Abstract
Biological materials typically display complex morphologies and hierarchical architectures, properties that are hardly matched by synthetic materials. Understanding the biological control of mineral properties will enable the development of new synthetic approaches toward biomimetic functional materials. Here, we combine biocombinatorial approaches with a proteome homology search and in vitro mineralization assays to assess the role of biological determinants in biomimetic magnetite mineralization. Our results suggest that the identified proteins and biomimetic polypeptides influence nucleation in vitro. Even though the in vivo role cannot be directly determined from our experiments, we can rationalize the following design principles: proteins, larger complexes, or membrane components that promote nucleation in vivo are likely to expose positively charged residues to a negatively charged crystal surface. In turn, components with acidic (negatively charged) functionality are nucleation inhibitors, which stabilize an amorphous structure through the coordination of iron.
1. Introduction
Nature has evolved biominerals with complex morphologies and hierarchical architectures that are hardly matched by synthetic materials so far.1 Understanding the exquisite control exerted by organisms over mineral properties might enable the exploitation of natural design principles for the development of biomimetic functional materials under physiological and environmentally friendly conditions.2−4 However, in many biomineralizing systems it is currently unclear which of the many biological determinants are critical in controlling particular material properties or steps in their formation such as synthesis, nucleation, growth, and morphogenesis. In cases where the molecular players are known, the mechanisms by which they interact with inorganic phases have often remained elusive.
A typical strategy for the identification of involved molecules is their extraction from an organism and the characterization of biomolecules that bind an isolated mineral phase.2 The effect of the identified molecules on mineralization can then be studied in vitro. Examples have been reported for biogenic silica,5 magnetite,6−8 and calcium carbonate.9
As an alternative synthetic approach to studying biomimetic molecular structures that interact with solids, the biocombinatorial selection of solid-binding peptides has developed into a powerful technique to identify short peptides with specific affinities for a large range of inorganic materials.10,11 Recent examples are selections for the binding of demosponge spicule silica,12 synthetic silica,13 ZnO,14 and GdO.15 Because the selections can be performed under close-to-physiological conditions, the question has arisen as to whether natural and synthetic selection evolves molecules with similar characteristics and whether the biomineralizing functionality might be encoded in homologue structures for materials also found in organisms.
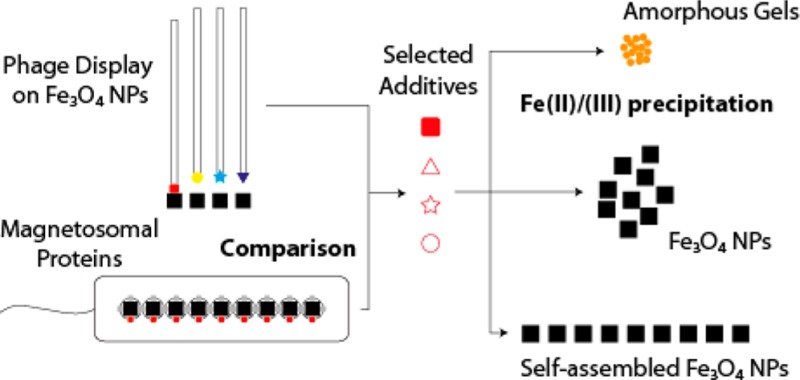
Here we investigated the example of the iron oxide mineral magnetite that is found in diverse organisms (bacteria, mollusks, birds, and fish) and where it serves geonavigational or mechanical purposes. Its biogenic formation is best studied in magnetotactic bacteria, which form chains of magnetic nanoparticles termed magnetosomes.16 Because of their size and high monodispersity, magnetosomes are envisioned for MRI contrast agents and cancer treatment applications.17 Furthermore, similarly structured synthetic magnetic nanoparticle assemblies have recently attracted much attention.18−20 Simple magnetotactic organisms have turned into a model system for iron oxide biomineralization because the genomes of several strains have been sequenced21 and because molecular techniques have been developed for their genetic manipulation.22,23 In particular, a whole set of deletion mutants has been studied in Magnetospirillum strains, with phenotypes ranging from size and morphology changes to the complete disappearance of biomineralization.24 It has been shown that about 20 genes are sufficient to restore magnetite formation in cells deficient of the whole magnetosome island, the gene cluster responsible for magnetite biomineralization.25,26 The encoded Mam, Mms, and Mtx proteins are therefore good potential candidates for comparison with synthetically selected molecules and subsequent in vitro mineralization studies. Furthermore, biocombinatorial peptide selection studies on magnetite have been reported earlier, which provide a basis for such a comparison (Figure 1). Using the biocombinatorial techniques of cell surface and phage display, Brown et al. and Barbas et al. had independently shown that polycationic polypeptides attach to magnetite or possibly to the very similar maghemite crystal surfaces.27,28
Figure 1.
Schematic method representation. A comparison of peptide sequences obtained by phage display and magnetosomal proteins affords proteins and peptides of interest for further study in Fe precipitation experiments. Depending on the additive characteristics, mineralization can be influenced to yield amorphous gels and magnetite in aggregates or self-assembled particle chains.
In this work, our idea is not to use phage display directly for the direct assessment of 12 amino acid sequences on mineralization but rather to provide an alternative route toward the identification of putative biomineralizing proteins without the need for in vivo mutant generation. We thus combine biocombinatorial approaches with a proteome homology search and assess in vitro the role of the identified proteins and associated biomimetic polypeptides in the mineralization of magnetite. Our results suggest that the macromolecules indeed influence nucleation in vitro.
2. Experimental Section
2.1. Phage Display
The Ph.D.-12 Phage Display Peptide Library (New England Biolabs) with approximately 2.7 × 109 random 12-mer peptide sequences was used for selections. Two independent selections were performed on magnetite powder. In a first experiment, ≤10 mg magnetite (Sigma-Aldrich, ≤5 μm particle size) was exposed to ∼4 × 1010 phages in 1 mL of Tris-buffered saline (TBS) and 0.1% Tween-20 (TBST, pH 7.5) for 30 min. The dispersed magnetite with bound phages was trapped on a magnetic flow-through column (MACS, Miltenyi Biotec) and washed 10 times with 1 mL of TBST. Bound phages were eluted with a low-pH buffer (0.2 M glycine-HCl, pH 2.2), neutralized with 1 M Tris-HCl (pH 9.1), and amplified by the infection of E. coli ER2738 in liquid culture. After 4.5 h, growth phages were isolated and concentrated by the removal of the bacterial cells through centrifugation and repeated precipitation with an aqueous PEG-8000 (20% w/v)/NaCl (2.5 M) solution. The amplified phages were used for the next round of selection on the mineral. In subsequent rounds, the surfactant concentration was raised by 0.1%. Individual phage clones were picked for sequencing after three and six panning rounds. Phage DNA was isolated with the M13 phage DNA isolation kit (Qiagen), sequenced (Eurofins MWG Operon), and translated into the encoded peptide. In a second experiment, the incubation time was reduced to 10 min to select for binders with a faster binding rate kon. The surfactant concentration was kept constant at 0.5% in both the incubation and washing phases to increase the initial stringency. Magnetite was kept in a 1.5 mL reaction tube, washed repeatedly (10 times) by precipitation with a magnet, resuspended in 1 mL of TBST, and transferred to a new reaction tube to prevent the potential selection of tube-material-binding peptides. Individual phages were picked and amplified for sequencing after four and six rounds of panning.
2.2. Peptide Characterization and Sequence Similarity Search
Sequence characteristics were determined using the ExPasy ProtParam tool.29 Peptides were compared to the M. magneticum (AMB-1), M. marinus (MC-1), M. magnetotacticum (MS-1), M. gryphiswaldense (MSR-1), and D. magneticus (RS-1) proteomes using the BLASTP 2.2.28+ tool on http://blast.ncbi.nlm.nih.gov/(30) using the default parameters (word size, 3; BLOSUM62 matrix; conditional compositional score matrix adjustment; gap costs, existence 11/extension 1; without low complexity filtering). Proteins for precipitation experiments were chosen on the basis of the lowest e values obtained for proteins unique to magnetotactic bacteria (MSR-1 MamJ, e = 1.1; MSR-1 MtxA, e = 0.12).
2.3. Proteins
Genes mamJ, mtxA, and mtxAΔ1–24 were amplified from M. gryphiswaldense MSR-1 genomic DNA by PCR (oligonucleotides in Table S1, produced by MWG Operon) using KOD polymerase (Novagen), purified, and ligated into a pET-51b(+)Ek/LIC vector (Novagen). mtxAΔ1–24 refers to the DNA sequence encoding for the MtxAΔ1–24 protein without the leading N-terminal 24 amino acids, which were identified as a signal peptide. MamJ was expressed in E. coli Rosetta 2 (DE3) (Novagen). MtxA and MtxΔ1–24 were expressed in E. coli BL21 (DE3) (Novagen). Precultures were grown in 5 mL of LB medium plus 100 μg mL–1 ampicillin overnight at 37 °C with 250 rpm stirring. Culture batches of 250 mL of autoinducing ZYM-5052 medium supplied with 100 μg mL–1 ampicillin were inoculated with 0.1% of the preculture and grown for 24 h at 30 °C with 250 rpm stirring.31 Cells were pelleted by centrifugation (4 °C, 4000 rpm, 15 min) and stored at −80 °C. Thawed cells were resuspended in 10 mL of Strep-Tactin wash buffer (Merck) supplied with 1 mg mL–1 lysozyme and 1 mM PMSF. After 30 min of incubation on ice, cells were lysed by sonication (10 × 15 s burst with 15 s pauses). The cell lysate was cleared by centrifugation (21 000 rpm, 4 °C, 45 min) and applied to a 5 mL bed Strep-Tactin SuperFlow agarose column (IBA). Washing and elution at 4 °C followed the manufacturer’s protocols using the respective buffers. Fractions were analyzed by SDS-PAGE using Coomassie-stained 4–20% precast linear gradient polyacrylamide gels (Bio-Rad). If the SDS-PAGE indicated impurities, then pooled His-tagged proteins were subjected to a second round of affinity chromatography on Ni-IDA matrix columns (Macherey-Nagel) at 4 °C. Generally proteins were stored at 4 °C in the respective elution buffers. The protein identity was verified by ESI-MS fingerprinting after trypsine digestion. Before magnetite coprecipitation experiments, protein solutions were dialyzed against Milli-Q water for 24 h with three solvent exchanges using ServaPor dialysis tubing with a 12–14 kDa molecular weight cutoff. Protein concentrations were determined by UV absorption at 280 nm on an Implen NanoPhotometer. Molecular weights and extinction coefficients were calculated on the basis of the sequence of the overexpressed proteins using the ProtParam tool on the ExPasy server.29 MamJ has a molecular weight of 48 481 Da and absorbs 40 115 M–1 cm–1. MtxAΔ1–24 has a mass of 34 774 Da and absorbs 33 920 M–1 cm–1. Concentrations were adjusted to 1 mg mL–1 by dilution with Milli-Q water or concentration on U-tube concentrator (30 kDa molecular weight cutoff, Novagen) tubes by centrifugation at 4 °C.
2.4. Precipitation Experiments
Reactions were performed with a computer-controlled titration system (Metrohm AG) consisting of a 776 Dosimat dosing device with an 806 exchange unit (1 mL dosing cylinder) and a 719 Titrino titration device with an 806 exchange unit (5 mL dosing cylinder). Microloader tips (Eppendorf) were used as inlets into the reaction vessel. Reactions were performed in a 50 mL vessel with a thermostated jacket kept at constant temperature (25.0 ± 0.1 °C) by a thermostat (Lauda M3). Solutions were stirred at 2050–2250 min–1 with a mechanical stirrer. The pH was measured using a Biotrode pH meter (Metrohm). All experiments were performed under a nitrogen atmosphere. Ten milliliters minus the additive volume of deionized water was initially set to the pH of interest with NaOH. In case of precipitation in the presence of an additive, the additive was then supplied and the pH was reset. Magnetite precipitation was initiated by the addition of an iron chloride solution (Fe3+/Fe2+ = 2/1) to the reactor at a rate of 1 μL min–1. The pH of the solution in the reaction vessel was simultaneously kept constant (ΔpH ±0.1) by the addition of sodium hydroxide.
2.5. Transmission Electron Microscopy
Particles were adsorbed from aqueous suspensions to carbon film Cu TEM grids for 15 min. After the removal of the liquid with Kimwipe paper, grids were washed with a drop of Milli-Q water to remove residual salt precipitates. Standard imaging was performed on a Zeiss EM 912 Omega at an acceleration voltage (U) of 120 kV. High-resolution imaging was performed on (i) a Jeol JEM 4010 transmission electron microscope (U = 400 kV) and on (ii) an FEI Titan 80/300 scanning transmission electron microscope (U = 300 kV) equipped with a probe corrector, an EDX detector, and an EELS spectrometer.
2.6. X-ray Diffraction
Precipitates were studied by synchrotron wide-angle X-ray diffraction at the μ-Spot beamline, BESSY II, Berlin. Samples were dried on a Kapton thin film (Breitlander GmbH) clamped on a custom-made sample holder. The beam size was set to 100 μm with an energy of 15 keV (λ = 0.82656 Å) defined by a Si(111) double-crystal monochromator. Diffraction data was acquired on a 3072 pixel × 3072 pixel MarMosaic 225 CCD camera (Mar USA) with a 73.242 μm pixel size. For data analysis, the beam center and the detector tilt were determined and corrected for using the respective routine of the Fit2D software.32 Peaks were fitted after baseline correction by a pseudo-Voigt function. Mean particle diameters were estimated under the neglect of strain-induced broadening with the Scherrer equation.
3. Results and Discussion
We first compared the published magnetite-adhering peptide sequences (RRTVKHHN, RRSRHH, RSKRGR, RSKKMR, and RFKRVRDR) to the proteomes of the well-studied magnetotactic bacterial strains Magnetospirillum magneticum AMB-1, Magnetococcus marinus MC-1, Magnetospirillum magnetotacticum MS-1, Magnetospirillum gryphiswaldense MSR-1, and Desulfovibrio magneticus RS-1.27,28 However, none of the sequences are similar to Mam-, Mms-, or Mtx-type proteins. We therefore performed new sequence pannings on magnetite particles using a randomized 12-mer phage display library to generate additional sequences (Table S2). In contrast to the earlier reported results, sequence alignments revealed neither a common motif among the retrieved peptides nor clear characteristics regarding amino acid composition, the isoelectric point (pI = 7.86 ± 2.00) or the hydrophobicity (average hydropathy = −0.77 ± 0.6833). We compared the resulting sequences by alignment with the proteomes of the aforementioned magnetotactic bacterial strains because our goal was to identify new putative biomineralizing macromolecules within the proteome of magnetotactic bacteria. Because of the short length of the peptides as compared to that of typical proteins, alignments yield a large number of random hits when searching entire genomes. Most sequences are dissimilar or only weakly similar to magnetosomal proteins. However, two sequences (out of 27) stood out because each one represented the best respective hits (lowest e values) within the search space encompassing the complete AMB-1, MC-1, MS-1, MSR-1, and RS-1 proteomes and could be attributed to proteins MamJ and MtxA of MSR-1 with known or hypothesized magnetosome or magnetotaxis functionality (Tables S3 and S4). We then overexpressed both proteins in E. coli for in vitro studies. The leading 24 N-terminal amino acids of MtxA represent a membrane translocation signal peptide (Figures S1–S3) that renders the protein insoluble when overexpressed in E. coli. Therefore, we used MtxAΔ1–24 without a signal peptide for in vitro assays.
We studied the influence of the selected proteins (MamJ and MtxAΔ1–24) as well as the two peptide polymers poly-l-arginine (polyR) and poly-l-glutamic acid (polyE) on magnetite formation. Apart from structural differences, the proteins/polymers differ primarily in the availability of differently charged groups that can interact with different iron or iron (oxyhydr)oxide species. Acid residues in MamJ and polyE provide binding moieties for cationic FeII/III, whereas the cationic guanidinium group of polyR is able to interact electrostatically with (in alkaline solution) negatively charged iron (oxyhydr)oxide crystal surfaces. The presence of the additives has a strong effect on the phase, crystallinity, particle size, morphology, and aggregation of the precipitates, in agreement with the interactions with soluble or solid iron species that occur either prior to or after the nucleation of the magnetite phase (Figures 2–4).
Figure 2.
Precipitation products in the presence of peptide polymer and protein additives. (A–C) Magnetite particles formed in the presence of poly-l-arginine. (Insets in C) FFTs of particles indexed as magnetite. (D) SDS-PAGE of MamJ. (E) Precipitation product in the presence of MamJ. (Inset in E) Electron diffraction reveals only amorphous scattering. (F) SDS-PAGE of MtxAΔ1–24. (G) Precipitation product in the presence of MtxAΔ1–24. (Inset in G) Electron diffraction shows diffraction consistent with magnetite.
Figure 4.
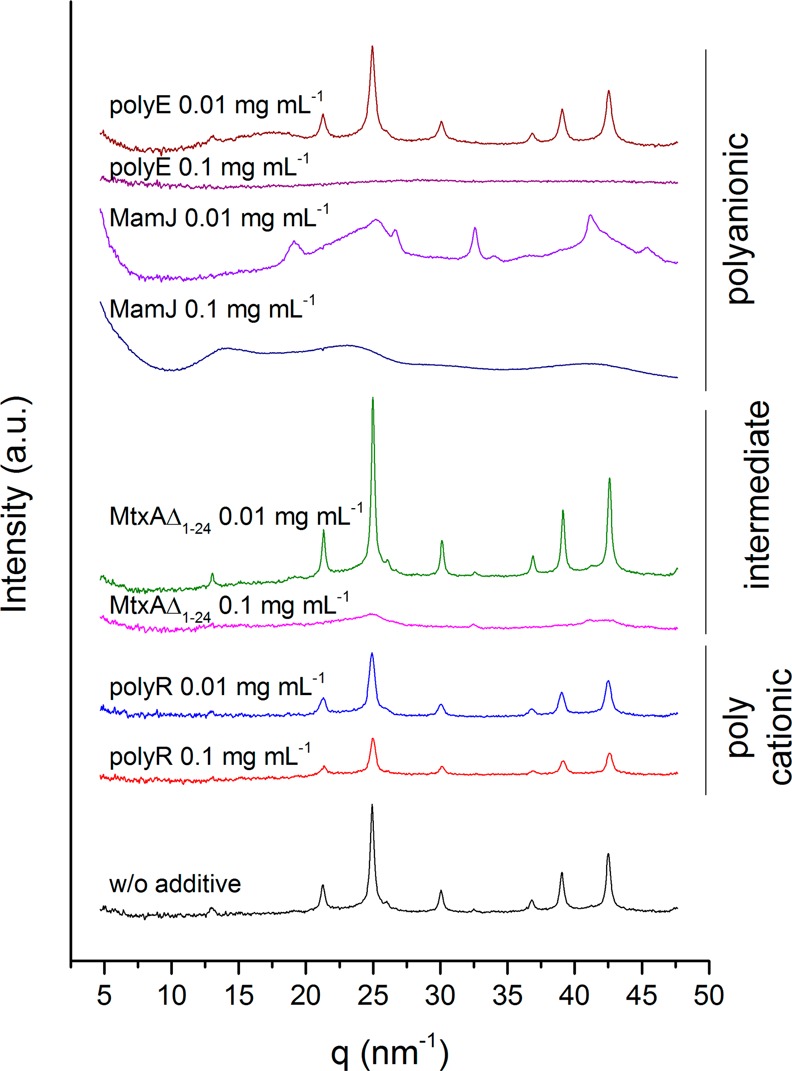
X-ray diffraction patterns of precipitations formed at pH 9 without additive and in the presence of protein and peptide polymers.
3.1. Polycationic Stabilization of Magnetite Nanoparticles by Poly-l-Arginine
polyR serves as a proxy for a potential polycationic biomolecular structure as inferred from the earlier literature reports by Barbas et al. and Brown et al.27,28 Coprecipitation of ferrous and ferric iron (at Fe3+/Fe2+ = 2/1) under alkaline conditions at pH ≥9 yields crystalline magnetite with grain sizes dependent on the alkalinity.34,35 Under these conditions, polyR affects the size, morphology, and aggregation behavior of the formed magnetite nanoparticles: in its presence, we obtained monodisperse, stable single-domain-sized nanoparticles of 35 ± 5 nm (Figure 3) that assemble to chain structures in solution (up to several micrometers; Figures 2A and S4–S6 and a video). Despite their irregular morphology, particles are mostly single-crystalline (Figure 2C). The nucleation and colloidal stabilization effects of polyR, leading to particle chain formation in vitro, are similar to the colloidal stabilization by magnetosome compartimentalization in the bacteria in vivo. This compartimentalization is provided by a lipid membrane containing diverse transmembrane proteins of yet mostly unknown functions. Interestingly, the lipid composition of the magnetosomes is dominated by phosphatidylethanolamines.36 Such lipid layers therefore expose mainly positively charged amines toward the intracellular magnetite crystals in line with our observation of a polyR-induced colloidal stabilization effect.
Figure 3.
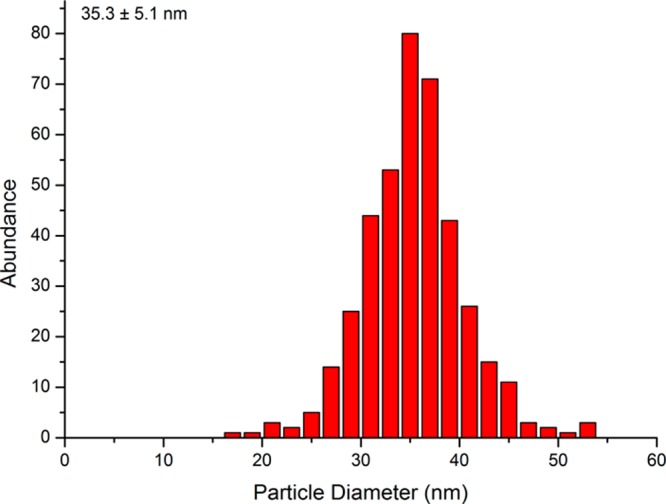
Particle size distribution of magnetite particles formed in the presence of polyR.
3.2. Polyanionic Inhibition of Magnetite Nucleation by MamJ and Poly-l-glutamic Acid
In contrast, MamJ and polyE (which resembles the polyanionic domain found in MamJ37,38) strongly affect the phase of the formed precipitates by the inhibition of magnetite nucleation with increasing additive concentration: in both cases, we obtained either amorphous gels or crystalline phases other than magnetite. Only at low additive concentrations (0.01 mg mL–1), magnetite could be obtained (Figure 4). At pH 11, in the presence of 0.01 mg mL–1 MamJ, diffraction patterns are consistent with an extremely small nanosized magnetite (Figure 5, line features are extremely broadened magnetite peaks, intensity increase toward low q). At pH 9, we obtained unidentifiable mixtures (Figure 4, peaks at q = 19.13, 26.66, 32.57, 34.03, 41.16, and 45.42 nm–1; d = 0.33, 0.24, 0.19, 0.18, 0.15, and 0.14 nm). With 0.1 mg mL–1 MamJ, we obtained a poorly ordered pattern at pH 9 (Figure 4, possibly ferrihydrite) and at pH 11 (Figure 5) a pattern with two distinct peaks at q = 15.51 nm–1 (d = 0.41 nm) and q = 25.25 nm–1 (d = 0.25 nm), which are consistent with goethite (α-FeOOH). Accordingly, in the presence of 0.1 mg mL–1 polyE we obtained no crystalline material within 1 h: at pH 9, we obtained an orange amorphous gel-like precipitate by centrifugation from a clear solution, whereas at pH 11 no pellet could be formed even by centrifugation. At low concentration (0.01 mg mL–1), polyE has no strong impact on the formed phase (Figure 4).
Figure 5.
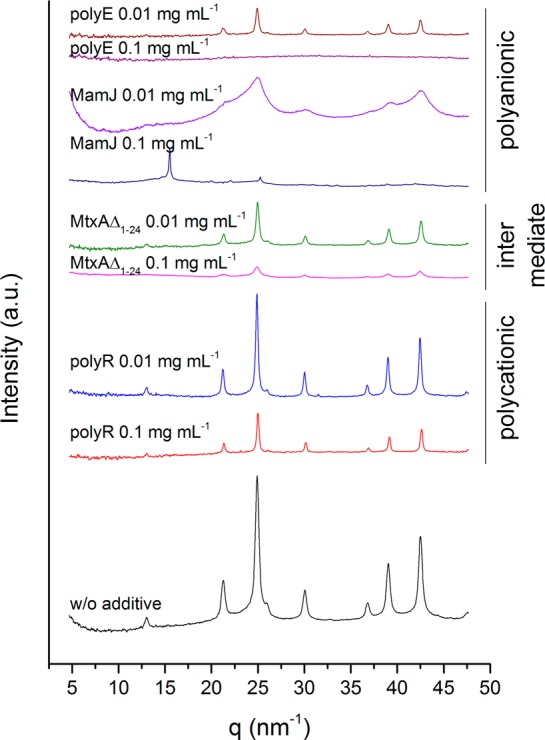
X-ray diffraction patterns of precipitates obtained at pH 11 from ferrous and ferric iron chloride solution mixtures without and in the presence of protein or polypeptide additives.
MamJ is known to be involved in the magnetosome chain assembly and has been suggested to serve as an anchor to the MamK filament in the cell.37 The anchoring function is provided exclusively by the N- and C-terminal domains without the involvement of a repetitive central sequence stretch rich in glutamic acid.38 The role of this polyanionic domain has remained unclear but was initially suspected to take part in biomineralization through the binding and accumulation of iron. The found consensus sequence PVA-LVNR can be located twice within this repetitive stretch of unknown function (Table S3; MamJ83–90 and MamJ171–178). However, the inhibitory function with respect to magnetite mineralization in vitro is inconsistent with the formation of the iron oxide both in and without the presence of the protein in vivo,37,38 unless a yet unknown regulatory function of mineral formation is required for magnetosome chain formation in the bacteria. The strong effects of the synthetic peptide polymer polyE on magnetite nucleation inhibition suggest that if such an inhibitor exists in the bacteria, it will likely act by iron binding through acidic moieties.
3.3. Weak Influence on Magnetite Mineralization by MtxAΔ1–24
Finally, MtxAΔ1–24 had only a minor influence on the phase of the formed precipitates: in the presence of 0.1 mg mL–1 MtxAΔ1–24 at pH 9, we obtained only an amorphous/poorly crystalline material with similar orange gel-like properties, whereas under all other conditions we obtained mixtures of magnetite and amorphous gels (Figure 2G).
MtxA was identified in or attached to the magnetosome membrane.39 It has been suggested to play a role in the magnetotaxis because of its gene location in an operon encoding signal transduction genes.40 However, no experimental evidence has shown the implication of the protein in such a mechanism so far. Our in vitro assays suggest that despite the possibility that the protein can bind to magnetite it will likely have no effect on the crystallization of the mineral in vivo.
3.4. Magnetite Particle Growth in the Presence of Additives
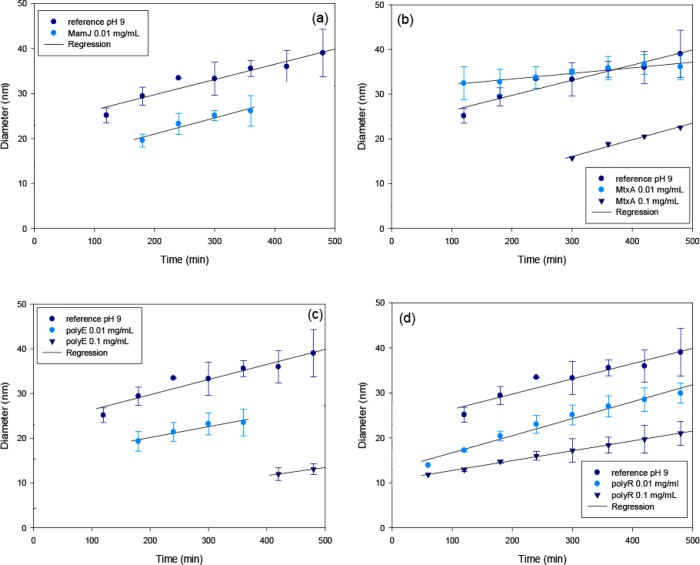
Growth experiments over several hours at pH 9, which without additives resembles the kinetics observed in the bacteria in vivo,35 indicate that generally the initial magnetite grain size becomes smaller with increasing additive concentration; however, the crystal growth rate after nucleation appears to be largely unaffected (Figure 6 and Table S5). This is consistent with an effect of ionic strength on the surface tension that in turn affects the nucleus size at the formation threshold.34,41 Furthermore, the nucleation is retarded in the presence of all investigated additives except polyR, in line with an interaction of these proteins/polymers with soluble iron species.
Figure 6.
Growth kinetics of magnetite nanoparticles formed in the presence of (a) MamJ, (b) MtxAΔ1–24, (c) polyE, and (d) polyR and control data. Error bars represent the standard deviation from three independent reactions.
3.5. Implications for Biomineral Formation
Both thermodynamic and kinetic arguments can be used to explain our observations of nucleation inhibition or phase stabilization. One can argue thermodynamically that the interaction with a charged additive will influence the surface energy of a nucleating particle. A positively charged polymer in interaction with the negatively charged magnetite particle will lower the surface tension and thereby facilitate nucleation. Accordingly, the interaction between equally charged polymers and particle surfaces would be energetically unfavorable with resulting higher surface tension and eventually nucleation inhibition. Kinetically, one can argue that in the inhibited case the interaction between a positively charged ionic iron precursor and an anionic polymer is likely more rapid than the formation of the crystalline iron oxide solid, therefore entrapping iron in a polymer-induced amorphous precursor state.42
Generally, synthetic polyelectrolytes by virtue of their electrostatics find extensive use as nucleation and crystal growth modifiers as well as in the stabilization or flocculation of colloidal suspensions.43 Equally, nature has evolved highly charged proteins (and other biomolecules) that can perform similar functions in vivo. In the context of biomineralization, this applies to proteins involved in the phase selection and growth modification of calcium carbonates and phosphates, silicate, and possibly iron oxides as investigated here. Similar to the effects observed here for the iron precipitates, the charge of interacting polyelectrolytic proteins determines the fate of the precursor species by stabilization or destabilization. For example, polyanionic aspartic acid-rich proteins play a presumably fundamental role in the stabilization of amorphous calcium carbonate (ACC) in mollusk shells.9 Whereas early-stage calcium carbonate precipitates possess slightly positive zeta potentials (for equimolar calcium and carbonate mixtures) facilitating the binding of polyanionic proteins,44 silica possesses negative surface charge throughout physiologically accessible pH ranges, favoring interaction with polycations.45 Thus, polycationic biopolymers have been shown to be involved in biomineralization by diatoms where they mediate the formation of SiO2.5
Although studying the mineralization role of proteins in vitro represents a promising alternative because deletion mutants are difficult to obtain and because their mechanistic role can be inferred more easily, most of the studies have focused on only one such magnetosomal protein (Mms6) in vitro so far.7,8,46 The Mms6 protein and even its 26 amino acids C-terminal peptide are supposed to impact the size of magnetite nanoparticles, although controversial results have been obtained so far. Mms6 was initially chosen because it was shown to be tightly bound to the magnetite mineral. Recently, the putative mechanistic role of MamP has also been inferred from an in vitro study.47 Although the in vivo effect of MamP had been recognized earlier,24 only the recent combination of protein structural and chemical reaction studies could assume its role of redox control in the oxidation of Fe(II).47
4. Conclusions
Although biocombinatorial approaches and in vitro mineralization assays alone cannot assume the role of proteins (or other biomolecules) in biomineralization in vivo, they can point toward possible molecular characteristics required by the involved biochemical machinery. Our experiments suggest that in the case of magnetite formation discussed here, proteins, larger complexes, or membrane components promoting the nucleation in vivo are likely to expose positively charged residues to a negatively charged crystal surface. Components with acidic (negatively charged) functionality are likely inhibitors by the stabilization of an amorphous structure through the coordination of iron.
Acknowledgments
We thank W. Schulze for ESI-MS peptide fingerprinting and C. Li and S. Siegel for support at the μ-Spot beamline, BESSY II. This research was supported in D.F.’s laboratory by the Max Planck Society, the DFG (SPP 1420, FA835/2-1; 1569 FA835/6-1), the European Union (Bio2MaN4MRI, no. 245542), and a starting grant from the European Research Council (project MB2, no. 256915).
Supporting Information Available
List of oligonucleotides for gene amplification. Summary of magnetite-adhering sequences and sequence alignment with magnetotactic bacteria-specific proteins. Sequence alignment of M. gryphiswaldense MSR-1 MamJ and phage display of selected peptide QFSLPVAKLVNR. Sequence alignment of M. gryphiswaldense MSR-1 MtxA and phage display of selected peptide VPSLTPSAQSRP. Fitted kinetic data assuming linear relation reaction-limited growth. TMHMM output from the MtxA sequence. SignalP-NN output for the MtxA sequence. SignalP-HMM output for MtxA sequence. Large and medium field of view transmission electron micrographs of magnetite particles formed in the presence of poly-l-arginine. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Dunlop J. W. C.; Fratzl P. Biological Composites. Ann. Rev. Mater. Res. 2010, 40, 1–24. [Google Scholar]
- Weiner S. Biomineralization: A Structural Perspective. J. Struct. Biol. 2008, 163, 229–234. [DOI] [PubMed] [Google Scholar]
- Fratzl P. Biomimetic Materials Research: What Can We Really Learn from Nature’S Structural Materials?. J. R. Soc. Interface 2007, 4, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman F.; Sommerdijk N. A. J. M. Biomineralization as an Inspiration for Materials Chemistry. Angew. Chem., Int. Ed. 2012, 51, 6582–6596. [DOI] [PubMed] [Google Scholar]
- Kröger N.; Deutzmann R.; Sumper M. Polycationic Peptides from Diatom Biosilica That Direct Silica Nanosphere Formation. Science 1999, 286, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Amemiya Y.; Arakaki A.; Staniland S. S.; Tanaka T.; Matsunaga T. Controlled Formation of Magnetite Crystal by Partial Oxidation of Ferrous Hydroxide in the Presence of Recombinant Magnetotactic Bacterial Protein Mms6. Biomaterials 2007, 28, 5381–5389. [DOI] [PubMed] [Google Scholar]
- Arakaki A.; Webbs J.; Matsunaga T. A Novel Protein Tightly Bound to Bacterial Magnetite particles in Magnetospirillum magnetotacticum Strain AMB-1. J. Biol. Chem. 2003, 278, 8745–8750. [DOI] [PubMed] [Google Scholar]
- Prozorov T.; Mallapragada S. K.; Narasimhan B.; Wang L.; Palo P.; Nilsen-Hamilton M.; Williams T. J.; Bazlinski D. A.; Prozorov R.; Canfield D. E. Protein-Mediated Synthesis of Uniform Superparamagnetic Magnetite Nanocrystals. Adv. Funct. Mater. 2007, 17, 951–957. [Google Scholar]
- Politi Y.; Mahamid J.; Goldberg H.; Weiner S.; Addadi L. Asprich Mollusk Shell Protein: In Vitro Experiments Aimed at Elucidating Function in CaCO3 Crystallization. CrystEngComm 2007, 9, 1171–1177. [Google Scholar]
- Chen C.-L.; Rosi N. L. Peptide-Based Methods for the Preparation of Nanostructured Inorganic Materials. Angew. Chem., Int. Ed. 2010, 49, 1924–1942. [DOI] [PubMed] [Google Scholar]
- Sarikaya M.; Tamerler C.; Schwartz D. T.; Baneyx F. Materials Assembly and Formation Using Engineered Polypeptides. Annu. Rev. Mater. Res. 2004, 34, 373–408. [Google Scholar]
- Müller W. E. G.; Schröder H. C.; Muth S.; Gietzen S.; Korzhev M.; Grebenjuk V. A.; Wiens M.; Schloßmacher U.; Wang X. The Silicatein Propeptide Acts as Inhibitor/Modulator of Self-Organization during Spicule Axial Filament formation. FEBS J. 2013, 280, 1693–1708. [DOI] [PubMed] [Google Scholar]
- Patwardhan S. V.; Emami F. S.; Berry R. J.; Jones S. E.; Naik R. R.; Deschaume O.; Heinz H.; Perry C. C. Chemistry of Aqueous Silica Nanoparticle Surfaces and the Mechanism of Selective Peptide Adsorption. J. Am. Chem. Soc. 2012, 134, 6244–6256. [DOI] [PubMed] [Google Scholar]
- Rothenstein D.; Claasen B.; Omiecienski B.; Lammel P.; Bill J. Isolation of ZnO-Binding 12-mer Peptides and Determination of Their Binding Epitopes by NMR Spectroscopy. J. Am. Chem. Soc. 2012, 134, 12547–12556. [DOI] [PubMed] [Google Scholar]
- Schwemmer T.; Baumgartner J.; Faivre D.; Börner H. G. Peptide-Mediated Nanoengineering of Inorganic Particle Surfaces: A General Route toward Surface Functionalization via Peptide Adhesion Domains. J. Am. Chem. Soc. 2012, 134, 2385–2391. [DOI] [PubMed] [Google Scholar]
- Faivre D.; Schüler D. Magnetotactic Bacteria and Magnetosomes. Chem. Rev. 2008, 108, 4875–4898. [DOI] [PubMed] [Google Scholar]
- Lisy M. R.; Hartung A.; Lang C.; Schüler D.; Richter W.; Reichenbach J. R.; Kaiser W. A.; Hilger I. Fluorescent Bacterial Magnetic Nanoparticles As Bimodal Contrast Agents. Invest. Radiol. 2007, 42, 235–241. [DOI] [PubMed] [Google Scholar]
- Hu M.-J.; Lu Y.; Zhang S.; Guo S.-R.; Lin B.; Zhang M.; Yu S.-H. High Yield Synthesis of Bracelet-like Hydrophilic Ni–Co Magnetic Alloy Flux-Closure Nanorings. J. Am. Chem. Soc. 2008, 130, 11606–11607. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Dong L.; Zhang L.-C.; Su Y.-D.; Yu S.-H. Biogenic and Biomimetic Magnetic Nanosized Assemblies. Nano Today 2012, 7, 297–315. [Google Scholar]
- Gao M.-R., Zhang S.-R., Xu Y.-F., Zheng Y.-R., Jiang J., Yu S.-H.Self-Assembled Platinum Nanochain Networks Driven by Induced Magnetic Dipoles. Adv. Funct. Mater. 2013, doi: 10.1002/adfm.201302262. [DOI] [Google Scholar]
- Jogler C.; Schüler D. Genomics, Genetics, and Cell Biology of Magnetosome Formation. Annu. Rev. Microbiol. 2009, 63, 501–521. [DOI] [PubMed] [Google Scholar]
- Schultheiss D.; Schüler D. Development of a Genetic System for Magnetospirillum gryphiswaldense. Arch. Microbiol. 2003, 179, 89–94. [DOI] [PubMed] [Google Scholar]
- Matsunaga T.; Nakamura C.; Burgess J. G.; Sode K. Gene Transfer in Magnetic Bacteria: Transposon Mutagenesis and Cloning of Genomic DNA Fragments Required for Magnetosome Synthesis. J. Bacteriol. 1992, 174, 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D.; Quinlan A.; Vali H.; Komeili A. Comprehensive Genetic Dissection of the Magnetosome Gene Island Reveals the Step-Wise Assembly of a Prokaryotic organelle. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohße A.; Ullrich S.; Katzmann E.; Borg S.; Wanner G.; Richter M.; Voigt B.; Schweder T.; Schüler D. Functional Analysis of the Magnetosome Island in Magnetospirillum gryphiswaldense: The mamAB Operon Is Sufficient for Magnetite Biomineralization. PLoS One 2011, 6, e25561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D.; Falahati V.; Bertinetti L.; Csencsits R.; Körnig A.; Downing K. H.; Faivre D.; Komeili A. The Magnetosome Membrane Protein, MmsF, Is a Major Regulator of Magnetite Biomineralization in Magnetospirillum magneticum AMB-1. Mol. Microbiol. 2012, 85, 684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. Engineered Iron Oxide-Adhesion Mutants of the Escherichia-Coli Phage-Lambda Receptor. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 8651–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C. F.; Rosenblum J. S.; Lerner R. A. Direct Selection of Antibodies That Coordinate Metals from Semisynthetic Combinatorial Libraries. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 6385–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E.; Hoogland C.; Gattiker A.; Duvaud S.; Wilkins M. R.; Appel R. D.; Bairoch A.. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker J. M., Ed.; Humana Press: Totowa, NJ, 2005; pp 571–607. [Google Scholar]
- Altschul S. F.; Madden T. L.; Schäffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Protein Production by Auto-Induction in High-Density Shaking Cultures. Protein Expression Purif. 2005, 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Hammersley A. The FIT2D Home Page. http://www.esrf.eu/computing/scientific/FIT2D/.
- Kyte J.; Doolittle R. F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Baumgartner J.; Dey A.; Bomans P. H. H.; Le Coadou C.; Fratzl P.; Sommdijk N. A. J. M.; Faivre D. Nucleation and Growth of Magnetite from Solution. Nat. Mater. 2013, 12, 310–314. [DOI] [PubMed] [Google Scholar]
- Baumgartner J.; Bertinetti L.; Widdrat M.; Hirt A. M.; Faivre D. Formation of Magnetite Nanoparticles at Low Temperature: From Superparamagnetic to Stable Single Domain Particles. PLoS One 2013, 8, e57070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K.; Müller E. C.; Otto A.; Reszka R.; Linder D.; Kube M.; Reinhardt R.; Schüler D. Biochemical and Proteomic Analysis of the Magnetosome Membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 2004, 70, 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A.; Gruska M.; Faivre D.; Linaroudis A.; Plitzko J. M.; Schüler D. An Acidic Protein Aligns Magnetosomes along a Filamentous Structure in Magnetotactic Bacteria. Nature 2006, 440, 110–115. [DOI] [PubMed] [Google Scholar]
- Scheffel A.; Schüler D. The Acidic Repetitive Domain of the Magnetospirillum gryphiswaldense MamJ Protein Displays Hypervariability but Is Not Required for Magnetosome Chain Assembly. J. Bacteriol. 2007, 189, 6437–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M.; Okamura Y.; Arakaki A.; Tanaka T.; Takeyama H.; Matsunaga T. Origin of Magnetosome Membrane: Proteomic Analysis of Magnetosome Membrane and Comparison with Cytoplasmic Membrane. Proteomics 2006, 6, 5234–5247. [DOI] [PubMed] [Google Scholar]
- Richter M.; Kube M.; Bazylinski D. A.; Lombardot T.; Glockner F. O.; Reinhardt R.; Schüler D. Comparative Genome Analysis of Four Magnetotactic Bacteria Reveals a Complex Set of Group-Specific Genes Implicated in Magnetosome Biomineralization and Function. J. Bacteriol. 2007, 189, 4899–4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssières L.; Chanéac C.; Tronc E.; Jolivet J. P. Size Tailoring or Magnetite Particles Formed by Aqueous Precipitation: An Example of thermodynamic Stability of Nanometric Oxide Particles. J. Colloid Interface Sci. 1998, 205, 205–212. [DOI] [PubMed] [Google Scholar]
- Gower L. B. Biomimetic Model Systems for Investigating the Amorphous Precursor Pathway and Its Role in Biomineralization. Chem. Rev. 2008, 108, 4551–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer D. A. Synthetic Polyelectrolytes—A Review. Polym. Int. 1991, 2529–41. [Google Scholar]
- Chibowski E.; Hotysz L.; Szcześ A. Time Dependent Changes in Zeta Potential of Freshly Precipitated Calcium Carbonate. Colloids Surf., A 2003, 222, 41–54. [Google Scholar]
- Behrens S. H.; Grier D. G. The Charge of Glass and Silica Surfaces. J. Chem. Phys. 2001, 115, 6716–6721. [Google Scholar]
- Galloway J. M.; Arakaki A.; Masuda F.; Tanaka T.; Matsunaga T.; Staniland S. S. Magnetic Bacterial Protein Mms6 Controls Morphology, Crystallinity and Magnetism of Cobalt-Doped Magnetite Nanoparticles in Vitro. J. Mater. Chem. 2011, 21, 15244–15254. [Google Scholar]
- Siponen M. I.; Legrand P.; Widdrat M.; Jones S. R.; Zhang W.-J.; Chang M. C. Y.; Faivre D.; Arnoux P.; Pignol D.. Structural Insight into Magnetochrome-Mediated Magnetite Biomineralization. Nature 2013, 502, 681–684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.