Abstract
Recent studies have suggested a potential role for wild birds in zoonotic transmission of Campylobacter jejuni, the leading cause of gastroenteritis in humans worldwide. In this study, we detected Campylobacter spp. in 66.9% (85/127) of free-ranging American crows (Corvus brachyrhyncos) sampled in the Sacramento Valley of California in 2012 and 2013. Biochemical testing and sequence analysis of 16S rRNA revealed that 93% of isolates (n = 70) were C. jejuni, with cytolethal distending toxin (CDT) and flagellin A genes detected by PCR in 20% and 46% of the C. jejuni isolates (n = 59), respectively. The high prevalence of C. jejuni, coupled with the occurrence of known virulence markers CDT and flagellin A, demonstrates that crows shed Campylobacter spp. in their feces that are potentially pathogenic to humans. Crows are abundant in urban, suburban, and agricultural settings, and thus further study to determine their role in zoonotic transmission of Campylobacter will inform public health.
INTRODUCTION
Campylobacter jejuni is a Gram-negative spiral rod bacterium that is commonly associated with human gastroenteritis (1, 2). In the United States, campylobacteriosis is estimated to affect 1.3 million people each year, with symptoms that included fever, abdominal cramping, and bloody diarrhea (1, 3, 4). In rare cases, infection can also lead to serious autoimmune disorders, such as Guillain-Barré syndrome (5). Routes of transmission and infection often involve consumption of feces-contaminated undercooked poultry and contaminated water, followed by invasion of the host gastrointestinal tract to cause disease (1, 2). Exact mechanistic details of how C. jejuni invades and causes damage are still unclear; however, several proteins, such as flagellin proteins, are vital for adherence to and invasion of human epithelial cells, and production of cytolethal distending toxin (CDT) has been shown to be involved with cytotoxicity, cell cycle arrest, and host cell death (6–9).
A number of recent studies have begun to reveal the importance of wildlife as reservoirs for Campylobacter (10–13). Although the most attention has been paid to transmission among animals within a facility (e.g., zoos [14, 15]) or an agricultural operation (e.g., chicken farms [16, 17]), realization is growing that wild birds might also be playing a role in zoonotic transmission of Campylobacter (11, 18, 19). Campylobacter bacteria are well suited to live in and be carried by birds because they grow well in thermophilic, microaerobic conditions (11, 13, 18, 19); however, presence of Campylobacter in an avian host does not necessarily indicate a significant role in human disease epidemiology. For example, human-commensal European starlings (Sturnus vulgaris) have been found to be infected with Campylobacter spp., but not with C. jejuni, and therefore pose little threat to human health (13). Similarly, greylag geese (Anser anser) carry C. jejuni, but they have been excluded as a major human health hazard because their gene profile suggests a high level of host specificity (12). In contrast, black-headed gulls (Larus ridibundus) have been found to carry C. jejuni species that are potentially pathogenic to humans (11). Because of these species-specific differences, the possible role of each species of wild bird in the spread of human disease must be considered on an individual basis.
The American crow (Corvus brachyrhynchos) is a widespread North American passerine that forages in a variety of settings, including dumps, animal feedlots, pastures, and urban areas. American crows therefore have potential to transfer pathogens from human waste or infected domestic animal manure to human food sources or uninfected domestic animals, acting both as reservoir and transport host, and they may enable zoonotic transmission. Previous studies have shown that different crow species found in Japan, Tanzania, New Zealand, and Malaysia carry Campylobacter spp. (20–23). For example, two species of crows (Corvus corone and Corvus levaillanti) sampled in a suburban population in Tokyo had a prevalence of C. jejuni approaching 34%, possibly because of their association with a municipal garbage dump (22, 23). Likewise, a study carried out in the mid-Atlantic United States reported that American crows had the highest prevalence of C. jejuni (3/7 samples; 43%) among 32 avian species sampled (24). No studies to date, however, have examined the likelihood that bacterial strains carried by American crows could be shared with and are pathogenic to humans.
Here, we assess the prevalence of Campylobacter in American crows and their potential role in the epidemiology of human campylobacteriosis using three lines of evidence: 16S rRNA sequencing, CDT gene presence, and flagellin A gene presence. We use phylogenetic analyses of 16S rRNA sequence data to distinguish C. jejuni from other species and to map strains found in crows with strains previously isolated from humans, livestock, and poultry. We assess CDT and flagellin A gene presence because they are necessary for cell pathology and virulence in humans. Specifically, we test the hypothesis that the C. jejuni strains in American crows in California have virulence characteristics consistent with those associated with human disease.
MATERIALS AND METHODS
Crow sampling.
Fecal or cloacal swab samples were collected from American crows (referred to as “crows” here) in Yolo County, California, between 8 May 2012 and 26 June 2013. Samples were collected using Amies clear gel collection and transport swabs (Remel BactiSwab; Thermo Fisher Scientific, Waltham, MA) and stored on ice (4 to 7°C) for 2 to 6 h prior to culture. Seventy-six samples were taken from crow nestlings (7 to 30 days after hatching) in May to June 2012, and 15 samples were taken from nestlings in May to June 2013 from either cloacal swabs or fresh feces. Forty additional samples (environmental samples) were collected from fresh feces under free-flying crow flocks, comprising adult, sexually mature birds (>2 years old) and sexually immature subadults (<2 years old). Twenty-six of these samples were collected in July of 2012 from single-species diurnal foraging flocks. We observed these foraging flocks prior to sampling to ascertain that each sample was fresh and originated from a unique crow. Fourteen environmental samples were collected on 26 February 2013 from a large communal crow roost. The >10,000 crows from this roost flew in circles above the roost for several minutes prior to their dawn departure. The likelihood is high that each sample originated from a different bird in the flock, because we collected fresh feces from these circling birds as they fell. To avoid collecting any background Campylobacter that might have been present in the environment, we swabbed only the top surface of each sample: we made no contact with the ground when swabbing droppings. All crow work was performed under protocols approved by the Institutional Animal Care and Use Committee of the University of California, Davis (IACUC 16897).
Campylobacter isolation and culture.
Fecal samples were inoculated onto 5% sheep blood agar containing cefoperazone, vancomycin, and amphotericin B (Campy CVA; Hardy Diagnostics, Santa Maria, CA), with incubation at 37°C under microaerophillic conditions (CampyGen; Oxoid Limited, Hampshire, United Kingdom) for 4 to 6 days. Bacterial colonies consistent with Campylobacter spp. were subjected to Gram staining, and those with Gram-negative curved rods observed were subcultured onto 5% sheep blood agar (Hardy Diagnostics) for further characterization.
Biochemical testing.
After Gram staining, each isolate was tested for the presence of catalase and the ability to hydrolyze hippurate (Dalynn Biologicals Inc., Calgary, Canada) using the manufacturer's instructions. Campylobacter isolates were also tested for their susceptibility to nalidixic acid and cephalothin by using 30-μg antibiotic disks (BD Biosciences) according to the manufacturer's instructions and analyzed for a zone of clearance, indicating bacterial susceptibility (Table 1). Isolates were stored at −80°C using Microbank freezer beads (Pro-Lab Diagnostics, Ontario, Canada) until further testing.
TABLE 1.
Campylobacter characterization based upon biochemical phenotype and 16S rRNA phylogenetic analyses
| Pattern no. or speciesa | No. of isolates with pattern | Catalase activity | Hippurate hydrolysis | Sensitivity tob: |
Biochemical phenotypec | No. of isolates amplified for 16S rRNAd | Isolate(s) identified by 16S rRNAe (%) | |
|---|---|---|---|---|---|---|---|---|
| Nalidixic acid | Cephalothin | |||||||
| 1 | 1 | − | − | S | R | Campylobacter spp. | 1 | Arcobacter (100 [n = 1]) |
| 2 | 3 | + | − | R | R | Campylobacter spp. | 3 | C. lari (100 [n = 3]) |
| 3 | 4 | + | − | S | R | Campylobacter spp. | 3 | C. jejuni (67), C. lari (33 [n = 3]) |
| 4 | 5 | + | + | R | R | C. jejuni | 4 | C. jejuni (100 [n = 4]) |
| 5 | 71 | + | + | S | R | C. jejuni | 59 | C. jejuni (100 [n = 59]) |
| C. jejuni (control) | 1 | + | + | S | R | C. jejuni | 1 | C. jejuni (100) |
| C. coli (control) | 1 | + | − | S | R | C. coli | 1 | C. coli (100) |
Five different patterns emerged from biochemical testing of isolates. Isolates were compared to known C. jejuni and C. coli controls.
S, susceptible; R, resistant.
Phenotypes were declared based on biochemical tests.
Some isolates could not be amplified by PCR.
The 16S rRNA gene was analyzed phylogenetically, and isolates were identified to the genus and species levels based upon their relationship (close proximity with support values) with known reference sequences.
DNA extraction, 16S rRNA, CDT, and flagellin A amplification analysis.
DNA for PCR amplification and sequencing was purified from bacterial strains grown on 5% sheep blood agar plates (Hardy Diagnostics, Santa Maria, CA). The boiling method for DNA extraction was used as previously described (10, 25). In brief, a generous loop of pure bacteria was suspended in 300 μl sterile molecular analysis-grade water and boiled at 100°C for 10 min. After boiling, tubes were chilled for 5 min, followed by centrifugation at 15,000 × g for 10 min. The supernatant was then transferred to a new 1.5-ml tube. DNA was further diluted as needed to a concentration of approximately 50 ng/μl.
For 16S rRNA amplification, universal eubacterial forward and reverse primers 8FPL (5′ CTG CAG AGT TTG ATC CTG GCT CAG 3′) and 1492RPL (5′ CGG GTT ACC TTG TTA CGA CTT 3′) were used to amplify the 16S rRNA genes. Each 20-μl reaction mixture contained 2 μl of 10× buffer, 0.4 μl of deoxynucleoside triphosphates (dNTPs), 0.5 μl of each 10 pM primer, 0.1 μl Qiagen HotStarTaq Plus (Venlo, Limburg, The Netherlands), and 14.5 μl water, and 2 μl DNA at 50 ng/μl was added to each reaction mixture. Amplification was performed on an Eppendorf Mastercycler thermal cycler (Hamburg, Germany) as follows: 95°C for 5 min followed by 35 cycles of 94°C for 1 min, 61°C for 1 min, and 72°C for 1 min 50 s followed by an elongation step at 72°C for 10 min. The 1,484-bp PCR product was separated on a 1.0% agarose gel by electrophoresis and visualized with ethidium bromide (EtBr). The PCRs were cleaned using an ExoSAP-IT system (USB Corporation) following the manufacturer's protocol by adding 2 μl of ExoSAP-IT to each 5 μl PCR product and incubating as follows: 37°C for 15 min and 80°C for 15 min and a hold at 4°C until sequencing. The DNA Sequencing Facility, Division of Biological Sciences, University of California, Davis, used Sanger technology to perform forward and reverse sequencing reactions on submitted amplicons.
Amplification of virulence genes was performed using protocols targeting CDT (cdtABC) and flagellin A (flaA) genes (1, 6, 26). The whole CDT gene cluster (cdtABC) was amplified using forward primer GNW (5′-GGA AAT TGG ATT TGG GGC TAT ACT-3′) and reverse primer LPF-X (5′-TTG CAC ATA ACC AAA AGG AAG-3′) with an amplicon of 1,215 bp (26). The same PCR mixtures and thermocycler parameters were used as with the 16S rRNA amplification. For flagellin A (flaA) amplification, the forward and reverse primers were flaA-F (5′-GGATTTCGTATTAACACAAATGGTGC-3′) and flaA-R (5′-CTGTAGTAATCTTAAAACATTTTG-3′), respectively, as previously published (1, 26). Each PCR used the following parameters: 95°C for 5 min, 95°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min (30 cycles) and 72°C for 10 min of final extension, producing a 1,728-bp flaA amplicon. Both cdtABC and flaA amplicons from each isolate were separated on a 1.0% agarose gel using electrophoresis and visualized with ethidium bromide (EtBr).
Phylogenetic analyses.
The 16S rRNA sequences were analyzed and compared to reference sequences from National Center for Biotechnology Information (NCBI) using the nucleotide database and Basic Local Alignment Search Tool (BLAST) (27). For alignment and tree building, Ab.1 files were uploaded to the Ribosome Database Project (RDP) (28) pipeline for trimming and quality check. Cleaned and processed reads were aligned using SSU-ALIGN (29) and subsequently merged using a custom perl script (http://figshare.com/articles/Prevalence_of_Campylobacter_/796458). To the merged alignment, Campylobacter reference sequences (see Table S2 in the supplemental material) available through RDP and NCBI were added. These alignments led to generation of a phylogenetic tree using FastTree2.1 (30) and default parameters with the “pseudo” option. Numbers at nodes in the tree are FastTree local support values based on a Shimodaira-Hasegawa (SH) test where 1.000 represents the strongest relationship and 0.000 indicates low support (30). The tree was visualized and annotated using Dendroscope 3 (version 3.2.8) (31) and Adobe Illustrator CS4.
Nucleotide sequence accession numbers.
Accession numbers for nucleotide sequences determined in this study can be found in Fig. 1 (see also Table S1 in the supplemental material).
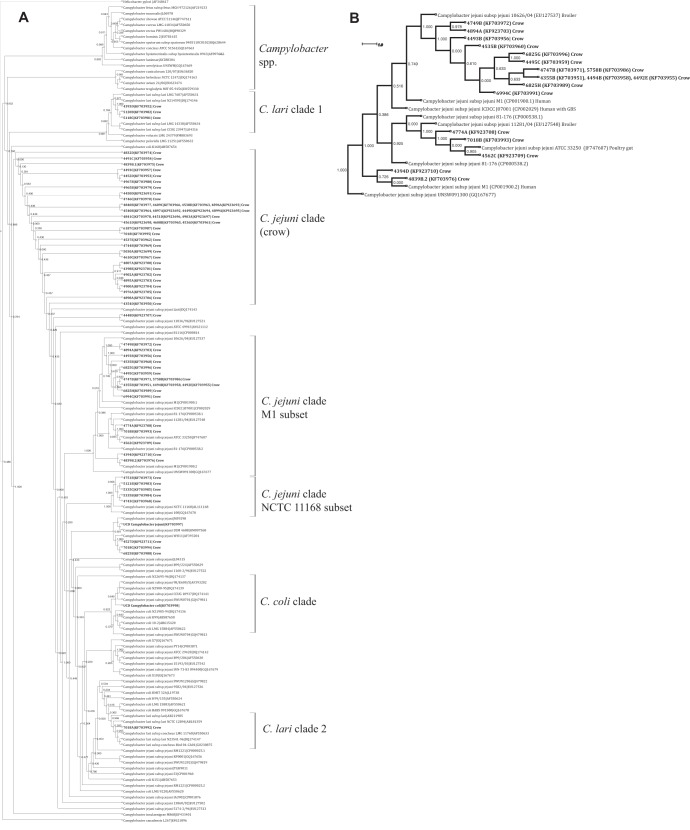
FIG 1.
16S rRNA phylogenetic analysis. (A) Annotated cladogram depicting relationships between the total Campylobacter isolates collected from American crows in this study and other known Campylobacter isolates. The tree is rooted to Helicobacter pylori. Campylobacter isolates from crows are indicated in bold. The species, strain, and accession number are listed for each publicly accessible sequence. The cluster corresponding to “Campylobacter spp.” contains non-C. jejuni Campylobacter sequences, including C. avium, C. rectus, and C. fetus along with the H. pylori root. Other clades are defined in the text. (B) Expanded phylogram depicting crow isolates clustered around known C. jejuni isolates, some of human origin (strain M1). Numbers at nodes are FastTree local support values based on the Shimodaira-Hasegawa (SH) test, where 1.000 represents the strongest relationship and 0.000 indicates low support. Distance is indicated by a scale bar.
RESULTS
Prevalence and biochemical findings.
Fecal culture with biochemical testing revealed that, overall, 66.9% (85/127) of the American crow samples were positive for Campylobacter spp. Sixty percent of samples (59/98) collected in 2012 were positive, whereas 89.6% (26/29) were positive in 2013. Of the crows sampled in 2012, 60.5% (46/76) of nestlings and 53.8% (14/26) of the adults and subadults were culture positive for Campylobacter spp. Of the nestlings sampled in 2013, 86.7% (13/15) were positive for Campylobacter spp., and 92.9% (13/14) of the adults and subadults were culture positive for Campylobacter spp.
Five distinct patterns emerged from the biochemical data, which were then compared to patterns from known C. jejuni and C. coli control bacteria (Table 1). A positive test for both catalase and hippurate hydrolysis was used to presumptively identify C. jejuni, whereas a positive catalase test and a negative hippurate test indicated non-jejuni Campylobacter spp. Campylobacter jejuni is typically sensitive to nalidixic acid and resistant to cephalothin as measured by a bacterial zone of clearing around the antibiotic disk; however, we occasionally detected isolates that were hippurate positive and also resistant to nalidixic acid (Table 1). It is typical that a few C. jejuni isolates would be resistant to nalidixic acid. Therefore, this phenotype, while rare, does not exclude those isolates from being members of the C. jejuni species. As expected, this abbreviated biochemical testing format could not fully categorize all isolates (Table 1).
16S rRNA gene sequence analysis.
Ninety-three percent (65/70) of Campylobacter isolates were identified as C. jejuni by 16S rRNA sequencing using phylogenetic analyses. Identification was based upon close association in clades with a high level of bootstrap support values with isolates previously identified as C. jejuni (Fig. 1). Species identification was further supported by their biochemical phenotype (described above and depicted in Table 1). Some C. jejuni isolates from crows clustered with samples originating from humans, chickens, and livestock in our phylogenetic analysis (Fig. 1A [C. jejuni, M1 clade, and C. jejuni, 1168 clade] and B). Two clades of C. lari emerged: four crow isolates (4393D, 5120B, 5118C, and 7018A) clustered with known reference C. lari isolates (Fig. 1A [C. lari clades 1 and 2]). One C. coli cluster emerged (Fig. 1A [C. coli clade]). None of the Campylobacter isolates from crows grouped in a clade with C. avium, a species recently found in poultry that is divergent from C. jejuni (32).
Four isolates were identified biochemically as hippurate negative; however, after three of the isolates were sequenced, two had 16S rRNA sequences most similar to those of other C. jejuni isolates and one isolate's sequence was most similar to that of C. lari. Due to a high degree of similarity between the 16S rRNA genes of C. coli and C. jejuni, the two C. jejuni isolates could be closer to the C. coli species than to the C. jejuni species, or those isolates could represent a strain of C. jejuni without the enzyme necessary to hydrolyze hippurate. Further work will continue to characterize those isolates.
CDT and flagellin A gene detection.
PCR amplification of extracted isolate DNA revealed that 20% (12/59) of the isolates classified as C. jejuni had all three CDT genes (cdtA, cdtB, and cdtC) and that 33% (1/3) of the isolates identified as C. lari had all three CDT genes. Forty-six percent (27/59) of the isolates, including one C. lari isolate, had the flagellin A gene (flaA) (Table 2).
TABLE 2.
Presence of flaA and cdtABC in Campylobacter isolates
| Species | % flaA presence (n/total no. of isolates) | % cdtABC presence (n/total no. of isolates) |
|---|---|---|
| Campylobacter jejuni (control) | 100 (1/1) | 100 (1/1) |
| Campylobacter coli (control) | 100 (1/1) | 100 (1/1) |
| Campylobacter jejuni | 46 (27/59) | 20 (12/59) |
| Campylobacter lari | 33 (1/3) | 33 (1/3) |
| Arcobacter | 0 (0/1) | 0 (0/1) |
DISCUSSION
Although high levels of infection have been reported for a number of species and populations in the genus Corvus (20–24), the prevalence of Campylobacter detected in this California American crow population (66.9%) was among the highest recorded in a wild bird population. Cytolethal distending toxin (CDT) and flagellin A (flaA) genes, both of which are associated with virulence, were detected in a subset of the C. jejuni and C. lari isolates. This study demonstrated that potentially virulent strains of Campylobacter are shed in the feces of American crows.
Phylogenetic analyses based on 16S rRNA sequence data were consistent with the hypothesis that some C. jejuni strains might be shared among crows, humans, poultry, and livestock and could be associated with disease (Fig. 1). For example, sequences from 18 (28%) of the isolates clustered in a large clade with C. jejuni M1 (Fig. 1B), a strain transmitted from poultry to humans that is associated with human gastroenteritis (32). Also in that large clade was C. jejuni ICDCCJ07001, isolated from a human with Guillain-Barré syndrome in Jilin, China (33) (Fig. 1B). Five C. jejuni isolates from this study clustered in the same clade as Campylobacter jejuni subsp. jejuni NCTC 11168, the first Campylobacter to be sequenced and of human origin (34). Likewise, several C. jejuni and C. coli isolates from diarrheal patients in Japan (35) and a highly virulent C. jejuni strain associated with sheep abortion in the United States (36) clustered with crow Campylobacter sequences (Fig. 1). Thirty-seven (57%) of the Campylobacter isolates collected from crows in this study seemed to form a separate, unique clade of C. jejuni, suggesting that some of the C. jejuni isolates could be crow specific or could come from a common parent strain (Fig. 1; see also Fig. S1 in the supplemental material). One of the sequences closest to this crow-specific clade was that of C. jejuni 81116 (37), isolated from a human case during a schoolhouse outbreak of campylobacteriosis. Wild birds were suspected to have contaminated the school's drinking water (38).
Cytolethal distending toxin (CDT) genes were detected in 20% (12/59) of C. jejuni isolates and 33% (1/3) of C. lari isolates. The expression of CDT genes is key to the pathogenicity of C. jejuni: production of all cdt genes (cdtA, cdtB, and cdtC) in C. jejuni has been shown to be necessary for the CDT toxin phenotype and cell pathology in humans cell in vitro (6). Likewise, the flaA gene was present in 46% (27/59) of C. jejuni isolates and 33% (1/3) of C. lari isolates. The flaA gene encodes the major flagellin protein in C. jejuni and has also been strongly associated with the ability of Campylobacter to infect an organism (8). In a study where the authors knocked out flagellin proteins, they demonstrated that a mutant C. jejuni strain lacking the flaA gene could no longer successfully invade cells (8). Note, however, that the presence of flaA genes or CDT genes does not necessary indicate their transcription or protein expression. Furthermore, C. jejuni strains may still have the ability to cause damage to human cells in the absence of flagellin A or in the absence of CDT toxin (8).
Campylobacter lari has previously been reported in low numbers in wild migratory birds (19) and aquatic mammals (e.g., northern elephant seals [10]) and has been associated with human disease (39, 40), although it might be less pathogenic than C. jejuni (39). The presence of CDT genes and flaA in one crow isolate suggests that C. lari from American crows could also be pathogenic.
Conclusions.
The high prevalence of C. jejuni, coupled with the occurrence of known virulence markers, suggests that crows shed Campylobacter spp. in their feces that are potentially pathogenic to humans. American crows are particularly relevant to the potential spread of pathogens because of their movement between human-dominated urban and agriculture landscapes (41). We note, however, that we did not directly test the extent to which crows and humans share Campylobacter strains or the potential transmission pathways linking crows and humans. Furthermore, it is possible that crows acquire the pathogen from humans (or domestic animals) but do not contribute to human disease epidemiology (i.e., a reverse zoonotic). Additional studies on the expression of each CDT protein and flagellin proteins, cytotoxicity, and other virulence factors, as well as antibiotic resistance markers, and full-genome sequencing to assess other virulence loci would further elucidate the potential role of the American crow as a transport host of Campylobacter in human disease epidemiology.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the UC Davis Veterinary Medicine Teaching Hospital for help with isolation and identification of Campylobacter isolates, particularly JoAnn Yee. Srijak Bhatnagar helped with 16S rRNA alignment and phylogenetic analysis.
Studies were funded by the University of California, Davis.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03393-13.
REFERENCES
- 1.Nachamkin I. 2003. Campylobacter and Arcobacter, p 902–914 In Murray PR, Baron EJ, Pfaller MA, Jorgensen JH, Yolken RH. (ed), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC [Google Scholar]
- 2.Coker AO, Isokpehi RD, Thomas BN, Amisu KO, Obi CL. 2002. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8:237–244. 10.3201/eid0803.010233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen EM. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85–89. 10.1046/j.1472-765X.2002.01143.x [DOI] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.09-1101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28–35. 10.3201/eid0501.990104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickett CL, Pesci EC, Cottle DL, Russell G, Erdem AN, Zeytin H. 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 64:2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elwell CA, Dreyfus LA. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37:952–963. 10.1046/j.1365-2958.2000.02070.x [DOI] [PubMed] [Google Scholar]
- 8.Wassenaar TM, Bleumink-Pluym NMC, van der Zeijst AMB. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sert V, Cans C, Tasca C, Bret-Bennis L, Oswald E, Ducommun B, De Rycke J. 1999. The bacterial cytolethal distending toxin (CDT) triggers a G2 cell cycle checkpoint in mammalian cells without preliminary induction of DNA strand breaks. Oncogene 18:6296–6304. 10.1038/sj.onc.1203007 [DOI] [PubMed] [Google Scholar]
- 10.Stoddard RA, Miller WG, Foley JE, Lawrence J, Gulland FM, Conrad PA, Byrne BA. 2007. Campylobacter insulaenigrae isolates from northern elephant seals (Mirounga angustirostris) in California. Appl. Environ. Microbiol. 73:1729–1735. 10.1128/AEM.01816-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broman T, Palmgren H, Bergstrom S, Sellin M, Waldenstrom J, Danielsson-Tham ML, Olsen B. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40:4594–4602. 10.1128/JCM.40.12.4594-4602.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colles FM, Dingle KE, Cody AJ, Maiden MC. 2008. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl. Environ. Microbiol. 74:3583–3590. 10.1128/AEM.02491-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colles FM, McCarthy ND, Howe JC, Devereux CL, Gosler AG, Maiden MC. 2009. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling). Environ. Microbiol. 11:258–267. 10.1111/j.1462-2920.2008.01773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misawa N, Shinohara S, Satoh H, Itoh H, Shinohara K, Shimomura K, Kondo F, Itoh K. 2000. Isolation of Campylobacter species from zoo animals and polymerase chain reaction-based random amplified polymorphism DNA analysis. Vet. Microbiol. 71:59–68. 10.1016/S0378-1135(99)00156-X [DOI] [PubMed] [Google Scholar]
- 15.Adesiyun AA, Caesar K, Inder L. 1998. Prevalence of Salmonella and Campylobacter species in animals at Emperor Valley Zoo, Trinidad. J. Zoo. Wildl. Med. 29:237–239 [PubMed] [Google Scholar]
- 16.Shreeve JE, Toszeghy M, Pattison M, Newell DG. 2000. Sequential spread of Campylobacter infection in a multipen broiler house. Avian Dis. 44:983–988. 10.2307/1593076 [DOI] [PubMed] [Google Scholar]
- 17.Petersen L, Nielsen EM, On SL. 2001. Serotype and genotype diversity and hatchery transmission of Campylobacter jejuni in commercial poultry flocks. Vet. Microbiol. 82:141–154. 10.1016/S0378-1135(01)00382-0 [DOI] [PubMed] [Google Scholar]
- 18.Broman T, Waldenström J, Dahlgren D, Carlsson I, Eliasson I, Olsen B. 2004. Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. J. Appl. Microbiol. 96:834–843. 10.1111/j.1365-2672.2004.02232.x [DOI] [PubMed] [Google Scholar]
- 19.Waldenström J, Broman T, Carlsson I, Hasselquist D, Achterberg RP, Wagenaar JA, Olsen B. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911–5917. 10.1128/AEM.68.12.5911-5917.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mdegela RH, Nonga HE, Ngowi HA, Kazwala RR. 2006. Prevalence of thermophilic Campylobacter infections in humans, chickens and crows in Morogoro, Tanzania. J. Vet. Med. B Infect. Dis. Vet. Public Health 53:116–121. 10.1111/j.1439-0450.2006.00926.x [DOI] [PubMed] [Google Scholar]
- 21.Ganapathy K, Saleha AA, Jaganathan M, Tan CG, Chong CT, Tang SC, Ideris A, Dare CM, Bradbury JM. 2007. Survey of Campylobacter, Salmonella and mycoplasmas in house crows (Corvus splendens) in Malaysia. Vet. Rec. 160:622–624. 10.1136/vr.160.18.622 [DOI] [PubMed] [Google Scholar]
- 22.Matsuda MST, Itoh Y, Takiguchi M, Furuhata K, Moore JE, Murayama O, Fukuyama M. 2002. First isolation of urease-positive thermophilic Campylobacter (UPTC) from crows (Corvus levaillantii) in Japan. Int. J. Hyg. Environ. Health 205:321–324. 10.1078/1438-4639-00157 [DOI] [PubMed] [Google Scholar]
- 23.Ito K, Kubokura Y, Kaneko K, Totake Y, Ogawa M. 1988. Occurrence of Campylobacter jejuni in free-living wild birds from Japan. J. Wildl. Dis. 24:467–470. 10.7589/0090-3558-24.3.467 [DOI] [PubMed] [Google Scholar]
- 24.Keller JI, Shriver WG, Waldenstrom J, Griekspoor P, Olsen B. 2011. Prevalence of Campylobacter in wild birds of the mid-Atlantic region, USA. J. Wildl. Dis. 47:750–754. 10.7589/0090-3558-47.3.750 [DOI] [PubMed] [Google Scholar]
- 25.Josefsen MH, Lübeck PS, Hansen F, Hoorfar J. 2004. Towards an international standard for PCR-based detection of foodborne thermotolerant campylobacters: interaction of enrichment media and pre-PCR treatment on carcass rinse samples. J. Microbiol. Methods 58:39–48. 10.1016/j.mimet.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 26.Bang DD, Nielsen EM, Scheutz F, Pedersen K, Handberg K, Madsen M. 2003. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 94:1003–1014. 10.1046/j.1365-2672.2003.01926.x [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 28.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawrocki EP. 2009. Structural RNA homology search and alignment using covariance models. Ph.D. thesis Washington University in Saint Louis, School of Medicine, Saint Louis, MO [Google Scholar]
- 30.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61:1061–1067. 10.1093/sysbio/sys062 [DOI] [PubMed] [Google Scholar]
- 32.Friis C, Wassenaar TM, Javed MA, Snipen L, Lagesen K, Hallin PF, Newell DG, Toszeghy M, Ridley A, Manning G, Ussery DW. 2010. Genomic characterization of Campylobacter jejuni strain M1. PLoS One 5:e12253. 10.1371/journal.pone.0012253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, He L, Li Q, Sun H, Gu Y, You Y, Meng F, Zhang J. 2010. Genomic characterization of the Guillain-Barre syndrome-associated Campylobacter jejuni ICDCCJ07001 isolate. PLoS One 5:e15060. 10.1371/journal.pone.0015060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- 35.Kabir SM, Kikuchi K, Asakura M, Shiramaru S, Tsuruoka N, Goto A, Hinenoya A, Yamasaki S. 2011. Evaluation of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the identification of Campylobacter strains isolated from diarrheal patients in Japan. Jpn. J. Infect. Dis. 64:19–27 [PubMed] [Google Scholar]
- 36.Burrough ER, Sahin O, Plummer PJ, Zhang Q, Yaeger MJ. 2009. Pathogenicity of an emergent, ovine abortifacient Campylobacter jejuni clone orally inoculated into pregnant guinea pigs. Am. J. Vet. Res. 70:1269–1276. 10.2460/ajvr.70.10.1269 [DOI] [PubMed] [Google Scholar]
- 37.Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. 2007. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J. Bacteriol. 189:8402–8403. 10.1128/JB.01404-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer SR, Gully PR, White JM, Pearson AD, Suckling WG, Jones DM, Rawes JC, Penner JL. 1983. Water-borne outbreak of Campylobacter gastroenteritis. Lancet i:287–290 [DOI] [PubMed] [Google Scholar]
- 39.Werno AM, Klena JD, Shaw GM, Murdoch DR. 2002. Fatal case of Campylobacter lari prosthetic joint infection and bacteremia in an immunocompetent patient. J. Clin. Microbiol. 40:1053–1055. 10.1128/JCM.40.3.1053-1055.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nachamkin I, Stowell C, Skalina D, Jones AM, Hoop RM, Smibert RM. 1984. Campylobacter laridis causing bacteremia in an immunosuppressed patient. Ann. Intern. Med. 101:55–57. 10.7326/0003-4819-101-1-55 [DOI] [PubMed] [Google Scholar]
- 41.Oravcova V, Zurek L, Townsend A, Clark AB, Ellis JC, Cizek A, Literak I. 16 July 2013. American crows as carriers of vancomycin-resistant enterococci with vanA gene. Environ. Microbiol.. 10.1111/1462-2920.12213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.