Abstract
Staphylococcus aureus USA300, the clonal type associated with epidemic community-acquired methicillin-resistant S. aureus (MRSA) infections, displays the giant protein Ebh on its surface. Mutations that disrupt the ebh reading frame increase the volume of staphylococcal cells and alter the cross wall, a membrane-enclosed peptidoglycan synthesis and assembly compartment. S. aureus ebh variants display increased sensitivity to oxacillin (methicillin) as well as susceptibility to complement-mediated killing. Mutations in ebh are associated with reduced survival of mutant staphylococci in blood and diminished virulence in mice. We propose that Ebh, following its secretion into the cross wall, contributes to the characteristic cell growth and envelope assembly pathways of S. aureus, thereby enabling complement resistance and the pathogenesis of staphylococcal infections.
INTRODUCTION
Staphylococcus aureus is both a commensal and an invasive pathogen that causes soft tissue infections, sepsis, endocarditis, and pneumonia (1). Owing to the frequent use of antibiotics, staphylococci have evolved resistance to many different drugs (2). These methicillin-resistant S. aureus (MRSA) strains are associated with therapeutic failure and increased mortality due to staphylococcal infections (3, 4). In the United States, clones of the USA300 lineage cause epidemics of community-acquired MRSA infections (2). Two drugs, daptomycin and linezolid, were recently licensed for the treatment of these infections (5, 6); however, MRSA strains evolve resistance against new antibiotics (7). Thus, there is a continued need for the identification of new drug targets and therapies that combat MRSA.
S. aureus is a spherical microbe with a thick cell wall envelope that is synthesized at the cross wall, a compartment formed during cell division from the plasma membranes of adjacent daughter cells (8). Upon completion of peptidoglycan synthesis, staphylococci split cross walls down the middle, thereby separating daughter cells that form new cross walls in a plane perpendicular to their previous division plane (9). Cell wall synthesis and cell separation at the cross wall require protein traffic into this compartment (10–13). Several precursor proteins with signal peptides that bear a YSIRK-G/S motif have been shown to be secreted at the cross wall, including sortase-anchored surface proteins (12), Geh (lipase) (14), and LytN, a murein hydrolase responsible for cross wall splitting (10). Although the mechanism of precursor secretion into the cross wall has not been revealed, the purpose of such traffic is either the display of cell wall proteins on the bacterial surface or a contribution of secreted proteins to cross wall synthesis and cell separation (15). Bioinformatic analysis identified Ebh, a giant precursor, as harboring the YSIRK-G/S signal peptide (Fig. 1A) (16, 17). A specific role for Ebh during the cell cycle of S. aureus was heretofore not known.
FIG 1.
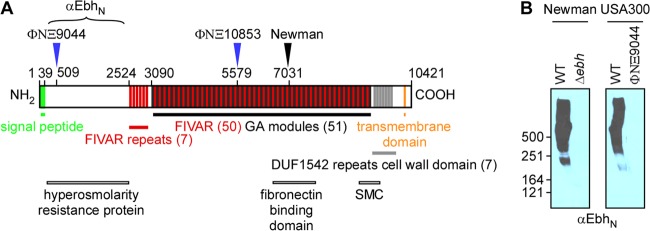
The giant Ebh protein of Staphylococcus aureus. (A) Schematic of Ebh in Staphylococcus aureus USA300 LAC and its variants. The diagram of Ebh domains includes the N-terminal YSIRK-G/S signal peptide, the hyperosmolarity resistance region, FIVAR and GA domain repeats, the SMC region, DUF1542, and the transmembrane and cytoplasmic domains. Blue arrows denote the positions of transposon insertions ΦNΞ9044 and ΦNΞ10853, which lead to truncated proteins of 509 and 5,579 residues, respectively. Compared to wild-type (WT) strain USA300 LAC, ΦNΞ9044 represents the ebh null mutant. S. aureus Newman harbors a truncated ebh gene with a termination codon after codon 7031 (black arrow). (B) Immunoblot experiments with staphylococcal cell lysates separated by 6% SDS-PAGE to detect Ebh expression in USA300 LAC compared with its ΦNΞ9044 ebh variant and in S. aureus Newman compared with its Δebh mutant. Rabbit immune serum against EbhN (residues 42 to 2524) was used to detect Ebh. Numbers indicate molecular mass markers (kDa).
Also described as encoding an extracellular matrix binding protein in Staphylococcus epidermidis (18, 19), ebh is conserved in S. aureus, S. epidermidis, Staphylococcus warneri, and Staphylococcus haemolyticus (20) but is not found in the Staphylococcus intermedius group and in Staphylococcus lugdunensis, species that do not colonize humans and are infrequently associated with human disease (21, 22). ebh appears to be transcribed as a monocistronic, 31,266-nucleotide gene in strain USA300. The predicted 10,421-amino-acid Ebh precursor comprises an N-terminal YSIRK-G/S signal peptide (amino acid [aa] residues 1 to 39), an N-terminal domain annotated as a hyperosmolarity resistance domain (EbhN; residues 179 to 2530) (23), 7 repeats of the 54-residue FIVAR domain (possibly associated with polysaccharide binding), 50 repeats of the 123-residue FIVAR-GA domain (albumin binding), 7 repeats of DUF1542 (a 72-residue domain of unknown function), a putative SMC domain (structural maintenance of chromosomes; residues 8976 to 9576), a transmembrane domain (residues 10227 to 10247), and, finally, the positively charged cytoplasmic domain (Fig. 1A) (24). Clones of the USA300 lineage, as well as several other clinical isolates, harbor intact ebh open reading frames (ORFs) (25). S. aureus isolates that have been maintained in research laboratories harbor nucleotide changes truncating the ebh ORF at coding sequences for the FIVAR-GA or DUF1542 repeats. For example, in strain Newman, Ebh is only 7,031 residues long (Fig. 1A) (26–28).
Recombinant purified FIVAR domains of Ebh (or its Embp homologue) have been reported to bind fibronectin, heparin, hyaluronate, and plasminogen; nevertheless, the physiological relevance of such associations is not yet appreciated (16, 18, 19). X-ray crystallography revealed FIVAR-GA as double (FIVAR)- and triple (GA)-helical bundle structures with an elongated shape (24, 29). Other work reported that S. aureus 8325-4 ebh mutants display increased sensitivity to teicoplanin (a glycopeptide antibiotic), osmotic stress, and Triton X-100 (23). To the best of our knowledge, previous studies on S. aureus ebh were performed with strains that carry truncating mutations in this gene. Previous work also left unresolved whether the giant Ebh protein contributes to methicillin resistance or to the pathogenesis of staphylococcal infections.
MATERIALS AND METHODS
Ethics statement.
Experiments with blood from human volunteers involved a protocol that was reviewed, approved, and performed under the regulatory supervision of the University of Chicago's Institutional Review Board (IRB). Written, informed consent was provided by all volunteers. Animal experiments involving S. aureus challenge followed protocols that were reviewed, approved, and performed under the regulatory supervision of the University of Chicago's Institutional Biosafety Committee (IBC) and the Institutional Animal Care and Use Committee (IACUC). Animals were handled by the University of Chicago Animal Resource Center, which is accredited by the American Association for Accreditation of Laboratory Animal Care and the Department of Health and Human Services. BALB/c mice and New Zealand White rabbits were purchased from Charles River Laboratories and Harlan Sprague Dawley, respectively. The statistical analysis of mouse survival data during staphylococcal sepsis was analyzed with the two-tailed log rank test. The results of all animal experiments were examined for reproducibility.
Bacterial strains and growth.
S. aureus strains were cultured on tryptic soy agar (TSA) or in tryptic soy broth (TSB) at 37°C. Escherichia coli strains DH5α and BL21(DE3) were cultured on Luria agar or in Luria broth at 37°C. Ampicillin (100 μg/ml) and erythromycin (10 μg/ml) were used for plasmid and transposon mutant selection, respectively.
Transposon mutagenesis.
Insertional mutations ΦNΞ9044 and ΦNΞ10853 from the Phoenix library were transduced into S. aureus USA300 LAC (30). Each mutant carries the bursa aurealis transposon, containing an erythromycin resistance cassette in the ebh gene (Fig. 1A); mutations were verified by DNA sequencing (30). Briefly, chromosomal DNA was extracted (Promega Wizard kit), digested with AciI, religated with T4 ligase to form individual episomes, and PCR amplified using Martn-F and Martn-R, primers specific to the bursa aurealis transposon (30). PCR products were sequenced to verify the site of transposon insertions in the ebh target of USA300 LAC.
Deletion mutagenesis.
DNA sequences 1 kb upstream and downstream of ebh were PCR amplified using the primers attB1_ebh (GGGGACAAGTTTGTACAAAAAAGCAGGCTGTTAGATCAAGGCTATTAACGC), ebh1_sacII (GGTTCCGCGGGGAGCACCGATTGACATCAC), ebh2_sacII (GGTTCCGCGGCTCCTTATCTTGTTGTTATGTC), and attbB2_ebh (GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAGAATTAGGTGTAACCTC). The fragments were cloned into pKOR1 by use of a BP clonase II kit (Invitrogen) (31). The resulting plasmid, pΔebh, was electroporated into S. aureus Newman (26). Transformants were subjected to a temperature shift, which induced allelic exchange to generate the corresponding Δebh deletion (31). Mutants were verified by PCR amplification of the gene locus, DNA sequencing, and immunoblot analysis (Fig. 1B).
Localizing Ebh to the staphylococcal cross wall.
Plasmid pEbhSP-mCherry was created via splicing by overhang extension PCR (SOE PCR). A fusion protein of the signal peptide for Ebh (residues 1 to 126; aa 1 to 42) and mCherry was created by amplification of the mCherry coding sequence with primers mCh_Ebh_f (AACAAATCAACCAGCAAGGGCGAGGAGGATAACATG) and mCh_NheI_rc (GATCGCTAGCTTACTTGTACAGCTCGTCCATGCC). The coding sequence for the Ebh signal peptide was amplified with primers EbhSP_EcoRI_f (GATCGAATTCGTGAATTATCGTGATAAAATTCAAAAG) and EbhSP_mCh_rc (GGTTGATTTGTTTCAGCAGCATGCTCGCCCTTGCT). PCR products were used as templates in the following amplification step, using primers EbhSP_EcoRI_f and mCh_NheI_rc, and the product from this reaction was purified, digested with EcoRI/NheI, and ligated into predigested pMF312 (a derivative plasmid of pOS1 containing the iTet promoter) (32) to generate plasmid pEbhSP-mCherry. Transformants were analyzed by DNA sequencing to confirm the correct sequence and subsequently transformed into S. aureus RN4220. Plasmid DNA was extracted from transformants and electroporated into S. aureus Newman or its Δebh variant. Expression was determined by fluorescence microscopy. Overnight cultures of S. aureus Newman and the Δebh mutant with empty vector or plasmid pEbhSP-mCherry were diluted in fresh culture medium, grown to mid-log phase, and induced with anhydrotetracycline. Cells were fixed, stained with BODIPY-vancomycin to delineate cell walls, and visualized by confocal fluorescence microscopy.
EbhN purification and rabbit antiserum.
The coding sequence of EbhN (codons 42 to 2524) was cloned as a PCR fragment with flanking XhoI and BglII sites into pET15b (33) cut with the same restriction enzymes to generate pT7-EbhN. PCR amplification used S. aureus USA300 LAC template DNA and primers Ebh-XhoI (gaaCTCGAGgctgaaacaaatcaaccagc) and Ebh-BglII (AGTAGATCTTTGTGGGAAATTAACCCAACG). Overnight cultures of E. coli BL21(DE3)(pT7-EbhN) were diluted 1:100 in fresh LB medium, grown at 37°C to an A600 of 0.5, and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. Cells were centrifuged at 6,000 × g, suspended in 1× column buffer (0.1 M Tris-HCl [pH 7.5], 0.5 M NaCl), and lysed in a French press at 14,000 lb/in2. Lysates were subjected to ultracentrifugation at 40,000 × g for 30 min, and supernatants were subjected to affinity chromatography on Ni-nitrilotriacetic acid (Ni-NTA) columns. Each column was washed with column buffer containing 25 mM imidazole, followed by elution with 500 mM imidazole. The eluate was dialyzed against 1× phosphate-buffered saline (PBS). To remove endotoxin, 1:1,000 Triton X-114 was added, and the solution was chilled for 5 min, incubated at 37°C for 10 min, and centrifuged at 13,000 × g. Supernatant was loaded onto a HiTrap desalting column to remove Triton X-114. A rabbit (6-month-old New Zealand White female) was immunized with 100 μg EbhN emulsified in complete Freund's adjuvant (Difco) for initial immunization or emulsified in incomplete Freund's adjuvant for booster immunizations on days 24 and 48. On day 60, the rabbit was bled. Serum was recovered after centrifugation of coagulated blood at 2,000 × g for 10 min.
Immunoblotting.
Overnight cultures of staphylococci in TSB were diluted 1:100 in fresh medium and grown with shaking at 37°C to an A600 of 0.4. One milliliter of each culture was lysed by adding 5 μl of lysostaphin from a 2-mg/ml stock for 30 min, followed by the addition of 75 μl of 100% (wt/vol) trichloroacetic acid (TCA) solution. Samples were incubated on ice for 10 min and centrifuged for 10 min at 13,000 × g. Protein sediment was washed with 1 ml ice-cold 100% acetone, air dried, and solubilized in 50 μl sample buffer (4% SDS, 50 mM Tris-HCl [pH 8.0], 10% glycerol, and bromophenol blue). Protein samples were separated by 6% SDS-PAGE and subjected to immunoblotting with EbhN antiserum and secondary mouse anti-rabbit Alexa Fluor 680 conjugate. Immunoreactive signals were quantified with a Li-Cor Odyssey instrument.
Immunofluorescence microscopy.
Overnight cultures of staphylococci in TSB were diluted 1:100 in fresh medium and grown to an A600 of 0.4. One milliliter of culture was centrifuged to sediment bacteria. Staphylococci were washed in 1× PBS and fixed (2.5% paraformaldehyde, 0.006% glutaraldehyde in 1× PBS, pH 7.4) for 20 min at room temperature. Bacteria were washed 3 times with PBS and suspended in 100 μl PBS, and a 30-μl droplet was added to a coverslip precoated with poly-l-lysine. For precoating, 60 μl of poly-l-lysine solution (Fisher) was placed on a glass coverslip for 5 min, followed by 3 washes with water. Cells adhering to the coverslip were washed 3 times with 60-μl droplets of PBS and then placed in blocking solution (3% bovine serum albumin [BSA], 1:200 human IgG [Sigma] in PBS) for 30 min. Blocking solution containing the specific rabbit antibody (1:1,000) was added to cells, and coverslips were incubated for 1 h. Coverslips were washed 10 times, and secondary antibody solution was added for 1 h (3% BSA, 1:200 Alexa Fluor 647-conjugated mouse anti-rabbit IgG). Slides were washed again in PBS, and 1:100 BODIPY-vancomycin and 1:1,000 Hoechst dye in PBS were added for 5 min. Coverslips were washed three times, mounted on glass slides with N-propylgallate, and sealed with nail polish. For visualization of mCherry, cells were processed with a similar protocol, but without antibody staining. Slides were stored at 4°C, and images were collected using a Leica SP5 AOBS spectral two-photon confocal microscope.
Transmission electron microscopy.
Staphylococcal cells were cultured in TSB to mid-log phase in the presence or absence of 2 μg/ml oxacillin, centrifuged, fixed in 3% paraformaldehyde, embedded in epoxy, and thin sectioned. Sections were stained with uranyl acetate and viewed using a Tecnai F30 (Philips/FEI) transmission electron microscope (field emission gun, 300-kV accelerating voltage, and magnification of ×10,000 to ×40,000) and a high-performance charge-coupled device camera with 4,000-by-4,000 resolution. Images were acquired using Gatan DigitalMicrograph software, contrast adjusted, and processed using Adobe Photoshop.
Oxacillin sensitivity.
Overnight cultures of S. aureus were normalized to an A600 of 4.0 and diluted 1:1,000 in TSB with or without oxacillin. The absorbance at 600 nm was measured for 18 h in a 96-well plate reader at 37°C with constant shaking.
Staphylococcal survival in blood or plasma.
Overnight cultures of S. aureus strains were diluted 1:100 into fresh TSB and grown at 37°C until they reached an A600 of 0.4. One milliliter of culture was washed and suspended in 1 ml PBS to obtain 1 × 108 CFU/ml stock. Whole blood from naive 6-week-old BALB/c mice was collected, and lepirudin (Refludan) was added to a final concentration of 50 μg/ml. To obtain fresh plasma, whole blood was centrifuged at 2,000 × g for 3 min to sediment blood cells, and the supernatant was transferred to a fresh tube. Plasma samples were either left untreated or heat inactivated at 60°C for 30 min with occasional mixing. Blood or plasma was aliquoted (450 μl) and mixed with 50 μl bacterial stock to a final concentration of 5 × 106 CFU/ml. Samples were incubated at 37°C with slow rotation. Aliquots (50 μl) were removed at timed intervals, mixed 1:1 with 2% saponin in PBS, and incubated on ice for 30 min. Five serial 10-fold dilutions were prepared, and 10-μl aliquots were spread on TSA for colony formation and enumeration.
Complement deposition assays.
S. aureus (1 × 107 CFU/ml stock) was incubated in PBS with 10% human plasma in a final volume of 1 ml. Reaction mixtures were incubated at 37°C with rotation, and aliquots of 100 μl were removed and quenched 1:10 in ice-cold PBS at 5-min intervals. Samples were washed three times with cold PBS, labeled with fluorescein isothiocyanate (FITC)-conjugated anti-C3 F(ab)2, washed three more times, fixed in 4% formalin, and analyzed by flow cytometry using an LSRII instrument. Aliquots that had been removed at 0, 5, and 15 min were also processed for immunoblotting and immunofluorescence microscopy. For immunoblotting, samples were quenched and then centrifuged at 13,000 × g, and the sediment was processed as described above. Briefly, the sediment was suspended in 50 μl sample buffer, boiled for 3 min, and centrifuged at 13,000 × g to remove insoluble material. Solubilized proteins were subjected to immunoblotting with goat anti-human C3 primary antibody and Alexa Fluor 680-conjugated donkey anti-goat antibody. SDS-PAGE gels were imaged using a Li-Cor Odyssey machine. For immunofluorescence microscopy, aliquots were removed at 0 and 5 min, quenched and washed as described above, fixed, and then stained with FITC-conjugated anti-C3 F(ab)2, using the aforementioned protocol for confocal fluorescence microscopy.
Mouse challenge experiments.
Overnight cultures of S. aureus were diluted 1:100 in fresh TSB and grown to an A600 of 0.4. Bacteria were sedimented by centrifugation at 7,500 × g, washed, and suspended in 1× PBS. Six-week-old female BALB/c mice (n = 10) were injected in the periorbital venous sinus with 5 × 106 CFU S. aureus USA300 LAC in 100 μl PBS. On day 5 postinfection, mice were euthanized (CO2 asphyxiation and cervical dislocation), and kidneys were removed during necropsy. All organs were examined for surface lesions, and 8 to 10 right kidneys were analyzed for histopathology by staining thin-sectioned paraffin-embedded tissues with hematoxylin-eosin. These slides were examined by light microscopy for abscess formation. For the lethal challenge model, all experimental conditions remained identical, except that 5 × 107 CFU S. aureus USA300 LAC were injected intravenously and mice were monitored for survival over 14 days postinfection.
RESULTS
S. aureus ebh mutants.
Compared with MRSA isolate USA300 LAC and its related clones (25, 34), S. aureus Newman harbors a nucleotide substitution in ebh that truncates the Ebh polypeptide at amino acid 7032 (26). Using pKOR1 allelic replacement technology (31), we deleted the entire ebh open reading frame of S. aureus Newman, thereby generating the Δebh variant. We also transduced two bursa aurealis alleles originally isolated from S. aureus strain Newman (30) into USA300 LAC, thereby terminating the open reading frame of ebh after codon 509 (ΦNΞ9044) or after codon 5579 (ΦNΞ10853) (Fig. 1A). Mutational lesions were verified by DNA sequencing. To analyze the wild-type and mutant strains for the synthesis of ebh gene products, we purified recombinant His-tagged EbhN (Ebh residues 42 to 2524) from E. coli lysates by affinity chromatography and raised rabbit immune serum (anti-EbhN). Mid-log-phase cultures of S. aureus Newman or USA300 LAC and ebh variants were centrifuged, the peptidoglycan envelope was digested with lysostaphin, and proteins were precipitated with TCA. Proteins were subjected to 6% SDS-PAGE and analyzed by immunoblotting with anti-EbhN. Lysates derived from S. aureus Newman and USA300 harbored anti-EbhN-immunoreactive species ranging in size from 250 kDa to >500 kDa (Fig. 1B). Neither the Δebh variant of S. aureus Newman nor ΦNΞ9044 generated EbhN-immunoreactive species, indicating that ebh expression had been abolished (Fig. 1B). Because of the very large size of the ebh gene, we were unable to complement the mutant phenotypes with a plasmid-borne copy of the wild-type ebh gene.
Ebh is displayed on the surface of S. aureus.
Immunofluorescence microscopy was used to localize Ebh in bacteria. Staphylococci were grown to mid-log phase, fixed with paraformaldehyde, and stained with either BODIPY-vancomycin (green fluorescence) or anti-EbhN and Alexa Fluor 647 conjugate (red fluorescence). BODIPY-vancomycin stains the cell wall envelope of S. aureus and is abundantly deposited at the cross wall (11). Anti-EbhN antibody staining was found on the surface of S. aureus USA300 LAC but not on S. aureus ΦNΞ9044 (Fig. 2A). Staphylococci displayed a spherical distribution of anti-EbhN staining, similar to that of sortase-anchored surface proteins secreted via YSIRK-G/S-type signal peptides (11). Anti-EbhN staining also generated immunofluorescence signals on the surface of S. aureus Newman, albeit that the fluorescence signals were less intense and less homogeneous than those for USA300 LAC (Fig. 2B). This phenotype may be due to the truncation of Ebh in S. aureus Newman, which eliminates the C-terminal transmembrane domain of the polypeptide (Fig. 1A). It is, however, not clear why truncated Ebh remains associated with the surface of S. aureus Newman. We observed occasional aggregates of anti-EbhN staining at cell contact sites of S. aureus Newman and its Δebh variant (Fig. 2B); these signals were interpreted as nonimmune binding of antibodies to staphylococcal protein A. Anti-EbhN staining on S. aureus USA300 and Newman did not occur at the cross wall, presumably because antibodies cannot access this compartment (Fig. 2A and B).
FIG 2.
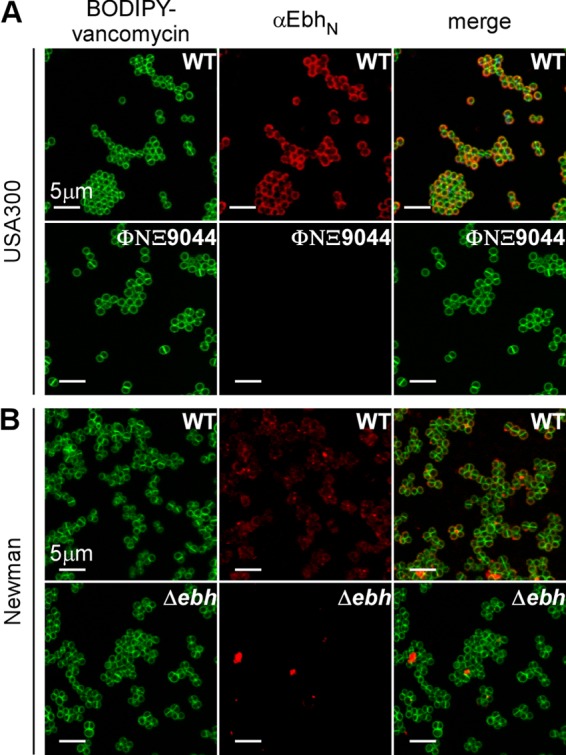
Ebh is displayed on the staphylococcal surface. Mid-log-phase staphylococcal cells were stained with BODIPY-vancomycin (green) to delineate peptidoglycan and with anti-EbhN and Alexa Fluor 647 to localize Ebh (red). Immunofluorescence microscopy images were acquired for wild-type strain USA300 LAC and its ΦNΞ9044 ebh variant (A) as well as for wild-type strain S. aureus Newman and its Δebh mutant (B). Ebh-specific staining was observed for USA300 LAC and S. aureus Newman but not for the ΦNΞ9044 and Δebh variants.
EbhSP-mCherry is secreted into the cross wall of S. aureus.
We wondered whether the YSIRK-G/S signal peptide of Ebh is sufficient to direct the secretion of a hybrid mCherry protein into the cross wall, as has been observed for LytN-mCherry and Geh-mCherry (10, 14). A hybrid gene comprising nucleic acid sequences for the tetracycline-inducible promoter and the ebh sequence (codons 1 to 62) were fused to the mCherry open reading frame, thereby generating pEbhSP-mCherry. This plasmid was transformed into S. aureus Newman or its Δebh variant. Staphylococci were grown to mid-log phase, induced for EbhSP-mCherry expression with anhydrotetracycline (35), fixed, and stained with BODIPY-vancomycin prior to fluorescence microscopy (Fig. 3). As expected, BODIPY-vancomycin (green) stained staphylococcal peptidoglycan, in particular the cross wall (Fig. 3). S. aureus Newman (without pEbhSP-mCherry) did not generate red fluorescence signals (Fig. 3). Staphylococci that had been transformed with pEbhSP-mCherry displayed mCherry signals at the cross wall for both wild-type and Δebh variant strains (Fig. 3). Cross wall staining of pEbhSP-mCherry cells is dependent on the Ebh signal peptide, as staphylococci expressing mCherry alone generated diffuse cytoplasmic staining (10, 14). These data suggest that the YSIRK-G/S signal peptide of Ebh is sufficient to direct reporter proteins to the cross wall, as has been observed for other YSIRK-G/S signal peptides (12, 14). Furthermore, the ebh gene is not essential for the secretion of YSIRK-G/S precursors into the cross wall, as EbhSP-mCherry trafficking was observed in both wild-type and Δebh mutant strains.
FIG 3.
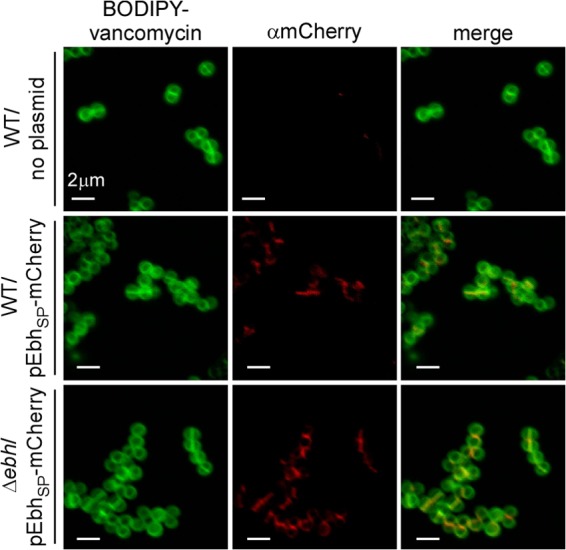
EbhSP-mCherry is secreted into the cross wall. S. aureus Newman and Δebh cells with or without pEbhSP-mCherry were grown to mid-log phase and induced with anhydrotetracycline. Cells were fixed and stained with BODIPY-vancomycin to delineate the cell wall (green). Samples were analyzed via confocal fluorescence microscopy to reveal mCherry staining (red).
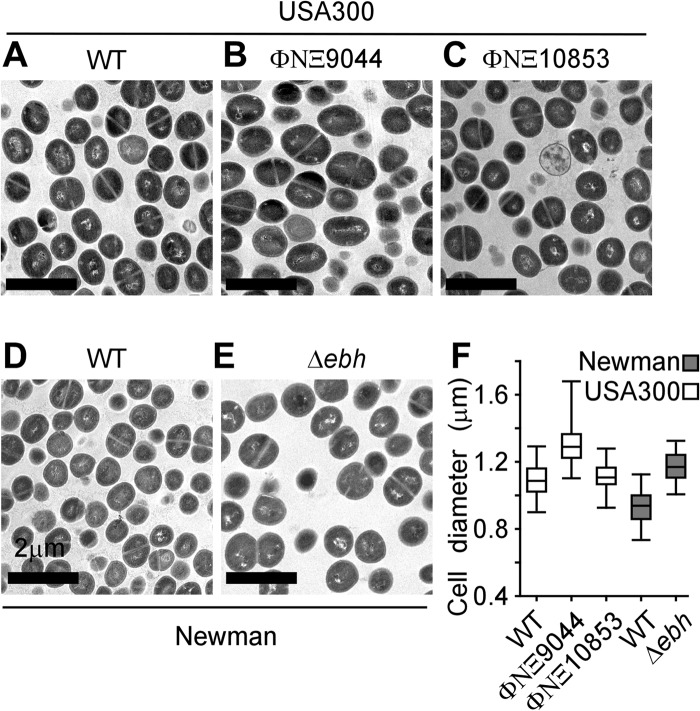
ebh is a cell size determinant of S. aureus.
Staphylococci from mid-log-phase cultures were sedimented by centrifugation, suspended in PBS, and fixed in glutaraldehyde. Samples were embedded in epoxy, thin sectioned, and stained with uranyl acetate prior to viewing by transmission electron microscopy (Fig. 4A to E). To quantify the sizes of individual staphylococci, we measured the diameters of thin-sectioned cells that had formed a cross wall and displayed two daughter cells with equal volumes, thereby ensuring that spherical cells selected for measurements had been cut across the middle. By calculating the average diameter (± standard error of the mean) for 80 to 100 cells per sample, we noted that S. aureus USA300 (diameter of 1.1 ± 0.2 μm) and S. aureus Newman (0.9 ± 0.3 μm) differed in size (Fig. 4F). Furthermore, cells of the ebh mutant strain ΦNΞ9044 (1.3 ± 0.4 μm) were, on average, 0.2 μm (20%) larger than those of wild-type parent strain USA300 LAC (Fig. 4D). Similar observations were made in determining the average cell diameter for the Δebh variant of S. aureus Newman (1.1 ± 0.3 μm) (Fig. 4F). Note that the ΦNΞ10853 insertion, which truncated ebh after codon 5579, did not affect the cell size of USA300 LAC (1.1 ± 0.2 μm) (Fig. 4F). Thus, mutations that abolish ebh expression (Δebh or ΦNΞ9044), but not mutations that truncate ebh in the FIVAR-GA module repeats, increase the cell size of staphylococci.
FIG 4.
The giant protein Ebh is a cell size determinant of S. aureus. (A to E) S. aureus USA300 LAC (A) and its ebh variants ΦNΞ9044 (B) and ΦNΞ10853 (C), as well as S. aureus Newman (D) and its Δebh mutant (E), were fixed, thin sectioned, and viewed by transmission electron microscopy. (F) The diameters of dividing cells were measured, and the results were plotted in a dot-and-whisker plot. The ebh mutant cells were larger than their wild-type parents: for Newman versus its Δebh variant, P < 1 × 10−4; and for USA300 LAC versus ΦNΞ9044, P < 1 × 10−4. Measurements represent the average diameters for 80 to 100 dividing cells. Statistical significance and P values were calculated with the Mann-Whitney U test.
ebh is required for methicillin (oxacillin) resistance.
S. aureus USA300 LAC, but not S. aureus Newman, is resistant to methicillin, a penicillinase-resistant β-lactam compound that is no longer commercially available (34). Over the past decade, oxacillin has been used as a surrogate for methicillin; this β-lactam is also resistant to cleavage by penicillinase. Expression of PBP2a (mecA) in S. aureus USA300 confers resistance to both methicillin and oxacillin (36). Following the addition of 2 μg ml−1 oxacillin to culture media, the growth of wild-type USA300 LAC was initially delayed but quickly resumed, as expression of PBP2a supports resistance to this antibiotic (37) (Fig. 5A and B). In contrast, oxacillin inhibited the growth of the ΦNΞ9044 ebh mutant, whereas ΦNΞ10853, encoding a longer Ebh variant, exhibited an intermediate phenotype of oxacillin sensitivity (Fig. 5A and B).
FIG 5.
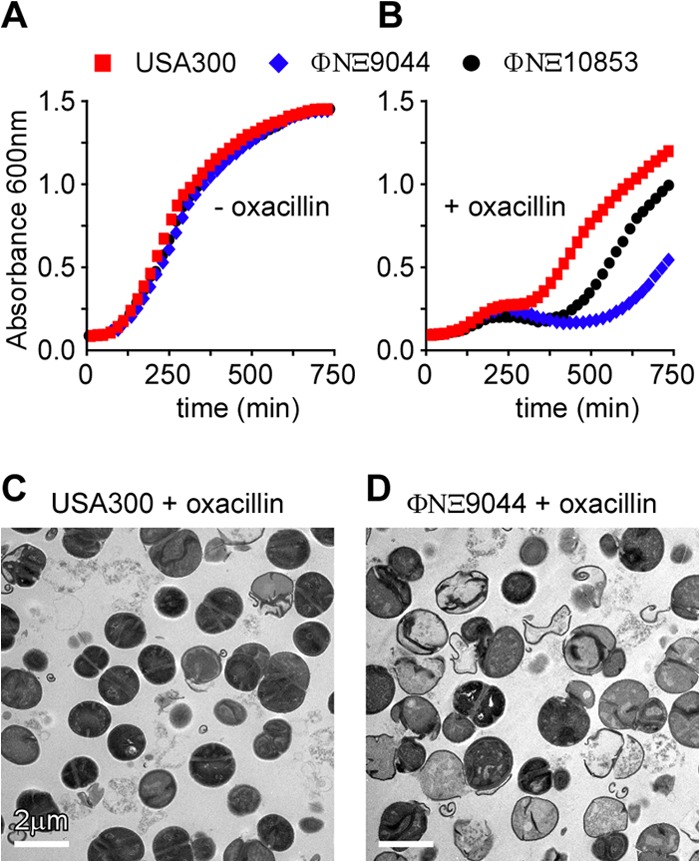
Ebh contributes to S. aureus methicillin resistance. Growth of S. aureus USA300 LAC and its ebh variants ΦNΞ9044 and ΦNΞ10853 was determined for samples diluted from overnight cultures in TSB without (A) or with (B) 2 μg/ml oxacillin. Growth was measured as increased absorbance at 600 nm (A600). S. aureus USA300 LAC (C) and its ebh variant ΦNΞ9044 (D) were grown to mid-log phase in 2 μg/ml oxacillin, fixed, thin sectioned, and viewed by transmission electron microscopy.
Electron microscopy of USA300 LAC and its ebh mutant ΦNΞ9044 grown in the presence of 2 μg ml−1 oxacillin revealed differences in cell wall integrity and cell structure. Wild-type USA300 LAC cells displayed the physiological cell and cell wall envelope architecture (Fig. 5C). In contrast, many cells of the ΦNΞ9044 variant had lysed: empty murein sacculi displayed defects, which occurred as small holes in the peripheral cell wall and as structural deformations of the cross wall (Fig. 5D). These data suggest that Ebh is involved in the physiological assembly, integrity, and separation of the cell wall when staphylococci synthesize peptidoglycan with PBP2a (MecA) in the presence of oxacillin.
ebh is a determinant of S. aureus complement resistance.
We wondered whether the structural changes to the envelope of ebh mutant staphylococci affect their ability to survive and replicate in host tissues. To begin to address this question, we first inoculated wild-type and ebh mutant staphylococci into fresh human blood (38). Studies with blood samples from many different volunteers had shown that the capacity of human blood to kill S. aureus USA300 LAC varies by more than 2 orders of magnitude (log CFU) (39). Such phenotypic variation may depend on hereditary factors; however, it is more likely to be influenced by adaptive immune responses that are based on individual encounters with the pathogen. To study the effects of the ebh gene, we selected blood samples from human volunteers that showed a moderate level (23.7%) of S. aureus USA300 LAC killing over a 30-min incubation period (Fig. 6A). Compared to USA300 LAC, the ebh variant ΦNΞ9044, but not ΦNΞ10853, was more susceptible to killing in blood, with 50.3% killing of ΦNΞ9044 (Fig. 6A). A similar phenotype was observed in comparing the survival of S. aureus Newman and its Δebh variant, with 16.8% and 32% killing of staphylococci in human blood, respectively (Fig. 6A). To determine whether ebh-mediated survival of staphylococci is specific for human blood, the experiment was performed with blood from naive mice that had been raised under pathogen-free conditions and lacked antibodies against staphylococci (38). Mouse blood did not affect the survival of USA300 LAC (1.3% killing) or the ΦNΞ10853 variant, whereas 41.8% of ΦNΞ9044 cells were killed during incubation with mouse blood (Fig. 6D). Bacterial killing in mouse blood may be a feature of phagocytic cells that recognize opsonized staphylococci and, following phagocytosis, derive oxygen radicals and lysosomal vesicles to eliminate the pathogen. At least for Gram-negative bacteria, complement deposition in the bacterial envelope triggers the formation of membrane attack complexes (MACs) that kill microbes without assistance from phagocytic cells (40). To distinguish between such possibilities, human blood was centrifuged, causing red blood cells and immune cells to sediment. Plasma samples were removed with the supernatant and used as a source of complement for incubation with staphylococci (Fig. 6B). Incubation in human plasma did not affect the survival of S. aureus USA300 LAC, in agreement with the general model that staphylococci cannot be killed by complement. In contrast, 38.9% of the ebh mutant ΦNΞ9044 cells were killed in human plasma (62.7% survival; P = 0.0317), whereas 19% of ΦNΞ10853 cells (P = 0.285) were killed. To verify whether the observed killing of ΦNΞ9044 in plasma was indeed caused by complement, human plasma was heat treated at 56°C to inactivate C3 convertases; heat treatment indeed abrogated killing of ΦNΞ9044 (Fig. 6C). These data suggest that the deposition of complement in the envelope of the ebh mutant may trigger staphylococcal killing in blood.
FIG 6.
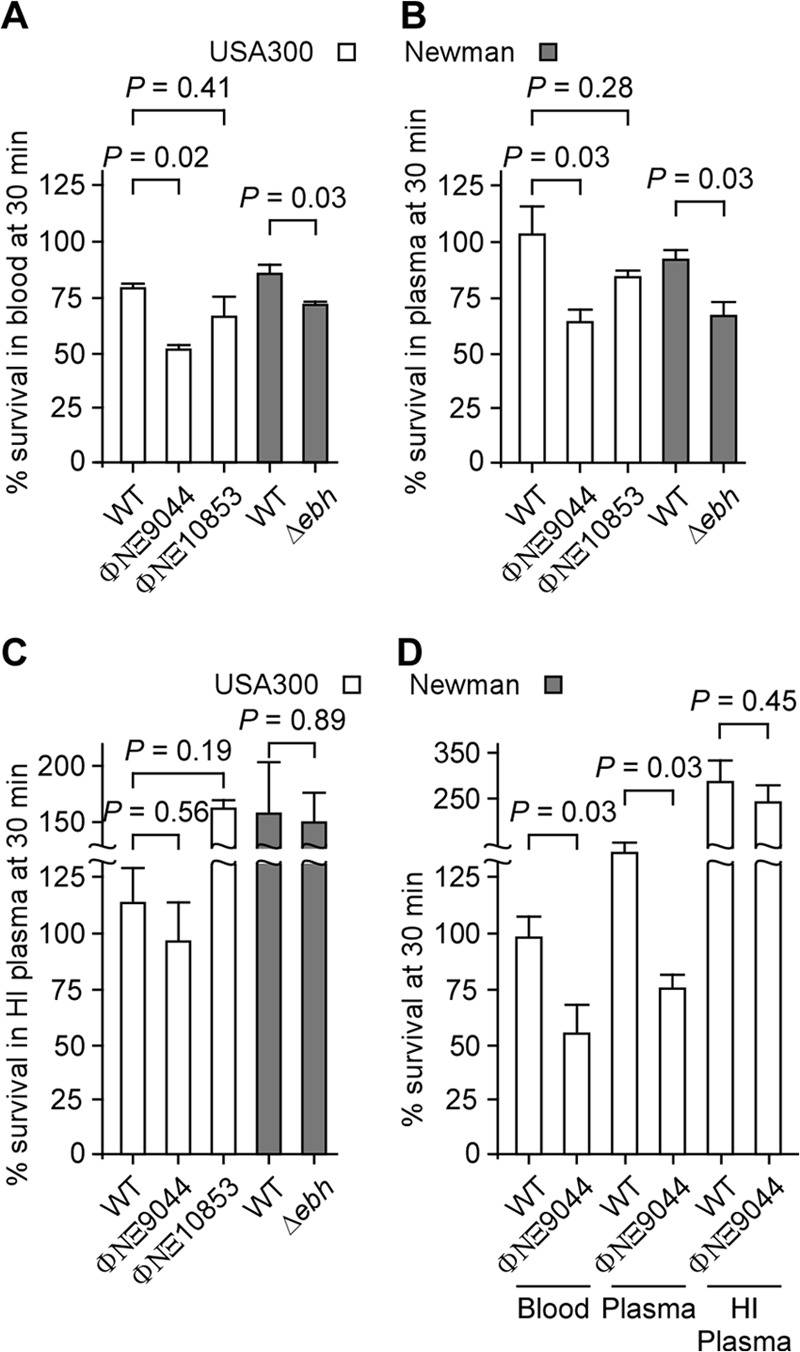
Ebh contributes to S. aureus resistance against complement. (A) S. aureus USA300 LAC and its ebh variants ΦNΞ9044 and ΦNΞ10853, as well as S. aureus Newman and its Δebh mutant, were incubated in human blood for 30 min. Bacterial survival was assessed by enumerating CFU on agar plates before and after incubation in blood and calculating the percent survival. (B) Staphylococci were incubated in human plasma for 30 min, and the percent survival was calculated. (C) Staphylococci were incubated in heat-inactivated (HI) human plasma for 30 min, and the percent survival was calculated. (D) Staphylococci were incubated in mouse blood, mouse plasma, or heat-inactivated mouse plasma for 30 min, and the percent survival was calculated. Data represent the averages for three or four independent trials. Statistical significance and P values were calculated with the Mann-Whitney U test.
Complement deposition on the staphylococcal surface.
Complement-mediated killing can be initiated via the deposition of antibody or lectin and complement convertase C3b or C5 binding to bacterial surfaces; this triggers a series of proteolytic cascades that result in the formation of a MAC (40). As alluded to above, complement proteins are effective at lysing Gram-negative bacteria, whereas the thick peptidoglycan envelope of Gram-positive pathogens prevents access of MACs to bacterial membranes (41). S. aureus isolates are particularly resistant to complement-mediated killing, as these microbes secrete SCIN, Sak, and CHIPS—proteins that neutralize or destroy complement (42–44). Furthermore, S. aureus uses cell wall-associated proteins (Eap, Efb, and Sbi) to sequester complement (45–47). To test whether Ebh protects staphylococci from complement deposition, we measured the abundance of C3b in the envelope of wild-type or ebh mutant cells by using both flow cytometry and immunofluorescence microscopy. Mid-log-phase wild-type and ebh mutant staphylococci were incubated with human plasma, and aliquots were analyzed at 5-min intervals. Samples were stained with anti-human C3b FITC-conjugated antibody and quantified by flow cytometry. We observed increased C3b deposition in the envelope of the ebh mutant ΦNΞ9044 compared to wild-type USA300 LAC (Fig. 7A). Wild-type and ebh mutant staphylococci were incubated with plasma for various times, washed with PBS, boiled in sample buffer, and subjected to immunoblotting with anti-C3. We observed increased deposition of C3 on ebh mutant surfaces (increase in anti-C3 from 0 to 15 min, 0.97% for USA300 LAC versus 69.19% for ΦNΞ9044; P = 0.0228), indicating that complement factors adhere to the surfaces of ebh mutant cells but not to wild-type staphylococci. We also visualized complement deposition via confocal microscopy (Fig. 7B). Staphylococcal cells were incubated in plasma, and aliquots were removed at timed intervals, stained with C3b FITC-conjugated antibody, and viewed under a confocal microscope. Cells of the ebh mutant ΦNΞ9044 were labeled with C3 within 10 s, whereas wild-type staphylococci exhibited very little C3 labeling even after 5 min of incubation. These data suggest that ebh expression is required for the assembly of a staphylococcal envelope that is refractory to C3 complement deposition and complement-mediated killing.
FIG 7.
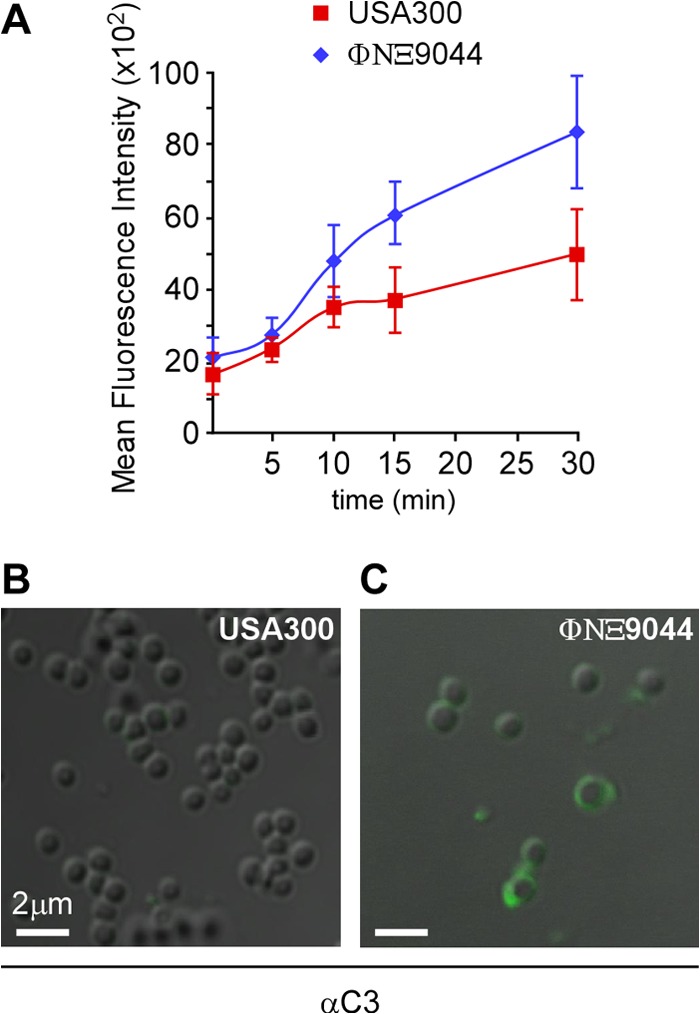
Ebh prevents C3 complement deposition in the envelope of S. aureus. (A) MRSA strain USA300 LAC and its ebh variant ΦNΞ9044 were incubated with human plasma. At timed intervals (0, 5, 10, 15, 20, 25, and 30 min), aliquots were removed, bacteria stained with FITC-conjugated anti-human C3, and fluorescence quantified via flow cytometry. Samples generated at the 5-min time point for panel A were viewed by confocal microscopy to reveal C3 deposition in the envelopes of S. aureus USA300 LAC (B) and ΦNΞ9044 (C).
Ebh and staphylococcal virulence.
To investigate whether ebh contributes to staphylococcal virulence, we infected cohorts of BALB/c mice via intravenous injection with either wild-type USA300 LAC or the ebh variant ΦNΞ9044 (Ebh1–509) or ΦNΞ10853 (Ebh1–5579) (Fig. 8A). At a challenge dose of 5 × 107 CFU, all BALB/c mice that had been infected with USA300 LAC or ΦNΞ10853 died within 24 to 48 h. In contrast, animals infected with the same dose of the ΦNΞ9044 ebh variant displayed a delayed time to death of up to 108 h (for USA300 LAC versus ΦNΞ9044, P < 0.005). Following intravenous challenge with 5 × 106 CFU, BALB/c mice infected for 5 days with USA300 LAC generated abscesses in kidneys and other internal organs (48). When renal tissues were analyzed for bacterial load, we observed an average of 6.2 log10 CFU USA300 LAC g−1 tissue (Fig. 8B). In contrast, the ΦNΞ9044 ebh variant displayed a 1.4-log10 reduction in CFU g−1 tissue (Fig. 8B). A similar defect, namely, a 1.2-log10 reduction in CFU g−1 tissue, was observed in comparing wild-type S. aureus Newman and its Δebh variant (Fig. 8B). Thus, mutations that abolish the expression of ebh in S. aureus Newman or USA300 LAC diminish staphylococcal virulence, likely because ebh mutant bacteria are subject to complement-mediated killing in infected host tissues.
FIG 8.
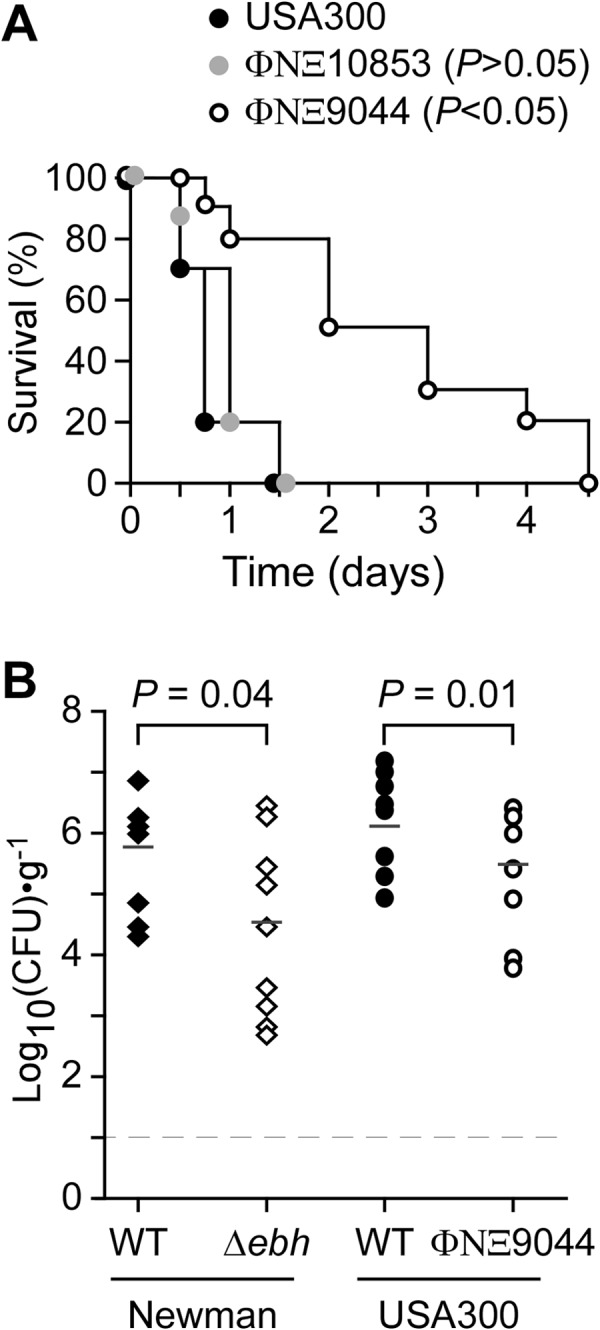
Ebh contributes to virulence of S. aureus in a mouse model. (A) BALB/c mice (n = 10) were injected into the periorbital venous plexus with 5 × 107 CFU of S. aureus USA300 LAC or its ebh variant ΦNΞ9044 or ΦNΞ10853. Animal survival was recorded over 14 days. When analyzed with the log rank test, the ebh variant ΦNΞ9044 displayed a significant delay in time to death compared to the wild type (for USA300 LAC versus ΦNΞ9044, P < 0.05). (B) BALB/c mice (n = 10) were injected into the periorbital venous plexus with 5 × 106 CFU S. aureus Newman or its Δebh variant and with USA300 LAC or its ebh variant ΦNΞ9044. At 5 days postinfection, animals were euthanized and necropsied and their kidneys removed. Renal tissues were homogenized, and staphylococcal loads were determined by plating aliquots on TSA and enumerating CFU. The mean bacterial load was calculated and analyzed for statistical significance by use of the Mann-Whitney U test to compare the wild-type and ebh variant strains: for USA300 LAC versus ΦNΞ9044, P = 0.0103; and for Newman versus the Δebh variant, P = 0.0409.
DISCUSSION
Enzymes responsible for lipid II synthesis (MurG) and polymerization into peptidoglycan (PBP1, PBP2, PBP2a, and PBP4) are localized to the cross wall (49–53). In the absence of cell division or under conditions of diminished abundance of cell division factors, PBPs cannot localize because the cross wall is not formed (54). Furthermore, depletion of lipid II from staphylococcal cells, as occurs when S. aureus is treated with murgocil or vancomycin, diminishes PBP localization to the cross wall (49, 52). PBP2 is the key enzyme of S. aureus responsible for the polymerization of glycan strands from lipid II (55). PBP2 cooperates with PBP4 to generate cross-linked peptidoglycan (51, 56). Indeed, most of the Gly5 cross bridges in the cell wall of S. aureus are linked to d-Ala of adjacent wall peptides (57, 58). For MRSA strains growing in the presence of β-lactams, PBP2a functions as the sole transpeptidase, replacing PBP1, PBP2, and PBP4, but not PBP2 transglycosylase (51). Sortase A, another enzyme requiring lipid II substrate (59), is also localized to the cross wall during cell division (60).
Recent work suggests a connection between the assembly of wall teichoic acid (WTA), which does not occur within the cross wall (13), and peptidoglycan synthesis at the cross wall (53). WTA synthesis is initiated by TagO and TagA, which link GlcNAc and ManNAc to bactoprenol (C55-PO4) (61–63). The product, C55-(PO4)2-GlcNAc-ManNAc, is extended with glycerol-3-phosphate (GroP) and poly-ribitol-5-phosphate (RboP) (64). Bactoprenol-linked WTA precursors are tethered to the C6-OH of glycan chains by three enzymes—LcpA, LcpB, and LcpC—that fulfill seemingly redundant functions (65, 66). S. aureus mutants lacking tagO display aberrant cross wall formation with parallel and incompletely formed structures (62, 67, 68). A similar phenotype is observed with lcpABC mutants (65). Finally, tagO mutants as well as wild-type staphylococci treated with tunicamycin, an inhibitor of TagO (69, 70), are susceptible to methicillin even when PBP2a is expressed, which implies that PBP2a may not be localized properly to the cross wall when teichoic acid synthesis is blocked (62, 71).
Biogenesis of the cross wall follows a series of temporal and spatially controlled cytokinesis events that are initiated with the polymerization of FtsZ, a tubulin-like GTPase (72). Once tethered to the membrane and associated with other cell division factors, FtsZ constricts as a ring structure, the divisome (72). The purpose of the divisome is the generation of two daughter cells, which is accomplished by separating the membranes of bacteria at a midcell position (73). EzrA, a negative regulator of the divisome in firmicutes (74), contributes to the proper positioning of FtsZ rings and PBPs at the staphylococcal cross wall (75). These observations can be integrated into a simple model. Divisome placement at midcell localizes MurG and lipid II synthesis, triggering accumulation of lipid II at the cell division site and directing PBPs and sortase to the cross wall. Perturbation of WTA synthesis (tagO or lcpABC) may increase the availability of bactoprenol and disturb the localized accumulation of lipid II at the cross wall, thereby perturbing the localization of PBPs and sortase A. Once the cross wall has formed, i.e., peptidoglycan is synthesized in the extracellular space derived from FtsZ-catalyzed membrane separation, other mechanisms provide for the trafficking of precursors with YSIRK-G/S signal peptides into the cross wall. These proteins are deposited in the cross wall and distributed over the staphylococcal surface (proteins with LPXTG sorting signals) (12). Other YSIRK-G/S precursors, for example, LytN and Ebh, traffic into the cross wall to shape the structure of its peptidoglycan and/or split the cross wall (10).
Experiments described herein reveal that Ebh is a surface protein of S. aureus Newman and USA300. The distribution pattern of Ebh is reminiscent of that observed for sortase-anchored surface proteins with YSIRK-G/S signal peptides, for example, protein A (SpA) (11). Unlike sortase A-anchored products, which are linked to peptidoglycan, the giant protein Ebh is thought to span the peptidoglycan layer and the plasma membrane (Fig. 1A). Ebh, similar to sortase A-anchored proteins, is presumably distributed over the staphylococcal surface once the cross wall is split (12). In the absence of ebh, mutant staphylococci display a surprising phenotype: the bacterial cell volume is nearly doubled, and cross walls assume irregular shapes and thicknesses. We suspect that the increased sensitivity of ebh variants to complement-mediated killing in plasma may be a feature of their cell wall separation defects. Weakened peptidoglycan within cross walls and increased cell size appear to increase the association with complement and may enable the deposition of MACs with membrane-disrupting attributes (76, 77). The aberrantly shaped ebh cells are also susceptible to treatment with oxacillin, a β-lactam that impairs the transpeptidation reaction of cell wall synthesis (78). These findings further corroborate our hypothesis that Ebh may enable physiological peptidoglycan synthesis and drug resistance at the cross wall. What might be the molecular mechanisms that lead to the unique phenotypes of ebh variants? We presume that Ebh may act as a cytoskeletal element for proper positioning of the cross wall and subsequent cross wall splitting and cell separation. Note that its predicted length (350 nm) would allow Ebh to span the calculated width of the cross wall. If it does so, perhaps staphylococci exploit the shape and repeat structures of Ebh for the proper assembly of peptidoglycan and/or other carbohydrate structures within the cross wall compartment.
Earlier work identified mecA (PBP2′ or PBP2a), encoding a transpeptidase that catalyzes the transpeptidation reaction of cell wall synthesis when MRSA cells are grown in the presence of methicillin or oxacillin (79, 80). Methicillin resistance in clinical isolates of S. aureus is typically heterogeneous in phenotypic expression, where most cells of a population show low resistance levels and a minor population displays high resistance (81). Reduced methicillin resistance (10- to 50-fold reduction in MIC) in MRSA strains is observed following inactivation of any one of 20 different fem genes (factors essential for methicillin resistance) (78). Several of these factors are involved in the synthesis of lipid II, i.e., C55-(PO4)2-MurNAc(l-Ala-d-iGln-l-Lys(NH2-Gly5)-d-Ala-d-Ala)-GlcNAc, for example, by synthesizing the pentaglycine (Gly5) cross bridge (FemAB and FmhB) (82), the di-amino acid l-Lys (FemF/MurE) (83), d-Gln (FemC) (84), or the GlcNAc moiety of lipid II (FemD [GlmM]) (85). Nevertheless, mutations in genes for cell wall- or membrane-associated proteins (FmtA, FmtB, and FmtC [MprF]) can also diminish methicillin resistance, although the underlying mechanisms are not yet understood (86–88). Deletion of the ebh gene does not affect the susceptibility of S. aureus Newman toward β-lactam antibiotics (MIC of oxacillin is 0.2 μg ml−1 for both wild-type and Δebh strains). However, β-lactam resistance of the MRSA strain USA300 LAC (MIC, 20 μg ml−1) is reduced by 2 orders of magnitude in the USA300 ebh mutant (ΦNΞ9044) (MIC, 0.2 μg ml−1).
We observed that the genomes of staphylococcal strains whose DNA sequences were determined in close temporal proximity to their clinical isolation may harbor intact (full-length) ebh genes (25). In contrast, strains that had been propagated for years in the laboratory prior to genome sequencing harbored truncated ebh variants with 3′ nonsense mutations. Our experiments involved mutants with bursa aurealis insertions in the ebh gene. Depending on the locations of the mutations, these variants display a gradient of defects, where the most severe phenotypes are associated with 5′ disruptions and nonsense mutations at the 3′ end cause minor phenotypes. From this information, we conclude that expression of full-length ebh is not required for staphylococcal growth in the laboratory. Nevertheless, secretion of full-length Ebh into the cross wall facilitates staphylococcal replication in host tissues by providing for envelope stability and protection against host defenses.
ACKNOWLEDGMENTS
We thank Michael Berry, Matthew Frankel, and Wade W. Williams Green for experimental assistance and members of our laboratory for discussions.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Diseases Branch (grants AI038897 and AI052474 to O.S. and grant AI075258 to D.M.). A.G.C. was a trainee of the NIH Medical Scientist Training Program at The University of Chicago (grant GM07281). D.M. and O.S. acknowledge membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH award 1-U54-AI-057153).
We declare a conflict of interest as inventors of patent applications that are related to the development of Staphylococcus aureus vaccines and are currently under commercial license.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 2.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens RM, Edwards JR, Gaynes RP. 2008. The impact of antimicrobial-resistant, health care-associated infections on mortality in the United States. Clin. Infect. Dis. 47:927–930. 10.1086/591698 [DOI] [PubMed] [Google Scholar]
- 4.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 5.Arbeit RD, Maki D, Tally FP, Campanaro E, Eisenstein BI. 2004. The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin. Infect. Dis. 38:1673–1681. 10.1086/420818 [DOI] [PubMed] [Google Scholar]
- 6.Stevens DH, Herr D, Campiris H, Hunt JL, Batts DH, Hafkin B. 2002. Linezolid versus vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin. Infect. Dis. 34:1481–1490. 10.1086/340353 [DOI] [PubMed] [Google Scholar]
- 7.van Hal SJ, Paterson DL. 2011. New Gram-positive antibiotics: better than vancomycin? Curr. Opin. Infect. Dis. 24:515–520. 10.1097/QCO.0b013e32834ab1de [DOI] [PubMed] [Google Scholar]
- 8.Giesbrecht P, Kersten T, Maidhof H, Wecke J. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzagoloff H, Novick R. 1977. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 129:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. 2011. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J. Biol. Chem. 286:32593–32605. 10.1074/jbc.M111.258863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeDent AC, McAdow M, Schneewind O. 2007. Distribution of protein A on the surface of Staphylococcus aureus. J. Bacteriol. 189:4473–4484. 10.1128/JB.00227-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeDent AC, Missiakas DM, Schneewind O. 2008. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 27:2656–2668. 10.1038/emboj.2008.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel MB, Schneewind O. 2012. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J. Biol. Chem. 287:10460–10471. 10.1074/jbc.M111.336404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu W, Götz F. 2012. Cell wall antibiotics provoke accumulation of anchored mCherry in the cross wall of Staphylococcus aureus. PLoS One 7:e30076. 10.1371/journal.pone.0030076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneewind O, Missiakas DM. 2012. Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1123–1139. 10.1098/rstb.2011.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke SR, Harris LG, Richards RG, Foster SJ. 2002. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70:6680–6687. 10.1128/IAI.70.12.6680-6687.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae T, Schneewind O. 2003. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185:2910–2919. 10.1128/JB.185.9.2910-2919.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams RJ, Henderson B, Sharp LJ, Nair SP. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70:6805–6810. 10.1128/IAI.70.12.6805-6810.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, Rohde H. 2010. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75:187–207. 10.1111/j.1365-2958.2009.06981.x [DOI] [PubMed] [Google Scholar]
- 20.McCarthy AJ, Lindsay JA. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 10:173. 10.1186/1471-2180-10-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben Zakour NL, Beatson SA, van den Broek AH, Thoday KL, Fitzgerald JR. 2012. Comparative genomics of the Staphylococcus intermedius group of animal pathogens. Front. Cell. Infect. Microbiol. 2:44. 10.3389/fcimb.2012.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilbronner S, Holden MT, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. 2011. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322:60–67. 10.1111/j.1574-6968.2011.02339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda M, Tanaka Y, Aoki R, Shu D, Tsumoto K, Ohta T. 2008. Staphylococcus aureus giant protein Ebh is involved in tolerance to transient hyperosmotic pressure. Biochem. Biophys. Res. Commun. 374:237–241. 10.1016/j.bbrc.2008.07.037 [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, Sakamoto S, Kuroda M, Goda S, Gao Y, Tsumoto K, Hiragi Y, Yao M, Watanabe N, Ohta T, Tanaka I. 2008. A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus. Structure 16:488–496. 10.1016/j.str.2007.12.018 [DOI] [PubMed] [Google Scholar]
- 25.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U. S. A. 105:1327–1332. 10.1073/pnas.0710217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300–310. 10.1128/JB.01000-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438. 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bæk KT, Frees D, Renzoni A, Barras C, Rodriguez N, Manzano C, Kelley WL. 2013. Genetic variation in the Staphylococcus aureus 8325 strain lineage revealed by whole-genome sequencing. PLoS One 8:e77122. 10.1371/journal.pone.0077122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto S, Tanaka Y, Tanaka I, Takei T, Yu J, Kuroda M, Yao M, Ohta T, Tsumoto K. 2008. Electron microscopy and computational studies of Ebh, a giant cell-wall-associated protein from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 376:261–266. 10.1016/j.bbrc.2008.08.117 [DOI] [PubMed] [Google Scholar]
- 30.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U. S. A. 101:12312–12317. 10.1073/pnas.0404728101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 32.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. 2010. ABI-domain containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol. Microbiol. 78:238–252. 10.1111/j.1365-2958.2010.07334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89 [DOI] [PubMed] [Google Scholar]
- 34.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 35.Gründling A, Schneewind O. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8478–8483. 10.1073/pnas.0701821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck WD, Berger-Bächi B, Kayser FH. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 165:373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. 1989. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 171:2882–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HK, Cheng AG, Kim H-Y, Missiakas DM, Schneewind O. 2010. Non-toxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections. J. Exp. Med. 207:1863–1870. 10.1084/jem.20092514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. 2009. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J. Exp. Med. 206:2417–2427. 10.1084/jem.20090097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gros P, Milder FJ, Janssen BJ. 2008. Complement driven by conformational changes. Nat. Rev. Immunol. 8:48–58. 10.1038/nri2231 [DOI] [PubMed] [Google Scholar]
- 41.Laarman A, Milder F, van Strijp JA, Rooijakkers SH. 2010. Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications. J. Mol. Med. 88:115–120. 10.1007/s00109-009-0572-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jongerius I, Köhl J, Pandey MK, Ruyken M, van Kessel KPM, van Strijp JAG, Rooijakkers SHM. 2007. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 204:2461–2471. 10.1084/jem.20070818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooijakkers SH, van Wamel WJ, Ruyken M, van Kessel KP, van Strijp JA. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect. 7:476–484. 10.1016/j.micinf.2004.12.014 [DOI] [PubMed] [Google Scholar]
- 44.Rooijakkers SHM, Wu J, Ruyken M, van Domselaar R, Planken KL, TZekou A, Ricklin D, Lambris JD, Janssen BJC, van Strijp JAG, Gros P. 2009. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 7:721–727. 10.1038/ni.1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harraghy N, Hussain M, Haggar A, Chavakis T, Sinha B, Herrmann M, Flock JI. 2003. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology 149:2701–2707. 10.1099/mic.0.26465-0 [DOI] [PubMed] [Google Scholar]
- 46.Lee LY, Liang X, Höök M, Brown EL. 2004. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb). J. Biol. Chem. 279:50710–50716. 10.1074/jbc.M408570200 [DOI] [PubMed] [Google Scholar]
- 47.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. 2007. A structural basis for complement inhibition by Staphylococcus aureus. Nat. Immunol. 8:430–437. 10.1038/ni1450 [DOI] [PubMed] [Google Scholar]
- 48.Anderson M, Chen YH, Butler EK, Missiakas DM. 2011. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J. Bacteriol. 193:1583–1589. 10.1128/JB.01096-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mann PA, Müller A, Xiao L, Pereira PM, Yang C, Ho Lee S, Wang H, Trzeciak J, Schneeweis J, Dos Santos MM, Murgolo N, She X, Gill C, Balibar CJ, Labroli M, Su J, Flattery A, Sherborne B, Maier R, Tan CM, Black T, Onder K, Kargman S, Monsma FJJ, Pinho MG, Schneider T, Roemer T. 2013. Murgocil is a highly bioactive staphylococcal-specific inhibitor of the peptidoglycan glycosyltransferase enzyme MurG. ACS Chem. Biol. 8:2442–2451. 10.1021/cb400487f [DOI] [PubMed] [Google Scholar]
- 50.Pereira SFF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189:3525–3531. 10.1128/JB.00044-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 187:1815–1824. 10.1128/JB.187.5.1815-1824.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799–807. 10.1111/j.1365-2958.2004.04420.x [DOI] [PubMed] [Google Scholar]
- 53.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho M, Filipe SR. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 107:18991–18996. 10.1073/pnas.1004304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinho MG, Errington J. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871–881. 10.1046/j.1365-2958.2003.03719.x [DOI] [PubMed] [Google Scholar]
- 55.Lovering AL, de Castro LH, Lim D, Strynadka NC. 2007. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405. 10.1126/science.1136611 [DOI] [PubMed] [Google Scholar]
- 56.Lovering AL, Safadi SS, Strynadka NC. 2012. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 81:451–478. 10.1146/annurev-biochem-061809-112742 [DOI] [PubMed] [Google Scholar]
- 57.de Jonge BLM, Chang YS, Gage D, Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 267:11248–11254 [PubMed] [Google Scholar]
- 58.de Jonge BLM, Chang YS, Gage D, Tomasz A. 1992. Peptidoglycan composition in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 267:11255–11259 [PubMed] [Google Scholar]
- 59.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241–16248. 10.1074/jbc.M109194200 [DOI] [PubMed] [Google Scholar]
- 60.Raz A, Fischetti VA. 2008. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. U. S. A. 105:18549–18554. 10.1073/pnas.0808301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung BC, Zhao J, Gillespie RA, Kwon DY, Guan Z, Hong J, Zhou P, Lee SY. 2013. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341:1012–1016. 10.1126/science.1236501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell J, Singh AK, Santa Maria JPJ, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6:106–116. 10.1021/cb100269f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown S, Zhang YH, Walker S. 2008. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 15:12–21. 10.1016/j.chembiol.2007.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia G, Peschel A. 2008. Toward the pathway of S. aureus WTA biosynthesis. Chem. Biol. 15:95–96. 10.1016/j.chembiol.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 65.Chan YGY, Frankel MB, Dengler V, Schneewind O, Missiakas DM. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr (LCP) family of enzymes release wall teichoic acids into the extracellular medium. J. Bacteriol. 195:4650–4659. 10.1128/JB.00544-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J. 30:4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183–4189. 10.1128/JB.00197-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243–245. 10.1038/nm991 [DOI] [PubMed] [Google Scholar]
- 69.Hancock IC, Wiseman G, Baddiley J. 1976. Biosynthesis of the unit that links teichoic acid to the bacterial wall: inhibition by tunicamycin. FEBS Lett. 69:75–80. 10.1016/0014-5793(76)80657-6 [DOI] [PubMed] [Google Scholar]
- 70.Soldo B, Lazarevic V, Karamata D. 2002. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148:2079–2087 http://mic.sgmjournals.org/content/148/7/2079.full.pdf+html [DOI] [PubMed] [Google Scholar]
- 71.Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 8:226–233. 10.1021/cb300413m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. 10.1038/354161a0 [DOI] [PubMed] [Google Scholar]
- 73.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653. 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- 74.Levin PA, Schwartz RL, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 96:9642–9647. 10.1073/pnas.96.17.9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorge AM, Hoiczyk E, Gomes JP, Pinho MG. 2011. EzrA contributes to the regulation of cell size in Staphylococcus aureus. PLoS One 6:e27542. 10.1371/journal.pone.0027542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hadders MA, Beringer DX, Gros P. 2007. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science 317:1552–1554. 10.1126/science.1147103 [DOI] [PubMed] [Google Scholar]
- 77.Podack ER. 2009. How to polymerize in order to survive. Immunity 30:668–670. 10.1016/j.immuni.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 78.Berger-Bächi B, Rohrer S. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165–171. 10.1007/s00203-002-0436-0 [DOI] [PubMed] [Google Scholar]
- 79.Utsui Y, Yokota T. 1985. Role of an altered penicillin binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397–403. 10.1128/AAC.28.3.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pinho MG, Filipe SR, De Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525–6531. 10.1128/JB.183.22.6525-6531.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bächi B. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175:1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bächi B. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. U. S. A. 96:9351–9356. 10.1073/pnas.96.16.9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ornelas-Soares A, de Lencastre H, de Jonge BLM, Tomasz A. 1994. Reduced methicillin resistance in a new Staphylococcus aureus transposon mutant that incorporates muramyl dipeptides into the cell wall peptidoglycan. J. Biol. Chem. 269:27246–27250 [PubMed] [Google Scholar]
- 84.Gustafson J, Strassle A, Hachler H, Kayser FH, Berger-Bächi B. 1994. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J. Bacteriol. 176:1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jolly L, Wu S, van Heijenoort J, de Lencastre H, Mengin-Lecreulx D, Tomasz A. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J. Bacteriol. 179:5321–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komatsuzawa H, Ohta K, Sugai M, Fujiwara T, Glanzmann P, Berger-Bächi B, Suginaka H. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 45:421–431. 10.1093/jac/45.4.421 [DOI] [PubMed] [Google Scholar]
- 87.Komatsuzawa H, Ohta K, Labischinski H, Sugai M, Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 203:49–54. 10.1111/j.1574-6968.2001.tb10819.x [DOI] [PubMed] [Google Scholar]