Abstract
Full genome sequences were determined for five Klebsiella pneumoniae strains belonging to the sequence type 512 (ST512) clone, producing KPC-3. Three strains were resistant to tigecycline, one showed an intermediate phenotype, and one was susceptible. Comparative analysis performed using the genome of the susceptible strain as a reference sequence identified genetic differences possibly associated with resistance to tigecycline. Results demonstrated that mutations in the ramR gene occurred in two of the three sequenced strains. Mutations in RamR were previously demonstrated to cause overexpression of the AcrAB-TolC efflux system and were implicated in tigecycline resistance in K. pneumoniae. The third strain showed a mutation located at the vertex of a very well conserved loop in the S10 ribosomal protein, which is located in close proximity to the tigecycline target site in the 30S ribosomal subunit. This mutation was previously shown to be associated with tetracycline resistance in Neisseria gonorrhoeae. A PCR-based approach was devised to amplify the potential resistance mechanisms identified by genomics and applied to two additional ST512 strains showing resistance to tigecycline, allowing us to identify mutations in the ramR gene.
INTRODUCTION
Carbapenem-resistant Klebsiella pneumoniae is an important nosocomial pathogen with limited treatment options. Antibiogram-based therapy of infections sustained by extremely resistant K. pneumoniae often combines drugs like colistin sulfate or polymyxin B with doripenem, ertapenem plus doripenem or meropenem, fosfomycin, or tigecycline (1). Clinical efficacy of tigecycline, the first-in-class glycylcycline, has been demonstrated in complicated skin and skin structure infections and intraabdominal infections, but it is also used (off-label) in combination with other drugs for the treatment of serious infections caused by carbapenem-resistant Enterobacteriaceae and Acinetobacter spp. (2). Tigecycline enters bacterial cells through passive diffusion or active transportation. Similarly to tetracyclines, tigecycline reversibly binds to the ribosome 30S subunit, interfering with accommodation of aminoacyl-tRNA in the ribosomal A site and blocks bacterial growth by inhibition of translation (3, 4). Structural data of the tigecycline/ribosome complex have been recently obtained for the Thermus thermophilus ribosome 30S subunit (5).
Recently reported studies revealed the emergence of tigecycline resistance in carbapenem-resistant K. pneumoniae (6–8). Since 2008, K. pneumoniae strains producing KPC carbapenemases have been detected in many different Italian hospitals. In particular, the clones designated sequence type 258 (ST258) and ST512 are the most frequently reported K. pneumoniae clones in this country, both belonging to the clonal group CC258 (6–18). In the period from 2010 to 2011, two surveillance studies were performed in 10 hospitals in Rome, collecting >100 KPC-3-producing K. pneumoniae strains belonging to CC258; several strains showed reduced susceptibility or resistance to tigecycline (6).
In laboratory-derived tigecycline-resistant K. pneumoniae strains, the increased expression of the AcrAB resistance-nodulation-division efflux system was described as one of the possible mechanisms of tigecycline resistance, causing the reduction of the intracellular concentration of tigecycline (19, 20). AcrAB is present in most Enterobacteriaceae, utilizes the outer membrane protein TolC, and reduces the susceptibility to fluoroquinolones, macrolides, chloramphenicol, trimethoprim, and tetracyclines (21). Besides K. pneumoniae, AcrAB-TolC has been implicated in reduced susceptibility to tigecycline in clinical isolates of Escherichia coli, Morganella morganii, Proteus mirabilis, and Enterobacter cloacae (22–24).
The transcription of the acrAB genes is controlled by several transcriptional activators of the AraC/XylS family, RamA, MarA, SoxS, and RobA, which interact with the acrAB promoter, increasing the production of AcrAB proteins and enhancing efflux (19, 22, 25). The transcriptional activators of the acrAB genes are themselves controlled by other trans-acting factors. RamA expression is controlled by the negative regulator RamR; the depletion of RamR has been implicated in the activation of RamA, causing the overexpression of the AcrAB efflux pump (19, 22, 25–29).
Recent studies demonstrated that tigecycline resistance can occur independently of AcrAB system upregulation (29, 30), suggesting the presence of different mechanisms of resistance. The recent availability of whole-genome sequencing permits the discovery of genetic differences between pairs of genetically related susceptible and resistant bacterial strains. In this study, we applied the genome sequencing approach to three tigecycline-resistant, one intermediate-resistant, and one susceptible K. pneumoniae strain belonging to the ST512 clone and producing KPC-3, with the aim of identifying genetic differences possibly associated with reduced susceptibility and resistance to tigecycline.
MATERIALS AND METHODS
Bacterial isolates.
Eight carbapenem-resistant, colistin-susceptible K. pneumoniae strains were selected from among those previously isolated and characterized in 2010 to 2011 (Table 1) (6). All strains showed identical pulsed-field gel electrophoresis (PFGE) patterns and were assigned to ST512 by multilocus sequence typing (MLST), carried the blaKPC-3 gene, and were positive for FIIK1 and FIIK2 replicons, as expected because of the presence of the pKpQIL-IT and pKPN-IT plasmids (6, 12). Among the selected strains, seven showed reduced susceptibility or resistance to tigecycline (KP1-I, KP2-R, KP4-R, KP5-R, KP6-R, KP7-R, and KP8-I) and one was susceptible to tigecycline (KP3-S) (Table 1). KP1-I and KP2-R were isolated in 2010 from the same patient under tigecycline therapy. KP4-R, KP5-R, KP6-R, KP7-R, and KP8-I were isolated in 2011 in the same hospital from different patients from two surgery wards. The KP3-S susceptible strain was isolated from the blood of a patient hospitalized in the intensive care unit. Strains KP1-I, KP2-R, KP3-S, KP4-R, and KP5-R were selected for complete genome sequencing.
TABLE 1.
Klebsiella pneumoniae ST512 strains analyzed in this study
| Strain | Yr | Patient | MIC (μg/ml) of: |
||||
|---|---|---|---|---|---|---|---|
| Tigecycline | Ertapenem | Imipenem | Meropenem | Colistin | |||
| KP1-I | 2010 | A | 2 | >32 | 8 | >32 | <0.5 |
| KP2-R | 2010 | A | 4 | >32 | >32 | >32 | <0.5 |
| KP3-S | 2011 | B | 1 | >32 | 24 | >32 | <0.5 |
| KP4-R | 2011 | C | 4 | >32 | >32 | >32 | <0.5 |
| KP5-R | 2011 | D | 4 | >32 | >32 | >32 | <0.5 |
| KP6-R | 2011 | E | 4 | >32 | >32 | >32 | <0.5 |
| KP7-R | 2011 | F | 4 | >32 | >32 | >32 | <0.5 |
| KP8-I | 2011 | G | 2 | >32 | 8 | >32 | <0.5 |
Tigecycline, ertapenem, meropenem, and imipenem MICs were determined for all the strains by Etest (bioMérieux, Italy), and colistin breakpoints were determined by using the Vitek 2 system (bioMérieux, Italy), following the manufacturer's procedures and interpreting the results as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 3.1 guidelines (breakpoint tables for interpretation of MICs and zone diameters [http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.pdf]).
Whole-genome DNA sequencing.
The complete DNA sequences of the five genomes were obtained by applying the 454-Genome Sequencer (454-GS) FLX procedure (see http://454.com/applications/whole-genome-sequencing/) to libraries constructed on total DNA purified with the NucleoSpin tissue kit (Macherey-Nagel, Inc., Bethlehem, PA), according to the manufacturer's procedure, producing >600-bp reads.
Genome analysis.
Draft genomes were assembled by using the GS De Novo Assembler v.2.6 software (Roche Diagnostics S.p.A., Monza, Italy), producing an average of 130 contigs per each genome, ranging from 543,722 bp to 101 bp, with at least 50-fold coverage. Each genome sequence was analyzed by BLASTN versus the complete genome of the K. pneumoniae strain HS11286 (GenBank accession no. CP003200) and against the other four fully sequenced genomes determined in this study (31, 32).
To detect single nucleotide polymorphisms (SNPs) occurring in specific loci of each genome under study, all of the 454-GS reads obtained for isolates KP1-I, KP2-R, KP4-R, and KP5-R were mapped against all contigs generated from the genome of the susceptible KP3-S, using the GS Reference Mapper v.2.6 software (Roche Diagnostics S.p.A., Monza, Italy). Each SNP identified in each specific locus was considered valid if confirmed in at least 80% of the reads mapped in that locus. Each SNP was classified using the ARTEMIS software (downloaded at the Sanger Institute, United Kingdom) as an intergenic SNP, silent SNP, or SNP changing a deduced coding sequence (CDS). Silent SNPs were not further analyzed. Intergenic SNPs and SNPs changing CDSs were further studied by manual inspection of the locus. In particular, the following genes, encoding porins and previously described efflux pumps and their respective promoters, were manually checked for SNPs, insertions, and deletions potentially implied in modification of the expression or alteration or depletion of the deduced protein sequences: ompK35, ompK36, rnfC, ramA, ramR, marA, marR, marC, soxR, yceE, norA, yceL, emrD, tolC, acrA, acrB, macA, macB, oqxA, and oqxB (33).
The 16S rRNA genes were amplified from the 8 copies of the rRNA gene cluster by PCR performed using the same 16S KPN RV primer in combination with 8 different forward primers, devised based on the first gene flanking each of the rRNA clusters (see Table S1 in the supplemental material).
The ResFinder software (see http://cge.cbs.dtu.dk/services/ResFinder) was used to identify acquired antimicrobial resistance genes in the sequenced genomes (34).
Plasmids were identified by BLASTN analysis performed against the complete nucleotide sequences of pKpQIL-IT (GenBank accession no. JN233705), pKPN-IT (JN233704), ColEST258 (JN247853), and IncX-SHV ST258 (JN247852) plasmids (12).
Protein comparative analysis.
Five pools of deduced CDSs with a size of >100 amino acid residues were obtained by using the ARTEMIS software provided by the Sanger Institute, using all contigs generated for each genome. The pool of CDSs obtained for the susceptible KP3-S genome was compared against the other four pools by the BLASTP program.
Draft genomes were submitted to the Prophage Finder website (http://bioinformatics.uwp.edu/∼phage/ProphageFinder.php) for detection of phage-related proteins (35).
Screening of tigecycline resistance candidate genes.
Mutations identified by genomics in the ramR and rpsJ genes were confirmed by PCR and DNA sequencing of the amplicons (using primers and protocols described in Table S1 in the supplemental material) of total genomic DNAs extracted with the NucleoSpin Tissue kit (Macherey-Nagel, Inc., Bethlehem, PA) and used as the templates.
Nucleotide sequence accession numbers.
The nucleotide sequences of genes reported in this work have been deposited in the GenBank nucleotide database under the following accession numbers: KC843628 (KP3-S barA), KC843629 (KP5-R barA), KC843631 (KP3-S HTH_AraC regulator), KC843630 (KP2-R HTH_AraC regulator), KC843632 (KP3-S yeaM-araC), KC843633 (KP5-R yeaM-araC), KC843636 (KP3-S rpsJ), KC686844 (KP4-R rpsJ), KC843634 (KP3-S ramAR operon), KC686846 (KP2-R ramAR operon-ISKpn18), KC686847 (KP5-R ramAR operon), KC843635 (KP3-S rcsC), and KC686845 (KP2-R rcsC).
The whole-genome shotgun project results have been deposited with DDBJ/EMBL/GenBank under the accession numbers AQCJ00000000 (KP1-S), AQCI00000000 (KP2-R), AOSJ00000000 (KP3-S), APMF00000000 (KP4-R), and APMG00000000 (KP5-R). The versions described in this paper are the first versions: AQCJ01000000, AQCI01000000, AOSJ01000000, APMF01000000, and APMG01000000, respectively.
RESULTS AND DISCUSSION
Common features of the Klebsiella pneumoniae ST512 genomes.
The five fully sequenced K. pneumoniae ST512 genomes were >99% identical at the nucleotide level, since no large deletions, inversions, or substitution events were observed by sequence alignments; they also showed >99% nucleotide identity with the ST11 K. pneumoniae strain HS11286, belonging to CC258 (GenBank accession no. CP003200) (31, 32). Previously described antimicrobial resistance genes, such as the plasmid-located blaKPC-3, the chromosomally located blaSHV-11, and the oqxA and oqxB genes, were identified in all five strains (12, 33). All strains also showed blaTEM-1, mph(A), sul1, dfrA12, aac(6′)-Ib, aadA2, aph(3′)-Ia, and catA1 genes and the operons encoding mercury, silver, and copper resistance. Defects in the outer membrane proteins OmpK35 and OmpK36 were identified, as previously described for an ST258 strain isolated in Rome in 2010 (12). Several allele variants were identified in each of the eight copies of the 16S rRNA genes. However, these variants were perfectly conserved in all five of the strains (data not shown).
The features identically conserved with respect to the sequence of the KP3-S susceptible strain were not further analyzed, as they were not implicated in tigecycline resistance.
Few genetic differences were identified in the tigecycline-intermediate KP1-I strain compared to the genome of the KP3-S-susceptible strain (see Table S2 in the supplemental material). The observed differences occurred in genes that were not previously implicated in tigecycline or tetracycline resistance mechanisms, and the demonstration that they are associated with the intermediate tigecycline phenotype deserves further investigation.
Amino acid substitutions identified in genes encoding adhesion proteins (the FimH protein, an outer membrane protein involved in fimbrial biogenesis and a pilin protein precursor), metabolic enzymes (protoheme, fumarate lyase, and quinone oxidoreductase) or hypothetical and phage-related proteins were not further characterized, since they were not considered good candidates as tigecycline resistance mechanisms (see Table S2 in the supplemental material).
The mutant RamR regulator of the AcrAB-TolC efflux system.
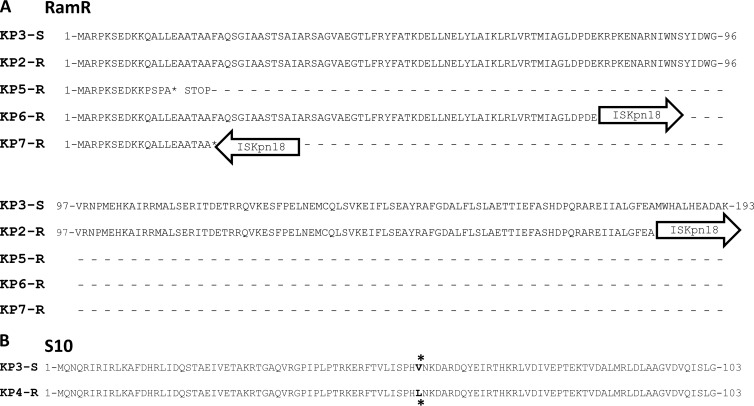
The genomic analysis contributed to clarification of the role of efflux in tigecycline resistance, since two resistant isolates (KP2-R and KP5-R) had different mutations in the ramR gene. In KP2-R, the insertion of the ISKpn18 element occurred at the 3′ end of the gene (duplication of the target site was identified at the boundaries of the integrated element; the ISKpn18 sequence was deposited with the IS Finder database (https://www-is.biotoul.fr), whereas in the KP5-R strain, a 13-bp deletion at the 5′ end caused a frameshift, creating a premature stop codon for RamR protein translation (Table 2). In detail, in KP2-R the last 12 amino acids of the predicted RamR reading frame were replaced by 24 random codons generated upon insertion of the ISKpn18 element, while in KP5-R a stop codon was generated after the first 20 amino acids of the predicted protein (Fig. 1A). Since RamR defects have been demonstrated to increase acrAB expression via RamA activation, the mutations observed in these two strains may account for their tigecycline resistance phenotype (19, 22, 25–29).
TABLE 2.
Mutations in efflux, regulatory systems, and the S10 ribosomal 30S protein possibly involved in tigecycline-intermediate and -resistant phenotypes identified by full-genome sequencing
| Protein product | Function | Mutation in strain |
Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| KP3-S (accession no.) | KP1-I (accession no.) | KP2-R (accession no.) | KP4-R (accession no.) | KP5-R (accession no.) | |||
| RamR | Regulator of RamA, positive regulator of the AcrAB efflux system | Functional (KC843634) | Functional | RamR mutation by ISKpn18 insertion (KC686846) | Functional | Premature termination of translation (KC686847) | 26 |
| RcsC | Hybrid sensory kinase in two-component regulatory system with RcsB and YojN, regulates capsule biosynthesis, cell division genes | Thr474 (KC843635) | Thr474 | Thr474→Ser474 (KC686845) | Thr474 | Thr474 | 35 |
| HTH-AraC | Bacterial regulatory helix-turn-helix protein, arabinose operon control protein | Conserved (KC843631) | Conserved | −33 promoter mutation (KC843630) | Conserved | Conserved | 36, 37 |
| BarA | Hybrid sensory histidine kinase, regulation of carbon metabolism | Val311 (KC843628) | Val311 | Val311 | Val311 | Val311→Gly311 (KC843629) | 38, 39 |
| YeaM | Putative AraC-type regulatory protein | Conserved (KC843632) | Conserved | Conserved | Conserved | Premature termination of translation (KC843633) | 40 |
| S10 | Architectural element in the 30S ribosomal subunit | Val57 (KC843636) | Val57 | Val57 | Val57→Leu57 (KC686844) | Val57 | 41 |
FIG 1.
Mutations identified in proteins potentially implicated in tigecycline resistance. (A) The entire RamR deduced protein sequence of the susceptible KP3-S strain is shown in comparison with the RamR mutation identified in KP5-R (by premature termination of translation) and KP2-R, KP6-R, and KP7-R (by insertion of the ISKpn18 element, represented by arrows). (B) The entire S10 deduced protein sequence of the susceptible KP3-S strain is shown in comparison with the mutated S10 identified in the KP4-R strain. The positions of the Val57→Leu57 mutation are indicated by asterisks.
However, KP2-R and KP5-R strains also showed mutations in other regulatory proteins. Strain KP2-R showed the Thr474→Ser474 mutation in the phosphorylation site of the hybrid sensory kinase RcsC and a mutation at the −33 position in the promoter of HTH-AraC, an arabinose operon regulatory helix-turn-helix protein (36–38). The mutation observed in RcsC occurred in a very well conserved domain of the protein, corresponding to the phosphorylation site. RcsC is the hybrid sensory kinase of a two-component regulatory system that regulates capsule biosynthesis, cell division genes, and osmC expression with RcsB and YojN (36). Strain KP5-R showed the Val311→Gly311 mutation in the hybrid sensory kinase BarA, which is involved in the regulation of carbon metabolism via the csrA/csrB regulatory system (39, 40), and a frameshift mutation causing the premature termination of translation of the YeaM protein, a putative AraC-type regulatory protein (41) (Table 2). In summary, both KP2-R and KP5-R genomes showed mutated ramR genes in association with mutations in the genes of two different hybrid sensory kinases and in two controls of the arabinose operon. The role of these latter systems in tigecycline resistance is not clear, and it deserves further investigation.
The ribosomal S10 protein.
The KP4-R tigecycline-resistant strain did not show any differences in efflux or other regulatory systems compared to KP3-S. However, an interesting mutation was observed in this strain that is probably involved in conferring tigecycline resistance. In fact, a mutation at codon 57, changing from GTC to CTC and leading to the Val57→Leu57 amino acid substitution, was identified in the rpsJ gene encoding the S10-30S ribosomal protein (Fig. 1B; Table 2).
The small S10 protein is a component of the 30S ribosomal subunit and, together with the NusB protein, participates in the formation of a BoxA binding module (42). The rpsJ gene encoding this protein is present in a single copy in the genome of K. pneumoniae and is conserved in all Enterobacteriaceae. An in silico analysis performed on 1,277 S10 protein sequences identified in GenBank from strains of several species and genera showed 98 to 100% amino acid identities with the sequences identified in the KP3-S, KP-1-I, KP2-R, and KP-5-R genomes, but no mutations were observed in the Val57 codon. An identical Val57→Leu57 mutation was identified in silico only in the genome of the K. pneumoniae KPNIH20 strain from the United States, which was reported to be tigecycline resistant, but the mechanism of resistance in that strain was not elucidated (43).
The X-ray crystallographic structure of the tigecycline antibiotic bound to the 70S ribosome of Thermus thermophilus demonstrated that the enhanced potency of tigecycline with respect to tetracycline resulted from a stacking interaction of tigecycline with the 16S rRNA and that this antibiotic coordinates two Mg2+ ions (Molecular Modeling Database [MMDB] identification [ID] no. 107830; Protein Data Bank [PDB] ID nos. 4G5T, 4G5U, 4G5V, and 4G5W [http://www.ncbi.nlm.nih.gov/structure]) (5). The Lys55 residue in T. thermophilus S10 is the equivalent of the Val57 in K. pneumoniae S10 and maps in the vertex of a loop that is located 8 Å from the tigecycline binding site in the 30S ribosomal subunit (see Fig. S1 in the supplemental material). S10 interacts with the 16S rRNA, the ribosomal proteins S14, S3, and S9, and Mg2+ ions. In the T. thermophilus model there are no peptides that are closer to the bound tigecycline than the S10 loop. Since the rpsJ gene is a single-copy gene in the chromosome of K. pneumoniae, the mutation causing the Val57→Leu57 substitution in S10 occurs in all the ribosomes of the KP4-R strain. This mutation may alter the ribosome structure near the tigecycline-binding site or disturb the coordination of the Mg2+ ion, leading to weaker binding of tigecycline to the 16S rRNA. To the best of our knowledge, such a target site-associated resistance mechanism has never been described for tigecycline, while it has been demonstrated for tetracycline in Neisseria gonorrhoeae (44). High levels of tetracycline resistance were described in that species due to a point mutation in the Val57 codon of the S10 protein, in combination with the mtrR and penB resistance determinants. In N. gonorrhoeae, site saturation mutagenesis of the codon Val57 identified three amino acids (Met, Leu, and Gln) that conferred identical levels of tetracycline resistance. This hypothesis opens the possibility that tetracycline and tigecycline can both select for this resistance mechanism.
Screening of tigecycline resistance candidate genes.
The rpsJ and ramR tigecycline resistance candidate genes were amplified and sequenced from three ST512 strains showing tigecycline-intermediate (KP8-I) or -resistant (KP6-R and KP7-R) phenotypes (Table 1). The two resistant strains showed the mutant RamR protein created by the ISKpn18 element insertion. Interestingly, in these strains the integration of the insertion sequence (IS) element occurred in different sites compared to KP2-R, producing truncated RamR deduced protein variants of different lengths (Fig. 1A). The strain KP8-I, which showed reduced susceptibility to tigecycline, carried a conserved ramR. These results enforced the idea that independent genetic events occurred in the resistant strains of our collection and demonstrated that no tigecycline-resistant clones were spread among different patients in the same hospital. Our data suggest that the inactivation of the ramR gene is the most common mechanism conferring resistance to tigecycline. These three strains showed a conserved rpsJ gene.
Conclusions.
Different events of mutation or insertion of mobile elements occurred in the ramR gene in four out of five tigecycline-resistant strains analyzed in this study. However, the structural alteration of the ribosomal protein S10 in the drug target site, which occurred in the non-RamR mutant tigecycline-resistant strain, is also a potential novel mechanism.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Antibacterial EUROPE ASPIRE 2012 Research Awards—a competitive grants program supported by Pfizer International Operations and by the Italian FLAGSHIP InterOmics project (PB.P05) funded by the MIUR and coordinated by the CNR.
A.C. is grateful to Daniela Meco and Andrea Di Mascio for their critical reading of the manuscript.
Footnotes
Published ahead of print 30 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01803-13.
REFERENCES
- 1.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Duin D, Kaye KS, Neuner EA, Bonomo RA. 2013. Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. Diagn. Microbiol. Infect. Dis. 75:115–120. 10.1016/j.diagmicrobio.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson MW, Ruzin A, Feyfant E, Rush TS, III, O'Connell J, Bradford PA. 2006. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob. Agents Chemother. 50:2156–2166. 10.1128/AAC.01499-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pankey GA. 2005. Tigecycline. J. Antimicrob. Chemother. 56:470–480. 10.1093/jac/dki248 [DOI] [PubMed] [Google Scholar]
- 5.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 110:3812–3816. 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capone A, Giannella M, Fortini D, Giordano A, Meledandri M, Ballardini M, Venditti M, Bordi E, Capozzi D, Balice MP, Tarasi A, Parisi G, Lappa A, Carattoli A, Petrosillo N, SEERBIO-GRAB Network 2013. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin. Microbiol Infect. 19:E23–E30. 10.1111/1469-0691.12070 [DOI] [PubMed] [Google Scholar]
- 7.Spanu T, De Angelis G, Cipriani M, Pedruzzi B, D'Inzeo T, Cataldo MA, Sganga G, Tacconelli E. 2012. In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 56:4516–4518. 10.1128/AAC.00234-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R; Survey Participants AMCLI-CRE, Pantosti A, Pagani L, Luzzaro F, Rossolini GM 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 18:pii:20489 [PubMed] [Google Scholar]
- 9.Agodi A, Voulgari E, Barchitta M, Politi L, Koumaki V, Spanakis N, Giaquinta L, Valenti G, Romeo MA, Tsakris A. 2011. Containment of an outbreak of KPC-3-producing Klebsiella pneumoniae in Italy. J. Clin. Microbiol. 49:3986–3989. 10.1128/JCM.01242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Carlo P, Pantuso G, Cusimano A, D'Arpa F, Giammanco A, Gulotta G, Latteri AM, Madonia S, Salamone G, Mammina C. 2011. Two cases of monomicrobial intraabdominal abscesses due to KPC-3 Klebsiella pneumoniae ST258 clone. BMC Gastroenterol. 11:103. 10.1186/1471-230X-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontana C, Favaro M, Sarmati L, Natoli S, Altieri A, Bossa MC, Minelli S, Leonardis F, Favalli C. 2010. Emergence of KPC-producing Klebsiella pneumoniae in Italy. BMC Res. Notes 3:40. 10.1186/1756-0500-3-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145. 10.1128/AAC.05308-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giani T, D'Andrea MM, Pecile P, Borgianni L, Nicoletti P, Tonelli F, Bartoloni A, Rossolini GM. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47:3793–3794. 10.1128/JCM.01773-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mammina C, Palma DM, Bonura C, Anna Plano MR, Monastero R, Sodano C, Calà C, Tetamo R. 2010. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae carbapenemase 3 in an intensive care unit in Italy. J. Clin. Microbiol. 48:1506–1507. 10.1128/JCM.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchese A, Coppo E, Barbieri R, Debbia E. 2010. Emergence of KPC-2 carbapenemase-producing Klebsiella pneumoniae strains and spread of an isolate of sequence type 258 in the neuro-rehabilitation unit of an Italian hospital. J. Chemother. 22:212–214. 10.1179/joc.2010.22.3.212 [DOI] [PubMed] [Google Scholar]
- 16.Mezzatesta ML, Gona F, Caio C, Petrolito V, Sciortino D, Sciacca A, Santangelo C, Stefani S. 2011. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin. Microbiol. Infect. 17:1444–1447. 10.1111/j.1469-0691.2011.03572.x [DOI] [PubMed] [Google Scholar]
- 17.Orsi GB, García-Fernández A, Giordano A, Venditti C, Bencardino A, Gianfreda R, Falcone M, Carattoli A, Venditti M. 2011. Risk factors and clinical significance of ertapenem-resistant Klebsiella pneumoniae in hospitalised patients. J. Hosp. Infect. 78:54–58. 10.1016/j.jhin.2011.01.014 [DOI] [PubMed] [Google Scholar]
- 18.Richter SN, Frasson I, Franchin E, Bergo C, Lavezzo E, Barzon L, Cavallaro A, Palù G. 2012. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog. 4:7. 10.1186/1757-4749-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruzin A, Immermann FW, Bradford PA. 2008. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3430–3432. 10.1128/AAC.00591-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruzin A, Visalli MA, Keeney D, Bradford PA. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017–1022. 10.1128/AAC.49.3.1017-1022.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido H. 1996. Multidrug efflux pumps of Gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeney D, Ruzin A, Bradford PA. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13:1–6. 10.1089/mdr.2006.9990 [DOI] [PubMed] [Google Scholar]
- 23.Ruzin A, Keeney D, Bradford PA. 2005. AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob. Agents Chemother. 49:791–793. 10.1128/AAC.49.2.791-793.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visalli MA, Murphy E, Projan SJ, Bradford PA. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-396) in Proteus mirabilis. Antimicrob. Agents Chemother. 47:665–669. 10.1128/AAC.47.2.665-669.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bratu S, Landman D, George A, Salvani J, Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J. Antimicrob. Chemother. 64:278–283. 10.1093/jac/dkp186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. 2010. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob. Agents Chemother. 54:2720–2723. 10.1128/AAC.00085-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. 2011. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38:39–45. 10.1016/j.ijantimicag.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veleba M, Schneiders T. 2012. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:4466–4467. 10.1128/AAC.06224-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez A, Poza M, Aranda J, Latasa C, Medrano FJ, Tomás M, Romero A, Lasa I, Bou G. 2012. Effect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob. Agents Chemother. 56:6256–6266. 10.1128/AAC.01085-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy S, Datta S, Viswanathan R, Singh AK, Basu S. 2013. Tigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia (2007–10) and role of an efflux pump in tigecycline non-susceptibility. J. Antimicrob. Chemother. 68:1036–1042. 10.1093/jac/dks535 [DOI] [PubMed] [Google Scholar]
- 31.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. 2012. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194:1841–1842. 10.1128/JB.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chmelnitsky I, Shklyar M, Hermesh O, Navon-Venezia S, Edgar R, Carmeli Y. 2013. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J. Antimicrob. Chemother. 68:74–83. 10.1093/jac/dks370 [DOI] [PubMed] [Google Scholar]
- 33.Kumar V, Sun P, Vamathevan J, Li Y, Ingraham K, Palmer L, Huang J, Brown JR. 2011. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob. Agents Chemother. 55:4267–4276. 10.1128/AAC.00052-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67:2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bose M, Barber R. 2006. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. 6:0020. [PubMed] [Google Scholar]
- 36.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385–400. 10.1046/j.1365-2958.2003.03455.x [DOI] [PubMed] [Google Scholar]
- 37.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405. 10.1146/annurev.micro.59.050405.101230 [DOI] [PubMed] [Google Scholar]
- 38.Schleif R. 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34:779–796. 10.1111/j.1574-6976.2010.00226.x [DOI] [PubMed] [Google Scholar]
- 39.Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, Zhu X, Mukhopadhyay S. 2012. BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PLoS One 7:e31348. 10.1371/journal.pone.0031348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, Normark S, Melefors O. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843–853. 10.1128/JB.185.3.843-853.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC. 2008. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell 32:791–802. 10.1016/j.molcel.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Comparative Sequencing Program Group NISC. Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4:148ra116. 10.1126/scitranslmed.3004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu M, Nandi S, Davies C, Nicholas RA. 2005. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob. Agents Chemother. 49:4327–4334. 10.1128/AAC.49.10.4327-4334.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.