Abstract
Mammalian kidney organogenesis involves reciprocal epithelial-mesenchymal interactions that drive iterative cycles of nephron formation. Recent studies have demonstrated that the Six2 transcription factor acts cell autonomously to maintain nephron progenitor cells, whereas canonical Wnt signaling induces nephron differentiation. How Six2 maintains the nephron progenitor cells against Wnt-directed commitment is not well understood, however. We report here that Six2 is required to maintain expression of Osr1, a homolog of the Drosophila odd-skipped zinc-finger transcription factor, in the undifferentiated cap mesenchyme. Tissue-specific inactivation of Osr1 in the cap mesenchyme caused premature depletion of nephron progenitor cells and severe renal hypoplasia. We show that Osr1 and Six2 act synergistically to prevent premature differentiation of the cap mesenchyme. Furthermore, although both Six2 and Osr1 could form protein interaction complexes with TCF proteins, Osr1, but not Six2, enhances TCF interaction with the Groucho family transcriptional co-repressors. Moreover, we demonstrate that loss of Osr1 results in β-catenin/TCF-mediated ectopic activation of Wnt4 enhancer-driven reporter gene expression in the undifferentiated nephron progenitor cells in vivo. Together, these data indicate that Osr1 plays crucial roles in Six2-dependent maintenance of nephron progenitors during mammalian nephrogenesis by stabilizing TCF-Groucho transcriptional repressor complexes to antagonize Wnt-directed nephrogenic differentiation.
Keywords: Kidney development, Nephron progenitor, Groucho, Odd-skipped, Osr1, Six2, Wnt signaling
INTRODUCTION
Kidneys play a crucial role in bodily health and homeostasis by filtering metabolic waste and excess fluid from the blood. In mammals, kidney morphogenesis begins with formation of the pronephros, which consists of simple tubules connecting to a nephric duct in the anterior intermediate mesoderm in the left and right side of the embryo. The pronephroi quickly regress, but the nephric ducts extend posteriorly and induce adjacent intermediate mesoderm to form mesonephroi, which are structurally more complex but are also transient in mammalian embryos. The definitive kidneys, the metanephroi, form at the posterior end of the intermediate mesoderm through several distinct processes: first, a unique population of nephrogenic cells, called metanephric mesenchyme (MM), is established in the posterior intermediate mesoderm; second, the MM induces outgrowth of ureteric bud (UB) from the nephric duct; third, the UB invades MM and induces MM cells to condense around the UB tip, forming the cap mesenchyme (CM); fourth, the CM cells induce UB to branch as a subset of the CM on the ventral side of the UB tips undergo mesenchymal-epithelial transformation to form renal vesicles, which subsequently differentiate into nephrons; fifth, reiterative reciprocal epithelial-mesenchymal interactions induce UB to branch repeatedly in a highly reproducible manner and new nephron formation from the mesenchyme adjacent to each new UB tip. The ureteric branches form the collecting duct tree that connects the nephrons to the ureter and drain urine into the bladder. The mature kidney contains ∼1,000,000 nephrons per kidney in humans and ∼11,000 nephrons per kidney in mice (Vainio and Lin, 2002; Dressler, 2006; Dressler, 2009; Costantini and Kopan, 2010).
Unlike other organs, such as the gut and skin, which have adult stem cells for maintaining homeostasis and injury recovery (Fuchs, 2008; Li and Clevers, 2010), all nephrogenic progenitors in the mammalian kidney are depleted by the final wave of nephrogenesis in the perinatal period (Little and Bertram, 2009). Thus, kidney organogenesis requires a balance between self-renewal and differentiation of the nephron progenitor cells to ensure the generation of sufficient numbers of nephrons (Little and McMahon, 2012). Premature depletion of progenitors during kidney development results in renal hypoplasia, a common cause of congenital kidney failure and a significant risk factor for hypertension in adults (Keller et al., 2003; Bertram et al., 2011).
Recent studies have identified Wnt9b as a primary inductive signal for nephrogenesis and Six2, a homeodomain transcription factor, as a key factor in regulating nephron progenitor cell self-renewal during metanephric kidney development. Wnt9b is expressed in the UB epithelium and mice lacking Wnt9b function showed complete lack of renal vesicle formation and arrest of metanephric kidney development at E11.5 (Carroll et al., 2005). Furthermore, heterologous Wnt9b-producing cells were sufficient to induce renal vesicle formation in isolated MM explants (Carroll et al., 2005). Loss- and gain-of-function studies indicate that canonical Wnt signaling mediated by β-catenin is necessary and sufficient for initiating epithelial transformation of the CM (Park et al., 2007). Constitutive stabilization of β-catenin in the mouse CM caused ectopic formation of pretubular aggregates and subsequently renal agenesis due to rapid depletion of CM (Park et al., 2007). Six2 is strongly expressed in the CM and is downregulated during renal vesicle formation (Self et al., 2006; Kobayashi et al., 2008). Genetic lineage tracing studies indicate that the Six2-expressing CM cells are maintained by self-renewal and give rise to all cell types of the main body of the nephrons (Kobayashi et al., 2008). Six2-/- mutant mouse embryos exhibited ectopic renal vesicle formation and premature depletion of CM cells (Self et al., 2006; Kobayashi et al., 2008). The differentiation of CM in Six2-/- mutant embryos still requires Wnt9b signaling, suggesting that Six2 acts to maintain the progenitor state of the CM against Wnt-induced nephrogenic differentiation (Kobayashi et al., 2008). However, Six2 protein is detected in the pretubular aggregates and renal vesicles, although at reduced levels in comparison with the CM (Park et al., 2012). In addition to their antagonistic activities, recent studies showed that Six2 and β-catenin had cooperative effects on expression of some shared target genes in the CM (Karner et al., 2011; Park et al., 2012). The molecular mechanisms that regulate the cooperative versus antagonistic actions of these pathways during kidney development remain to be elucidated.
Odd-skipped related 1 (Osr1) encodes a zinc-finger protein homologous to the Drosophila odd-skipped transcription factor (So and Danielian, 1999; Lan et al., 2001). During mouse embryogenesis, Osr1 mRNA expression is first activated in the nascent intermediate mesoderm at E7.5 and remains strong throughout the intermediate mesoderm at E8.5 (So and Danielian, 1999; Wang et al., 2005). Although early Osr1-expressing intermediate mesoderm cells give rise to the majority of cell types in the metanephric kidney, Osr1 mRNA expression itself is progressively restricted to the CM and is downregulated upon mesenchymal-epithelial transition during renal vesicle formation (James et al., 2006; Mugford et al., 2008). Although generation and analyses of mice carrying targeted null mutations revealed a crucial role of Osr1 in MM specification (Wang et al., 2005; James et al., 2006; Mugford et al., 2008), whether Osr1 is required for CM maintenance during metanephric kidney development has not been addressed because the Osr1-/- mutant mouse embryos exhibit aberrant intermediate mesoderm apoptosis starting at E9.5 and lack UB induction (Wang et al., 2005; James et al., 2006). Here, we report that tissue-specific inactivation of Osr1 in the CM caused premature differentiation of nephron progenitor cells, resulting in renal hypoplasia. Our data indicate that Osr1 and Six2 interact synergistically to maintain the nephron progenitor cell pool during metanephric kidney organogenesis.
RESULTS
Tissue-specific inactivation of Osr1 in the cap mesenchyme results in hypoplastic kidneys
We examined the pattern of Osr1 expression during metanephric kidney development using the Osr1tm1Jian/+ mice, which express lacZ from the endogenous Osr1 locus (Wang et al., 2005). At E11.5, when the UB had just invaded the MM, Osr1 was strongly expressed throughout MM cells while no expression was detected in UB or nephric duct (Fig. 1A). As development proceeds, Osr1 expression was restricted mainly in undifferentiated CM cells (Fig. 1B-D), and absent from renal vesicle (Fig. 1C), S-shape body (Fig. 1D) or further differentiated nephron structures. As recent reports indicated that Cited1+ Six2+ doubly positive immunoreactivity marks the nephron progenitor cell population, we compared expression of eGFP from the Osr1GCE allele (Mugford et al., 2008) with the expression of endogenous Cited1 and Six2 proteins, respectively, in Osr1GCE/+ embryos. Direct comparison of Osr1-eGFP with Cited1 protein immunostaining patterns identified a Cited1-Osr1+ domain of CM in between the Cited1+Osr1+ undifferentiated CM and Cited1-Osr1- pretubular aggregates (Fig. 1E). However, although Osr1 and Six2 are co-expressed in all undifferentiated CM cells (Fig. 1F), Six2, but not Osr1, persists in cells forming pretubular aggregates (Fig. 1F). To further confirm that Six2 protein persists in the early differentiating cells, we directly compared the distribution of eGFP and Six2 with Jag1, a marker of proximal part of the renal vesicle (Cheng et al., 2007), and found that Six2 protein was present in the Jag1- cells in the newly formed renal vesicle (Fig. 1F). These results indicate that the CM progresses from the Cited1+Osr1+Six2+ progenitor state through Cited1-Osr1+Six2+ and Cited1-Osr1-Six2+ stages during renal vesicle induction.
Fig. 1.
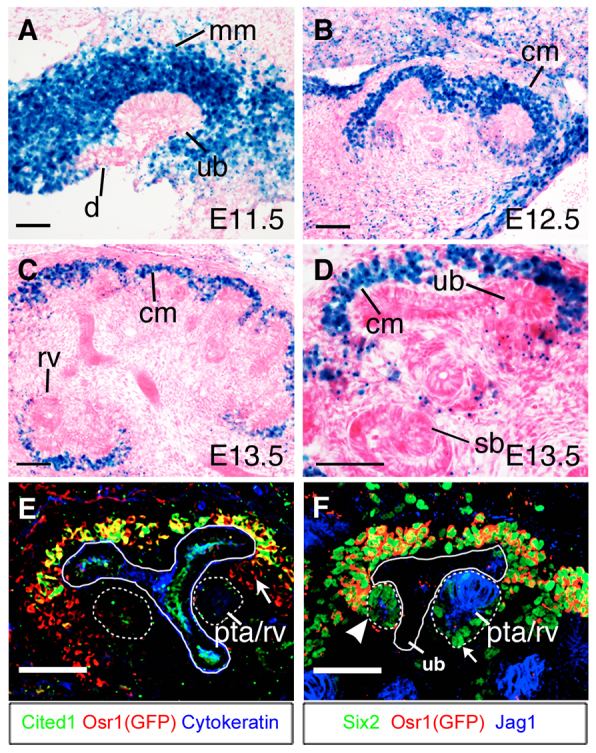
Expression of Osr1 during metanephric kidney development. (A-C) Osr1-lacZ expression in developing kidneys in Osr1tm1Jian/+ embryos at E11.5 (A), E12.5 (B) and E13.5 (C). (D) High-magnification view of the cortical region of an E13.5 Osr1tm1Jian/+ kidney section. (E) Immunofluorescent staining of eGFP (red), Cited1 (green) and pan-cytokeratin (blue) in section of E13.5 Osr1GCE/+ kidney. Arrow indicates Cited1-Osr1+ cells between the Cited1+ cap mesenchyme and pretubular aggregate (outlined with white dashed circle). (F) Immunofluorescent staining of eGFP (red), Six2 (green) and Jag1 (blue) in E13.5 Osr1GCE/+ kidney section. Arrowhead indicates early pretubular aggregate cells that have begun to express Jag1 but still have high levels of Six2 proteins. Arrow indicates cells in the distal part of the renal vesicle showing high levels of Six2 proteins. cm, cap mesenchyme; pta, pretubular aggregate; rv, renal vesicle; sb, S-shaped body. Scale bars: 50 μm.
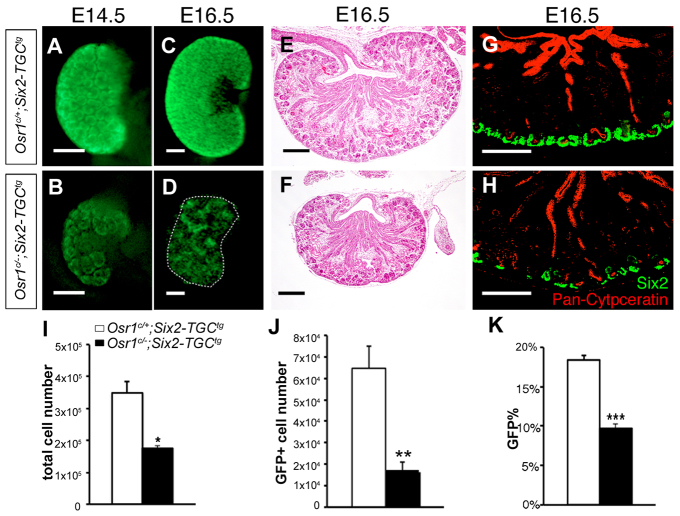
To investigate the role of Osr1 in metanephric kidney development, we first used the Six2-TGCtg BAC transgenic mice, which express an eGFP-Cre fusion protein in the CM (Kobayashi et al., 2008), in combination with Osr1c/c mice (Lan et al., 2011) to specifically inactivate Osr1 in the CM at the onset of metanephric kidney development. We initially analyzed kidneys from Osr1c/+;Six2-TGCtg control and Osr1c/-;Six2-TGCtg mutant embryos at E14.5 and E16.5. Morphologically, Osr1c/-;Six2-TGCtg kidneys (Fig. 2B,D) were significantly smaller than those of control littermates (Fig. 2A,C). In Osr1c/-;Six2-TGCtg kidneys, eGFP levels were obviously reduced by E14.5 (Fig. 2B), in comparison with control littermates (Fig. 2A). HE staining of frontal sections of E16.5 kidneys showed that Osr1c/-;Six2-TGCtg kidneys had differentiated glomeruli structures (Fig. 2F), although the mutant kidney was much smaller than the Osr1c/+;Six2-TGCtg control kidneys (Fig. 2E). Immunofluorescent detection of Six2 and pan-cytokeratin showed that the Osr1c/-;Six2-TGCtg kidneys had significantly reduced Six2+ CM cells (Fig. 2H) compared with littermate control kidneys (Fig. 2G). Quantification of GFP+ cells by using fluorescence-activated cell sorting (FACS) showed that the total cell number in Osr1c/-;Six2-TGCtg kidneys was reduced to ∼50% of that in Osr1c/+;Six2-TGCtg/+ kidneys at E14.5 (Fig. 2I). Moreover, whereas ∼18% of the cells were GFP+ in the E14.5 Osr1c/+;Six2-TGCtg control kidneys, <10% of the cells in the E14.5 Osr1c/-;Six2-TGCtg kidneys were GFP+ (Fig. 2J,K). These results indicate that Osr1 is required for cap mesenchyme maintenance and normal kidney development.
Fig. 2.
Tissue-specific inactivation of Osr1 in the cap mesenchyme causes renal hypoplasia. (A-D) Whole-mount images of GFP expression in Osr1c/-;Six2-TGCtg (B,D) and Osr1c/+;Six2-TGCtg (A,C) kidneys. (E,F) Frontal sections of E16.5 kidneys from Osr1c/-;Six2-TGCtg embryos (F) and Osr1c/+;Six2-TGCtg littermates (E). (G,H) Immunofluorescence detection of Six2 (green) and pan-cytokeratin (red) in Osr1c/-;Six2-TGCtg (H) and Osr1c/+;Six2-TGCtg (G) kidneys. Scale bars: 200 μm. (I-K) Comparison of total cell number (I), GFP-positive cell number (J) and percentage of GFP-positive cells (K) in E14.5 Osr1c/-;Six2-TGCtg and Osr1c/+;Six2-TGCtg/+ control kidneys. Error bars indicate s.e.m. *P<0.05; **P<0.01; ***P<0.001.
Osr1 plays an important role in maintaining the progenitor state of the CM cells
To define the onset and progression of kidney developmental defects in Osr1c/-;Six2-TGCtg mutant embryos, we analyzed the patterns of UB branching morphogenesis and nephrogenic differentiation from E11.5 to E13.5 in Osr1c/+;Six2-TGCtg (Fig. 3A,C,E) and Osr1c/-;Six2-TGCtg (Fig. 3B,D,F) embryos. At E11.5 and E12.5, the CM cells were similarly condensed around the UBs in Osr1c/+;Six2-TGCtg (Fig. 3A,C) and Osr1c/-;Six2-TGCtg (Fig. 3B,D) kidneys. At E13.5, the UB had branched multiple times and renal vesicles appeared on the medullary side of the UB branches in Osr1c/+;Six2-TGCtg control embryos (Fig. 3E). The Osr1c/-;Six2-TGCtg kidneys were smaller and had fewer ureteric bud branches (Fig. 3E,F). Moreover, Six2+ CM cells formed aggregates in some cortical regions in Osr1c/-;Six2-TGCtg kidneys (Fig. 3F). Histological analysis also detected aberrant aggregates of CM cells distal or cortical to the UB tips in the E13.5 Osr1c/-;Six2-TGCtg kidneys (supplementary material Fig. S1). These results suggest that the Osr1-deficient CM cells undergo premature differentiation.
Fig. 3.
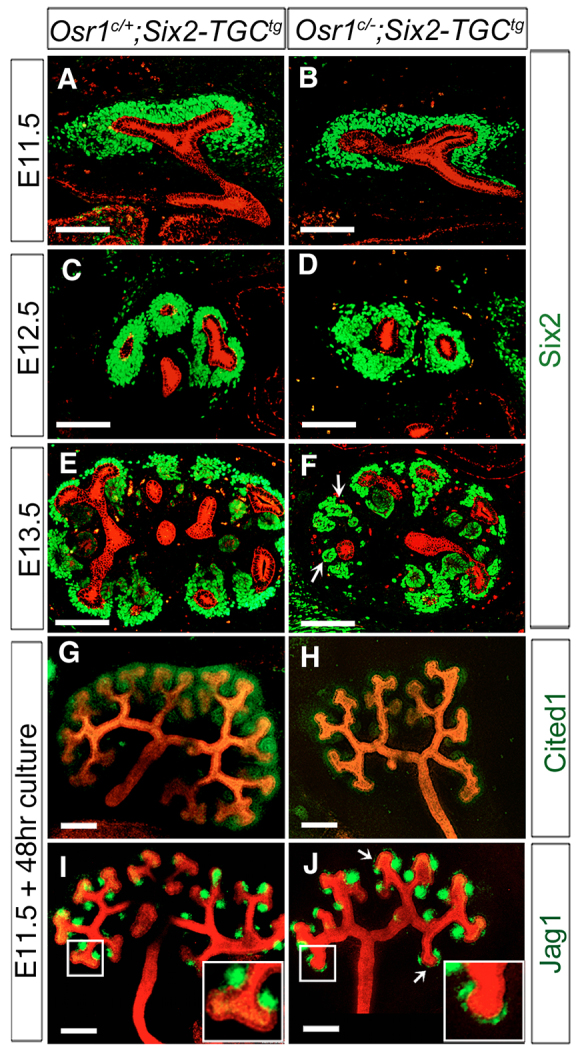
Tissue-specific inactivation of Osr1 in the cap mesenchyme causes premature differentiation. (A-F) Immunofluorescent detection of Six2 (green) and pan-cytokeratin (red) in developing kidneys of Osr1c/+;Six2-TGCtg (A,C,E) and Osr1c/-;Six2-TGCtg (B,D,F) embryos at E11.5 (A,B), E12.5 (C,D) and E13.5 (E,F). Arrows in F indicate abnormal mesenchymal aggregates in Osr1c/-;Six2-TGCtg kidneys. Scale bars: 100 μm. (G-J) Whole-mount immunofluorescence detection of Cited1 (green, G,H) or Jag1 (green, I,J) and pan-cytokeratin (red) in cultured kidney explants. Arrows in J indicate ectopic Jag1 expression in the CM distal to the UB tips. Insets in the lower right-hand corners of I and J show higher magnification pictures of the respective boxed region. Scale bars: 200 μm.
To further investigate the defects in kidney organogenesis in Osr1c/-;Six2-TGCtg mutant embryos, we analyzed kidney morphogenesis using organ cultures. The UBs underwent repeated branching in culture, similar to their branching morphogenesis in vivo, but the explants appear relatively flat and provide easy visualization of the pattern of the collecting duct tree. After 48 hours of culture, explants from E11.5 Osr1c/+;Six2-TGCtg control embryos averaged six to seven rounds of UB branching, with Cited1+ undifferentiated CM maintained at the cortical region of the developing kidneys and Jag1+ renal vesicles forming on the medullary side of each new UB branch (Fig. 3G,I). In explants from Osr1c/-;Six2-TGCtg embryos, there were fewer UB branches and significantly reduced Cited1+ immunostaining of the CM cells after 48 hours of culture (Fig. 3H). Consistent with the observation of aberrant cell aggregates in the cortical regions of the Osr1c/-;Six2-TGCtg kidneys in vivo (Fig. 3F; supplementary material Fig. S1), ectopic Jag1+ aggregates are detected in the CM cortical to the UB tips in Osr1c/-;Six2-TGCtg mutant kidney explants (Fig. 3J). These results indicate that Osr1 function is required for maintenance of the progenitor state of the CM cells during kidney organogenesis.
Osr1 acts downstream of Six2 in the CM
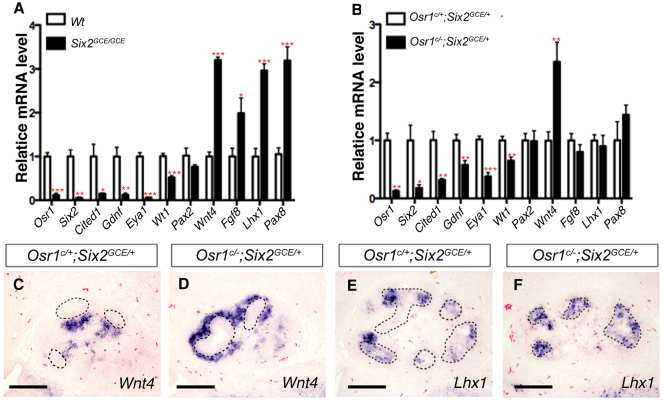
Previous studies have shown that Six2 is required for CM cell maintenance and self-renewal (Self et al., 2006; Kobayashi et al., 2008). Expression of a number of well-characterized genes required for normal kidney development, including Gdnf, Eya1, Wt1, Pax2, Pax8, Wnt4, Fgf8 and Lhx1, was altered at E11.5 and E12.5 in Six2 mutant embryos (Kreidberg et al., 1993; Stark et al., 1994; Moore et al., 1996; Pichel et al., 1996; Sánchez et al., 1996; Xu et al., 1999; Brophy et al., 2001; Grieshammer et al., 2005; Kobayashi et al., 2005; Self et al., 2006; Park et al., 2007; Kobayashi et al., 2008). To investigate whether the kidney developmental defects in the Osr1c/-;Six2-TGCtg embryos was due to loss of Six2 function, we carried out quantitative real-time RT-PCR analysis of expression of Osr1, Six2 and several other important kidney developmental regulators at E12.5, prior to the observed morphological defect in the mutant kidney. As shown in Fig. 4, the level of Osr1 mRNAs in the Osr1c/-;Six2-TGCtg mutant kidney was reduced to 12% of that in the Osr1c/+;Six2-TGCtg control kidney by E12.5 when analyzed using total RNAs isolated from the whole-kidney samples. Moreover, full-length Osr1 mRNA was not detectable in FACS-isolated GFP+ cells from E12.5 Osr1c/-;Six2-TGCtg mutant kidneys, confirming that the Osr1 gene was effectively inactivated in the Osr1c/-;Six2-TGCtg CM cells (Fig. 4A,B). Interestingly, whereas Cited1 mRNA level was significantly reduced in the Osr1c/-;Six2-TGCtg kidneys, especially in the FACS-isolated GFP+ cells (Fig. 4A,B), Six2 mRNA level was not significantly reduced in Osr1c/-;Six2-TGCtg whole kidneys or in FACS-isolated GFP+ cells compared with the Osr1c/+;Six2-TGCtg control kidneys at E12.5 (Fig. 4A,B). Remarkably, we found that expression of Wnt4 mRNAs was significantly increased in the FACS-isolated GFP+ cells from the E12.5 mutant kidneys in comparison with the Osr1c/+;Six2-TGCtg control kidneys (Fig. 4A,B). In situ hybridization assays confirmed that expression of Wnt4 mRNAs was ectopically activated in the CM in Osr1c/-;Six2-TGCtg mutant kidneys by E12.5, in comparison with the control littermates (Fig. 4C,D). These data indicate that Osr1 function is required for maintenance of the progenitor state of the CM even in the presence of normal levels of Six2 expression.
Fig. 4.
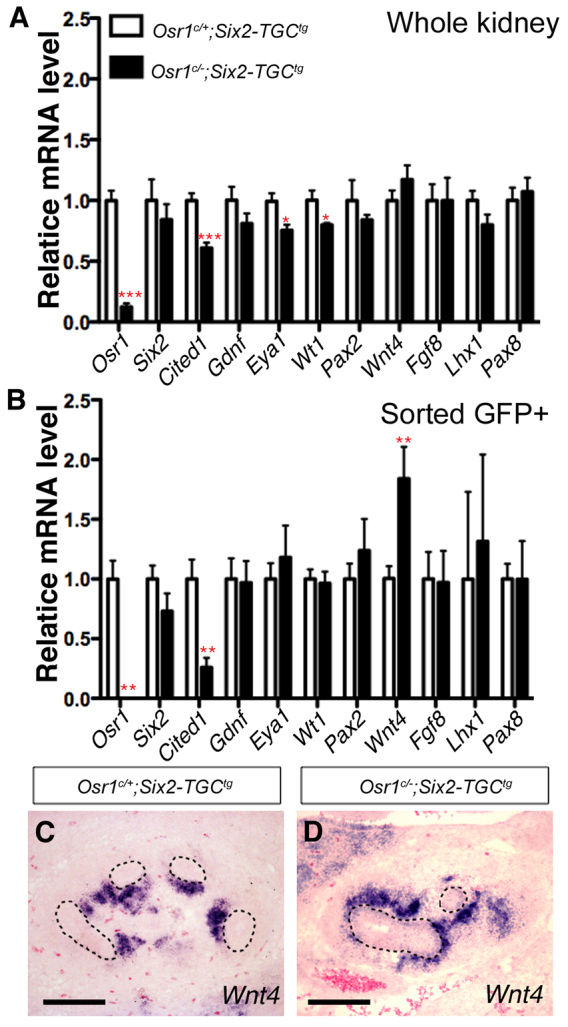
Comparison of gene expression in E12.5 Osr1c/-;Six2-TGCtg mutant and control kidney samples. (A,B) Real-time RT-PCR analysis of cap mesenchyme and nephron differentiation markers in E12.5 Osr1c/-;Six2-TGCtg whole kidney (A) and FACS-isolated GFP+ cells (B). Error bars indicate s.e.m. *P<0.05; **P<0.01; ***P<0.001. (C,D) Section in situ hybridization detection of Wnt4 mRNA in the E12.5 Osr1c/+;Six2-TGCtg (C) and Osr1c/-;Six2-TGCtg (D) kidneys. UB epithelial structures are circled with dashed lines. Scale bars: 100 μm.
To investigate further the relationship between Osr1 and Six2 in the maintenance of the progenitor state of the CM cells during kidney development, we examined Osr1 expression in Six2-null mutant embryos by section in situ hybridization. For this study, we used mice carrying the Six2GCE allele, which contains an eGFPCreERT2 fusion protein expression cassette inserted at the translation initiation codon position in the Six2 locus and thus represents a Six2-null allele (Kobayashi et al., 2008). As shown in Fig. 5, we found that Osr1 mRNA expression was dramatically downregulated in Six2GCE/GCE mutant embryos as early as E11.5 when the UB was undergoing the first round of branching (Fig. 5A,B). These data indicate that Six2 is required for the maintenance of Osr1 mRNA expression in the CM.
Fig. 5.
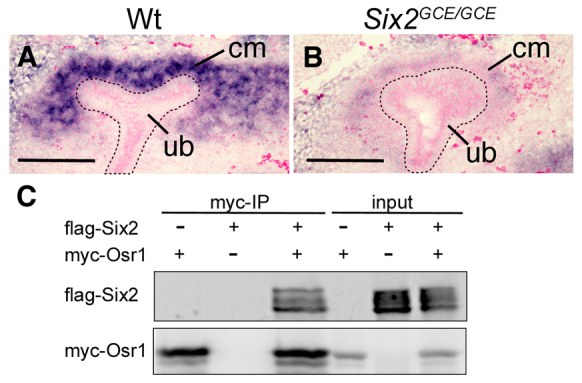
Six2 is required to maintain expression of Osr1 in metanephric mesenchyme. (A,B) Section in situ hybridization detection of Osr1 mRNA expression in E11.5 wild-type (A) and Six2GCE/GCE (B) kidneys. Scale bars: 100 μm. (C) Co-immunoprecipitation analysis of Osr1 and Six2 using co-transfected HEK297T cells.
Osr1 interacts and acts synergistically with Six2 to maintain the progenitor state of the CM
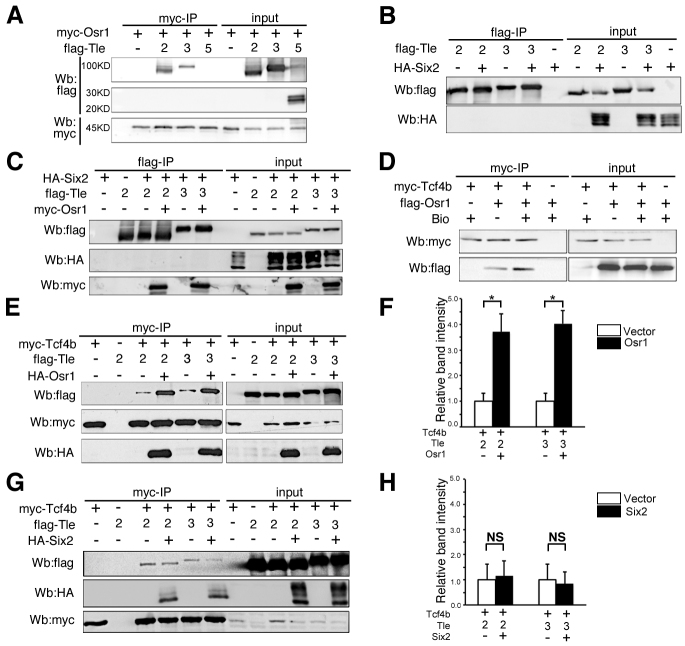
As Osr1 and Six2 are co-expressed in the undifferentiated CM compartment and both are required for maintenance of the nephron progenitor cells, we analyzed whether Osr1 and Six2 proteins interact with each other. Owing to lack of a working antibody against the endogenous Osr1 protein, we performed co-immunoprecipitation assays using co-transfected HEK293T human embryonic kidney cells. Immunoprecipitation of Myc-Osr1 indeed pulled down Flag-Six2 from the co-transfected cells (Fig. 5C), indicating that Osr1 and Six2 could form interactive protein complexes when co-expressed.
Previous studies showed that Six2-/- mutant embryos exhibit premature depletion of nephron progenitor cells as early as E12.5 but Six2+/- heterozygous mice develop normal kidneys (Self et al., 2006; Kobayashi et al., 2008). With our findings that Osr1 also plays a crucial role in the maintenance of nephron progenitors and that Osr1 expression is downregulated in the Six2-/- CM, we next tested whether Osr1 and Six2 interact genetically to regulate kidney organogenesis. We crossed Osr1+/- mice to Six2GCE/+ mice and found that Osr1+/-;Six2GCE/+ double heterozygous mice had normal kidneys. We then crossed Osr1+/-;Six2GCE/+ double heterozygous male mice with Osr1c/c female mice and injected the pregnant female mice with 2 mg tamoxifen at E10.5 to induce Cre-mediated inactivation of the Osr1c allele in the CM cells in embryos carrying the Six2GCE allele. Analysis of embryonic kidneys at E16.5 showed that Osr1c/-;Six2GCE/+ embryos had severely hypoplastic kidneys (Fig. 6B,D), in comparison with Osr1c/+;Six2GCE/+ control embryos (Fig. 6A,C). Histological analysis showed that the nephrogenic zone was absent in E16.5 Osr1c/-;Six2GCE/+ kidneys (Fig. 6E,F) Immunofluorescence staining using antibodies against Six2 and pan-cytokeratin antibodies showed that Six2+ CM cells were absent and UB branching was disrupted in E16.5 Osr1c/-;Six2GCE/+ kidneys (Fig. 6G,H). These results indicate that nephrogenic progenitor cells were prematurely depleted during early metanephric kidney organogenesis in Osr1c/-;Six2GCE/+ mutant embryos.
Fig. 6.
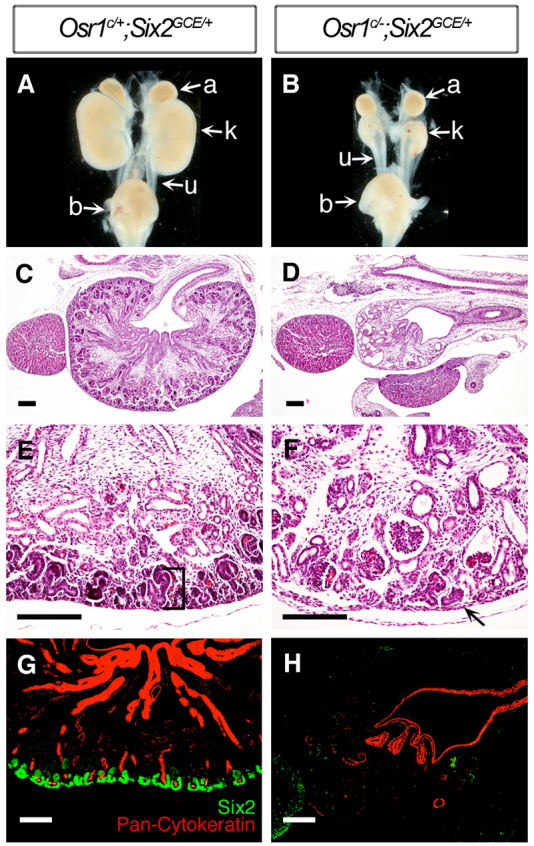
The Osr1c/-;Six2GCE/+ embryos exhibit premature depletion of nephron progenitor cells. (A,B) Whole-mount view of the kidneys from E16.5 Osr1c/+;Six2GCE/+ (A) and Osr1c/-;Six2GCE/+ (B) littermates. (C-F) Histological analysis of E16.5 kidney sections of Osr1c/-;Six2GCE/+ embryos (D,F) and Osr1c/+;Six2GCE/+ littermates (C,E). Osr1c/-;Six2GCE/+ mutant kidney had lost the nephrogenic zone (F, arrow). (G,H) Immunofluorescence detection of Six2 (green) and pan-cytokeratin (red) in E16.5 Osr1c/+;Six2GCE/+ (G) and Osr1c/-;Six2GCE/+ (H) kidneys. Scale bars: 100 μm.
We analyzed the patterns of UB branching and nephrogenic differentiation in Osr1c/-;Six2GCE/+ mutant embryos from E11.5 to E13.5, in comparison with Osr1c/+;Six2GCE/+ littermates. At E11.5, the first round of UB branching and the condensation of CM cells around the UB tips occurred normally in Osr1c/-;Six2GCE/+ embryos (Fig. 7A,B). At E12.5, whereas the CM cells were condensed around the UBs in the controls (Fig. 7C), the CM cells formed multiple pretubular aggregates around the UBs in Osr1c/-;Six2GCE/+ embryos (Fig. 7D, arrows). By E13.5, the UB had branched multiple times and renal vesicles formed on the medullary side of the UB branches in control embryos (Fig. 7E). The E13.5 Osr1c/-;Six2GCE/+ kidneys were much smaller than the controls and almost all Six2+ cells appeared in large epithelialized aggregates around the UBs (Fig. 7F). Using explant culture assays, we found that Osr1c/-;Six2GCE/+ mutant kidneys had very limited UB branching and the CM cells lost Cited1 expression and differentiated into several large Jag1-positive epithelialized structures within 48 hours of culture (Fig. 7G-J).
Fig. 7.
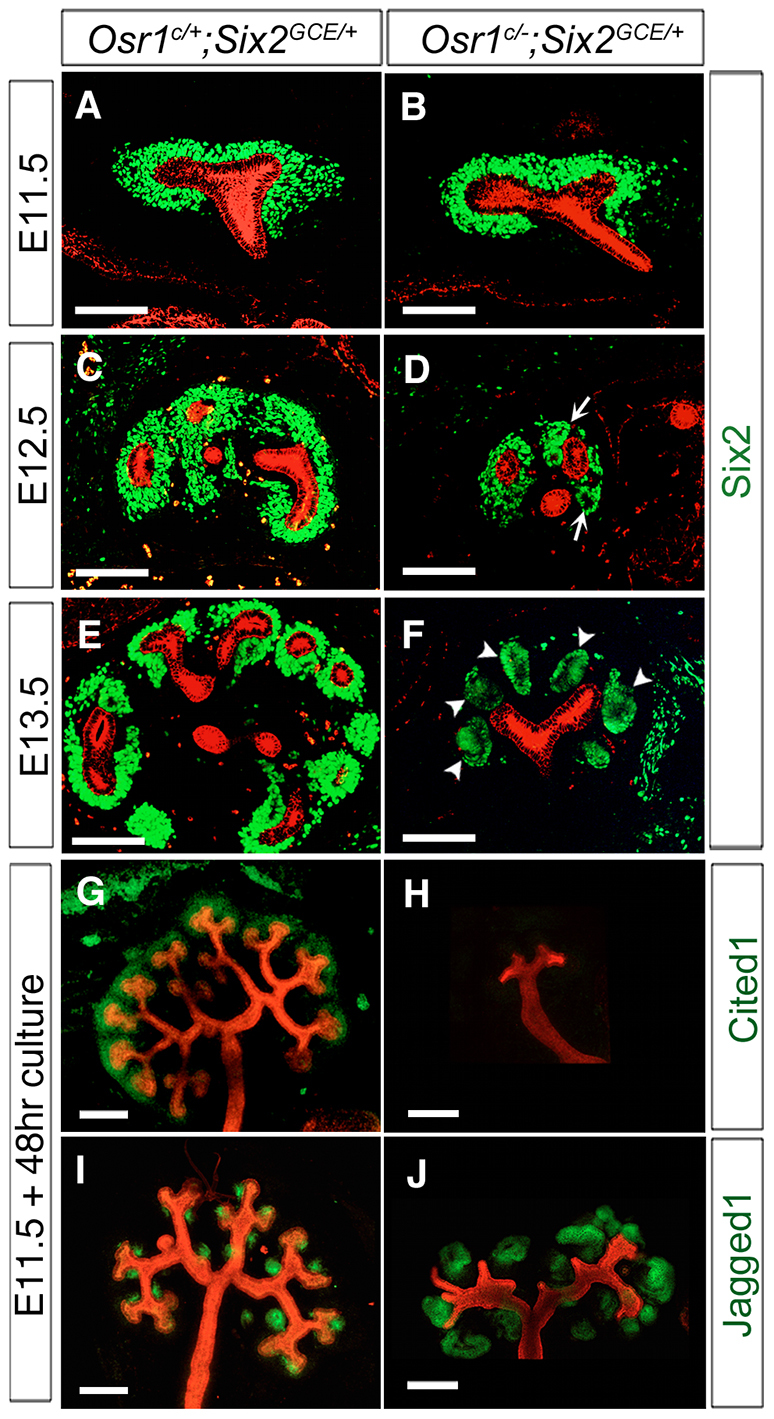
Analysis of cap mesenchyme and UB branching in the Osr1c/-;Six2GCE/+ mutant embryos. (A-F) Immunofluorescence detection of Six2 (green) and pan-cytokeratin (red) in developing kidneys of Osr1c/+;Six2GCE/+ (A,C,E) and Osr1c/-;Six2GCE/+ (B,D,F) embryos at E11.5 (A,B), E12.5 (C,D) and E13.5 (E,F). Arrows in D indicate abnormal mesenchymal aggregates surrounding the UB in the E12.5 Osr1c/-;Six2GCE/+ mutant kidney. Arrowheads in F indicate large epithelializing aggregates in place of the CM surrounding the UBs. Scale bars: 100 μm. (G-J) Whole-mount immunofluorescence detection of Cited1 (green, G,H) or Jag1 (green, I,J) and pan-cytokeratin (red) in cultured Osr1c/+;Six2GCE/+ (G,I) and Osr1c/-;Six2GCE/+ (H,J) kidney explants. Scale bars: 200 μm.
The precocious differentiation of the CM and defects in UB branching in Osr1c/-;Six2GCE/+mutant kidneys are remarkably similar to the kidney developmental defects reported previously in Six2-/- mutant embryos (Self et al., 2006; Kobayashi et al., 2008). We compared the changes in expression of molecular markers for the CM as well as early differentiating nephron precursor cells in E12.5 Six2GCE/GCE and Osr1c/-;Six2GCE/+mutant kidneys by quantitative real-time RT-PCR. As shown in Fig. 8A, the mRNA levels of most of the genes normally expressed in the CM, including Osr1, Cited1, Gdnf, Eya1 and Wt1, were significantly reduced, whereas the mRNA levels of genes that are upregulated during nephrogenic differentiation, including Wnt4, Fgf8, Lhx1 and Pax8, were significantly increased in E12.5 Six2GCE/GCE kidneys (Fig. 8A), consistent with previous in situ hybridization studies (Self et al., 2006; Kobayashi et al., 2008). In E12.5 Osr1c/-;Six2GCE/+mutant kidneys, the level of full-length Osr1 mRNAs was reduced to 13% of that in the Osr1c/+;Six2GCE/+ control kidneys, confirming efficient inactivation of the Osr1c allele. The mRNA levels of the CM marker genes, including Cited1, Six2, Gdnf, Eya1 and Wt1, were all significantly reduced in the Osr1c/-;Six2GCE/+mutant kidneys in comparison with the Osr1c/+;Six2GCE/+controls (Fig. 8B). In particular, the level of Six2 mRNAs in the Osr1c/-;Six2GCE/+mutant kidneys was reduced by ∼80% in comparison with Osr1c/+;Six2GCE/+control kidneys. In contrast to Six2GCE/GCE mutants, however, of the nephrogenic differentiation markers examined, only Wnt4 expression was significantly upregulated in E12.5 Osr1c/-;Six2GCE/+ kidneys in comparison with the control kidneys (Fig. 8B). In situ hybridization analysis confirmed that Wnt4 mRNAs were ectopically expressed in the CM cells cortical to UB tips in E12.5 Osr1c/-;Six2GCE/+mutant kidneys (Fig. 8C,D). No ectopic expression of Lhx1 mRNAs was detected in the same embryonic kidneys (Fig. 8E,F). Thus, the precocious differentiation of the CM in Osr1c/-;Six2GCE/+mutant kidneys starts at about E12.5, later than that in Six2-null kidneys, which exhibit precocious differentiation of the CM at E11.5 (Self et al., 2006; Kobayashi et al., 2008), but earlier than that in Osr1c/-;Six2-TGCtg embryos. These data indicate that Osr1 and Six2 act synergistically to maintain the nephron progenitor cells.
Fig. 8.
Comparison of kidney gene expression in Six2GCE/GCE and Osr1c/-;Six2GCE/+ mutant embryos. (A,B) Real-time RT-PCR analysis of cap mesenchyme and nephron differentiation markers in E12.5 Six2GCE/GCE (A) and Osr1c/-;Six2GCE/+ mutant (B) kidneys. Error bars indicate s.e.m. *P<0.05; **P<0.01; ***P<0.001. (C-F) In situ hybridization detection of Wnt4 (C,D) and Lhx1 (E,F) mRNA expression in E12.5 Osr1c/+;Six2GCE/+ (C,E) and Osr1c/-;Six2GCE/+ (D,F) kidneys. Scale bars: 100 μm.
Osr1 interacts with Tcf4 and enhances Tcf4 interaction with Groucho family transcriptional co-repressors
It has been reported that the Drosophila odd-skipped protein repressed target gene expression during embryonic segmentation by interacting with the transcriptional co-repressor Groucho through an engrailed homology domain, eh1 (Goldstein et al., 2005). The vertebrate Osr1 and Osr2 proteins have a similar eh1 domain and overexpression studies in Xenopus embryos suggest that Osr1 and Osr2 also act as transcriptional repressors (Tena et al., 2007). The human and mouse genomes each contains six Groucho-related genes, which are also called transducin-like enhancer-of-split (Tle) genes, Tle1-Tle4, Tle6 and Aes/Tle5 (Buscarlet and Stifani, 2007). Of these, the protein products of the Tle1-Tle4 genes contain all structural motifs homologous to the Drosophila Groucho protein, whereas the Aes protein contains only the N-terminal regions while Tle6 protein is structurally significantly divergent. Aes and Tle1-Tle3 mRNAs are expressed in the cap mesenchyme of developing mouse kidneys (www.GUDMAP.org), whereas Tle4 expression is low in the mesenchyme cells and is increased in polarized renal vesicles and S-shaped bodies (Cai et al., 2003). Thus, we tested whether Osr1 was able to interact with Aes, Tle2 or Tle3. By using co-immunoprecipitation of co-transfected HEK293T cells, we detected robust interactions of Osr1 with Tle2 and Tle3, but not with Aes (Fig. 9A). By contrast, we did not detect any protein complex containing Six2 and Tle2 or Tle3 in co-transfected HEK293T cells (Fig. 9B). When co-transfected with both Osr1 and Six2 expression constructs, both Tle2 and Tle3 were co-immunoprecipitated with Osr1 but not Six2 (Fig. 9C).
Fig. 9.
Osr1 interacts with both TCF and Groucho/Tle proteins and stabilizes the repressor complex. (A) Osr1 forms complexes with Tle2 and Tle3, but not with Aes. (B) Six2 was not co-immunoprecipitated with Tle2 or Tle3. (C) When co-expressed with Osr1 and Six2, Tle2 and Tle3 were detected in complexes with Osr1 but not Six2. (D) Osr1 formed complexes with Tcf4b. (E,F) Addition of Osr1 significantly increased the amount of Tle2/3 proteins co-immunoprecipitated with Tcf4b. Error bars indicate s.e.m. *P<0.05. (G,H) Six2 interacts with Tcf4b but does not enhance Tcf4b interaction with Tle2/3. NS, not significantly different.
The TCF/Lef family proteins have been shown to interact with Groucho-related proteins to repress Wnt target gene expression (Cavallo et al., 1998; Roose et al., 1998). We found that Osr1 was co-immunoprecipitated with Tcf4b in co-transfected HEK293T cells (Fig. 9D). Moreover, whereas Tcf4b interacted weakly with either Tle2 or Tle3 in co-transfected HEK293T cells, addition of Osr1 significantly increased the amount of Tle2/3 protein co-immunoprecipitated with Tcf4b (Fig. 9E,F). By contrast, although Six2 has also been shown to interaction with Tcf4b (Park et al., 2012), co-transfection of Six2 and Tcf4b did not have any significant effect on Tcf4b interaction with either Tle2 or Tle3 (Fig. 9G,H). Together, these data suggest that, when co-expressed with TCF/Lef (e.g. as in the CM in developing kidneys), Osr1 can stabilize interactions between TCF/Lef and the Groucho family co-repressors.
Evidence that Osr1 inhibits Wnt/β-catenin mediated activation of Wnt4 enhancer in the CM cells in vivo
By performing chromatin immunoprecipitation followed by high-throughput sequencing, Park et al. (Park et al., 2012) found that Six2 and β-catenin could co-occupy 130 cis-regulatory elements in the developing kidney, including an enhancer element ∼60 kb upstream of the Wnt4 gene. Remarkably, whereas this 350 bp Wnt4 enhancer directed specific reporter gene expression in the developing renal vesicles in the Wnt4e-lacZ transgenic mouse embryos, mutating the two TCF-binding sites in the enhancer completely abolished reporter expression in the developing kidneys (Park et al., 2012), indicating that Wnt/β-catenin activation of this Wnt4 enhancer during renal vesicle formation is mediated by TCF. As our in vitro co-immuoprecipitation data suggested that Osr1, TCF and Groucho family members form stable repressor complexes, we hypothesized that the Wnt4e-lacZ reporter would be ectopically activated in the CM cells in Osr1-deficient kidneys. To test this hypothesis, we generated Wnt4e-lacZ transgenic mice and crossed the Wnt4e-lacZ reporter into the Osr1c/c;Six2GCE/+ mice. As shown in Fig. 10, whereas the Wnt4e-lacZ reporter was strongly expressed in the newly formed renal vesicles but absent from the CM in Osr1c/+;Six2GCE/+;Wnt4e-lacZtg mouse kidney (Fig. 10A,C), it was ectopically activated throughout the CM and renal vesicles in Osr1c/c;Six2GCE/+;Wnt4e-lacZtg mutant kidneys by E12.5 (Fig. 10B,D). Together, these data indicate that Osr1 prevents premature differentiation of CM cells through enhancing TCF/Tle-mediated inhibition of Wnt/β-catenin target genes, including Wnt4.
Fig. 10.
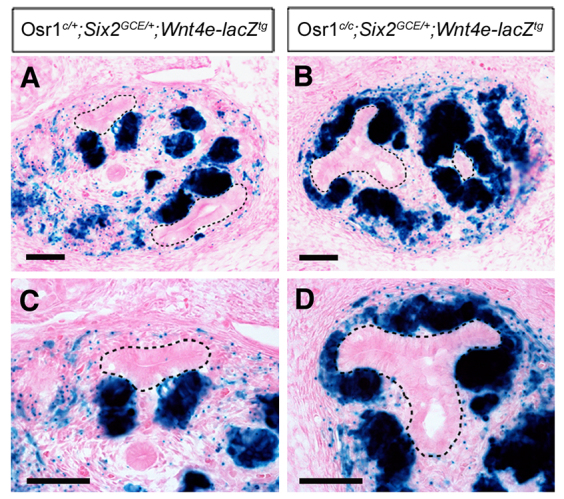
Osr1 maintains nephron progenitor cells by inhibiting β-catenin/TCF-mediated activation of Wnt4 expression. (A,B) X-gal staining of frozen sections of E12.5 Osr1c/+;Six2GCE/+;Wnt4e-lacZtg (A) and Osr1c/c;Six2GCE/+;Wnt4e-lacZtg (B) embryos. Black dashes outline UBs. (C,D) Higher magnification views of kidney sections showing restricted lacZ expression in the renal vesicles in Osr1c/+;Six2GCE/+;Wnt4e-lacZtg control (C) and ectopic lacZ expression in the cap mesenchyme in Osr1c/+;Six2GCE/+;Wnt4e-lacZtg mutant (D) kidneys. Scale bars: 50 μm.
DISCUSSION
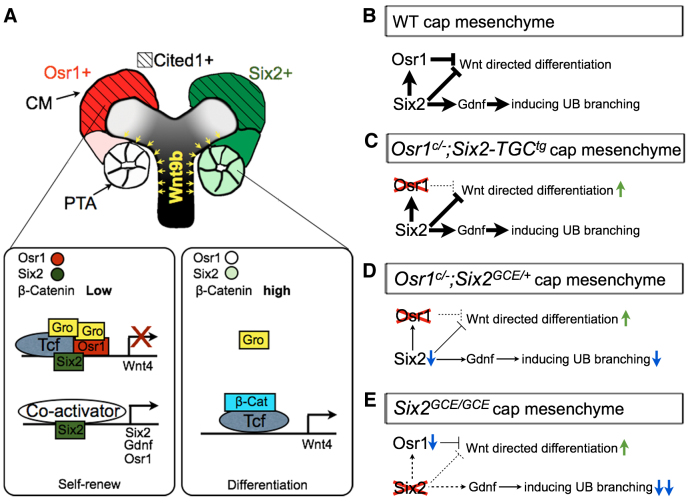
Wnt9b induces nephrogenic differentiation through the canonical Wnt signaling pathway in which activation of Wnt receptors leads to stabilization and nuclear translocation of β-catenin, which binds to the TCF/Lef family of transcription factors to regulate downstream target genes (Cadigan and Nusse, 1997; Carroll et al., 2005; Park et al., 2007; Angers and Moon, 2009; Karner et al., 2011). The TCF/Lef transcription factors interact with a number of transcriptional co-repressors, including CtBP and members of the Groucho/Tle family (Cavallo et al., 1998; Levanon et al., 1998; Roose et al., 1998; Waltzer and Bienz, 1998; Brannon et al., 1999; Brantjes et al., 2001; Park et al., 2012). As these co-repressors are broadly expressed, TCF/Lef factors function primarily as transcriptional repressors in the absence of β-catenin (Willert and Jones, 2006). Conversion of TCF/Lef to transcriptional activator involves functional competition between β-catenin and Groucho for binding to TCF (Cavallo et al., 1998; Roose et al., 1998; Range et al., 2005). Roose et al. (Roose et al., 1998) demonstrated that high levels of Groucho overexpression were able to overcome β-catenin-mediated activation even when the transactivation domain of β-catenin was fused directly to the TCF molecule, indicating that the outcome of canonical Wnt signaling depends on relative levels of TCF/Groucho repressor and TCF/β-catenin activator complexes (Roose et al., 1998; Range et al., 2005). During kidney organogenesis, the regions where CM cells undergo mesenchymal-epithelial transition correlates with highest levels of Wnt9b activity as Wnt9b is expressed at higher levels in the UB stalk than in UB tip cells (Carroll et al., 2005). In this study, we found that Osr1 interacts with both TCF and Groucho family proteins and enhanced TCF/Groucho complex formation. Together with the findings of premature differentiation of the CM in Osr1c/-;Six2-TGCtg and Osr1c/-;Six2GCE/+ mutant kidneys, these data indicate that Osr1 acts to maintain the nephron progenitor cell pool in the CM by stabilizing the TCF/Groucho repressor complexes to antagonize Wnt/β-catenin-driven differentiation (Fig. 11A). Our finding that the Wnt4e-lacZ transgene is ectopically activated in the CM in Osr1c/c;Six2GCE/+;Wnt4e-lacZtg mutant kidneys further supports this conclusion.
Fig. 11.
A model depicting the molecular mechanism involving Osr1, Six2 and Wnt9b/β-catenin signaling in the regulation of nephron progenitor renewal and differentiation. (A,B) In wild-type kidney, Osr1 and Six2 interact and form a strong repressor complex with TCF and Groucho/Tle proteins to prevent the activation of Wnt/β-catenin target genes such as Wnt4 in the CM. Six2 also acts independently of Osr1 to activate expression of many CM genes, including itself, Gdnf and Osr1. The CM cells in the ‘armpit’ of new UB branches receive the highest level of Wnt9b signaling, which converts TCF from repressor to activator and activates expression of differentiation genes, including Wnt4. (C) In Osr1c/-;Six2-TGCtg mutant embryos, Osr1 deficiency in the CM causes premature differentiation but Six2 is still expressed at high levels and maintains expression of many CM-specific genes, including Gdnf. (D) In Osr1c/-;Six2GCE/+ mutant embryos, loss of Osr1 and significant downregulation of Six2 cause earlier onset of premature nephrogenic differentiation and reduced UB branching than in Osr1c/-;Six2-TGCtg mutant embryos. (E) In Six2GCE/GCE mutant embryos, the complete lack of Six2 causes rapid loss of both Gdnf and Osr1, resulting in rapid depletion of CM cells as well as lack of UB branching.
Whereas both Osr1c/-;Six2-TGCtg and Osr1c/-;Six2GCE/+ mutant embryos exhibit premature nephrogenic differentiation accompanied by ectopic Wnt4 mRNA expression, the phenotypic onset is earlier and more synchronized in Osr1c/-;Six2GCE/+ embryos than in Osr1c/-;Six2-TGCtg embryos (compare Fig. 7 with Fig. 3). Two factors probably contributed to these results. First, the Six2-TGCtg transgenic embryos exhibit slight mosaicism in Cre expression (Kobayashi et al., 2008). It is likely that some CM cells retained a functional Osr1 allele in the early stages of kidney development in Osr1c/-;Six2-TGCtg embryos. Second, whereas Osr1c/-;Six2-TGCtg embryos had nearly normal levels of Six2 mRNA expression at E12.5 (Fig. 4), the Osr1c/-;Six2GCE/+ mutant embryos showed dramatic downregulation of Six2 mRNA expression by E12.5 (Fig. 8). The dramatic downregulation of Six2 mRNA expression might have caused the amount of Six2 protein to fall below a threshold required for CM self-renewal. The significant decrease in expression of a number of CM marker genes in E12.5 Osr1c/-;Six2GCE/+ mutant kidneys compared with Osr1c/+;Six2GCE/+ littermates support this explanation. However, several nephron induction genes that were ectopically activated in E12.5 Six2-/- kidneys, including Fgf8, Lhx1 and Pax8, were not significantly increased in E12.5 Osr1c/-;Six2GCE/+ mutant kidneys, indicating that other Six2-dependent factors also contribute to suppressing nephrogenic differentiation. Thus, in wild-type embryos, Six2 maintains expression of Osr1 and other important effectors to maintain the nephron progenitor cells against Wnt-directed differentiation as well as induces UB branching though positive regulation of Gdnf (Fig. 11B). In Osr1c/-;Six2-TGCtg embryos, even though Six2 expression in the CM is maintained at high levels, Osr1 deficiency causes premature activation of some Wnt target genes, such as Wnt4, and results in accelerated differentiation of nephron progenitors (Fig. 11C). In Osr1c/-;Six2GCE/+ mutant embryos, Osr1 deficiency in combination with significant downregulation of Six2 expression causes both reduced UB branching and premature CM differentiation (Fig. 11D). In Six2-/- embryos, expression of both Gdnf and Osr1 in the CM is downregulated, resulting in lack of UB branching and synchronous premature differentiation of the CM (Fig. 11E).
By direct comparison of the expression patterns of Osr1-eGFP with Cited1, Six2 and Jag1 in the E13.5 kidney, we found a Cited1-Osr1+Six2+ transition zone between the Cited1+Osr1+Six2+ progenitor cells and the pretubular aggregates (Fig. 1E,F). Our results confirm the recent finding of Brown et al. (Brown et al., 2013), who showed that a Six2+-only CM compartment is present in between the Cited1+Six2+ CM cells and Lef1+ pretubular aggregates in the developing mouse kidneys. Brown et al. (Brown et al., 2013) suggested that the Cited1+Six2+ nephron progenitor cells are first primed by Bmp7 signaling and transition to the Six2+-only compartment where they become inducible by Wnt/β-catenin signaling (Brown et al., 2013). As Osr1 interacts with Tcf/Tle to repress β-catenin/TCF-driven Wnt4 gene expression in the CM, downregulation of Osr1 expression is an important part of the priming mechanism during nephron formation. Whereas our data demonstrate that Osr1 acts downstream of Six2 to maintain the progenitor state of the CM, further studies are necessary to elucidate the molecular mechanism that turns off Osr1 expression at the onset of pretubular aggregate formation.
MATERIALS AND METHODS
Mouse strains
The Osr1+/- (Osr1tm1Jian), Osr1c/c, Osr1GCE/+, Six2-TGCtg BAC transgenic, Six2GCE/+ and Wnt4e-lacZ transgenic mice have been described previously (Wang et al., 2005; Kobayashi et al., 2008; Mugford et al., 2008; Lan et al., 2011; Park et al., 2012) and were maintained by crossing to C57BL/6J mice. To inactivate Osr1 in the CM, Osr1+/- mice were first crossed with either Six2GCE/+ or Six2-TGCtg mice to generate Osr1+/-;Six2GCE/+ or Osr1+/-;Six2-TGCtg double heterozygous mice, which were then crossed with Osr1c/c mice to generate Osr1c/-;Six2GCE/+ and Osr1c/-;Six2-TGCtg mutant embryos, respectively. Noon of the day a vaginal plug was identified was designated as embryonic day (E) 0.5. To activate Cre recombinase in embryos carrying the Six2GCE allele, pregnant dams were injected intraperitoneally at E10.5 with 2 mg of tamoxifen (Sigma, T5648) (Kobayashi et al., 2008).
Histology, X-gal and immunofluorescent staining
For histological analysis, embryos were dissected at desired stages from timed pregnant mice. Embryos were fixed in 4% paraformaldehyde (PFA), dehydrated through an ethanol series, embedded in paraffin, sectioned at 7 μm, and stained with Hematoxylin and Eosin. Immunofluorescent staining was performed using paraffin sections or frozen sections following standard protocols. Antibodies used include rabbit anti-Six2 (ProteinTech, 11562-1-AP; 1:200), mouse anti-Pan cytokeratin (Sigma; 1:100), rabbit anti-Jag1 (Santa Cruz, sc-8303; 1:50), rabbit anti-Cited1 (Thermo, RB-9219-P0; 1:50) and chicken anti-GFP (Aves labs, GFP-1010; 1:50).
For X-gal staining, embryos were dissected at pre-determined stages and fixed in 1% PFA at 4°C overnight. After fixation, embryos were washed with PBS three times, soaked in 30% sucrose (Sigma), embedded in Neg-50 (Thermo), and sectioned at 14 μm using a cryostat microtome. Sections were post-fixed in 0.2% PFA, washed in LacZ rinse buffer (0.1 M phosphate buffer, PH 7.3, 0.02% NP-40, 0.01% sodium deoxycholate, 2 mM MgCl2), and stained by immersion in X-gal staining solution (0.1 M NaPO4 buffer, PH 7.3, 2 mM MgCl2, 0.02% NP-40, 0.01% sodium deoxycholate, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 0.1% X-gal) overnight at 37°C in the dark. The X-gal-stained sections were counterstained with Eosin.
Kidney explant culture and whole-mount immunofluorescence staining
The metanephric rudiments were dissected from E11.5 mouse embryos and positioned on a filter membrane (1.0 μm pore size, BD Falcon, 353102) in individual wells of a six-well tissue culture plate and cultured in DMEM/F12 media plus 10% fetal bovine serum (Invitrogen). The explant cultures were maintained at 37°C at an atmosphere of 5% CO2 and 100% humidity.
After 48 hours, the kidney explants were fixed in methanol for 10 minutes, washed with PBST (PBS with 0.1% Tween 20) and blocked with 2.5% goat serum, 5% BSA in PBST for 1 hour. The explants were incubated in primary antibodies diluted in blocking buffer overnight at 4°C. Explants were washed three times in PBST, with the final wash extending overnight at 4°C. Cultures were incubated with secondary antibodies for 3 hours at room temperature and washed three times in PBST. All specimens were examined and photographed using a Nikon inverted confocal microscope.
Quantitative real-time RT-PCR
The metanephric kidneys were dissected from E12.5 mouse embryos in cold DEPC-treated PBS and immediately frozen -70°C. Total RNAs were isolated using the Qiagen RNeasy Micro kit (Qiagen, 74004). First-strand cDNAs were synthesized using SuperScript First-Strand Synthesis System (Invitrogen, 11904-018). Primers for specific transcripts were designed for real-time RT-PCR (SYBR). β-Actin was used as internal control in each reaction. Real-time PCR was performed using a Bio-Rad CFX96 Real-Time System using conditions recommended by the manufacturer. Each reaction was performed in triplicate. The quantity of each mRNA was first determined using a standard curve method and normalized to the internal control. The primers used for real-time RT-PCR are listed in supplementary material Table S1.
In situ hybridization
In situ hybridization was performed as previously described (Zhang et al., 1999). At least three embryos of each genotype were hybridized to each probe and only probes that detected consistent patterns of expression in all samples were considered as valid.
Co-immunoprecipitation
The Osr1-coding sequence was subcloned into the pCS2, pCMV7.1 (Sigma) or pCMV-HA (Clontech) vectors to express Osr1 with Myc-, Flag- or HA-epitope tag. Tle2-, Tle3- and Aes-coding sequences were subcloned into pCMV7.1 (Sigma) vector with a Flag-tag. Flag- or Myc-tagged Six2 and Tcf4b expression vectors have been reported previously (Park et al., 2012).
For immunoprecipitation assays, HEK293T cells were co-transfected with plasmids as indicated. After transfection, cells were cultured in DMEM supplemented with 10% fetal bovine serum for 48 hours. The cells were lysed in RIPA buffer containing proteinase inhibitors (Santa Cruz, SC-24948). Whole cell lysate was incubated with anti-c-Myc antibody (4A6, Millipore, 16-219) conjugated to protein-G agarose beads, and rotated at 4°C overnight. The beads were washed three times with RIPA buffer. Western blot was performed using anti-Flag (M2, Sigma F3165), anti-c-Myc (4A6, Millipore 05-724) or anti-HA (Clontech 631207) antibodies. The intensity of detected bands on western blots was quantified using Photoshop Histogram Analysis from three independent experiments.
Fluorescence-activating cell sorting (FACS)
The metanephric kidneys of E12.5 and E14.5 embryos from Osr1c/c females crossed with Osr1+/-;Six2-TGCtg males were harvested and digested with the trypsin-EDTA solution (Invitrogen) at 37°C for 4 minutes. After inactivation of trypsin with DMEM containing 10% FBS, cells were dissociated by pipetting. The dissociated kidney cells were resuspended in PBS with 2% FBS and 10 mM EDTA, and filtered through a 40 μm nylon cell strainer (BD Falcon, 352340). GFP+ cells were isolated using BD FACSAria II.
Statistics
All results are presented as mean±s.e.m. All statistical analyses were performed using GraphPad Prism5 software. Two-tailed Student’s t-tests were used for comparisons between two groups. P-values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Steve Potter and Eric Brunskill for comments and discussions.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
R.J., Y.L. and J.X. conceived the project and designed experiments; J.X., H.L. and Y.L. performed experiments; J.-S.P. provided new reagents; J.X., H.L., J.-S.P., Y.L. and R.J analyzed and discussed the data; J.X. and R.J. wrote and revised the manuscript.
Funding
This work was supported by a National Institutes of Health/National Institute of Dental and Craniofacial Research grant [DE013681 to R.J.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.103283/-/DC1
References
- Angers S., Moon R. T. (2009). Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- Bertram J. F., Douglas-Denton R. N., Diouf B., Hughson M. D., Hoy W. E. (2011). Human nephron number: implications for health and disease. Pediatr. Nephrol. 26, 1529–1533 [DOI] [PubMed] [Google Scholar]
- Brannon M., Brown J. D., Bates R., Kimelman D., Moon R. T. (1999). XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126, 3159–3170 [DOI] [PubMed] [Google Scholar]
- Brantjes H., Roose J., van De Wetering M., Clevers H. (2001). All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29, 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy P. D., Ostrom L., Lang K. M., Dressler G. R. (2001). Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128, 4747–4756 [DOI] [PubMed] [Google Scholar]
- Brown A. C., Muthukrishnan S. D., Guay J. A., Adams D. C., Schafer D. A., Fetting J. L., Oxburgh L. (2013). Role for compartmentalization in nephron progenitor differentiation. Proc. Natl. Acad. Sci. USA 110, 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M., Stifani S. (2007). The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17, 353–361 [DOI] [PubMed] [Google Scholar]
- Cadigan K. M., Nusse R. (1997). Wnt signaling: a common theme in animal development. Genes Dev. 11, 3286–3305 [DOI] [PubMed] [Google Scholar]
- Cai Y., Brophy P. D., Levitan I., Stifani S., Dressler G. R. (2003). Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 22, 5522–5529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T. J., Park J. S., Hayashi S., Majumdar A., McMahon A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283–292 [DOI] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. (1998). Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395, 604–608 [DOI] [PubMed] [Google Scholar]
- Cheng H. T., Kim M., Valerius M. T., Surendran K., Schuster-Gossler K., Gossler A., McMahon A. P., Kopan R. (2007). Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development 134, 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F., Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R. (2006). The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22, 509–529 [DOI] [PubMed] [Google Scholar]
- Dressler G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. (2008). Skin stem cells: rising to the surface. J. Cell Biol. 180, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. E., Cook O., Dinur T., Pisanté A., Karandikar U. C., Bidwai A., Paroush Z. (2005). An eh1-like motif in odd-skipped mediates recruitment of Groucho and repression in vivo. Mol. Cell. Biol. 25, 10711–10720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U., Cebrián C., Ilagan R., Meyers E., Herzlinger D., Martin G. R. (2005). FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development 132, 3847–3857 [DOI] [PubMed] [Google Scholar]
- James R. G., Kamei C. N., Wang Q., Jiang R., Schultheiss T. M. (2006). Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development 133, 2995–3004 [DOI] [PubMed] [Google Scholar]
- Karner C. M., Das A., Ma Z., Self M., Chen C., Lum L., Oliver G., Carroll T. J. (2011). Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 138, 1247–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G., Zimmer G., Mall G., Ritz E., Amann K. (2003). Nephron number in patients with primary hypertension. N. Engl. J. Med. 348, 101–108 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Kwan K. M., Carroll T. J., McMahon A. P., Mendelsohn C. L., Behringer R. R. (2005). Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 132, 2809–2823 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G., McMahon A. P. (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R. (1993). WT-1 is required for early kidney development. Cell 74, 679–691 [DOI] [PubMed] [Google Scholar]
- Lan Y., Kingsley P. D., Cho E. S., Jiang R. (2001). Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech. Dev. 107, 175–179 [DOI] [PubMed] [Google Scholar]
- Lan Y., Liu H., Ovitt C. E., Jiang R. (2011). Generation of Osr1 conditional mutant mice. Genesis 49, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Goldstein R. E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., Groner Y. (1998). Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95, 11590–11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Clevers H. (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. H., Bertram J. F. (2009). Is there such a thing as a renal stem cell? J. Am. Soc. Nephrol. 20, 2112–2117 [DOI] [PubMed] [Google Scholar]
- Little M. H., McMahon A. P. (2012). Mammalian kidney development: principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 4, a008300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. W., Klein R. D., Fariñas I., Sauer H., Armanini M., Phillips H., Reichardt L. F., Ryan A. M., Carver-Moore K., Rosenthal A. (1996). Renal and neuronal abnormalities in mice lacking GDNF. Nature 382, 76–79 [DOI] [PubMed] [Google Scholar]
- Mugford J. W., Sipilä P., McMahon J. A., McMahon A. P. (2008). Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 324, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Valerius M. T., McMahon A. P. (2007). Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development 134, 2533–2539 [DOI] [PubMed] [Google Scholar]
- Park J. S., Ma W., O’Brien L. L., Chung E., Guo J. J., Cheng J. G., Valerius M. T., McMahon J. A., Wong W. H., McMahon A. P. (2012). Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev. Cell 23, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichel J. G., Shen L., Sheng H. Z., Granholm A. C., Drago J., Grinberg A., Lee E. J., Huang S. P., Saarma M., Hoffer B. J., et al. (1996). Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382, 73–76 [DOI] [PubMed] [Google Scholar]
- Range R. C., Venuti J. M., McClay D. R. (2005). LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev. Biol. 279, 252–267 [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. (1998). The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395, 608–612 [DOI] [PubMed] [Google Scholar]
- Sánchez M. P., Silos-Santiago I., Frisén J., He B., Lira S. A., Barbacid M. (1996). Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382, 70–73 [DOI] [PubMed] [Google Scholar]
- Self M., Lagutin O. V., Bowling B., Hendrix J., Cai Y., Dressler G. R., Oliver G. (2006). Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 25, 5214–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So P. L., Danielian P. S. (1999). Cloning and expression analysis of a mouse gene related to Drosophila odd-skipped. Mech. Dev. 84, 157–160 [DOI] [PubMed] [Google Scholar]
- Stark K., Vainio S., Vassileva G., McMahon A. P. (1994). Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372, 679–683 [DOI] [PubMed] [Google Scholar]
- Tena J. J., Neto A., de la Calle-Mustienes E., Bras-Pereira C., Casares F., Gómez-Skarmeta J. L. (2007). Odd-skipped genes encode repressors that control kidney development. Dev. Biol. 301, 518–531 [DOI] [PubMed] [Google Scholar]
- Vainio S., Lin Y. (2002). Coordinating early kidney development: lessons from gene targeting. Nat. Rev. Genet. 3, 533–543 [DOI] [PubMed] [Google Scholar]
- Waltzer L., Bienz M. (1998). Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395, 521–525 [DOI] [PubMed] [Google Scholar]
- Wang Q., Lan Y., Cho E. S., Maltby K. M., Jiang R. (2005). Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev. Biol. 288, 582–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K., Jones K. A. (2006). Wnt signaling: is the party in the nucleus? Genes Dev. 20, 1394–1404 [DOI] [PubMed] [Google Scholar]
- Xu P. X., Adams J., Peters H., Brown M. C., Heaney S., Maas R. (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23, 113–117 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao X., Hu Y., St Amand T., Zhang M., Ramamurthy R., Qiu M., Chen Y. (1999). Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev. Dyn. 215, 45–53 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.