Abstract
Soil microbial communities are extremely complex, being composed of thousands of low-abundance species (<0.1% of total). How such complex communities respond to natural or human-induced fluctuations, including major perturbations such as global climate change, remains poorly understood, severely limiting our predictive ability for soil ecosystem functioning and resilience. In this study, we compared 12 whole-community shotgun metagenomic data sets from a grassland soil in the Midwestern United States, half representing soil that had undergone infrared warming by 2°C for 10 years, which simulated the effects of climate change, and the other half representing the adjacent soil that received no warming and thus, served as controls. Our analyses revealed that the heated communities showed significant shifts in composition and predicted metabolism, and these shifts were community wide as opposed to being attributable to a few taxa. Key metabolic pathways related to carbon turnover, such as cellulose degradation (∼13%) and CO2 production (∼10%), and to nitrogen cycling, including denitrification (∼12%), were enriched under warming, which was consistent with independent physicochemical measurements. These community shifts were interlinked, in part, with higher primary productivity of the aboveground plant communities stimulated by warming, revealing that most of the additional, plant-derived soil carbon was likely respired by microbial activity. Warming also enriched for a higher abundance of sporulation genes and genomes with higher G+C content. Collectively, our results indicate that microbial communities of temperate grassland soils play important roles in mediating feedback responses to climate change and advance the understanding of the molecular mechanisms of community adaptation to environmental perturbations.
INTRODUCTION
Extant biodiversity, i.e., the total organisms and their genes present in a habitat, has been recognized as telling the evolutionary history of life while also providing an evolutionary scaffold for the future. Consequently, one of the great challenges in the natural sciences is to better understand how the inventory of biodiversity determines the evolutionary path(s) that will shape the future (1, 2). Soil prokaryotic communities, in particular, are composed of thousands of distinct species (3–6), each of which typically makes up a rather small fraction (i.e., <0.1%) of the total community and encodes hundreds of species-specific genes of unknown function (7, 8). How such complex communities respond to natural and anthropogenic fluctuations in the environment, including major perturbations such as global climate change, is poorly understood. For instance, little is known about what genomic adaptations, interactions, and feedback mechanisms occur among members of the community during perturbations such as increases in ambient temperatures and carbon dioxide (CO2) concentrations (9, 10). Advancing the understanding of these issues will also lead to a more predictable understanding of the role of the soil ecosystem and its biota for models of climate change.
The recent advancements in sequencing technologies provide an opportunity to comprehensively assess community-wide shifts in response to environmental perturbations. Several studies have recently attempted to quantify the impacts of elevated temperatures (11) and CO2 levels (12, 13), input of exogenous organic matter of varied degrees of recalcitrance (14, 15), a substantial reduction in soil organic matter (16), and different regimes of nitrogen fertilization (17) on soil microbial communities. Most of these studies analyzed small subunit rRNA (16S rRNA) gene sequences recovered from the indigenous communities and revealed important differences in community composition in response to the perturbations. Although the 16S rRNA gene successfully serves as the best phylogenetic marker to identify the taxa present in a sample, it has several important limitations. Most importantly, it represents just one of the genes in the genome and is highly conserved in terms of sequence similarity; hence, important levels of functional and/or ecological differentiation frequently distinguish closely related organisms with identical or almost identical 16S rRNA genes (18). Therefore, in order to better understand and model the functional significance of the observed shifts in species composition, it is important to perform whole-genome-level analysis. A recent study highlighted the power of whole-genome approaches by linking methane (CH4) emissions from a thawed permafrost soil to specific genes and species of the indigenous communities (19).
In this study, we report on the whole-genome shotgun metagenomic analysis of microbial communities of temperate grassland soils (well-aerated soil in Oklahoma, United States) that experienced 2°C infrared heating for 10 years (measured soil temperature). Our previous analysis of samples collected from the same soils but in different years, using the GeoChip microarray technology, revealed that even such mild and relatively short-lived perturbations induce significant changes in the functional potential of the indigenous microbial community, related to carbon and nitrogen cycling (20). However, whether the observed changes were community wide as opposed to being attributable to a few taxa, which taxa responded to warming, and what genomic adaptations underlined the responses remained essentially unknown in our previous study. The specific objectives of the present study were to provide a high-resolution picture of the genes and taxa responding to warming and their effects on the total soil carbon balance and to uncover potential interactions among taxa, as well as to assess the molecular signatures of individual-genome and whole-community acclimation to warming. In the process, novel bioinformatics approaches to analyze and statistically compare unassembled metagenomic reads were also developed.
MATERIALS AND METHODS
Experimental setup and sampling.
This study was conducted at the Kessler Farm Field Laboratory (KFFL), located at the Great Plain Apiaries in McClain County, Oklahoma, United States (34°58′54″N, 97°31′14″W). This is an old field tallgrass prairie that had been abandoned from agriculture for more than 30 years (see the supplemental material). The experiment was established in November 1999 with a blocked split-plot design in which warming is a primary factor. Two levels of warming (ambient and +2°C) were set for six pairs of 1-m by 1-m subplots by utilizing a real or dummy infrared radiator (Kalglo Electronics, Bethlehem, PA) as the heating device, suspended 1.5 m above the ground in warming plots. In control plots, a dummy infrared radiator is also suspended (but not functional), to exclude the shading effect of the device itself. The 12 soil samples were taken from the 0-to-15-cm layer in 6 warming and 6 control plots in October 2010. Each sample was composited from two soil cores (2.5 cm in diameter by 15 cm deep) and was sieved with 2-mm sieves prior to being transported to the laboratory and stored at −80°C. The annual temperature measured on actual soil samples was, on average, 1.2°C higher in heated than in control plots at a 15-cm depth (see Table S3 in the supplemental material), confirming that our heating strategy was effective. The pH, moisture, total C and N, labile and recalcitrant C, microbial biomass, ammonium and nitrate content, temperature, and nitrification and denitrification potentials of soil samples were measured as previously described (21–25).
DNA extraction, sequencing, and preprocessing.
Soil DNA was extracted by freeze-grinding mechanical lysis as described previously (26), purified using a low-melting-point agarose gel followed by phenol extraction, and sequenced on an Illumina HiSeq 2000 instrument as described previously (27). The resulting sequencing reads were processed and trimmed as described previously (28). Read trimming was also applied on the publicly available metagenomes used in this study, for purposes of consistency (see Table S1 in the supplemental material).
Metagenome assembly and gene annotation.
The assembly of metagenomes was carried out using a hybrid protocol that combines Velvet (29), SOAPdenovo (30), and Newbler 2.0, as described previously (28). The protein-coding genes included in the assembled contigs were identified by using MetaGeneMark (31) and functionally annotated based on BLASTX searches against the nr database. The protein-coding genes in individual reads were identified by using FragGeneScan (32) with the Illumina 0.5% error model and default settings. The amino acid sequences coded by these genes were searched against the SEED database (33) by using BLAT (34) with the default settings. The best match for each read, using a cutoff E value of <1e−10, an alignment length of >20 amino acids, and an amino acid identity of >30% against the SEED genes, was recorded, and the number of best-matching reads was taken as a proxy for the abundance of the SEED genes and subsystems in each sample after normalizing for the posttrimming size of the sample. The relative abundances of domains in the metagenomes (e.g., bacteria versus eukaryotes) were estimated based on the best match of amino acid sequences using the MG-RAST server (35).
Analysis of SSU rRNA gene reads.
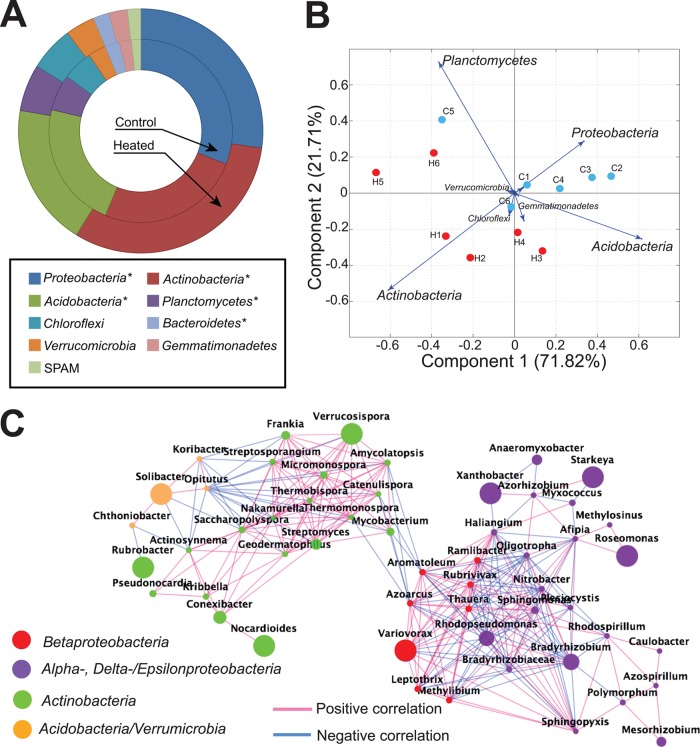
To identify reads that included prokaryotic 16S rRNA gene fragments, we clustered the full-length sequences available in the GreenGenes database, August 2012 release (36), at the 79% nucleotide sequence identity level and used one representative sequence from each resulting cluster as a reference. Reads were identified using a BLASTN (37) search (settings: −v 1 −b 1 −X 150 −q −1 −e 1e−12; remaining parameters at default settings) with the GreenGenes sequences as a reference database and a cutoff for a match of at least 70% nucleotide sequence identity and 50-bp alignment length. The matching metagenomic reads were extracted and searched against a nonredundant version of the GreenGenes database, in which all GreenGenes sequences were first preclustered to operational taxonomic units (OTUs) at the 99% nucleotide sequence identity level. Pair-ended (PE) reads with both ends matching the same OTU with higher than 97% nucleotide identity were assigned to that OTU. The relative abundances of different genera/phyla in each sample were quantified by the number of reads assigned to each taxon, normalized by the sample size (assuming each community/sample is characterized by the same rRNA copy number per genome, on average). Reads containing fungal 18S rRNA genes were identified by searching all the reads against the SILVA 18S rRNA gene database (38) and the Ribosomal Database Project 23S rRNA gene database (39) using the same BLASTN settings and cutoffs as described above, except that an 80% nucleotide identity cutoff was used to assign reads to phylum level and reads that had a better match in the 16S rRNA gene reference database were excluded. The normalized counts for genera/phyla were subjected to principal component analysis (PCA) as implemented in MatLab using default settings and Euclidean distance. The contributions of the genera/phyla in separating the samples were visualized as biplot vectors in the principal component space (see Fig. 2B). Genera/phyla that were significantly differentially present were identified using the paired t test from statlib in Python (Benjamini-Hochberg [B-H] adjusted).
FIG 2.
Shifts in taxon abundance and cooccurrence network as effects of warming. (A) Rings represent the average abundances (from six replicate samples) of phyla that made up at least 1% of the whole community; phyla that were significantly different in abundance between heated and control samples are marked by asterisks (P < 0.05, two-tailed paired t test). SPAM is a candidate phylum (55). (B) PCA biplot of the phylum abundance values separated heated from control samples. The contribution of the phyla to each principal component is represented by the arrows. (C) Cooccurrence network, based on Pearson correlation analysis, of the relative abundance of genera in the 12 soil samples (only genera with a correlation coefficient of >0.7 and P value of <0.01 are shown). Each node represents a genus and is color coded for the phylum the genus is assigned to; the size of the node is proportional to the average relative abundance of the genus across the 12 samples. Each line represents a significant correlation between the two genera it connects and is color coded for positive or negative correlation.
Abundance of genera and cooccurrence networks.
Reads not containing SSU rRNA gene fragments were searched against all complete and draft bacterial genomes available from NCBI in August 2012 (www.ncbi.nlm.nih.gov) using BLAT (cutoff for a match, E value of <1e−10, alignment length of >50 bp, and nucleotide identity of >80%). Only paired ends that had the same genome sequence as their best match were considered for further analysis. Reads were assigned to a genus based on the taxonomic classification of the genome that provided the best match. The number of reads recruited by each genome was normalized for the sample size by dividing by the total number of reads of the sample. The normalized read counts were used as a proxy of the genus abundance in the corresponding sample. The correlation of the abundances of any two genera between all samples was calculated using the Pearson correlation (statlib in Python), and a genus cooccurrence network was built using pairs of genera with correlation coefficients of >0.7 and P values of <0.01 (see Fig. 2C).
Differentially present pathways.
To identify pathways that were significantly differentially present between the control and the heated samples, we employed an approach combining resampling techniques, the DESeq package (40), and binomial testing (see Fig. S4 in the supplemental material). A jackknife method was used to generate all combinations of three control and three heated samples (from a total of six samples in each set). For each combination, a count table was generated. Each row of the table represented a SEED subsystem, each column represented a sample, and each element was the number of reads from the sample assigned to the SEED subsystem from the previous BLAT search (counting reads that were assigned to all genes that constitute the subsystem). DESeq 1.12 was then used, with the default settings, to detect the difference between heated and control samples for each SEED subsystem. For the same SEED subsystem, the log2-fold changes from the DESeq analysis of all combinations of samples followed a distribution whose mean represented the best estimate of fold change, while the variance reflected the reliability of the estimate. A binomial test was carried out to test the significance of the log2-fold changes, and the P value was adjusted for false discovery rate using the Benjamini-Hochberg method. SEED subsystems that recruited at least 100 reads in one sample with a P value of <0.01 and a fold change of >5% are reported in Table S6 in the supplemental material.
Data accession numbers.
The supplemental material includes additional information about procedures and analytical techniques. The sequence data are available in NCBI's Sequence Read Archive under BioProject PRJNA219368, accession number SRP029969.
RESULTS AND DISCUSSION
Community complexity and sequence-discrete populations.
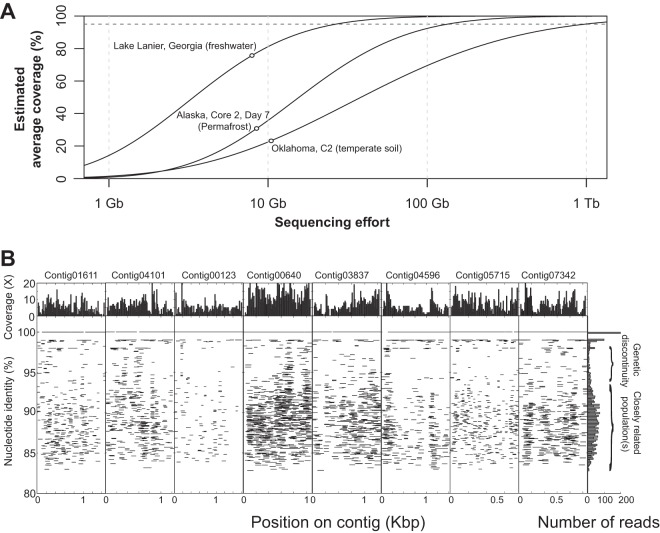
Community DNA was extracted and sequenced from six replicate samples representing the heated soils (H1 to H6) and six samples representing the adjacent unheated soils (controls; C1 to C6), yielding about 10 to 15 gigabytes (Gb) of short pair-ended (PE) sequence data per sample (100 by 100 bp) (see Tables S1 and S2 in the supplemental material). Prokaryotes represented the great majority of each community sampled, since an average of 92% of the total genes recovered had best matches (average amino acid identity, ∼35%) against bacterial and archaeal genomes in MG-RAST (35). Due to the high complexity of the soil communities in terms of species richness, the assembly of the metagenomes using established algorithms, such as Velvet (29) or our recently described hybrid protocol (28), yielded only short contigs (e.g., N50 of 500 to ∼1,000 bp), while the majority of the reads remained unassembled (see Table S1). The community complexity was quantitatively evaluated based on the fraction of unique reads in randomly drawn subsets of the data using Nonpareil, an algorithm recently developed for these purposes (41), and was compared to the complexity of other available metagenomes. The soil community was estimated to be about eight times more complex in terms of species richness than the planktonic communities of the open ocean or Lake Lanier (Atlanta, GA), assuming an average genome size three times larger in the soil than in the aquatic communities (42) and two and a half times more complex than the permafrost soil community reported recently (assuming similar genome sizes) (Fig. 1A) (19). Lake Lanier was previously estimated to contain about 1,000 OTUs (defined at the 97% 16S rRNA gene sequence identity level; Chao1 index) and to show diversity comparable to that of the open ocean (43). Therefore, the number of OTUs in each soil sample sequenced was extrapolated to be about 8,000, which is in agreement with previous estimates (44) but likely represents an underestimate of species richness due to the high conservation of 16S rRNA gene sequences (45), the more even distribution of abundance in soils with respect to the distribution of abundance in freshwater communities, and the nature of the Chao1 index as a “lower boundary” estimation of richness. We also estimated that about 4.58 terabytes (Tb) of sequencing would be required to cover 99% of the sequence diversity within each sample used in the study (interquartile range, 1.5 to 6.4 Tb) (Fig. 1A) or 530 Gb at the 95% diversity coverage level (interquartile range, 247 to 732 Gb). Although the average amounts of sequence diversity (or community complexity) were comparable between heated and control samples, heated samples showed significantly less variability in their diversity estimates (t test, P < 0.05) (see Fig. S1), indicating that warming reduced stochastic variations in terms of the amount of total diversity within the corresponding communities. Shannon and inverse Simpson alpha diversity indices based on 16S rRNA gene-containing reads recovered in the metagenomes also showed no significant differences between heated and control samples (P values ranged from 0.35 to 0.72 based on Shannon or inverse Simpson indices at both the genus and phylum levels; paired t test) (see Table S6).
FIG 1.
Soil community complexity and dominance of sequence-discrete populations. (A) The average coverage, estimated from the portion of nonunique reads (defined as reads with at least one match at the 95% nucleotide identity level; y axis) as a function of the size of subsamples randomly drawn from metagenomes of different habitats (x axis), is shown. The solid lines indicate the fitted model based on subsampling, the empty circles mark the actual size and estimated coverage of the metagenome data sets, and the horizontal dashed line denotes the 95% average coverage level. (B) Eight contig sequences assembled from a control metagenome (C5) were used as references to recruit reads, essentially as described previously (54). The graph shows the identity of each read against the reference sequence (y axes) plotted against the position of the read on the reference sequence (x axes). The histogram on the top represents the read coverage across the length of the contigs; the histogram on the right represents the number of reads recruited per unit of nucleotide identity. Note the genetic discontinuity typically observed in the 95-to-98% nucleotide identity range.
A recent synthesis of the findings from previous metagenomic studies has revealed that microbial communities of many habitats, such as the open ocean, freshwater ecosystems, the human gut, iron-reducing biofilms, and phosphorus-removing bioreactors, are predominantly composed of sequence-discrete populations and these populations may represent the important units of microbial diversity (46). Due to the unavailability of appropriate data sets, it has not yet been possible to test the applicability of these findings to soil ecosystems. Fragment recruitment plots using the contigs assembled from the temperate soil metagenomes as references revealed that sequence-discrete populations dominate the soil microbial communities, similar to other habitats (Fig. 1B). Using the number of reads recruited by each contig (at the >95% nucleotide identity level) as a proxy for in situ abundance and the phylogenetic affiliation of the housekeeping genes encoded on the contigs, we found that Burkholderia species, Conexibacter species, and Rhizobacter species were the most abundant species in the samples and that no single species recruited more than 0.1% of the total reads.
Taxon distribution and cooccurrence patterns as an effect of warming.
At a sequencing depth of about 100 million PE reads per sample, no domain-level differences in abundance were observed as an effect of warming (see Table S1 in the supplemental material). Within the bacterial and fungal domains, however, several significant differences were observed. Based on the 16S rRNA gene fragments recovered in the metagenomes, the most abundant bacterial phyla, i.e., Actinobacteria, Proteobacteria, Acidobacteria, Planctomycetes, and Bacteroidetes, were significantly differentially present between heated and control data sets, albeit the difference was not dramatic, 2% on average (P < 0.05, paired t test, Benjamini-Hochberg [B-H] adjusted for false discovery in multitesting) (Fig. 2A); similar results were obtained based on DESeq analysis (data not shown). PCA projection using phylum relative abundance confirmed that the samples from the two different treatments clustered separately, which was primarily attributable to the differences in the five phyla mentioned above (Fig. 2B). These results were reproducible (see Fig. S2) when the analysis was performed using the FastUniFrac algorithm (see the supplemental material) and for different regions of the 16S rRNA gene sequence (47). Significant differences in abundance were also observed for the two most abundant fungal phyla, Ascomycota (comprising about 70% of total fungal 18S rRNA gene sequences) and Basidiomycota (about 15% of the total), which were enriched in the control samples; consistent results were also observed with 23S rRNA reads (data not shown). These findings indicated that some of the above-mentioned bacterial taxa might have been more competitive than the dominant fungal taxa under conditions of higher abundance of labile carbon in heated soils (see also below). Alternatively, the patchy distribution of fungi in soils and the small volume used for sampling might have somewhat biased the results on the relative abundance of fungi. Clearly, more research is needed to test these hypotheses.
Interestingly, a significant increase in the G+C content of the sequences containing 16S rRNA genes (∼0.3% on average) or all reads (∼1% on average) assigned to four of the five major bacterial phyla was observed in the heated metagenomes (P < 0.05 paired t test; B-H adjusted) (see Fig. S3 in the supplemental material). The increased G+C content likely reflects the selection pressure of elevated temperatures, as suggested in previous comparative genomics studies based on the higher thermostability of GC bonds relative to AT ones (48). Other factors, such as shifts in organic nitrogen availability in heated samples (see below), might also have contributed to the higher G+C content of the heated data sets (49).
To determine the taxa whose abundance was correlated (potential synergistic interactions) or anticorrelated (potential antagonistic interactions) as an effect of warming, a genus cooccurrence network was constructed based on the abundances of all bacterial genera present in all 12 samples, the latter defined by the number of reads recruited by available representative genomes of each genus. The resulting network was composed of four major well-connected subgraphs representing Betaproteobacteria, Alpha-, Delta-, and Epsilonproteobacteria, Actinobacteria, and Acidobacteria/Verrucomicrobia (Fig. 2C). We observed mostly positive correlations within a subgraph, whereas only negative correlations were typically observed between genera from different subgraphs. These results indicated that, upon environmental perturbation, genera of the same subgraph (corresponding usually to the phylum or order levels) act synergistically, as a cohesive unit, among themselves and antagonistically to genera of different subgraphs.
Relative abundances of metabolic pathways in heated versus control metagenomes.
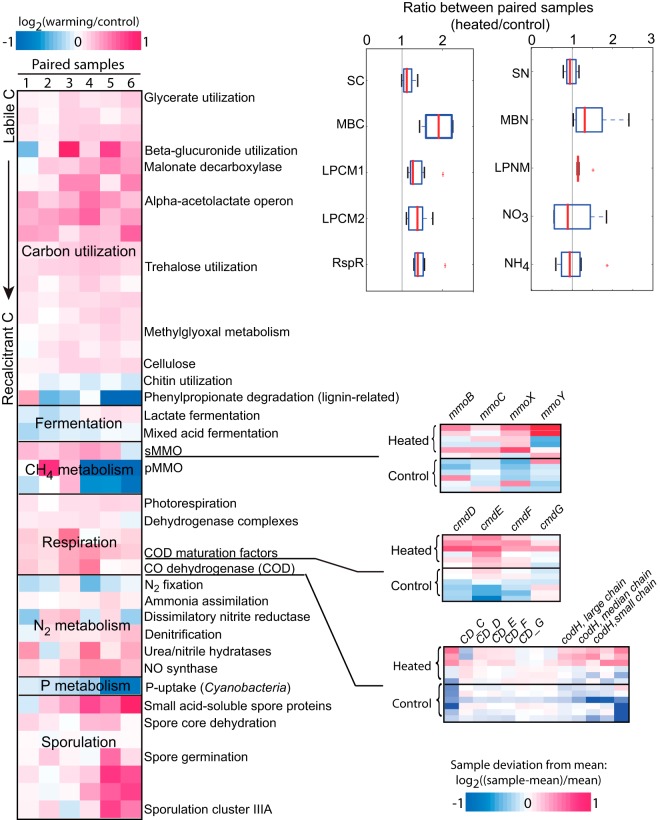
The protein-encoding PE reads were assigned to pathways in the SEED database (33) based on homology searches of the protein sequences encoded by the reads, and the number of reads was taken as a proxy of the relative abundance of the pathway in the corresponding sample (between 30 and 40% of the total reads in each sample could be assigned to the SEED database). A statistical approach was developed that employed jackknife resampling (to account for the large heterogeneity of soil samples) and the B-H method for adjusting P values to identify pathways presented differentially between the heated and control data sets (see Fig. S7 in the supplemental material). Consistent with the low-impact perturbation applied (2°C warming for 10 years), the differences in pathway abundances between heated and control samples were small (e.g., typically <5% change). Nonetheless, several significant changes were also noted, and these were reproducible in biological and technical replicates (see Fig. S5). A large portion of pathways involved in carbon source utilization and degradation and nitrogen cycling showed significant changes in relative abundance (Fig. 3). In particular, several pathways that are involved in labile carbon source metabolism were enriched in heated samples, such as glycerate metabolism (+13%), cellulose degradation (+13%), and β-glucuronide utilization (+22%). The opposite trend was observed for pathways related to (more) recalcitrant carbon sources, such as chitin utilization (−9%) and lignin degradation (−18%). These observations were consistent with field physicochemical measurements (Fig. 3), which showed on average higher labile carbon content (8%; albeit not statistically significant) (see Table S3) and higher primary production in heated soils, driven mostly by aboveground plant communities (P = 0.004 for forbs and P = 0.006 for grass, paired t test) (see Table S4). Recalcitrant carbon was also higher in heated samples, consistent with a larger contribution of plant residues, and was apparently not preferred by bacteria relative to the labile carbon. No significant difference in plant community composition was observed between treatment and control samples (P value = 0.74; permuted test on the first two principles of PCA) (see Table S5). These observations were also consistent with previous results based on the GeoChip analysis of samples taken from the same site but in earlier years (20), indicating that the trends observed were robust. Furthermore, the enriched pathways in heated samples included carbon monoxide dehydrogenases (COD), their maturation factors, and various respiratory pathways. These findings were in agreement with the higher respiration (31% on average) and elevated carbon dioxide (CO2) emissions measured in the heated versus the control soils (20). In contrast, fermentation pathways, e.g., lactate and mixed-acid fermentation, were typically less abundant in the heated metagenomes, apparently due to the prevalence of aerobic metabolism and the availability of additional, plant-derived labile organic soil carbon as an effect of warming.
FIG 3.
Changes in relative abundance of pathways as an effect of warming. The heat map on the left represents changes in the abundance of different pathways (rows) for each pair of samples (columns), color coded based on the magnitude of the change (see scale on the top left). For selected pathways related to the emission of greenhouse gases, the relative abundances of the individual genes that constitute the pathways are shown on the right (small heat maps; rows represent samples, and columns represent genes). In this case, changes in abundance represent deviations from the average abundance of the gene in all 12 samples, are color coded based on the magnitude of the difference (see scale on the bottom right), and are generally consistent with the results for the whole pathway. The results of physicochemical measurements are represented by box plots on the top right. The vertical lines at ratio 1 indicate no change between heated and control samples; the medians of six paired replicate samples are marked by the red bars; the first and third quartiles are represented by the left and right boundaries of the boxes, respectively; the left and right whiskers represent the 1.5 interquartile range; outliers are marked by red asterisks. Abbreviations: SC, soil carbon; MBC, microbial carbon; LPCM1 or -2, labile pool 1 or 2 of carbon, microbial; RspR, respiration rate; SN, soil nitrogen, MBN, microbial nitrogen, LPNM, labile pool of nitrogen, microbial; s- or pMMO, soluble or particulate methane monooxygenase, respectively.
Taken together, our results indicated that warming induced higher primary production and microbial respiration rates in the temperate soils studied here; microbial respiration appeared to release most if not all of the soil organic carbon fixed by (primarily) aboveground plant activity to the atmosphere. In agreement with these interpretations, we found that although the aboveground plant biomass was on average 10 to 30% greater in heated than in control sites, depending on the sample considered (see Table S4 in the supplemental material), the total soil carbon concentration, including soil organic matter and microbial carbon, was not significantly different between the treatments (Fig. 3).
With respect to nitrogen metabolism, a significantly higher abundance of denitrification genes was observed in heated samples, consistent with the GeoChip results (20). However, unlike the GeoChip results, nitrogen fixation genes did not differ significantly in relative abundance (Fig. 3). These observations indicated higher turnover and decreased content of available inorganic nitrogen in heated soils, in agreement with the higher respiration (microbial activity) and physicochemical measurements (Fig. 3; see also Table S3 in the supplemental material). It should be also mentioned that soil moisture, which is typically positively associated with the prevalence of anaerobic conditions and processes (such as denitrification), was relatively lower in heated than in control samples, by about 4% (see Table S3), although the difference was not statistically significant. The majority of genes related to the cycling of other nutrients, such as phosphorus, did not show significant differences in abundance as an effect of warming, with the exception of a few lineage-specific genes like those encoding the cyanobacterial phosphorus uptake system (decreased in warming [see Table S7]; the overall abundance of Cyanobacteria was not affected by warming). Finally, a higher abundance of sporulation-related genes and pathways, e.g., spore core dehydration (5% difference) and spore germination (12% difference), was observed in the communities that underwent warming (Fig. 3; see also Table S7), which was consistent with our expectations for the latter communities based on the higher ambient temperature (high temperatures induce sporulation, in general).
Community-wide versus taxon-specific shifts.
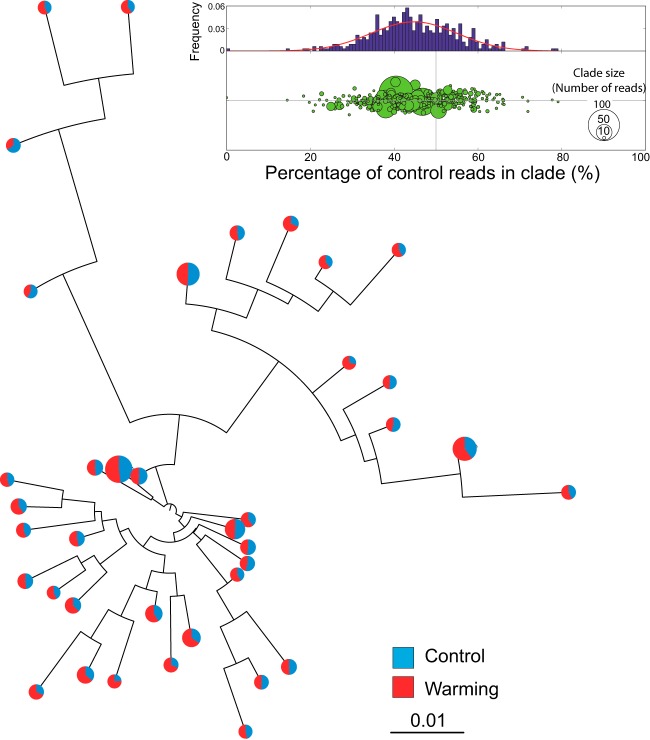
We also evaluated whether the shifts observed between heated and control data sets were due to systematic community-wide adaptations or instead to the differential presence of a few taxa. To this end, all (control and heated) overlapping PE reads encoding a gene that was found to be differentially abundant in heated versus control data sets were clustered at the 80% sequence identity level, providing the OTUs present in the samples for each gene. The percentages of heated and control reads constituting each OTU were compared to determine the OTU(s) that contributed to the higher abundance of genes and pathways in the heated samples. Overall, most of the gene content shifts were attributable to many OTUs, typically more than 50% of the total OTUs observed for each gene analyzed, revealing that warming induced community-wide adaptations (Fig. 4; see also Fig. S6).
FIG 4.
Changes in pathway abundance are community wide and not attributable to only a few taxa. Representative sequences from all OTUs (or clades) of a specific gene (in this case, a CO2 dehydrogenase, CD_D) were analyzed to produce the distance-based phylogenetic tree shown. The pie charts at the tips of the tree represent the percentage of heated versus control reads that made up each OTU, and the size of each chart is proportional to the number of reads in the OTU; only OTUs with at least 50 reads are shown, for simplicity. Note that no OTU was heat or control specific and about ∼60% of the pie charts had a higher number of heated than of control reads, revealing that many distinct taxa are responsible for the higher abundance of CO2 dehydrogenase in heated metagenomes. This is also evident in the graph shown at the top (inset). In the circle graph, each circle represents an OTU; the x coordinate represents the percentage of the total reads of the OTU that are control reads, and the y coordinate represents a random value for visualization purposes. Note that more OTUs have less than 50% control reads than have more than 50% control reads. The histogram at the top shows the distribution of the percentages of control reads in all OTUs, with a fitted Gaussian curve shown by the solid line.
Conclusions and perspectives.
For long-term perturbations, during which the responding microorganisms undergo several replications (grow), like the one performed here and in contrast to pulse-like, short-term perturbations, DNA-based data can provide robust views of the responses of the indigenous organisms and are typically consistent with transcriptomic, proteomic, or other functional data (50, 51). In this study, the metagenomic data obtained from replicate samples were quantitative (as seen in Fig. 3), highly reproducible (e.g., see Fig. S5 in the supplemental material), and consistent with macroscopic, biochemical, and physicochemical measurements of soil and aboveground plant biota. Therefore, the results and methods reported here offer meaningful insights into the genomic adaptations and functional responses of complex soil microbial communities to experimental warming and, hence, the predicted effects of climate change. It should be noted, however, that the specific activities performed by the microorganisms sampled cannot be captured by our methodology, but only indirectly inferred.
These data revealed that soil microbial communities adapt fast to perturbations, even low-impact ones, perhaps faster than previously anticipated. In the case of (mild) warming, adaptation was evident, for instance, by significant shifts in the metabolic pathway abundances and G+C contents in the genomes of the indigenous microbes. These adaptations apparently took place in less than 10 years, and we find it remarkable that features such as G+C content, which are thought to represent stable properties of the genome and community, can change in such a (relatively) short period of time.
The systematic responses and the shifts in G+C and gene content mentioned above indicated that the differences between heated and control samples were likely attributable to long-term adaptations of preexisting, abundant taxa as opposed to short-term, pulse-like responses of a few, perhaps newly emerged, genotypes. Consistent with this interpretation, similar patterns were observed when OTUs based on functional genes (Fig. 4; see also Fig. S6 in the supplemental material) were clustered at the 95% nucleotide sequence identity level, which corresponds to the sequence-discrete populations observed (Fig. 1B) and the species demarcation standards (46). Thus, if newly emerged genotypes that were dormant and/or not abundant before the incubation responded to warming and, thus, accounted for the results observed in the case of the 95% identity cutoff (as opposed to preexisting, abundant genotypes), these would have been genotypes of the same population (or species) as the abundant genotypes. However, this is unlikely, given that the phenotypic differentiation among highly similar genotypes (i.e., within species) is generally considered to be low. Nonetheless, time-series metagenomic data will be required to fully resolve the matter and determine the relative contributions of preexisting taxa to community adaptation.
We also noted several hypothetical genes and mobile elements to be differentially abundant as an effect of warming (e.g., see Table S7 in the supplemental material). While at least a few of these genes likely represent important (uncharacterized) adaptations to warming, we also expect many of the lineage-specific genes differentially present between heated and control samples, such as the cyanobacterial phosphate uptake system mentioned above, to represent hitchhiking events, i.e., the corresponding taxon was favored by warming for other reasons (e.g., available carbon sources).
Our study also indicated that complex interactions between community members probably played a role in community acclimation to warming, and it is notable that mostly negative correlations in terms of abundance were observed between organisms assigned to different broad taxonomic ranks, such as different phyla (Fig. 2B). Similar patterns were reported previously for soil communities, using different approaches. For example, based on bromodeoxyuridine (BrdU)-labeled 16S rRNA gene quantification by PhyloChip, Goldfarb and colleagues reported antagonistic interactions among bacterial phyla in response to carbon substrate addition (14), and Barberán and colleagues concluded that taxa within the same phylum tend to cooccur more often than expected based on the analysis of 151 soil 16S rRNA gene amplicon data sets (52). These observations across different soil and data types collectively reveal a hierarchical structure within soil microbial communities, consistent with what was suggested previously (53). It is important to note, however, that cooccurrence does not necessarily indicate direct interactions between the taxa, as cooccurrence may be due to hidden (indirect) factors. For instance, r (copiotrophic) versus K (oligotrophic) ecological strategies may underlie some of the taxon cooccurrence patterns observed; we expect a higher relative abundance of K strategists in the control samples due to the relative higher concentration of available recalcitrant organic carbon and genes for its degradation. Consistent with these interpretations, taxa that are thought to represent K strategists, such as Acidobacteria, were more abundant in control samples.
Nonetheless, functional experiments will be necessary to elucidate the mechanisms that underlie the correlation patterns revealed here. Furthermore, disentangling the direct effect of warming on the belowground microbial communities from the indirect effect of warming due to the stimulation of aboveground plant communities remains challenging. Additional samples across time and soils of different types and latitudes need to be examined before more robust conclusions can emerge with respect to the importance of the belowground microbial communities for mitigating or exacerbating the effects of climate change.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy (award DE-SC0004601).
We thank the personnel of the Los Alamos National Laboratory Genomics Facility for their assistance with DNA sequencing.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03712-13.
REFERENCES
- 1.Allison SD, Martiny JB. 2008. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105(Suppl 1):11512–11519. 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shade A, Peter H, Allison SD, Baho DL, Berga M, Burgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JB, Matulich KL, Schmidt TM, Handelsman J. 2012. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. 10.3389/fmicb.2012.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torsvik V, Goksoyr J, Daae FL. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578–6583. 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494–10499. 10.1073/pnas.142680199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handelsman J, Tiedje J, Alvarez-Cohen L, Ashburner M, Cann I, Delong E, Doolittle W, Fraser-Liggett C, Godzik A, Gordon J, Riley M, Schmidt T. 2007. The new science of metagenomics: revealing the secrets of our microbial planet. The National Academies Press, Washington, DC: [PubMed] [Google Scholar]
- 7.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson KE, Paulsen IT, Heidelberg JF, Fraser CM. 2000. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 18:1049–1054. 10.1038/80235 [DOI] [PubMed] [Google Scholar]
- 9.Bond-Lamberty B, Thomson A. 2010. Temperature-associated increases in the global soil respiration record. Nature 464:579–582. 10.1038/nature08930 [DOI] [PubMed] [Google Scholar]
- 10.Heimann M, Reichstein M. 2008. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451:289–292. 10.1038/nature06591 [DOI] [PubMed] [Google Scholar]
- 11.Deslippe JR, Hartmann M, Simard SW, Mohn WW. 2012. Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol. Ecol. 82:303–315. 10.1111/j.1574-6941.2012.01350.x [DOI] [PubMed] [Google Scholar]
- 12.Deng Y, He Z, Xu M, Qin Y, Van Nostrand JD, Wu L, Roe BA, Wiley G, Hobbie SE, Reich PB, Zhou J. 2012. Elevated carbon dioxide alters the structure of soil microbial communities. Appl. Environ. Microbiol. 78:2991–2995. 10.1128/AEM.06924-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, Van Nostrand JD, Hobbie SE, Reich PB, Zhou J. 2010. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol. Lett. 13:564–575. 10.1111/j.1461-0248.2010.01453.x [DOI] [PubMed] [Google Scholar]
- 14.Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2:94. 10.3389/fmicb.2011.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar J, Eichorst SA, Gallegos-Graves LV, Silva S, Xie G, Hengartner NW, Evans RD, Hungate BA, Jackson RB, Megonigal JP, Schadt CW, Vilgalys R, Zak DR, Kuske CR. 2012. Common bacterial responses in six ecosystems exposed to 10 years of elevated atmospheric carbon dioxide. Environ. Microbiol. 14:1145–1158. 10.1111/j.1462-2920.2011.02695.x [DOI] [PubMed] [Google Scholar]
- 16.Sul WJ, Asuming-Brempong S, Wang Q, Tourlousse DM, Penton R, Deng Y, Rodrigues JLM, Adiku SGK, Jones JW, Zhou J, Cole JR, Tiedje JM. 2013. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol. Biochem. 65:33–58. 10.1016/j.soilbio.2013.05.007 [DOI] [Google Scholar]
- 17.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6:1007–1017. 10.1038/ismej.2011.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10:504–509. 10.1016/j.mib.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 19.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. 2011. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371. 10.1038/nature10576 [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, Fei S, Deng S, He Z, Van Nostrand JD, Luo Y. 2012. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Change 2:106–110. 10.1038/nclimate1331 [DOI] [Google Scholar]
- 21.Belay-Tedla A, Zhou XH, Su B, Wan SQ, Luo YQ. 2009. Labile, recalcitrant, and microbial carbon and nitrogen pools of a tallgrass prairie soil in the US Great Plains subjected to experimental warming and clipping. Soil Biol. Biochem. 41:110–116. 10.1016/j.soilbio.2008.10.003 [DOI] [Google Scholar]
- 22.Luo Y, Wan S, Hui D, Wallace LL. 2001. Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413:622–625. 10.1038/35098065 [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, White L, Hui D. 2004. Comment on “Impacts of fine root turnover on forest NPP and soil C sequestration potential.” Science 304:1745. 10.1126/science.1098080 [DOI] [PubMed] [Google Scholar]
- 24.Sherry RA, Zhou X, Gu S, Arnone JA, III, Schimel DS, Verburg PS, Wallace LL, Luo Y. 2007. Divergence of reproductive phenology under climate warming. Proc. Natl. Acad. Sci. U. S. A. 104:198–202. 10.1073/pnas.0605642104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Wan SQ, Luo YQ. 2007. Source components and interannual variability of soil CO2 efflux under experimental warming and clipping in a grassland ecosystem. Glob. Chang. Biol. 13:761–775. 10.1111/j.1365-2486.2007.01333.x [DOI] [Google Scholar]
- 26.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh S, Tandukar M, Pavlostathis SG, Chain PSG, Konstantinidis KT. 2013. Microbial community adaptation to quaternary ammonium biocides as revealed by metagenomics. Environ. Microbiol. 15:2850–2864. 10.1111/1462-2920.12154 [DOI] [PubMed] [Google Scholar]
- 28.Luo C, Tsementzi D, Kyrpides NC, Konstantinidis KT. 2012. Individual genome assembly from complex community short-read metagenomic datasets. ISME J. 6:898–901. 10.1038/ismej.2011.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38:e132. 10.1093/nar/gkq275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rho M, Tang H, Ye Y. 2010. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 38:e191. 10.1093/nar/gkq747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-R LM, Konstantinidis KT. 5 November 2013. Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics. 10.1093/bioinformatics/btt584 [DOI] [PubMed] [Google Scholar]
- 42.Konstantinidis KT, Braff J, Karl DM, DeLong EF. 2009. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl. Environ. Microbiol. 75:5345–5355. 10.1128/AEM.00473-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh S, Caro-Quintero A, Tsementzi D, DeLeon-Rodriguez N, Luo C, Poretsky R, Konstantinidis KT. 2011. Metagenomic insights into the evolution, function, and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Appl. Environ. Microbiol. 77:6000–6011. 10.1128/AEM.00107-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U. S. A. 109:21390–21395. 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole J, Konstantinidis KT, Farris RJ, Tiedje JM. 2010. Microbial diversity and phylogeny: extending from rRNAs to genomes, p 1–20 In Liu W-T, Jansson J. (ed), Environmental molecular biology. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 46.Caro-Quintero A, Konstantinidis KT. 2012. Bacterial species may exist, metagenomics reveal. Environ. Microbiol. 14:347–355. 10.1111/j.1462-2920.2011.02668.x [DOI] [PubMed] [Google Scholar]
- 47.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakashima H, Fukuchi S, Nishikawa K. 2003. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J. Biochem. 133:507–513. 10.1093/jb/mvg067 [DOI] [PubMed] [Google Scholar]
- 49.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolonen A, Webb EA, Zinser ER, Chisholm SW. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047. 10.1038/nature01947 [DOI] [PubMed] [Google Scholar]
- 50.Turnbaugh PJ, Gordon JI. 2008. An invitation to the marriage of metagenomics and metabolomics. Cell 134:708–713. 10.1016/j.cell.2008.08.025 [DOI] [PubMed] [Google Scholar]
- 51.Hettich RL, Pan C, Chourey K, Giannone RJ. 2013. Metaproteomics: harnessing the power of high performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal. Chem. 85:4203–4214. 10.1021/ac303053e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6:343–351. 10.1038/ismej.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8:523–529. 10.1038/nrmicro2367 [DOI] [PubMed] [Google Scholar]
- 54.Konstantinidis KT, DeLong EF. 2008. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2:1052–1065. 10.1038/ismej.2008.62 [DOI] [PubMed] [Google Scholar]
- 55.Lipson DA, Schmidt SK. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70:2867–2879. 10.1128/AEM.70.5.2867-2879.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.