Abstract
Streptococcus suis is a major bacterial pathogen of young pigs causing worldwide economic problems for the pig industry. S. suis is also an emerging pathogen of humans. Colonization of porcine oropharynx by S. suis is considered to be a high risk factor for invasive disease. In the oropharyngeal cavity, where glucose is rapidly absorbed but dietary α-glucans persist, there is a profound effect of carbohydrate availability on the expression of virulence genes. Nineteen predicted or confirmed S. suis virulence genes that promote adhesion to and invasion of epithelial cells were expressed at higher levels when S. suis was supplied with the α-glucan starch/pullulan compared to glucose as the single carbon source. Additionally the production of suilysin, a toxin that damages epithelial cells, was increased more than ten-fold when glucose levels were low and S. suis was growing on pullulan. Based on biochemical, bioinformatics and in vitro and in vivo gene expression studies, we developed a biological model that postulates the effect of carbon catabolite repression on expression of virulence genes in the mucosa, organs and blood. This research increases our understanding of S. suis virulence mechanisms and has important implications for the design of future control strategies including the development of anti-infective strategies by modulating animal feed composition.
Introduction
Streptococcus suis is a major bacterial pathogen of young pigs and a worldwide economic problem for the pig industry. Furthermore, S. suis is emerging as a zoonotic pathogen associated with meningitis and septicaemia in humans [1], [2]. In pigs, invasive disease is thought to be caused by translocation of S. suis across the mucosal epithelium in the upper respiratory tract [3], [4]. The ecological conditions that promote adhesion to, and invasion of the host mucosa by S. suis are unknown and probably depend on the environmental conditions and the bacterial genotype including presence of virulence genes. In vitro studies on adhesion and invasion by S. suis have often been performed in medium containing glucose as a carbon source which does not accurately reflect the situation in vivo. In the oropharyngeal cavity including the saliva, glucose may be present but concentrations usually diminish rapidly (within 30 min) after ingestion [5], [6] as glucose is readily absorbed by the host and metabolized by commensal bacteria. In contrast, starch α-glucans, large polymers of D-glucose that are present in large amounts in animal feeds [7] can persist in high concentrations in the oropharynx of humans and pigs [8]–[11]. A second type of carbohydrate that may promote proliferation of pathogenic bacteria is animal glycogen released from damaged or lysed cells. Suilysin, a hemolytic toxin encoded by the S. suis sly gene, may release, through its cytotoxic effect on epithelial cells, cellular glycogen [12], [13] that may serve as an important substrate for pathogen growth during the early stages of infection.
Currently, little is known about the expression and regulation of S. suis virulence factors such as suilysin. Streptococcal pathogens contain genes required for efficient utilization of α-glucans including amylases and/or pullulanases which cleave α-1,4 and α-1,6 glycosidic bonds in starch or glycogen [14]–[18]. Previously, we have shown that a cell wall anchored amylopullulanase of S. suis serotype 2 (apuA-SSU1849) was necessary to support bacterial proliferation on the α-glucan starch/pullulan (an α-1,6; α-1,4 linked glucose polymer) as a single carbon source and to promote adhesion of S. suis to porcine tracheal epithelial cells [19].
As in all Gram-positive bacteria, in S. suis the expression of carbohydrate metabolic enzymes are under the control of the global transcriptional regulator, catabolite control protein A (CcpA) that mediates carbon catabolite control (CCC) in presence of a preferred sugar, usually glucose. CcpA can repress or activate transcription by binding to cis-acting catabolite response element (cre) sites in the gene promoter regions [20], [21].
Given the relative abundance of dietary complex carbohydrates such as α-glucans and low level of glucose in the porcine upper oropharyngeal niche occupied by S. suis, we compared the genome-wide effects of growth on glucose or the α-glucan starch/pullulan on S. suis metabolism and virulence gene expression. We first constructed a metabolic map for S. suis and used it to analyze the key metabolic pathways altered by growth on glucose or pullulan. In addition to genes that play roles in (carbohydrate) metabolic pathways, 19 virulence genes were differentially expressed, seven of which were strongly induced (ratios>10) by growth in pullulan compared to glucose and nine were shown to contain a consensus cre site in their promoter sequences. The regulation of apuA, the virulence factor most strongly induced in pullulan compared to glucose, was investigated in detail using qPCR analysis of gene transcripts in bacteria grown in different carbon sources, promoter mapping and binding studies with CcpA and a newly identified transcriptional regulator, ApuR. The biological consequences of carbohydrate metabolism and virulence gene expression were also assessed in an in vitro porcine tracheal cell model using qPCR, hemolytic assays and adhesion and invasion assays. Based on these findings we propose a model for the transcriptional regulation of production of virulence factors during different stages of infection dependent on a CcpA-mediated, carbon catabolite control-dependent mechanism. To verify the predictions of this model, the in vivo expression of apuA and sly was measured for S. suis serotype 2 recovered from the blood and organs of pigs infected under controlled conditions.
Materials and Methods
Bacterial strains, plasmids and culture conditions
The virulent S. suis serotype 2 strain S10 [22] and S735-pCOM1-V10 [23] were used in this study. The genome of S. suis S10 is more than 99% identical to the genome of S. suis 2 strain P1/7, a sequenced reference strain of which the genome had been annotated previously (NCBI Genome and NCBI BioProject http://www.ncbi.nlm.nih.gov/genome/?term=Streptococcus%20suis) [24]. S. suis was grown in Todd-Hewitt broth (THB) (Difco) or on Columbia agar plates with 6% sheep blood (Oxoid) at 37°C under 5% CO2 for 18 hr. A complex medium (CM) (Text S1) was prepared as previously described [19], [25]. The carbohydrates were added separately and sterilized by autoclaving at 100°C for 10 min (pullulan) or filtration with 0.45 µM pore size filter (glucose, lactose and maltotriose). We previously demonstrated that S. suis only grows to high density in CM when exogenous carbohydrates are added [19]. Growth in complex medium was determined by measurement of turbidity at OD600 using a SpectraMax M5 reader (Molecular Devices LLC).
RNA extraction from in vitro grown S. suis and quantitative PCR (qPCR)
For RNA extraction, S. suis S10 was grown to exponential (e) and early stationary (s) phase as indicated in Figure S1. Ten ml of culture was collected and centrifugated for each time point. The pellet was immediately frozen in liquid nitrogen until further handling. The frozen pellet was dissolved in 600 µl RA1 reagent (Macherey-Nagel) plus β-mercaptoethanol and lysed using a FastPrep-24 (MP. Biomedicals, Solon, OH) for 6.0 m/sec at 20 sec. Total RNA was purified using NucleoSpin RNA II (Macherey-Nagel). The quality and the concentration of RNA were assessed with an Experion System (Bio-Rad) and by analysis of the A260/A280 ratio (NanoDrop 8000 UV-Vis Spectrophotometer). For qPCR, cDNA was synthesized using SuperScript VILO (Invitrogen). Primers were designed using Oligo Program version 6 (MedProbe, Oslo, Norway) (Table S1). Quantitative PCR was performed using a LightCycler 4.0 V and the LightCycler FastStart DNA Master SYBR Green I Kit (Roche). Constitutive gene expression in complex media was determined as a ratio of target gene vs reference gene proS (Text S1) [26]. The level of proS expression was constant at all the time points analyzed (data not shown). Two replicates of all samples and primer pairs were included and the experiment was performed in triplicate. Non-template controls were included for each gene in each run.
Microarray transcriptome analysis
An S. suis oligoarray (8×15 K) containing in situ synthesized 60-mers was produced by Agilent Technologies (Santa Clara, USA), based on the genome sequence of S. suis P1/7 [24]. A total of 7651 unique 60-mers having a theoretical melting temperature of approximately 81°C and representing 1960 ORFs were selected as described [27]. Genes were represented by 4 (91%), 3 (4%), 2 (2%), or 1 probe (3%). Twenty-five putative genes were not represented on the array because no unique probe satisfying the selection criteria could be selected. RNA (1 µg) from S. suis samples was labeled using the Cyanine 5 (Cy5) labeling reaction (Amersham Biosciences, Buckinghamshire, UK). Co-hybridization with labeled cDNA probes was performed on these oligonucleotide arrays at 42°C for 16 h in Slidehyb#1 (Ambion, Austin, USA). The data were normalized using Lowess normalization [28] as available in MicroPrep [29] and corrected for inter-slide differences on the basis of total signal intensity per slide using Postprep [29]. Significance of differential gene expression was based on FDR values lower than 0.05. All microarray data are MIAME compliant and available in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE40658. Details of the custom-made 60-mer oligonucleotide array design (Agilent Biotechnologies, Amstelveen, The Netherlands) are deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession number A-MEXP-1671.
Bioinformatic tools: microarray analysis and cre motifs search
Differential gene expression of S. suis bacteria grown in CM supplemented with pullulan (Pul) or glucose (Glc) and harvested at early exponential (e) or early stationary (s) phase was cross-compared in different combinations (Pul_e vs Glc_s, Pul_s vs Glc_s, Pul_e vs Pul_s and Glc_e vs Glc_s). Overlapping and unique differentials were visualized using Venn diagrams at http://bioinfogp.cnb.csic.es/tools/venny/index.html.
For all genes and proteins identified in the S. suis P1/7 genome, Gene Ontology (GO, http://www.geneontology.org) and KEGG pathway annotations were obtained using the BLAST2GO software (www.blast2go.org) [30] including annotations based on terms obtained from EBI using the InterPROScan feature [31] that is part of BLAST2GO.
BLAST2GO was used to annotate all known S. suis genes and proteins according to standard Gene Ontology (GO) (www.geneontology.org) nomenclature. BLAST2GO uses the integrated Gossip package [30] for statistical assessment of differences in GO term abundance between two sets of sequences. The GO enrichment analysis feature of BLAST2GO was then used to identify functional GO terms that were statistically over- or underrepresented in the set of genes differentially expressed in pullulan vs glucose in exponential and early stationary phases of growth (Fisher's Exact Test p<0.05).
The MEME (http://meme.sdsc.edu/meme/meme.html) software suite (version 4.1.0) was used for the identification of motifs OM1 and OM2 [32]. As input, we used the promoter sequences of the MdxE and MalE genes, with well-characterized cre sites, from Bacillus and Listeria (TGWAARCGYTWNCW [W = A or T; R = A or G; Y = C or T; N = any base]). A range of motif widths (15 nt in length) and zero or one motif per sequence were specified in our queries and FIMO, part of MEME, was used to search for this motif in the genome of S. suis P1/7. The algorithm in MAST, also part of the MEME suite, calculates position scores for the motif at each possible position within a sequence [33]; motif hits with a position-specific goodness-of-fit P value below 10−4 were considered to identify putative CcpA binding sites. In silico searches and comparisons of predicted cre sites within the S. suis P1/7 genome sequence and reconstruction of cre locations in the predicted operons were conducted using the corresponding databases provided by the MicrobesOnline database web server (http://microbesonline.org) and RegPrecise database [34].
5′-Rapid Amplification of cDNA Ends (5′-RACE)
The 5′-rapid amplification of cDNA ends (RACE) system (Invitrogen) was used to determine the transcription start site of the apuA gene. Briefly, the first strand cDNA was reverse transcribed from RNA from 1 µg of total RNA S. suis grown in CM plus pullulan using the specific primer ASP1 (Table S1). A homopolymeric tail was added to the 3′-end of the cDNA using terminal deoxynucleotidyl transferase (TdT) and the deoxynucleotide dCTP. The tailed cDNA was amplified in nested-PCR with Abridged Anchor Primer (AAP) and a second apuA specific primer ASP2 primer upstream ASP1. The resulting 5′-RACE product of ∼380 bp was sequenced and analysed by Vector NTI software (Invitrogen).
Recombinant regulators production and infrared EMSA
The genes encoding the regulators ApuR (amino acids 2 to 312) and CcpA (amino acids 2 to 333) were amplified by PCR from S. suis 2 S10 genomic DNA using GoTaq (Promega) with primers ApuR_F/R and CcpA_F/R respectively (Table S1). The recombinant ApuR (rApuR) and CcpA (rCcpA) proteins were cloned in pTrcHis TOPO2 TA and purified by HPLC affinity chromatography (HisTrap affinity column, Amersham Pharmacia Biotech) as previously described (Text S1) [19]. Fractions containing purified fusion proteins of the expected size (approx. 38 kDa for ApuR and 40 kDa for CcpA) were collected and dialyzed against buffer (500 mM NaCl, 50 mM Tris-HCl, pH 7.4) and stored at −80°C with 10% of glycerol. Protein concentrations were measured using a BCA Protein Assay kit (Thermo Scientific). Further details can be found in the Text S1.
Infrared electrophoretic mobility shift assays (EMSA) were performed utilizing three pairs of fluorescent InfraRed-dye 800 (IRdye 800) labelled primer on the 5′ ends (Biolegio, The Netherlands). The IRdye-primers used to PCR-amplify three DNA fragments of ca. 120 bp (Pr1-Pr3) contained overlapping regions to cover the full length of the S. suis P apuA promoter sequence (Table S1). The IRdye-PCR fragments were purified with QIAquick PCR Purification Kit (Qiagen) and used for the binding reaction at a concentration of around 50 nM. DNA binding reactions were performed in 20 µl of binding buffer containing 10 mM Tris-HCl pH 8.5, 50 mM NaCl, 10 mM EDTA, 0.5% Tween-20, 10 mM DTT, and 1 mg of poly(dI-dC)-labelled IRdye-PCR fragments at room temperature for 30 min. For the specific and non-specific competition assays, D(+)-glucose 6-phosphate (30 mM) (Sigma) was added to the binding buffer in the EMSA as CcpA cofactor [35]. Purified rApuR and rCcpA proteins (from 0.5 to 4.0 µM) were incubated, in separate experiments, with the fragment Pr2 and a non-specific competitor fragment that was obtained by PCR amplification of the gene SSU0879 (from 25 to 150 nM). Two 95 bp oligonucleotides complementary to Pr2 fragment lacking the two predicted binding sites (ΔOM1 and ΔOM2/cre) were synthesized (Eurogentec, The Netherlands), PCR-amplified with labelled Pr2F/R primers and incubated with the proteins. Free and bound DNAs were separated on 5% Tris-Borate-EDTA (TBE) native gels for 30 min at 10 cm/v. The gels were visualized using a LI-COR Odyssey Imager and scanned at 800 nm wavelengths.
Titration of hemolytic activity
The hemolytic activity was assayed as previously described [36]. Two different independent assays were carried out in triplicate. Briefly, S. suis bacteria were grown in CM+Glc and CM+Pul at three different growth stages: lag (OD600 0.1–0.2), exponential (OD600 0.2–0.5) and stationary (OD600 0.5–0.7). The supernatant was collected from 1 ml for each culture by centrifugation at 12000 g for 1 min. Serial twofold dilutions (150 µl) of test samples were prepared in polystyrene deep-well titer plates (Beckman) with 10 mM Tris-buffered saline (PBS, pH 7.4). Subsequently, 150 µl of a 2% washed horse erythrocyte suspension in 10 mM Tris-buffered saline containing 0.5% BSA was added to each well. After the wells were sealed, the plates were incubated on a Coulter mixer for 2 h at 37°C. Unlysed erythrocytes were sedimented by centrifugation (1500 g for 10 min), 150 µl portions of the supernatant were transferred to a polystyrene flat-bottom microtiter plate and measured at 540 nm with a microELISA (enzyme-linked immunosorbent assay) reader (SpectraMax M5, Molecular Devices LLC). A 100% lysis reference sample was obtained by lysing cells with 1% Triton-X and the background lysis was subtracted before calculation of hemolytic activity.
Adherence and invasion assays using NPTr cell line
Culturing of Newborn pig tracheal cells (NPTr) [37] and adhesion assays were performed as previously described [19]. For the invasion assays, S. suis was added to the cell culture at an multiplicity of infection (m.o.i.) of ∼50 and incubated for 2 h at 37°C with 5% CO2 to allow cellular invasion by the bacteria. The monolayers were then washed three times with PBS; 1 ml of cell culture medium containing 100 µl/ml gentamycin and 5 µg/ml penicillin G was added to each well, and the plates were incubated for 2 h at 37°C, 5% CO2 to kill extracellular and surface-adherent bacteria. The monolayers were washed three times with PBS, and cells were disrupted by the addition of 800 µl of ice-cold Milli-Q water and repeated up-and-down pipetting to release intracellular bacteria. To enumerate the viable bacteria, serial dilutions of each cell lysate were plated in triplicate on Columbia sheep blood agar plates and incubated at 37°C for 24 h. The rate of invasion was expressed as percentage of initial inoculum that was recovered per well. Two independent assays were performed in triplicate.
Experimental infection of pigs
The animal experiment described in this paper was approved by the ethical committee of the Central Veterinary Institute of Wageningen UR, Lelystad, The Netherlands under proposal number 2010113c in accordance with Dutch legislation (The Experiments on Animals Act, 1997) and the general principles governing the use of animals in experiments of the European Communities (Directive 86/609/EEC). To reduce unnecessary suffering of animals, humane end points were used: piglets were euthanized as soon as S. suis-specific symptoms occurred. Caesarean-derived, colostrum-deprived (CDCD) piglets were infected i.v. with 1×106 CFU of S735-pCOM1-orf2 [23]. The strain used to experimentally infect pigs was different to that used in our in vitro qPCR assays but was selected because it was known to be effectively recovered from the blood and organs in experimental pig infections. Genes encoding apuA and sly are both present in the genome of this strain (not shown). Animals were euthanized when specific S. suis symptoms (meningitis, arthritis, sepsis) were observed, or at the end of the experiment (6 days post infection). After animals were euthanized; organs and specific sites of S. suis infection were examined macroscopically and bacteriologically. Tissue samples were collected during post-mortem observation and snap-frozen in liquid nitrogen.
Quantitative PCR using bacterial-enriched RNA isolated from in vivo tissues and blood
Blood collected in PAXgene tubes was treated as recommended by the manufacturer (PreAnalytix/Qiagen). Samples taken from heart, brain and joints consisted of a mixed pellet of porcine cells combined with bacteria. All pellets from in vivo samples were thawed on ice, resuspended in 600 µl of Trizol (Invitrogen, Carlsbad, CA, USA) and subjected to 40 seconds of 6.5 m s−2 in the Fastprep-24 (MP Biomedicals, Solon, OH, USA) to disrupt bacteria. The mixture was extracted with 120 µl of chloroform, mixed for 15 seconds, incubated for 3 min at RT and centrifuged for 15 min at 20.000 x g. Supernatant was removed and extracted with 1 volume of chloroform. RNA in the supernatant was precipitated with 1 volume of isopropanol. After incubation for 30 min at RT or 16 h at −20°C, RNA was collected by centrifugation, and washed with 70% ethanol. The pellet was resuspended in water. Subsequently, RNA was purified as described above. To remove eukaryotic RNA all samples were treated using the MicrobEnrich kit (Ambion, Austin, Tx, USA). Bacterial-enriched RNA was amplified using the Ovation PicoSl WTA system v2 (Nugen, San Carlos, CA, USA). cDNA was purified using MinElute spin columns (Qiagen, Hilden, Germany) and diluted 25 times for qPCR analysis. Primers were designed using PrimerExpress software (Applied Biosystems, Foster City, CA, USA) (Table S1). Each reaction contained 12.5 pmol forward primer, 12.5 pmol reverse primer and POWR SYBR Green PCR Master Mix (Applied Biosystems). qPCR was performed using an ABI7500 (Applied Biosystems). As the bacterial RNA was amplified we used a more stringent GeNorm method [38] for normalization of the real-time qPCR data. GeNorm utilizes multiple internal control genes for normalization, in this case gyrA, proS, and mutS that were most stably expressed among 7 tested genes (data not shown). In each run a standard curve was incorporated consisting of seven 10-fold dilutions of a vector containing the cloned target of the PCR. In this way, both target genes and the reference genes could be related to a standard line. For each reaction, negative water controls were included. Analysis was performed using the ABI7500 Software (Applied Biosystems).
Statistical analysis
The results obtained in the in vitro studies were analyzed using GraphPad Prism version 5.0 software (San Diego California, USA). All qPCR experiments and hemolytic activity assay were reproduced at least two times in triplicates and, where indicated, representative experiments are shown. Two-way ANOVA tests were carried out using Bonferroni's post hoc test. The adhesion and invasion assays were performed at least two times using triplicate samples. All numerical data presented here are expressed as means ± standard error of the mean (SEM). Statistical significance was determined using a two-tailed unpaired Student's t test. Differences were considered significant at p<0.05. Statistical significance was indicated as follows: * p<0.05; ** p<0.01; *** p<0.001.
Results
The switch from glucose to α-glucan starch fermentation has pleiotropic effects on gene expression
To investigate the global effects of a shift in carbohydrate metabolism on S. suis gene expression we compared genome-wide transcriptomic data from exponential (e) and stationary phase (s) cultures of S. suis in complex media (CM) supplemented with 1% of starch/pullulan (α-1,4; α-1,6 glucan) (Pul) or glucose (Glc) as carbon sources (Figure S1 and Table S2).
The numbers of genes differentially expressed during growth in pullulan versus glucose were 1028 (52% of annotated genes) during exponential growth and 1015 (51% of annotated genes) during early stationary growth (Figure 1A). In total 738 (37% of annotated genes) genes were differentially regulated in pullulan compared to glucose, irrespective of the growth phase. In starch/pullulan, 209 genes were differentially regulated between the exponential and early stationary phases of growth; in glucose, 432 genes were differentially regulated for the same comparison (Figure 1A).
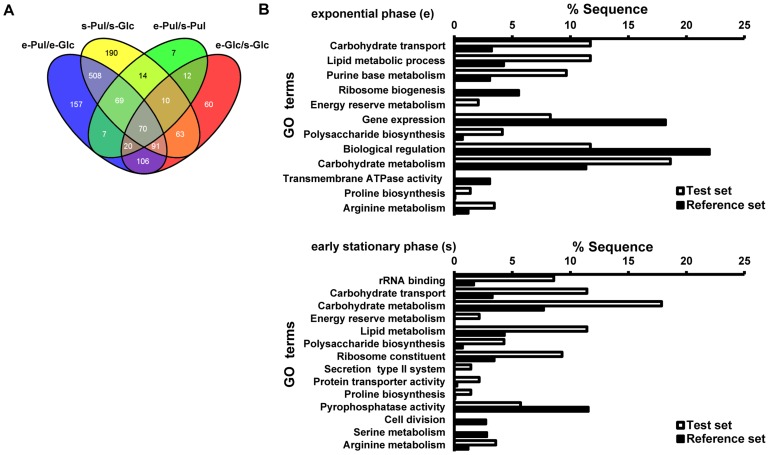
Figure 1. Comparison of genome-wide transcriptomic data for exponential (e) and early stationary phase (s) cultures of S. suis grown in pullulan (Pul) or glucose (Glc).
A. Venn diagram of S. suis genes differentially regulated during growth in pullulan (Pul) vs glucose (Glc) in the exponential (e) or stationary (s) phase. In each sector the numbers of shared or unique differentially expressed genes is indicated. B. GO term distribution of S. suis genes differentially regulated in pullulan vs glucose in early exponential and early stationary phase. GO Enrichment analysis was performed using BLAST2GO (P = 0.05, two-tailed Fisher's Exact test).
To link gene expression data to changes in bacterial metabolic and physiological pathways, we obtained gene ontology (GO) functional gene annotations of all differentially expressed S. suis P1/7 genes using BLAST2GO (see Methods) (Figure 1B). Similar analyses were also performed for a subset of differentials, namely all upregulated genes within the GO category “carbohydrate metabolism” (Figure S2). The highest number of genes differentially expressed in both growth phases participated in “carbohydrate metabolism” (18%) and “carbohydrate transport” (11%). Genes in the GO category “energy reserve metabolism” (2% for both growth phases) and lipid metabolism that generates the precursors of lipoteichoic acids and membrane phospholipids (11% for both growth phases) were also enriched when bacteria were grown in starch/pullulan vs glucose (Figure 1B). Other enriched GO categories included amino acid metabolic pathways for arginine and proline.
These analyses showed that culturing S. suis in presence of these different carbon sources leads to changes of 35–50% of the S. suis transcriptome, affecting not only carbohydrate metabolism but also basal metabolic and stress survival pathways. To summarize these broad changes in global transcriptomes, we generated a visual representation of the main carbohydrate pathway genes for six growth conditions comparisons according to their expression ratios (see Methods) (Figure 2A).
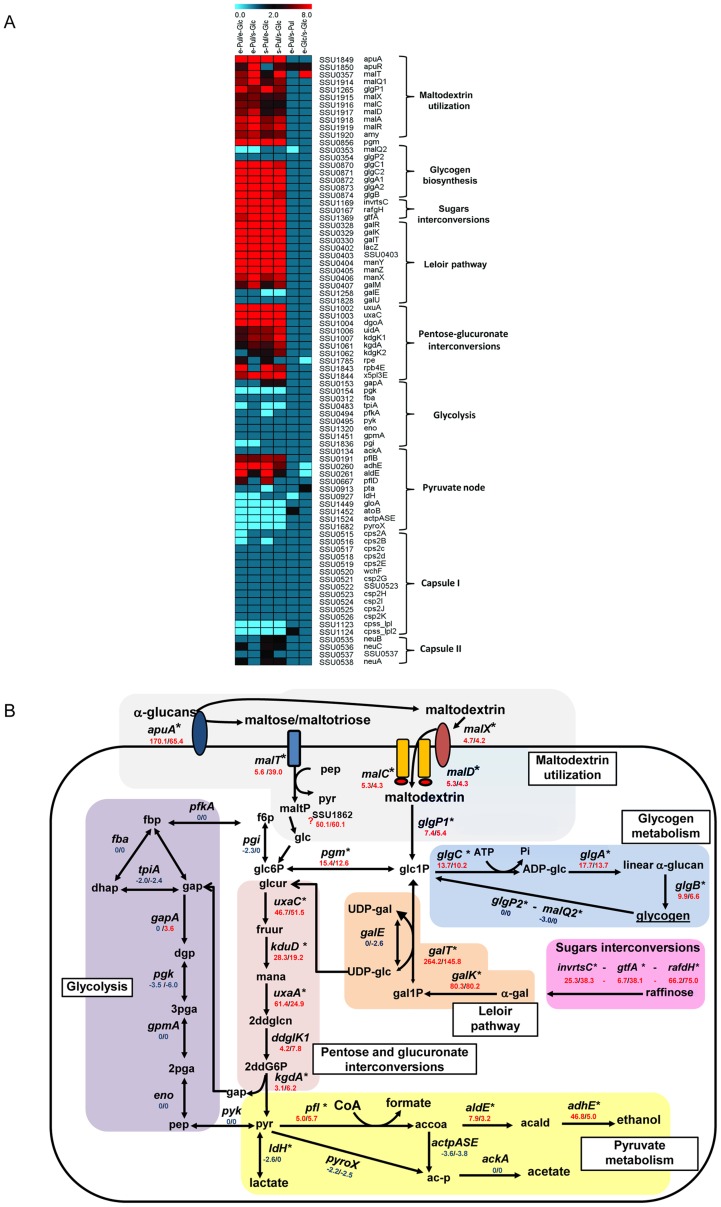
Figure 2. Transcriptome analysis of S. suis metabolism in presence of pullulan.
A. Heatmap showing the effect of starch/pullulan on the transcription of genes involved in carbohydrate metabolism and capsule production. Expression (ratios) of genes participating in different pathways (indicated at the right of the heatmap) are shown for 6 different comparisons (indicated at the top of the heatmap). At the top of the Figure 2A, a color scale depicts the ratio of expression during growth in pullulan vs. glucose. Red indicates induction (upregulation) of the respective genes and blue indicates repression (downregulation) of the respective genes for each comparison. For each gene, the S. suis P1/7 locus tag and the gene name is depicted on the right. B. Schematic representation of S. suis metabolic pathways differentially regulated in pullulan vs glucose. α-glucans (i.e. starch/pullulan) are degraded by extracellular amylopullulanase (apuA) and the end degradation products, maltose/maltotriose and maltodextrins, are transported by PTS for maltose/maltotriose (malT) and maltodextrin ABC transport inside the bacteria (malX, malC and malD). Maltodextrins and maltose are most lilkely converted to glucose-1-phosphate (Glc1P) or α-glucose by 4-α-glucanotransferase and maltodextrin phosphorylase (malQ1 and glgP1 respectively). Glc1P can be metabolized in different pathways: phosphoglucomutase (pgm) isomerize glc1P to glucose-6-phosphate (glc6P) which may enter glycolysis (violet box) where it is consequently oxidated to pyruvate (pyr). Homolactic fermentation reduces pyruvate into lactate, whereas heterofermentative growth leads to other products, such as formate, acetate and ethanol (pyruvate metabolism, yellow box). The excess of glc1P that cannot enter in glycolysis may be used for synthesis of glycogen as an energy reserve (light blue box). The genome of S. suis is predicted to encode the enzymes sucrose phosphorylase gtfA, α-fructofuranosidase (interconvertase) invrtsC, and raffinose galactohydrolase, rafgH for the interconversion of raffinose- like sugars. These enzymes participate in the starch and galactose Leloir pathway. Part of Leloir pathway (e.g. galactose-1-phosphate uridylyltransferase, galT, and galactokinase, galK) was induced more strongly in starch/pullulan. GalT interconverts galactose-1-phosphate (gal1-P) and UDP-Glucose (UDP-glc) to UDP-galactose (UDP-gal) and glc1P. Alternatively, UDP-glc may be converted into glucuronic acid (glcur) by UDP-D-glucuronate (UDP-glcur) to enter in an alternative (to glycolysis) pathway for pyruvate (pyr) production. Pathway predictions were reconstructed based on genome information, literature and database surveys (KEGG, MetaCyc). The following gene annotation was downloaded from NCBI: galM, aldose 1-epimerase; galK, galactokinase; galE, UDP-glucose 4-epimerase; galT, galactose 1-phosphate uridylyltransferase; pgm, Phosphoglucomutase/phosphomannomutase; pfkA, 6-phosphofructokinase; fba, fructose bisphosphate aldolase; tpiA, triosephosphate isomerase; gapA, glyceraldehyde-3-phosphate dehydrogenase; pgk, phosphoglycerate kinase; gpmA, phosphoglyceromutase; eno, phosphopyruvate hydratase; pyk, pyruvate kinase; ldh, L-lactate dehydrogenase; pyroX, pyruvate oxidase; ackA, acetate kinase; pfl, pyruvate formate-lyase; adlE acetaldehyde-CoA dehydrogenase; adhE alcohol dehydrogenase; glgB, glgA glycogen synthase; glgC glucose-1-phosphate adenylyltransferase; glgP glycogen phosphorylase.
Comparative transcriptome analysis of S. suis grown in starch/pullulan vs glucose revealed that growth in pullulan induced expression of the maltodextrins utilization gene cluster that includes apuA and the corresponding phosphotransferase systems (PTS) and ATP-binding cassette (ABC) transporters. The expression of the glycolysis pathway genes did not change significantly during growth in pullulan compared to glucose, suggesting that activity of this pathway was not altered during growth in both carbon sources. Rather, it appeared that excess intracellular glucose was converted in glycogen as energy reserves (glycogen biosynthesis pathways) or had entered into hexose-pentose carbohydrate pathways as an alternative to glycolysis (sugar interconversion and Leloir pathways) (Figure 2A). The biochemical links between the differentially expressed metabolic pathways are shown in Figure 2B; for each pathway, an extensive overview of the main genes involved and their functions can be found in the SI Text S2.
Links between carbohydrate metabolism and virulence gene regulation
We analyzed the transcriptome data to see whether the differential fermentation of starch/pullulan and glucose affected expression of known or predicted S. suis virulence genes. Seven genes possibly involved in the invasion of mucosal tissues or avoidance of host defenses by streptococcal pathogens were highly upregulated (expression ratio>10) in starch/pullulan compared to glucose (Table 1, Figure S3, Text S2). As CcpA in Gram-positive bacteria controls carbohydrate metabolism, one of the most profoundly altered categories in our microarray experiment, we investigated the potential role of CcpA in regulating virulence gene expression. We first used the MEME [39] and MAST [33] algorithms to mine the genome sequence of S. suis P1/7 for catabolite-responsive element (cre) sites, short DNA regions that can be bound by CcpA, using the consensus B. subtilis cre sequence [40]–[42].
Table 1. Confirmed and proposed S. suis virulence factors differentially expressed in pullulan (Pul) compared to glucose (Glc).
| Annotation S. suis P 1/7 | Protein | Function | Virulence | Pul/Glc1 | Biblio |
| Galactosyl/rhamnosyl transferase - SSU0520 | CpsE/F | CPS biosynthesis | Attenuated-pig | [71] | |
| Tyrosine-protein kinase Wze - SSU0517 | Cps2C | CPS biosynthesis | Attenuated-pig | [71] | |
| N-acetylneuraminic acid synthase - SSU0535 | NeuB | Sialic acid synthesis | Attenuated-pig | U | [72] |
| Peptidoglycan GlcNAc deacetylase - SSU1448 | PgdA | Peptidoglycan | Attenuated-pig | D | [73] |
| D-alanine-poly ligase - SSU0554 | DltA | LTA D-alanylation | Attenuated-pig | [74] | |
| Fibronectin-fibrinogen binding - SSU1311 | FbpS | Adhesion ECM | Attenuated-pig | [75] | |
| Enolase - SSU1320 | Eno | Adhesion ECM | no Mutant | [76] | |
| Glyceraldehyde-3-P-dehydrog - SSU0153 | GAPDH | Adhesion ECM | no Mutant | U | [77] |
| Di-peptidyl peptidase IV - SSU0187 | DppIV | Adhesion ECM | Attenuated-mouse | U | [78] |
| Amynoacyl histidine peptidase - SSU1215 | PepD | Subtilisin- protease | No Mutant | U>10 | [79] |
| 6-phosphogluconate-dehydrogen - SSU1541 | 6-PGD | Adhesion epithelium | No Mutant | [80] | |
| Amylopullulanase - SSU1849 | ApuA | Adhesion epithelium | Not tested | U>10 | [19] |
| Glutamine synthetase - SSU0157 | GlnA | Adhesion epithelium | Attenuated-mouse | D | [81] |
| Streptococcal adhesin P - SSU0253 | SadP | Adhesion epithelium | no Mutant | U | [82] |
| Arginine deaminase - SSU0580 | ArcB | Resistance to acidity | Not tested | U>10 | [83] |
| Anchored DNA nuclease - SSU1760 | SsnA | DNA degradation | Not tested | U | [84] |
| Cell envelope proteinase - SSU0757 | SspA | Subtilisin- protease | Attenuated-mouse | U | [85] |
| Metallo-serine protease - SSU1773 | IgAP | IgA1 protease | Attenuated-pig | U>10 | [86] |
| Suilysin - SSU1231 | Sly | Haemolysin | Unaffected-pig | U>10 | [36] |
| Hyaluronate lyase - SSU1050 | Hyl | hyaluronidase | Not tested | U>10 | [87] |
| putative oligohyaluronate lyase - SSU1048 | HepI/III | hyaluronidase | Not tested | U>10 | [88] |
| Sortase A - SSU0925 | SrtA | Protein sorting | Attenuated-pig | [89] | |
| Serum opacity-like factor - SSU1474 | OFS | Serum opacification | Attenuated-pig | D | [90] |
| S-ribosyl homocysteinase - SSU0376 | LuxS | Quorum sensing | Attenuated zebrafish | D | [91] |
| Muramidase released protein - SSU0706 | MRP | Unknown | Unaffected-pig | U | [22] |
| Extracellular protein factor - SSU0171 | Ef | Unknown | Unaffected-pig | U | [92] |
Pul/Glc1 upregulated (U) or downregulated (D) expression when S. suis was grown in pullulan (Pul) compared to glucose (Glc).
38 potential cre sites located upstream of the start codon of predicted proteins (P) or in proximity of the gene transcription start site (G) were predicted to control expression of 172 genes through interaction with CcpA (Table S3). Of the 172 genes in the predicted CcpA regulon, 145 (84%) were differentially regulated during growth in starch/pullulan compared to glucose (Table S3). In Table S3 we show 38 cre sites and their downstream genes or operons and if these were differentially expressed in pullulan compared to glucose (our study) or in an S. suis serotype 2 ΔccpA mutant compared to wild-type [43].
As expected, cre sites were commonly associated with the predicted promoters of carbohydrate PTS and ABC transporters (17%) and enzymes for carbohydrate metabolism (25%). Additionally, cre sites were identified in the promoters of regulators (10%) and 9 (8%) out of the 19 virulence genes (47%) that were differentially expressed in pullulan compared to glucose, including sly and apuA (Table S3). As apuA is essential for growth on pullulan and was shown to play a role in colonization of mucosal epithelia in vitro [19] and suilysin is a major virulence factor, we sought to understand the regulation of the encoding genes in more detail and explore the role of CcpA in transcriptional regulation during the switch from glucose to starch fermentation.
Expression of apuA and sly is repressed by CCR
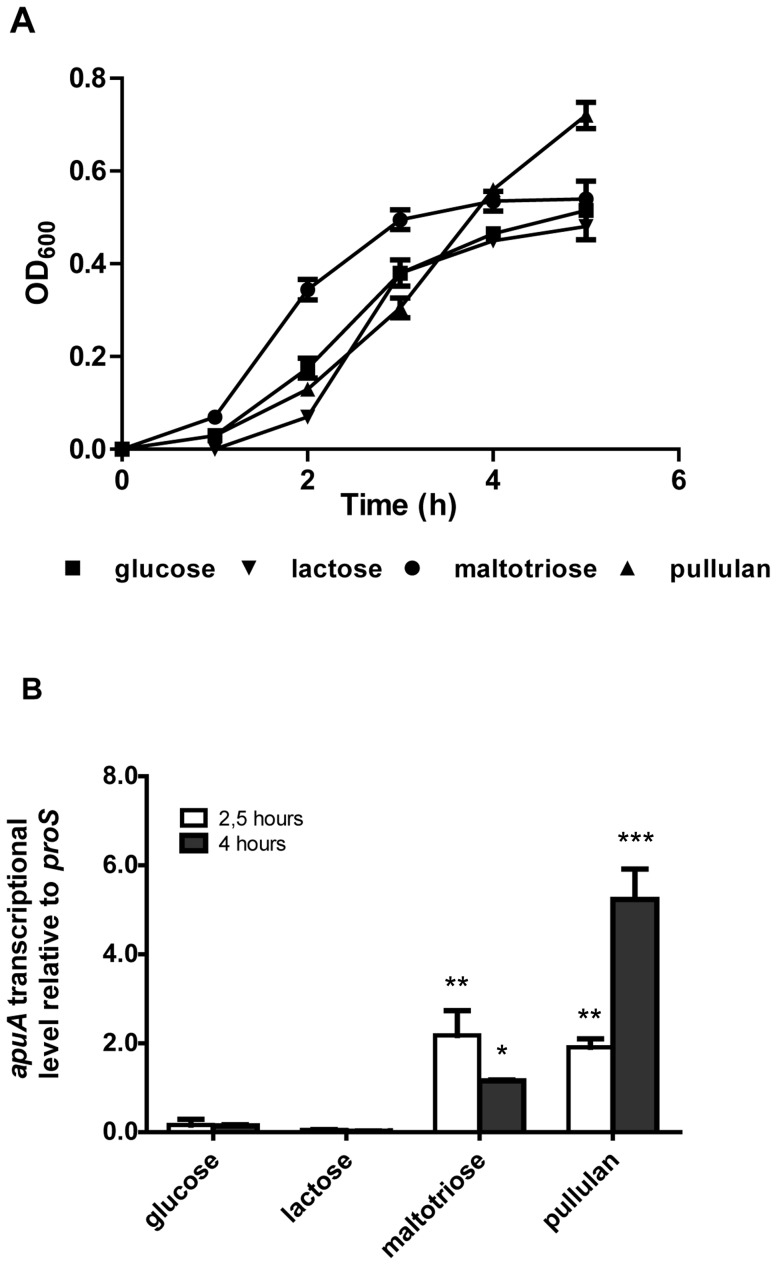
We assessed the relative expression of apuA and sly by qPCR when S. suis was grown in complex media supplemented with glucose, lactose, starch/pullulan or maltotriose, all of which supported efficient growth (Figure 3 panels A). Compared to growth in glucose or lactose, apuA and sly transcription was induced by growth in pullulan (up to 5.2 and 1.3 fold after 4 hours respectively; P<0.001) and maltotriose (up to 2.2 and 0.23 after two 2.5 hours respectively; P<0.01) (Figure 3 panels B and C).
Figure 3. Transcriptional regulation of apuA and sly grown in the presence of different carbon sources.
A. S. suis S10 growth curve at 37°C in CM containing 1% (w/v) of different sugars as indicated. The graph shows the means and standard deviations from two independent experiments. B. and C. Relative expression of apuA and sly in S. suis grown in CM containing 1% (w/v) different sugars was determined by qPCR. The transcript levels of apuA were measured after 2.5 hours and 4 hours of growth relative to the reference gene proS, which is constitutively expressed at similar levels during growth in different sugars (data not shown). The height of the bars represent mean values for the relative expression data ± SEM from 2 independent experiments (n = 3). Statistical significance was calculated using a two-way ANOVA test followed by Bonferroni's post hoc test (* p<0.05; ** p<0.01; *** p<0.001.).
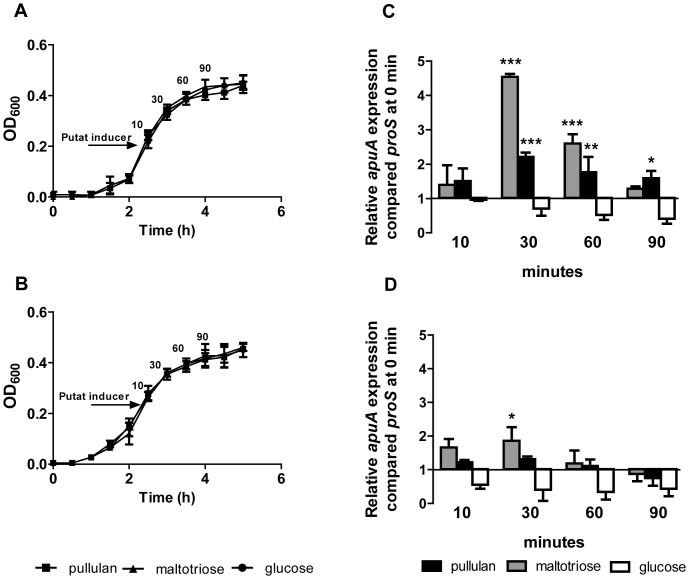
As apuA and sly contain a conserved cre in the promoter region (Table S3), we predicted that their transcription might be repressed during growth on glucose but not lactose. However, the relative level of apuA transcription in lactose was comparable to that of transcription in glucose suggesting possible regulation by a second transcriptional regulator. To test this hypothesis we added starch/pullulan or maltotriose to S. suis growing exponentially in either lactose (Figure 4A) or glucose (Figure 4B) and quantified apuA transcription at different time points (Figure 4 panels C and D). In medium containing lactose, both maltotriose and pullulan strongly induced expression of apuA after 30 min (4.5 fold and 2.2 fold respectively; P<0.001; Figure 4C). In medium containing glucose the addition of maltotriose or starch/pullulan had a small effect on the expression of apuA; only a slight increase (1.6 fold; P<0.05) of apuA expression was observed 30 min after addition of maltotriose (Figure 4D). These results show that when S. suis is growing in glucose for 30 min, apuA transcription is hardly induced by pullulan or maltotriose but when grown in lactose for 30 min, apuA expression is strongly induced by pullulan and maltotriose.
Figure 4. Induction of apuA expression by putative inducers in the presence of glucose or lactose.
A. Growth curves of S. suis S10 in CM containing 1% w/v lactose or B. 1% w/v glucose before and after addition of 0.25% w/v putative inducers (arrow) i.e. maltotriose, pullulan or glucose. The graphs show the means and standard deviations from two independent experiments. C. Relative expression of apuA genes following addition of putative inducers to S. suis growing in CM plus lactose. D. Relative expression of apuA genes following addition of putative inducers. The relative expression of apuA, was measured by qPCR 10, 30, 60 and 90 minutes after addition of the putative inducers, The height of the bars shows the mean (n = 3) fold change in expression ± SEM from two independent experiments. Statistical significance was calculated using a two-way ANOVA test followed by Bonferroni's post hoc test (* p<0.05; ** p<0.01; *** p<0.001.).
CcpA and ApuR bind to conserved operator motifs in the apuA promoter
We hypothesized that the induction of apuA expression by maltotriose was due to ApuR, a putative transcriptional regulator upstream of apuA that possesses its own promoter and a predicted rho-independent downstream terminator (Figure 5A). Homology searches indicated that ApuR was a LacI/GalR type regulator containing an N-terminus DNA-binding and a C-terminus ligand-binding domain that can be bound by a specific sugar. Comparison of ApuR with protein sequences in the UniProt database revealed similarities to several transcriptional regulators of operons (Figure S4) which comprising maltodextrin utilization gene clusters. In the top ranking were listed for highest similarities: BL23 YvdE (Lactobacillus casei; 53% identity) [44], MdxR (syn. YyvdE; B. subtilis 168; 49% identity) [45], [46], the activator EGD-e Lmo2128 (Listeria monocytogenes; 47% identity) [47] (Figure 5 E–F) and the activator MdxR (Enterococcus faecium E1162; 49% identity) [48].
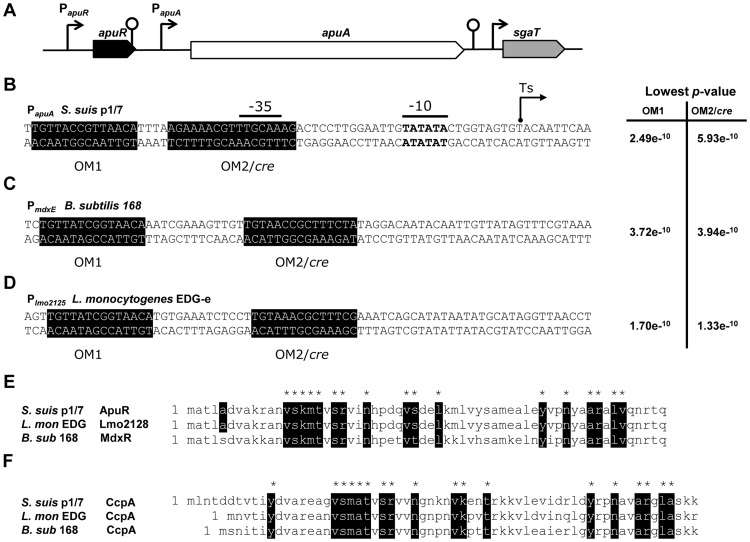
Figure 5. Identification of conserved operator binding motifs for ApuR (OM1) and CcpA (OM2) in S. suis P1/7, B. subtilis 168 and L. monocytogenes EDG-e.
A. The 6 kb amylopullulanase gene apuA is located downstream of apuR which encodes a putative transcriptional regulator of the LacI/GalR family. Located downstream of apuA are a cluster of genes predicted to be involved in uptake and fermentation of ascorbate (sgaT, sgaB). For each gene, the direction of transcription is indicated by an arrow, the size of which is proportional to the length of the corresponding open reading frame. Putative promoters are represented by arrows and transcription terminators by loops. The operator motifs OM1 and OM2/cre (shaded sequences in B, C and D) were determined using the MEME software suite and their relative probability p-values are indicated. B. The apuA promoter based on the experimentally determined transcription start site (Ts arrow) C. The B. subtilis mdxE (BSU34610) promoter D. The L. monocytogenes lmo2125 promoter E. and F. Sequence alignment of the DNA binding domains of the ApuR and CcpA proteins of S. suis, L. monocytogenes and B. subtilis. Conserved amino acid sequences are indicated in black.
As apuA is considered a relevant virulence factor and its regulation by different carbohydrates could be of biologic relevance, we decided to investigate the binding of CcpA and ApuR to operator sites in the apuA promoter. To elucidate the apuA promoter regulatory modules we determined the transcription start site, conducted homology searches for binding motifs of known regulators in silico, and performed in vitro promoter binding assays with purified transcription factors (TFs). The transcriptional start site of the apuA transcript was determined experimentally by 5′-RACE to be 31 nt upstream of the start codon. The consensus −35 element (5′-TTGCAA-3′) for RpoD (sigma 70) and the −10 element (5′-TATATA-3′) required for interaction with the RNA polymerase and transcription initiation were found near the expected positions upstream of the transcription start site (Figure 5B). Visual inspection of the apuA promoter region revealed the presence of two potential operator motifs (OMs) that were also listed in the RegPrecise database [34]. One mdxR-like operator motif, designated here as OM1, was found 13 bp upstream of the −35 element; a second operator motif predicted cre site designated OM2/cre, located at position −30 nt, overlapping the conserved −35 element; this overlap could interfere with binding of the RNA polymerase (Figure 5 C–F).
To demonstrate binding of ApuR and CcpA to the predicted operator motifs, we expressed and purified these proteins in E. coli with a C-terminal His-tag (Figure S5 and B) and tested DNA binding in electrophoretic mobility shift assays (EMSA) using three overlapping fragments of the PapuA promoter region (Pr 1–3; approx. 120 bp in length). A single rApuR-DNA complex was observed with promoter fragments Pr2 and Pr3 suggesting the binding motif for ApuR lies within the 64 bp overlapping region (Figure S6 panels A and B). The binding of ApuR to Pr2 was shown to be concentration dependent in the range from 0.5 to 4.0 µM of rApuR (Figure S6C). The specificity of rApuR binding to Pr2 was shown in a competition EMSA using unlabeled competitor DNA (i.e. fragment Pr2) or an unlabeled non-specific competitor lacking the two predicted OM1 and OM2/cre operator binding motifs. The non-specific competitor DNA fragment had no effect on Pr2 complex formation whereas the specific competitor (Pr2) substantially reduced rApuR binding (Figure S6 panels D and E). To investigate whether ApuR binds to OM1 or OM2/cre, promoter fragments lacking these motifs were synthesized and tested in the EMSA (i.e. ΔOM1 and ΔOM2/cre, Table S3 and S4). In three independent experiments we observed a lower amount of the rApuR-DNA complex with fragments lacking ΔOM1 suggesting that ApuR binds most strongly to this motif. In some experiments, complex formation was slightly reduced when ΔOM2/cre was deleted, possibly due to low affinity binding of ApuR to ΔOM2/cre at higher protein concentrations (Figure S6F).
Similarly, binding of CcpA to the PapuA OM2/cre that overlaps with the predicted −35 promoter element was demonstrated by EMSA using a fluorescent IRdye-Pr2 fragment containing the putative cre site (Figure S6 panels G–J). An increasing amount of an rCcpA-DNA complex was observed with an increasing concentration of purified rCcpA (1.5 to 5 µM) (Figure S6G). The complex could be outcompeted by addition of unlabeled Pr2 but not with non-specific competitor DNA indicating that CcpA binds specifically to PapuA (Figure S6 panels H and I). Recombinant CcpA also appears to bind to the ΔOM2/cre fragment although less DNA/transcription factor complex is observed suggesting that CcpA may also bind OM1 or other sequences in Pr2 with lower affinity (Figure S6J).
Taken together, these results show that apuA expression may be regulated via repression through CcpA-mediated carbon catabolite control and via transcriptional activation through a dedicated regulator encoded by the apuR gene.
Relief from CCR increases adhesion and invasion of S. suis to porcine epithelial cells
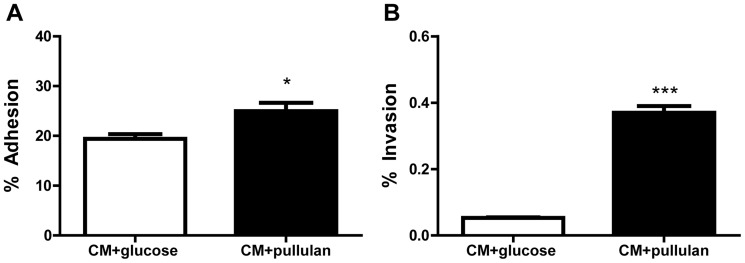
As our microarray data showed increased transcription of apuA and seven other genes predicted to play a role in S. suis adhesion and invasion in starch/pullulan compared to glucose, we hypothesized that culturing S. suis in pullulan as single carbon source, thus in absence of glucose might increase its adhesion and invasion capacity. Exponentially growing S. suis grown in CM supplemented with 1% of glucose or pullulan were incubated with NPTr cells for 2 h at m.o.i. of ∼50 bacteria/cell. To maintain similar conditions during the period of co-culture, the cell culture medium was replaced with glucose-free DMEM supplemented with either 1% glucose or pullulan during the 2 h incubation with S. suis. In agreement with previous adhesion studies using the NPTr cell line [19], we found that adherence of S. suis bacteria grown in CM plus glucose was 19.4±1.0% of original inoculum (averaged over 3 independent replicates). The adherence of S. suis was significantly increased (24.9±1.7%; P<0.05; 3 replicates) after growth in 1% pullulan (Figure 6A). In accordance with previous studies using a human Hep-2 cell line [49], S. suis 2 S10 showed low invasion capacity (0.05% of original inoculum) in glucose. Invasiveness of S. suis was nearly 9-fold higher when grown in pullulan (0.40%±0.01) (P<0.01) (Figure 6B), corresponding to approximately 8.0×102 cfu/ml when grown in glucose, to 7.2×103 cfu/ml in pullulan. In conclusion, S. suis grown in starch/pullulan showed a small but significant increase in adherence to, and a strongly increased invasiveness of NPTr cells compared to S. suis grown in glucose.
Figure 6. Comparison of adherence A. and invasion B. of S. suis after growth in CM+1% w/v pullulan (black bars) vs. CM+1% w/v glucose (white bars).
NPTr confluent monolayers were co-cultivated for 2 h with S. suis S10 bacteria grown in CM plus pullulan or glucose. Adherence and invasion are shown as mean % values of the initial inoculum from two independent experiments in triplicate. Error bars indicate the SD.
Carbon catabolite control of suilysin expression
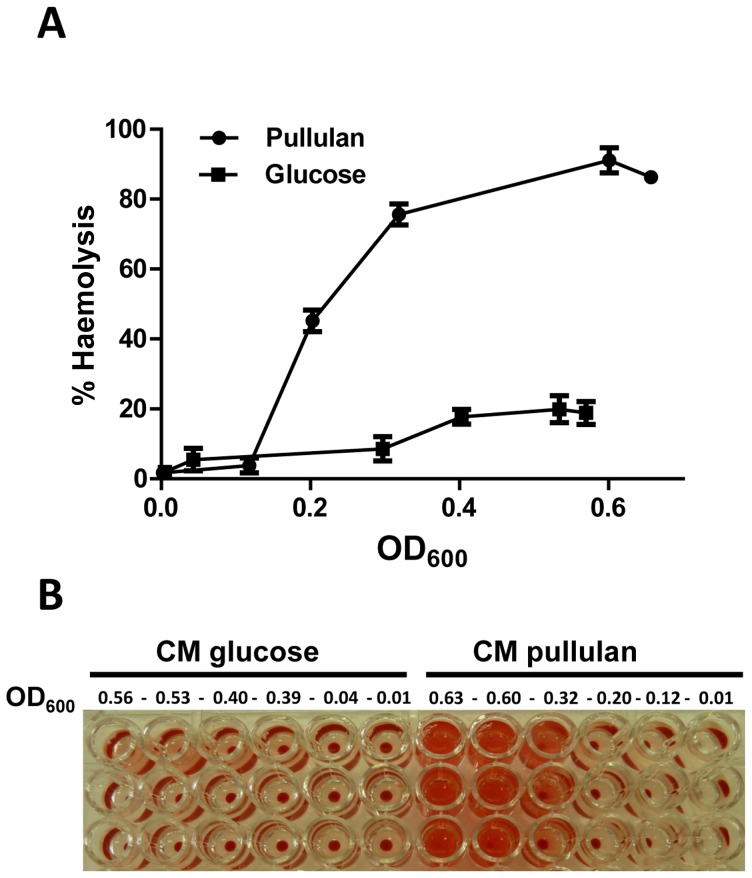
The expression of sly was strongly induced in pullulan compared to glucose, both in exponential and stationary phase (+18.1/+17.9 ratio in pullulan vs glucose). The presence of a cre site in the sly promoter region suggested that the induction of sly during growth in pullulan was due to relief from CcpA-mediated carbon catabolite repression rather than a specific induction by starch/pullulan. As suilysin has been proposed to compromise the integrity of the host epithelium and facilitate entry into the body we measured erythrocyte hemolytic activity (HA) of culture supernatants of S. suis grown in glucose or pullulan. HA was significantly higher in supernatants of S. suis grown in pullulan compared to glucose over a wide range (ca. 0.5) of OD600 values (P<0.001; Figure 7). In early stationary phase, HA for pullulan and glucose cultures was approx. 91% and 18%, respectively (Figure 7).
Figure 7. Hemolysis assay of S. suis growing in two different carbon sources.
A. The hemolysis production was measured by analyzing the supernatant of S. suis grown in CM plus 1% w/v of glucose or pullulan in the lag, exponential and stationary phase (OD600 values 0 to 0.56). B. Deep-well titer plate showing hemolytic activity of supernatants collected from S. suis grown in CM supplemented with glucose or pullulan.
In vivo expression of the virulence factors apuA and sly
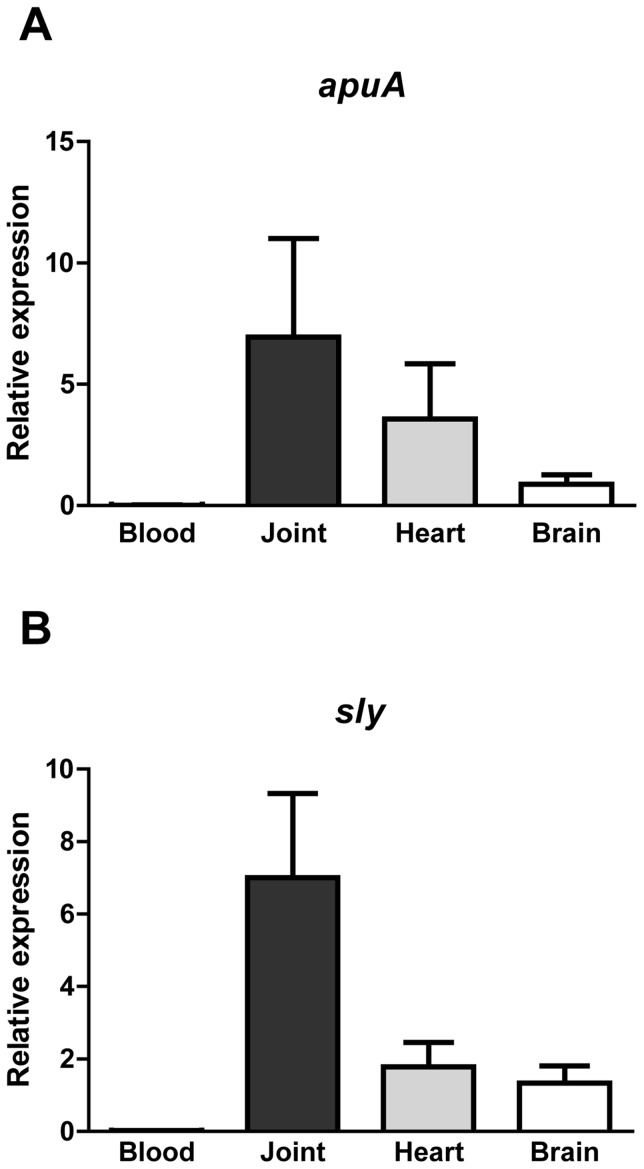
Based on the results from this study so far, we predicted that expression of apuA and sly would be substantially higher in the mucosal colonization stage of the infection when glucose is scarce than in the bloodstream where glucose levels are sufficient to support growth and induce CCR. S. suis invasive disease often leads to infection of the joints, heart and brain. To test our hypothesis S. suis was recovered from the blood, synovial joints, heart and brains of infected piglets immediately after euthanasia and RNA was extracted from the blood and organs. As hypothesized we found transcription of sly and apuA to be significantly higher in the synovial joints, heart and brain than in the blood (Figure 8). These results show that our regulatory model (Figure 9) was informative for some aspects of porcine infection with S. suis.
Figure 8. Expression of apuA and sly in S. suis recovered from blood and tissues of experimentally infected piglets.
The relative expression of apuA and sly in S. suis blood, joints, heart and brain recovered from intravenously infected piglets calculated using the GeNorm method [38] using three housekeeping genes for data normalization. A. The relative expression of apuA (×107) are shown for S. suis recovered from blood and different organs. B. Relative expression of sly (×105) in blood and different organs.
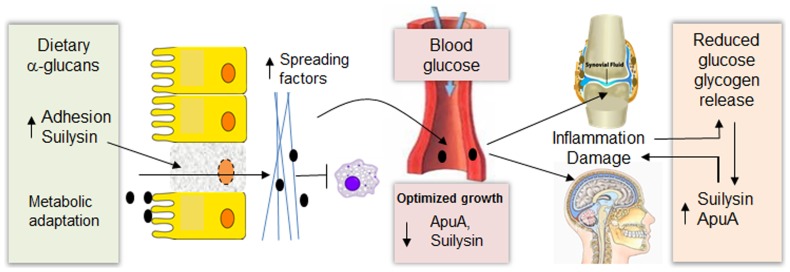
Figure 9. Links between carbohydrate metabolism and virulence in Streptococcus suis.
At the mucosal surfaces a high ratio of a-glucans to glucose upregulates expression of several sugar transport systems and metabolic pathways associated with starch metabolism. Additionally, several virulence factors involved in adherence to host cells, degradation of connective tissue (spreading factors), and avoidance of phagocytic killing, including ApuA and suilysin are upregulated when glucose is diminished. Suilysin may facilitate dispersion of bacteria in mucosal tissues due to loss of barrier integrity. Once S. suis reaches the bloodstream metabolism is adapted for optimal growth on glucose and the expression of virulence factors is reduced by CcpA mediated-repression. In infected organs glucose levels are lower than in the blood and are further reduced by inflammation and utilization by S. suis leading to upregulation of ApuA, suilysin and other virulence factors. In the organs and tissues, glycogen released from damaged cells is degraded by ApuA to generate maltodextrins which sustain growth of S. suis.
Discussion
Asymptomatic carriage of S. suis is common in adult pigs but young piglets that become colonized by S. suis may develop invasive disease. Host susceptibility to S. suis infection may occur due to inadequate host immune responses but may also be due to environmental factors such as the availability of glucose or other carbohydrates. Here we investigated whether the availability of different types of carbohydrates, notably starch and glucose, could influence the switch from an asymptomatic to a pathogenic association of S. suis with the host. To search for the mechanism controlling such a switch, we initially performed an in vitro transcriptomics study on S. suis grown in CM medium supplemented with pullulan or glucose; the latter mediates carbon catabolite control in streptococci via catabolite control protein A (CcpA). Pullulan, a starch, was chosen since we had previously found that growth of S. suis in presence of this carbohydrate induces expression of apuA, a gene known to be involved in carbohydrate metabolism [19].
A switch from growth in glucose to starch/pullulan resulted in a change of ca. 50% of its transcriptome, involving multiple GO function categories (Figure 1). As anticipated many genes annotated with GO term “carbohydrate metabolism” were differentially expressed, including the α-glucan-starch-degrading amylopullulanase apuA [19] and downstream pathway genes required for maltodextrin metabolism (Figure 2).
In other streptococcal pathogens CcpA and CCC have been shown to play a role in virulence gene expression as well as colonization [50], [51] and virulence in an animal model [51]. To predict which S. suis genes might be regulated by CCC we searched for conserved cre sites in the genome of S. suis P1/7 and identified 172 genes, some of which were organized in operons (Table S3). Of these 172 genes, 145 (84%) were differentially regulated during growth in pullulan compared to glucose. The difference in these two values may be due to incorrect cre site predictions or to the fact that expression of certain genes or operons are controlled by additional transcription factors, for example suilysin may be also co-regulated by the two-component system CovS/CovR (SSU1190-SSU1191) [52]. The 145 genes that appeared to be controlled by CCC were also compared to a published microarray data set generated by comparison of an S. suis serotype 2 ΔccpA mutant with the corresponding wild-type strain [43]. In total, 99 genes comprising 38 operons were differentially regulated in both datasets (68% overlap; Table S3).
The CCC-regulon included genes encoding transcriptional regulators, carbohydrate metabolic enzymes and, importantly, putative or known virulence factors (8%) (Table S3). The most highly up-regulated virulence gene in pullulan compared to glucose was apuA encoding a cell-surface amylopullulanase required for growth on starch or glycogen that also contributes to adherence to porcine epithelial cells in vitro and thus may be relevant to mucosal colonization [19]. Expression of apuA was highly induced during growth in starch/pullulan or maltotriose, the most abundant degradation product of pullulan, and repressed during growth in glucose (Figure 3 and 4). An analysis of the promoter region of apuA revealed the presence of two transcription binding motifs (Figure 5) and EMSA studies showed that apuA expression was co-regulated by CcpA and a second regulator, ApuR (Figure S6). The similar location of the conserved OM in the promoters regulated by ApuR and the known transcriptional activator of maltodextrin utilization cluster Lmo2128 in L. monocytogenes [47] and MdxR in E. faecium [48], with high sequence similarity to ApuR, suggested that ApuR might also be a transcriptional activator. Addition of maltotriose to S. suis exponentially growing in medium containing lactose as a carbon source, significantly induced expression of apuA and, to a lesser extent, apuR suggesting that ApuR, like Lmo2128, might be allosterically regulated by maltotriose (Figure 4). In contrast, CcpA binding repressed apuA transcription, most likely through binding to an OM2/cre site overlapping with the −35 promoter element, since addition of maltotriose to S. suis bacteria precultured in glucose as sole carbon source did not lead to significant induction of apuA. This dual regulatory model postulates that apuA is maximally expressed when glucose level is low, thus allowing relief from CcpA-mediated catabolite repression, and when substrates that can be degraded by apuA are present.
In addition to apuA, eighteen other predicted or known virulence genes were differentially regulated in pullulan vs glucose, seven of which were highly upregulated in pullulan compared to glucose (Table 1, Figure S3). One of these genes was sly encoding suilysin, a pore-forming toxin which was secreted in 5-fold higher amounts in pullulan compared to glucose (Figure 7); its increased expression was confirmed in qPCR assays (Figure 3). Suilysin plays an important role in damaging host epithelial [12], [13], [53], endothelial [54], [55] and immune cells [56], suggesting that suilysin could play roles in vivo in damaging and penetration of different cell and tissue types, promoting tissue invasion and inhibition or killing of leukocytes [3], [57]. Of note, virulent S. suis strains that do not produce Sly may still be invasive in pigs, possibly best exemplified by Allen et al (2001) [58] who reported that an isogenic S. suis sly mutant, lacking of the hemolytic characteristics, was nearly as invasive as the wild-type parental strain in a pig infection studies after bacteria intravenous injection. The authors of the study by Allen et al (2001) [58] proposed that Sly is relevant for translocation across epithelia and during the infection stages prior to dispersion via the blood, and proposed that production of Sly may correlate with increased severity of clinical symptoms and the capacity to reach higher colonization of organs. This notion is supported by the study of King et al (2001) [59] who reported that sly was present in a significantly higher number of isolates from pigs with meningitis, septicemia, and arthritis than isolates from pigs with pneumonia. Conversely, sly is also significantly overrepresented in asymptomatic carriage strains of pigs [59], showing that presence of sly does not exclusively correlate with invasive isolates. In addition to suilysin, it is likely that other virulence factors are relevant for the invasion of connective tissue, such as the hyaluronidase and heparinase II/III-like proteins. Note that the function of these putative virulence proteins still has to be demonstrated. In contrast to our results, sly expression was not increased in a recent study which compared gene expression of a wild-type strain with a ΔccpA mutant grown to stationary phase [43]. The reason for these different findings may be related to the use of THB growth medium [43], a rich medium containing glucose, instead of the supplemented CM medium that we used, or different growth stages or different physiological states of the bacteria in the different media. Lastly, it is also possible that expression of sly is controlled by a CcpA-independent carbon catabolite repression mechanism although a cre site is present in the sly promoter region.
We propose that at mucosal surfaces the high ratios of starch to glucose promotes production of extracellular ApuA to support growth of S. suis in the oropharyngeal cavity. Additionally relief from CCR leads to higher expression of other virulence genes involved in mucosal infection (Figure 9). Indeed, in an in vitro experiment employing tracheal epithelial cells and S. suis bacteria grown with starch/pullulan or glucose as sole carbon source, a nearly 10-fold increase of epithelial cell invasion was observed when bacteria were grown with pullulan. We observed that in presence of starch, S. suis induced expression of the metabolic pathway enzymes required for transport and metabolism of maltodextrins and the glycogen biosynthesis enzymes which generate bacterial glycogen energy reserves. Several studies have linked bacterial glycogen metabolism to environmental survival, symbiotic performance, and colonization and virulence [60]–[67]. Glycogen reserves may be important for survival and fitness of S. suis when carbon sources are scarce. During colonization, relief from CCR would increase production of suilysin and may facilitate dispersion of S. suis into the deeper tissues due to loss of barrier integrity [4], [53], [58]. We hypothesized that once S. suis would reach the bloodstream, higher glucose concentrations (from 4.4 to 6.6 mM) would repress expression of apuA and sly by CCR (Figure 9). Indeed, gene expression analysis of bacteria isolated from the blood or organs of acutely infected pigs showed significantly lower expression of apuA and sly in blood than in the organs (Figure 8) supporting occurrence of CCR. In contrast, S. suis isolated from infected joints, heart or brain tissue expressed significantly higher levels of sly and apuA than bacteria isolated from blood. These results are probably due to the lower levels of glucose in the tissues (0.25–0.55 mM in normal synovial fluid and joint cavities) than in the blood (from 4.4 to 6.6 mM) [68], [69] and thus, relief from CCR when bacteria were located in the organs. In inflamed tissues, glucose levels may have been reduced further due to the consumption of glucose by neutrophils and macrophages. It is tempting to speculate that induction of suilysin expression in the tissues will release host glycogen from damaged cells that would be degraded by ApuA and metabolized to further support growth of S. suis (Figure 9). Additionally S. suis may be able to metabolize host glucans such as hyaluronan which is present in high concentrations (3–4 mg/ml) in synovial fluid [70].
Taken together, our results clearly demonstrate that the availability of glucose and other carbohydrates such as starch serves as an environmental cue to regulate the expression of apuA and other virulence genes. We propose a schematic model of how carbohydrate content of different tissues could modulate S. suis metabolism at the different stages of infection (Figure 9). Our novel findings demonstrate a regulatory mechanism dependent on relief from CcpA repression that links carbohydrate metabolism and virulence at least in vitro and that might play roles in vivo, at different stages of S. suis infection. Awareness of the role of carbohydrate content of the S. suis environment may lead to new strategies for combating this important disease, for instance via modulation of carbohydrate content and composition of animal feeds, or the inhibition of S. suis enzymes required for metabolism of starch.
Supporting Information
S. suis S10 growth curve at 37°C in CM containing pullulan or glucose at 1% w/v.
(TIF)
GO term distribution of S. suis genes with the annotation “carbohydrate metabolism” that were differentially upregulated in starch/pullulan vs glucose. A. early exponential (e) and B. early stationary phase (s) Enrichment analysis performed using BLAST2GO (P = 0.05, two-tailed Fisher's Exact test).
(TIF)
Putative and characterized virulence gene expression ratios in pullulan vs. glucose. The genes are grouped according to their predicted or described function in S. suis pathogenesis and expression ratios are shown for exponential (white bar) and early stationary (black bar) growth phases. Envelope: cps2E*-SS0519 putative galactosyl transferase; wchF*-SSU0520 putative rhamnosyl transferase; cps2C-SSU0517 tyrosine-protein kinase; cpss_lpl-SSU1123 putative glycosyltransferase; cpss_lpl2-SSU1124 putative rhamnosyl transferase pgdA-SSU1448 peptidoglycan GlcNAc deacetylase, dltA-SSU0596 D-alanine-poly(phosphoribitol) ligase subunit1; Envelope/Adhesion neuB-SSU0535 putative N-acetylneuraminic acid synthase; neuC-SSU0536 putative UDP-N acetylglucosamine 2-epimerase; neuA-SSU0538 N-acylneuraminate cytidylyltransferase; Adhesion: apuA*-SSU1849 amylopullulanase; sadP-SSU0253 putative surface-anchored protein receptor; gnd-SSU1541 6-phosphogluconate dehydrogenase; Adhesion/Invasion: srtA-SSU0925 sortase; fbpS-SSU1311 fibronectin-fibrinogen binding protein; gapdH-SSU0153 glyceraldehyde-3-phosphate dehydrogenase; eno-SSU1320 enolase; pepD-SSU1215 putative surface-anchored dipeptidase; dpp IV-SSU0187 Xaa-Pro dipeptidyl-peptidase; Invasion: sly* SSU1231 suilysin (haemolysin); hepII/III*-SSU1048 heparinase II/III-like protein; hyl*-SSU1050 hyaluronidase precursor; ssnA*-SSU1760 surface-anchored DNA nuclease; arcB*-SSU0580 arginine deaminase; igaP-SSU1773 putative surface-anchored serine protease; sspA-SSU0757 cell envelope proteinase; ofs-SSU1474 serum opacity factor; luxS-SSU0376 S-ribosyl homocysteinase; Marker: mrp-SSU0706 muramidase-released protein precursor; ef-SSU0171 putative surface-anchored protein. * Indicates the presence of a predicted cre in the virulence gene promoter region.
(TIF)
Gene homologues in Gram-positive bacteria that share ≥45% of protein identity with the S. suis apuR gene (black arrows). The annotations of the genes downstream of apuR are also indicated and colored to show functional relatedness. Gene names are indicated above the arrows.
(TIF)
SDS polyacrylamide electrophoresis of purified transcriptional regulators. A. Coomassie stained SDS-PAGE gel (12%), showing purified fraction His-ApuR at the expected size of 38 kDa and B. Western Blot of the same gel with a monoclonal His-tag antibody protein. C. Coomassie stained SDS-PAGE gel (12%) of purified His-CcpA at expected size of 40 kDa.
(TIF)
EMSA with purified ApuA and CcpA. A. Schematic representation of the apuA promoter (PapuA) and fluorescently labeled PCR amplified DNA fragments (Pr1, Pr2 and Pr3) used for EMSAs. B. to F. DNA amplicons and DNA/rApuR protein complexes visualized in native 5% acrylamide gels using the Odyssey Imager. In these EMSA assays the concentration of each DNA amplicon was around 6 ng (∼50 nM) B. EMSA of 100 nM rApuR binding to Pr1, Pr2 and Pr3. C. Increasing DNA/rApuR complex formation in presence of 4 ng Pr2 DNA amplicon and an increasing amount of rApuR (∼0,5–4 µM as indicated) D. Competitive EMSA using increasing concentrations of non-fluorescent non-specific competitor DNA (lacking OM1 binding motifs). E. Competitive EMSA using increasing concentrations of non-fluorescent Pr2 as a specific competitor. The amounts of competitor DNA added are indicated (25–150 nM) F. Identification of specific ApuR binding sites in PapuA. PR2: native promoter region fragment 2. ΔOM1 and ΔOM2/cre are synthetic DNA fragments of PapuA that lack the predicted binding sites. + rApuR recombinant present - rApuR recombinant absent. G. to J. DNA amplicons and DNA/rCcpA protein complexes visualized in native 5% acrylamide gels using the Odyssey Imager. G. DNA/rCcpA complexes in the presence of increasing amounts of rCcpA as indicated. H. Competitive EMSA using increasing concentrations of non-fluorescent specific competitor I. Competitive EMSA using increasing concentrations of non-fluorescent non-specific competitor DNA (lacking OM2/cre binding motifs) J. DNA/rCcpA complex formation with fluorescent Pr2 and two synthetic promoter Pr2 fragments lacking either OM2/cre or OM1.
(TIF)
Oligonucleotide primers used in this study.
(DOCX)
Microarray data (xl file): S. suis genes differentially regulated during growth in pullulan (Pul) vs glucose (Glc) in the exponential (e) or stationary (s) phase.
(XLSX)
cre-site prediction in the genome of S. suis P1/7.
(DOCX)
Motif OM1-OM2/cre like motifs identified in S. suis 2 P1/7 and other Gram positive bacteria.
(DOCX)
Material and methods.
(DOCX)
Results (Supporting text on differentially expressed genes, pathways and their predicted or known functions).
(DOCX)
Acknowledgments
The authors are grateful to Michiel Kleerebezem for valuable discussions and insights on heterofermentative metabolism.
Funding Statement
The work was funded by The Netherlands Top Institute for Food and Nutrition. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lun ZR, Wang QP, Chen XG, Li AX, Zhu XQ (2007) Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis 7: 201–209. [DOI] [PubMed] [Google Scholar]
- 2. Wertheim HF, Nghia HD, Taylor W, Schultsz C (2009) Streptococcus suis: an emerging human pathogen. Clin Infect Dis 48: 617–625. [DOI] [PubMed] [Google Scholar]
- 3. Gottschalk M, Segura M (2000) The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol 76: 259–272. [DOI] [PubMed] [Google Scholar]
- 4. Fittipaldi N, Segura M, Grenier D, Gottschalk M (2012) Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis . Future Microbiol 7: 259–279. [DOI] [PubMed] [Google Scholar]
- 5. Meurman JH, Rytomaa I, Kari K, Laakso T, Murtomaa H (1987) Salivary pH and glucose after consuming various beverages, including sugar-containing drinks. Caries Res 21: 353–359. [DOI] [PubMed] [Google Scholar]
- 6. Gough H, Luke GA, Beeley JA, Geddes DA (1996) Human salivary glucose analysis by high-performance ion-exchange chromatography and pulsed amperometric detection. Arch Oral Biol 41: 141–145. [DOI] [PubMed] [Google Scholar]
- 7. Bakker GC, Dekker RA, Jongbloed R, Jongbloed AW (1998) Non-starch polysaccharides in pig feeding. Vet Q 20 Suppl 3: S59–64. [PubMed] [Google Scholar]
- 8. Mormann JE, Muhlemann HR (1981) Oral starch degradation and its influence on acid production in human dental plaque. Caries Res 15: 166–175. [DOI] [PubMed] [Google Scholar]
- 9. Mormann JE, Amadò R, Neukom H (1982) Comparative Studies on the in vitro alpha-Amylolysis of Different Wheat Starch Products. Starch - Stärke 34: 121–124. [Google Scholar]
- 10. Taravel FR, Datema R, Woloszczuk W, Marshall JJ, Whelan WJ (1983) Purification and characterization of a pig intestinal alpha-limit dextrinase. Eur J Biochem 130: 147–153. [DOI] [PubMed] [Google Scholar]
- 11. Shelburne SA 3rd, Granville C, Tokuyama M, Sitkiewicz I, Patel P, et al. (2005) Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun 73: 4723–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Norton PM, Rolph C, Ward PN, Bentley RW, Leigh JA (1999) Epithelial invasion and cell lysis by virulent strains of Streptococcus suis is enhanced by the presence of suilysin. FEMS Immunol Med Microbiol 26: 25–35. [DOI] [PubMed] [Google Scholar]
- 13. Lalonde M, Segura M, Lacouture S, Gottschalk M (2000) Interactions between Streptococcus suis serotype 2 and different epithelial cell lines. Microbiology 146 Pt 8: 1913–1921. [DOI] [PubMed] [Google Scholar]
- 14. Walker GJ, Builder JE (1971) Metabolism of the reserve polysaccharide of Streptococcus mitis. Properties of branching enzyme, and its effect on the activity of glycogen synthetase. Eur J Biochem 20: 14–21. [DOI] [PubMed] [Google Scholar]
- 15. Eisenberg RJ, Elchisak M, Lai C (1974) Glycogen accumulation by pleomorphic cells of Streptococcus sanguis. Biochem Biophys Res Commun 57: 959–966. [DOI] [PubMed] [Google Scholar]
- 16. Bongaerts RJ, Heinz HP, Hadding U, Zysk G (2000) Antigenicity, expression, and molecular characterization of surface-located pullulanase of Streptococcus pneumoniae . Infect Immun 68: 7141–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shelburne SA 3rd, Keith DB, Davenport MT, Horstmann N, Brennan RG, et al. (2008) Molecular characterization of group A Streptococcus maltodextrin catabolism and its role in pharyngitis. Mol Microbiol 69: 436–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbott DW, Higgins MA, Hyrnuik S, Pluvinage B, Lammerts van Bueren A, et al. (2010) The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae . Mol Microbiol 77: 183–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferrando ML, Fuentes S, de Greeff A, Smith H, Wells JM (2010) ApuA, a multifunctional alpha-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology 156: 2818–2828. [DOI] [PubMed] [Google Scholar]
- 20. Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH Jr (2001) Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol 39: 1366–1381. [DOI] [PubMed] [Google Scholar]
- 21. Deutscher J, Francke C, Postma PW (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70: 939–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vecht U, Arends JP, van der Molen EJ, van Leengoed LA (1989) Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am J Vet Res 50: 1037–1043. [PubMed] [Google Scholar]
- 23. Smith HE, Buijs H, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA (2001) Selection of virulence-associated determinants of Streptococcus suis serotype 2 by in vivo complementation. Infect Immun 69: 1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Greeff A, Wisselink HJ, de Bree FM, Schultsz C, Baums CG, et al. (2011) Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC Microbiol 11: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santi I, Pezzicoli A, Bosello M, Berti F, Mariani M, et al. (2008) Functional characterization of a newly identified group B Streptococcus pullulanase eliciting antibodies able to prevent alpha-glucans degradation. PLoS One 3: e3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saulnier DM, Santos F, Roos S, Mistretta TA, Spinler JK, et al. (2011) Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One 6: e18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP (2003) MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2: 241–244. [PubMed] [Google Scholar]
- 30. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 31. Zdobnov EM, Apweiler R (2001) InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. [DOI] [PubMed] [Google Scholar]
- 32. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54. [DOI] [PubMed] [Google Scholar]
- 34. Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, et al. (2010) RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res 38: W299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaptal V, Gueguen-Chaignon V, Poncet S, Lecampion C, Meyer P, et al. (2006) Structural analysis of B. subtilis CcpA effector binding site. Proteins 64: 814–816. [DOI] [PubMed] [Google Scholar]
- 36. Jacobs AA, Loeffen PL, van den Berg AJ, Storm PK (1994) Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis . Infect Immun 62: 1742–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferrari M, Losio MN, Bernori E, Lingeri R (1993) Established thyroid cell line of newborn pig (NPTh). New Microbiol 16: 381–384. [PubMed] [Google Scholar]
- 38. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- 40. Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y (2000) Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis . Nucleic Acids Res 28: 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lorca GL, Chung YJ, Barabote RD, Weyler W, Schilling CH, et al. (2005) Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J Bacteriol 187: 7826–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujita Y (2009) Carbon catabolite control of the metabolic network in Bacillus subtilis . Biosci Biotechnol Biochem 73: 245–259. [DOI] [PubMed] [Google Scholar]
- 43. Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, et al. (2011) Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis . Microbiology 157: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 44. Monedero V, Yebra MJ, Poncet S, Deutscher J (2008) Maltose transport in Lactobacillus casei and its regulation by inducer exclusion. Research in microbiology 159: 94–102. [DOI] [PubMed] [Google Scholar]
- 45. Schonert S, Seitz S, Krafft H, Feuerbaum EA, Andernach I, et al. (2006) Maltose and maltodextrin utilization by Bacillus subtilis . Journal of bacteriology 188: 3911–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shim JH, Park JT, Hong JS, Kim KW, Kim MJ, et al. (2009) Role of maltogenic amylase and pullulanase in maltodextrin and glycogen metabolism of Bacillus subtilis 168. Journal of bacteriology 191: 4835–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gopal S, Berg D, Hagen N, Schriefer EM, Stoll R, et al. (2010) Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PloS one 5: e10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X, Rogers M, Bierschenk D, Bonten MJ, Willems RJ, et al. (2013) A LacI-family regulator activates maltodextrin metabolism of Enterococcus faecium . PLoS One 8: e72285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benga L, Goethe R, Rohde M, Valentin-Weigand P (2004) Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol 6: 867–881. [DOI] [PubMed] [Google Scholar]
- 50. Iyer R, Baliga NS, Camilli A (2005) Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae . J Bacteriol 187: 8340–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shelburne SA 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, et al. (2008) A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus . Proc Natl Acad Sci U S A 105: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pan X, Ge J, Li M, Wu B, Wang C, et al. (2009) The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J Bacteriol 191: 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seitz M, Baums CG, Neis C, Benga L, Fulde M, et al. (2013) Subcytolytic effects of suilysin on interaction of Streptococcus suis with epithelial cells. Vet Microbiol 167: 584–591. [DOI] [PubMed] [Google Scholar]
- 54. Vanier G, Segura M, Gottschalk M (2007) Characterization of the invasion of porcine endothelial cells by Streptococcus suis serotype 2. Can J Vet Res 71: 81–89. [PMC free article] [PubMed] [Google Scholar]
- 55. Charland N, Nizet V, Rubens CE, Kim KS, Lacouture S, et al. (2000) Streptococcus suis serotype 2 interactions with human brain microvascular endothelial cells. Infect Immun 68: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lecours MP, Gottschalk M, Houde M, Lemire P, Fittipaldi N, et al. (2011) Critical role for Streptococcus suis cell wall modifications and suilysin in resistance to complement-dependent killing by dendritic cells. J Infect Dis 204: 919–929. [DOI] [PubMed] [Google Scholar]
- 57. Gottschalk M, Xu J, Calzas C, Segura M (2011) Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5: 371–391. [DOI] [PubMed] [Google Scholar]
- 58. Allen AG, Bolitho S, Lindsay H, Khan S, Bryant C, et al. (2001) Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect Immun 69: 2732–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. King SJ, Heath PJ, Luque I, Tarradas C, Dowson CG, et al. (2001) Distribution and genetic diversity of suilysin in Streptococcus suis isolated from different diseases of pigs and characterization of the genetic basis of suilysin absence. Infect Immun 69: 7572–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bonafonte MA, Solano C, Sesma B, Alvarez M, Montuenga L, et al. (2000) The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis . FEMS Microbiol Lett 191: 31–36. [DOI] [PubMed] [Google Scholar]
- 61. Bourassa L, Camilli A (2009) Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae . Mol Microbiol 72: 124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, et al. (2004) Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101: 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, et al. (2008) Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Henrissat B, Deleury E, Coutinho PM (2002) Glycogen metabolism loss: a common marker of parasitic behaviour in bacteria? Trends Genet 18: 437–440. [DOI] [PubMed] [Google Scholar]
- 65. Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, et al. (2008) Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun 76: 2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sambou T, Dinadayala P, Stadthagen G, Barilone N, Bordat Y, et al. (2008) Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Mol Microbiol 70: 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilson WA, Roach PJ, Montero M, Baroja-Fernandez E, Munoz FJ, et al. (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 34: 952–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pitsillides AA, Skerry TM, Edwards JC (1999) Joint immobilization reduces synovial fluid hyaluronan concentration and is accompanied by changes in the synovial intimal cell populations. Rheumatology (Oxford) 38: 1108–1112. [DOI] [PubMed] [Google Scholar]
- 69. Praest BM, Greiling H, Kock R (1997) Assay of synovial fluid parameters: hyaluronan concentration as a potential marker for joint diseases. Clin Chim Acta 266: 117–128. [DOI] [PubMed] [Google Scholar]
- 70. Hui AY, McCarty WJ, Masuda K, Firestein GS, Sah RL (2012) A systems biology approach to synovial joint lubrication in health, injury, and disease. Wiley Interdiscip Rev Syst Biol Med 4: 15–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, et al. (1999) Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67: 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith HE, de Vries R, van't Slot R, Smits MA (2000) The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb Pathog 29: 127–134. [DOI] [PubMed] [Google Scholar]
- 73. Fittipaldi N, Sekizaki T, Takamatsu D, de la Cruz Dominguez-Punaro M, Harel J, et al. (2008) Significant contribution of the pgdA gene to the virulence of Streptococcus suis . Mol Microbiol 70: 1120–1135. [DOI] [PubMed] [Google Scholar]
- 74. Fittipaldi N, Sekizaki T, Takamatsu D, Harel J, Dominguez-Punaro Mde L, et al. (2008) D-alanylation of lipoteichoic acid contributes to the virulence of Streptococcus suis . Infect Immun 76: 3587–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. de Greeff A, Buys H, Verhaar R, Dijkstra J, van Alphen L, et al. (2002) Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect Immun 70: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, et al. (2008) Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis . Microbiology 154: 2668–2679. [DOI] [PubMed] [Google Scholar]
- 77. Brassard J, Gottschalk M, Quessy S (2004) Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet Microbiol 102: 87–94. [DOI] [PubMed] [Google Scholar]
- 78. Ge J, Feng Y, Ji H, Zhang H, Zheng F, et al. (2009) Inactivation of dipeptidyl peptidase IV attenuates the virulence of Streptococcus suis serotype 2 that causes streptococcal toxic shock syndrome. Curr Microbiol 59: 248–255. [DOI] [PubMed] [Google Scholar]
- 79. Jobin MC, Grenier D (2003) Identification and characterization of four proteases produced by Streptococcus suis . FEMS Microbiol Lett 220: 113–119. [DOI] [PubMed] [Google Scholar]
- 80. Tan C, Fu S, Liu M, Jin M, Liu J, et al. (2008) Cloning, expression and characterization of a cell wall surface protein, 6-phosphogluconate-dehydrogenase, of Streptococcus suis serotype 2. Vet Microbiol 130: 363–370. [DOI] [PubMed] [Google Scholar]
- 81. Si Y, Yuan F, Chang H, Liu X, Li H, et al. (2009) Contribution of glutamine synthetase to the virulence of Streptococcus suis serotype 2. Vet Microbiol 139: 80–88. [DOI] [PubMed] [Google Scholar]
- 82. Kouki A, Haataja S, Loimaranta V, Pulliainen AT, Nilsson UJ, et al. (2011) Identification of a novel streptococcal adhesin P (SadP) protein recognizing galactosyl-alpha1-4-galactose-containing glycoconjugates: convergent evolution of bacterial pathogens to binding of the same host receptor. J Biol Chem 286: 38854–38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Winterhoff N, Goethe R, Gruening P, Rohde M, Kalisz H, et al. (2002) Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the Arginine Deiminase system of Streptococcus pyogenes . J Bacteriol 184: 6768–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fontaine MC, Perez-Casal J, Willson PJ (2004) Investigation of a novel DNase of Streptococcus suis serotype 2. Infect Immun 72: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bonifait L, Vaillancourt K, Gottschalk M, Frenette M, Grenier D (2010) Purification and characterization of the subtilisin-like protease of Streptococcus suis that contributes to its virulence. Vet Microbiol 148: 333–340. [DOI] [PubMed] [Google Scholar]
- 86. Zhang A, Mu X, Chen B, Han L, Chen H, et al. (2010) IgA1 protease contributes to the virulence of Streptococcus suis . Vet Microbiol 148: 436–439. [DOI] [PubMed] [Google Scholar]
- 87. Allen AG, Lindsay H, Seilly D, Bolitho S, Peters SE, et al. (2004) Identification and characterisation of hyaluronate lyase from Streptococcus suis . Microb Pathog 36: 327–335. [DOI] [PubMed] [Google Scholar]
- 88. Tonnaer EL, Hafmans TG, Van Kuppevelt TH, Sanders EA, Verweij PE, et al. (2006) Involvement of glycosaminoglycans in the attachment of pneumococci to nasopharyngeal epithelial cells. Microbes Infect 8: 316–322. [DOI] [PubMed] [Google Scholar]
- 89. Vanier G, Sekizaki T, Dominguez-Punaro MC, Esgleas M, Osaki M, et al. (2007) Disruption of srtA gene in Streptococcus suis results in decreased interactions with endothelial cells and extracellular matrix proteins. Vet Microbiol [DOI] [PubMed] [Google Scholar]
- 90. Baums CG, Kaim U, Fulde M, Ramachandran G, Goethe R, et al. (2006) Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis . Infect Immun 74: 6154–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wen ZT, Burne RA (2002) Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans . Appl Environ Microbiol 68: 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Smith HE, Vecht U, Wisselink HJ, Stockhofe-Zurwieden N, Biermann Y, et al. (1996) Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect Immun 64: 4409–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. suis S10 growth curve at 37°C in CM containing pullulan or glucose at 1% w/v.
(TIF)
GO term distribution of S. suis genes with the annotation “carbohydrate metabolism” that were differentially upregulated in starch/pullulan vs glucose. A. early exponential (e) and B. early stationary phase (s) Enrichment analysis performed using BLAST2GO (P = 0.05, two-tailed Fisher's Exact test).
(TIF)
Putative and characterized virulence gene expression ratios in pullulan vs. glucose. The genes are grouped according to their predicted or described function in S. suis pathogenesis and expression ratios are shown for exponential (white bar) and early stationary (black bar) growth phases. Envelope: cps2E*-SS0519 putative galactosyl transferase; wchF*-SSU0520 putative rhamnosyl transferase; cps2C-SSU0517 tyrosine-protein kinase; cpss_lpl-SSU1123 putative glycosyltransferase; cpss_lpl2-SSU1124 putative rhamnosyl transferase pgdA-SSU1448 peptidoglycan GlcNAc deacetylase, dltA-SSU0596 D-alanine-poly(phosphoribitol) ligase subunit1; Envelope/Adhesion neuB-SSU0535 putative N-acetylneuraminic acid synthase; neuC-SSU0536 putative UDP-N acetylglucosamine 2-epimerase; neuA-SSU0538 N-acylneuraminate cytidylyltransferase; Adhesion: apuA*-SSU1849 amylopullulanase; sadP-SSU0253 putative surface-anchored protein receptor; gnd-SSU1541 6-phosphogluconate dehydrogenase; Adhesion/Invasion: srtA-SSU0925 sortase; fbpS-SSU1311 fibronectin-fibrinogen binding protein; gapdH-SSU0153 glyceraldehyde-3-phosphate dehydrogenase; eno-SSU1320 enolase; pepD-SSU1215 putative surface-anchored dipeptidase; dpp IV-SSU0187 Xaa-Pro dipeptidyl-peptidase; Invasion: sly* SSU1231 suilysin (haemolysin); hepII/III*-SSU1048 heparinase II/III-like protein; hyl*-SSU1050 hyaluronidase precursor; ssnA*-SSU1760 surface-anchored DNA nuclease; arcB*-SSU0580 arginine deaminase; igaP-SSU1773 putative surface-anchored serine protease; sspA-SSU0757 cell envelope proteinase; ofs-SSU1474 serum opacity factor; luxS-SSU0376 S-ribosyl homocysteinase; Marker: mrp-SSU0706 muramidase-released protein precursor; ef-SSU0171 putative surface-anchored protein. * Indicates the presence of a predicted cre in the virulence gene promoter region.
(TIF)
Gene homologues in Gram-positive bacteria that share ≥45% of protein identity with the S. suis apuR gene (black arrows). The annotations of the genes downstream of apuR are also indicated and colored to show functional relatedness. Gene names are indicated above the arrows.
(TIF)
SDS polyacrylamide electrophoresis of purified transcriptional regulators. A. Coomassie stained SDS-PAGE gel (12%), showing purified fraction His-ApuR at the expected size of 38 kDa and B. Western Blot of the same gel with a monoclonal His-tag antibody protein. C. Coomassie stained SDS-PAGE gel (12%) of purified His-CcpA at expected size of 40 kDa.
(TIF)
EMSA with purified ApuA and CcpA. A. Schematic representation of the apuA promoter (PapuA) and fluorescently labeled PCR amplified DNA fragments (Pr1, Pr2 and Pr3) used for EMSAs. B. to F. DNA amplicons and DNA/rApuR protein complexes visualized in native 5% acrylamide gels using the Odyssey Imager. In these EMSA assays the concentration of each DNA amplicon was around 6 ng (∼50 nM) B. EMSA of 100 nM rApuR binding to Pr1, Pr2 and Pr3. C. Increasing DNA/rApuR complex formation in presence of 4 ng Pr2 DNA amplicon and an increasing amount of rApuR (∼0,5–4 µM as indicated) D. Competitive EMSA using increasing concentrations of non-fluorescent non-specific competitor DNA (lacking OM1 binding motifs). E. Competitive EMSA using increasing concentrations of non-fluorescent Pr2 as a specific competitor. The amounts of competitor DNA added are indicated (25–150 nM) F. Identification of specific ApuR binding sites in PapuA. PR2: native promoter region fragment 2. ΔOM1 and ΔOM2/cre are synthetic DNA fragments of PapuA that lack the predicted binding sites. + rApuR recombinant present - rApuR recombinant absent. G. to J. DNA amplicons and DNA/rCcpA protein complexes visualized in native 5% acrylamide gels using the Odyssey Imager. G. DNA/rCcpA complexes in the presence of increasing amounts of rCcpA as indicated. H. Competitive EMSA using increasing concentrations of non-fluorescent specific competitor I. Competitive EMSA using increasing concentrations of non-fluorescent non-specific competitor DNA (lacking OM2/cre binding motifs) J. DNA/rCcpA complex formation with fluorescent Pr2 and two synthetic promoter Pr2 fragments lacking either OM2/cre or OM1.
(TIF)
Oligonucleotide primers used in this study.
(DOCX)
Microarray data (xl file): S. suis genes differentially regulated during growth in pullulan (Pul) vs glucose (Glc) in the exponential (e) or stationary (s) phase.
(XLSX)
cre-site prediction in the genome of S. suis P1/7.
(DOCX)
Motif OM1-OM2/cre like motifs identified in S. suis 2 P1/7 and other Gram positive bacteria.
(DOCX)
Material and methods.
(DOCX)
Results (Supporting text on differentially expressed genes, pathways and their predicted or known functions).
(DOCX)