Abstract
Colistin resistance remains rare among clinical isolates of Acinetobacter species. We noted the emergence of colistin-resistant bloodstream isolates of the Acinetobacter genomic species (GS) 13BJ/14TU from patients at a university hospital between 2003 and 2011. We report here, for the first time, the microbiological and molecular characteristics of these isolates, with clinical features of Acinetobacter GS 13BJ/14TU bacteremia. All 11 available patient isolates were correctly identified as Acinetobacter GS 13BJ/14TU using partial rpoB gene sequencing but were misidentified using the phenotypic methods Vitek 2 (mostly as Acinetobacter baumannii), MicroScan (mostly as A. baumannii/Acinetobacter haemolyticus), and the API 20 NE system (all as A. haemolyticus). Most isolates were susceptible to commonly used antibiotics, including carbapenems, but all were resistant to colistin, for which it is unknown whether the resistance is acquired or intrinsic. However, the fact that none of the patients had a history of colistin therapy strongly suggests that Acinetobacter GS 13BJ/14TU is innately resistant to colistin. The phylogenetic tree of multilocus sequence typing (MLST) showed that all 11 isolates formed a separate cluster from other Acinetobacter species and yielded five sequence types. However, pulsed-field gel electrophoresis (PFGE) revealed 11 distinct patterns, suggesting that the bacteremia had occurred sporadically. Four patients showed persistent bacteremia (6 to 17 days), and all 11 patients had excellent outcomes with cleared bacteremia, suggesting that patients with Acinetobacter GS 13BJ/14TU-associated bacteremia show a favorable outcome. These results emphasize the importance of precise species identification, especially regarding colistin resistance in Acinetobacter species. In addition, MLST offers another approach to the identification of Acinetobacter GS 13BJ/14TU, whereas PFGE is useful for genotyping for this species.
INTRODUCTION
Acinetobacter species have emerged as important causative pathogens of nosocomial infections, including ventilator-associated pneumonia, bacteremia, and urinary tract infections (1, 2). Among these infections, the most common species isolated from clinical specimens is Acinetobacter baumannii, and a number of other Acinetobacter species are also frequently observed to be clinically relevant (1). To date, more than 32 Acinetobacter species have been reported, including at least 17 named species; the remaining species are generally referred to as genomic species (GS) pending further characterization (2–6). For species other than A. baumannii, the majority of Acinetobacter clinical isolates are isolated from blood and are associated with severe bacteremia or septicemia (1, 7), highlighting the increasing clinical importance of non-baumannii Acinetobacter species in bloodstream infections.
As the prevalence of multidrug-resistant (MDR) Acinetobacter infections has increased, the polymyxin antibiotic colistin has reemerged as a first-line therapy for the treatment of MDR Acinetobacter infections (2, 8, 9). While colistin-resistant Acinetobacter strains remain rare among clinical isolates of Acinetobacter species, colistin resistance has been documented in Acinetobacter GS 13BJ/14TU, an uncommon Acinetobacter species that rarely causes disease in humans (1, 8). Acinetobacter GS 13BJ and Acinetobacter GS 14TU were independently described in 1989 by two groups based on DNA-DNA hybridization (10, 32); however, these two isolates were later found to represent a single species (11). In our previous study, Acinetobacter GS 13BJ/14TU accounted for 4.7% of all Acinetobacter isolates from blood cultures at Chonnam National University Hospital (a 1,000-bed tertiary care hospital in Gwangju, South Korea) over a 4-year period (12). However, despite the frequency of these infections, little is known regarding the microbiological, genotypic, and clinical characteristics of Acinetobacter GS 13BJ/14TU bloodstream isolates. In this study, we examined the results of the identification and antimicrobial susceptibilities for all 11 available bloodstream isolates of Acinetobacter GS 13BJ/14TU. Additionally, we evaluated the genetic relationships of these isolates using both multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) and the clinical characteristics of the patients with Acinetobacter GS 13BJ/14TU bacteremia.
MATERIALS AND METHODS
Bacterial isolates and identification.
In this study, we analyzed all 11 available Acinetobacter GS 13BJ/14TU bloodstream isolates, recovered from 11 patients at Chonnam National University Hospital (a 1,000-bed tertiary care hospital in Gwangju, South Korea) between January 2003 and December 2011; two duplicate isolates were obtained from two patients (patients 8 and 11) during this period. Laboratory strains Acinetobacter GS 13BJ/14TU ATCC 17905 and A. baumannii ATCC 19606 were used as controls. All bloodstream isolates were identified by partial rpoB gene analysis using a 450-bp sequence (zone 2) in the rpoB gene region (13). Primers Ac1055F (5′-GTGATAARATGGCBGGTCGT-3′) and Ac1598R (5′-CGBGCRTGCATYTTGTCRT-3′) were used to amplify the rpoB zone 2 sequence. All loci were sequenced in both the forward and reverse directions with the same primers as used for the amplification. Sequences were assembled and compared with reference sequences using the BLAST tool at the NCBI database (http://www.ncbi.nlm.nih.gov/blast). Phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis version 5 (MEGA5) (14) comparing the 450-bp rpoB gene sequences (zone 2) obtained in this study to Acinetobacter reference sequences obtained from GenBank. A dendrogram was constructed using the neighbor-joining method and bootstrap analysis with 1,000 replicates (15, 16). Phenotypic identification of bloodstream isolates was performed using the Vitek 2 XL (Gram-negative identification [GN-ID] card; bioMérieux, France), MicroScan WalkAway 96 plus (Neg Breakpoint Combo Panel; Siemens Healthcare Diagnostics), and API 20 NE systems (bioMérieux, France) according to the manufacturers' instructions. API 20 NE identification results were interpreted using the apiweb identification software with a 7-digit numerical profile (code number). Microbiological features were investigated by streaking of isolates on blood agar plates (bioMérieux, France) and by reading the reaction of the API 20 NE strip.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing of bloodstream isolates was performed using the disk diffusion method (Becton, Dickinson and Company) on Mueller-Hinton agar for ampicillin-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid, ceftazidime, cefepime, ceftriaxone, imipenem, meropenem, gentamicin, tobramycin, amikacin, tetracycline, and ciprofloxacin; colistin susceptibility was determined using the agar dilution method according to the Clinical Laboratory and Standards Institute guidelines (17–19).
Multilocus sequence typing.
MLST was performed for all Acinetobacter GS 13BJ/14TU bloodstream isolates and control strains (Acinetobacter GS 13BJ/14TU ATCC 17905 and A. baumannii ATCC 19606) using the Institut Pasteur MLST scheme (http://www.pasteur.fr/mlst). The MLST scheme uses internal fragments of the following seven housekeeping genes: cpn60 (60-kDa chaperonin), fusA (elongation factor EF-G), gltA (citrate synthase), pyrG (CTP synthase), recA (homologous recombination factor), rplB (50S ribosomal protein L2), and rpoB (RNA polymerase subunit B). The rpoB region used for MLST is distinct from the partial rpoB gene region (zone 2) used for identification. A phylogenetic tree was inferred from 2,976-bp concatenated sequences of seven housekeeping genes for the 11 bloodstream isolates and two control strains tested in this study, along with reference Acinetobacter species sequences obtained from the MLST database and previous reports (20, 21). Phylogenetic relationships were determined using MEGA5 (14), using a 1,000-replicate bootstrap analysis; a phylogenetic tree was constructed using the neighbor-joining method (15).
Pulsed-field gel electrophoresis.
Restriction endonuclease analysis of ApaI-digested genomic DNA was performed using PFGE, as described previously (22). The CHEF DNA size standard (Bio-Rad Laboratories) was used as a molecular weight standard. PFGE patterns were analyzed visually, and the epidemiological relatedness inferred from the number of band differences in the PFGE pattern, as described previously (23). Acinetobacter GS 13BJ/14TU ATCC 17905 and A. baumannii ATCC 19606 isolates were used as control strains.
Clinical characteristics.
Demographic and clinical data for the 11 patients whose blood isolates were identified as Acinetobacter GS 13BJ/14TU were obtained from electronic medical records. Data were collected regarding clinical variables, including comorbidities, number of blood culture-positive days, stays in the intensive care unit (ICU), previous antibiotics usage, invasive procedures at the time of bacteremia onset, and outcomes. Neutropenia was defined as a neutrophil count <1,500/mm3 at the onset of infection (24). Previous use of antimicrobial agents or steroids was defined as administration within 30 days prior to onset of bacteremia (25). Recent operation was defined as an operation occurring within 30 days prior to the onset of bacteremia. Mortality was defined as Acinetobacter-related death in the absence of another definite cause of death (26).
RESULTS
Molecular identification.
A BLAST search using the 450-bp partial rpoB gene (zone 2) sequences from all 11 isolates included in this study identified Acinetobacter GS 13BJ/14TU as the closest match, which had 97 to 100% identity with the reference strain Acinetobacter GS 13BJ/14TU CIP 64.2 (GenBank accession number DQ207478.1) (Table 1).
TABLE 1.
Identification results obtained using rpoB gene sequencing and three commercial systems, antimicrobial susceptibility, and genotyping results for the 11 bloodstream isolates of Acinetobacter GS 13BJ/14TU
| Isolate no. | Date of isolation (yr-mo-day) | Species identified by: |
Antimicrobial susceptibilityb | MLSTc |
PFGEg pattern | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| rpoB gene sequence analysisa | Vitek 2a | MicroScana | API 20 NE |
|||||||
| Resulta | Code no. | Allele profiled | STf | |||||||
| 1 | 2003-10-07 | Acinetobacter GS 13BJ/14TU (97) | A. baumannii complex (95) | A. baumannii/haemolyticus (99.99) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 18-22-42e −11-21-14-21 | 198e | P1 |
| 2 | 2004-03-15 | Acinetobacter GS 13BJ/14TU (100) | A. baumannii complex (96) | A. lwoffii (99.99) | A. haemolyticus (99.9) | 0010051 | All S, except colistin (R) | 21-25-24-15-24-17-24 | 69 | P2 |
| 3 | 2005-10-18 | Acinetobacter GS 13BJ/14TU (97) | A. baumannii complex (99) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except tetracycline (R) and colistin (R) | 18-22-42e-11-21-14-21 | 198e | P3 |
| 4 | 2008-08-03 | Acinetobacter GS 13BJ/14TU (100) | A. baumannii complex (98) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 21-25-24-28e-46e-17-24 | 200e | P4 |
| 5 | 2009-07-24 | Acinetobacter GS 13BJ/14TU (99) | A. baumannii complex (99) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 21-25-42e-15-24-17-24 | 201e | P5 |
| 6 | 2009-10-01 | Acinetobacter GS 13BJ/14TU (97) | A. baumannii complex (98) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (99.9) | 0010051 | All S, except colistin (R) | 18-22-42e-11-21-14-21 | 198e | P6 |
| 7 | 2009-11-03 | Acinetobacter GS 13BJ/14TU (97) | A. baumannii complex (99) | A. baumannii/haemolyticus (66.99) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 18-22-42e-11-21-14-21 | 198e | P7 |
| 8 | 2010-05-11 | Acinetobacter GS 13BJ/14TU (99) | A. baumannii complex (99) | A. baumannii/haemolyticus (95.16) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 18-22-42e-11-21-14-24 | 199e | P8 |
| 9 | 2010-07-03 | Acinetobacter GS 13BJ/14TU (97) | A. lwoffii (97) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 18-22-42e-11-21-14-21 | 198e | P9 |
| 10 | 2011-07-11 | Acinetobacter GS 13BJ/14TU (97) | A. baumannii complex (99) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except ciprofloxacin (I) and colistin (R) | 18-22-42e-11-21-14-21 | 198e | P10 |
| 11 | 2011-08-09 | Acinetobacter GS 13BJ/14TU (99) | A. baumannii complex (99) | P. stuartii (75.88) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 21-25-24-28e-46e-17-24 | 200e | P11 |
| ATCC 17905 | Acinetobacter GS 13BJ/14TU (100) | A. baumannii complex (98) | A. baumannii/haemolyticus (98.18) | A. haemolyticus (95.2) | 0010053 | All S, except colistin (R) | 21-25-24-15-24-17-24 | 69 | P12 | |
Numbers in parentheses are the probabilities of correct identification (as percentages).
Antimicrobial susceptibility testing was performed using the disk diffusion method for ampicillin-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid, ceftazidime, cefepime, ceftriaxone, imipenem, meropenem, gentamicin, tobramycin, amikacin, tetracycline, and ciprofloxacin; colistin susceptibility was determined using the agar dilution method. S, susceptible; I, intermediate; R, resistant.
MLST, multilocus sequence typing.
Allele profiles for cpn60-fusA-gltA-pyrG-recA-rplB-rpoB.
Three new alleles and four new STs identified in this study were added to the Institut Pasteur MLST database (http://www.pasteur.fr/mlst).
ST, sequence type.
PFGE, pulsed-field gel electrophoresis.
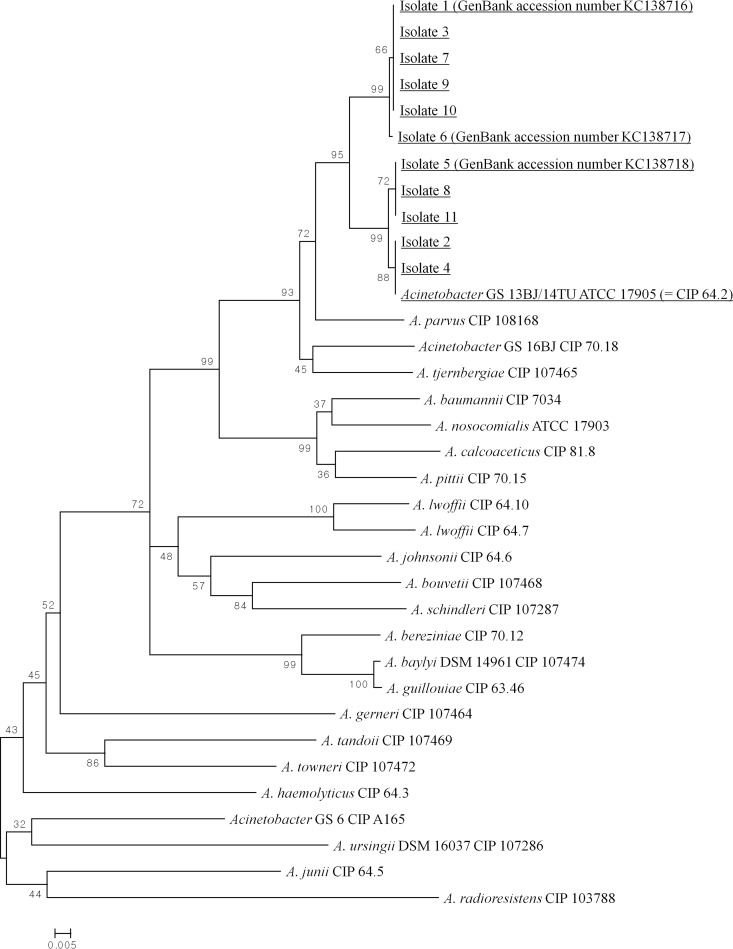
A phylogenetic tree was generated using the 450-bp partial rpoB gene sequences to identify relationships among the 11 Acinetobacter GS 13BJ/14TU bloodstream isolates (isolates 1 to 11), along with the Acinetobacter species reference strains available in GenBank (Fig. 1). In this analysis, all 11 bloodstream isolates were well differentiated from other Acinetobacter species, clearly clustering with Acinetobacter GS 13BJ/14TU ATCC 17905 (CIP 64.2). The partial rpoB gene sequences of isolates 1, 3, 7, 9, and 10 were identical, as were those of isolates 5, 8, and 11.
FIG 1.
Phylogenetic tree of the 11 Acinetobacter GS 13BJ/14TU bloodstream isolates (isolates 1 through 11) from Chonnam National University Hospital and reference strains of Acinetobacter species from the GenBank database, constructed from 450-bp zone 2 sequences of the rpoB gene. This tree was inferred using the neighbor-joining method and a 1,000-replicate bootstrap analysis. The sequences tested in this study are underlined. New rpoB gene sequences (submitted to GenBank) identified in this study include accession no. KC138716 (isolate 1), KC138717 (isolate 6), and KC138718 (isolate 5). The scale bar indicates the number of base substitutions per site.
Phenotypic identification and antimicrobial susceptibilities.
All 11 Acinetobacter GS 13BJ/14TU bloodstream isolates were characterized using three commercial methods: Vitek 2, MicroScan, and the API 20 NE system (Table 1). Vitek 2 misidentified 10 of 11 isolates as A. baumannii, with the remaining isolate identified as Acinetobacter lwoffii. Similarly, MicroScan misidentified 9 of 11 isolates as A. baumannii/A. haemolyticus, with the remaining two isolates identified as Providencia stuartii and A. lwoffii. The API 20 NE system identified all isolates as A. haemolyticus with high predictive values. The code numbers obtained by the API 20 NE system were 0010053 for nine isolates and 0010051 for two isolates. Using the API 20 NE system, 100% of isolates (11/11) showed gelatin hydrolysis, and assimilation of capric acid, malate, and citrate; 82% (9/11) showed assimilation of phenylacetic acid. All 11 bloodstream isolates exhibited beta hemolysis on blood agar plates.
Antimicrobial susceptibility testing revealed that all (100%) bloodstream isolates were susceptible to the following 11 antimicrobial agents: ampicillin-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid, ceftazidime, cefepime, ceftriaxone, imipenem, meropenem, gentamicin, tobramycin, and amikacin. Ten of 11 isolates (91%) were also susceptible to tetracycline and ciprofloxacin. However, despite this widespread susceptibility to common antimicrobial agents, all 11 isolates were resistant to colistin as determined by the agar dilution method (Table 1).
MLST and PFGE analysis.
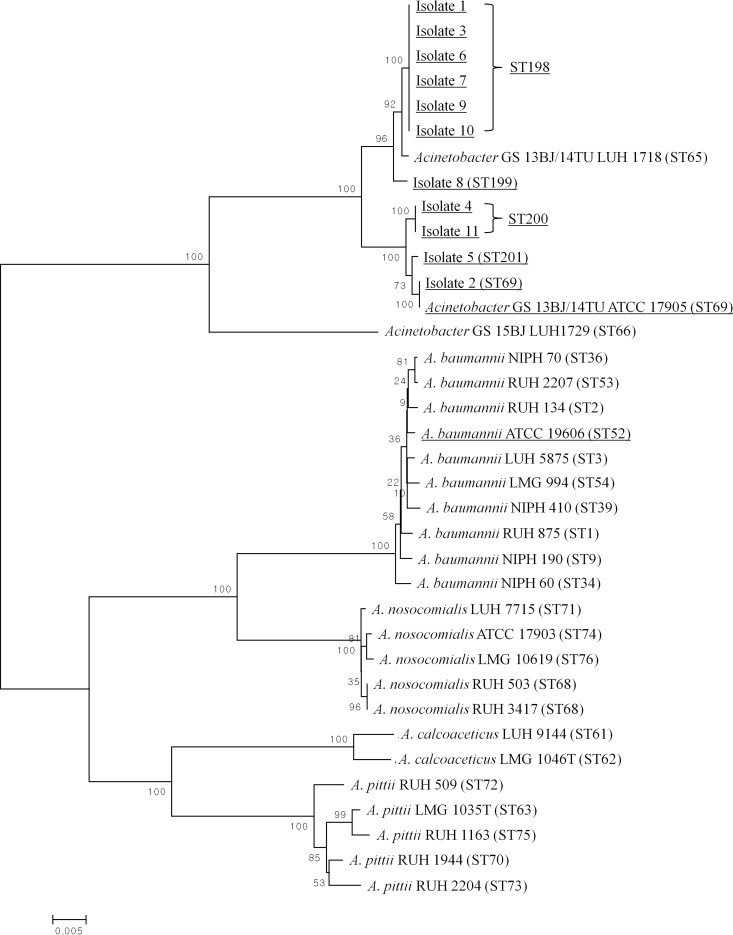
Three new alleles (gltA 42, pyrG 28, and recA 46) and four new sequence types (STs) (ST198, ST199, ST200, and ST201) were identified by MLST and were added to the MLST database (see http://www.pasteur.fr/mlst). Overall, five distinct STs were obtained from a combination of seven housekeeping genes (Table 1). Of the five STs, ST198 and ST200 were shared by six and two isolates, respectively, with the remaining three STs (ST69, ST199, and ST201) found in one isolate each. The control strain, Acinetobacter GS 13BJ/14TU ATCC 17905, was classified as ST69. Within the phylogenetic tree, all 11 bloodstream isolates clustered with the reference strains Acinetobacter GS 13BJ/14TU ATCC 17905 and Acinetobacter GS 13BJ/14TU LUH 1718 (Fig. 2). This cluster was distinct from the Acinetobacter calcoaceticus-A. baumannii complex that comprised A. baumannii, A. calcoaceticus, A. nosocomialis, and A. pittii. Acinetobacter GS 15BJ LUH 1729, previously described as ST66, was also distinct from the cluster of Acinetobacter GS 13BJ/14TU.
FIG 2.
Dendrogram of the MLST analysis of 11 bloodstream isolates (isolates 1 to 11) and reference strains of Acinetobacter species. A 2,976-bp concatenated sequence consisting of seven housekeeping genes was inferred using the neighbor-joining method and a 1,000-replicate bootstrap analysis. Corresponding MLST profiles present in the Institut Pasteur MLST database (see http://www.pasteur.fr/mlst) are shown in parentheses. The sequences tested in this study are underlined. New STs recovered in this study include ST198, ST199, ST200, and ST201. The scale bar indicates the number of base substitutions per site.
The representative PFGE patterns for each of the 11 bloodstream isolates are shown in Fig. 3. PFGE of ApaI-digested genomic DNA revealed 11 distinct PFGE patterns (Table 1). In the two patients (patients 8 and 11) from whom serial bloodstream isolates were collected, all strains identified from each patient exhibited identical MLST and PFGE patterns.
FIG 3.
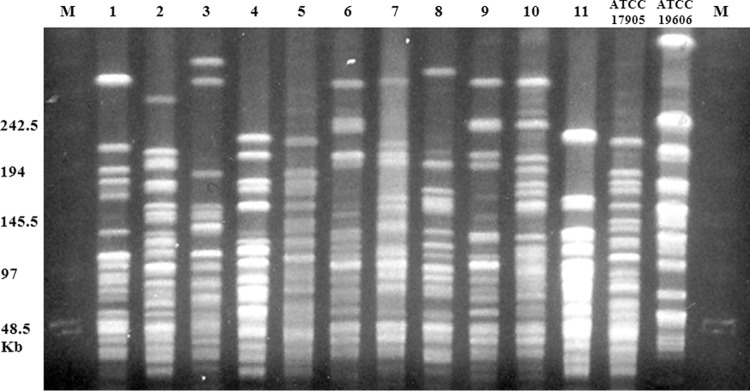
Restriction endonuclease analysis of genomic ApaI-digested DNA using the PFGE method for 11 nonduplicate Acinetobacter GS 13BJ/14TU bloodstream isolates; all 11 isolates showed different band patterns. Details are summarized in Table 1. Lanes 1 and 15, lambda ladder markers; lanes 2 to 12, isolates 1 to 11; lane 13, Acinetobacter GS 13BJ/14TU ATCC 17905; lane 14, A. baumannii ATCC 19606.
Clinical characteristics.
The clinical characteristics of the 11 patients with Acinetobacter GS 13BJ/14TU bacteremia are summarized in Table 2. Most patients had severe comorbidities, such as leukemia, infective endocarditis, and acute myocardial infarction, or underwent invasive procedures, such as tracheal intubation, central venous catheterization, coronary artery intervention, and peritoneal dialysis. No previous use of colistin was identified in any of the 11 patients. Ten patients were deemed to have nosocomial infections, while the remaining patient was an outpatient undergoing regular peritoneal dialysis. While all patients with Acinetobacter GS 13BJ/14TU bacteremia had excellent outcomes with cleared bacteremia, four patients (patients 1, 5, 8, and 11) were repeatedly culture positive (two to seven times), with continuous, persistent bacteremia (6 to 17 days). These four patients with continuous bacteremia were implicated in necrotizing pneumonia, infective endocarditis, primary bacteremia, and peritonitis.
TABLE 2.
Clinical characteristics of the 11 patients with Acinetobacter GS 13BJ/14TU bacteremia
| Patient clinical characteristic | Patient no.a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Age (yr)/gender | 12/F | 76/F | 57/M | 74/F | 80/M | 68/F | 85/F | 75/M | 83/F | 48/M | 42/M |
| Department/ward | CS/SICU | GI/ER | ID/7822 | ID/7862 | CV/8766 | RHD/B767 | CV/8761 | CV/MICU | HPBS/B866 | NR/U866 | KD/outpatient |
| Hospital days from admission to first bacteremia | 46 | 5 | 8 | 7 | 8 | 32 | 16 | 7 | 12 | 11 | Outpatient |
| Underlying diseases | Leukemia | Duodenal ulcer bleeding and perforation, coronary artery disease | Renal impairment, gout | Liver cirrhosis, gastric ulcer | Mitral regurgitation, heart failure | Dermatomyositis, DM | COPD, heart failure | Acute myocardial infarction, DM, renal impairment | Choledocholithiasis | Cerebral vascular accident | End-stage renal disease, DM |
| No. of blood cultures (days)b positive | 7 (0, 1, 2, 9, 11, 14, 17) | 1 | 1 | 1 | 6 (0, 2, 3, 4, 7, 8) | 1 | 1 | 4 (0, 4, 7, 8) | 1 | 1 | 2 (0, 6) |
| ICU stay | Y | N | N | N | N | N | N | Y | N | Y | N |
| Clinical status at positive culture | |||||||||||
| Previous use of colistin/other antibiotics | N/Y | N/Y | N/Y | N/Y | N/N | N/N | N/N | N/N | N/Y | N/Y | N/N |
| Previous use of steroid/neutropenia | N/N | N/N | Y/N | N/N | N/N | Y/N | N/N | N/N | N/N | N/N | N/N |
| Recent operation | N | N | N | N | N | N | N | N | Y | N | N |
| Probable primary site of infection | Necrotizing pneumonia | Primary bacteremia | Catheter-related infection | Pneumonia | Infective endocarditis | Pneumonia | Pneumonia | Primary bacteremia | Intraabdominal infection | Primary bacteremia | CAPD peritonitis |
| Invasive procedures at bacteremia onset | Tracheal intubation, CVC | CVC, TPN, GI endoscopy, NG tube, IVI, Foley catheter | CVC, hemodialysis | TPN | TPN | None | Coronary angiography | Coronary angiography/intervention, TPN | TPN | Tracheal intubation, arterial catheter, TPN, NG tube, IVI, Foley catheter | Peritoneal dialysis |
| Antibiotic therapy | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y |
| Outcome of bacteremia/mortality | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive | Cleared/alive |
CS, cardiothoracic surgery; SICU, surgical intensive care unit; GI, gastrointestinal; ER, emergency room; ID, infectious disease; CV, cardiovascular; RHD, rheumatic disease; MICU, medical intensive care unit; HPBS, hepatopancreaticobiliary surgery; NR, neurology; KD, kidney disease; DM, diabetes mellitus; COPD, chronic obstructive pulmonary disease; CAPD, continuous ambulatory peritoneal dialysis; CVC, central venous catheterization; TPN, total parenteral nutrition; NG, nasogastric; IVI, intravascular intervention; Y, yes; N, no.
Days are numbered relative to the time of first positive blood culture.
Nucleotide sequence accession numbers.
The zone 2 rpoB sequences of isolates 1, 5, and 6 have been deposited in GenBank under accession numbers KC138716, KC138718, and KC138717, respectively.
DISCUSSION
Although infrequently isolated from clinical samples, Acinetobacter GS 13BJ/14TU isolates exhibit resistance to colistin, the last line of defense against MDR Acinetobacter infections (1, 8). It is unknown whether this resistance is the result of secondary mutations conferring acquired resistance, or if Acinetobacter GS 13BJ/14TU is intrinsically resistant to colistin. In this study, all 11 bloodstream isolates of Acinetobacter GS 13BJ/14TU were susceptible to a wide range of clinically relevant antibiotics, but all exhibited significant in vitro resistance to colistin. As none of the isolates included in this study were recovered from patients with a history of colistin therapy, these data strongly suggest that Acinetobacter GS 13BJ/14TU is intrinsically resistant to colistin. In addition, this report is noteworthy from a clinical standpoint in that it represents the first description of Acinetobacter GS 13BJ/14TU as an opportunistic pathogen capable of causing health care-associated bacterial infections with favorable outcomes. These data highlight the importance of proper identification of Acinetobacter at the species level, and the need for physicians to be aware of the antimicrobial susceptibility profiles of different Acinetobacter species, particularly those causing bloodstream infections.
Although all 11 bloodstream isolates were misidentified by the phenotypic identification methods using the Vitek 2, MicroScan, and API 20 NE systems, all were correctly identified as Acinetobacter GS 13BJ/14TU by rpoB gene (zone 2) sequencing, confirming the suitability of this region for species identification, as reported previously (13, 27, 28). Misidentification by commercial systems may have arisen due to limitations in existing databases and the lack of nonspecialized substrates for Acinetobacter GS 13BJ/14TU species identification, as reported previously (2, 29–31).
All of our isolates produced beta hemolysis on blood agar plates, a phenotype not observed in A. calcoaceticus-A. baumannii complex species (2, 10, 32). Further characterization of these isolates using the API 20 NE system revealed gelatin hydrolysis and assimilation of capric acid, malate, and citrate, similar to A. haemolyticus (33). These phenotypic characteristics likely explain why the API 20 NE system misidentified these isolates as A. haemolyticus with high predictive values. However, further characterization revealed assimilation of phenylacetic acid in 9 of 11 isolates (82%), a finding that is inconsistent with identification of A. haemolyticus because <1% of all A. haemolyticus isolates exhibit such a phenotype, information which is present in the manufacturer's database and a previous report (33). A previous report also showed 100% susceptibility (10/10 isolates) of A. haemolyticus to colistin (8), in contrast to the results described here. Taken together, our study identified three characteristic features that can be used to diagnose Acinetobacter GS 13BJ/14TU infections in clinical microbiology laboratories. These features include (i) evidence of beta hemolysis on blood agar plates, (ii) colistin resistance, and (iii) positive identification as A. haemolyticus by the API 20 NE system, especially when combined with positive assimilation of phenylacetic acid. Molecular methods remain necessary for definitive species identification; however, these criteria may allow early detection of Acinetobacter GS 13BJ/14TU infections.
Two MLST schemes for Acinetobacter species identification are currently available, those of Bartual et al. (6) and the Institut Pasteur. Of these two, only the Institut Pasteur MLST scheme provided a successful performance of MLST testing for all isolates of Acinetobacter GS 13BJ/14TU, suggesting that the scheme of the Institut Pasteur may be a more suitable MLST method for this species. All bloodstream Acinetobacter GS 13BJ/14TU isolates are well clustered with the ST 65 of Acinetobacter GS 13BJ/14TU LUH 1718 (SEIP 5.84) and the ST 69 of Acinetobacter GS 13BJ/14TU LUH 1717 (ATCC 17905) reference strains of previous reports (20, 21), suggesting that MLST is suitable for data transfer and comparison and thus providing a tool for long-term or global epidemiological studies (6, 34). Furthermore, all MLST concatenated sequences for Acinetobacter GS 13BJ/14TU were easily differentiated from other Acinetobacter species (Fig. 2). These findings suggest that STs 65, 69, 198, 199, 200, and 201 may be indicative of Acinetobacter GS 13BJ/14TU strains, and the MLST can be used as genetic identification tool for this species.
This study represents the first report of the genotypic relationships among Acinetobacter GS 13BJ/14TU clinical isolates using both MLST and PFGE analysis. In the current study, MLST differentiated 11 isolates into 5 STs, whereas PFGE revealed 11 distinct patterns, suggesting that PFGE is a more discriminatory method than MLST for this species. Despite the apparent divergence among PFGE patterns of isolates from different patients, each of the duplicate isolates from patients 8 and 11 exhibited identical MLST and PFGE patterns, indicating a high degree of reproducibility for these two methods. In the present study, none of the bloodstream isolates were highly related spatiotemporally, and PFGE showed 11 distinct patterns, suggesting that each bacteremia occurred sporadically. However, as PFGE patterns are likely to reflect rapidly evolving genetic markers, isolates with identical STs but different PFGE patterns may have been derived from a recent common ancestor (35). These data suggest that the PFGE method may be useful for epidemiological typing of Acinetobacter GS 13BJ/14TU isolates, while MLST is more effective for species identification.
Although Acinetobacter GS 13BJ/14TU has been implicated in cases of septic shock, endocarditis, and pyrexia (1), little is known about the clinical characteristics of Acinetobacter GS 13BJ/14TU bacteremia. Our data suggest that Acinetobacter GS 13BJ/14TU can cause health care-associated bacteremia in patients with severe underlying diseases or those who had undergone invasive medical procedures. The isolates from seven patients were recovered from blood just once, and the isolate from one patient disappeared without antibiotic therapy. This finding suggests that this species may cause transient bacteremia. However, four patients who had severe underlying diseases such as leukemia, mitral regurgitation, uncontrolled diabetes mellitus, or end-stage renal disease showed persistent bacteremia (6 to 17 days), demonstrating this species may be implicated in necrotizing pneumonia, infective endocarditis, primary bacteremia, or peritonitis. However, despite the persistent bacteremia in some patients, all Acinetobacter GS 13BJ/14TU bacteremias were eventually cleared, with positive outcomes in all 11 patients. These observations are consistent with previous reports of significant differences in the clinical features, including mortality, among Acinetobacter species, with A. baumannii associated with significantly worse outcomes compared to other Acinetobacter species (36–41). This work therefore highlights the importance of correct GS identification, which may facilitate prediction of the outcome of Acinetobacter bacteremia.
ACKNOWLEDGMENTS
We thank platform Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles (available at www.pasteur.fr/mlst).
Footnotes
Published ahead of print 8 January 2014
REFERENCES
- 1.Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48:1445–1449. 10.1128/JCM.02467-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doughari HJ, Ndakidemi PA, Human IS, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ. 26:101–112. 10.1264/jsme2.ME10179 [DOI] [PubMed] [Google Scholar]
- 4.Karah N, Haldorsen B, Hegstad K, Simonsen GS, Sundsfjord A, Samuelsen O. 2011. Species identification and molecular characterization of Acinetobacter spp. blood culture isolates from Norway. J. Antimicrob. Chemother. 66:738–744. 10.1093/jac/dkq521 [DOI] [PubMed] [Google Scholar]
- 5.Nemec A, Musilek M, Maixnerova M, De Baere T, van der Reijden TJ, Vaneechoutte M, Dijkshoorn L. 2009. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans. Int. J. Syst. Evol. Microbiol. 59:118–124. 10.1099/ijs.0.001230-0 [DOI] [PubMed] [Google Scholar]
- 6.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter spp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114–4123. 10.1128/AAC.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemec A, Dijkshoorn L. 2010. Variations in colistin susceptibility among different species of the genus Acinetobacter. J. Antimicrob. Chemother. 65:367–369. 10.1093/jac/dkp440 [DOI] [PubMed] [Google Scholar]
- 9.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 10.Bouvet PJ, Jeanjean S. 1989. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res. Microbiol. 140:291–299. 10.1016/0923-2508(89)90021-1 [DOI] [PubMed] [Google Scholar]
- 11.Gerner-Smidt P, Tjernberg I, Ursing J. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29:277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SY, Shin JH, Lee K, Joo MY, Park KH, Shin MG, Suh SP, Ryang DW, Kim SH. 2013. Comparison of the Vitek 2, MicroScan, and Etest methods with the agar dilution method in assessing colistin susceptibility of bloodstream isolates of Acinetobacter species from a Korean University Hospital. J. Clin. Microbiol. 51:1924–1926. 10.1128/JCM.00427-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832. 10.1128/JCM.44.3.827-832.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 16.Zharkikh A, Li WH. 1995. Estimation of confidence in phylogeny: the complete-and-partial bootstrap technique. Mol. Phylogenet. Evol. 4:44–63. 10.1006/mpev.1995.1005 [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial disk susceptibility tests; approved standard, 10th ed. (M02-A10) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. (M07-A8) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 19.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement (M100-S21). Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 20.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162:393–404. 10.1016/j.resmic.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 21.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335. 10.1128/JCM.43.9.4328-4335.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh MM, Everhart JE, Byrd-Holt DD, Tisdale JF, Rodgers GP. 2007. Prevalence of neutropenia in the U.S. population: age, sex, smoking status, and ethnic differences. Ann. Intern. Med. 146:486–492. 10.7326/0003-4819-146-7-200704030-00004 [DOI] [PubMed] [Google Scholar]
- 25.Huang ST, Chiang MC, Kuo SC, Lee YT, Chiang TH, Yang SP, Ti Y, Chen TL, Fung CP. 2012. Risk factors and clinical outcomes of patients with carbapenem-resistant Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. 45:356–362. 10.1016/j.jmii.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 26.Jang HC, Kim SH, Kim KH, Kim CJ, Lee S, Song KH, Jeon JH, Park WB, Kim HB, Park SW, Kim NJ, Kim EC, Oh MD, Choe KW. 2009. Salvage treatment for persistent methicillin-resistant Staphylococcus aureus bacteremia: efficacy of linezolid with or without carbapenem. Clin. Infect. Dis. 49:395–401. 10.1086/600295 [DOI] [PubMed] [Google Scholar]
- 27.Lee Y, Lee J, Jeong SH, Bae IK, Lee K. 2011. Carbapenem-non-susceptible Acinetobacter baumannii of sequence type 92 or its single-locus variants with a G428T substitution in zone 2 of the rpoB gene. J. Antimicrob. Chemother. 66:66–72. 10.1093/jac/dkq402 [DOI] [PubMed] [Google Scholar]
- 28.Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333–2341. 10.1099/mic.0.026054-0 [DOI] [PubMed] [Google Scholar]
- 29.Bernards AT, Dijkshoorn L, Van der Toorn J, Bochner BR, Van Boven CP. 1995. Phenotypic characterisation of Acinetobacter strains of 13 DNA-DNA hybridisation groups by means of the biolog system. J. Med. Microbiol. 42:113–119. 10.1099/00222615-42-2-113 [DOI] [PubMed] [Google Scholar]
- 30.Bernards AT, van der Toorn J, van Boven CP, Dijkshoorn L. 1996. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur. J. Clin. Microbiol. Infect. Dis. 15:303–308. 10.1007/BF01695662 [DOI] [PubMed] [Google Scholar]
- 31.Horrevorts A, Bergman K, Kollee L, Breuker I, Tjernberg I, Dijkshoorn L. 1995. Clinical and epidemiological investigations of Acinetobacter genomospecies 3 in a neonatal intensive care unit. J. Clin. Microbiol. 33:1567–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tjernberg I, Ursing J. 1989. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS 97:595–605. 10.1111/j.1699-0463.1989.tb00449.x [DOI] [PubMed] [Google Scholar]
- 33.Dortet L, Legrand P, Soussy CJ, Cattoir V. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471–4478. 10.1128/JCM.01535-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, Doi Y. 2011. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J. Clin. Microbiol. 49:3849-3854. 10.1128/JCM.00619-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin JH, Bougnoux ME, d'Enfert C, Kim SH, Moon CJ, Joo MY, Lee K, Kim MN, Lee HS, Shin MG, Suh SP, Ryang DW. 2011. Genetic diversity among Korean Candida albicans bloodstream isolates: assessment by multilocus sequence typing and restriction endonuclease analysis of genomic DNA by use of BssHII. J. Clin. Microbiol. 49:2572–2577. 10.1128/JCM.02153-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuang YC, Sheng WH, Li SY, Lin YC, Wang JT, Chen YC, Chang SC. 2011. Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with acinetobacter bacteremia. Clin. Infect. Dis. 52:352–360. 10.1093/cid/ciq154 [DOI] [PubMed] [Google Scholar]
- 37.Lee YC, Huang YT, Tan CK, Kuo YW, Liao CH, Lee PI, Hsueh PR. 2011. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J. Antimicrob. Chemother. 66:1839–1846. 10.1093/jac/dkr200 [DOI] [PubMed] [Google Scholar]
- 38.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, Wenzel RP, Seifert H. 2012. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 64:282–290. 10.1016/j.jinf.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 39.Houang ET, Chu YW, Chu KY, Ng KC, Leung CM, Cheng AF. 2003. Significance of genomic DNA group delineation in comparative studies of antimicrobial susceptibility of Acinetobacter spp. Antimicrob. Agents Chemother. 47:1472–1475. 10.1128/AAC.47.4.1472-1475.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim YM, Shin KS, Kim J. 2007. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genomic species of Acinetobacter isolates. J. Clin. Microbiol. 45:902–905. 10.1128/JCM.01573-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park KH, Shin JH, Lee SY, Kim SH, Jang MO, Kang SJ, Jung SI, Chung EK, Ko KS, Jang HC. 2013. The clinical characteristics, carbapenem resistance, and outcome of acinetobacter bacteremia according to genospecies. PLoS One 8:e65026. 10.1371/journal.pone.0065026 [DOI] [PMC free article] [PubMed] [Google Scholar]