ABSTRACT
Bovine spongiform encephalopathy (BSE) can be efficiently transmitted to small ruminants (sheep and goats) with certain prion protein (PrP) genotypes. Polymorphisms in PrP of both the host and donor influence the transmission efficiency of transmissible spongiform encephalopathies (TSEs) in general. These polymorphisms in PrP also modulate the PrP conversion underlying TSE agent replication. Here we demonstrate that single-round protein misfolding cyclic amplification (PMCA) can be used to assess species and polymorphism barriers at the molecular level. We assessed those within and between the ovine and bovine species in vitro using a variety of natural scrapie and experimentally generated cross-species BSE agents. These BSE agents include ovBSE-ARQ isolates (BSE derived from sheep having the ARQ/ARQ PrP genotype), and two unique BSE-derived variants: BSE passaged in VRQ/VRQ sheep and a cow BSE agent isolate generated by back-transmission of ovBSE-ARQ into its original host. PMCA allowed us to quantitatively determine PrP conversion profiles that correlated with known in vivo transmissibility and susceptibility in the two ruminant species in which strain-specific molecular signatures, like its molecular weight after protease digestion, were maintained. Furthermore, both BSE agent isolates from ARQ and VRQ sheep demonstrated a surprising transmission profile in which efficient transmissions to both sheep and bovine variants was combined. Finally, all data support the notion that ARQ-derived sheep BSE points to a significant increase in virulence compared to all other tested scrapie- and BSE-derived variants reflected by the increased conversion efficiencies of previously inefficient convertible PrP variants (including the so-called “resistant” sheep ARR variant).
IMPORTANCE Prion diseases such as scrapie in sheep and goats, BSE in cattle, and Creutzfeldt-Jakob disease (CJD) in humans are fatal neurodegenerative diseases caused by prions. BSE is known to be transmissible to a variety of hosts, including sheep and humans. Based on the typical BSE agent strain signatures and epidemiological data, the occurrence of a novel variant of CJD in humans was linked to BSE occurrence in the United Kingdom. Measures, including genetic selection of sheep toward less susceptible PrP genotypes, have been implemented to lower the risk of BSE transmission into sheep, since the disease could potentially spread into a natural reservoir. In this study, we demonstrated using molecular PrP conversion studies that when BSE is first transmitted through sheep, the host range is modified significantly and the PrP converting potency increased, allowing the ovine BSE to transmit more efficiently than cow BSE into supposedly less susceptible hosts.
INTRODUCTION
Prion diseases, or transmissible spongiform encephalopathies (TSEs), include scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, and Creutzfeldt-Jakob disease (CJD) in humans. TSEs in sheep and cattle are naturally occurring infectious and fatal neurological disorders characterized by the accumulation of pathologically folded prion protein (PrPSc) mainly in tissues of the central nervous system. Pathogenesis of both scrapie and experimental BSE in sheep starts after uptake in the gut via the gut-associated lymphoid tissues into the enteric nervous system toward the central nervous system (1–3). PrPSc formation is the key event in prion disease, in which it is converted from normal cellular PrP (PrPC) by a template-assisted seeding reaction in which cellular cofactors can play a role (4–6). Therefore, PrPSc (the seed) is a primary component of the infectious agent in TSEs.
Transmission of TSEs between and within species usually encounters a species or polymorphism barrier that is largely determined by the PrP amino acid sequence of both the donor and recipient. For sheep, over 55 PrP polymorphisms have been described (7), of which several have been associated with a particular disease phenotype (8–10). Most important are the variants having a polymorphism at codon 136 (alanine [A] to valine [V]), usually designated VRQ, which has been associated with a high risk of disease and short survival times (sheep PrP alleles are indicated by amino acids in single-letter code at positions 136, 154, and 171 only). In contrast, the ARR allele having a glutamine (Q)-to-arginine (R) polymorphism at codon 171 renders sheep almost resistant to most kinds of (experimental) TSEs. The wild type (ARQ) is considered intermediately susceptible to several forms of scrapie but seems thus far one of the most sensitive variants for experimental BSE transmissions (11). We have shown earlier that mutations in either PrPC or PrPSc can directly determine the rate of conversion of PrPC into PrPSc, reflecting the observed susceptibility and transmissibility of sheep scrapie in vivo (12, 13). Even though the mechanisms behind ovine prion protein conversion are still poorly understood, PrP self-interaction domains and polymorphic regions involved in a proposed molecular switch correlate with the impact polymorphisms have on prion protein conversion (14, 15). Surprisingly, there seems to be no measurable effect on the primary interaction between PrPC and PrPSc molecules (16, 17).
BSE from cattle can be transmitted to a variety of species, including ruminants, mice, and humans (10, 11, 18–20). Typical for BSE during cross-species transmission are the conservation of some key characteristics that can be measured in mouse strain-typing studies or by molecular phenotyping (mouse brain lesion profiles and the typical molecular weight and predominant diglycosylated species on Western blots). These signatures usually remain preserved for several passages in foreign species (21). Based on the typical signatures and epidemiological data, the occurrence of a novel variant of CJD in humans was linked to BSE (22). While no “natural” BSE has been found in sheep thus far, BSE in goats in two isolated cases has been found in France and the United Kingdom recently (23–25).
Contrasting natural scrapie in small ruminants, BSE in cattle is predominantly restricted to the central nervous system (26, 27), while ovine BSE efficiently spreads through lymphoid tissues, similar to sheep scrapie and variant CJD in humans (2, 28–30). This indicator of increased pathogenicity was underlined by studies of transmission of ovine BSE into transgenic mice expressing bovine, porcine, elk, or human PrP (31–33). Whereas in our view, pathogenicity reflects the ability to cause disease in host organisms and/or specific organs, the transmission into known less susceptible “species” like sheep ARR indicates that ovine BSE exhibits different virulence properties. Even though ovine ARQ BSE is transmitted more efficiently than classical cow BSE into several lines of transgenic mice, the underlying quantitative molecular mechanism of cow BSE adapting to sheep PrP variants and simultaneously increasing its potency for many PrP variants and species is still poorly understood.
Several types of cell-free conversion reactions have been developed in model species like the hamster and mouse that allow the investigation of PrP conversion and/or sensitive detection in diagnostic assays (34–38). In some of these systems, it was demonstrated that these can be used for assessing species and polymorphism barriers in nonmodel species like cattle and sheep (12, 13, 39–41). Moreover, newer systems have been developed with the primary aim of ultrasensitive species-independent diagnostic detection of TSEs in, for instance, blood and animal products (42–46). All these systems use the discriminative power of proteinase K, where PrPC is entirely protease sensitive while a large fragment of PrPSc is protease resistant (PrPres). The tissue homogenate-driven conversion, or protein misfolding cyclic amplification (PMCA), assay has proven itself to be a very effective conversion system that allowed the ultrasensitive detection of prions (34, 47). PMCA has also been used to assess the hamster and mouse model species barrier, the barriers limiting transmission of various CWD (chronic wasting disease) strains, and the barriers involved in human CJD (48–52). Thus far, PMCA is the only published conversion reaction that allows the detectable amplification of “classical TSE-associated” infectivity (43, 52). PMCA is based on the conversion of large quantities of PrPC by minute quantities of PrPSc—both isolated from brain tissues—generating more aggregated PrPSc. These aggregates are subsequently repeatedly sheared by sonication to generate more seeds, allowing subsequent exponential amplification of the PrP (47).
In this study, we demonstrated that protein misfolding cyclic amplification (PMCA) using a variety of PrP sources can be used to quantify and study strain, species, and polymorphism barriers at the molecular level. We assessed these barriers within and between ruminant species in vitro using a variety of experimentally generated cross-species BSE agents. These included not only cow BSE and ovBSE-ARQ (ovine BSE derived from sheep having the ARQ/ARQ PrP genotype) but also two unique BSE-derived variants: one isolate of BSE agent passaged in VRQ/VRQ sheep and one ovBSE-ARQ isolate that was back-transmitted into cattle (its original host).
MATERIALS AND METHODS
Ethic statement.
All animal experiments have been performed in compliance with our institutional and Dutch national guidelines, in accordance with European Community Council Directive 86/609/EEC. Any experimental protocol was approved by the WUR-ASG ethics committee.
Animals and brain tissues.
Confirmed scrapie-negative materials of various PrP genotypes were obtained from a well-defined scrapie-free flock kept on the Central Veterinary Institute of Wageningen UR (CVI) premises. Confirmed BSE-negative bovine materials were obtained individually from a healthy pool of control animals. Directly after euthanization, the brain materials (mainly brain stem) were washed in ice-cold phosphate-buffered saline (PBS) to remove possible blood clots and subsequently snap-frozen in liquid nitrogen and stored at −80°C until use.
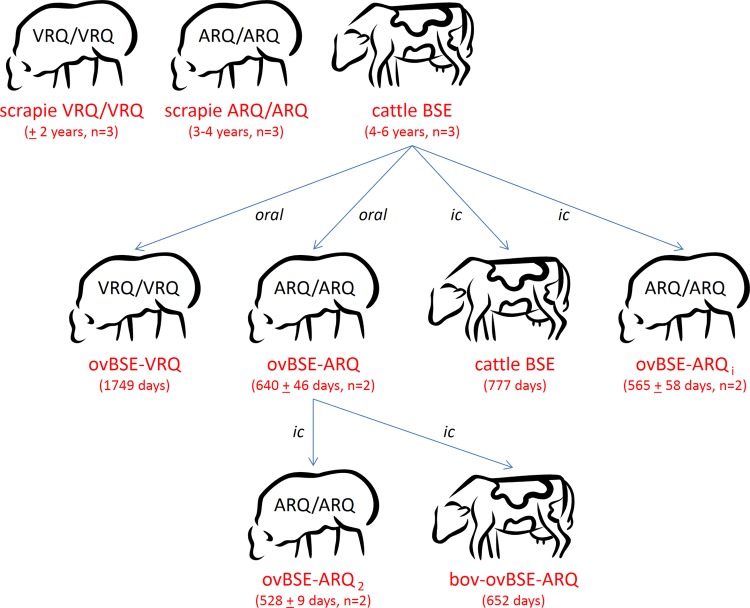
Classical BSE and scrapie materials were derived from various natural Dutch or United Kingdom cases. A sheep-derived BSE agent (ovBSE-ARQ) was obtained from three individual ARQ/ARQ sheep of which two were orally and one was intracerebrally (i.c.) inoculated with classical BSE agent in the context of European BSE transmission studies (53, 54). In addition, a VRQ-derived ovine BSE (ovBSE-VRQ) agent was obtained from a single clinical VRQ/VRQ sheep after oral inoculation with classical cow BSE agent (1,749-day incubation time). The second rare isolate, a BSE agent from a cow challenged with ovBSE-ARQ (bov-ovBSE-ARQ), was obtained from a clinically affected animal 652 days after intracerebral injection. TSE-positive brain tissues (mainly brain stem and cerebrum) were directly stored at −20°C and lower. Because of the complexity of this set of sheep and cow TSE isolates, Fig. 1 was included to assist in understanding the conditions and incubation times for the used samples from ruminant experiments.
FIG 1.
Origins of the various TSE-positive isolates used within this study (naturally acquired or generated). The top line represents the three “natural” TSE agent types that we started from (natural scrapie from either VRQ/VRQ ([n = 3] or ARQ/ARQ [n = 3] sheep or cattle BSE [n = 3]). Their respective expected incubation times under natural conditions are specified in years. For cow BSE and BSE-derived isolates, several experimental passages to cattle and sheep (either ARQ/ARQ or VRQ/VRQ) have been performed by various transmission routes (intracerebral or oral). Their averaged incubation times are given in days (±standard deviation) after inoculation until clinical disease. Data for oral transmission of cow BSE to ARQ/ARQ sheep are from Thuring et al. (53). Other transmissions are new (L. J. M. van Keulen, unpublished data).
All negative and positive isolates were fully PrP genotyped by Sanger sequencing of the entire open reading frame using routinely established methods.
Preparation of tissue homogenates.
Brain homogenates (10% [wt/vol]) were prepared in homogenization buffer (PBS containing 150 mM NaCl, 1% Triton X-100 [pH 7.4], and complete protease inhibitor cocktail with EDTA [Roche; final concentration, EDTA 1 mM]). Briefly, 0.5 g of positive or negative brain tissue was macerated and homogenized with 5 ml of ice-cold homogenization buffer for 30 to 60 s on ice at medium speed with Omnitip plastic disposable rotor stator generator probes. The homogenates were then precleared by low-speed centrifugation (440 × g for 2 min). BSE-negative bovine homogenates were precleared by low-speed centrifugation for 10 min at 4°C to clear large debris and to minimize nonspecific reactions. Supernatants of the negative and positive homogenates were frozen as small aliquots at −80°C; subsequent freezing and thawing cycles were avoided prior to PMCA. PrPres levels in TSE samples were checked after digestion with proteinase K (PK) by Western blotting (see below for digestion and Western blotting procedures).
PMCA procedure.
PMCA was performed initially as described by Morales et al. (55) with some adjustments. Briefly, PMCA samples consisted of equal volumes of negative isolates (PrPC containing substrate) and TSE-positive isolates (inoculum). Concentrations of PrPres in the various TSE inocula were matched by diluting homogenates in PBS containing 1% (wt/vol) Triton X-100 to such a level that the PrPres signal was just visible and quantifiable on a Western blot. Conversion experiments were performed with 50 μl of these mixture samples in 0.2-ml PCR vials (Nunc; 230895). Usually inocula were diluted at least 10-fold before mixing, but where necessary for sufficiently accurate quantifications, additional less diluted fractions were also analyzed up to 2-fold dilutions.
For PMCA of samples, vials were immersed in the sonicator water bath and subjected to 48 sonication cycles during a 24-h period. Each PMCA cycle consisted of 29 min of incubation at 37°C and 1 min of sonication at 70% of sonicator power output, 200 to 204 W (S-3000; Misonix, Farmingdale, NY). To avoid systemic errors between similar experiments, the order of sample position in the water bath was systematically varied.
Nonamplified control aliquots were always taken, directly briefly centrifuged, and frozen at −80°C to be used as a reference for starter PrPres content. After PMCA, sonicated samples were centrifuged briefly and frozen at −80°C or directly subjected to PK digestion (100 μg/ml for 60 min at 37°C). The reactions were stopped by adding 30 μl of inhibitors (Pefabloc, Roche) and 25 μl of loading buffer (LDS4x, no reducing agent added; Invitrogen), and then samples were heated at 96°C for 10 min and briefly centrifuged prior to Western blotting.
Western blotting.
Samples were separated on 12% bis-tris NuPAGE gels (Invitrogen), transferred to polyvinylidene difluoride (PVDF) membranes, and subsequently immunostained using monoclonal antibody 9A2 (56) at 0.5 μg of IgG/ml. PrPres signal development was performed with rabbit anti-mouse IgG-alkaline phosphatase conjugate and fluorescent ECF substrate (Molecular Dynamics). The 9A2 antibody was quantified by Western blotting to react equally well with a variety of PrP species—sheep, cattle, and cervid—and different prions, including scrapie, BSE, and CWD agents. The fluorescent signal was visualized after a maximum of 30 min of substrate incubation on a Typhoon Trio imager (Molecular Dynamics) and quantified with ImageQuant 5.2 software (Molecular Dynamics).
PMCA quantification and statistical analysis.
PMCA-induced conversion efficiencies were calculated from the quantified signals in the molecular mass range of 21 to 26 kDa as ratios of sonicated divided by the corresponding directly frozen control samples. To emphasize the relative conversion efficiencies, conversion ratios were normalized within each series against the averaged homologous conversion reaction. A series/profile of experiments is defined as the complete panel of PrPC variants (VRQ, ARQ, ARR, and bovine [bov]) used for each individual PrPSc isolate. Statistically significant differences between conversion efficiencies were determined by the double-sided (unpaired) Welch-corrected Student t test. Possible linear correlation between sets of conversions were assessed by the Pearson product-moment correlation efficient and tested for significance using the nondirectional t-distribution (N − 2, where N is the number of data points).
RESULTS
We adapted the PMCA technique into a semiquantitative reaction for sheep- and cow-derived materials, thereby allowing the system to assess species and polymorphism barriers under in vitro conditions. To quantify polymorphism and ovine and bovine species barriers, we performed our studies in the still-linear kinetic range of a one-round PMCA reaction (contrasting serial PMCA used to detect ultrasmall amounts of PrPSc). PMCA-obtained conversion efficiencies, which follow an entirely different principle from that of denatured cell-free conversion reactions, confirmed and extended established sheep scrapie conversion rates and reflected the observed survival times of cattle and sheep suffering from BSE and natural scrapie, respectively. Ovine BSE contrasted these “natural” isolates by exposing an unequivocal increase in conversion potency and was able to convert less-susceptible PrP variants to large extents.
One-round PMCA of natural TSE sources.
To investigate the potential of (one-round) PMCA for quantification of conversion barriers, we used TSE-positive materials from naturally infected animals, which were scrapie-infected sheep with ARQ/ARQ and VRQ/VRQ PrP genotypes and BSE-infected cattle identified at routine surveillance. Healthy control animals were sourced from histopathologically confirmed negative sheep and cattle. Initial amplification experiments under standard conditions of serial PMCA (eight rounds of subsequent PMCA reactions) revealed high background and/or spontaneous conversion of PrPC from brain homogenates into protease-resistant PrP isoforms different from the characteristic PrPSc molecular profiles (data not shown). We therefore optimized and standardized the procedure (preparation of the homogenates, sonication conditions, tubes, and temperature control) as well as the collection of fresh substrates. Using these conditions we could detect highly specific conversion reactions (characteristic PrPres migration in the 19- to 30-kDa molecular mass region and the typical three PrP glycoform variants (di-, mono-, and nonglycosylated) by incubation both with and without sonication (Fig. 2A). PK conditions were standardized such that all input PrPC was completely digested. Undigested bands would be readily visible as bands with slower migration (corresponding to 3 to 4 kDa larger than PrPres). The sonicated reactions were more efficient than the incubation-only reactions for a variety of tested PrPSc and PrPC combinations. Thus, VRQ PrPC converted best in PMCA, around 10-fold, while with incubation alone, an approximately 4- to 5-fold maximum could be achieved. Under these conditions, bovine PrPC achieved roughly 1.5- and 6-fold conversion ratios, respectively, for incubated and sonicated conditions (Fig. 2A, incubated versus sonicated). Differences in the amounts of host PrPC present in the negative brain homogenates were minimal (as assessed by Western blotting), and dilution series of PrPC only revealed a linear decline in absolute conversion ratios while maintaining its specific profile of conversion over different PrPC variants (data not shown).
FIG 2.
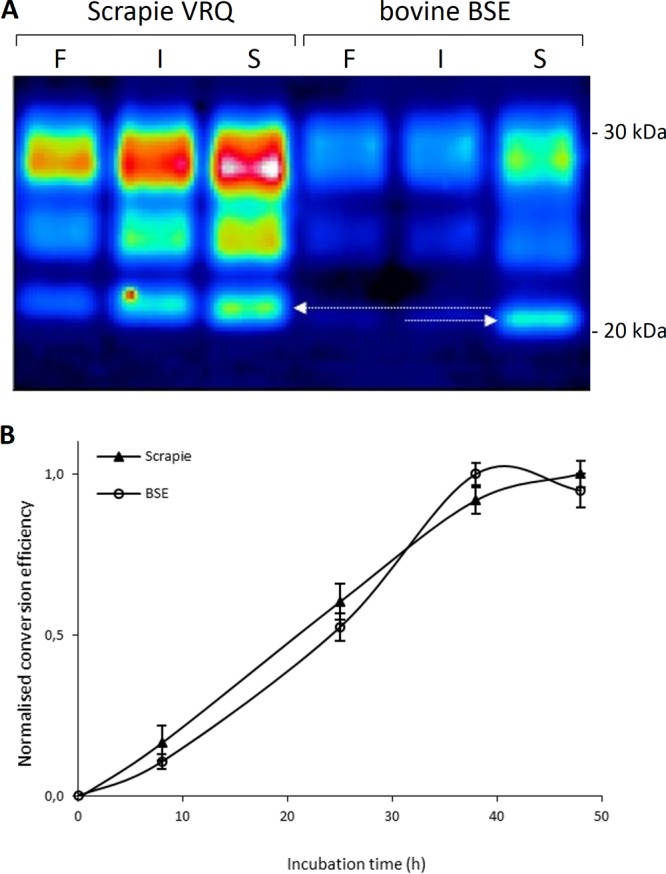
Homologous PMCA conversion reactions of ovine scrapie (VRQ/VRQ) and BSE. (A) A representative Western blot of the typical three banding patterns (di-, mono-, or nonglycosylated PrP) is shown in pseudointensity colors after PK digestion. Detection was performed with monoclonal antibody 9A2. Arrows indicate the molecular mass difference between sheep scrapie and cow BSE which is most apparent in the nonglycosylated PrP fraction (71). Lanes before treatment (frozen fraction [F]), after incubation without sonication (only incubated and not sonicated [I]), and after PMCA (incubated and sonicated [S]) are indicated. (B) Incubation time curves of homologous PMCA reactions as shown in panel A. Conversion efficiencies are indicated on a normalized scale. Linear regression lines for scrapie and BSE had R2 of 0.98 and 0.95, respectively (including the last measurement near its plateau). Excluding the last measurements at 48 h results in R2 of 1.00 and 0.97 for scrapie and BSE, respectively.
Obvious was the reproduction of the 1.5-kDa molecular mass difference of the nonglycosylated fraction of PrPres between scrapie and BSE as reported (arrows in Fig. 2A). Negative-control reactions (TSE-positive inoculum was replaced with negative brain homogenate of the same PrP genetic background) were always negative under the currently used conditions, indicating that no detectable spontaneous PrP conversion occurred. The inverse experiment, in which the negative brain homogenate was replaced either with conversion buffer or with PrP knockout mouse brain homogenate, also did not result in the increase of PrPres signal on a Western blot, excluding an impeded PK reactivity due to, e.g., aggregates newly formed in the sonication process.
Even though conversion ratios in single-round PMCA were relatively low, using the same conditions in serial PMCA, we could reproduce conversion efficiencies for hamster PrP making around 108-fold dilutions positive in only 4 rounds, while 106-fold dilutions could be reproducibly obtained positive for sheep scrapie VRQ-homologous reactions in only 5 rounds (data not shown).
Time course series of efficient homologous conversions (VRQ with VRQ or BSE with bovine) in single-round PMCA demonstrated that saturation of the reactions due to, for instance, depletion of PrPC occurred only after 40 h (Fig. 2B), with near linear correlation between time and conversion efficiency in the 0- to 40-h range (linear regression coefficient [R2] better than 0.97). Therefore, in this study, our PMCA experiments were standardized at 24 h to stay amply in the linear range, allowing adequate quantification of conversion reactions.
Species and polymorphism specificity in natural TSE isolates.
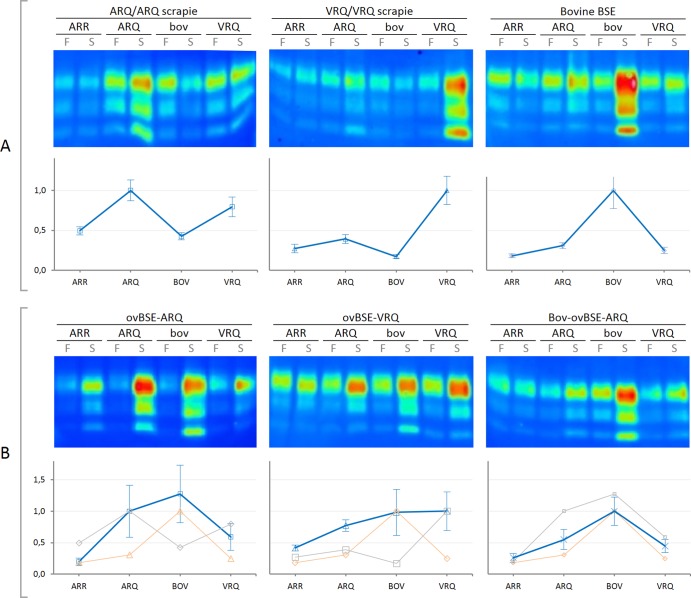
As PrP substrates, TSE-negative brain tissue from sheep homozygous for the ARR, ARQ, and VRQ PrP alleles and from cattle (6 octarepeats) were used throughout the studies. Quantification of the results (Fig. 3A) confirmed previous observations in guanidine-based cell-free conversions that in general, homologous conversion reactions were the most efficient reactions (12, 13, 41). In addition, we confirmed the apparent more conversion-resistant variant ARR by each of the natural TSE-positive materials.
FIG 3.
One-round PMCA conversion reactions of various ovine and bovine isolates. Shown are PMCA reactions of bovine (bov) and ovine (ARQ, VRQ, and ARR) PrP variants for conversion series using various TSE agent types (each of the six panels). All reactions were performed in triplicate for each PrPC, and each of these triplicates was carried out on three different PrPSc isolates (thus leading to a total of 9 analyses) for scrapie and cow BSE, duplicate for ovBSE-ARQ (n = 6), and single for both ovBSE-VRQ and bov-ovBSE-ARQ (n = 3). Panel A shows conversion results induced by natural sheep scrapie (VRQ/VRQ and ARQ/ARQ) and cow BSE agent isolates, while panel B displays conversion results of experimental sheep-derived BSE (ovBSE-ARQ or ovBSE-VRQ), as well as a unique ovBSE-ARQ isolate back-transmitted into cattle (bov-ovBSE-ARQ). Within each of the six sets of images the top portion shows a representative Western blot of the single-round PMCA prior to (frozen fraction [F]) or after (sonicated [S]) PMCA. The bottom portion of each set of images shows the corresponding normalized data as a relative conversion profile where the homologous (usually most efficient) reaction was set at unity and each point represents average value ± standard error of the mean. For reference, the cow BSE-induced profile is shown in orange, while for each B graph the gray line shows the corresponding profile of sheep scrapie agent isolate conversions of panel A.
Each of the conversion reactions was repeated for at least three independent animal-derived substrates to incorporate possible animal-animal variation as has been observed previously. For all natural TSE variants (ovine VRQ/VRQ and ARQ/ARQ scrapie and cow BSE) and ovBSE-ARQ, at least three animal-independent isolates per TSE-positive type have been used to drive the conversions (total, n = 9).
All heterologous conversion reactions (Fig. 3A) were significantly different (P < 0.01) from their homologous counterparts, except for ARQ PrPSc in combination with VRQ PrPC (P = 0.27). This relatively high conversion rate of VRQ is in agreement with earlier guanidine-hydrochloride (Gdn-HCl) denaturation-based cell-free conversion studies (Fig. 4). Overall, Fig. 3 displays that each type of PrPSc (except bov-ovBSE-ARQ) generated a quite distinct and unique conversion profile. Each profile was reproducible for various PrPC and PrPSc isolates and reflected within each set of PrPSc conversions the known biological parameters of classical TSE susceptibility and transmission studies. For example, it is known that transmission to ARR-homozygous sheep is limited for both classical scrapie as well as BSE, which is reflected with a relative low conversion rate. In addition to the most efficient homologous reactions, cow BSE converts ovine ARQ PrPC efficiently, albeit far less efficient than bovine PrPC.
FIG 4.
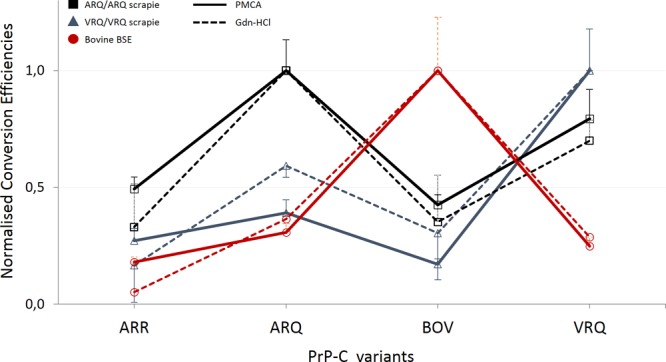
Correlation of the two in vitro PrP conversion systems. Conversion efficiencies are shown on a relative scale of the Gdn-HCl-based or the PMCA conversion system. A clear correlation exists between the Gdn-HCl and PMCA system for both scrapie types as well as cow BSE. Data for the scrapie-driven Gdn-HCl system came from Bossers et al. (13). Every measurement consisted of at least three independent conversion reactions.
Ovine BSE-driven conversions.
Surprisingly, altered convertibility profiles were observed for ARQ/ARQ sheep-derived BSE (ovBSE-ARQ) (Fig. 1), which converted sheep ARQ and bovine PrPC equally well (compare the first image in Fig. 3B with that in Fig. 3A). The conversion profile seems to represent a merger of the susceptible features of these profiles of both sheep scrapie ARQ and cow BSE (reflected by the highly efficient conversions of ARQ as well as bovine PrPC), while also showing an increased but relative limited conversion of ARR. The conversion in absolute efficiencies of ARR by ovBSE-ARQ was higher than observed for scrapie ARQ/ARQ or cow BSE (Fig. 3). The ovBSE-ARQ-driven relative conversion efficiency of bovine PrPC was significantly different (P < 0.05) from a sheep scrapie-driven conversion. Furthermore, the bovine BSE-driven conversions of sheep ARQ PrPC were significantly different from the conversions driven by ovBSE-ARQ (P < 0.001). It should be noted that there was considerable difference between the absolute conversion efficiencies induced by the two different ARQ-derived ovBSE variants (from oral inoculation). However, after normalization, relative conversion efficiencies were highly reproducible for all measured PrPC variants within one substrate panel.
For study of transmission of cow BSE to VRQ/VRQ sheep, only a single animal TSE isolate was available. To compensate for experimental variation, three different homogenates from the same tissue source were used for PMCA experiments on the four PrPC variants (therefore, n = 3 per PrPC variant). Compared to VRQ/VRQ scrapie, ovBSE-VRQ (Fig. 1) also yielded an altered conversion profile (Fig. 3B, second image). In addition, when comparing the two types of ovBSE agent variants (ovBSE-ARQ and ovBSE-VRQ), the genetic background of the sheep—where the ovBSE agent originated from (ARQ versus VRQ, respectively)—also appeared to have its effect on the molecular conversion profile, respectively, driving either the ARQ or VRQ PrPC reaction more efficiently (Fig. 3B, first or second image, respectively). Both variants, however, exerted a strikingly efficient conversion of bovine PrPC as well contrasting them from the classical scrapie variants.
PMCA of BSE agent variants from sheep and cow transmission and cow back-transmission.
Within this study, a variety of sheep- and cow-derived BSE agent isolates were used (Fig. 1). These isolates and strains include “naturally occurring” cow BSE agent isolates as well as some rare experimentally generated ovBSE agent variants. In addition, it was assessed what would happen to the conversion profiles for ovBSE-ARQ when back-transmitted into cattle (bov-ovBSE-ARQ) compared to a cow BSE agent from i.c. challenged animals (Fig. 1). Even though only a single transmission could be performed for both isolates, the incubation period was slightly shorter than that of a transmission with cow BSE within the same experimental setup (652 days compared to 777 days, respectively). From these two BSE back-into-cattle transmissions, three different homogenates from the same isolate were investigated for PMCA combined with the various PrPC variants (totals, n = 3 per variant on the animal level but n = 9 on the total experimental level). Natural and i.c. challenged cow BSE produced similar conversion profiles (data not shown), but the conversion profile of bov-ovBSE-ARQ (Fig. 3B, third image) was different from that of ovBSE-ARQ as well as the cow BSE agent variant. It thus appeared that the profile of bov-ovBSE-ARQ restored features of the natural cow BSE profile. This was also reflected by the statistics in which the bov-ovBSE-ARQ variant was not significantly different from either ovBSE-ARQ or the cow BSE agent for any of the tested PrPC variants (P > 0.1 for all). The conversion profile of bov-ovBSE-ARQ correlated best with bovBSE (Pearson coefficient [r] = 0.967; P < 0.001) but also with ovBSE-ARQ (r = 0.927; P < 0.01), while all other conversion profiles did not show any clear correlation (r < 0.8; P > 0.1).
Conversion potency of ovine BSE.
The relative (normalized) conversion profiles demonstrated the differential convertibility of various PrPC variants for each TSE agent type. However, the ovBSE-ARQ variant was the only TSE agent type having a significant increase in potency to convert almost any sheep and cattle PrPC variant (albeit limited for the less susceptible ARR PrPC). Therefore, all the conversion ratios of each PrPC variant (including ARR) per type of PrPSc used were combined in a whisker plot (Fig. 5). From Fig. 5 it is clear that the three independent ovBSE-ARQ isolates demonstrated a high potency to convert any PrPC. This is significantly different for any comparison of ovBSE-ARQ with any of the other PrPSc isolates (P < 0.001), even taking into account the large spread in the data due to individual PrPSc isolate differences and due to the inclusion of the poorly converting ARR variant.
FIG 5.
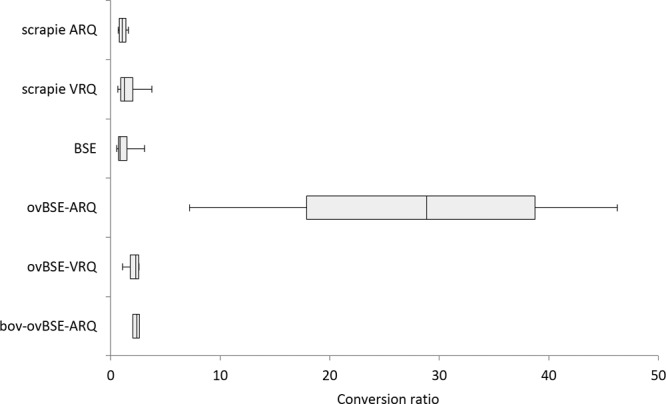
Whisker plot demonstrating the high conversion rates obtained using ovBSE-ARQ compared to all other TSE agent types. Indicated are the median nonnormalized conversion ratios of all PrPC variants (VRQ, ARQ, and bovine but also ARR) subjected to conversion by a particular TSE agent isolate type (vertical axis).
DISCUSSION
PMCA has a different conversion principle than denatured cell-free conversion (thereby allowing potential prion adaptation). It quantitatively confirms some of the earlier established PrP conversion profiles of classical sheep scrapie and cow BSE in vitro. These profiles correlate with known in vivo transmissibility and susceptibility in the two ruminant species. Strain-specific molecular signatures, like its molecular weight after protease digestion, were maintained. Furthermore, both ovine BSE agent isolates from ARQ and VRQ sheep demonstrated a conversion profile in which the efficient transmissions to both sheep and bovine PrP variants were maintained. Our data reveal that ovBSE-ARQ displays a significant increase in virulence as reflected by an increased conversion potency for all PrPC allelic variants studied (ovine VRQ, ARQ, and ARR as well as bovine PrP).
The observed increased conversion efficiency of the ARQ-derived ovine BSE agent in a variety of sheep PrP genotypes and cattle recipients is in agreement with data for BSE transmissions into bovinised, porcinized, or humanized PrP transgenic mice (31, 32, 57–59). These bioassay data show that sheep-passaged BSE can transmit efficiently in bovinized mice with stable incubation times between serial passages (31) and can be more readily transmitted to human transgenic mice (32, 57). Our data clearly add that the ARQ-derived ovine BSE transmits to other hosts (in our case, cattle) with efficiencies similar to that of within-species transmissions. This increased efficiency seems comparable to the measured effect that sheep-derived BSE agent titers were comparable as determined in two different hosts, ARQ sheep and RIII mice (60). Finally, the conversion of the sheep ARR PrPC was also significantly increased (Fig. 3B and 5), which seems in agreement with efficient intracerebral and intraperitoneal infections of ARR/ARR sheep with the ovBSE agent (60). Furthermore, similar observations can be made from transmissions depicted in Fig. 1, where the incubation period of bovine BSE to cattle tended to be longer than the back-transmission of ARQ-derived ovine BSE into cattle. Even though that these transmission data need to be considered with care due to the single animal examples used in these expensive studies, they display the same trend of loss of virulence after back-transmission into cattle (Fig. 3B and Fig. 5). This is in agreement with data for ovine BSE agent passage directly in transgenic humanized mice compared with data for first passing it through bovinized transgenic mice, restoring the human species barrier (57). Probably the primary bovine amino acid sequence of PrP directs the ovine BSE strain back into the classical BSE conformer and/or the bovine PrP sequence somehow specifically selects from a quasispecies mixture of conformers of the classical BSE agent type. It is probable that host factors other than the PrP structure play a role in such selection.
It is generally accepted that prion protein conversion studies, similar to transgenic-mouse studies, resemble the trend of incubation periods in intracerebral inoculations, and that for sheep (and some other species) they also seem to be biologically relevant for oral and natural transmission routes (61, 62). Even though PrPSc accumulation in the brain is considered the prime indicator for successful TSE agent spread, other peripheral organs like the liver, spleen, and gut can also significantly contribute to agent spread (2, 59, 63). In this study, all materials had a natural origin or were experimentally generated by oral or intracerebral inoculation. We were also able to generate some preliminary PMCA data demonstrating the absence of potential differences in infection routes as well as the absence of a potential difference in primary versus serial passages of ovine BSE. These isolates, a serial passage of an ARQ-derived ovine BSE agent (designated ovBSE-ARQ_2) and a primary passage of a cow BSE agent to ARQ/ARQ sheep by the intracerebral instead of the oral route designated ovBSE-ARQ_i (Fig. 1), gave conversion profiles similar to those of the materials coming from primary oral passage of BSE into ARQ/ARQ sheep (data not shown). Neither of these two isolates was by PMCA thus far statistically significantly different in Pearson correlation tests from the orally transmitted BSE agent. This could be expected since it has been demonstrated that specific diagnostic features characteristic for BSE in sheep remain conserved over at least three serial passages in sheep (21).
We observed the preservation of the specific molecular weight signatures of the isolates but noticed that for BSE-derived isolates, the PMCA was slightly more efficient in nonglycosylated species. This is probably a characteristic feature for BSE transmission in heterologous hosts itself rather than an in vitro artifact, since transmission experiments using BSE-derived strains in transgenic mice also favored less glycosylated PrP isoforms (58).
In vitro cell-free conversion efficiencies have been demonstrated to inversely correlate with natural or experimental scrapie or BSE transmission incubation times in sheep of homozygous or heterozygous PrP genotypes (64, 65). Extrapolating relative susceptibilities to homozygous counterparts from these measurements using heterozygous animals might be risky since one allelic variant might contribute more or less significantly to the conversion process. This is demonstrated by, for instance, experimental transmission studies of VRQ scrapie (SSBP/1) to VRQ/VRQ, VRQ/ARQ, VRQ/ARR, ARQ/ARQ, and ARR/ARR animals where all VRQ carriers succumb to disease with a clear inverse correlation and a short (170 to 364 days) incubation time (9). However, the homozygous counterparts of the susceptible heterozygous animals (ARQ/ARQ for VRQ/ARQ and the ARR/ARR for VRQ/ARR) did not succumb to disease at all at incubation times exceeding 1,200 days, suggesting a complex interplay of each of the allelic contributions in the PrP conversion and disease process, where it has been demonstrated that PrP heterozygosity by itself can even be a protective factor (17, 66). In another study, it was also demonstrated that transmission of ARQ-derived scrapie has different kinetics than VRQ-derived scrapie when transmitted to either homozygous or heterozygous (ARQ or VRQ) recipients (67). In PrPres ARQ/ARR sheep with natural scrapie or experimental BSE PrPres, the ARQ/ARR ratio appeared differently expressed, indicating that strain differences can influence allelic involvement in prion formation (68). Not every PrP allelic variant-negative substrate can be freshly stored routinely mostly due to their rare presence in the population, thereby restricting studies involving many (rare) PrP allelic variants. For such studies, alternate sources from, for instance, transgenic mice or transformed cell lines might be more beneficial (study in progress).
Within this study, we tried to fine-tune parameters and use a PrP conversion system that generates physiologically relevant data. Incubation time series ensured that measurements were performed under conditions that allowed the assessment of conversion efficiency differences without the risk of complete substrate depletion or reaction saturation. These conditions were, however, not the conditions that produced the highest conversion ratios. Higher conversion ratios could be obtained by adding, for instance, poly(A) to the reaction mixture (45). In our experiments, we found that adding poly(A) completely abolished species and polymorphism specificity (data not shown). Probably it modifies conditions such that they resemble conditions, for instance, similar to the conditions of the QuIC PrP conversion system that is deliberately deprived of species and polymorphism specificity (38, 44). The removal of any strain and species specificity is useful for sensitive routine diagnostics but not for species barrier measurements, for which such specificity should be retained.
Another way to significantly improve the conversion efficiency (up to 106-fold improvement) is by using serial PMCA (sPMCA) (69). We were also able to increase the absolute conversion ratio of the current system to allow detection of PrPSc after at least 106-fold dilutions of sheep scrapie inoculum. Even though we did see species specificity being preserved when using sPMCA over at least 3 rounds, relative conversion ratios altered after more than 4 or 5 rounds in sPMCA, especially for the combinations of TSE donor and recipient that are known to allow some adaptation in vivo during experimental serial transmission of isolates in transgenic mice (subject of future studies). This potential adaptation within multiple rounds of PMCA opens up experiments to study biologically relevant TSE adaptation in vitro, pushing assessments to the very limit. This adaptation has been demonstrated to occur, for instance, for hamster and mouse as well as for chronic wasting disease prions. In the last case, adaptation for certain types of TSE donor and recipient combinations only required three to six rounds of PMCA (52, 70). Future studies of sPMCA utilizing the “resistant” ARR PrP allelic variant of sheep that is currently used to genetically control natural scrapie in sheep in Europe might elucidate whether some TSE agent might be more prone to adapt to this particular PrP genotype in sheep.
Our study focused primarily on readily transmittable classical TSE agent types for both the natural scrapie from sheep and BSE agent isolates from cattle. The nonnatural isolates we used are the ones originating from the experimental transmissions as described for Fig. 1. Future studies should assess the effects of, for instance, the recently discovered L-type or H-type forms of BSE. Due to the limited availability of such materials, and the costs to generate such materials experimentally, only some preliminary data could be generated for two atypical scrapie agent isolates (data not shown). One isolate from an ARR/ARR (so-called resistant) sheep and two isolates from atypical scrapie type Nor98 (atypical scrapie is usually limited to single index cases in flocks, contrasting classical scrapie transmissions). The ARR scrapie-driven conversion reactions were very inefficient, and the generated conversion profile (that included additional concentration steps by centrifugation) resembled mostly that of ARQ scrapie. The preliminary results of the Nor98-driven conversion reactions were initially all negative for the standardize conditions we have used in our system as described in the current paper; however, by significantly lowering the PK conditions, we could generate some preliminary data for the conversion of mainly those variants that are associated with Nor98 susceptibility (namely, the sheep PrP ARQ and AHQ alleles). The AHQ allelic variant was not further assessed in this study, but previous conversion studies demonstrated an inefficient conversion of AHQ using classical scrapie agent isolates, albeit not as inefficient as that with the resistance-associated ARR PrP allelic variant (13). Since we needed to modify the conversion and analysis conditions at several points, the data should be interpreted with care since no full correlation study of atypical scrapie has been performed using those new conditions yet.
Since sheep ARR was limitedly, but clearly, converted by the ovine BSE agent (Fig. 3B, first image, lanes 1 and 2), it remains a critical question what impact these slightly elevated ARR conversion rates have on, for instance, scrapie control programs based on selective breeding for ARR. Even though PMCA using various ovine and bovine isolates did clearly demonstrate that biologically relevant conversion and/or adaptation data were generated, it still reflects at best experimental intracerebral transmissions, which are generally far more efficient than oral transmissions or transmissions under natural conditions. Under natural conditions, the limited conversion of ARR might easily prolong the incubation time beyond the normal commercial life span of sheep. Furthermore, it might be expected that the limited conversion of ARR PrP will reflect only a limited susceptibility under field conditions, even for a potent BSE strain in sheep. Whether and when BSE might transmit spontaneously to sheep remains to be established, as well as the potential impact of such a transmission for human health from animal and animal-derived products. The level of spread of the agent through the periphery and excreta also are determinants of spread of infection in the field. It remains to be seen how virulent the BSE materials would be in another small-ruminant species, namely, goat, as reported in the few field BSE cases in small ruminants (23, 24, 25).
ACKNOWLEDGMENTS
We thank Alice Wiefferink and Ruth Bossers for technical assistance and Freddy de Bree for assistance with statistics.
This study was supported by European project “BSE in sheep” QLK3-CT-2002-01309 and EU project “goatBSE” FOOD-CT-2006-36353, as well as several grants from the Dutch Ministry of Economic Affairs (WOT-01-002-01).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Andréoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen L, Schelcher F, Elsen JM, Lantier F. 2000. Early accumulation of PrP(Sc) in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J. Gen. Virol. 81:3115–3126 http://vir.sgmjournals.org/content/81/12/3115 [DOI] [PubMed] [Google Scholar]
- 2.van Keulen LJ, Bossers A, van Zijderveld F. 2008. TSE pathogenesis in cattle and sheep. Vet. Res. 39:24. 10.1051/vetres:2007061 [DOI] [PubMed] [Google Scholar]
- 3.van Keulen LJ, Schreuder BE, Vromans ME, Langeveld JP, Smits MA. 2000. Pathogenesis of natural scrapie in sheep. Arch. Virol. Suppl. 2000(16):57–71 [DOI] [PubMed] [Google Scholar]
- 4.Caughey B, Baron GS, Chesebro B, Jeffrey M. 2009. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu. Rev. Biochem. 78:177–204. 10.1146/annurev.biochem.78.082907.145410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. 2012. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U. S. A. 109:E1938–E1946. 10.1073/pnas.1206999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Supattapone S. 2004. Prion protein conversion in vitro. J. Mol. Med. 82:348–356. 10.1007/s00109-004-0534-3 [DOI] [PubMed] [Google Scholar]
- 7.Hunter N, Bossers A. 2006. The PrP genotype as a marker for scrapie susceptibility in sheep, p 640–647 In Hörnlimann B, Riesner D, Kretzschmar H. (ed), Prions in humans and animals. de Gruyter, Berlin, Germany [Google Scholar]
- 8.Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA. 1996. PrP genotype contributes to determining survival times of sheep with natural scrapie. J. Gen. Virol. 77(Part 10):2669–2673 [DOI] [PubMed] [Google Scholar]
- 9.Goldmann W, Hunter N, Smith G, Foster J, Hope J. 1994. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 75(Part 5):989–995 [DOI] [PubMed] [Google Scholar]
- 10.Hunter N. 2003. Scrapie and experimental BSE in sheep. Br. Med. Bull. 66:171–183. 10.1093/bmb/66.1.171 [DOI] [PubMed] [Google Scholar]
- 11.Foster JD, Hope J, Fraser H. 1993. Transmission of bovine spongiform encephalopathy to sheep and goats. Vet. Rec. 133:339–341. 10.1136/vr.133.14.339 [DOI] [PubMed] [Google Scholar]
- 12.Bossers A, Belt P, Raymond GJ, Caughey B, de Vries R, Smits MA. 1997. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U. S. A. 94:4931–4936. 10.1073/pnas.94.10.4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossers A, de Vries R, Smits MA. 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 74:1407–1414. 10.1128/JVI.74.3.1407-1414.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigter A, Langeveld JP, Timmers-Parohi D, Jacobs JG, Moonen PL, Bossers A. 2007. Mapping of possible prion protein self-interaction domains using peptide arrays. BMC Biochem. 8:6. 10.1186/1471-2091-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Fernandez-Borges N, Schwarz P, Castilla J, Wuthrich K, Aguzzi A. 2010. A molecular switch controls interspecies prion disease transmission in mice. J. Clin. Invest. 120:2590–2599. 10.1172/JCI42051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigter A, Bossers A. 2005. Sheep scrapie susceptibility-linked polymorphisms do not modulate the initial binding of cellular to disease-associated prion protein prior to conversion. J. Gen. Virol. 86:2627–2634. 10.1099/vir.0.80901-0 [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi M, Priola SA, Chabry J, Caughey B. 2000. Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers. Proc. Natl. Acad. Sci. U. S. A. 97:5836–5841. 10.1073/pnas.110523897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellworthy SJ, Dexter G, Stack M, Chaplin M, Hawkins SA, Simmons MM, Jeffrey M, Martin S, Gonzalez L, Hill P. 2008. Oral transmission of BSE to VRQ/VRQ sheep in an experimental flock. Vet. Rec. 162:130–131. 10.1136/vr.162.4.130 [DOI] [PubMed] [Google Scholar]
- 19.Ridley RM, Baker HF. 1996. Oral transmission of BSE to primates. Lancet 348:1174. 10.1016/S0140-6736(05)65312-3 [DOI] [PubMed] [Google Scholar]
- 20.Béringue V, Andreoletti O, Le Dur A, Essalmani R, Vilotte JL, Lacroux C, Reine F, Herzog L, Biacabe AG, Baron T, Caramelli M, Casalone C, Laude H. 2007. A bovine prion acquires an epidemic bovine spongiform encephalopathy strain-like phenotype on interspecies transmission. J. Neurosci. 27:6965–6971. 10.1523/JNEUROSCI.0693-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stack M, Gonzalez L, Jeffrey M, Martin S, Macaldowie C, Chaplin M, Thorne J, Sayers R, Davis L, Bramwell J, Grimmer S, Bellworthy S. 2009. Three serial passages of bovine spongiform encephalopathy in sheep do not significantly affect discriminatory test results. J. Gen. Virol. 90:764–768. 10.1099/vir.0.005983-0 [DOI] [PubMed] [Google Scholar]
- 22.Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, Collinge J, Doey LJ, Lantos P. 1997. The same prion strain causes vCJD and BSE. Nature 389:448–450, 526 [DOI] [PubMed] [Google Scholar]
- 23.Spiropoulos J, Lockey R, Sallis RE, Terry LA, Thorne L, Holder TM, Beck KE, Simmons MM. 2011. Isolation of prion with BSE properties from farmed goat. Emerg. Infect. Dis. 17:2253–2261. 10.3201/eid1712.110333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eloit M, Adjou K, Coulpier M, Fontaine JJ, Hamel R, Lilin T, Messiaen S, Andreoletti O, Baron T, Bencsik A, Biacabe AG, Beringue V, Laude H, Le Dur A, Vilotte JL, Comoy E, Deslys JP, Grassi J, Simon S, Lantier F, Sarradin P. 2005. BSE agent signatures in a goat. Vet. Rec. 156:523–524 [DOI] [PubMed] [Google Scholar]
- 25.Jeffrey M, Martin S, Gonzalez L, Foster J, Langeveld JP, van Zijderveld FG, Grassi J, Hunter N. 2006. Immunohistochemical features of PrP(d) accumulation in natural and experimental goat transmissible spongiform encephalopathies. J. Comp. Pathol. 134:171–181. 10.1016/j.jcpa.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 26.Wells GA, Hawkins SA, Green RB, Austin AR, Dexter I, Spencer YI, Chaplin MJ, Stack MJ, Dawson M. 1998. Preliminary observations on the pathogenesis of experimental bovine spongiform encephalopathy (BSE): an update. Vet. Rec. 142:103–106. 10.1136/vr.142.5.103 [DOI] [PubMed] [Google Scholar]
- 27.Buschmann A, Groschup MH. 2005. Highly bovine spongiform encephalopathy-sensitive transgenic mice confirm the essential restriction of infectivity to the nervous system in clinically diseased cattle. J. Infect. Dis. 192:934–942. 10.1086/431602 [DOI] [PubMed] [Google Scholar]
- 28.Bellworthy SJ, Hawkins SA, Green RB, Blamire I, Dexter G, Dexter I, Lockey R, Jeffrey M, Ryder S, Berthelin-Baker C, Simmons MM. 2005. Tissue distribution of bovine spongiform encephalopathy infectivity in Romney sheep up to the onset of clinical disease after oral challenge. Vet. Rec. 156:197–202. 10.1136/vr.156.7.197 [DOI] [PubMed] [Google Scholar]
- 29.van Keulen LJ, Schreuder BE, Vromans ME, Langeveld JP, Smits MA. 1999. Scrapie-associated prion protein in the gastrointestinal tract of sheep with natural scrapie. J. Comp. Pathol. 121:55–63. 10.1053/jcpa.1998.0300 [DOI] [PubMed] [Google Scholar]
- 30.Bruce ME, McConnell I, Will RG, Ironside JW. 2001. Detection of variant Creutzfeldt-Jakob disease infectivity in extraneural tissues. Lancet 358:208–209. 10.1016/S0140-6736(01)05411-3 [DOI] [PubMed] [Google Scholar]
- 31.Espinosa JC, Andreoletti O, Castilla J, Herva ME, Morales M, Alamillo E, San-Segundo FD, Lacroux C, Lugan S, Salguero FJ, Langeveld J, Torres JM. 2007. Sheep-passaged bovine spongiform encephalopathy agent exhibits altered pathobiological properties in bovine-PrP transgenic mice. J. Virol. 81:835–843. 10.1128/JVI.01356-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plinston C, Hart P, Chong A, Hunter N, Foster J, Piccardo P, Manson JC, Barron RM. 2011. Increased susceptibility of human-PrP transgenic mice to bovine spongiform encephalopathy infection following passage in sheep. J. Virol. 85:1174–1181. 10.1128/JVI.01578-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamgüney G, Miller MW, Giles K, Lemus A, Glidden DV, DeArmond SJ, Prusiner SB. 2009. Transmission of scrapie and sheep-passaged bovine spongiform encephalopathy prions to transgenic mice expressing elk prion protein. J. Gen. Virol. 90:1035–1047. 10.1099/vir.0.007500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saborío GP, Soto C, Kascsak RJ, Levy E, Kascsak R, Harris DA, Frangione B. 1999. Cell-lysate conversion of prion protein into its protease-resistant isoform suggests the participation of a cellular chaperone. Biochem. Biophys. Res. Commun. 258:470–475. 10.1006/bbrc.1999.0660 [DOI] [PubMed] [Google Scholar]
- 35.Lucassen R, Nishina K, Supattapone S. 2003. In vitro amplification of protease-resistant prion protein requires free sulfhydryl groups. Biochemistry 42:4127–4135. 10.1021/bi027218d [DOI] [PubMed] [Google Scholar]
- 36.Kocisko DA, Priola SA, Raymond GJ, Chesebro B, Lansbury PT, Jr, Caughey B. 1995. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc. Natl. Acad. Sci. U. S. A. 92:3923–3927. 10.1073/pnas.92.9.3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eiden M, Palm GJ, Hinrichs W, Matthey U, Zahn R, Groschup MH. 2006. Synergistic and strain-specific effects of bovine spongiform encephalopathy and scrapie prions in the cell-free conversion of recombinant prion protein. J. Gen. Virol. 87:3753–3761. 10.1099/vir.0.81590-0 [DOI] [PubMed] [Google Scholar]
- 38.Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA, Caughey B. 2008. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat. Methods 5:211–212. 10.1038/nmeth0308-211 [DOI] [PubMed] [Google Scholar]
- 39.Kirby L, Birkett CR, Rudyk H, Gilbert IH, Hope J. 2003. In vitro cell-free conversion of bacterial recombinant PrP to PrPres as a model for conversion. J. Gen. Virol. 84:1013–1020. 10.1099/vir.0.18903-0 [DOI] [PubMed] [Google Scholar]
- 40.Raymond GJ, Bossers A, Raymond LD, O'Rourke KI, McHolland LE, Bryant PK, III, Miller MW, Williams ES, Smits M, Caughey B. 2000. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 19:4425–4430. 10.1093/emboj/19.17.4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymond GJ, Hope J, Kocisko DA, Priola SA, Raymond LD, Bossers A, Ironside J, Will RG, Chen SG, Petersen RB, Gambetti P, Rubenstein R, Smits MA, Lansbury PT, Jr, Caughey B. 1997. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature 388:285–288. 10.1038/40876 [DOI] [PubMed] [Google Scholar]
- 42.Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. 2008. Cell-free propagation of prion strains. EMBO J. 27:2557–2566. 10.1038/emboj.2008.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castilla J, Saa P, Hetz C, Soto C. 2005. In vitro generation of infectious scrapie prions. Cell 121:195–206. 10.1016/j.cell.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 44.Orrú CD, Wilham JM, Hughson AG, Raymond LD, McNally KL, Bossers A, Ligios C, Caughey B. 2009. Human variant Creutzfeldt-Jakob disease and sheep scrapie PrP(res) detection using seeded conversion of recombinant prion protein. Protein Eng. Des. Sel. 22:515–521. 10.1093/protein/gzp031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorne L, Terry LA. 2008. In vitro amplification of PrPSc derived from the brain and blood of sheep infected with scrapie. J. Gen. Virol. 89:3177–3184. 10.1099/vir.0.2008/004226-0 [DOI] [PubMed] [Google Scholar]
- 46.Orrú CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. 2011. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio 2(3):e00078–11. 10.1128/mBio.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saborio GP, Permanne B, Soto C. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810–813. 10.1038/35081095 [DOI] [PubMed] [Google Scholar]
- 48.Jones M, Peden AH, Head MW, Ironside JW. 2011. The application of in vitro cell-free conversion systems to human prion diseases. Acta Neuropathol. 121:135–143. 10.1007/s00401-010-0708-8 [DOI] [PubMed] [Google Scholar]
- 49.Fernández-Borges N, de Castro J, Castilla J. 2009. In vitro studies of the transmission barrier. Prion 3:220–223. 10.4161/pri.3.4.10500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurt TD, Telling GC, Zabel MD, Hoover EA. 2009. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology 387:235–243. 10.1016/j.virol.2009.02.025 [DOI] [PubMed] [Google Scholar]
- 51.Meyerett C, Michel B, Pulford B, Spraker TR, Nichols TA, Johnson T, Kurt T, Hoover EA, Telling GC, Zabel MD. 2008. In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology 382:267–276. 10.1016/j.virol.2008.09.023 [DOI] [PubMed] [Google Scholar]
- 52.Castilla J, Gonzalez-Romero D, Saa P, Morales R, De Castro J, Soto C. 2008. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 134:757–768. 10.1016/j.cell.2008.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thuring CM, van Keulen LJ, Langeveld JP, Vromans ME, van Zijderveld FG, Sweeney T. 2005. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J. Comp. Pathol. 132:59–69. 10.1016/j.jcpa.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 54.Andreoletti O. 2003. European FP5 project QLK3-CT-2002-01309 to study BSE strain in sheep. http://cordis.europa.eu/projects/rcn/67194_en.html [Google Scholar]
- 55.Morales R, Duran-Aniotz C, Diaz-Espinoza R, Camacho MV, Soto C. 2012. Protein misfolding cyclic amplification of infectious prions. Nat. Protoc. 7:1397–1409. 10.1038/nprot.2012.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langeveld JP, Jacobs JG, Erkens JH, Bossers A, van Zijderveld FG, van Keulen LJ. 2006. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet. Res. 2:19. 10.1186/1746-6148-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Padilla D, Beringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, Herzog L, Gutierrez-Adan A, Pintado B, Laude H, Torres JM. 2011. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 7:e1001319. 10.1371/journal.ppat.1001319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Espinosa JC, Herva ME, Andreoletti O, Padilla D, Lacroux C, Cassard H, Lantier I, Castilla J, Torres JM. 2009. Transgenic mice expressing porcine prion protein resistant to classical scrapie but susceptible to sheep bovine spongiform encephalopathy and atypical scrapie. Emerg. Infect. Dis. 15:1214–1221. 10.3201/eid1508.081218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Béringue V, Herzog L, Jaumain E, Reine F, Sibille P, Le Dur A, Vilotte JL, Laude H. 2012. Facilitated cross-species transmission of prions in extraneural tissue. Science 335:472–475. 10.1126/science.1215659 [DOI] [PubMed] [Google Scholar]
- 60.González L, Chianini F, Martin S, Siso S, Gibbard L, Reid HW, Jeffrey M. 2007. Comparative titration of experimental ovine BSE infectivity in sheep and mice. J. Gen. Virol. 88:714–717. 10.1099/vir.0.82426-0 [DOI] [PubMed] [Google Scholar]
- 61.Beck KE, Thorne L, Lockey R, Vickery CM, Terry LA, Bujdoso R, Spiropoulos J. 2013. Strain typing of classical scrapie by transgenic mouse bioassay using protein misfolding cyclic amplification to replace primary passage. PLoS One 8:e57851. 10.1371/journal.pone.0057851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorne L, Holder T, Ramsay A, Edwards J, Taema MM, Windl O, Maddison BC, Gough KC, Terry LA. 2012. In vitro amplification of ovine prions from scrapie-infected sheep from Great Britain reveals distinct patterns of propagation. BMC Vet. Res. 8:223. 10.1186/1746-6148-8-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Everest SJ, Ramsay AM, Chaplin MJ, Everitt S, Stack MJ, Neale MH, Jeffrey M, Moore SJ, Bellworthy SJ, Terry LA. 2011. Detection and localisation of PrP(Sc) in the liver of sheep infected with scrapie and bovine spongiform encephalopathy. PLoS One 6:e19737. 10.1371/journal.pone.0019737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bossers A, Rigter A, de Vries R, Smits MA. 2003. In vitro conversion of normal prion protein into pathologic isoforms. Clin. Lab. Med. 23:227–247. 10.1016/S0272-2712(02)00063-X [DOI] [PubMed] [Google Scholar]
- 65.Bucalossi C, Cosseddu G, D'Agostino C, Di Bari MA, Chiappini B, Conte M, Rosone F, De Grossi L, Scavia G, Agrimi U, Nonno R, Vaccari G. 2011. Assessment of the genetic susceptibility of sheep to scrapie by protein misfolding cyclic amplification and comparison with experimental scrapie transmission studies. J. Virol. 85:8386–8392. 10.1128/JVI.00241-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Priola SA, Caughey B, Race RE, Chesebro B. 1994. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68:4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.González L, Jeffrey M, Dagleish MP, Goldmann W, Siso S, Eaton SL, Martin S, Finlayson J, Stewart P, Steele P, Pang Y, Hamilton S, Reid HW, Chianini F. 2012. Susceptibility to scrapie and disease phenotype in sheep: cross-Prnp genotype experimental transmissions with natural sources. Vet. Res. 43:55. 10.1186/1297-9716-43-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs JG, Bossers A, Rezaei H, van Keulen LJ, McCutcheon S, Sklaviadis T, Lantier I, Berthon P, Lantier F, van Zijderveld FG, Langeveld JP. 2011. Proteinase K-resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein. J. Virol. 85:12537–12546. 10.1128/JVI.00448-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saá P, Castilla J, Soto C. 2006. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J. Biol. Chem. 281:35245–35252. 10.1074/jbc.M603964200 [DOI] [PubMed] [Google Scholar]
- 70.Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, Telling GC. 2008. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog. 4:e1000139. 10.1371/journal.ppat.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thuring CM, Erkens JH, Jacobs JG, Bossers A, Van Keulen LJ, Garssen GJ, Van Zijderveld FG, Ryder SJ, Groschup MH, Sweeney T, Langeveld JP. 2004. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J. Clin. Microbiol. 42:972–980. 10.1128/JCM.42.3.972-980.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]