Abstract
The Staphylococcus aureus Agr system regulates virulence gene expression by responding to cell population density (quorum sensing). When an extracellular peptide signal (AIP-III in strain UAMS-1, used for these experiments) reaches a concentration threshold, the AgrC-AgrA two-component regulatory system is activated through a cascade of phosphorylation events, leading to induction of the divergently transcribed agrBDCA operon and the RNAIII gene. RNAIII is a posttranscriptional regulator of numerous metabolic and pathogenesis genes. CodY, a global regulatory protein, is known to repress agrBDCA and RNAIII transcription during exponential growth in rich medium, but the mechanism of this regulation has remained elusive. Here we report that phosphorylation of AgrA by the AgrC protein kinase is required for the overexpression of the agrBDCA operon and the RNAIII gene in a codY mutant during the exponential-growth phase, suggesting that the quorum-sensing system, which normally controls AgrC activation, is active even in exponential-phase cells in the absence of CodY. In part, such premature expression of RNAIII was attributable to higher-than-normal accumulation of AIP-III in a codY mutant strain, as determined using ultrahigh-performance liquid chromatography coupled to mass spectrometry. Although CodY is a strong repressor of the agr locus, CodY bound only weakly to the agrBDCA-RNAIII promoter region, suggesting that direct regulation by CodY is unlikely to be the principal mechanism by which CodY regulates agr and RNAIII expression. Taken together, these results strongly suggest that cell population density signals inducing virulence gene expression can be overridden by nutrient availability, a condition monitored by CodY.
INTRODUCTION
Bacteria can adapt to complex and rapidly changing environments. They have diverse and sophisticated systems for sensing their surroundings and regulating their gene expression accordingly. Because most pathogens can obtain nutrients from their host organisms, at least in part by producing virulence factors whose activities can also be deleterious to the host, synthesis of such factors is tightly regulated. For instance, the opportunistic pathogen Staphylococcus aureus, which produces a plethora of secreted and cell wall-associated pathogenicity factors, can sense and respond to environmental cues to optimize the expression of virulence genes. Indeed, the synthesis of staphylococcal pathogenicity factors changes in response to variations in growth phase (1–3), cell population density (4), pH (5, 6), glucose availability (7, 8), NaCl concentration (9), and exposure to antibiotics (10).
S. aureus virulence is centrally regulated by the accessory gene regulator (Agr) system (4). The agr locus consists of two divergent transcription units (Fig. 1). The P2 promoter drives the transcription of the agrBDCA operon, producing an mRNA called RNAII, whereas the P3 promoter drives the synthesis of RNAIII, an RNA molecule that is both an mRNA for delta-hemolysin and a regulatory factor for a large number of genes. RNAIII reduces the expression of genes encoding surface adhesins and increases the synthesis of capsule, toxins, and proteases (4). The agrB and agrD gene products are involved in the synthesis, transport, and maturation of a cyclic thiolactone that acts as an autoinducing peptide (AIP), whereas the agrC and agrA genes encode, respectively, the AIP-sensing membrane kinase and response regulator of a two-component transcriptional regulatory system. When the extracellular concentration of AIP reaches a threshold level, AIP binds to AgrC, inducing autophosphorylation and leading to the activation-by-phosphorylation of AgrA. Phosphorylated AgrA stimulates both the transcription of the operon that encodes it and the transcription of RNAIII by binding to the P2-P3 promoter region. SarA, a small DNA-binding protein, also regulates the expression of many S. aureus virulence genes (11) and functions in part by activating the transcription of the agr locus (12–14). The detailed molecular mechanism by which SarA regulates virulence gene expression is unclear (15–19).
FIG 1.
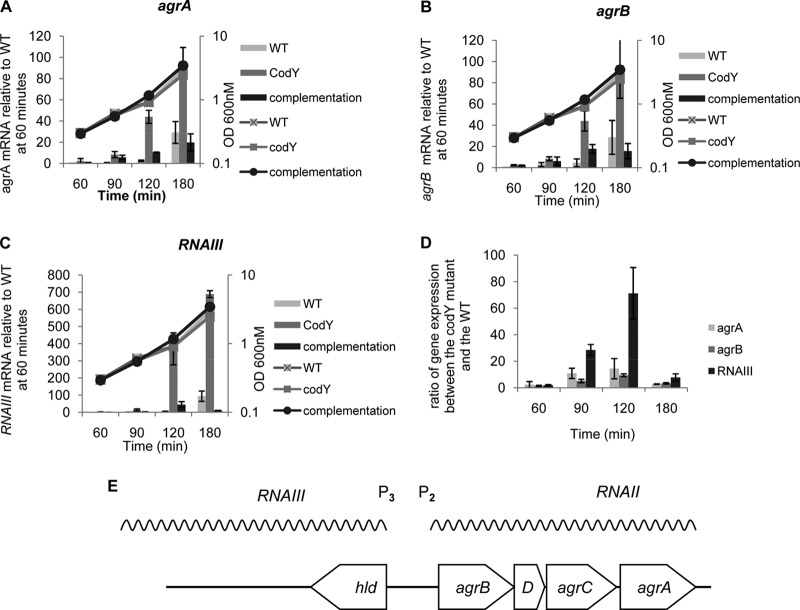
CodY is a negative regulator of the agr genes during the exponential-growth phase. (A through C) Two biological replicates of exponential-phase culture samples of UAMS-1 and MS-1 (ΔcodY) stains were collected after 60, 90, 120, and 180 min of growth in TSB medium. RNA samples were prepared, and levels of agrA (A), agrB (B), and RNAIII (C) RNAs were determined by real-time quantitative PCR. For each target gene at each time point, the ratio of the transcript to 16S rRNA was determined and was normalized to the transcript/16S rRNA ratio for the wild-type strain at 60 min. (D) Data from panels A through C were converted to ratios of expression in the codY mutant relative to expression in the parent strain at each time point. Error bars correspond to the standard errors of the means.
Many staphylococcal virulence genes are regulated by nutritional signals. CodY, a global transcriptional regulator present in most low-G+C Gram-positive bacteria, senses directly the intracellular concentrations of branched-chain amino acids (BCAAs) and GTP (20). These effectors couple CodY activity to the availability of rapidly metabolized carbon and nitrogen sources and, specifically, to amino acid pools (21). CodY is most active during the exponential-growth phase in rich medium, when the intracellular concentrations of BCAAs and GTP are high (20). In S. aureus, as well as in several other pathogenic bacteria, CodY regulates both metabolic and virulence genes (22–37). Interestingly, the S. aureus agr locus is overexpressed in a codY mutant during exponential phase, suggesting that CodY is a repressor of transcription from the agr P2 and P3 promoters (25).
Although CodY represses agr expression, we report here that CodY regulates agr transcription even though it binds at best weakly to the P2-P3 promoter region and to a putative binding site within agrC. Additionally, we found that the overexpression of the entire agr locus seen in exponential-phase cells of a codY mutant is correlated with increased accumulation of AIP and depends on the presence of the phosphorylated form of the response regulator AgrA as well as on the kinase AgrC. On the other hand, CodY-dependent regulation of the agr locus was independent of SarA and of any other regulatory protein known to affect agr expression. These results suggest that CodY regulates the expression of the agr genes indirectly. Moreover, these results indicate that the Agr quorum-sensing system is active under conditions of relatively low cell population density but that the effects of intercellular signaling are masked by CodY-mediated repression under nutrient-replete conditions. Thus, CodY-dependent regulation of the agr genes serves to integrate cell population density and nutritional signals that control virulence.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strain DH5α (recA) was used for plasmid construction and was cultivated in Luria-Bertani (LB) medium. S. aureus strains were grown in tryptic soy broth (TSB; Becton, Dickinson Co.). Cultures were incubated with shaking (250 rpm) at 30°C, 37°C, or 42°C, with a flask-to-medium volume ratio of 10:1. Chloramphenicol (10 μg/ml) and ampicillin (50 μg/ml) were added for plasmid maintenance when needed. Chloramphenicol was used at 2.5 μg/ml when a chloramphenicol resistance gene was integrated into the chromosome. Growth was monitored as an increase in the absorbance at 600 nm in an Ultraspec II UV-visible spectrophotometer (LKB Biochrom).
TABLE 1.
Plasmids and strains used in the study
| Plasmid or strain | Relevant genotype and/or characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pTS1 | Shuttle vector; pE194ori(Ts); ColE1 bla cat | 74 |
| pNL9164 | Shuttle vector; pT181 cop-634(Ts) repC4 ori | 75 |
| pKM1 | S. aureus codY cloned into pBAD30 with 5 additional histidine codons at the C terminus | K. McCarty and A. Sonenshein, unpublished data |
| pCL15 | Expression shuttle vector, derivative of pSI-1; PSPAC cat | 44, 46 |
| pAR12 | DNA regions surrounding agrA cloned in pST1 | This study |
| pAR23 | DNA regions surrounding the P2-P3 agr promoters cloned in pST1 | This study |
| pAR28 | Cloned intron DNA targeted to SACOL2585 in pNL9164 | This study |
| pAR31 | DNA regions surrounding agrC cloned into pST1 | This study |
| pTL6936 | pCL15 PSPAC-codY | 26 |
| PSPAC-agrA | pCL15 PSPAC-agrA | This study |
| S. aureus strains | ||
| UAMS-1 | Clinical isolate | 76 |
| MS1 | UAMS-1 ΔcodY::ermC | 26 |
| AR87 | UAMS-1 SA2585::intron | This study |
| AR89 | UAMS-1 ΔcodY::ermC SA2585::intron | This study |
| CM18 | UAMS-1 agr::tetM | 25 |
| CM20 | UAMS-1 sarA::kan | 25 |
| CM22 | UAMS-1 ΔcodY::ermC sarA::kan | 25 |
| AR45 | UAMS-1 ΔagrA | This study |
| AR48 | UAMS-1 ΔcodY::ermC ΔagrA | This study |
| AR70 | UAMS-1 ΔP2-P3 | This study |
| AR71 | UAMS-1 ΔcodY::ermC ΔP2-P3 | This study |
| AR145 | UAMS-1 ΔagrC | This study |
| AR147 | UAMS-1 ΔcodY::ermC ΔagrC | This study |
DNA manipulations and transformations.
All molecular biology techniques, including E. coli transformations, were performed as described by Sambrook and Russell (38). Oligonucleotide primers were obtained from Integrated DNA Technologies and are listed in Table 2. All cloned fragments were verified by sequencing by the Tufts University Nucleic Acids and Protein Core Facility. A rapid boiling procedure was used to prepare template DNA from S. aureus as described previously (39).
TABLE 2.
Primers used in the study
| Primer name | Sequence (5′→3′) |
|---|---|
| oAR39 | AATAATCCCGGGCGCAAGTTCCGTCATGATTA |
| oAR69 | GTCATGCTTACGAATTTCAC |
| oAR70 | GAAATTCGTAAGCATGACCAAAATCTCACAGACTCATTGC |
| oAR71 | GGGGAGCTCCATTATGGGATAACGCAGAAG |
| oAR95 | GATTGGTAGAACTATAAACACGC |
| oAR97 | GGGCCCGGGCACCTGCAGCTACTAAGAAG |
| oAR96 | GCGTGTTTATAGTTCTACCAATCGATGCATCAACAATCAAACAG |
| oAR98 | GGGGAGCTCATTCACATCCTTATGGCTAG |
| oAR88 | GGGCCCGGGGTATTAAATTAGATTTGTATAAATAAAAAGCAC |
| oAR85 | CGACACAGTGAACAAATTCAC |
| oAR87 | GGGGAGCTCGCACAATATAAAATGATTTGAGTAAC |
| oAR86 | GTGAATTTGTTCACTGTGTCGGAGGAGAGTGGTGTAAAATTG |
| oAR134 | AAAAGCTTTTGCAACCCACGTCGATCGTGAAGAAATAATAATAGTGCGCCCAGATAGGGTG |
| oAR135 | CAGATTGTACAAATGTGGTGATAACAGATAAGTCATAATAAGTAACTTACCTTTCTTTGT |
| oAR136 | CGCAAGTTTCTAATTTCGGTTATTTCTCGATAGAGGAAAGTGTCT |
| oAR174 | CTATTTTCCATCACATCTCTGTG |
| oCM43 | TGAATTTGTTCACTGTGTCGAT |
| oCM114 | AGTAAGGATCCTTTGGATCGTCTTCGCAAAT |
| oAR154 | CAATTTTACACCACTCTCCTC |
| p-ilvD2 | CAGAAATAGGACTTAAAGCGTTTAG |
| oCM123 | ATTGAATTCCGCAAGTTCCGTCATGATTA |
| oAR187 | TGACCAGTTTGCCACGTATCTTCA |
| oAR188 | GCTAAGACCTGCATCCCTAATCGT |
| oAR52 | CCTATGGAAATTGCCCTCGC |
| oAR64 | GCCTAATTTGATACCATTAATATCAG |
| oAR185 | GTGAATTTGTTCACTGTGTCGATAATCC |
| oAR186 | GGAAGGAGTGATTTCAATGGCACA |
| oAR16 | CGTGTCTCAGTTCCAGTGTG |
| oAR17 | CTTCTCTGATGTTAGCGGCG |
| oAR157 | GGGGGATCCAGAGGAGAAATTAACTATGAAAATTTTCATTTGCGAAGACG |
| oAR158 | GGGGAGCTCAATTGAATACGCCGTTAAC |
Deletion mutants.
Three unmarked deletion mutants were constructed: one with a deletion of P2 and P3 (ΔP2-P3), one with an in-frame deletion (ΔagrC), and one with an out-of-frame deletion (ΔagrA). PCR-based mutagenesis followed by splicing overlap extension (SOEing) (40) was carried out using UAMS-1 genomic DNA and primer sets oAR39/oAR69 and oAR70/oAR71 for ΔagrA, oAR95/oAR97 and oAR96/oAR98 for ΔagrC, and oAR88/oAR85 and oAR87/oAR86 for the ΔP2-P3 promoters. The fragments were cloned in the thermosensitive integrative plasmid pTS1 (26) by using the restriction sites SacI and XmaI. The resulting plasmids, pAR12, pAR31, and pAR23, were introduced by electroporation into the restriction-deficient S. aureus strain RN4220 according to methods described previously (41), but with incubation of the transformants at 30°C. The plasmids were then transferred to strain UAMS-1 using the transducing phage ϕ11, as described previously (42). Transductants were grown in TSB medium supplemented with chloramphenicol for 12 h at 30°C. A 1:100 dilution of the culture was then grown in TSB containing chloramphenicol for 12 h at 42°C, a nonpermissive temperature for pTS1 replication, thereby selecting for integration into the chromosome by homologous recombination. The cultures were then shifted to 30°C, and the bacteria were passaged four times in TSB without antibiotics. Bacteria were finally plated on tryptic soy agar (TSA) and were screened for chloramphenicol-sensitive clones.
The SACOL2585 insertion mutation was constructed using the S. aureus TargeTron gene knockout system from Sigma-Aldrich according to the manufacturer's instructions. Briefly, primers oAR134, oAR135, and oAR136 were designed using the TargeTron design website (Sigma-Aldrich) and were used to amplify a PCR fragment encoding a mutated intron designed to specifically target the SACOL2585 gene. The fragment was then cloned into the thermosensitive TargeTron vector pNL9164 by using the HindIII and BsrGI restriction sites. The resulting plasmid was first introduced by electroporation in RN4220 according to methods described previously (41) and then transferred to strain UAMS-1 by using the transducing phage ϕ11 (42). The transformants and transductants were selected and cultivated at 30°C. Insertion mutants were finally cultivated at 42°C, and plasmid-free, erythromycin-sensitive mutants were selected.
The chromosomal mutation ΔcodY::erm was transferred into appropriate strain backgrounds by using transducing phage ϕ11, as described previously (42).
Detection of AIP-III.
S. aureus strains UAMS-1, MS1 (ΔcodY), and CM18 (Δagr) were grown overnight in TSB medium, diluted in 10 ml fresh TSB to an initial optical density at 600 nm (OD600) of 0.05, and incubated with shaking at 37°C until they reached an OD600 of about 0.5. The cultures were then diluted in 60 ml of TSB to give an initial OD600 of 0.05 and were incubated with shaking at 37°C. Ten-milliliter samples were removed at 60-min intervals and were subjected to centrifugation at 10,000 × g for 10 min at 4°C. The supernatants were sterilized by passage through a 0.22-μm filter and were frozen on dry ice.
An approach used previously for the detection of AIP-I (43) was modified to facilitate measurements of the accumulation of AIP-III. AIP-III was analyzed in the filtered culture fluids using reversed-phase liquid chromatography (LC) (C18 column with T3 endcapping; Waters Corporation) in an Acquity ultrahigh-performance liquid chromatograph (UPLC; Waters Corporation) coupled to an Orbitrap mass spectrometer (MS) with an electrospray source (LTQ Orbitrap XL; Thermo Scientific) over a scan range of m/z 300 to 2,000. The mass spectrometer settings were as follows: tube lens voltage, 110 V; source voltage, 4.50 kV; source current, 100 μA; heated capillary voltage, 20.0 V; heated capillary temperature, 300.0°C; sheath gas flow rate, 20.0; auxiliary gas flow rate, 0.00. Samples (5 μl) were injected and eluted at a flow rate of 0.25 ml per min with the following binary gradient, where solvent A is 0.1% formic acid in H2O and solvent B is 0.1% formic acid in acetonitrile: 0 to 6 min, from 80% A to 20% A; 6.0 to 6.5 min, from 20% A to 80% A; 6.5 to 7.0 min, 80% A (isocratic). For tandem MS (MS-MS) analysis, the precursor mass of 819.41 (the predicted mass of AIP-III) was subjected to collision-induced dissociation with an activation energy of 35%.
PSPAC-agrA plasmid construction.
The agrA gene was amplified from UAMS-1 genomic DNA using primers oAR157, containing the strong ribosome binding site (RBS) of the pQE plasmid (Qiagen), and oAR158. The PCR fragment was introduced into pCL15 (44) using the BamHI and SstI restriction sites. In the resulting plasmid, PSPAC-agrA, the agrA gene is under the control of the PSPAC promoter.
Gel mobility shift assay.
To generate a 32P-end-labeled probe, the P2-P3, P2-P3-plus-CodY motif, ilvD, and agrC regions were PCR amplified from strain UAMS-1 genomic DNA using 32P-labeled oAR174, oCM43, oAR175, and oCM114, respectively, and unlabeled oAR154 (for both P2-P3 region versions), p-ilvD2, and oCM123, respectively. The 32P-labeled primer was 5′ end labeled with [γ-32P]ATP (PerkinElmer) using T4 polynucleotide kinase (New England BioLabs) according to the manufacturer's recommendations and was then purified with the QIAquick nucleotide removal kit (Qiagen). Following amplification, 3,000 cpm of the end-labeled PCR product was used for each gel shift reaction. CodY with a 6-histidine tag at the C terminus was purified as described previously (25) and was mixed at various concentrations (25 to 200 nM) with the 32P-end-labeled promoter DNA in binding buffer (20 mM Tris-HCl [pH 8], 50 mM KCl, 2 mM MgCl2, 5% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol [DTT], 0.05% Nonidet P-40, 25 μg/ml salmon sperm DNA) supplemented with 10 mM (each) isoleucine, leucine, and valine and 2 mM GTP. After 20 min of incubation at room temperature, the reaction mixtures were loaded onto an 8% nondenaturing polyacrylamide gel prepared in a buffer (10 mM Tris-HCl [pH 8], 77 mM glycine, 0.2 mM EDTA) supplemented with 10 mM (each) isoleucine, leucine, and valine and were subjected to electrophoresis for 2 h at room temperature. The electrophoresis buffer contained 3.5 mM HEPES and 4.3 mM imidazole. Following electrophoresis, the gel was dried under a vacuum, exposed to a phosphorimager screen, and analyzed using an Applied Biosystems phosphorimager and Image Quant software (GE Healthcare).
DNase I protection experiment.
To generate a 32P-end-labeled probe, the P2-P3, P2-P3-plus-CodY motif, and ilvD regions were PCR amplified from strain UAMS-1 genomic DNA using the same primers as those used for the gel mobility shift experiment with oAR174, 0AR154, and pilvD2 as 32P-labeled primers. CodY was incubated with the 32P-end-labeled promoter DNA fragments in binding buffer (see above) supplemented with 10 mM (each) isoleucine, leucine and valine and 2 mM GTP. The total volume was 10 μl. After incubation for 20 min at room temperature, 1 μl of binding buffer containing 0.06 U of RQ1 DNase I (Promega), 10 mM MgCl2, and 20 mM CaCl2 was added to the samples. After 1 min of incubation, 4 μl of stop solution (20 mM EDTA and 95% formamide dye solution) was added, and the samples were heated at 80°C for 5 min. The samples were then loaded without further purification onto 7 M urea–5% polyacrylamide DNA sequencing gels. G+A sequence ladders were created by incubating 80,000 cpm of each radioactive fragment in 20 μl of 60% stop solution containing 1 μl of a 5% formic acid solution and boiling for 20 min. Following electrophoresis, the gel was dried under a vacuum, exposed to a phosphorimager screen, and analyzed using an Applied Biosystems phosphorimager and Image Quant software (GE Healthcare).
RNA sample collection and preparation.
S. aureus colonies from TSA plates were used to inoculate 3-ml overnight cultures in TSB. These cultures were then used to inoculate 10 ml of fresh TSB to a starting OD600 of 0.05. After the bacteria reached an OD600 between 0.5 and 0.8, they were diluted again in fresh TSB to an OD600 of 0.05. During subsequent incubation, samples (5 ml) of exponentially growing cultures (OD600, between 0.5 and 0.8) were rapidly withdrawn from each flask and were transferred to 15-ml conical tubes (Becton, Dickinson) containing an equal volume of an ice-cold (1:1 [vol/vol]) mixture of ethanol and acetone. The tubes were agitated vigorously and were immediately stored at −80°C until RNA extraction. Thawed samples were spun for 10 min at 4°C and 3,200 × g, and the pellets were washed twice with 0.5 ml TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA) with centrifugation at 3,200 × g and 4°C for 10 min. The pellets were then resuspended in 1 ml of Qiagen RLT buffer supplemented with 1% (vol/vol) 2-mercaptoethanol, mixed with 0.25 ml of 0.1-mm-diameter silica beads (Biospec Products, Inc.), and disrupted in a Mini-Beadbeater (Biospec Products, Inc.) for two 60-s intervals at the maximum disruption frequency (setting, 48) separated by a 5-min incubation on ice. The suspensions were then centrifuged at 13,000 × g for 15 min at 4°C, and the supernatant fluid was used for RNA isolation by using RNeasy minicolumns (Qiagen) according to the manufacturer's recommendations, but without the on-column DNase treatment. Genomic DNA was eliminated by using the Turbo DNA-free DNase treatment and removal kit (Ambion) according to the manufacturer's instructions, using 5 μg of nucleic acid. RNA concentrations were determined by absorbance at 260 nm using a computer-controlled NanoDrop ND-1000 spectrophotometer.
cDNA synthesis and quantitative real-time reverse transcriptase PCR (RT-PCR).
A 1-μg sample of total RNA was used to synthesize cDNA by using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer's instructions. Primers for quantitative PCR (qPCR) were designed using the online PrimerQuest tool from Integrated DNA Technologies and are listed in Table 2. We performed qPCR using the LightCycler 480 system and associated SYBR green I chemistry (Roche Applied Science) to analyze transcript abundance from prepared cDNA samples. Each 20-μl reaction mixture contained 600 nM specific primers (oAR187 and oAR188 for agrB, oAR64 and oAR52 for agrA, oAR185 and oAR186 for RNAIII, and oAR16 and oAR17 for 16S rRNA). Thermal cycling was performed according to the LightCycler 480 SYBR green I template protocol, except that we used annealing temperatures of 50°C and 55°C as the minimum temperatures for melting curve analysis. Standard curves were generated for each target by using purified DNA fragments amplified by PCR from the S. aureus chromosome. Serial dilutions spanning at least 4 orders of magnitude were analyzed. Standard curves as well as standard PCR controls (including no-template and no-reverse-transcriptase reactions) were run on each plate along with test reactions. Single amplification products were verified by melting curve analysis (melting temperature [Tm]-calling algorithm). The results of quantitative PCRs were calculated using the second-derivative maximum analysis algorithm. Data are presented either as numbers of copies of target transcripts per 106 copies of 16S rRNA or as the target/rRNA ratio of a mutant sample relative to the target/rRNA ratio of the wild-type (WT) sample cultivated under the same conditions. We used 16S rRNA for normalization because its abundance was not expected to change significantly under the conditions tested.
RESULTS
CodY-dependent regulation of the agr locus.
The Agr quorum-sensing system directly or indirectly regulates a large number of virulence genes (4). Interestingly, microarray analysis demonstrated that the expression of the agr genes is derepressed in a codY mutant, compared to that in its parent strain, during the exponential-growth phase of strains UAMS-1 and Newman (25, 27), suggesting that CodY is a repressor of agr gene expression. To confirm and quantify this regulatory effect for strain UAMS-1, we performed quantitative, real-time RT-PCR experiments at several time points during the exponential-growth phase to compare the levels of agrA and agrB mRNAs (reflecting P2 activity) and RNAIII (reflecting P3 activity) in the parental strain and the codY mutant strain (MS-1). As expected, expression of the agr locus in the wild-type strain increased significantly only at 180 min, i.e., near the end of exponential phase, but the codY-null mutation led to levels of all three RNAs significantly higher than those in the parent strain starting at 90 min (Fig. 1). Additionally, introduction into the codY mutant strain of the wild-type codY gene cloned in a low-copy-number plasmid (44) suppressed the overexpression phenotype (Fig. 1A). As reported by others (45), we found that the level of RNAIII was higher than the levels of agrA and agrB mRNAs. Interestingly, we found that CodY regulates P3 promoter expression to a greater extent than P2 promoter expression, suggesting that CodY regulates the two promoters differentially (Fig. 1D). These results confirm that CodY is a negative regulator of the agr genes and also show that agr transcription is not strictly dependent on high cell population density; when CodY is inactive, the agr locus is expressed even early in the exponential-growth phase.
AgrA is required for hyperexpression of the agr locus in a codY mutant.
To determine whether the overexpression of the agr locus in a codY mutant depends on the normal Agr autoregulatory system, we constructed an agrA deletion mutant strain and measured the effect of a codY mutation on agrB and RNAIII expression in this strain. As expected, deletion of agrA prevented the activation of agr gene expression in both the exponential (Fig. 2) and the stationary (data not shown) phase. Interestingly, the introduction of a codY mutation did not cause any derepression of the agr locus in the agrA mutant background (Fig. 2), showing that the overexpression of the agr locus in the codY mutant during the exponential-growth phase requires the presence of AgrA. To confirm this result, we cloned agrA under the control of the PSPAC promoter in a low-copy-number plasmid (44, 46). To limit the expression of agrA in trans and to prevent the induction of the P2 and P3 promoters in the codY+ strain, we took advantage of the leakiness of the PSPAC promoter and cultivated the strains in the absence of an inducer. The expression of agrA from the plasmid induced greater increases in agrB and RNAIII expression in the agrA codY double mutant than in the agrA single mutant (Fig. 2). (In fact, we may have underestimated the effect of the codY mutation in this case, because the transcription of agrA from the plasmid was unexpectedly lower in the codY mutant strain than in the wild-type strain [data not shown].) These results suggest that the effect of the CodY protein on agr locus expression depends on the presence of the AgrA response regulator.
FIG 2.
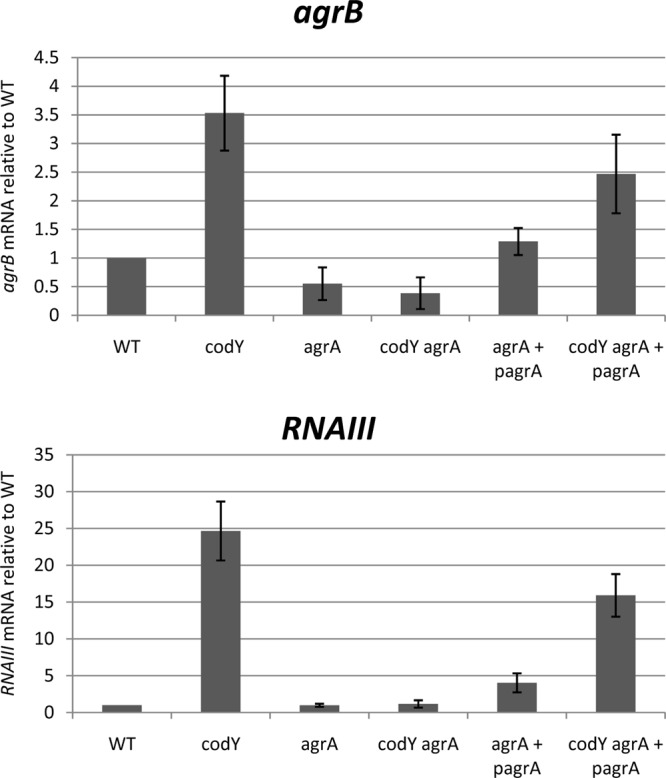
AgrA is required for overexpression of the agr locus in a codY mutant. Three biological replicates of exponential-phase culture samples (OD600, 0.5 to 0.8) of strains UAMS-1, UAMS-1 ΔagrA, UAMS-1 ΔagrA (PSPAC-agrA), and their respective codY mutants were collected, and RNAs were prepared. The relative levels of agrB and RNAIII RNAs were determined by real-time quantitative PCR. For each target gene, the ratio of the transcript to 16S rRNA was normalized to the corresponding ratio obtained for wild-type cells. Error bars correspond to the standard errors of the means.
Phosphorylation of AgrA by AgrC is required for agr overexpression in the codY mutant.
Phosphorylation of AgrA is required for the activation of the P2 and P3 promoters in stationary-phase cells (45, 47). Both the phosphorylated and unphosphorylated forms of AgrA can bind to the agr promoter region with high affinity in vitro, but the affinity and stability of the binding are higher for the phosphorylated form (45). Because CodY is active mostly during the exponential phase of growth in rich media, we investigated whether AgrA phosphorylation is required for agr overexpression in a codY mutant during exponential phase. To do so, we constructed an in-frame deletion of the histidine kinase-encoding gene agrC. Such a strain should synthesize AgrA but should not be able to mediate its phosphorylation. To check that the deletion of agrC was not affecting the expression of the downstream agrA gene, we compared, by RT-PCR, the expression of agrA in the constructed strain UAMS-1 ΔagrC and in the P2-P3 promoter deletion mutant strain, where agrA expression is not driven by the P2 promoter. We found that whereas the deletion of the P2-P3 promoter region leads to very low expression of agrA, the deletion of agrC allows a basal level of agrA expression identical to the level seen in the wild-type strain (data not shown). This result confirms that agrA is expressed at a wild-type level in the UAMS-1 ΔagrC strain. We then measured the effect of codY deletion on the levels of agrB, agrA, and RNAIII transcripts using RT-PCR analysis in exponential-phase cells of strains UAMS-1, MS-1 (ΔcodY), UAMS-1 ΔagrC, and UAMS-1 ΔagrC ΔcodY. We found that the deletion of codY led to the overexpression of RNAII (agrB and agrA) and RNAIII in agrC+ cells, as expected, but not in agrC mutant strains (Fig. 3). Therefore, we infer that phosphorylation of AgrA by AgrC is required for the overexpression of agr in a codY mutant. These results also imply that no kinase other than AgrC and no metabolite found in exponential-phase cells is able to phosphorylate AgrA sufficiently to stimulate agr gene expression in exponential-phase cells.
FIG 3.
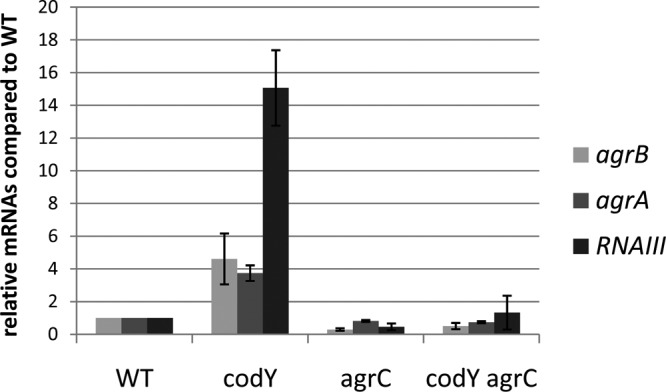
AgrC is required for overexpression of the agr locus in a codY mutant. Three biological replicates of exponential-phase culture samples (OD600, 0.5 to 0.8) of strains UAMS-1, MS-1 (ΔcodY), UAMS-1 ΔagrC, and UAMS-1 ΔcodY ΔagrC were collected, and RNAs were prepared. The relative levels of agrA, agrB, and RNAIII RNAs were determined by real-time quantitative PCR. For each target gene, the ratio of the transcript to 16S rRNA was normalized to the corresponding ratio obtained for wild-type cells. Error bars correspond to the standard errors of the means.
AIP-III accumulation in a codY mutant strain.
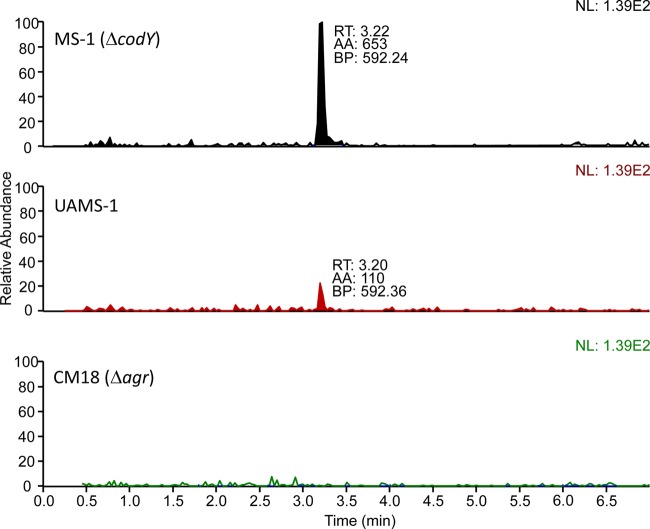
The dependence on AgrA and AgrC of agr locus hyperexpression in a codY mutant strain (Fig. 2 and 3) implies that an autoinducing peptide is involved in the derepression. Moreover, this result suggests that, even in exponential phase, when the cell density is relatively low, there is a sufficient level of autoinducing peptide to promote agr expression. To compare the levels of autoinducing peptide in the parental strain and the codY mutant, we prepared culture fluid samples from exponential-phase and early-stationary-phase cells grown in TSB. The samples were subjected to coupled liquid chromatography-mass spectrometry. There are at least four different types of AIPs produced by different S. aureus strains (48). All are short peptides (7 to 9 amino acids) that assume a thiolactone configuration (48). Strain UAMS-1 has been shown to encode AIP-III (49). To verify that the isolate of UAMS-1 used for our experiments did indeed code for AIP-III, the agrD gene was sequenced and found to encode within it the heptapeptide INCDFLL, as predicted (S. R. Brinsmade, personal communication). Figure 4 shows that we detected by mass spectrometry a heptapeptide thiolactone with the anticipated sequence and structure. As far as we can determine, this represents the first physicochemical assay for AIP-III in culture fluid. The amount of AIP-III in the culture fluid increased during growth for both the parental strain and the codY mutant, but at early times, the amount was considerably higher in the codY mutant (Table 3). At 180 min of growth, the codY mutant had accumulated about 15 times more AIP-III in the culture fluid than the parental strain (Table 3 and Fig. 5). The control strain, CM18 (Δagr), did not accumulate any detectable AIP-III at any time tested.
FIG 4.
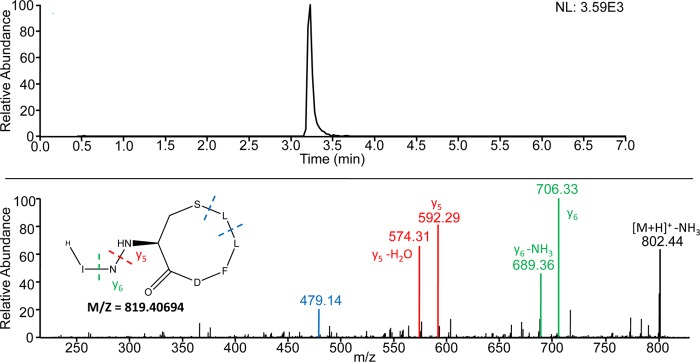
Detection of AIP-III. The filter-sterilized culture fluid of strain MS1 (ΔcodY) was subjected to reversed-phase liquid chromatography coupled to mass spectrometry. (Top) Extracted ion chromatogram for the transition from m/z 819.4 to 592.2, the y5 fragment of AIP-III. Filtering the data in this way enabled the peak corresponding to AIP-III to be clearly visualized at a retention time of 3.23 min. (Bottom) The [M+H]+ ion for AIP-III (m/z 819.4) was subjected to collision-induced dissociation to generate the MS-MS spectrum shown. The predicted structure and calculated mass of AIP-III are shown on the left. Product ion y6 corresponds to loss of the N-terminal isoleucine residue and y5 to loss of the N-terminal isoleucine-asparagine dipeptide. The product ion with a mass of 479.14 corresponds to cleavage on both sides of the leucine residue at position 7.
TABLE 3.
Accumulation of AIP-III in culture fluid of parental and mutant strainsa
| Time of sampling (min) | Amt of AIP-III (arbitrary units)b in culture fluid of strain: |
||
|---|---|---|---|
| UAMS-1 | MS1 (ΔcodY) | CM18 (Δagr) | |
| 60 | ND | ND | ND |
| 120 | ND | ND | ND |
| 180 | 30 ± 30 | 503 ± 186 | ND |
| 240 | 1,728 ± 788 | 4,365 ± 1,241 | ND |
| 360 | 7,449 ± 322 | 11,007 ± 1,321 | ND |
All strains were grown in TSB medium in three biological replicates, and samples were removed for centrifugation at the times indicated. The amount of accumulated AIP-III in the culture fluid was determined by LC-MS, as described in Materials and Methods.
ND, no peak detected for the ion corresponding to AIP-III (with the threshold for detection set at a signal/noise ratio of 3:1).
FIG 5.
Comparison of AIP-III accumulation in the parental and codY mutant strains. Filter-sterilized culture fluid from 180-min cultures of the parent strain UAMS-1, the codY-null mutant MS1, and the agr deletion mutant CM18 were subjected to coupled LC-MS, as described in Materials and Methods. Extracted ion chromatograms for the 819.4-to-592.2 transition of the [M+H]+ ion of AIP-III are shown for the three different cultures. All chromatograms were normalized to the same value (1.39 × 102) to facilitate comparison of the relative abundances of AIP-III. A peak representing AIP-III was detected at a retention time (RT) of 3.20 min, with base peak (BP) at m/z 592.36 and area (AA) of 110 (arbitrary units).
CodY integrates cell density and nutritional signals.
To assess the impact of overaccumulation of AIP-III, we measured RNAIII levels at several points during growth in the parental strain and the codY mutant (harvested at an OD600 of 0.05), which were resuspended in filtered culture fluids of later-exponential-phase cultures (180 min) of strain UAMS-1, CM18 (Δagr), or MS-1 (ΔcodY). (The culture fluids were harvested at 180 min so as to allow the resuspended cells to continue growing for several generations.) We found that growth in the culture fluid from the parental strain led to an increase in agr expression even in the parental strain (compare Fig. 6A and C), presumably due to the presence of some AIP-III in the 180-min culture fluid. In parental culture fluid, a codY-null mutant showed a statistically significant higher level of expression of RNAIII than the parental strain (Fig. 6A). As expected, resuspension of parental strain or codY mutant cells in the culture fluid of a codY mutant caused even higher levels of RNAIII expression (Fig. 6B). These results suggest that overproduction of AIP-III in a codY mutant is responsible, at least in part, for the hyperexpression of the agr operon and the RNAIII gene. Moreover, CodY normally prevents the expression of RNAIII even after the population density threshold has been reached. Interestingly, we could not detect this effect for the P2 promoter (data not shown), confirming that the P2 and P3 promoters are differentially regulated by CodY.
FIG 6.
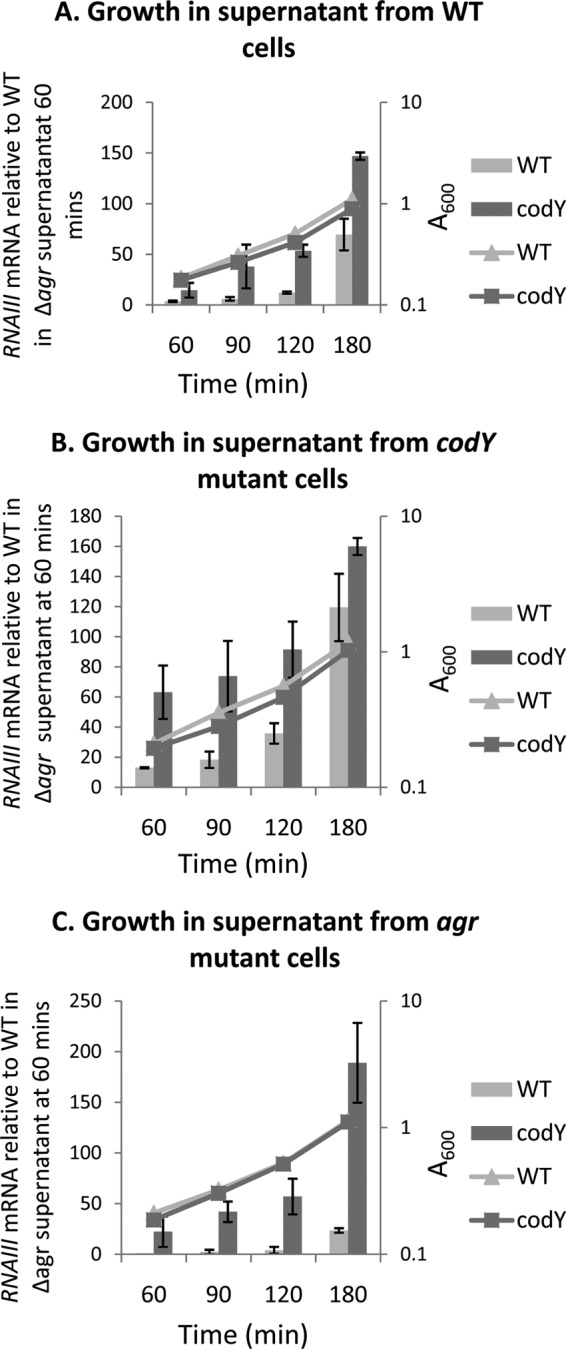
CodY integrates cell density and nutritional signals. Cultures of strains UAMS-1 (A), MS-1 (ΔcodY) (B), and CM18 (Δagr) (C) were harvested at 180 min of growth in TSB medium (late-exponential phase), filtered, and used to resuspend exponential-phase cells of strain UAMS-1 or MS-1 (ΔcodY) at an initial A600 of 0.05. Samples of the latter cultures were harvested at 60, 90, 120, and 180 min of growth in the culture fluids, RNA samples were prepared, and relative levels of RNAIII were determined by real-time quantitative PCR. For each target gene, the ratio of the transcript to 16S rRNA was normalized to the corresponding ratio obtained for wild-type cell RNA harvested at 60 min of growth in culture fluid from Δagr cells. The data represent the results of two biological replicates. Error bars correspond to the standard errors of the means.
CodY binds weakly to the agr P2-P3 promoter region in vitro.
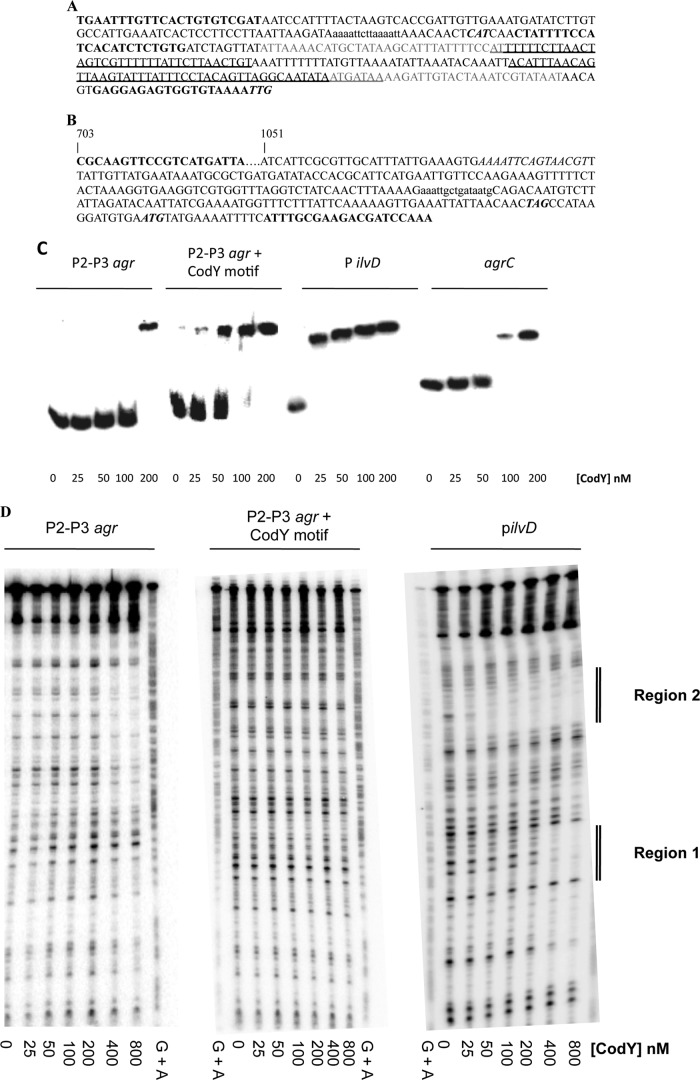
A consensus motif, AATTTTCWGAAAATT (“CodY box”), has been shown to play an important, but not necessarily sufficient, role in the interaction of CodY with DNA in Lactococcus lactis and Bacillus subtilis (50, 51). In S. aureus, a conserved motif highly reminiscent of the canonical CodY box was strongly associated with sites of CodY binding (25). Sequence analysis showed the presence of one CodY box with 3 mismatches near the beginning of the RNAIII gene, whereas no CodY box with fewer than 5 mismatches was identified in the intergenic region that includes the P2 and P3 promoters of the agr locus (Fig. 7A). Moreover, unlike hundreds of bona fide CodY binding sites, neither P2 nor P3 was copurified with CodY in a genomewide screen for CodY targets (25). To analyze further the ability of CodY to bind within the agr locus in vitro, we performed an electrophoretic mobility shift experiment using DNA fragments containing either the P2-P3 promoter intergenic region alone (P2-P3 agr) or both the P2-P3 promoter region and the putative CodY motif present within RNAIII (P2-P3 agr plus CodY motif) (Fig. 7A). As shown in Fig. 7C, S. aureus CodY was able to bind with high affinity to the ilvD promoter region (KD [equilibrium dissociation constant], ≤25 nM; KD is estimated as the concentration of CodY that causes 50% of the DNA fragments to be shifted), used here as a positive control, but bound with low affinity to the agr P2-P3 promoter region (KD, 100 to 200 nM). Interestingly, we found that CodY was able to bind with higher affinity to the DNA fragment containing the putative CodY box present within RNAIII in addition to the P2-P3 promoter region (KD, 50 to 100 nM) (Fig. 7C). This result raised the possibility that the putative binding motif present within RNAIII is a physiologically relevant binding site for CodY. To try to verify the apparent binding, we performed DNase I footprinting analyses of the various DNA fragments. As expected, we found that CodY protected two regions in the ilvD promoter fragment, corresponding to the regions identified in a genome-wide pulldown assay (25) (Fig. 7D). However, we were unable to detect protection with the P2-P3-containing fragments, even the fragment with the apparent CodY box, at CodY concentrations below 400 nM (Fig. 7D). The relatively weak interaction of CodY with the P2-P3 promoter region and the absence of a footprint for this region strongly suggest that the relatively weak binding observed in vitro does not reflect meaningful interaction in vivo and that CodY is likely to regulate the transcription of the agr genes indirectly.
FIG 7.
CodY binds with low affinity to the agr locus in vitro. (A and B) Sequences of the RNAIII-agrB intergenic region probes (A) and the agrC gene internal probe (B). The agrC probe is a 640-bp fragment that starts at position 703 of the agrC coding sequence, extends through the agrC stop codon, and ends at the 36th base of the agrA coding sequence. The oligonucleotides used to amplify each region for use as a probe are in boldface. The 3′ oligonucleotides at the downstream ends of each sequence are the complements of the actual oligonucleotides used. Putative CodY binding motifs are in lowercase. (A) The start codons for agrB and the transcription start site for RNAIII are in bold italics; the P2 and P3 promoter regions are indicated by gray letters; and the regions protected by AgrA in DNase footprinting experiments (45) are underlined. (B) The stop codon of agrC and the start codon of agrA are in bold italics. (C) Electrophoretic mobility shift experiment for S. aureus CodY binding to the P2-P3, agrC, and ilvD promoter regions. (D) DNase I protection assays of S. aureus CodY binding to the P2-P3 and ilvD promoter regions. Regions 1 and 2 correspond to CodY-protected sites in the ilvD promoter region, as described in reference 25. The lanes marked “G + A” provide sequence markers based on cleavages specific for dGMP and dAMP residues.
CodY does not regulate the expression of agrA through binding within agrC.
Given that CodY binds weakly to the P2 promoter region, which normally drives agrA transcription, a possible explanation for the AgrA dependence of agr overexpression in codY mutant cells would be that, in such cells, CodY controls the expression of agrA by binding to a DNA region located inside the agrBDCA operon. Such a mechanism would also explain the derepression of P3-dependent transcription in codY mutant cells. Interestingly, in vitro binding experiments showed previously that CodY binds to a site located within agrC (25). To confirm this result, we performed a gel shift experiment using a fragment of the agrC region containing the putative CodY binding site (Fig. 7B) in the presence of GTP and BCAAs. As shown in Fig. 7C, CodY bound with intermediate affinity to the agrC region (KD, 50 to 100 nM). Because agrC is located upstream of agrA, CodY bound at that site might function as a transcription roadblock, preventing the transcription of agrA from P2. To test that hypothesis, we compared the absolute levels of agrB and agrA mRNAs in the wild-type strain during exponential growth. We found that agrB and agrA were expressed at the same basal level (Table 4). Thus, we could find no evidence that any transcriptional roadblock exists downstream of agrB.
TABLE 4.
CodY does not regulate the expression of agrA through binding within agrCa
| Strain | Level of mRNAb |
|
|---|---|---|
| agrB | agrA | |
| WT (UAMS-1) | 17.17 ± 0.49 | 18.15 ± 2.19 |
| UAMS-1 ΔP2-P3 | ND | 0.52 ± 0.02 |
| UAMS-1 ΔcodY ΔP2-P3 | ND | 0.51 ± 0.01 |
Exponential-phase culture samples (OD600, between 0.5 and 0.8) of UAMS-1, UAMS-1 ΔP2-P3, and UAMS-1 ΔcodY ΔP2-P3 were collected, and RNAs were prepared. The absolute levels of agrB and agrA mRNAs were determined by real-time quantitative PCR.
Expressed as the number of copies of agr mRNA per 106 copies of 16S rRNA. ND, not detectable.
Another possible explanation for the CodY-dependent regulation of agr expression would be that an additional promoter, controlled by CodY, was driving agrA expression. In fact, the initial characterization of the agr locus reported the presence of a promoter, P1, that lies within the agrC coding sequence and could, in principle, contribute to the expression of agrA (52). The role, if any, played by this promoter in the regulation of the agr locus has never been determined. Interestingly, the agrC region bound by CodY in vitro overlaps with the putative P1 promoter (25). To examine the potential regulatory effect of CodY on transcription from P1, we measured the effect of a codY deletion on agrA gene expression in a strain that lacks the P2-P3 promoter region and therefore can express agrA only from P1. To that end, we constructed a mutant strain carrying a deletion of the P2-P3 promoter region and monitored the expression levels of agrA in the ΔP2-P3 and ΔcodY ΔP2-P3 mutant strains by quantitative RT-PCR. Very low expression of agrA was detected in the codY+ strain, and agrA expression did not increase in the codY mutant strain lacking the P2-P3 promoter region (Table 4). Thus, P1 is not likely to contribute significantly to agrA expression under the conditions tested and therefore is not likely to be the target of regulation by CodY.
Neither SarA nor SACOL2585 is the mediator of the CodY effect on agr gene expression.
CodY might control agr expression indirectly by controlling the expression of another regulatory protein that acts directly on the agr operon. In addition to CodY, at least 15 other proteins have been reported to regulate the agr locus, including AgrA, SarA (11), SarT (53), SarU (54), SarX (55), SarZ (56), MgrA (55–57), CcpA (58), ArlRS (59), ClpP (60, 61), CvfA (62, 63), Msa (64), Rsr (65), SrrAB (66), and RpiR homologs (67). Other than AgrA, however, none of the known transcriptional regulators of the agr operon was found to be differentially expressed more than 2-fold in a codY mutant by microarray analysis (25, 27) or by transcriptome sequencing (RNA-seq) analysis (A. Roux, unpublished data). Since SarA is a particularly well-studied regulator of agr and binds directly to the P2 and P3 promoter regions (12, 13, 68), we pursued further the possibility that the activity of SarA, rather than its synthesis, might mediate the CodY effect. Previous results, obtained using semiquantitative RT-PCR analysis, showed that RNAII and RNAIII were overexpressed in both the codY single mutant and the codY sarA double mutant (25). To address this question more rigorously, we used quantitative RT-PCR to compare the accumulation of agrA and agrB mRNAs (from P2) and RNAIII (from P3) during the exponential-growth phase in strains UAMS-1, MS-1 (ΔcodY), UAMS-1 ΔsarA, and UAMS-1 ΔcodY ΔsarA. As expected, the levels of all three RNAs were significantly higher in the codY mutant and significantly lower in the sarA mutant than in the parental strain. In a sarA mutant, however, there was still significant derepression when a codY mutation was introduced (Fig. 8). These results demonstrate that while SarA is required for maximal agr expression, the derepressing effect of a codY mutation does not depend on SarA. Thus, SarA and CodY appear to be independent regulators of the agr promoters.
FIG 8.
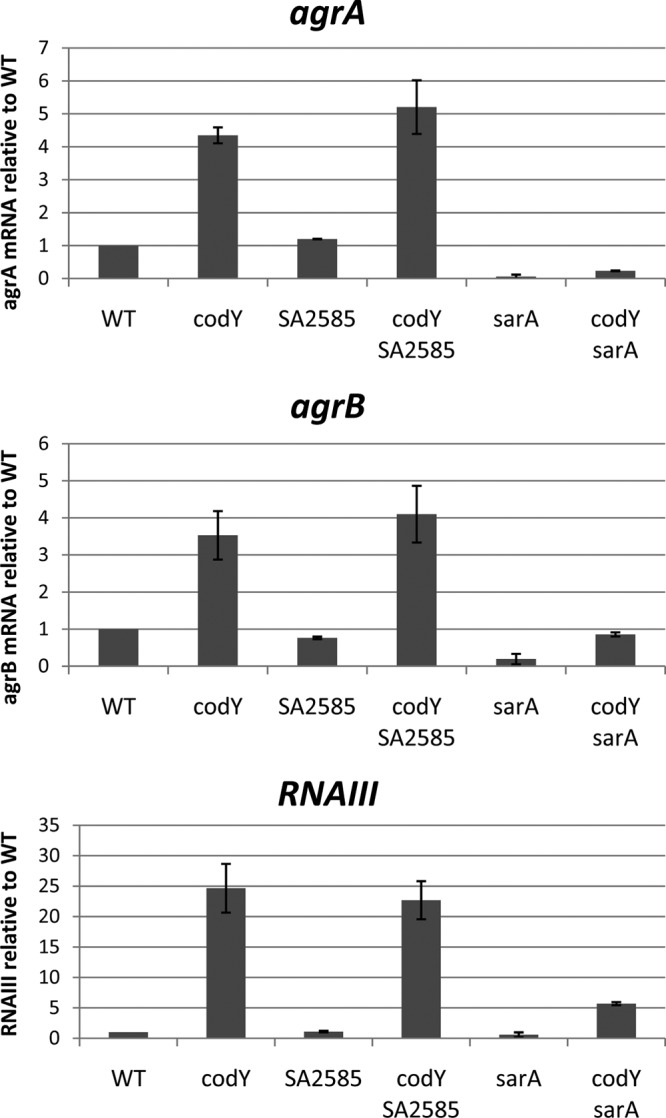
Neither SarA nor SACOL2585 is a mediator of CodY-dependent agr regulation. Three biological replicates of exponential-phase culture samples (OD600, 0.5 to 0.8) of strains UAMS-1, UAMS-1 SACOL2585::intron, UAMS-1 ΔsarA, and their respective codY mutants were collected, and RNAs were prepared. The relative levels of agrA, agrB, and RNAIII RNAs were determined by real-time quantitative PCR. For each target gene, the ratio of the transcript to 16S rRNA was normalized to the corresponding ratio obtained for wild-type cells. Error bars correspond to the standard errors of the means.
Two other transcription factor-encoding genes, saeR and SACOL2585, were significantly overexpressed in a codY mutant strain (25). The saeR gene encodes the response regulator protein of the SaeRS two-component system and is known to act on virulence gene expression in S. aureus, but several studies have shown that SaeR is not an activator of agr expression, seeming to rule out SaeR as the regulator that links CodY to the regulation of the agr locus (69–71). SACOL2585 encodes a potential transcriptional regulator of the PfoR family. To assess the role of this gene in the regulation of agr in a codY mutant, we constructed a SACOL2585 insertion mutation and measured its effect on agr expression. Interruption of SACOL2585 did not affect the expression of agr in the parental strain or in a codY mutant (Fig. 8), making it unlikely that this protein mediates CodY-dependent regulation of agr.
To test the possibility that a previously unidentified regulatory protein mediates the effect of CodY on the agr locus, we purified proteins that bind to the P2-P3 promoter region by streptavidin-affinity chromatography using biotinylated P2-P3 promoter region DNA as the bait. The same four polypeptides in the same relative amounts were found to bind to the P2-P3 region in extracts of parental and codY mutant cells (data not shown). Mass spectrometry of the bands excised from SDS-polyacrylamide gels identified the polypeptides as MgrA, SarZ, SarA, and enolase (note that CodY was not among the proteins in wild-type extracts that copurified with the regulatory region). Except for enolase, a glycolytic enzyme that binds nonspecifically to DNA, all of these proteins are known regulators of the agr locus (12, 13, 57, 68, 72). The fact that expression of the genes that encode these proteins is not affected by a codY mutation and that they were present in the same amounts in wild-type and codY mutant extracts argues against (but does not rule out) their serving as mediators of the CodY effect.
DISCUSSION
In the present study, we have analyzed the regulation of the agr genes by CodY and found that this mechanism is likely to be multifaceted. CodY binds to the P2 and P3 promoter regions in vitro but does so with low affinity. Therefore, CodY binding to the P2-P3 promoter region, if physiologically relevant, is unlikely to be the primary mode by which CodY controls agr expression. It is still possible that the binding of CodY to the P2-P3 promoter region is enhanced in vivo through simultaneous binding of an unidentified cofactor, but there is no evidence for such a cofactor in any of the dozens of other cases where CodY binding to a regulatory site has been studied. Also, in accord with a previous result (25), we showed that CodY binds with moderate affinity in vitro within the agrC gene. We found that this binding has no effect on agrA transcription in vivo, however. In fact, we could find no evidence that CodY acts as a roadblock to transcription from P2 and no evidence that CodY represses agrA transcription from the previously described, poorly active P1 promoter, which also resides within agrC (52). Taken together, these results strongly suggest that CodY does not control agr gene expression through direct binding within the agr locus.
The most surprising aspect of our results is that AgrA can be activated by AgrC-mediated phosphorylation even when the cells are at relatively low population density. Thus, our results imply that the autoinducing peptide accumulates to a sufficient level to signal to AgrC at an earlier stage of growth than previously assumed. In wild-type cells, this signaling has a minimal effect, because the agr locus is independently repressed by CodY. Only when CodY loses activity (that is, when the intracellular pools of the BCAAs or GTP, or both, drop sufficiently) does the release of CodY-mediated repression allow the activated AgrA to stimulate agr gene expression. Therefore, the apparent role of CodY is to prevent inappropriate autoinduction of agr until the nutrient limitation and the population density thresholds have both been reached. Thus, expression of the quorum-sensing system is itself dependent on nutritional signals that indicate amino acid availability. The fact many other proteins also contribute both positively and negatively to regulation of agr locus expression emphasizes the extraordinary capacity of S. aureus to integrate multiple signals simultaneously and to regulate Agr-dependent gene expression accordingly.
Although our conclusion is that CodY regulates the agr system indirectly, we have not been able to determine the mechanism of such regulation. Several hypotheses can be offered. First, the synthesis of a direct regulator of agr could be under CodY control, but we could find no evidence that the expression of any known or previously unknown transcriptional regulator of agr, other than AgrA, is controlled by CodY. In fact, overexpression of the agr locus in a codY mutant depends on the same kinase (AgrC) and the same response regulator (AgrA) as does the induction that normally occurs when cells reach high population density. Alternatively, CodY could regulate the activity of AgrA, AgrC, or another regulator through a posttranscriptional mechanism. Such a regulatory mechanism has been inferred for Bacillus anthracis, in which CodY has been shown to control the stability of AtxA, the master regulator of virulence (30). Thus, S. aureus CodY might positively regulate the synthesis of a protease that degrades AgrA or AgrC during the exponential-growth phase. In a codY-null mutant, the protease would not be made, and active AgrA or AgrC would accumulate to higher-than-normal levels. Alternatively, since the activity of AgrA is modulated by disulfide bond formation under oxidizing conditions (73), lack of CodY might affect the cell's redox balance. Given that the activity of AgrC is thought to depend on sensing a peptide signal outside the cell, we cannot rule out a posttranscriptional effect of CodY on AgrD or AgrB as well. In summary, the direct target of CodY that mediates its effect on the expression of the Agr system remains elusive; identifying this target will undoubtedly reveal a novel pathway of regulation that affects most virulence genes of S. aureus.
ACKNOWLEDGMENTS
We thank Chia Lee for providing plasmids for complementation, Alex Horswill and Mark Smeltzer for advice about assays for AIP-III, and Boris Belitsky, Laurent Bouillaut, Shaun Brinsmade, Claudette Gardel, and Kathryn Kerstein for helpful discussions. Mass spectrometry data were collected in the Triad Mass Spectrometry Facility (UNC Greensboro) under the direction of Brandie Ehrmann.
The research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award R01 GM042219.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Vandenesch F, Kornblum J, Novick RP. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jelsbak L, Hemmingsen L, Donat S, Ohlsen K, Boye K, Westh H, Ingmer H, Frees D. 2010. Growth phase-dependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Int. J. Med. Microbiol. 300:229–236. 10.1016/j.ijmm.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 3.Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. 2004. The influence of agr and σB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4:3034–3047. 10.1002/pmic.200400937 [DOI] [PubMed] [Google Scholar]
- 4.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 5.Regassa LB, Betley MJ. 1992. Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 174:5095–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407–8423. 10.1128/JB.186.24.8407-8423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regassa LB, Novick RP, Betley MJ. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60:3381–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regassa LB, Couch JL, Betley MJ. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect. Immun. 59:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan PF, Foster SJ. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144(Part 9):2469–2479. 10.1099/00221287-144-9-2469 [DOI] [PubMed] [Google Scholar]
- 10.Reiss S, Pane-Farre J, Fuchs S, Francois P, Liebeke M, Schrenzel J, Lindequist U, Lalk M, Wolz C, Hecker M, Engelmann S. 2012. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 56:787–804. 10.1128/AAC.05363-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341–7353. 10.1128/JB.183.24.7341-7353.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung AL, Projan SJ. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien Y, Manna AC, Cheung AL. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991–1001. 10.1046/j.1365-2958.1998.01126.x [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien Y, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169–37176. 10.1074/jbc.274.52.37169 [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto DF, Brunskill EW, Bayles KW. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822–4828. 10.1128/JB.182.17.4822-4828.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Manna AC, Pan CH, Kriksunov IA, Thiel DJ, Cheung AL, Zhang G. 2006. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103:2392–2397. 10.1073/pnas.0510439103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, Hurlburt B, Smeltzer M, Overbeek R, Disz T, Dunman PM. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188:2593–2603. 10.1128/JB.188.7.2593-2603.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterba KM, Mackintosh SG, Blevins JS, Hurlburt BK, Smeltzer MS. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410–4417. 10.1128/JB.185.15.4410-4417.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207. 10.1016/j.mib.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 21.Inaoka T, Ochi K. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923–3930. 10.1128/JB.184.14.3923-3930.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malke H, Ferretti JJ. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56:707–714. 10.1099/jmm.0.46984-0 [DOI] [PubMed] [Google Scholar]
- 23.Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453–1467. 10.1111/j.1365-2958.2007.05597.x [DOI] [PubMed] [Google Scholar]
- 24.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206–219. 10.1111/j.1365-2958.2007.05906.x [DOI] [PubMed] [Google Scholar]
- 25.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877. 10.1128/JB.00220-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265. 10.1128/JB.01545-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953–2963. 10.1128/JB.01492-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frenzel E, Doll V, Pauthner M, Lucking G, Scherer S, Ehling-Schulz M. 2012. CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol. Microbiol. 85:67–88. 10.1111/j.1365-2958.2012.08090.x [DOI] [PubMed] [Google Scholar]
- 29.Lindback T, Mols M, Basset C, Granum PE, Kuipers OP, Kovacs AT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14:2233–2246. 10.1111/j.1462-2920.2012.02766.x [DOI] [PubMed] [Google Scholar]
- 30.van Schaik W, Chateau A, Dillies MA, Coppee JY, Sonenshein AL, Fouet A. 2009. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect. Immun. 77:4437–4445. 10.1128/IAI.00716-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chateau A, van Schaik W, Six A, Aucher W, Fouet A. 2011. CodY regulation is required for full virulence and heme iron acquisition in Bacillus anthracis. FASEB J. 25:4445–4456. 10.1096/fj.11-188912 [DOI] [PubMed] [Google Scholar]
- 32.McDowell EJ, Callegari EA, Malke H, Chaussee MS. 2012. CodY-mediated regulation of Streptococcus pyogenes exoproteins. BMC Microbiol. 12:114. 10.1186/1471-2180-12-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291–5299. 10.1128/JB.00288-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PW. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590–601. 10.1128/JB.00917-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caymaris S, Bootsma HJ, Martin B, Hermans PW, Prudhomme M, Claverys JP. 2010. The global nutritional regulator CodY is an essential protein in the human pathogen Streptococcus pneumoniae. Mol. Microbiol. 78:344–360. 10.1111/j.1365-2958.2010.07339.x [DOI] [PubMed] [Google Scholar]
- 36.Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. 2012. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80:2382–2389. 10.1128/IAI.06172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera FE, Miller HK, Kolar SL, Stevens SM, Jr, Shaw LN. 2012. The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12:263–268. 10.1002/pmic.201100298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell DW. 2006. Transformation of E. coli by electroporation. CSH Protoc. 2006:pdb.prot3933. 10.1101/pdb.prot3933 [DOI] [PubMed] [Google Scholar]
- 39.Skow A, Mangold KA, Tajuddin M, Huntington A, Fritz B, Thomson RB, Jr, Kaul KL. 2005. Species-level identification of staphylococcal isolates by real-time PCR and melt curve analysis. J. Clin. Microbiol. 43:2876–2880. 10.1128/JCM.43.6.2876-2880.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chastanet A, Derre I, Nair S, Msadek T. 2004. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 186:1165–1174. 10.1128/JB.186.4.1165-1174.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 42.Nedelmann M, Sabottke A, Laufs R, Mack D. 1998. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentralbl. Bakteriol. 287:85–92. 10.1016/S0934-8840(98)80151-5 [DOI] [PubMed] [Google Scholar]
- 43.Junio HA, Todd DA, Ettefagh KA, Ehrmann BM, Kavanaugh JS, Horswill AR, Cech NB. 2013. Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography-high resolving power mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 930:7–12. 10.1016/j.jchromb.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luong TT, Lee CY. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123–3131. 10.1099/mic.0.29177-0 [DOI] [PubMed] [Google Scholar]
- 45.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186:7549–7555. 10.1128/JB.186.22.7549-7555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144(Part 11):3097–3104. 10.1099/00221287-144-11-3097 [DOI] [PubMed] [Google Scholar]
- 47.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick RP, Vandenesch F. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655–662. 10.1046/j.1365-2958.1998.00830.x [DOI] [PubMed] [Google Scholar]
- 48.Malone CL, Boles BR, Horswill AR. 2007. Biosynthesis of Staphylococcus aureus autoinducing peptides by using the Synechocystis DnaB mini-intein. Appl. Environ. Microbiol. 73:6036–6044. 10.1128/AEM.00912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cassat JE, Dunman PM, McAleese F, Murphy E, Projan SJ, Smeltzer MS. 2005. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 187:576–592. 10.1128/JB.187.2.576-592.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.den Hengst CD, Curley P, Larsen R, Buist G, Nauta A, van Sinderen D, Kuipers OP, Kok J. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512–521. 10.1128/JB.187.2.512-521.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belitsky BR, Sonenshein AL. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224–1236. 10.1128/JB.01780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt KA, Manna AC, Gill S, Cheung AL. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749–4758. 10.1128/IAI.69.8.4749-4758.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manna AC, Cheung AL. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343–353. 10.1128/IAI.71.1.343-353.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manna AC, Cheung AL. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188:4288–4299. 10.1128/JB.00297-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamber S, Cheung AL. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 77:419–428. 10.1128/IAI.00859-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423–1431. 10.1128/IAI.73.3.1423-1431.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183–1194. 10.1128/AAC.50.4.1183-1194.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486–5492. 10.1128/JB.187.15.5486-5492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frees D, Andersen JH, Hemmingsen L, Koskenniemi K, Baek KT, Muhammed MK, Gudeta DD, Nyman TA, Sukura A, Varmanen P, Savijoki K. 2012. New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J. Proteome Res. 11:95–108. 10.1021/pr200956s [DOI] [PubMed] [Google Scholar]
- 61.Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, Hacker J, Ohlsen K. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783–5796. 10.1128/JB.00074-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagata M, Kaito C, Sekimizu K. 2008. Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem. 283:2176–2184. 10.1074/jbc.M705309200 [DOI] [PubMed] [Google Scholar]
- 63.Kaito C, Kurokawa K, Matsumoto Y, Terao Y, Kawabata S, Hamada S, Sekimizu K. 2005. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56:934–944. 10.1111/j.1365-2958.2005.04596.x [DOI] [PubMed] [Google Scholar]
- 64.Sambanthamoorthy K, Smeltzer MS, Elasri MO. 2006. Identification and characterization of msa (SA1233), a gene involved in expression of SarA and several virulence factors in Staphylococcus aureus. Microbiology 152:2559–2572. 10.1099/mic.0.29071-0 [DOI] [PubMed] [Google Scholar]
- 65.Donegan NP, Thompson ET, Fu Z, Cheung AL. 2010. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 192:1416–1422. 10.1128/JB.00233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123. 10.1128/JB.183.4.1113-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y, Nandakumar R, Sadykov MR, Madayiputhiya N, Luong TT, Gaupp R, Lee CY, Somerville GA. 2011. RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 193:6187–6196. 10.1128/JB.05930-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinrichs JH, Bayer MG, Cheung AL. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742–7758. 10.1128/JB.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Novick RP, Jiang D. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709–2717. 10.1099/mic.0.26575-0 [DOI] [PubMed] [Google Scholar]
- 71.Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74:4655–4665. 10.1128/IAI.00322-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ballal A, Manna AC. 2009. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J. Bacteriol. 191:3301–3310. 10.1128/JB.01496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun F, Liang H, Kong X, Xie S, Cho H, Deng X, Ji Q, Zhang H, Alvarez S, Hicks LM, Bae T, Luo C, Jiang H, He C. 2012. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. U. S. A. 109:9095–9100. 10.1073/pnas.1200603109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greene C, McDevitt D, Francois P, Vaudaux PE, Lew DP, Foster TJ. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143–1152. 10.1111/j.1365-2958.1995.mmi_17061143.x [DOI] [PubMed] [Google Scholar]
- 75.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281. 10.1261/rna.68706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]