Abstract
The I222K, I222R, and I222T substitutions in neuraminidase (NA) have been found in clinically derived 2009 pandemic influenza A/H1N1 viruses with altered susceptibilities to NA inhibitors (NAIs). The effects of these substitutions, together with the most frequently observed resistance-related substitution, H274Y, on viral fitness and resistance mechanisms were further investigated in this study. Reduced sensitivities to oseltamivir were observed in all three mutants (I222K, I222R, and I222T). Furthermore, the I222K and I222T substitutions had a combinational effect of further increasing resistance in the presence of H274Y, which might result from a conformational restriction in the NA binding site. Of note, by using molecular dynamics simulations, R152, the neighbor of T222, was observed to translate to a position closer to T222, resulting in the narrowing of the binding pocket, which otherwise only subtends the residue substitution of H274Y. Moreover, significantly attenuated NA function and viral growth abilities were found in the I222K+H274Y double mutant, while the I222T+H274Y double mutant exhibited slightly delayed growth but had a peak viral titer similar to that of the wild-type virus in MDCK cells. The relative growth advantage of the I222T mutant versus the I222K mutant and the higher frequency of I222T emerging in N1 subtype influenza viruses raise concerns necessitating close monitoring of the dual substitutions I222T and H274Y.
INTRODUCTION
Influenza viruses are highly contagious in the human population and result in acute respiratory infectious diseases ranging from mild to severe. Since most of currently circulating influenza viruses have been found to be resistant to M2 ion channel blockers (1), neuraminidase inhibitors (NAIs), such as oseltamivir and zanamivir, which target the NA glycoproteins of influenza A and B viruses, are widely used in the prophylaxis and treatment of influenza virus infections. In 2009, a novel triple reassortant swine-origin influenza A(H1N1) virus that was naturally resistant to adamantanes emerged and quickly spread worldwide (2). Although NAIs are effective against A(H1N1)pdm09, and <2% of the oseltamivir-resistant viruses harboring an H274Y substitution in NA were detected (3), the outbreak of a cluster infection of H274Y A(H1N1)pdm09 in New South Wales in 2011, as well as the emergence of multidrug-resistant clinical isolates with novel genotypes, raised global concerns (4, 5).
NAI resistance is mostly related to influenza NA mutations in or around the active site (6). The active site is composed of 8 functional residues (R118, D151, R152, R224, E276, R292, R371, and Y406) and 11 framework residues (E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425) (7). To our knowledge, the amino acid substitutions in the functional residues are rare, and only substitutions at D151, R292, and R371 have been detected in clinic specimens or field isolates (8, 9). Nevertheless, those substitutions usually result in decreased NA activity or impaired fitness in MDCK cells (10–12). Substitutions in the framework site of NA, such as residues 119, 198, 222, 274, and 294, are relatively more common and diverse in their presentation (8, 13). Those framework substitutions usually reduce the NAI susceptibilities of the viruses by interrupting the binding of NAIs and NA. Some of the substitutions spread widely due to their minor effects on NA activity or viral fitness (6, 14, 15). For example, the well-studied substitution H274Y was found to confer oseltamivir resistance in the seasonal H1N1 virus in 2008 to 2009 (16). The substitutions on residue 222 have also been regarded to be key markers in the monitoring of the ability of a strain to increase drug resistance by combining with H274Y (17). Until now, at least seven types of NA substitutions in A(H1N1)pdm09 have been identified to be drug resistant in response to NAI treatment (see Table S1 in the supplemental material).
Some of the NAI resistance-related substitutions are subtype specific and drug specific. It has been reported that the H274Y substitution is oseltamivir resistant in the N1 subtype, and D151V (8, 9) is zanamivir resistant in the N2 subtype. Some substitutions, however, are not type/subtype specific. For example, substitutions at residue 222 confer reduced susceptibility in N1, N2, and type B viruses (18, 19). The amino acid substitutions at residue 222 of NA are varied, including substitutions of I222T/V in type B, I222V/M/T/R in N1, and I222V in N2 (18–25), and these exhibit varied effects on the enzymatic properties of NA and NAI susceptibility. They emerged either naturally or after oseltamivir treatment or prophylaxis. I222R increases the 50% inhibitory concentration (IC50) of oseltamivir and zanamivir relative to the wild-type (WT) strain in A(H1N1)pdm09 and has a combinational effect on resistance to oseltamivir and zanamivir when combined with the H274Y substitution (5, 21, 26, 27). The I222T substitution results in moderately reduced susceptibility of influenza B virus to oseltamivir but not zanamivir (18). I222T can be found in a wide host range, which has been detected in A(H1N1)pdm09, H5N1, and type B viruses (see Tables S2 and 3 in the supplemental material). It has been reported that most B viruses with the I222T substitution were isolated from untreated patients, which indicates that I222T may occur naturally without NAI selective pressure (18). The I222K substitution was detected in A(H1N1)pdm09, resulting in significant oseltamivir resistance and moderate zanamivir resistance (28). Although the viral fitness and NA enzymatic properties of A(H1N1)pdm09 viruses have been studied in several single substitutions (such as I222V or I222R) and double substitutions (I222V/R combined with H274Y) (29, 30), the features of I222T and I222K mutants have not yet been determined. To address this issue, in this work, we generated recombinant A(H1N1)pdm09 viruses containing I222T and I222K NA substitutions with or without H274Y and then compared their NAI resistance phenotypes and in vitro replicative capacities. Furthermore, we studied the resistance mechanisms involved in the various impacts of I222T substitutions on NA biological functions of A(H1N1)pdm09 using molecular dynamics simulations.
MATERIALS AND METHODS
Cell culture.
Madin-Darby canine kidney (MDCK) and human embryonic kidney (293T) cells were obtained from the American Type Culture Collection as reported previously (31). The 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA, USA), and MDCK was maintained in minimum essential medium (MEM) (Invitrogen), supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 2 mM glutamine (Invitrogen).
Generation of recombinant viruses and virus titrations.
All eight gene segments of influenza virus strain A/California/04/2009 (H1N1; CA04) were amplified by real-time PCR (RT-PCR) and cloned into a modified version of the bidirectional expression plasmid pCQI, derived from pHW2K. Mutations were introduced into the pCQI plasmid containing the NA segment using the appropriate primers and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), according to the manufacturer's instructions. The nucleotide changes were I/ATA to K/AAA (I222K), I/ATA to R/AGA (I222R), I/ATA to T/ACA (I222T), and H/CAC to Y/TAC (H274Y). Based on the H274Y-NA gene, we introduced a second mutation. All plasmids were sequenced to ensure the presence of the introduced mutations and the absence of undesired mutations. The eight bidirectional plasmids were cotransfected into a 293T/MDCK coculture monolayer using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. All the recombinant viruses were sequenced and then titrated by the 50% tissue culture infective dose (TCID50). As previously reported (31), the TCID50 was measured on MDCK cells. A log0.5 dilution of the virus was inoculated on monolayer MDCK cells. After virus adsorption for 1 h at 35°C, unadsorbed virus was removed by washing with phosphate-buffered saline (PBS). The infection medium, containing 2 mg/liter N-tosyl-l-phenylalanine chloromethyl ketone-treated (TPCK) trypsin (Sigma, St. Louis, MO, USA), was added. The cytopathogenic effect (CPE) was observed daily and confirmed by a hemagglutination assay with 0.5% turkey red blood at 96 h postinfection. The TCID50 was calculated by the Reed-Muench formula (32).
NA inhibitor assays.
The rescued viruses were tested for susceptibility to oseltamivir and zanamivir by NA inhibition assays using the 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid (MUNANA) (Sigma, St. Louis, MO) substrate (23), with minor modifications. Briefly, recombinant viruses were standardized to an NA activity level 10-fold higher than that of the background. Forty microliters of virus was mixed with various concentrations of inhibitor (final concentrations of NA inhibitors, 0.01 nM to 10,000 nM) in black microtiter plates. The virus-inhibitor mixture was incubated at 37°C for 45 min prior to the addition of 40 μl of MUNANA substrate (final concentration, 100 μM) and then incubated at 37°C for 60 min. The reaction was terminated by the addition of 80 μl of the stop solution. The IC50 for each drug was performed using the GraphPad Prism software version 5.
Michaelis constant measurement.
The NA enzyme kinetics was measured at pH 6.5 with 33 mM 2-(N-morpholino)ethanesulfonic acid hydrate (MES) (Sigma-Aldrich, St. Louis, MO, USA), 4 mM CaCl2, and the fluorogenic substrate MUNANA (final substrate concentration, 1.5625 to 200 nM). The same titer of 105.8 TCID50/ml of the virus particles was used in the measurement of the Michaelis constant (Km). The reaction was conducted at 37°C in a total volume of 100 μl, and the fluorescence of released 4-methylumbelliferone was measured every 60 s for 90 min by Infinite 200 PRO (Tecan, Medford, MA), using excitation and emission wavelengths of 360 and 465 nm, respectively. The Michaelis constant (Km) kinetic parameters were calculated by fitting the data to the appropriate Michaelis-Menten equations by using nonlinear regression in the GraphPad Prism 5 software (GraphPad Software, La Jolla, CA).
Expression of NA at the surface of virus-infected cells, and total NA activity.
The expression of NA at the surface of virus-infected MDCK cells was determined as previously described (6). In brief, the rescued viruses were diluted in appropriate medium containing 2 mg/liter TPCK-trypsin to infect confluent MDCK 48-well plates at a multiplicity of infection (MOI) of 1 50% tissue culture infectious dose (TCID50) per cell. All media and viruses were maintained at 4°C and then incubated with MDCK cells for 1 h at 35°C to allow the viruses to bind to the surface of the cells. Then inocula were disposed and placed in freshly working medium (200 μl/well). The plates were incubated at 35°C under 5% CO2. At 12 h postinfection, the cells were digested with 0.25% EDTA-trypsin and collected together. The cells were washed three times with PBS and then divided into 3 parts: one for NA surface staining, one for the detection of total NA activity, and one for neuropilin (NP) staining. The cells for the total NA activity assay were lysed by an appropriate volume of passive lysis buffer and then mixed with 400 μM MUNANA. The mixtures were incubated at 37°C. The fluorescence value of released 4-methylumbelliferone was detected 30 min later. Specific A/New Caledonia/20/1999 strain NA monoclonal antibody (Immunotech, Suzhou, China) was added at a dilution of 1:100 in PBS, and the plates were incubated 1 h at room temperature. The cells were washed three times as previously described. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (ZSGB-Bio, Beijing, China) was added at a dilution of 1:100 in PBS, and the tubes were incubated 1 h at room temperature. After a last washing step, the cells were suspended by 500 μl PBS and were detected by flow cytometry. Blank corrected data were normalized against the quantity of infected cells reflected by the relative quantity of NP expressed in the cells, determined by the same assays performed in parallel after permeabilization of the infected cells.
Growth curves.
In vitro MDCK cells were infected with recombinant viruses at a multiplicity of infection (MOI) of 0.001 TCID50 per cell and incubated in the appropriate medium containing 2 μg/ml TPCK-trypsin at 35°C. The supernatants were collected at 12, 24, 48, and 72 h postinoculation (hpi), and the virus titers in the supernatants were determined by titration in MDCK cells by daily observation for cytopathogenic effect (CPE) after incubation for 96 h at 35°C; these were confirmed by a hemagglutination assay performed with 0.5% turkey chicken red blood. The TCID50 was calculated by the Reed-Muench formula (32).
Bioinformatics analysis.
The wild-type NA crystal structure (PDB code 4B7R) was obtained from the Protein Data Bank (33). NA mutants were first produced by using the side-chain modeling software CISRR. Next, the modeling structures were optimized by molecular dynamics simulations using NAMD (34) (version 2.9) and CHARMM22 force fields (35). The parameters for oseltamivir were generated by SwissParam (36). The molecules were solvated with TIP3P water in a cubic box, where the minimal distance between the atoms in the complex and the edges of the box was set to 12 Å. A switch distance of 10 Å and a cutoff distance of 12 Å were used for nonbonded interactions. Particle mesh Ewald (PME) was chosen to calculate the long-range electronic interactions. The simulations were conducted at 310 K and a constant pressure of 1 bar. They were first minimized for 2,000 steps and then equilibrated in the isothermal-isobaric (NPT) ensemble for 1.8 ns, with an integration time step of 1 fs. The simulations were repeated twice.
Statistical analysis.
The IC50s, Kms, NA surface expression levels, NA activities, and viral titers of the mutants were compared with those of the wild type (WT) or H274Y mutant by using an unpaired two-tailed t test in the GraphPad Prism 5 software package (GraphPad Software, USA). A P value of <0.05 was considered to indicate statistical significance.
RESULTS
Enzymatic properties of the recombinant viruses.
To determine the impact of the substitutions of H274Y, I222K, and I222T on the enzymatic properties of the viruses, we generated the recombinant WT and five variants (I222K, I222T, H274Y, I222K+H274Y, and I222T+H274Y) by reverse genetics in the context of the A/California/04/2009 virus. The I222R and I222R+H274Y mutants were also generated for comparison. To characterize the NA enzymatic parameters of these viruses, the Michaelis constant (Km), which reflects NA affinity for the substrate, was determined using MUNANA as a substrate. Except for I222K, all single substitutions significantly decreased enzymatic affinity (2.2 to 3.4-fold increase in Km; P < 0.05). The substitution I222K resulted only in a minor reduction of affinity (1.4-fold increase in Km; P = 0.072) compared with the WT. The enzymatic affinities of all double mutants decreased further, especially for the I222R+H274Y mutant (9-fold increase; P = 0.0001) (Table 1).
TABLE 1.
Enzymatic characteristics of the NA of recombinant A(H1N1)pdm09 viruses
| NA substitution(s)a | Km (μM) (mean ± SDb [fold changec]) | IC50 (nM) for indicated drug |
|
|---|---|---|---|
| Oseltamivir | Zanamivir | ||
| Mean ± SDb (fold changec) | Mean ± SDb (fold changec) | ||
| WT | 19.14 ± 2.50 (1.0) | 0.69 ± 0.05 (1.0) | 0.28 ± 0.01 (1.0) |
| H274Y | 41.65 ± 4.37d (2.2) | 396.2 ± 15.56d (574.2) | 0.3 ± 0.03 (1.1) |
| I222K | 26.73 ± 2.05 (1.4) | 20.48 ± 1.73e (29.7) | 0.87 ± 0.15 (3.1) |
| I222T | 47.61 ± 5.72d (2.5) | 4.42 ± 0.86d (6.4) | 0.55 ± 0.03 (2.0) |
| I222R | 64.66 ± 12.83d (3.4) | 22.07 ± 2.27e (32) | 2.27 ± 0.18 (8.1) |
| I222K+H274Y | 148.64 ± 20.35e,g (7.8) | 3,974 ± 477.58e,g (5,759) | 1.99 ± 0.03e (7.1) |
| I222T+H274Y | 97.44 ± 16.9d,f (5.1) | 950.15 ± 47.73e,f (1,377) | 0.57 ± 0.09 (2.0) |
| I222R+H274Y | 174.63 ± 3.18e,g (9.1) | 3,936 ± 336.7e,g (5,704) | 3.74 ± 1.09d (13.4) |
All substitutions are in N2 numbering.
Data from three independent experiments.
Compared to that of WT (wild type).
P value of <0.05, unpaired t test against wild type.
P value of <0.01, unpaired t test against wild type.
P value of <0.05, unpaired t test against the H274Y mutant.
P value of <0.01, unpaired t test against the H274Y mutant.
Susceptibilities of the recombinant viruses to neuraminidase inhibitors.
As expected, the recombinant WT virus was sensitive to the two NAIs tested (oseltamivir and zanamivir), while the H274Y mutant was resistant to oseltamivir (mean 574-fold increase in IC50) but susceptible to zanamivir. The I222T mutant showed only a 6-fold reduction in susceptibility to oseltamivir and had almost no impact on the susceptibility to zanamivir. Interestingly, the combination of I222T and H274Y induced a mean 1,377-fold increase in the IC50 of oseltamivir compared to that of the WT virus (P < 0.01). I222K was also associated with a low level of resistance to oseltamivir and with a small reduction in zanamivir susceptibility (29- and 3-fold increases in the IC50, respectively). The I222R substitution conferred moderate resistance to oseltamivir and reduced susceptibility to zanamivir (32- and 8-fold increases in the IC50, respectively). The IC50s of the double mutants I222K+H274Y and I222R+H274Y were further increased >5,000-fold in oseltamivir and >7-fold in zanamivir compared to the recombinant WT virus (Table 1).
Replicative capacity of recombinant viruses in vitro in MDCK cells.
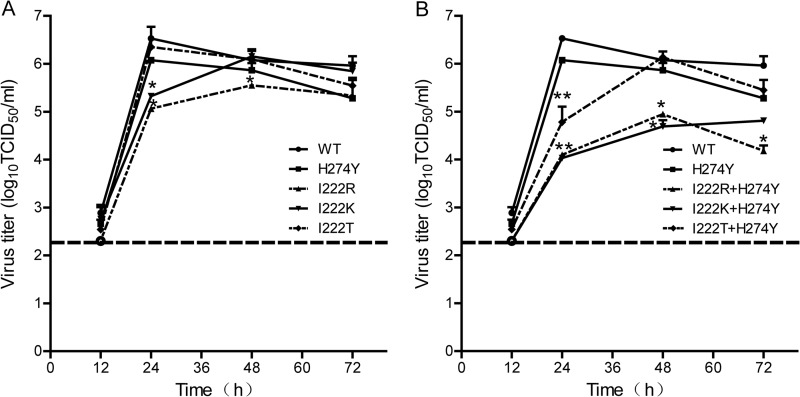
A multicycle growth assay of recombinant viruses was performed in MDCK cells at an MOI of 0.001 TCID50 per cell, the supernatants were harvested at 12, 24, 48, and 72 h after infection, and the viral titers were determined on MDCK cells. Viral growth was slightly impaired for the H274Y mutant, while there was almost no difference in growth for the I222T mutant. The initial virus replication rates were similar for the single-residue-substitution variants, except for I222R, whose titer was below the lower limit of detection. The peak points of replication of the I222R and I222K mutants were delayed to 48 hpi, with lower peak titers than that of the WT virus (Fig. 1A). A significantly lower level of viral growth was seen for the I222R+H274Y and I222K+H274Y mutants, with peak titers of ∼101 to 102.5 TCID50/ml lower than that of WT (P < 0.05). Their viral titers maintained the lower levels throughout the time points. Although the I222T+H274Y mutant reached its peak at 48 hpi, 24 h later than the WT virus, their peak titers were comparable, with 106.53 TCID50/ml for the WT virus and 106.14 TCID50/ml for the I222T+H274Y mutant (Fig. 1B).
FIG 1.
Replication kinetics of recombinant influenza A(H1N1)pdm09 viruses in MDCK cells. Confluent MDCK cells were inoculated with rescued viruses at an MOI of 0.001. Supernatants were collected at 12, 24, 48, and 72 hpi at 35°C and titrated by a TCID50 assay using MDCK cells. (A) WT, I222R, I222K, I222T, and H274Y mutants. (B) WT, I222R+H274Y, I222K+H274Y, and I222T+H274Y mutants. Each data point represents the yield of a virus tested in three separated experiments (mean ± SD of TCID50/ml). *, P value of <0.05, and **, P value of <0.01 for differences in viral titers between the mutant and WT viruses as determined by an unpaired t test. The dotted line indicates the lower limit of detection.
NA total activity and NA surface expression.
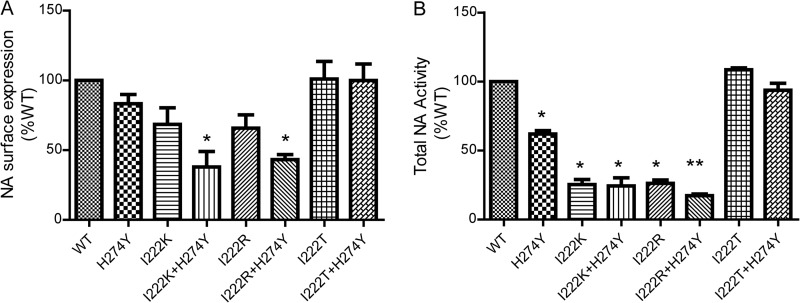
To understand what is responsible for the varied viral fitness of different amino acid substitutions at the NA 222 residue, we detected the levels of NA activity and surface expression in the mutants. The surface-expressed NA protein levels on the infected cells of the I222K, I222R, and H274Y mutants were decreased but without a significant difference relative to that of the wild type (P > 0.05) (Fig. 2A). The I222R and I222K mutants had very low total NA activity, which represented 25.4% and 26.2% of that of the wild type, respectively (P = 0.02 and 0.017, respectively). The H274Y mutant also showed decreased NA activity, with 61% of that of the wild type (P = 0.03) (Fig. 2B). Surprisingly, the I222T substitution, alone or with the H274Y substitution, had little impact on total NA activity and NA surface expression compared to that of the wild type (P > 0.07), while the I222R+H274Y and I222K+H274Y mutants showed significantly reduced total NA activity (P < 0.02) and NA surface expression (P < 0.05).
FIG 2.
Expression of NA at the surface of infected MDCK cells, and total NA activity of recombinant A(H1N1)pdm09 virus. MDCK cells were infected with rescued viruses at an MOI of 1 and collected at 12 hpi. The level of NA surface expression and NA total activity were expressed as a percentage of that of the wild-type viruses. (A) Expression of all the recombinant virus NAs at the surface of infected cell surface. (B) Total NA activity of all the recombinant viruses determined by fluorometric assays using MUNANA as a substrate. The mean ± SD values are from three independent assays. *, P value of <0.05, and **, P value of <0.01 versus the wild type.
Molecular simulations for the inhibitor resistance mutants.
The above results show that the I222K, I222T, I222K+H274Y, and I222T+H274Y substitutions have obvious impacts on oseltamivir susceptibility, but what is responsible for this phenomenon? Crystal structure studies have demonstrated that I222R results in shrinkage at the edge of the active site pocket that accommodates the hydrophobic pentoxyl group of oseltamivir (37). The H274Y substitution perturbs the position and reorientation of the side chain of E276, which further restricts the binding of oseltamivir in the opposite direction (38, 39). We suppose the mechanism for the I222K and I222K+H274 substitutions is similar, because lysine (K) is similar to arginine (R) in its size and biochemical properties (Fig. 3A and B). However, unlike R or K, threonine (T) does not possess a bulky side chain, so why does it perform similarly regarding resistance to NAI? If it is because of the loss of hydrophobic effects for polar threonine (T) replacing hydrophobic isoleucine (I), how does the combinational effect take place? To investigate its mechanism, we employed the molecular dynamics simulations for three systems, the wild-type NA complexed with oseltamivir and the I222K and I222T mutants each complexed with oseltamivir. The three systems equilibrated in the NPT ensemble for 1.8 ns. We analyzed the conformational changes of residue 222 and discovered that there is an obvious difference at their surrounding residues. Compared to the wild type and I222K mutant, R152, the neighbor of T222, tends to translate to a closer position to T222 (Fig. 3C), driven by the forming of hydrogen bonds between two residues and waters. If we use the average distances between the terminal nitrogen (NH1) of R152 and carbon (CB) of I/K/T222 as a descriptor, it is 5.8 Å for the WT, 5.6 Å for I222K, and 4.8 Å for I222T in the simulations (Fig. 3D). By superimposing the modeling structures on the crystal structure, we found that the terminal group of R152 translated much closer to the hydrophobic pentoxyl group of oseltamivir (Fig. 3C). Their distance decreases from 4.1 Å in the wild type to 3.3 Å in the I222K mutant, which means there is steric hindrance to the binding of oseltamivir to the mutant NA. With reference to the cooperative restricting effect of the I222R+H274Y substitutions, we suggest that the double substitution perturbs the side chain of R152 by the smaller threonine (T) and of E276 by the bulkier tyrosine (Y), resulting in shrinkage at the edge of the active site pocket in two opposite directions. Thus, the double mutant has a combinational effect that has a much lower IC50 than either of the two single mutants.
FIG 3.
Structure modeling of I222K and I222T mutants. I222K and I222T mutants were generated from the crystal structure of the NA of H1N1 2009 pandemic influenza A virus (PDB code 4B7R) by using the side-chain modeling software CISRR. The structures of the wild-type and mutant viruses were further performed in 1.8-ns molecular dynamics simulation. The side-chain conformations of Y274 and E276 were obtained from the crystal structure of an NA mutant (PDB code 3CL0) after the structural alignment. NAs binding with oseltamivir are represented in green (wild-type residues) and light gray (mutant residues). The distances between the terminal nitrogen (NH1) of R152 and the carbon (CB) of I/T/K222 are shown by the dotted lines. The structural figures were generated by PyMOL (Delano Scientific, Palo Alto, CA). (A) Wild-type conformation of residue 222 and oseltamivir. (B) Conformation of K222 and oseltamivir. The substitution of K for I results in relatively close contact (represented as a yellow spark) between the terminal atom of K222 and oseltamivir, which restricts the reorientation of oseltamivir upon H274Y substitution. (C) Conformation of T222 and oseltamivir. The substitution of T for I leads to a conformational change in R152, which turns to contact T222 closely. Meanwhile, the distance between the terminal atom of R152 and oseltamivir is shortened, restricting the reorientation of oseltamivir. (D) Distances between the terminal nitrogen (NH1) of R152 and the carbon (CB) of I/T/K 222 in the 1.8-ns simulations. The x axis shows the time (ps) of the molecular dynamic simulation, and the y axis shows the distance (Å) between NH1 and CB. The dark yellow, blue, and red lines are the distance values between NH1 and CB of the I222, K222, and T222 mutants, respectively. The average distance of T222 (blue) is shorter than those of I222 (dark yellow) and K222 (red).
DISCUSSION
Influenza A virus has led to significant morbidity and mortality in humans and global societal costs every year. To combat the burden of disease caused by influenza virus, vaccination and antiviral drugs are the main strategies. However, when vaccine efficacy is low or a vaccine is unavailable, antiviral drugs are an important line of defense. Two classes of anti-influenza drugs, M2 ion channel blockers and neuraminidase inhibitors, were approved for the treatment of influenza. NAIs were the first-line drugs available for combating 2009 pandemic A(H1N1) virus for its natural resistance to adamantine. Early administration of oseltamivir was shown to improve clinical outcomes in infant, adult, and pregnant woman patients infected by A(H1N1)pdm09 (40). Apart from oseltamivir, inhaled zanamivir and intravenous peramivir were also used in patients who experienced failed oseltamivir treatment (41). Since the NAIs are more widely used, antiviral resistance to these drugs is monitored and a cause for concern.
The amino acid isoleucine (I) at residue 222 around the NA active site interacts with the pentoxyl group of oseltamivir or the N-acetyl and glycerol groups of zanamivir to facilitate NA binding to the drugs (37). Substitutions at residue 222 may result in reduced binding of NA to oseltamivir or/and zanamivir. The A(H1N1)pdm09 I222R or I222R+H274Y variants were already detected in immunocompromised patients after treatment with oseltamivir (5, 21, 22). Consistent with earlier reports, we found that a single substitution of I222R conferred only moderate resistance to oseltamivir and zanamivir, while its combination with H274Y led to a sharply increased NAI IC50 (Table 1). The impaired growth of both I222R and I222R/H274Y variants in MDCK cells detected in this study (see Fig. 1) and LeGoff et al. (26) indicates that the I222R substitution may be harmful to the viral fitness of the variants. However, contradictions were also reported. The discrepancies might originate from the different strains used in the experiments (30). The I222K and I222K+H274Y mutants were observed in a clinical case (28). This study and previous reports show that the I222K substitution confers resistance to oseltamivir and zanamivir similar to that conferred by the I222R substitution, and it enhances resistance to oseltamivir and zanamivir when combined with the H274Y substitution. Our study also indicates that the I222K substitution decreased the affinity of NA to substrate and impaired viral fitness; especially, the I222K+H274Y dual substitutions had a marked negative effect on the replicative kinetics of A(H1N1)pdm2009.
Our molecular dynamics simulations further indicate that the varied combinational effect of the I222T substitution might attribute to a distinct resistance mechanism. We found that the average distance between the terminal atoms of R152 and T222 is shorter than that in wild type, which leads to the narrowing of the binding pocket. As the conformational change of R152 is in the opposite direction of residue 274, the double substitutions I222T and H274Y have a combinational effect on oseltamivir binding (Fig. 3).
Furthermore, the replication capacities of the mutants with I222T or I222T+H274Y were comparable to that of the recombinant WT virus, indicating that the substitution I222T is not harmful to the viral fitness of A(H1N1)pdm09. It is generally accepted that the immune status of infected hosts correlates with the emergence and persistence of NAI-resistant mutants with fitness deficits, such as I222R (5). Unlike I222R or I222K, the I222T substitution seemed to occur naturally, as most of the viruses harboring this substitution were isolated from patients not treated with NAIs (18). Based on the fact that the I222T mutant has been frequently detected in current subtype viruses infecting humans, this substitution in A(H1N1)pdm09 should cause much concern.
In summary, the substitutions of I to R or K at residue 222 conferred a multidrug-resistant phenotype in A(H1N1)pdm09 viruses with significantly impaired viral fitness. However, the I222T substitution exhibited a combinational effect with H274Y with regard to oseltamivir resistance, without compromising viral fitness. Our molecular dynamics simulations suggest that the displacement of R152 toward T222 plays a key role in NA resistance to NAIs. Coupled with its higher frequency in N1 and natural emergence, the potential emergence or possible community transmission of I222T+H274Y requires close monitoring and investigation regarding the clinical usage of antiviral drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lu Yi of the Institute of Biophysics, Chinese Academy of Sciences, for providing technical support.
This work was supported by the major research project for infectious disease (project no. 2013ZX10004611), National Basic Research Program 973 of China (grant no. 2011CB504704), and Sichuan University to Y.C. (grant no. 2011SCU11104).
NAMD was developed by the Theoretical and Computational Biophysics Group at the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign.
Footnotes
Published ahead of print 23 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01848-13.
REFERENCES
- 1.Okomo-Adhiambo M, Sheu TG, Gubareva LV. 2013. Assays for monitoring susceptibility of influenza viruses to neuraminidase inhibitors. Influenza Other Respir. Viruses 7(Suppl 1):44–49. 10.1111/irv.12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605–2615. 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- 3.Meijer A, Jonges M, Abbink F, Ang W, van Beek J, Beersma M, Bloembergen P, Boucher C, Claas E, Donker G, van Gageldonk-Lafeber R, Isken L, de Jong A, Kroes A, Leenders S, van der Lubben M, Mascini E, Niesters B, Oosterheert JJ, Osterhaus A, Riesmeijer R, Riezebos-Brilman A, Schutten M, Sebens F, Stelma F, Swaan C, Timen A, van 't Veen A, van der Vries E, te Wierik M, Koopmans M. 2011. Oseltamivir-resistant pandemic A(H1N1) 2009 influenza viruses detected through enhanced surveillance in the Netherlands, 2009–2010. Antiviral Res. 92:81–89. 10.1016/j.antiviral.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 4.Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Gehrig N, Kelso A. 2011. Community transmission of oseltamivir-resistant A(H1N1)pdm09 influenza. N. Engl. J. Med. 365:2541–2542. 10.1056/NEJMc1111078 [DOI] [PubMed] [Google Scholar]
- 5.van der Vries E, Stelma FF, Boucher CA. 2010. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N. Engl. J. Med. 363:1381–1382. 10.1056/NEJMc1003749 [DOI] [PubMed] [Google Scholar]
- 6.Richard M, Ferraris O, Erny A, Barthélémy M, Traversier A, Sabatier M, Hay A, Lin YP, Russell RJ, Lina B. 2011. Combinatorial effect of two framework mutations (E119V and I222L) in the neuraminidase active site of H3N2 influenza virus on resistance to oseltamivir. Antimicrob. Agents Chemother. 55:2942–2952. 10.1128/AAC.01699-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman PM, Hoyne PA, Lawrence MC. 1993. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J. Virol. 67:2972–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HT, Fry AM, Gubareva LV. 2012. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir. Ther. 17:159–173. 10.3851/IMP2067 [DOI] [PubMed] [Google Scholar]
- 9.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284–3292. 10.1128/AAC.00555-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr J, Ives J, Kelly L, Lambkin R, Oxford J, Mendel D, Tai L, Roberts N. 2002. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 54:79–88. 10.1016/S0166-3542(01)00215-7 [DOI] [PubMed] [Google Scholar]
- 11.Herlocher ML, Carr J, Ives J, Elias S, Truscon R, Roberts N, Monto AS. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54:99–111. 10.1016/S0166-3542(01)00214-5 [DOI] [PubMed] [Google Scholar]
- 12.Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, Govorkova EA. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J. Virol. 80:8787–8795. 10.1128/JVI.00477-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferraris O, Lina B. 2008. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J. Clin. Virol. 41:13–19. 10.1016/j.jcv.2007.10.020 [DOI] [PubMed] [Google Scholar]
- 14.Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 4:e1000103. 10.1371/journal.ppat.1000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275. 10.1126/science.1187816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzaki Y, Mizuta K, Aoki Y, Suto A, Abiko C, Sanjoh K, Sugawara K, Takashita E, Itagaki T, Katsushima Y, Ujike M, Obuchi M, Odagiri T, Tashiro M. 2010. A two-year survey of the oseltamivir-resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamivir. Virol. J. 7:53. 10.1186/1743-422X-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKimm-Breschkin JL. 2013. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 7(Suppl 1):25–36. 10.1111/irv.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatakeyama S, Sugaya N, Ito M, Yamazaki M, Ichikawa M, Kimura K, Kiso M, Shimizu H, Kawakami C, Koike K, Mitamura K, Kawaoka Y. 2007. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA 297:1435–1442. 10.1001/jama.297.13.1435 [DOI] [PubMed] [Google Scholar]
- 19.Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395–2402. 10.1128/AAC.01339-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CDC 2009. Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:969–972 [PubMed] [Google Scholar]
- 21.Nguyen HT, Fry AM, Loveless PA, Klimov AI, Gubareva LV. 2010. Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin. Infect. Dis. 51:983–984. 10.1086/656439 [DOI] [PubMed] [Google Scholar]
- 22.Eshaghi A, Patel SN, Sarabia A, Higgins RR, Savchenko A, Stojios PJ, Li Y, Bastien N, Alexander DC, Low DE, Gubbay JB. 2011. Multidrug-resistant pandemic (H1N1) 2009 infection in immunocompetent child. Emerg. Infect. Dis. 17:1472–1474. 10.3201/eid1708.102004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurt AC, Holien JK, Barr IG. 2009. In vitro generation of neuraminidase inhibitor resistance in A(H5N1) influenza viruses. Antimicrob. Agents Chemother. 53:4433–4440. 10.1128/AAC.00334-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garg S, Moore Z, Lee N, McKenna J, Bishop A, Fleischauer A, Springs CB, Nguyen HT, Sheu TG, Sleeman K, Finelli L, Gubareva L, Fry AM. 2012. A cluster of patients infected with I221V influenza b virus variants with reduced oseltamivir susceptibility–North Carolina and South Carolina, 2010–2011. J. Infect. Dis. 207:966–973. 10.1093/infdis/jis776 [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Sleeman K, Huang W, Nguyen HT, Levine M, Cheng Y, Li X, Tan M, Xing X, Xu X, Klimov AI, Gubareva LV, Shu Y. 2013. Neuraminidase inhibitor susceptibility testing of influenza type B viruses in China during 2010 and 2011 identifies viruses with reduced susceptibility to oseltamivir and zanamivir. Antiviral Res. 97:240–244. 10.1016/j.antiviral.2012.12.013 [DOI] [PubMed] [Google Scholar]
- 26.LeGoff J, Rousset D, Abou-Jaoudé G, Scemla A, Ribaud P, Mercier-Delarue S, Caro V, Enouf V, Simon F, Molina JM, van der Werf S. 2012. I223R mutation in influenza A(H1N1)pdm09 neuraminidase confers reduced susceptibility to oseltamivir and zanamivir and enhanced resistance with H275Y. PLoS One 7:e37095. 10.1371/journal.pone.0037095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Vries E, Veldhuis Kroeze EJ, Stittelaar KJ, Linster M, Van der Linden A, Schrauwen EJ, Leijten LM, van Amerongen G, Schutten M, Kuiken T, Osterhaus AD, Fouchier RA, Boucher CA, Herfst S. 2011. Multidrug resistant 2009 A/H1N1 influenza clinical isolate with a neuraminidase I223R mutation retains its virulence and transmissibility in ferrets. PLoS Pathog. 7:e1002276. 10.1371/journal.ppat.1002276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen HT, Trujillo AA, Sheu TG, Levine M, Mishin VP, Shaw M, Ades EW, Klimov AI, Fry AM, Gubareva LV. 2012. Analysis of influenza viruses from patients clinically suspected of infection with an oseltamivir resistant virus during the 2009 pandemic in the United States. Antiviral Res. 93:381–386. 10.1016/j.antiviral.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 29.Pizzorno A, Abed Y, Bouhy X, Beaulieu E, Mallett C, Russell R, Boivin G. 2009. Impact of mutations at residue I223 of the neuraminidase protein on the resistance profile, replication level, and virulence of the pandemic influenza virus. Antimicrob. Agents Chemother. 56:1208–1214. 10.1128/AAC.05994-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizzorno A, Bouhy X, Abed Y, Boivin G. 2011. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J. Infect. Dis. 203:25–31. 10.1093/infdis/jiq010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W, Zhu Y, Qin K, Yu Z, Gao R, Yu H, Zhou J, Shu Y. 2012. Mutations in polymerase genes enhanced the virulence of 2009 pandemic H1N1 influenza virus in mice. PLoS One 7:e33383. 10.1371/journal.pone.0033383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493–497 [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217. 10.1002/jcc.540040211 [DOI] [Google Scholar]
- 36.Zoete V, Cuendet MA, Grosdidier A, Michielin O. 2011. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 32:2359–2368. 10.1002/jcc.21816 [DOI] [PubMed] [Google Scholar]
- 37.van der Vries E, Collins PJ, Vachieri SG, Xiong X, Liu J, Walker PA, Haire LF, Hay AJ, Schutten M, Osterhaus AD, Martin SR, Boucher CA, Skehel JJ, Gamblin SJ. 2009. H1N1 2009 pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog. 8:e1002914. 10.1371/journal.ppat.1002914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ. 2008. Crystal structures of oseltamivir-resistant influenzavirus neuraminidase mutants. Nature 453:1258–1261. 10.1038/nature06956 [DOI] [PubMed] [Google Scholar]
- 39.Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Martin SR, Daniels RS, Gregory V, Skehel JJ, Gamblin SJ, Hay AJ. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27:6317–6323. 10.1016/j.vaccine.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 40.Muthuri SG, Myles PR, Venkatesan S, Leonardi-Bee J, Nguyen-Van-Tam JS. 2013. Impact of neuraminidase inhibitor treatment on outcomes of public health importance during the 2009-10 influenza A(H1N1) pandemic: a systematic review and meta-analysis in hospitalized patients. J. Infect. Dis. 15:553–563. 10.1093/infdis/jis726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birnkrant D, Cox E. 2009. The Emergency Use Authorization of peramivir for treatment of 2009 H1N1 influenza. N. Engl. J. Med. 361:2204–2207. 10.1056/NEJMp0910479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.