient detectable signal to visualize HDAC binding with slow-on inhibitors.
These parameters are not required for therapeutic agents (e.g., fast uptake may not be necessary a drug) or for in vitro tracers (most nonspecific binding can be eliminated with extended wash times). To this end, it should be appreciated that desirable properties of radiotracers are usually different from those of drugs. For example, slow clearance of drugs from brain may maintain effective receptor occupancy for a long period of time and result in a beneficial therapeutic effect. However, for a radiotracer, slowing clearance may induce high nonspecific binding in brain. This contributes to the reality that only a small percentage of in vitro tracers and therapeutic agents are useful as in vivo imaging agents. In this case, a compound with sub-optimal properties as a drug could still be a useful PET radiotracer and vice versa.
(d) Metabolites
Besides the challenging design factors described above, a tracer candidate can still be rendered unusable in vivo if it is metabolized rapidly and those metabolites pervade regions of interest (e.g. the brain). If tracer metabolism generates lipophilic radioactive metabolites, they may enter the brain in significant concentration. If they do not bind to the target, they will increase nonspecific binding and thereby decrease the signal-to-background measurement of the target. On the other hand, if the radioactive metabolites bind to the target, quantification is highly confounded because the measured signal represents undetermined proportions of parent tracer and metabolite, each of which may have a different affinity for the target. The problem of lipophilic radioactive metabolites may sometimes be avoided by selecting a labeling position in the chemical structure with a built in liability such that labeled metabolites will be sufficiently polar to minimize brain uptake and retention. If the uptake and washout of the parent tracer are fast relative to the production of radioactive metabolites, then their contribution to the total measured activity may be negligible. In summary, PET radiotracers for brain imaging can be designed by labeled at a metabolically labile position (e.g., methylation at nitrogen site) in the molecule to produce hydrophilic radiolabeled metabolites in the peripheral organs which are not able to cross over the BBB; therefore, the nonspecific binding can be minimized by careful design.
One example of a potential 18F-labelled PET radiotracer for HDAC imaging is [18F]-suberoylanilide hydroxamic acid ([18F]SAHA), which is a close analogue of the clinically relevant HDAC inhibitor SAHA and was reported (Hendricks et al., 2011). Reported findings from pharmacokinetic studies indicate that [18F]SAHA has near-identical biochemical activity profiles to that of SAHA. Using a murine ovarian cancer model, Hendricks and colleagues demonstrated that HDAC inhibitor target binding efficacy can be quantitated within 24 h of administration [18F]SAHA. However, this case, the brain penetrance was also low, indicating that the utility of [18F]SAHA may not be ideal for evaluating HDAC expression in the brain.
Radiolabeled HDAC inhibitors for PET imaging
There are two ways to radiolabel drugs for PET imaging: one method is by isotopic substitution, such as 11C for 12C or 18F for 19F, wherein the physicochemical characteristics of the drug remain constant; the other method is bio-isosteric substitution (for example, 18F for a proton or 11C-labelled methyl group or 18F-labelled fluoroalkyl for a proton). By a careful design of the substitutions, it is possible for a radiolabeled drug to retain similar characteristics to the parent drug. (Willmann et al., 2008)
One published example investigating a radiolabeled HDAC inhibitor for PET imaging features MS-275, an HDAC inhibitor in clinical trials for the treatment of several types of cancer. Recent studies have indicated that MS-275 can cross the BBB and cause region-specific changes in rodent brain histone acetylation (Simonini et al., 2006). However, using PET, [11C]MS-275 showed low uptake in brain tissue when administered intravenously to nonhuman primates, and pharmacokinetics and brain accumulation of [11C]MS-275 were not changed by the co-administration of large doses of unlabeled MS-275 in rodent (Hooker et al., 2010). These results indicated that the efficacy of MS-275 for the treatment of neurological disorders by targeting HDACs in the CNS may be limited unless administered at high doses - which may lead to undesired off-target and toxicity-related effects. There is significant value in radiolabeling HDAC modulating drugs; however, we must re-emphasize that these efforts are often distinct from the development of a radiotracer designed to visualize HDAC in the brain.
Function: visualizing the role of a protein in regulating biological activity
Invasive measurement of enzyme function
Chromatin modifying enzymes each function to catalyze posttranslational changes on proteins or nucleic acids. Visualizing this function is more difficult than imaging static protein presence but provides insight into how changes in brain activity could be regulated by epigenetic change. After methylation of DNA, modification of nucleosomal histone proteins is arguably the best characterized evidence of epigenetic function. HDAC enzymes catalyze the removal of acetyl-groups from the amino-terminal tails of core histone proteins as well as non-histone proteins. Using similar techniques we have reviewed for measuring protein expression, antibodies raised against modified (acetylated) protein targets can be used to demonstrate a change in HDAC activity. Methodologically, the same limitations apply to the semi-quantitative western blotting and immunohistochemistry. Using a labeled substrate, HDAC activity can be assayed and measured in brain homogenates. This method can provide a relative measure of HDAC activity, but as the substrate selectivity for HDACs is limited, the activity contributed by each subtype currently remains an open question. (Faraco et al., 2006).
Activatable fluorescent HDAC probe
Kazuya Kikuchi and colleagues described a one-pot fluorogenic HDAC probe, a nine-residue piece of the histone H3 N-terminus with acetylation on the fourth amino acid and has an acylated coumarin at the C-terminus. An acyl group functions to “quench” the fluorescence under normal conditions, and when a transesterification reaction transfers the acyl group from the dye to the lysine in the presence of HDAC, the reaction works to “switch-on” the fluorophore. This unique probe will undoubtedly provide a tool for epigenetic research and the discovery of HDAC-targeted drugs in vitro. (Baba et al., 2012). A similar strategy may be explored in developing reagents to probe methylation and demethylation reactions which are relevant to chromatin modifications of histones as well as DNA.
Measuring transcriptional effects
A well-described role of chromatin modifying enzymes is to regulate transcriptional activity by modulating chromatin structure. Thus, one technique to visualize the impact of HDAC enzyme function is to use chromatin immunoprecipitation (ChIP) using a modified histone antibody. Co-precipitated DNA can then be assayed by quantitative PCR, microarray or deep sequencing to visualize the local enrichment of histone acetylation - the result of altered enzyme function - as well as changes in transcription. (Peleg et al., 2010, McFarland et al., 2013). These methods can reveal transcriptional changes that are the result of changes in promoter region chromatin structure. Such changes are likely direct effects of epigenetic enzymes, as well as those genes whose expression is altered downstream of primary response genes. These methods can be used to understand the impact of HDAC function using treatment with small molecule HDAC inhibitor compounds.
Impact of chromatin modifying drugs
The early identification that small molecule inhibitors that can block HDAC activity, such as trichostatin A (Korzus et al., 2004) has led to a vast literature base using HDAC inhibitors to understanding the impact of inhibited function. Small molecules that block the activity of a number of class I, II and IV HDAC enzymes (TSA, butyrate, crebinostat) have been used to clarify the role of HDAC activity in modulating synaptic growth, cellular differentiation, and onward to diverse, complex behavioral response in rodent models. As mentioned above, disruption of enzyme expression can also be interpreted in terms of a change in enzyme activity. Efforts to resolve small molecules that can inhibit a single HDAC subtype are emerging (Malvaez et al., 2013, Wagner et al., 2013) and will be invaluable in further resolving the impact of suppressing a specific HDAC subtype on brain function.
Noninvasive measurement of HDAC activity
PET imaging with a radiolabeled HDAC substrate [18F]FAHA
6-([18F]fluoroacetamido)-1-hexanoicanilide ([18F]FAHA), a labeled HDAC substrate, was used to show accumulation of the major metabolite, [18F]fluoroacetate, in rat brain. This compound was further used to visualize target binding in non-human primate brain which was modulated in a dose-dependent manner by administration of SAHA, indicating that [18F]FAHA and SAHA share the same biological targets (Reid et al., 2009). Very recently, Yeh and colleagues clarified these results, demonstrating that FAHA has substrate selectivity for class IIa HDAC enzyme subtypes (HDAC 4, 5, 7, and 9) compared to other HDAC classes (Yeh et al., 2013). Further, using the radiolabeled metabolite of [18F]FAHA, [18F]fluoroacetate (FACE), Yeh showed that the accumulation of radioactivity in the brain following [18F]FAHA administration was the result of its catabolism by class IIa HDAC to [18F]FACE, and further, to [18F]flurocitrate (Yeh et al., 2013). Importantly, both studies examining [18F]FAHA highlight its rapid metabolism in vivo and, thus, that accumulated radioactive signal attributed to binding of labeled parent probe to HDAC targets is difficult to distinguish from metabolites. The primary challenge, however, is that systemic blockade of HDAC to determine specific binding leads to an increase in uptake of [18F]FAHA. While this may seem counterintuitive, it is expected that brain uptake will increase when peripheral HDAC is blocked (and thus more radiotracer is available in the blood for brain binding). Deconvolving and interpreting the uptake of [18F]FAHA is not trivial and could limit its utility for brain HDAC imaging. This tool is, however, a remarkable advance for PET HDAC imaging. Given that the HDAC selectivity of SAHA has been shown to be class I HDAC subtypes HDAC1–3, 8 and the class IIb HDAC, HDAC6, it is possible that the SAHA-mediated depletion of signal from [18F]FAHA may result from a compensatory increase in the activity of class IIa HDAC enzymes following inhibition of class I and IIb subtypes by SAHA.
Together, these studies provide evidence of a tool to image HDAC activity in vivo. Importantly, [18F]FAHA is a substrate that is targeted specifically by class IIa HDAC enzymes (HDAC 4,5,7,9). Additionally, [18F]FAHA can facilitate the development and clinical translation of novel class-IIa HDAC inhibitors (Yeh et al., 2013). Given the importance implicated in class I HDAC enzymes (HDAC 1,2,3, 8) in CNS disease-related behaviors, there remains a great need to develop tools to measure the activity of class I HDACs or to synthesize radiolabeled ligands to visualize the expression of these enzymes in normal and diseased brain.
Even without a dedicated radiolabeled substrate like [18F]FAHA, the downstream impact of chromatin modifying enzymes can be visualized using existing PET tracers or other imaging modalities, such as MRI. For example, uptake of the glucose analog [18F]FDG can be measured by PET and is an established surrogate for evaluating changes in brain activity. (Mosconi et al., 2008) Differential binding of the D2-antagonist radiotracer, [11C]raclopride, can provide insight into the dopaminergic impact of a genetic model or drug-treatment regimen.
These techniques are currently used in clinical medicine and represent part of continued investigation to clarify brain activity changes associated with drug treatment and behavioral response in humans and rodent models. (Mayberg et al., 2000, Patel et al., 2008, Busto et al., 2009, Jang et al., 2011, Williams et al., 2011)
MRI measurement of drug effects
MRI is a noninvasive medical imaging technique used in radiology to visualize detailed internal structures and limited functions of the body. MRI is used to create images of the body with extraordinary detail, including the brain, by applying the nuclear magnetic resonance phenomena. The distribution of hydrogen nuclei found in water depends on the tissue type and whether or not the tissue is healthy or diseased. At the most basic level, MRI measures and records changes in the magnetic properties of protons in water.
Despite a vast and growing literature base on the behavioral effects of HDAC inhibitor treatment in rodents, few studies have utilized in vivo MRI imaging to assess the impact of HDAC inhibition on brain activity. Circumventing the challenges of PET-probe development, established techniques in MRI – and variations thereof - can provide detailed information on the impact of disease and drug treatment on changes in brain structure, blood flow and metabolite density. In the context of addiction, blood-oxygen level dependent (BOLD) activation of cortico-limbic circuitry in rat was shown to be increased after subchronic treatment with the HDAC inhibitor, sodium butyrate (Febo et al., 2009). This study was the first of its kind to couple chromatin modifying drug treatment with in vivo brain imaging. While the result of the study advanced that HDAC inhibition may functionally activate cortio-limbic neurocircuitry and enhance brain activation following co-treatment with cocaine, it also provided insight that neuroimaging tools could be effectively applied to investigate epigenetic mechanisms in animal models and, as discussed, provide a forward view on the therapeutic potential of HDAC inhibitors. Related, functional MRI (fMRI) and functional connectivity MRI (fcMRI) relates the BOLD signal between different parts of the brain to resolve putative connections between brain nuclei. This is most often coupled to a change in brain function based on a visual or cognitive task in awake subjects – a decided challenge for research animals, though analogies can be made for connectivity using pharmacological (ph) MRI. The impact of chromatin modifying drugs can be investigated using pharmacological MRI (phMRI), which couples the functional changes in cerebral blood flow to pharmacological drug challenge. As recently reviewed, this method provides high temporal resolution and, like PET, has virtually identical methods that can be translated to human phMRI imaging experiments (Jenkins, 2012).
High resolution structural information can be used to investigate the impact of chromatin modifying drugs on parameters such as hippocampal volume, a phenotype linked to a rodent model of schizophrenia (Sandner et al., 2011). Likewise, in a model of ischemic stroke, MRI was used to measure infarct size induced by medial cerebral artery occlusion as well as the functional recovery promoted by chronic treatment with the HDAC inhibitor sodium valproate (Wang et al., 2012). These studies represent important milestones in epigenetics research in which imaging tools are applied to understand the functional and structural changes in the brain regulated in part by HDAC activity.
Proton (1H) MRS utilizes spectroscopy and the exquisite temporal resolution of MRI to link functional changes in tissues including brain to concentrations of metabolic biomarkers. Altered levels of N-acetyl aspartate, choline, and myoinositol have been identified in the brain of patients with neurodegenerative diseases, cognitive deficits and psychiatric phenotypes. HDAC inhibitor treatment has been linked to restoration of MRS-measured levels of choline and N-acetyl-aspartate in cellular and animal tumor models (Beloueche-Babari et al., 2012, Wei et al., 2012, Ward et al., 2013) although in each case these are indirect measures of changes in HDAC activity.
Advancing on this limitation, an 19F-labeled HDAC substrate was developed which allowed the activity of HDAC enzymes to be visualized using MRS (Sankaranarayanapillai et al., 2006, Sankaranarayanapillai et al., 2008). 19F-MRS of the targeted molecular imaging agent fluorinated lysine derivative Boc-Lys-(Tfa)-OH (BLT) can be used to monitor delivery and activity of HDACis at the tumor site, whereas 31P-MRS can be used to monitor the downstream metabolic consequences of HDAC inhibition. Together, these two MRS methods provide both a direct marker of HDAC inhibition and a downstream biomarker of cellular response to the inhibition. The combination of 19F and 31P-MRS has the potential to serve as a reliable noninvasive modality to assess HDAC inhibition. Importantly, the acetyl substrates of most class I and II HDAC enzymes are not unique to any one HDAC subtype. Therefore, the specific HDAC subtypes responsible for any observed changes using 19F-BLT or another labeled HDAC substrate by MRI or PET would be difficult or impossible to interpret.
Using multiple modalities in concert presents new imaging frontiers. The simultaneous acquisition of different functional parameters using PET, fMRI or MRS, in addition to high-resolution anatomic MRI information, creates enormous possibilities and provides completely new opportunities to study pathology and biochemical processes in vivo. With this combined imaging system, both modalities preserve their functionality we simultaneously acquired functional and morphological PET-MRI data from living subjects. MR-PET will improve the diagnostic relevance and functional consequence of PET imaging. The simultaneous scanning will not only resolve many of the impediments to precise coregistration of anatomo-molecular information, but also be used to develop receptor occupancy and function correlations; this new imaging tool will lead us to discover a new MR-PET imaging technology to ask roles of chromatin modifications in brain disorders and explore the correlation between the therapeutic treatment and brain’s responses and might have strong potential for drug development.
Conclusion
In this review, we have provided an overview of methodology used to visualize chromatin modifying enzymes and their function in the brain. We highlight the tools used so far to extend these findings to visualize the brain in vivo and the powerful translational aspect of neuroimaging in animals and humans. Integrating the knowledge base from biochemical, in vitro and ex vivo studies with an understanding of what is happening in the living brain is inextricably linked to techniques in noninvasive neuroimaging. Beyond providing a better understanding of how to interpret results from basic research, developing in vivo imaging tools will give a chance to intervene in brain disease.
Identification and approval of the beta amyloid radiotracer, [18F]fluorbetapir was a major breakthrough in linking what basic science and ex vivo studies have shown regarding amyloid accumulation and what changes are evident in the brains from patients living with frontotemporal dementia, including Alzheimer’s disease (AD). This stands as a powerful example of how neuroimaging could change the trajectory of a disease. Nevertheless, treatment options for AD remain limited. However, like the number of tools available for imaging the dopaminergic system especially the D2-antagonist radiotracer, [11C]raclopride, dedicated in vivo imaging tools will help us understand protein expression and function in the context of many diseases.
Understanding of the role of chromatin modifying enzymes such as DNA and histone methyltransferases, lysine specific demethylases and is emerging, however evaluating how the expression and activity of these enzymes is distributed throughout the healthy and diseased brain in vivo remains an open question. Similar to the evidence we presented for the HDAC family of enzymes, many additional chromatin modifying enzymes are already well positioned to be investigated using neuroimaging techniques as specific chemical activators and inhibitors have been described and provide an important foothold to initiate characterization using in vivo imaging tools.
Highlights.
Invasive measurements of epigenetics can be conceptually adapted for human brain imaging.
Noninvasive measurements of HDAC expression and function are being developed.
Functional consequences of epigenetic modulation can be imaged with PET and fMRI.
Acknowledgments
The authors would like to thank Drs. Nicole R. Zurcher and Genevieve Van de Bittner for their help in figure preparation.
Abbreviations
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- BLT
fluorinated lysine derivative Boc-Lys-(Tfa)-OH
- BOLD
Blood Oxygenation Level Dependent
- ChIP
chromatin immunoprecipitation
- CNS
central nervous system
- DNA
Deoxyribonucleic acid
- ELISA
enzyme-linked immunosorbent assays
- FACE
fluoroacetate
- FAHA
6-(fluoroacetamido)-1-hexanoicanilide
- FDG
Fluorodeoxyglucose
- fMRI
Functional magnetic resonance imaging
- HDAC
histone deacetylase
- IHC
immunohistochemistry
- I.P
immunoprecipitation
- MRI
magnetic resonance imaging
- mRNA
messenger RNA
- MRS
magnetic resonance spectroscopy
- NAD+
Nicotinamide adenine dinucleotide
- PCR
polymerase chain reaction
- PET
positron emission tomography
- phMRI
pharmacologic magnetic resonance imaging
- PPB
plasma protein binding
- RNA
Ribonucleic acid
- SAHA
suberoylanilide hydroxamic acid
- SIRT
sirtuins
- TSA
trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atwal JK, Chen Y, Chiu C, Mortensen DL, Meilandt WJ, Liu Y, Heise CE, Hoyte K, Luk W, Lu Y, Peng K, Wu P, Rouge L, Zhang Y, Lazarus RA, Scearce-Levie K, Wang W, Wu Y, Tessier-Lavigne M, Watts RJ. A therapeutic antibody targeting BACE1 inhibits amyloid-beta production in vivo. Sci Transl Med. 2011;3:84ra43. doi: 10.1126/scitranslmed.3002254. [DOI] [PubMed] [Google Scholar]
- Baba R, Hori Y, Mizukami S, Kikuchi K. Development of a fluorogenic probe with a transesterification switch for detection of histone deacetylase activity. J Am Chem Soc. 2012;134:14310–14313. doi: 10.1021/ja306045j. [DOI] [PubMed] [Google Scholar]
- Bacskai BJ, Kajdasz ST, Christie RH, Carter C, Games D, Seubert P, Schenk D, Hyman BT. Imaging of amyloid-beta deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy. Nat Med. 2001;7:369–372. doi: 10.1038/85525. [DOI] [PubMed] [Google Scholar]
- Beloueche-Babari M, Arunan V, Troy H, te Poele RH, te Fong AC, Jackson LE, Payne GS, Griffiths JR, Judson IR, Workman P, Leach MO, Chung YL. Histone deacetylase inhibition increases levels of choline kinase alpha and phosphocholine facilitating noninvasive imaging in human cancers. Cancer research. 2012;72:990–1000. doi: 10.1158/0008-5472.CAN-11-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. Journal of molecular neuroscience: MN. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Busto UE, Redden L, Mayberg H, Kapur S, Houle S, Zawertailo LA. Dopaminergic activity in depressed smokers: a positron emission tomography study. Synapse. 2009;63:681–689. doi: 10.1002/syn.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- Fass DM, Kemp MM, Schroeder FA, Wagner FF, Wang Q, Holson EB. Histone Acetylation and Deacetylation. In: Meyers RA, editor. Meyers: Encyclopedia of Molecular Cell Biology and Molecular Medicine: Epigenetic Regulation and Epigenomics. Wiley-VCH Verlag GmbH & Co; KGaA: 2012. [Google Scholar]
- Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. Epigenetic mechanisms in mood disorders: Targeting neuroplasticity. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febo M, Akbarian S, Schroeder FA, Ferris CF. Cocaine-induced metabolic activation in cortico-limbic circuitry is increased after exposure to the histone deacetylase inhibitor, sodium butyrate. Neurosci Lett. 2009;465:267–271. doi: 10.1016/j.neulet.2009.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti SB, Chou A, Moll AD, Roskams AJ. HDAC inhibitors dysregulate neural stem cell activity in the postnatal mouse brain. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2013 doi: 10.1016/j.ijdevneu.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Solomon B. Filamentous phage as vector-mediated antibody delivery to the brain. Proc Natl Acad Sci U S A. 2002;99:5675–5679. doi: 10.1073/pnas.072027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Mitchell A, Schneider A, Halene T, Akbarian S. Epigenetic dysregulation in schizophrenia: molecular and clinical aspects of histone deacetylase inhibitors. Eur Arch Psychiatry Clin Neurosci. 2013 doi: 10.1007/s00406-013-0395-2. [DOI] [PubMed] [Google Scholar]
- Hendricks JA, Keliher EJ, Marinelli B, Reiner T, Weissleder R, Mazitschek R. In vivo PET imaging of histone deacetylases by 18F-suberoylanilide hydroxamic acid (18F-SAHA) Journal of medicinal chemistry. 2011;54:5576–5582. doi: 10.1021/jm200620f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker JM, Kim SW, Alexoff D, Xu Y, Shea C, Reid A, Volkow N, Fowler JS. Histone deacetylase inhibitor, MS-275, exhibits poor brain penetration: PK studies of [C]MS-275 using Positron Emission Tomography. ACS Chem Neurosci. 2010;1:65–73. doi: 10.1021/cn9000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Jang M, Kim SH, Um KB, Kang YK, Kim HJ, Chung S, Park MK. Regulation of dopaminergic neuron firing by heterogeneous dopamine autoreceptors in the substantia nigra pars compacta. J Neurochem. 2011;116:966–974. doi: 10.1111/j.1471-4159.2010.07107.x. [DOI] [PubMed] [Google Scholar]
- Janssen C, Schmalbach S, Boeselt S, Sarlette A, Dengler R, Petri S. Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis. Journal of neuropathology and experimental neurology. 2010;69:573–581. doi: 10.1097/NEN.0b013e3181ddd404. [DOI] [PubMed] [Google Scholar]
- Jenkins BG. Pharmacologic magnetic resonance imaging (phMRI): imaging drug action in the brain. NeuroImage. 2012;62:1072–1085. doi: 10.1016/j.neuroimage.2012.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han MH, Bassel-Duby R, Olson EN, Nestler EJ. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Magklara A, Smith CL. HDAC activity is required for efficient core promoter function at the mouse mammary tumor virus promoter. J Biomed Biotechnol. 2011;2011:416905. doi: 10.1155/2011/416905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nature biotechnology. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Li H, Sun J, Sui X, Liu J, Yan Z, Liu X, Sun Y, He Z. First-principle, structure-based prediction of hepatic metabolic clearance values in human. Eur J Med Chem. 2009;44:1600–1606. doi: 10.1016/j.ejmech.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Liu H, Hu Q, Kaufman A, D’Ercole AJ, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J Neurosci Res. 2008;86:537–543. doi: 10.1002/jnr.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Developmental dynamics: an official publication of the American Association of Anatomists. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS, Brannan SK, Tekell JL, Silva JA, Mahurin RK, McGinnis S, Jerabek PA. Regional metabolic effects of fluoxetine in major depression: serial changes and relationship to clinical response. Biol Psychiatry. 2000;48:830–843. doi: 10.1016/s0006-3223(00)01036-2. [DOI] [PubMed] [Google Scholar]
- McFarland BC, Gray GK, Nozell SE, Hong SW, Benveniste EN. Activation of the NF-kappaB Pathway by the STAT3 Inhibitor JSI-124 in Human Glioblastoma Cells. Mol Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-12-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M, Benn CL, Franklin SA, Smith DL, Woodman B, Marks PA, Bates GP. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLoS One. 2011;6:e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Monteggia LM. Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int J Dev Neurosci. 2013 doi: 10.1016/j.ijdevneu.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Tsui WH, Herholz K, Pupi A, Drzezga A, Lucignani G, Reiman EM, Holthoff V, Kalbe E, Sorbi S, Diehl-Schmid J, Perneczky R, Clerici F, Caselli R, Beuthien-Baumann B, Kurz A, Minoshima S, de Leon MJ. Multicenter standardized 18F-FDG PET diagnosis of mild cognitive impairment, Alzheimer’s disease, and other dementias. J Nucl Med. 2008;49:390–398. doi: 10.2967/jnumed.107.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VD, Lee DE, Alexoff DL, Dewey SL, Schiffer WK. Imaging dopamine release with Positron Emission Tomography (PET) and (11)C-raclopride in freely moving animals. Neuroimage. 2008;41:1051–1066. doi: 10.1016/j.neuroimage.2008.02.065. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Poulin P, Haddad S. Microsome composition-based model as a mechanistic tool to predict nonspecific binding of drugs in liver microsomes. J Pharm Sci. 2011 doi: 10.1002/jps.22619. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, Adkins NL, Stavreva DA, Wiench M, Georgel PT, Schiltz RL, Hager GL. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Reid AE, Hooker J, Shumay E, Logan J, Shea C, Kim SW, Collins S, Xu Y, Volkow N, Fowler JS. Evaluation of 6-([(18)F]fluoroacetamido)-1-hexanoicanilide for PET imaging of histone deacetylase in the baboon brain. Nucl Med Biol. 2009;36:247–258. doi: 10.1016/j.nucmedbio.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandner G, Host L, Angst MJ, Guiberteau T, Guignard B, Zwiller J. The HDAC Inhibitor Phenylbutyrate Reverses Effects of Neonatal Ventral Hippocampal Lesion in Rats. Frontiers in psychiatry. 2011;1:153. doi: 10.3389/fpsyt.2010.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanapillai M, Tong WP, Maxwell DS, Pal A, Pang J, Bornmann WG, Gelovani JG, Ronen SM. Detection of histone deacetylase inhibition by noninvasive magnetic resonance spectroscopy. Molecular cancer therapeutics. 2006;5:1325–1334. doi: 10.1158/1535-7163.MCT-05-0494. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanapillai M, Tong WP, Yuan Q, Bankson JA, Dafni H, Bornmann WG, Soghomonyan S, Pal A, Ramirez MS, Webb D, Kaluarachchi K, Gelovani JG, Ronen SM. Monitoring histone deacetylase inhibition in vivo: noninvasive magnetic resonance spectroscopy method. Molecular imaging. 2008;7:92–100. [PubMed] [Google Scholar]
- Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, Guidotti A. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci U S A. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase K, Oda S, Kuroda M, Funato H. Monoaminergic and neuropeptidergic neurons have distinct expression profiles of histone deacetylases. PLoS One. 2013;8:e58473. doi: 10.1371/journal.pone.0058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner FF, Olson DE, Gale JP, Kaya T, Weiwer M, Aidoud N, Thomas M, Davoine EL, Lemercier BC, Zhang YL, Holson EB. Potent and Selective Inhibition of Histone Deacetylase 6 (HDAC6) Does Not Require a Surface-Binding Motif. J Med Chem. 2013 doi: 10.1021/jm301355j. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Onestep generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013a;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang YL, Hennig K, Gale JP, Hong Y, Cha A, Riley M, Wagner F, Haggarty SJ, Holson E, Hooker J. Class I HDAC imaging using [ ( 3) H]CI-994 autoradiography. Epigenetics: official journal of the DNA Methylation Society. 2013b;8 doi: 10.4161/epi.25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tsai LK, Munasinghe J, Leng Y, Fessler EB, Chibane F, Leeds P, Chuang DM. Chronic valproate treatment enhances postischemic angiogenesis and promotes functional recovery in a rat model of ischemic stroke. Stroke; a journal of cerebral circulation. 2012;43:2430–2436. doi: 10.1161/STROKEAHA.112.652545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CS, Eriksson P, Izquierdo-Garcia JL, Brandes AH, Ronen SM. HDAC inhibition induces increased choline uptake and elevated phosphocholine levels in MCF7 breast cancer cells. PloS one. 2013;8:e62610. doi: 10.1371/journal.pone.0062610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Hong S, Yoon Y, Hwang SN, Park JC, Zhang Z, Olson JJ, Hu XP, Shim H. Early prediction of response to Vorinostat in an orthotopic rat glioma model. NMR in biomedicine. 2012;25:1104–1111. doi: 10.1002/nbm.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rush AJ, Koslow SH, Wisniewski SR, Cooper NJ, Nemeroff CB, Schatzberg AF, Gordon E. International Study to Predict Optimized Treatment for Depression (iSPOT-D), a randomized clinical trial: rationale and protocol. Trials. 2011;12:4. doi: 10.1186/1745-6215-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- Yeh HH, Tian M, Hinz R, Young D, Shavrin A, Mukhapadhyay U, Flores LG, Balatoni J, Soghomonyan S, Jeong HJ, Pal A, Uthamanthil R, Jackson JN, Nishii R, Mizuma H, Onoe H, Kagawa S, Higashi T, Fukumitsu N, Alauddin M, Tong W, Herholz K, Gelovani JG. Imaging epigenetic regulation by histone deacetylases in the brain using PET/MRI with (1)(8)F-FAHA. Neuroimage. 2013;64:630–639. doi: 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci U S A. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]