Abstract
Antifungal azoles are widely used for controlling fungal infections. Fungi are able to change the expression of many genes when they adapt to azole stress, and increased expression of some of these genes can elevate resistance to azoles. However, the regulatory mechanisms behind transcriptional adaption to azoles in filamentous fungi are poorly understood. In this study, we found that deletion of the transcription factor gene ccg-8, which is known to be a clock-controlled gene, made Neurospora crassa hypersensitive to azoles. A comparative genome-wide analysis of the responses to ketoconazole of the wild type and the ccg-8 mutant revealed that the transcriptional responses to ketoconazole of 78 of the 488 transcriptionally ketoconazole-upregulated genes and the 427 transcriptionally ketoconazole-downregulated genes in the wild type were regulated by CCG-8. Ketoconazole sensitivity testing of all available knockout mutants for CCG-8-regulated genes revealed that CCG-8 contributed to azole adaption by regulating the ketoconazole responses of many genes, including the target gene (erg11), an azole transporter gene (cdr4), a hexose transporter gene (hxt13), a stress response gene (locus number NCU06317, named kts-1), two transcription factor genes (NCU01386 [named kts-2] and fsd-1/ndt80), four enzyme-encoding genes, and six unknown-function genes. CCG-8 also regulated phospholipid synthesis in N. crassa in a manner similar to that of its homolog in Saccharomyces cerevisiae, Opi1p. However, there was no cross talk between phospholipid synthesis and azole resistance in N. crassa. CCG-8 homologs are conserved and are common in filamentous fungi. Deletion of the CCG-8 homolog-encoding gene in Fusarium verticillioides (Fvccg-8) also made this fungus hypersensitive to antifungal azoles.
INTRODUCTION
Antifungal azoles, which inhibit ergosterol biosynthesis by disrupting the essential P450 superfamily protein lanosterol 14α-demethylase CYP51 (also called ERG11), are the most widely used drugs for controlling fungal infections. However, when subjected to antifungal azole stress, fungi are able to transcriptionally change the expression of a number of genes (1–4). For example, Candida albicans can elevate the transcriptional levels of the azole efflux pump genes CDR1 and CDR2 and the azole drug target gene ERG11 in response to ketoconazole treatment (5, 6). Similar responses were found in Saccharomyces cerevisiae (4), Aspergillus fumigatus (3), Trichophyton rubrum (1), and Fusarium graminearum (2, 7). Overexpression of these genes increases resistance to azoles in many fungal species (8–14). In addition to these genes, several other genes that are transcriptionally elevated by antifungal azoles have been found. For example, in S. cerevisiae, many genes downstream of ERG11 in ergosterol biosynthesis and genes involved in lipid and cell wall biosynthesis are also transcriptionally elevated by ketoconazole treatment (4). However, the roles of the majority of azole-responsive genes in azole adaption are unknown.
Transcriptional responses to azole stress are known to be regulated by several transcription factors in S. cerevisiae and C. albicans. In S. cerevisiae, the transcription factors Pdr1p and Pdr3p regulate the transcription of multidrug efflux pump genes, such as PDR5, SNQ2, YOR1, and FLR1 (15). Their C. albicans homologs, Fcr1p and Fcr3p, have similar roles (16). Our BLASTp search found that filamentous fungi do not contain proteins that are homologous to Pdr1p or Pdr3p. A zinc finger domain containing transcription factor Upc2p in C. albicans and its paralog in S. cerevisiae, Ecm22p, regulate ergosterol synthesis genes (17–20). Upc2p can directly bind to the promoters of ergosterol biosynthesis genes, such as NCP1, ERG11, and ERG2, and to multidrug efflux genes, including CDR1, MDR1, and YOR1, in C. albicans (21). Although Upc2p homologs are present in filamentous fungi, their functions in the azole response are unknown. To date, only a bzip-type transcription factor, AP-1, is known to be important for azole resistance in both yeasts and filamentous fungi (22–24). Thus, compared with those of C. albicans and S. cerevisiae, the regulatory mechanisms behind azole responses in filamentous fungi are poorly understood. The unknown transcription factors that regulate azole responses need to be identified in filamentous fungi.
Neurospora crassa has transcriptional responses to ketoconazole that are similar to those of pathogenic fungi for both ergosterol biosynthesis genes and the Pdr5p-like ABC transporter gene cdr4 (25, 26), and about 70% of the genes in N. crassa have knockout mutants, which means that N. crassa may be an excellent model for identifying regulatory genes in drug resistance. Using this model, we have demonstrated that sterol C-22 desaturase ERG5 is required for wild-type azole resistance in N. crassa and Fusarium verticillioides (26). In this study, we found that the transcription factor CCG-8 positively regulates the ketoconazole responses of many genes in N. crassa and is required for wild-type resistance to antifungal azoles in N. crassa and F. verticillioides.
MATERIALS AND METHODS
Strains and culture conditions.
Most of the N. crassa strains, including FGSC 4200 (wild-type strain), FGSC 20378 (ccg-8; mating type a), and knockout mutants for genes regulated by ccg-8 deletion, were obtained from the Fungal Genetics Stock Center (www.fgsc.net/; University of Kansas Medical Center). Vogel's minimum medium (27), supplemented with 2% (wt/vol) sucrose for slants or 2% glucose for plates and the liquid medium, was used for culturing the strains. All cultures were grown at 28°C. Antifungal compounds were added as needed.
F. verticillioides wild-type strain 7600 and its transformants were maintained on potato dextrose agar (PDA) (Difco, Detroit, MI).
Drug susceptibility test.
Ketoconazole, fluconazole, and itraconazole were dissolved in dimethyl sulfoxide (DMSO) and then aseptically added to the autoclaved medium before the agar plates were made. The final concentrations of ketoconazole, fluconazole, and itraconazole in the agar plates were 2, 15, and 6 μg/ml, respectively. The final DMSO concentration was below 0.25% (vol/vol). Two microliters of conidial suspension was inoculated onto individual plates (diameter [Φ] = 9 cm) with or without antifungal drugs and incubated at 28°C.
Complementation of the N. crassa ccg-8 mutant.
To complement the ccg-8 knockout mutant, a complementary plasmid, pCB1532-ccg8, was created by inserting a 4,301-bp DNA fragment containing the ccg-8 gene (2,015 bp) flanked by a 1,383-bp upstream regulatory region and a 903-bp downstream region into the pCB1532 plasmid, which also contained a sulfonylurea resistance allele of the Magnaporthe grisea ILV1 gene as a selective marker (28). The insert was amplified from the wild-type strain, FGSC 4200, using primers ccg8(p)F-SmaI (TCCCCCGGGCTTCTTACATAGGTAGTCGGATTGG) and ccg8(3)R-EcoRI (CGGAATTCGCTCCAAGTTGTTTGCCAT), digested by SmaI and EcoRI, and then ligated into the pCB1532 plasmid. The pCB1532-ccg8 construct was transformed into the ccg-8 deletion mutant, FGSC 20378 (ccg-8; a), using the previously reported protoplast transformation method (29). Chlorimuron ethyl (15 μg/ml; Sigma) was added to the top of the agar to inhibit the growth of nontransformed protoplasts.
RNA extraction and transcriptional analysis by reverse transcription-quantitative PCR (qRT-PCR).
Mycelia were harvested, immediately frozen, and then ground into a fine powder in liquid nitrogen. Total RNA was extracted and treated with DNase I to remove genomic DNA according to the standard TRIzol protocol (Invitrogen, Carlsbad, CA, USA). The cDNAs were synthesized from total cellular RNA using a cDNA synthesis kit (Fermentas, Burlington, Canada) according to the manufacturer's protocol.
Gene-specific primers (Table 1) were designed using online tools from PrimerQuest. The qRT-PCR was carried out using the iQ5 multicolor real-time PCR detection system (Bio-Rad, Hercules, CA) with SYBR green detection (SYBR PrimeScript RT-PCR kit; TaKaRa) according to the manufacturer's instructions. Each cDNA sample was analyzed in triplicate, and the average threshold cycle (CT) was calculated. Relative expression levels were calculated using the 2−ΔΔCT method (30). The results were normalized to the expression level of β-tubulin.
TABLE 1.
Gene-specific primer pairs used for reverse transcription-quantitative PCR assay
| Gene | Primer name | Nucleotide sequence (5′→3′) |
|---|---|---|
| NCU09686 (ccg-8) | QN9686F | AAGGTGGCTCTCTCCTTTA |
| QN9686R | GGTCATTTGGTTCATCTTCTTG | |
| NCU02624 (erg11) | QN2624F | CCGCCATTGTCAAGGAAA |
| QN2624R | ACGTGATCGGTCGGAATA | |
| NCU05591 (cdr4) | QN5591F | ACGCTTTGGAAATGGATGGTGACG |
| QN5591R | ATGAACAAGGCGACGGAAATGCAG | |
| NCU06666 (ino1) | QN6666F | CACACCGTTGTGATCAAGTA |
| QN6666R | CACGTTGAAGAGCGAGATG | |
| NCU06317 (kts-1) | QN6317F | CACTACCACCACCAACAAG |
| QN6317R | GGACCAGCATGTGCAATA | |
| NCU01386 (kts-2) | QN1386F | ATGGCACCCTTTGTGATG |
| QN1386R | CAGTCCATCTCTCTTGGAAAC | |
| NCU09915 (fsd-1/ndt80) | QN9915F | ATCCGAGAGGTGGCTATG |
| QN9915R | GTAGATCGTTGCAGGGAAAT | |
| NCU09691 (kts-3) | QN9691F | GATTGCGACCCGAAGAAG |
| QN9691R | GGATCATCCACACAAGTCAG | |
| NCU02084 (arginase) | QN2084F | GATAGTCACATCGACACATGG |
| QN2084R | ATGGAGGTGTTGTGGATTAAG | |
| NCU09635 (kts-4) | QN9635F | CATCCCTCCTAACAACAAGAC |
| QN9635R | CCGTTGGCATAATTGACATAAG | |
| NCU01315 (gt41-1) | QN1315F | CTGATTGCTGGCGATGATTA |
| QN1315R | CACCACCAGCACCATTATAC | |
| NCU08957 (kts-5) | QN8957F | CTCTGACATCTCGGACTTTG |
| QN8957R | TTGGAGGAGGAGGAAGATT | |
| NCU01633 (hxt13) | QN1633F | GACTCCATTGGCTACTTCTATG |
| QN1633R | TACATGGTCTCGATCTCCTC | |
| NCU00247 (kts-6) | QN0247F | CGCCTTCTTCAGCTTCTTT |
| QN0247R | GATGATGGTGGGATGAATGAG | |
| NCU01302 (hydrolase) | QN1302F | CATTCCCTACCAAGAGATCAAG |
| QN1302R | GTTGCAGATAGGACGAGTATG | |
| NCU00611 (kts-7) | QN0611F | CATATCTCCATGCCACTCTTG |
| QN0611R | CAAGGAGTTCAGCTGGTAAG | |
| NCU00248 (kts-8) | QN0248F | GAGGTAGGAGGAACGACAT |
| QN0248R | GGGCATCCCTCTGATAAGTA | |
| NCU04334 (cpn10) | QN4334F | CCCGAGTCCTCCGTTAAG |
| QN4334R | GCCGTACTGAGGGATAAGAA | |
| NCU04923 (gcy-1) | QN4923F | GAGAGCAACTTCCAGATTCC |
| QN4923R | CCAGACATCGTAGCCAAAG | |
| β-tub | QbtubF | CCCAAGAACATGATGGCTGCTTCT |
| QbtubR | TTGTTCTGAACGTTGCGCATCTGG |
Transcriptomic profiling analysis.
Genome-wide transcriptional profiles for the wild type and the ccg-8 deletion mutant in Vogel's medium with or without ketoconazole treatment (2.5 μg/ml for 24 h) were obtained by digital gene expression (DGE) profiling (31). Briefly, conidia from the wild-type strain and the ccg-8 deletion mutant were added to 20 ml liquid medium in a plate (Φ = 9 cm) and incubated for 24 h at 28°C in the dark until they formed a mycelial mat on the surface of the liquid medium. The mycelial mat was then cut into small pieces (Φ = 10 mm) and transferred to Vogel's liquid medium (two pieces/100 ml) in 250-ml flasks. The cultures were shaken at 180 rpm and incubated at 28°C for 12 h. Ketoconazole was then added to the medium to reach a final concentration of 2.5 μg/ml. After 24 h of incubation, mycelia were harvested and total RNA was extracted and subjected to DGE analysis as described by Sun et al. (32). In this study, the transcriptional ratios between two samples of more than 2 and less than 0.5 were used as the thresholds for defining differentially expressed genes.
Deletion of the ccg-8 homolog gene in F. verticillioides.
The vector for deleting the ccg-8-homologous gene in F. verticillioides, Fvccg8 (FVEG_06643), was constructed as follows. The 5′-end-flanking sequences (1,351 bp) of FVEG_06643 were amplified using primers Fv06643(p)F-KpnI (TGGGGTACCCTTCTCCTCAACGCACCCT) and Fv06643(p)R-XhoI (CCGCTCGAGCGCAAGACGATAGCCCAC), and the 3′-end-flanking sequences (1,597 bp) of FVEG_06643 were amplified using primers Fv06643(3)F-SmaI (TCCCCCGGGCGTTTGGTGGGCTTTGTA) and Fv06643(3)R-NotI (ATAAGAATGCGGCCGCGAATGACGGGAGGCGATGA). The resulting PCR products were cloned into the pCSN44 plasmid (33) to create the knockout construct pCSN44-ΔFv06643, in which the PCR product was ligated with the hygromycin phosphotransferase (hph) gene. Then, the deletion cassette was transformed into F. verticillioides 7600 to create the deletion mutants. Fungal transformation followed the protocol reported by Miller et al. (34), with minor modifications, as described by Li et al. (35).
RESULTS
N. crassa ccg-8 deletion elevates azole sensitivity.
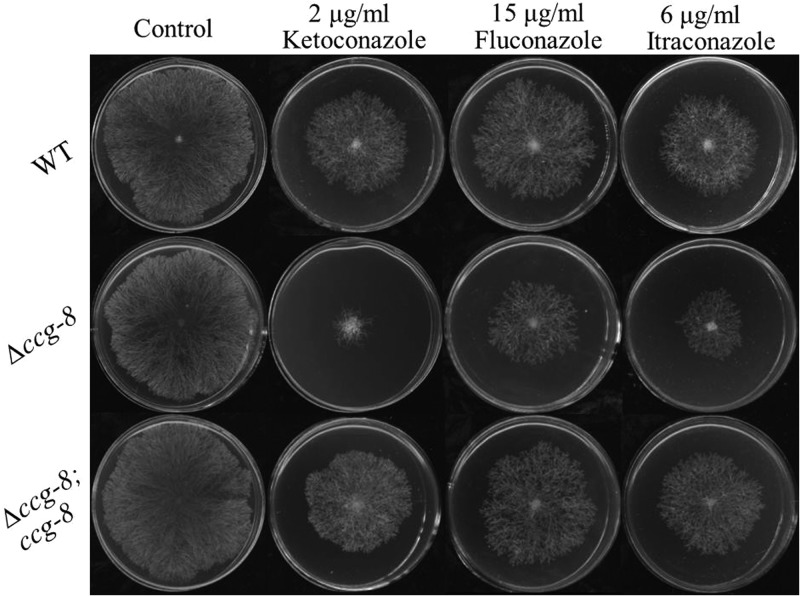
By screening the knockout mutants of the transcription factor-encoding genes, we successfully isolated a mutant, FGSC 20378, that displayed hypersensitivity to multiple antifungal azoles. FGSC 20378 lacks the ccg-8 (locus number NCU09686) gene, which has previously been identified as a clock-controlled gene (36). Without antifungal drugs, the differences in the growth rates (Fig. 1) and conidial yields (data not shown) between the ccg-8 deletion mutant and the wild-type strain were not significant. However, when the strains were grown on agar plates with ketoconazole, fluconazole, or itraconazole, the growth of the ccg-8 deletion mutant was more inhibited by these azoles than was that of the wild-type strain (Fig. 1). The rates of inhibition of the ccg-8 deletion mutant by ketoconazole (2 μg/ml), fluconazole (15 μg/ml), and itraconazole (6 μg/ml) were 80% ± 3%, 44% ± 2%, and 58% ± 2%, respectively. In contrast, the rates of inhibition of the wild type by these drugs were only 30% ± 2%, 22% ± 2%, and 34% ± 3%, respectively.
FIG 1.
Drug susceptibility analysis of the N. crassa wild type (WT), the ccg-8 knockout mutant (Δccg-8), and the ccg-8-complemented strain (Δccg-8; ccg-8). Two microliters of conidial suspension (1 × 104 conidia/ml) was inoculated onto the center of each plate (Φ = 9 cm) with or without antifungal drugs; the final concentrations of ketoconazole, fluconazole, and itraconazole were 2 μg/ml, 15 μg/ml, and 6 μg/ml, respectively. The plates were incubated at 28°C in the dark for about 66 h. Each test had three replicates, and the experiment was independently repeated twice.
N. crassa ccg-8 deletion does not affect sensitivities to other stresses.
In addition to testing sensitivities to antifungal drugs, we tested the sensitivity of the ccg-8 mutant to other stresses, including high temperature (40°C), osmotic stress (1 M NaCl), and oxidative stress (18 μg/ml menadione). The ccg-8 mutant displayed wild-type levels of sensitivity to these stresses (data not shown), which suggested that CCG-8 is not involved in adaptation to these stresses.
Complementation of the ccg-8 deletion mutant.
To complement the ccg-8 deletion mutant, we transformed the complementary pCB1532-ccg8 plasmid into the ccg-8 deletion mutant. Ten transformants were obtained, and six transformants displayed wild-type azole sensitivity (the phenotypic characteristics of a representative transformant are shown in Fig. 1), which confirmed that the hypersensitivity to azoles in the ccg-8 mutant was caused by ccg-8 deletion.
Transcription of ccg-8 is induced by ketoconazole.
Genome-wide transcriptional responses to ketoconazole treatment were analyzed by the transcriptome sequencing (RNA-seq) method, and the transcriptional level of ccg-8 increased by at least three times after ketoconazole (2.5 μg/ml) treatment for 24 h (Table 2). The transcriptional levels of ccg-8 in the wild type treated with or without ketoconazole were further analyzed by qRT-PCR. The ccg-8 transcriptional level in the wild type increased 3.6 times after 24 h of ketoconazole treatment (2.5 μg/ml) relative to the levels without treatment (Fig. 2).
TABLE 2.
Comparison of the transcriptional responses by the wild type and the ccg-8 mutant to ketoconazole stress by gene seta
| Locus and/or categoryb | Gene | Function | TPM in WT | TPM in WT(k) | Fold change [WT(k)/WT] | P value | TPM in Δccg-8 | TPM in Δccg-8(k) | Fold change [Δccg-8(k)/Δccg-8] | P value | Fold change [Δccg-8(k)/WT(k)] | Sensitivity to ketoconazole compared to WT's |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phospholipid synthesis pathways | ||||||||||||
| NCU06666 | ino1 | Inositol-3-phosphate synthase | 38.86 | 21.55 | 0.55 | 2.65E−05 | 180.9 | 17.25 | 0.09 | 4.7E−117 | 0.80 | Similar |
| NCU08045 | cho2 | Phosphatidylethanolamine N-methyltransferase | 11.22 | 6.16 | 0.55 | 0.022589 | 110 | 27.06 | 0.25 | 3.34E−41 | 4.39 | NA |
| NCU03695 | psd1 | Phosphatidylserine decarboxylase proenzyme 1 | 28.78 | 51.28 | 1.78 | 1.87E−14 | 69.7 | 115.08 | 1.65 | 7.63E−10 | 2.26 | Similar |
| NCU02381 | cho1 | Phosphatidylserine synthase | 18.13 | 8.12 | 0.45 | 0.000217 | 29.96 | 13.08 | 0.44 | 1.91E−06 | 1.61 | NA |
| NCU09643 | cds1 | Phosphatidate cytidylyltransferase | 17.56 | 13.43 | 0.76 | 0.165267 | 24.02 | 18.73 | 0.78 | 0.138875 | 1.39 | NA |
| Kinases participating in Opi1p (CCG-8 homolog in S. cerevisiae) phosphorylation | ||||||||||||
| NCU06240 | pkac-1 | Catalytic subunit protein kinase A | 26.48 | 24.63 | 0.93 | 0.6266 | 40.04 | 9.81 | 0.25 | 5.40E−16 | 0.40 | Similar |
| NCU06544 | pkc | Protein kinase C | 24.75 | 67.16 | 2.71 | 0 | 52.79 | 24.98 | 0.47 | 5.28E−09 | 0.37 | Similar |
| NCU03124 | ck2 | Catalytic subunit of protein kinase CK2 | 68.21 | 155.87 | 2.29 | 1.61E−13 | 52.2 | 60.07 | 1.15 | 0.173376 | 0.39 | Similar |
| Target enzyme of azoles and multidrug transporters | ||||||||||||
| NCU02624 | erg11 | Cytochrome P450 lanosterol 14a-demethylase | 69.08 | 676.66 | 9.80 | 0 | 113.9 | 254.84 | 2.24 | 4.01E−13 | 0.38 | NA |
| NCU05591 | cdr4 | ABC transporter CDR4 | 3.74 | 259.97 | 69.51 | 2.17E−07 | 4.75 | 54.42 | 11.46 | 4.10E−08 | 0.21 | Hypersensitive |
| NCU04161 | Multidrug resistance-associated protein 5 | 2.59 | 20.99 | 8.10 | 2.57E−06 | 4.75 | 6.84 | 1.44 | 0.264586 | 0.33 | Similar | |
| Stress responses | ||||||||||||
| NCU01499 | Related to hsp70 | 16.12 | 39.74 | 2.47 | 1.97E−09 | 35.29 | 147.49 | 4.18 | 0 | 3.71 | Similar | |
| NCU06317 | kts-1 | Stress response RCI peptide | 123.19 | 1142.04 | 9.27 | 1.56E−13 | 323.3 | 412.45 | 1.28 | 1.56E−09 | 0.36 | Hypersensitive |
| NCU09873 | pck-1/acu-6 | Phosphoenolpyruvate carboxykinase | 4.89 | 31.06 | 6.35 | 1.68E−08 | 300.1 | 275.96 | 0.92 | 0.064493 | 8.88 | Similar |
| NCU00355 | cat-3 | Catalase-3 | 135.85 | 252.42 | 1.86 | 0 | 54.28 | 24.98 | 0.46 | 1.08E−09 | 0.10 | Similar |
| Transcription factors | ||||||||||||
| NCU09686 | ccg-8 | Clock-controlled protein 8 | 33.97 | 127.89 | 3.76 | 3.94E−13 | 0 | 0 | Hypersensitive | |||
| NCU01386 | kts-2 | Zn(II)2Cys6 domain-containing protein | 0.58 | 26.86 | 46.31 | 0.000112 | 1.78 | 5.95 | 3.34 | 0.00582 | 0.22 | Hypersensitive |
| NCU01629 | C2H2 finger domain-containing protein | 4.03 | 53.45 | 13.26 | 1.15E−07 | 28.77 | 25.57 | 0.89 | 0.427838 | 0.48 | Similar | |
| NCU05536 | Zn(II)2Cys6 domain-containing protein | 6.04 | 48.97 | 8.11 | 1.25E−09 | 6.23 | 8.33 | 1.34 | 0.317684 | 0.17 | Similar | |
| NCU09496 | C2H2 transcription factor | 10.07 | 97.94 | 9.73 | 2.66E−13 | 37.37 | 47.58 | 1.27 | 0.042249 | 0.49 | NA | |
| NCU04022 | C2H2 finger domain-containing protein | 12.66 | 120.89 | 9.55 | 6.62E−14 | 33.22 | 29.44 | 0.89 | 0.38236 | 0.24 | NA | |
| NCU09915 | fsd-1/ndt80 | Female sexual development-1 protein | 7.77 | 46.17 | 5.94 | 2.51E−11 | 46.56 | 21.41 | 0.46 | 1.60E−08 | 0.46 | Hypersensitive |
WT, wild type; WT(k) or Δccg-8(k), wild type or Δccg-8 mutant treated with ketoconazole, respectively; TPM, number of locus tags per million; NA, the mutant was not available.
Locus numbers and function were annotated according to the N. crassa genome assembly (http://www.broadinstitute.org/annotation/genome/neurospora/).
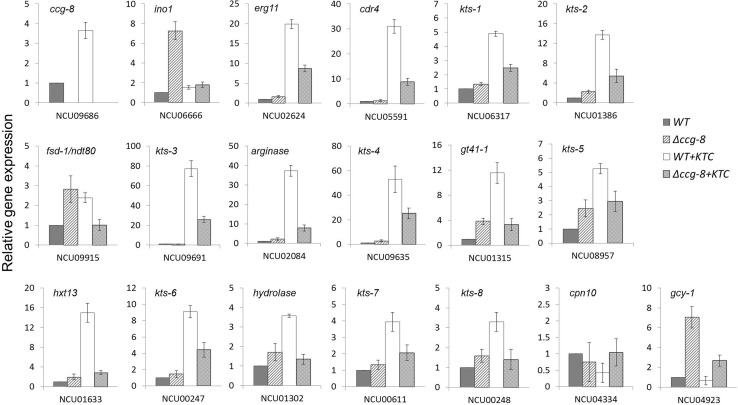
FIG 2.
Differential expression of genes in the ccg-8 deletion mutant relative to that in the wild-type strain in response to azole stress as determined by qRT-PCR. Values shown are means of results from three independent replicates. Standard deviations are indicated with error bars. The wild-type strain and the ccg-8 deletion mutant cultures were inoculated into Vogel's liquid medium and incubated at 28°C with shaking at 180 rpm for 12 h. Ketoconazole (KTC) was then added into the medium to reach a final concentration of 2.5 μg/ml. After incubation for 24 h, mycelia were harvested and total RNA was extracted and subjected to qRT-PCR analysis.
CCG-8 regulates transcriptional responses to ketoconazole stress.
We analyzed genome-wide transcriptional profiles in the wild type and the ccg-8 deletion mutant in Vogel's medium with or without ketoconazole (2.5 μg/ml for 24 h) using the DGE method in order to understand how CCG-8 influences azole sensitivity.
After ketoconazole treatment, 488 genes were upregulated and 427 genes were downregulated in the wild-type strain (see Table S1 in the supplemental material). The transcriptional levels for 70 of the 488 genes upregulated by ketoconazole stress in the wild type were significantly lower in the ccg-8 mutant than in the wild type (Tables S1 and S2 in the supplemental material), and transcriptional levels for 8 (NCU07332, NCU00999, NCU07454, NCU02579, NCU01219, NCU04334, NCU04720, and NCU04923) of the 427 genes downregulated by ketoconazole stress in the wild type were significantly higher in the ccg-8 mutant than in the wild-type strain (Tables S1 and S2). These results indicated that ccg-8 deletion can affect the normal transcriptional responses to ketoconazole.
The DEG data also showed that without ketoconazole treatment, ccg-8 deletion upregulated 570 genes and downregulated 386 genes relative to transcriptional levels in the wild type (see Table S3 in the supplemental material). There were no obvious defects in growth and development under the tested culture conditions without azole, so the transcriptional changes caused by ccg-8 deletion should not affect growth and development.
Roles of genes regulated by CCG-8 in azole resistance.
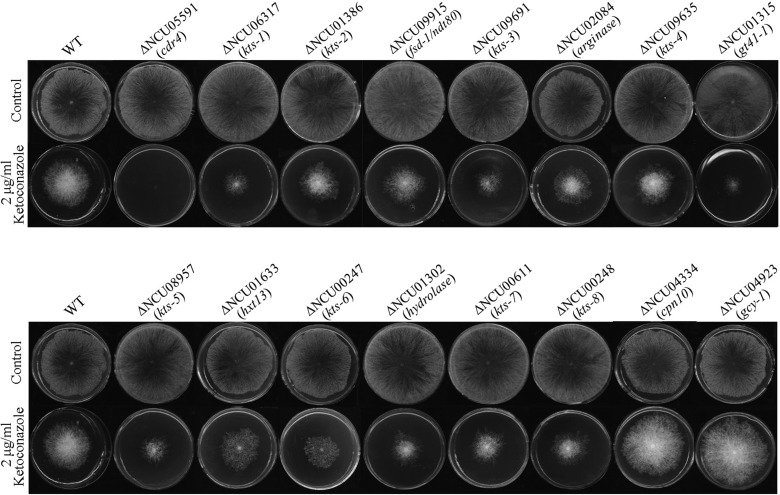
Seventy-eight genes are differentially expressed between the ccg-8 mutant and the wild type during ketoconazole stress, and overexpression of either of two of these genes, cdr4 (the PDR5 homolog) and erg11, has previously been shown to confer azole resistance in many fungi (8–14). The sensitivity to ketoconazole of their knockout mutants was tested in order to understand the roles of the remaining CCG-8-regulated genes in azole resistance. For 70 genes positively regulated by CCG-8, 56 single-gene-knockout mutants are available from the Fungal Genetics Stock Center. Our test showed that 14 mutants with single-gene deletions were hypersensitive to ketoconazole compared to the wild type (Fig. 3; Tables 2 and 3). Among the knocked-out genes were two transcription factor-encoding genes, fsd-1 or ndt80 (NCU09915) and NCU01386. Transcription factor FSD-1 is essential for female fertility in N. crassa (37). An FSD-1 homolog in S. cerevisiae, Ndt80, controls the expression of genes that regulate meiotic progression and spore formation (38, 39). NCU01386 encodes a Zn(II)Cys6-type transcription factor, and its highly conserved homologs are present in filamentous fungi, including the human pathogen A. fumigatus, but not in yeasts. To date, none of its homologs has previously been characterized. Mutants for the hexose transporter HXT13 gene hxt13 (NCU01633), the azole transporter gene cdr4 (NCU05591), a stress-responsive gene (NCU06317), a putative UDP-N-acetylglucosaminyltransferase gene, gt41-1 (NCU01315), an arginase-encoding gene (NCU02084), and a hydrolase-encoding gene (NCU01302) were also hypersensitive to ketoconazole (Fig. 3). In addition, 6 mutants with deletions of genes encoding hypothetical proteins with unknown functions, NCU09691, NCU09635, NCU08957, NCU00247, NCU00611, and NCU00248, were also hypersensitive to ketoconazole (Fig. 3). We named all unannotated genes as shown in Tables 2 and 3, based on the ketoconazole-hypersensitive phenotypes of the knockout mutants. For example, NCU06317 was named the ketoconazole-sensitive 1 (kts-1) gene, and NCU01386 was named the ketoconazole-sensitive 2 (kts-2) gene.
FIG 3.
Ketoconazole susceptibility analyses of N. crassa knockout mutants for genes regulated by CCG-8. Two microliters of conidial suspension (1 × 104 conidia/ml) was inoculated onto the center of each plate (Φ = 9 cm) with or without ketoconazole (2 μg/ml), and the plates were incubated at 28°C in the dark for about 66 h.
TABLE 3.
Significantly differently expressed genes between the wild type and the ccg-8 mutant in response to ketoconazole stressa
| Category and locusb | Gene | Function | TPM in WT | TPM in WT(k) | Fold change [WT(k)/WT] | P value | TPM in Δccg-8 | TPM in Δccg-8(k) | Fold change [Δccg-8(k)/Δccg-8] | P value | Fold change [Δccg-8(k)/WT(k)] | Sensitivity to ketoconazole compared to WT's |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upregulated genes | ||||||||||||
| NCU09691 | kts-3 | Hypothetical protein | 0.01 | 14.83 | 1483 | 8.88E−16 | 0.01 | 3.87 | 387 | 0.00012 | 0.26 | Hypersensitive |
| NCU02084 | Arginase | 13.82 | 1007.43 | 72.90 | 9.08E−14 | 67.92 | 304.5 | 4.48 | 2.78E−13 | 0.30 | Hypersensitive | |
| NCU09635 | kts-4 | Hypothetical protein | 3.17 | 216.04 | 68.15 | 7.57E−07 | 8.3 | 69.88 | 8.42 | 2.00E−11 | 0.32 | Hypersensitive |
| NCU01315 | gt41-1 | UDP-N-acetylglucosaminyltransferase | 2.59 | 38.9 | 15.02 | 2.57E−06 | 20.46 | 11.3 | 0.55 | 0.002807 | 0.29 | Hypersensitive |
| NCU08957 | kts-5 | Hypothetical protein | 5.47 | 69.4 | 12.69 | 4.61E−09 | 16.31 | 27.06 | 1.66 | 0.002712 | 0.39 | Hypersensitive |
| NCU01633 | hxt13 | Hexose transporter HXT13 | 377.04 | 4679.24 | 12.41 | 1.73E−12 | 622.5 | 1352.73 | 2.17 | 0 | 0.29 | Hypersensitive |
| NCU00247 | kts-6 | Hypothetical protein | 6.62 | 64.36 | 9.72 | 3.39E−10 | 7.71 | 13.98 | 1.81 | 0.013643 | 0.22 | Hypersensitive |
| NCU01302 | Hydrolase | 8.63 | 41.98 | 4.86 | 3.78E−12 | 17.2 | 9.81 | 0.57 | 0.009111 | 0.23 | Hypersensitive | |
| NCU00611 | kts-7 | Hypothetical protein | 8.06 | 30.78 | 3.82 | 1.54E−11 | 6.52 | 14.57 | 2.23 | 0.001244 | 0.47 | Hypersensitive |
| NCU00248 | kts-8 | Hypothetical protein | 59 | 216.04 | 3.66 | 3.92E−13 | 92.24 | 85.05 | 0.92 | 0.322136 | 0.39 | Hypersensitive |
| Downregulated genes | ||||||||||||
| NCU04334 | cpn10 | Chaperonin | 45.76 | 14.55 | 0.32 | 1.18E−14 | 18.09 | 31.22 | 1.73 | 0.000581 | 2.15 | Resistant |
| NCU04923 | gcy-1 | Glycerol dehydrogenase | 83.76 | 33.86 | 0.40 | 1.77E−18 | 462.4 | 163.85 | 0.35 | 1.7E−110 | 4.84 | Resistant |
WT, wild type; WT(k) or Δccg-8(k), wild type or Δccg-8 mutant treated with ketoconazole, respectively; TPM, number of locus tags per million; NA, the mutant was not available.
Locus numbers and function were annotated according to the N. crassa genome assembly (http://www.broadinstitute.org/annotation/genome/neurospora/).
The positive regulation of these genes by CCG-8 under ketoconazole stress was confirmed by qRT-PCR (Fig. 3). In response to ketoconazole, the expressions of these genes in both the wild type and the ccg-8 mutant were induced. However, their expression levels were lower in the ccg-8 mutant than in the wild type under the ketoconazole treatment condition. Since deletion of these genes results in increased sensitivity to ketoconazole, lower levels of expression of these genes may be the reason behind the hypersensitivity to antifungal azoles in the ccg-8 mutant.
Of the eight genes negatively regulated by CCG-8 (see Table S2 in the supplemental material), six currently have knockout mutants available. Two mutants with single genes (NCU04334 and NCU04923) knocked out were more resistant to ketoconazole than the wild type (Fig. 3). NCU04334 is a chaperonin-encoding gene, cpn10 (http://www.broadinstitute.org/annotation/genome/neurospora/GeneDetails.html?sp=S7000007580545759), and NCU04923 is a glycerol dehydrogenase-encoding gene, gcy-1 (http://www.broadinstitute.org/annotation/genome/neurospora/GeneDetails.html?sp=S7000007580553568). The negative regulation of NCU04923 (gcy-1) by CCG-8 under ketoconazole stress was confirmed by qRT-PCR (Fig. 2). Since their deletion is able to elevate resistance to ketoconazole, negative regulation of their expression by CCG-8 should make N. crassa resistant to azoles.
CCG-8 regulates phospholipid synthesis that is not involved in azole resistance.
The homolog of CCG-8 in S. cerevisiae is Opi1p (40), which plays a negative regulatory role in the expression of inositol-3-phosphate synthase upstream activation sequence (UASINO)-containing genes involved in phospholipid synthesis (41). In N. crassa, CCG-8 also regulates phospholipid synthesis because most UASINO-containing genes were upregulated in response to ccg-8 deletion. For example, in response to ccg-8 deletion, the inositol-3-phosphate synthase gene ino1 (NCU06666), phosphatidylserine decarboxylase proenzyme gene psd1 (NCU03695), and phosphatidylethanolamine N-methyltransferase gene cho2 (NCU08045) were transcriptionally increased by 4.6, 2.4, and 9.8 times, respectively (Table 2). However, the UASINO-containing genes showed no transcriptional response to ketoconazole stress in the wild type (Table 2). Furthermore, all available mutants for UASINO-containing genes displayed wild-type ketoconazole susceptibility (Table 2). These results indicated that phospholipid synthesis was not involved in azole adaption. The regulation of phospholipid synthesis and the regulation of azole responses are two independent functions of CCG-8.
Deletion of the ccg-8 homolog gene increases azole susceptibility in F. verticillioides.
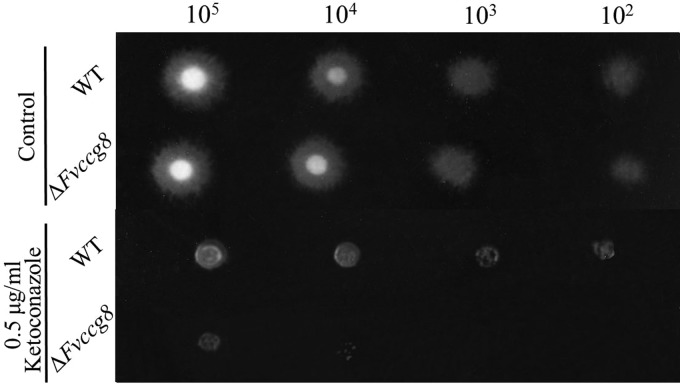
CCG-8 homologs are widely distributed in filamentous fungi (40). We chose the plant-pathogenic fungus F. verticillioides as a test species to investigate whether CCG-8 in other filamentous fungi also mediates azole sensitivity. F. verticillioides not only is a primary pathogen of maize but also can cause diseases in other crops. Moreover, ingestion of food contaminated by this fungus can cause esophageal cancer and neural tube defects in animal and human diseases, likely due to toxic metabolites produced by F. verticillioides, such as fumonisins, fusarins, and bikaverin (42). The ccg-8-homologous gene FVEG_06643 (Fvccg-8 in this study) was deleted in F. verticillioides. On the medium without the drug, the growth rate of the Fvccg-8 deletion mutant was similar to that of the wild-type strain (Fig. 4) and no conidiation defects were observed in the mutant (data not shown). When inoculated onto the medium with 0.5 μg/ml ketoconazole, the Fvccg-8 deletion mutant showed greater growth inhibition than the wild-type strain, which indicated that the CCG-8 homologs have similar azole resistance roles in different fungal species.
FIG 4.
Ketoconazole susceptibility analysis of the Fvccg-8 (FVEG_06643) knockout mutant. Two-microliter specimens of conidial suspensions with different concentrations (1 × 105, 1 × 104, 1 × 103, or 1 × 102 conidia/ml) were inoculated onto the plates (Φ = 9 cm), with or without ketoconazole, and incubated at 28°C for 72 h. Each test had three replicates, and the experiment was independently repeated twice.
DISCUSSION
Fungi are able to adjust the transcriptional levels of many genes when they adapt to azole stresses. In filamentous fungi, only transcription factor AP-1 was previously known as a regulator in azole adaption (24). In this study, we have shown that a second transcription factor, CCG-8, was also involved in the regulatory mechanism behind azole adaption in filamentous fungi by demonstrating that CCG-8 regulates transcriptional responses to ketoconazole in N. crassa and that CCG-8 is required for wild-type azole resistance in both N. crassa and F. verticillioides. Although CCG-8 in N. crassa has been identified as a clock-controlled gene (36) and its homolog Opi1p has previously been shown to be a negative regulator of UASINO-containing genes, which are involved in phospholipid synthesis (40, 41), the role of CCG-8 or its homologs in antifungal drug resistance has not previously been identified. In addition, this study has provided the first report of transcriptomic responses to antifungal azoles in the model fungus N. crassa and tested the ketoconazole sensitivities of 62 ketoconazole-responsive genes, which resulted in the identification of 15 new genes involved in azole adaption.
CCG-8 activates the transcriptional responses of the azole target gene erg11 and the azole pump gene cdr4 to ketoconazole stress in N. crassa. In C. albicans, the responses of the azole target gene ERG11 and the azole pump gene CDR1 (the cdr4 homolog) to azoles are regulated by transcription factor Upc2p, which can directly bind to the promoters of ERG11 and the azole pump gene CDR1 (21). Deletion of Upc2p resulted in hypersensitivity to antifungal azoles in C. albicans (18, 19). Although it is unknown whether a Upc2p homolog binds to the promoters of the azole target gene erg11 and azole pump gene cdr4, by screening knockout mutants, we found that a mutant with a deletion of the Upc2p-homologous gene tah-3 (NCU03686), FGSC 11076, was not hypersensitive to ketoconazole (data not shown). This suggests that the role of the Upc2p homolog in N. crassa is not as important as that of Upc2p in C. albicans. In addition, as mentioned in the introduction, filamentous fungi do not have homologs for the transcription factors Pdr1p and Pdr3p, which regulate the transcription of the azole pump gene in S. cerevisiae, PDR5 (15). These results suggested that filamentous fungi and yeasts may have different sets of transcription factors that regulate the responses to azole stress. However, since it is already known that transcription factor AP-1 is important for azole resistance in both yeasts and filamentous fungi (22–24), filamentous fungi and yeasts might share some common transcription factors in regulating azole adaptation. Homologs of CCG-8 are also found in C. albicans and other human-pathogenic Candida spp. (40). It is necessary to test the roles of CCG-8 homologs of Candida spp. in azole response and adaptation. CCG-8 activates the transcriptional responses of the azole target gene erg11 and the azole pump gene cdr4. Overexpression of either the cdr4 or erg11 homolog is able to confer azole resistance in many fungi. Thus, CCG-8 probably positively regulates resistance to azoles by transcriptional activation of cdr4 and erg11 during azole stress. This study has shown that 13 of the 70 genes positively regulated by CCG-8, in addition to erg11 and cdr4, made a significant contribution to azole resistance. Thus, CCG-8 may also positively regulate resistance to azoles by transcriptional activation of these genes during azole stress. Moreover, the reduced expression of two genes among the eight negatively regulated by CCG-8 was able to increase the organism's resistance to ketoconazole. Therefore, CCG-8 may elevate resistance to azoles by transcriptionally repressing these genes during azole stress. In addition to the genes that made significant contributions to azole resistance, many other CCG-8-regulated genes may contribute to azole resistance. For example, a multidrug resistance-associated protein 5-encoding gene, NCU04161 (http://www.broadinstitute.org/annotation/genome/neurospora/GeneDetails.html?sp=S7000007580550095), and three stress-responsive genes, NCU01499, pck-1 (NCU09873), and catalase-3 (NCU00355), were transcriptionally responsive to ketoconazole stress and were regulated by CCG-8 (Table 2). Increased expression of these genes may improve fungal survival under azole stress, although their roles may be detected by single-gene deletion. Not all genes differentially expressed between the ccg-8 mutant and the wild type were directly regulated by CCG-8. We showed that transcription factors FSD-1 and KTS-2 (NCU01386) were regulated by CCG-8 and were involved in azole resistance. In addition to these two transcription factors, four other transcription factors (NCU01629, NCU05536, NCU09496, and NCU04022) responded to ketoconazole stress, and their transcriptional responses to ketoconazole were regulated by CCG-8 (Table 2). Thus, many of the genes differentially expressed between the ccg-8 mutant and the wild type may be regulated by these transcription factors.
With the exceptions of those of erg11 and cdr4, links to azole resistance for the CCG-8-regulated genes identified in this study have not been previously reported. Among them, stress-responsive gene NCU06317 (kts-1), UDP-N-acetylglucosaminyltransferase-encoding gene gt41-1 (NCU01315), arginase-encoding gene NCU02084, and hydrolase-encoding gene NCU01302 may be generally required for adaptation to stresses because their transcriptional levels were also induced by oxidative stress (43). The ketoconazole-hypersensitive phenotype of the mutant with a deletion of the hexose transporter HXT13 gene hxt13 (NCU01633) suggested that this transporter may be involved in azole transportation or that the transportation of hexose might contribute to azole resistance under azole stress. Deletion of the glycerol dehydrogenase-encoding gene gcy-1 caused an increase in ketoconazole resistance. This suggested that glycerol metabolism may be involved in azole adaption. It has been reported that treatment by the fungicide fludioxonil, a derivative of the phenylpyrroles, which are structurally different from azoles, caused an accumulation of glycerol in N. crassa (44). Deletion of gcy-1 theoretically reduces the conversion from glycerol to glycerone, which probably results in the accumulation of glycerol in cells. However, the role of glycerol in azole resistance still needs to be investigated. For the rest of the genes identified in this study, more functional information is required to explain how they contribute to azole sensitivity. The discovery of their roles in azole resistance is important when it comes to identifying the genetic causes of drug resistance in clinical or agricultural isolates of fungal pathogens.
In the future, the consensus DNA motifs bound by CCG-8 need to be identified, as does whether CCG-8 homologs in yeasts also regulate transcriptional responses to azoles. In addition, since CCG-8 is a clock-controlled protein (36), the relationship between the circadian clock and azole resistance needs to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
This project is supported by grants 31000047 (to Hanxing Zhang), 31000551, and 31370024 (to Xianyun Sun) from the National Natural Science Foundation of China and a 100 Talent Program grant from the Chinese Academy of Sciences (to Shaojie Li).
Footnotes
Published ahead of print 16 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02244-13.
REFERENCES
- 1.Yu L, Zhang W, Wang L, Yang J, Liu T, Peng J, Leng W, Chen L, Li R, Jin Q. 2007. Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum. Antimicrob. Agents Chemother. 51:144–153. 10.1128/AAC.00755-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Jiang J, Shao J, Yin Y, Ma Z. 2010. Gene transcription profiling of Fusarium graminearum treated with an azole fungicide, tebuconazole. Appl. Microbiol. Biotechnol. 85:1105–1114. 10.1007/s00253-009-2273-4 [DOI] [PubMed] [Google Scholar]
- 3.da Silva Ferreira ME, Malavazi I, Savoldi M, Brakhage AA, Goldman MH, Kim HS, Nierman WC, Goldman GH. 2006. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50:32–44. 10.1007/s00294-006-0073-2 [DOI] [PubMed] [Google Scholar]
- 4.Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998–35015. 10.1074/jbc.M306291200 [DOI] [PubMed] [Google Scholar]
- 5.Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer DJ, Ward DJ, Marsden K, Bennett JE. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T, Lee R, Barker K, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226–2236. 10.1128/AAC.49.6.2226-2236.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher R, Weihmann F, Deising H, Wirsel S. 2011. Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics 12:52. 10.1186/1471-2164-12-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion JP, Konings WN, Goffeau A. 1996. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271:31543–31548. 10.1074/jbc.271.49.31543 [DOI] [PubMed] [Google Scholar]
- 9.Niimi M, Niimi K, Takano Y, Holmes AR, Fischer FJ, Uehara Y, Cannon RD. 2004. Regulated overexpression of CDR1 in Candida albicans confers multidrug resistance. J. Antimicrob. Chemother. 54:999–1006. 10.1093/jac/dkh456 [DOI] [PubMed] [Google Scholar]
- 10.Schuetzer-Muehlbauer M, Willinger B, Egner R, Ecker G, Kuchler K. 2003. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int. J. Antimicrob. Agents 22:291–300. 10.1016/S0924-8579(03)00213-9 [DOI] [PubMed] [Google Scholar]
- 11.Hamamoto H, Hasegawa K, Nakaune R, Lee YJ, Makizumi Y, Akutsu K, Hibi T. 2000. Tandem repeat of a transcriptional enhancer upstream of the sterol 14α-demethylase gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 66:3421–3426. 10.1128/AEM.66.8.3421-3426.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnabel G, Jones AL. 2001. The 14α-demethylase (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistant to myclobutanil. Phytopathology 91:102–110. 10.1094/PHYTO.2001.91.1.102 [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Proffer TJ, Jacobs JL, Sundin GW. 2006. Overexpression of the 14α-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 72:2581–2585. 10.1128/AEM.72.4.2581-2585.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du W, Coaker M, Sobel J, Akins R. 2004. Shuttle vectors for Candida albicans: control of plasmid copy number and elevated expression of cloned genes. Curr. Genet. 45:390–398. 10.1007/s00294-004-0499-3 [DOI] [PubMed] [Google Scholar]
- 15.Gulshan K, Moye-Rowley WS. 2007. Multidrug resistance in fungi. Eukaryot. Cell 6:1933–1942. 10.1128/EC.00254-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talibi D, Raymond M. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vik Å, Rine J. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395–6405. 10.1128/MCB.21.19.6395-6405.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silver PM, Oliver BG, White TC. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391–1397. 10.1128/EC.3.6.1391-1397.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacPherson S, Akache B, Weber S, de Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745–1752. 10.1128/AAC.49.5.1745-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhäuser J, Rogers PD. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180–1190. 10.1128/EC.00103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, de Deken X, Robert F, Raymond M. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836–847. 10.1128/EC.00070-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarco AM, Balan I, Talibi D, Mainville N, Raymond M. 1997. AP-1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304–19313. 10.1074/jbc.272.31.19304 [DOI] [PubMed] [Google Scholar]
- 23.Chen KH, Miyazaki T, Tsai HF, Bennett JE. 2007. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386:63–72. 10.1016/j.gene.2006.08.010 [DOI] [PubMed] [Google Scholar]
- 24.Qiao J, Liu W, Li R. 2010. Truncated Afyap1 attenuates antifungal susceptibility of Aspergillus fumigatus to voriconazole and confers adaptation of the fungus to oxidative stress. Mycopathologia 170:155–160. 10.1007/s11046-010-9309-2 [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Zhang Z, Zhang X, Zhang H, Sun X, Hu C, Li S. 2012. CDR4 is the major contributor to azole resistance among four Pdr5p-like ABC transporters in Neurospora crassa. Fungal Biol. 116:848–854. 10.1016/j.funbio.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 26.Sun X, Wang W, Wang K, Yu X, Liu J, Zhou F, Xie B, Li S. 2013. Sterol C-22 desaturase ERG5 mediates the sensitivity to antifungal azoles in Neurospora crassa and Fusarium verticillioides. Front. Microbiol. 4:127. 10.3389/fmicb.2013.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel HJ. 1956. A convenient growth medium for Neurospora (medium N). Microbiol. Genet. Bull. 13:42–43 [Google Scholar]
- 28.Sweigard J, Chumley FG, Carroll A, Farrall L, Valent B. 1997. A series of vectors for fungal transformation. Fungal Genet. Newsl. 44:52–53 [Google Scholar]
- 29.Royer JC, Yamashiro CT. 1992. Generation of transformable spheroplasts from mycelia, macroconidia, microconidia and germinating ascospores of Neurospora crassa. Fungal Genet. Newsl. 39:76–79 [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 31.Nielsen K, Høgh AL, Emmersen J. 2006. DeepSAGE-digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 34:e133. 10.1093/nar/gkl714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X, Zhang H, Zhang Z, Wang Y, Li S. 2011. Involvement of a helix-loop-helix transcription factor CHC-1 in CO2-mediated conidiation suppression in Neurospora crassa. Fungal Genet. Biol. 48:1077–1086. 10.1016/j.fgb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 33.Staben C, Jensen B, Singer M, Pollock J, Schechtman M, Kinsey J, Selker E. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36:79–81 [Google Scholar]
- 34.Miller B, Miller K, Timberlake W. 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5:1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Myung K, Guse D, Donkin B, Proctor RH, Grayburn WS, Calvo AM. 2006. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol. Microbiol. 62:1418–1432. 10.1111/j.1365-2958.2006.05447.x [DOI] [PubMed] [Google Scholar]
- 36.Bell-Pedersen D, Shinohara ML, Loros JJ, Dunlap JC. 1996. Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific. Proc. Natl. Acad. Sci. U. S. A. 93:13096–13101. 10.1073/pnas.93.23.13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutchison EA, Glass NL. 2010. Meiotic regulators Ndt80 and ime2 have different roles in Saccharomyces and Neurospora. Genetics 185:1271–1282. 10.1534/genetics.110.117184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu S, Herskowitz I. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685–696. 10.1016/S1097-2765(00)80068-4 [DOI] [PubMed] [Google Scholar]
- 39.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705. 10.1126/science.282.5389.699 [DOI] [PubMed] [Google Scholar]
- 40.Lombardi LM, Brody S. 2005. Circadian rhythms in Neurospora crassa: clock gene homologues in fungi. Fungal Genet. Biol. 42:887–892. 10.1016/j.fgb.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 41.White MJ, Hirsch JP, Henry SA. 1991. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem. 266:863–872 [PubMed] [Google Scholar]
- 42.Nelson PE, Desjardins AE, Plattner RD. 1993. Fumonisins, mycotoxins produced by Fusarium species: biology, chemistry and significance. Annu. Rev. Phytopathol. 31:233–252. 10.1146/annurev.py.31.090193.001313 [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Yu X, Xie B, Gu X, Zhang Z, Li S. 2013. Transcriptomic profiling-based mutant screen reveals three new transcription factors mediating menadione resistance in Neurospora crassa. Fungal Biol. 117:422–430. 10.1016/j.funbio.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 44.Fujimura M, Ochiai N, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. 2000. Fungicide resistance and osmotic stress sensitivity in os mutants of Neurospora crassa. Pestic. Biochem. Physiol. 67:125–133. 10.1006/pest.2000.2479 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.