Abstract
The past decade has seen a proliferation of new species of Miniopterus bats (family Miniopteridae) recognized from Madagascar and the neighboring Comoros archipelago. The interspecific relationships of these taxa, their colonization history, and the evolution of this presumed adaptive radiation have not been sufficiently explored. Using the mitochondrial cytochrome-b gene, we present a phylogeny of the Malagasy members of this widespread Old World genus, based on 218 sequences, of which 82 are new and 136 derived from previous studies. Phylogenetic analyses recovered 18 clades, which divide into five primary lineages: (1) M. griveaudi; (2) M. mahafaliensis, M. sororculus and X3; (3) M. majori, M. gleni and M. griffithsi; (4) M. brachytragos; M. aelleniA, and M. aelleniB; and (5) M. manavi and M. petersoni recovered as sister species, which were in turn linked to a group comprising M. egeri and five genetically distinct populations referred to herein as P3, P4, P5, P6 and P7. Beast analysis indicated that the initial divergence within the Malagasy Miniopterus radiation took place 4.5 Myr; most species diverged between 4 and 2.5 Myr, and a secondary period was between 1.25 and 1 Myr. DNA K2P-distances between recognized taxa ranged from 12.9% to 2.5% and intraspecific variation was less than 1.8%. Of the 18 identified clades, Latin binomials are only associated with 11, which indicates much greater differentiation than currently recognized for Malagasy Miniopterus. These data are placed in a context of the dispersal history of this genus on the island and patterns of ecological diversity.
Introduction
Madagascar is well known as a center of endemism for a wide assortment of plant and animal taxa. This is directly associated with the island's considerable ecological and topographic diversity, as well as isolation in deep geological time [1], [2], [3]. In contrast to other areas of the Old World tropics, Madagascar's distinctive biota contains numerous endemic groups at higher taxonomic levels, representing distinct radiations. In some cases, such as certain reptiles [4], these endemic groups are best explained as vicariant relicts originating before the break-up of Gondwana some 165 million years ago [5], [6]. However, much more common are plant and animal groups that successfully colonized the island by over-water dispersal in more recent geological time [7]. These post-Gondwana-split colonizations occurred across multiple geological periods, resulting in levels of differentiation ranging from endemic orders to genera [7]–[9].
Recent molecular research has provided considerable new insight into these different evolutionary events, levels of taxonomic diversity, and the complexity of various Malagasy radiations. These studies have uncovered cryptic species belonging to previously unrecognized taxa that are largely indiscernible using more classic taxonomic characters. As such, the results of these studies provide the means to differentiate shared evolutionary history versus convergence. The recent recognition of an endemic Malagasy bird family, the Bernieridae, is an excellent example. It comprises 11 species that share no defining morphological characters and formerly were placed in three different songbird families [10], [11]. Members of the endemic family Vangidae were also previously placed in three separate songbird families [12], [13]. Finally, although the island holds a considerable diversity of land mammals, all existing groups (carnivorans, lemurs, rodents, and tenrecs), which show extraordinary morphological variation, can be explained by four colonization events [14]. Study of the extant fauna has therefore shown that successful colonization of Madagascar by land mammals has been rare and accompanied by subsequent adaptive radiations. While several different hypotheses have been presented to explain patterns of endemism and micro-endemism in the island's biota [2], [3], [15], recent research has shown that a single model cannot explain the different patterns observed in the living biota of the island.
In the present study, we explore the complex micro-radiation of a widespread Old World group of bats, the family Miniopteridae. While their wing structure is not designed for high speed, they are relatively strong flyers [16], attested by their capacity to colonize offshore and oceanic islands. In a review of Madagascar's chiropteran fauna, Peterson et al. [17] reported four species from the island: one endemic, two shared with the nearby Comoro archipelago, and one in common with continental Africa. Less than two decades later, largely based on insights from molecular genetics and to a lesser extent morphology and bioacoustics, 11 species are recognized today from the island, all endemic with the exception of two shared with the Comoros [18], [19].
To date, systematic research on Malagasy Miniopterus has concentrated on the delimitation of species. Little attention has been given to the evolutionary relationships of the different taxa relative to Madagascar or nearby islands and continental areas. The purposes of this paper are to apply molecular phylogenetic data to explore primarily the patterns of diversification of members of this genus in Madagascar, within an ecological context. Secondarily, to explore aspects of their colonization history and patterns of dispersal.
Methods
Bat sampling and specimens examined
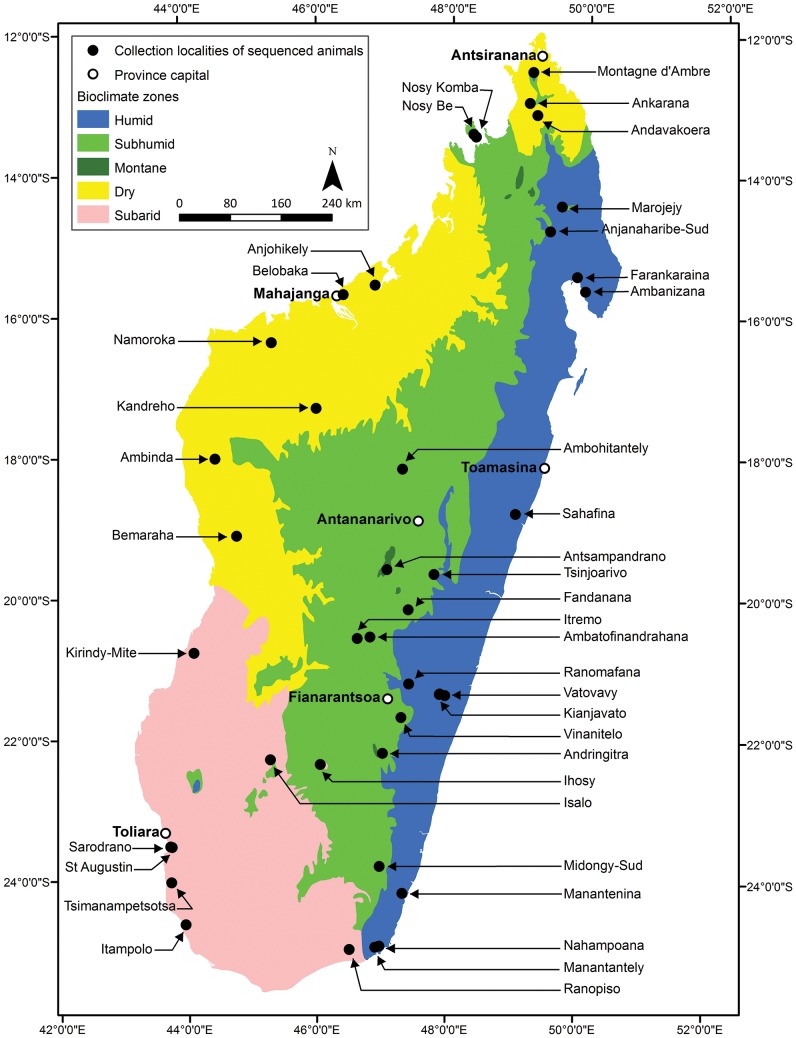
Specimens were captured from diverse areas and habitats for this study, essentially covering the entire range of Miniopterus spp. on Madagascar (Figure 1), using mist nets and harp traps most often placed at cave entrances. This study was conducted in strict accordance with the terms of research permits issued by national authorities in Madagascar (Direction du Système des Aires Protégées, Direction Générale de l’Environnement et des Forêts, and Madagascar National Parks; and in the Union of the Comoros (Centre National de Documentation et de Recherche Scientifique), following the laws of these countries, and the associated research permit numbers are listed in the acknowledgements. Seventy-five animals were captured, manipulated and euthnanized in accordance with guidelines accepted by these different national authorities and the scientific community for the handling of wild animals [20]. Voucher specimens are housed in the Field Museum of Natural History (FMNH), Chicago, and the Université d’Antananarivo, Département de Biologie Animale (UADBA), Antananarivo.
Figure 1. Bioclimatic map of Madagascar with collection localities of all specimens sequenced in this study (see Table S1).
The mitochondrial cytochrome-b (cyt-b: 1140 bp) has previously been shown to be informative at the species level in the study of miniopterine bats [21]–[24], which is our primary focus herein. The dataset we have employed includes all recently published work on Malagasy miniopterine species and incorporates new sequences from specimens previously defined as M. manavi [17]. In total, 264 sequences have been employed herein, 75 acquired for this study and 189 previously used in different taxonomic studies (Table S1). The dataset also incorporates sequences from islands in the Comoros, including Grande Comore and Anjouan [25]. Due to the reliance on pre-existing published sequences to build a complete taxonomy, the study was limited to the use of cyt-b alone, specifically as a number of tissue samples amomgst the 264 samples are not available to the authors for sequencing nuclear or microsatellite markers.
Cyt-b sequences of African, European, Asian and Australasian Miniopterus spp. were also included from Genbank records (Table S1). With no clear sister group to the genus Miniopterus or the family Miniopteridae, we chose Myotis ricketti (EF530349) as the outgroup. The use of outgroup sequences from other chiropteran families did not alter the relationships between the Miniopterus spp. [23], [24], [26], [27] Analysis using M. ricketti as the outgroup resulted in two fully supported (posterior probability 1.00) Miniopterus clades: one consisting of Malagasy, African and European taxa and another consisting of Asian and Australasian taxa. For reasons detailed below and to improve resolution, the Asian and Australasian clade was then used as the outgroup for determining relationships between the Malagasy, African and European taxa.
Molecular analysis
Production of the sequences was achieved using the same methods described in previous studies on Malagasy Miniopterus [27].
Sequences were assembled and aligned using Sequencher version 4.6 (Gene Codes Corporation, Ann Arbor, MI). Analysis using DNA strider [28] showed that sequences did not contain insertions, deletions or stop codons. All new sequences were deposited in GenBank (Accession numbers listed in Table S1). The program jModeltest v2.1.4 [29], [30] reported HKY + G as the optimal nucleotide substitution model for the dataset according to Hierarchical Likelihood Ratio tests, Aikake Information Criterion and Bayesian Information Criterion. This model was applied to the Bayesian and molecular clock analyses.
Bayesian analyses were conducted using MrBayes v3.2 [31] under uniform priors. Four chains were run under MrBayes for 2,000,000 generations with a sampling frequency of 1,000. Burn-in was set at 25% of initial trees. The deviation of split frequencies was below 0.01 at the conclusion of the analysis. Maximum likelihood analyses were run using RaXML Black Box workbench [32], [33], using the GTRGAMMA model. Bootstrap values were estimated using 1000 pseudoreplicates.
Bayesian and ML analyses were initially run with the full dataset in order to confirm fine-scale topology (Figure S1); however, due to the influence of wide variations in sequence divergences on the gamma distribution and increased branch length, these analyses were repeated using only two to four sequences from each major clade and with the removal of the highly divergent outgroup. Myotis proved to be more than 24% divergent in cyt-b (Kimura 2-parameter, K2P) [34] from Miniopterus, significantly altering the shape of the tree and resulting in difficulties in the estimation of rate heterogeneity parameters. As a consequence, Myotis was removed from the analysis in order to aid in the resolution of the tree and to avoid the extensive branch length difficulties reported by recent studies of the phenomenon [35], [36]. The overall topology was unaffected by the removal of the additional individuals and the outgroup.
Molecular clock analyses were conducted using BEAST 1.7.4 [37], [38], incorporating a Yule tree model under a uniform speciation prior. A relaxed uncorrelated lognormal molecular clock [39] was applied using a variable rate of 2.0% sequence evolution per lineage per million years [40]. No further calibration was possible due to the paucity of the fossil record with regard to this group.
All posterior parameter distributions for analysis were checked in Tracer v1.5 [41] for modality and effective sample size (ESS).
Genetic divergence between and within clades were computed as pairwise Kimura 2-parameter distances (K2P) with the software MEGA version 3.1 [42]. The K2P model was chosen to be comparable with previous studies reporting taxonomic inferences on miniopterid bat species based on genetic distances [22], [24], [25], [27], [43].
Results
Complete or near complete cyt-b sequences (1100 to 1140 bp) were obtained for most of the 82 samples sequenced in this study, as well as some critical specimens used in previous taxonomic studies. Exceptions to this were: (1) the paratype of Miniopterus manavi (FMNH 5650), a museum skin collected in 1896, and from which 220 bp were obtained; and (2) a tissue sample of FMNH 151718 from which only 710 bp were obtained. All available cyt-b sequences, including pre-existing sequences sourced from Genbank, are provided by region and taxon in Table 1. Full specimen details, including Genbank references, are provided in Table S1.
Table 1. Number of cyt-b sequences by taxon and region included in the present study; with one exception all belong to the genus Miniopterus.
| Region | Species/clade | Number of sequences |
| Madagascar | M. sororculus | 17 |
| X3 | 1 | |
| M. mahafaliensis | 19 | |
| M. griveaudi | 47 | |
| M. brachytragos | 12 | |
| M. manavi | 5 | |
| M. petersoni | 11 | |
| P6 | 10 | |
| P7 | 2 | |
| P5 | 3 | |
| P4 | 5 | |
| P3 | 2 | |
| M. egeri | 13 | |
| M. majori | 38 | |
| M. griffithsi | 6 | |
| M. gleni | 28 | |
| M. aelleni A | 15 | |
| M. aelleni B | 24 | |
| Africa | M. minor | 9 |
| M. fraterculus | 10 | |
| M. natalensis | 14 | |
| M. newtoni | 4 | |
| Europe | M. schreibersii | 10 |
| Australasia/Asia | M. australis | 1 |
| M. macrocneme | 1 | |
| M. oceanensis bassanii | 1 | |
| M. oceanensis orianae | 1 | |
| M. blepotis | 1 | |
| M. fuliginosus | 3 | |
| Myotis ricketti | 1 |
Full details including Genbank numbers and literature references are included in Table S1.
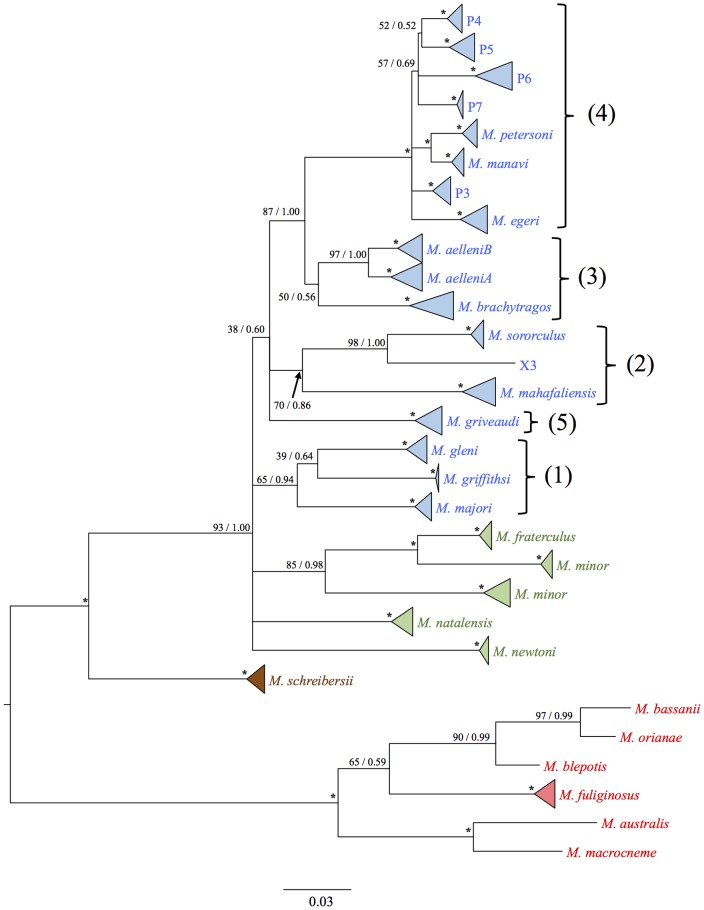
The initial ML and Bayesian analyses recovered the 264 specimens of Malagasy Minioptserus included in this study, as 18 clades (Figure S1). The X3 clade is represented by a single individual. The two to four most divergent haplotypes in each clade were then used for more extensive Bayesian, BEAST and ML analyses. The resulting ML and Bayesian phylogenetic trees produced broadly similar tree topologies, which recovered the reduced set of 54 of Malagasy Miniopterus included in this study as 18 clades (Figure 2). Each of these clades received 100% bootstrap (ML) and 1.00 posterior probability (Bayesian) support.
Figure 2. Bayesian majority consensus tree based on cvt-b sequence data and according to a HKY + G nucleotide substitution model.
The first number at each node represents bootstrap support according to the Maximum Likelihood analysis; the second represents Bayesian posterior probability. An asterisk (*) at a node indicates full support from both analyses, i.e. 100/1.00. Where clades contain more than a single individual, these have been collapsed into triangles. Colour coding refers to the origin of the species, as follows: Blue = Madagascar; Green = Africa; Brown = Europe; Red = Asia and Australasia. Large bold numbers beside lineages indicate the five primary lineages referred to in the text.
The 18 clades further clustered into five primary lineages. One of these, M. griveaudi, encompassed a single species. The remaining four sub-clades were comprised as follows: 1) the three taxa M. gleni, M. griffithsi and M. majori, supported with 0.94 posterior probability; 2) the three taxa M. sororculus, X3 and M. mahafaliensis, supported with 0.86 posterior probability; 3) the three taxa M. aelleniA, M. aelleniB and M. brachytragos with a lower support of 0.56; and 4) a sub-clade including M. petersoni, M. manavi, M. egeri and the genetically distinct populations referred to herein as P3, P4, P5, P6 and P7, with an overall support of 1.00. Sister relationships between 1) M. petersoni and M. manavi; 2) M. aelleniA and M. aelleniB; 3) and M. sororculus and X3 were all supported within their respective lineages at 1.00. The African taxa including M. fraterculus and M. minor, as well as the Malagasy M. gleni/M. griffithsi/M. majori lineage, formed a polytomy with M. natalensis, M. newtoni and the remainder of the Malagasy species. This is most likely due to the effect of a rapid radiation combined with the fast rate of evolution and fixation of the mitochondrial genome.
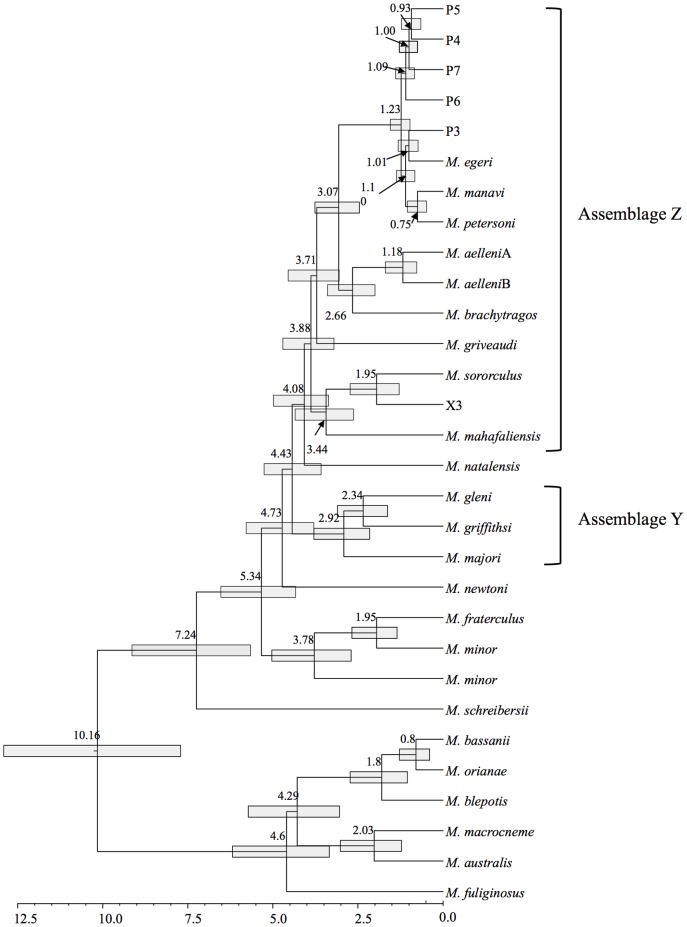
The Beast analysis (Figure 3) revealed that the initial divergence within the Malagasy Miniopterus radiation took place at about 4.5 million years ago (Mya). This is, therefore, the minimum date proposed for colonization of Madagascar by Miniopterus. Most of the species level divergences are recorded from the period between 4 and 2 Mya. A second set of diversifications took place 1.25 to 0.75 Mya, although uncertainty around these estimates allows some minor overlap between these two diversification periods.
Figure 3. BEAST molecular clock analysis of representative cvt-b sequences, incorporating a HKY + G nucleotide substitution model and a Yule model of speciation.
Molecular evolutionary rates were calibrated at 2% per million years under a relaxed lognormal clock. Numbers at nodes refer to the age of the node in millions of years (my); the scale bar indicates branch length in my. Grey bars represent 95% highest posterior distributions around node age estimates. Assemblages Y and Z are indicated as noted in the text.
Levels of DNA divergences between the recognized species of Malagasy Miniopterus ranged from 12.9% to 2.5% Kimura 2-parameter (K2P) (Table 1). Levels of within species variation were less than 1.8% K2P.
Discussion
Origins of Malagasy Miniopterus
The DNA phylogenetic analyses recovered Malagasy and African Miniopterus spp. as a monophyletic clade relative to Asian and Australasian taxa (Figure 2). Although some Malagasy bird species, which have similar capacity to bats for flight dispersal, appear to have originated through colonization events from Asia and Australasia across the Indian Ocean [44], [45], the cyt-b data of the present study clearly supports an African origin for Malagasy Miniopterus. With the recent taxonomic revision of Malagasy members of the tribe Emballonurni [46], the only remaining bat genus occurring on Madagascar that is demonstrably Asiatic in origin is the large and strong-flying Pteropus.
The available mtDNA data did not resolve conclusively whether there were one or multiple colonization events from Africa into Madagascar. The 18 clades identified among the Malagasy Miniopterus clustered into five primary lineages, but these were not recovered as a monophyletic assemblage, relative to African taxa (Figures 2 and 3). The prevailing winds in the nearly 400 km stretch of water separating Madagascar from Africa are westerly since the early Cenozoic [47] and well before the evolution of Miniopterus, indicating that colonizations in an easterly direction (i.e. Africa to Madagascar) would be against the prevailing winds. Under this scenario and based on extrapolation from a variety of flying and terrestrial vertebrates [10]–[13], [48], it is unlikely that the five identified Malagasy lineages of Miniopterus would each represent a separate colonization event.
Although the five primary lineages were not recovered as a monophyletic group, two major assemblages were identifiable among the Malagasy species: (1) M. gleni, M. griffithsi and M. majori; and (2) the remaining taxa. These two assemblages, referred to as Y and Z (Figure 3), diverged approximately 4.5 Myr ago. They represent either two separate colonization events or a single event that was followed by early divergence. Current data does not allow for the rejection of either of these hypotheses.
The BEAST analysis (Figure 3) also indicated that the five primary Malagasy lineages arose over a period of 3 to 4 Myr ago. There was a pulse of diversification in both assemblages Y and Z around 2 to 3 Myr ago and a further one in assemblage Z within the last million years. As Madagascar's fossil record has a major gap from the Late Cretaceous to Late Quaternary, little is known about the existing habitats and climatic regime on the island during the inferred Pliocene-Pleistocene period of Miniopterus diversification.
A caveat to the preceding discussion is that only a single MtDNA gene (cyt-b) was analyzed in this study. Although this gene has proven highly useful in identifying cryptic taxonomic diversity in Miniopterus [21]–[24] it is clear that further mtDNA augmented with nuclear sequence data is required to better resolve the early radiation and colonization history of the genus in Madagascar. The clarification of the number of taxa as discussed further below will set the framework for more detailed sequencing analyses.
Diversification of Malagasy Miniopterus
The various phylogenetic analyses (ML, Bayesian) all recovered the same 18 clades (Figure 2) of Malagasy Miniopterus. Eleven of these correspond directly with currently recognized species [18], [19], [24], [27], [49]–[51]. The other seven clades may represent additional species level diversity, but in certain cases other markers will be needed to resolve relationships.
In terms of DNA distances, the lowest recorded level between recognized sister species involved M. petersoni and M. manavi, where DNA distances ranged from 2.5% to 3.3% across the different haplotypes (Table 2). Distances between these two species and M. egeri ranged from 3.7% to 5.3%, while those involving comparisons between M. majori, M. gleni and M. griffithsi were higher still, ranging from 7.3% to 8.7% (Table 2). These relationships provide context for assessing the level of cyt-b differentiation recorded between M. sororculus and taxon X3. Although recovered as sister taxa, the two differed by a DNA distance of 7.2%, which is consistent with species level differentiation. As currently understood, M. sororculus is restricted to the central and southern portions of the Central Highlands and the single known individual referred to the X3 clade is from the foothills (810 m) of the central portion of the Central Highlands (Table 3). These two clades are not known to occur in sympatry and most likely represent an example of allopatric speciation that occurred 2 Myr ago based on the BEAST analysis (Figure 3). If this relationship is supported with additional specimens and sequence data, a taxonomic diagnosis for X3 will be required.
Table 2. mtDNA distances between Malagasy taxa belonging to the genus Miniopterus based on Kimura distances [34].
| Comparison between taxa | Distance range % | Comparison within taxa | Maximum distance % |
| majori vs griffithsi | 7.6 – 8.5 | majori | 1.1 |
| majori vs gleni | 8.3 – 8.7 | griffithsi | 0.3 |
| griffithsi vs gleni | 7.3 – 7.9 | gleni | 1.0 |
| petersoni vs manavi | 2.5 – 3.3 | manavi | 1.8 |
| manavi-petersoni vs P complex | 3.2 – 5.3 | petersoni | 1.1 |
| manavi-petersoni vs egeri | 3.7 – 5.3 | egeri | 1.7 |
| egeri vs P7 | 4.0 – 5.3 | P7 | 0.3 |
| egeri vs P6 | 4.6 – 6.9 | P6 | 1.6 |
| egeri vs P5 | 3.8 – 5.3 | P5 | 1.6 |
| egeri vs P4 | 3.3 – 4.5 | P4 | 0.4 |
| egeri vs P3 | 2.9 – 4.1 | P3 | 0.7 |
| within the P complex | 2.5 – 6.9 | ||
| aelleniA vs aelleniB | 3.1 – 3.6 | aelleniA | 1.7 |
| aelleniB | 1.5 | ||
| sororculus vs X3 | 7.2 | sororculus | 0.5 |
Table 3. Summary of different size and life-history parameters of Malagasy (M) and Comorian (C) Miniopterus spp. [19], [52].
| Taxon | Body Size | Elevation (m) | Distribution | Habitat |
| sororculus | MB | 950–2200 | C, S | mhf, oh, sbf |
| X3 | SB | 810 | E | lhf |
| mahafaliensis | SB | 0–950 | C, S, W | ddf, oh, sbf |
| griveaudi (M) | SB | 0–600 | N, W | ddf |
| griveaudi (C) | SB | 0–900 | Grande Comore, Anjouan | lhf, oh |
| majori | MB | 0–1550 | C, N, S | lhf, mhf, oh |
| griffithsi | LB | 25–110 | S | sbf |
| gleni | LB | 0–1200 | E, N, W, S | ddf, lhf, mhf, oh, sbf |
| brachytragos | SB | 0–600 | E, N, W | ddf, lhf |
| aelleniA (M) | SB | 40–500 | N, W | ddf |
| aelleniA (C) | SB | 220–700 | Anjouan | lhf |
| aelleniB | SB | 810–1340 | N, C | lhf, mhf |
| manavi | SB | 900–1500 | E, C | mhf, oh |
| petersoni | MB | 10–550 | S, E | lhf, oh |
| egeri | SB | 0–550 | N, E | lhf |
| P3 | SB | 810–1340 | N, C | lhf, mhf |
| P4 | SB | 800–875 | E | lhf |
| P5 | SB | 50–1340 | E, C | lhf, mhf |
| P6 | SB | 60–1425 | C, W | ddf, mhf, oh |
| P7 | SB | 1340–1425 | C | mhf, oh |
Body size: based on mean forearm length (FA), and animals are designated as small-bodied (SB), medium-bodied (MB) and large-bodied (LB); Distribution: E = east, N = north, W = west, S = south, C = central and for the Comoros the name of the island is presented; Habitat: lhf = lowland humid forest, mhf = montane humid forest, oh = open habitat (anthropogenic), ddf = dry deciduous forest, sbf = spiny bush forest.
The M. aelleniA and M. aelleniB clades were less differentiated with distances ranging from 3.1% to 3.6% across the different haplotypes. Nevertheless, this is comparable to levels recorded between closely related and morphologically distinct species such as M. petersoni and M. manavi, as well as between these two species and M. egeri (Table 2). In this context, the two M. aelleni clades are best treated as separate species. The type series of M. aelleni includes individuals from the M. aelleniA clade [27]. Consequently, a taxonomic diagnosis for M. aelleniB is required. The M. aelleniA clade includes individuals taken in dry deciduous forests, three of the four being from Ankarana, while those in the M. aelleniB clade are from humid forest formations, three being from Montagne d’Ambre (Table 3). The sites of Ankarana and Montagne d’Ambre are in close geographical proximity (about 40 km) and share numerous faunistic elements [52].
The five taxa assigned to the P-group were genetically differentiated at levels comparable to those separating M. manavi and M. petersoni (Table 2). However, additional markers, specifically based on nuclear DNA, are needed to differentiate between a single genetically variable taxon or several distinct species. It is important to note that the clade assigned to M. manavi is based on sequence data from a paratype of this species [24]. The most closely related forms were P3, P4, and P7 with genetic distances between them ranging from 2.5% to 3.3% (Table 2). Within each taxon, haplotypes differed by 0.3% to 0.7%. Genetic distances involving comparisons with P6 ranged from 3.2% to 5.3%, while those involving P5 ranged from 2.7% to 5.1%. Within both P5 and P6, haplotype variation did not exceed 1.6%. Distances between M. egeri and the various P taxa ranged from 2.9% to 6.9%.
The geographical distribution of the P clades provides further insights into the patterns of taxonomic and genetic diversity, although the following conclusions will need to be verified with further field data. P3 is largely restricted to the central portions of the Central Highlands, notably at Ambohitantely, where it shares a day roost with members of the M. manavi, P7, M. aelleniB, and P5 clades. It also occurs at sites in the Northern Highlands [53], specifically the Anjanaharibe-Sud and Marojejy Massifs. The form P4 is restricted to a relatively well-defined region at the foot of the Central Highlands (three individuals are from Midongy Sud and the fourth individual is from Andringitra) and all taken between 800 and 875 m. The single known individual of the X3 clade was also collected at Andringitra at 810 m. The form P5 shows a relatively broad distribution, with two individuals taken at Ambohitantely in the Central Highlands and at same day roost site as M. manavi, M. aelleniB, P7 and P3, as well as at 50 m elevation in the eastern lowlands. P7 also occurs in the Central Highlands (Ambohitantely) and Fanadana co-occurring with M. manavi, P6 and M. majori. The form P6 is widespread and includes individuals from the central western site of Namoroka, the northwestern offshore island of Nosy Be, and Fanandana.
The fact that each P clade was recovered as monophyletic, combined with the levels of genetic divergences between clades, and the co-occurrence at Ambohitantely and Fanandana of several of the P3 to P7 clades, requires further comment. One interpretation is that the P clades represent distinct but closely related cryptic species. The previous identification of additional species diversity in the M. manavi complex (e.g. M. petersoni, M. egeri) has been defined by both DNA and morphological evidence, such as tragus shape [19], [27], [49]. An examination of the tragi of animals from Ambohitantely belonging to clades P5 and P7 did not disclose noticeable differences in tragus structure. It may be that the P forms are incipient species and although genetic separation has occurred, obvious morphological differences have not yet evolved.
The P clade may comprise cases of incomplete lineage sorting of the mitochondrial genome, introgression and hybridization of closely related taxa or possibly a combination of these processes [54], [55]. The source taxon, however, is not clear from the available data: none of the P haplotypes were associated with any named species. Further analysis of this complex will require multiple nuclear markers in order to resolve their taxonomic status and relationship.
One further aspect that may have confounded our analyses of genetic relationships in the manavi group, including M. petersoni, M. egeri and the P group of clades, is that we used a mitochondrial marker that is only transmitted by females [56]. Hence, our evaluations here are only of the genetic relationships passed on by females, which may not accurately reflect patterns of overall gene flow. In bats, females tend to be notably more philopatric than males [57] and the use of bi-parentally inherited genes might provide further insight into the phylogenetic relationships of these different clades. However, we come to the same question as above: to which species do the P haplotypes align? Clearly, further morphological, ecological and behavioral work is required to understand better the taxonomic status of the five P taxa. At the very least, they may represent a single cryptic species with high levels of haplotype diversity to five currently unrecognized taxa requiring formal description.
Colonization of the Comoros
It has been demonstrated that two species of Miniopterus are shared between Madagascar and the Comoros [58], [59] and include M. griveaudi and M. aelleni [27]. Using mtDNA and microsatellites, it was concluded that M. griveaudi colonized the Comoros from Madagascar some 180 000 years ago [58]. Although this suggests a similar colonization history for M. aelleni, DNA differentiation was not compared between Malagasy and Comorian populations of the latter because of low sample sizes [58].
Herein, our study reveals that individuals of M. aelleni from Comoros are nested within the M. aelleniA clade (Figure 2). This indicates a recent colonization of the Comoros from a member of this apparent species complex. As the principal four islands of the Comoros have never been connected to other landmasses, dispersal would have been over-water in a westerly direction and following the prevailing winds.
Although M. griveaudi and M. aelleniA are not phylogenetically closely related, they are similar in their habitats, distribution and morphology (Table 3). Both taxa occur in dry deciduous forests in Madagascar. The ability to colonize islands in these two taxa is likely linked to similar ecological parameters. Interestingly, the populations in the Comoros of both M. griveaudi and M. aelleniA occur in lowland humid forest, the former also occurring in open habitat (anthropogenic).
Patterns of ecological diversification and speciation in Madagascar
All of the individuals in the P3 to P7 clades occur in the eastern humid forests at high (Central Highlands) to low (Sahafina) elevations (Table 3), with only a few exceptions: P6 includes two individuals from the dry deciduous forest site of Namoroka and one individual from the transitional dry deciduous/humid forest Sambirano formation of Nosy Be. Thus, most of the genetic diversity has been generated in the east. Based on the Beast analysis for extant species, the differentiation of the Malagasy Miniopterus radiation commenced about 4.5 Myr ago, with two periods of cladogenesis: 4–2 Myr and 0.75–1.25 Myr. These are periods for which little hard data exist for environmental conditions on the island and an interpretation of the ecological and evolutionary forces that gave rise to this differentiation is not evident. However, given the high level of syntopic occurrence of members of the P group, specifically at Ambohitantely and Fanandana, it is possible that past fragmentation of populations followed by range expansion are related, for example, to the cyclic climate changes of the Late Pleistocene/Early Holocene [60], [61], particularly in more montane zones, such as the Central Highlands, where Ambohitantely and Fandanana are found.
Using measures of species diversity of Miniopterus on Madagascar and the Comoros, the number of recognized species has gone from four in 1995 [17] to eleven currently recognized species [18], [19]. When the current genetic data are analyzed together, there are indications that, at least from the perspective of phylogenetic species, something approaching 18 taxa occur on Madagascar. Similar studies of continental African Miniopterus reveal that levels of species richness are higher than current estimates would indicate [62] and that, for example M. minor is paraphyletic (Figures 2 and 3). We suspect the same pattern will be found in other portions of the Old World range of this genus. Before the recent wave of molecular studies of members of this genus, something approaching 20 species were recognized across its Old World distribution [63]. If the patterns of cryptic species richness on Madagascar hold for other areas, it is conceivable that over a hundred taxa comprise this genus, making it an example of one of the most successful adaptive radiations amongst bats.
Supporting Information
Maximum Likelihood (ML) tree generated using RaXML Blackbox (see main text for citation) and derived from all available cyt -b sequences, corroborated with Bayesian analysis using MrBayes 3.2 (see main text for citation).
(DOCX)
Specimen data for individuals included in the study.
(DOCX)
Acknowledgments
We thank the Direction du Système des Aires Protégées, Direction Générale de l’Environnement et des Forêts; Madagascar National Parks (Madagascar); and Centre National de Documentation et de Recherche Scientifique (Union of the Comoros) for kindly providing authorizations for the capture, collection, and exportation of animals (No. 194/12/MEF/SG/DGF/DCB.SAP/SCB, No. 032/12/MEF/SG/DGF/DCB.SAP/SCBSE, No. 283/11/MEF/SG/DGF/DCB.SAP/SCB, No. 067/12/MEF/SG/DGF/DCB.SAP/SCBSE). The specimen material from Madagascar was obtained under a protocol of collaboration between the Département de Biologie Animale (Université d’Antananarivo), Association Vahatra and the Field Museum of Natural History. We acknowledge Scott G. Cardiff, Julie Ranivo, Zafimahery Rakotomalala, Eddy N. Rakotonandrasana, Beza Ramasindrazana, Fanja Ratrimomanarivo, Ishaka Saïd, M. Corrie Schoeman, William T. Stanley, Voahangy Soarimalala, and Peter Taylor for their aid with fieldwork or access to tissues. We are grateful to three anonymous reviewers for their comments on an earlier version of this paper.
Funding Statement
Financial support for the fieldwork associated with this project was graciously given by CABS of Conservation International, The John D. and Catherine T. MacArthur Foundation and Volkswagen Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Goodman SM, Benstead JP (2005) Updated estimates of biotic diversity and endemism for Madagascar. Oryx 39: 73–77. [Google Scholar]
- 2. Wilmé L, Goodman SM, Ganzhorn J (2006) Biogeographic evolution of Madagascar's microendemic biota. Science 312: 1063–1065. [DOI] [PubMed] [Google Scholar]
- 3. Vences M, Wollenberg KC, Vieites DR, Lees DC (2009) Madagascar as a model region of species diversification. Trends Ecol Evol 24: 456–465. [DOI] [PubMed] [Google Scholar]
- 4. Gaffney E, Forster C (2003) Side-necked turtle lower jaws (Podocnemididae, Bothremydidae) from the Late Cretaceous Maevarano Formation of Madagascar. Am Mus Novit 3397: 1–13. [Google Scholar]
- 5. Ali JR, Aitchison JC (2008) Gondwana to Asia: plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth-Science Reviews 88: 145–166. [Google Scholar]
- 6. Eagles G, Konig M (2008) A model of plate kinematics in Gondwana breakup. Geophys J Int 173: 703–717. [Google Scholar]
- 7. Samonds KE, Godfrey LR, Ali JR, Goodman SM, Vences M, et al. (2013) Imperfect isolation: factors and filters shaping Madagascar's extant vertebrate fauna. Plos One 8(4): e62086 doi:10.1371/journal.pone.0062086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poux C, Madsen O, Glos J, de Johng WW, Vences M (2008) Molecular phylogeny and divergence times of Malagasy tenrecs: influence of data partitioning and taxon sampling on dating analyses. BMC Evol Biol 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samonds KE, Godfrey LR, Ali JR, Goodman SM, Vences M, et al. (2012) Spatial and temporal arrival patterns of Madagascar's vertebrate fauna explained by distance, ocean currents, and ancestor type. Proc Natl Acad Sci, USA 109: 5352–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cibois A, Slikas B, Schulenberg TS, Pasquet E (2001) An endemic radiation of Malagasy songbirds is revealed by mitochondrial DNA sequence data. Evol 55: 1198–1206. [DOI] [PubMed] [Google Scholar]
- 11. Cibois A, Davis N, Gregory SMS, Pasquet E (2010) Bernieridae (Aves: Passeriformes): a family group name for the Malagasy sylvioid radiation. Zootaxa 2554: 65–68. [Google Scholar]
- 12. Jønsson KA, Fabre P-H, Fritz SA, Etienne RS, Ricklefs RE, et al. (2012) Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc Natl Acad Sci USA 109: 6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reddy S, Driskell A, Rabosky DL, Hackett SJ, Schulenberg TS (2012) Diversification and the adaptive radiation of the vangas of Madagascar. Proc Roy Soc Lond B: Biological Sciences 279: 2062–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poux C, Madsen O, Marquard E, Vieites DR, De Jong WW, et al. (2005) Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst Biol 54: 719–730. [DOI] [PubMed] [Google Scholar]
- 15. Pearson RG, Raxworthy CJ (2009) The evolution of local endemism in Madagascar: watershed versus climatic gradient hypotheses evaluated by null biogeographic models. Evol 63: 959–967. [DOI] [PubMed] [Google Scholar]
- 16. Norberg UM, Rayner JMV (1987) Ecological morphology and flight in bats (Mammalia: Chiroptera): wing adaptations, flight performance, foraging strategy and echolocation. Phil Trans Roy Soc B 316: 335–427. [Google Scholar]
- 17.Peterson RL, Eger JL, Mitchell L (1995) Chiroptères. Vol. 84. Faune de Madagascar. Paris: Muséum national d’Histoire naturelle.
- 18.Goodman SM (2011) Les chauves-souris de Madagascar. Antananarivo: Association Vahatra. 129 p. [Google Scholar]
- 19. Goodman SM, Ramasindrazana B, Maminirina CP, Schoeman MC, Appleton B (2011) Morphological, bioacoustical, and genetic variation in Miniopterus bats from eastern Madagascar, with the description of a new species. Zootaxa 2880: 1–19. [Google Scholar]
- 20. Sikes RS, Gannon WL (2011) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mam 92: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cardinal BR, Christidis L (2000) Mitochondrial DNA and morphology reveal three geographically distinct lineages of the large bentwing bat (Miniopterus schreibersii) in Australia. Aust J Zool 48: 1–19. [Google Scholar]
- 22. Appleton BR, McKenzie JA, Christidis L (2004) Molecular systematics and biogeography of the bent-wing bat complex Miniopterus schreibersii (Kuhl, 1817) (Chiroptera: Vespertilionidae). Mol Phylo Evol 31: 431–439. [DOI] [PubMed] [Google Scholar]
- 23. Miller-Butterworth CM, Eick G, Jacobs DS, Schoeman MC, Harley EH (2005) Genetic and phenotypic differences between South African long-fingered bats, with a global miniopterine phylogeny. J Mam 86: 1121–1135. [Google Scholar]
- 24.Goodman SM, Ryan KE, Maminirina CP, Fahr J, Christidis L, et al. (2007) The specific status of populations on Madagascar referred to Miniopterus fraterculus (Chiroptera: Vespertilionidae), with description of a new species. J Mam: 1216–1229.
- 25. Weyeneth N, Goodman SM, Stanley WT, Ruedi M (2008) The biogeography of Miniopterus bats (Chiroptera: Miniopteridae) from the Comoro Archipelago inferred from mitochondrial DNA. Mol Ecol 17: 5205–5219. [DOI] [PubMed] [Google Scholar]
- 26. Miller-Butterworth CM, Murphy WJ, O’Brien SJ, Jacobs DS, Springer MS, et al. (2007) A family matter: conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. . Mol Biol Evol 24: 1553–1561. [DOI] [PubMed] [Google Scholar]
- 27. Goodman SM, Maminirina CP, Weyeneth N, Bradman HM, Christidis L, et al. (2009a) The use of molecular and morphological characters to resolve the taxonomic identity of cryptic species: the case of Miniopterus manavi (Chiroptera, Miniopteridae). Zool Scripta 38: 339–363. [Google Scholar]
- 28.Marck C (1990) DNA Strider: A C program for DNA and protein sequence analysis. Gif Sur Yvette: Service de Biochimie et de Génétique et Moléculaire, direction des Sciences de la vie.
- 29. Guindon S, Gascuel O (2003) A simple, fast and accurate method to estimate large phylogenies by maximum likelihood. Sys Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 30. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 32. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 33. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web Servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- 34. Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 35. Marshall DC (2009) Cryptic failure of partitioned Bayesian phylogenetic analyses: lost in the land of the long trees. Sys Biol 59: 108–117. [DOI] [PubMed] [Google Scholar]
- 36. Brown JM, Hedtke SM, Lemmon AR, Lemmon EM (2010) When trees grow too long: investigating the causes of highly inaccurate Bayesian branch-length estimates. Sys Biol 59: 145–161. [DOI] [PubMed] [Google Scholar]
- 37. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Drummond AJ, Suchard MA, Xie D, Rambaut A (2013) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol and Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. Plos Biol 4: e88 doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bermingham E, Lessios HA (1993) Rate variation of protein and mitochondrial DNA evolution as revealed by sea urchins separated by the Isthmus of Panama. Proc Natl Acad Sci USA 90: 2734–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut A, Drummond AJ (2007) Tracer v1.5. Available at: http://beast.bio.ed.ac.uk/Tracer.
- 42. Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–63. [DOI] [PubMed] [Google Scholar]
- 43. Juste J, Ferrández A, Fa JE, Masefield W, Ibáyez C (2007) Taxonomy of little bent-winged bats (Miniopterus, Miniopteridae) from the African islands of Sao Tomé, Grand Comoro and Madagascar, based on mtDNA. Acta Chiropterol 9: 27–37. [Google Scholar]
- 44. Kundu S, Jones CG, Prys-Jones RP, Groombridge JJ (2012) The evolution of the 21 Indian Ocean parrots (Psittaciformes): extinction, adaptive radiation and eustacy. Mol Phylo Evol 62: 296–305. [DOI] [PubMed] [Google Scholar]
- 45. Jønsson KA, Irestedt M, Fuchs J, Ericson PGP, Christidis L, et al. (2008) Explosive avian radiations and multi-directional dispersal across Wallacea: evidence from the Campephagidae and other Crown Corvida (Aves). Mol Phylo Evol 47: 221–236. [DOI] [PubMed] [Google Scholar]
- 46. Goodman SM, Puechmaille SJ, Friedli-Weyeneth N, Gerlach J, Ruedi M, et al. (2012) Phylogeny of the Emballonurini (Emballonuridae) with descriptions of a new genus and species from Madagascar. J Mam 93: 1440–1455. [Google Scholar]
- 47. Ali JR, Huber M (2010) Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463: 653–656. [DOI] [PubMed] [Google Scholar]
- 48. Yoder AD, Burns MM, Zehr S, Delefosse T, Veron G, et al. (2003) Single origin of Malagasy Carnivora from an African ancestor. Nature 421: 734–737. [DOI] [PubMed] [Google Scholar]
- 49. Goodman SM, Bradman HM, Maminirina CP, Ryan KE, Christidis L, et al. (2008) A new species of Miniopterus (Chiroptera: Miniopteridae) from lowland southeastern Madagascar. Mam Biol 73: 199–213. [Google Scholar]
- 50. Goodman SM, Maminirina CP, Bradman HM, Christidis L, Appleton B (2009b) The use of molecular phylogenetic and morphological tools to identify cryptic and paraphyletic species: examples from the diminutive long-fingered bats (Chiroptera: Miniopteridae: Miniopterus) on Madagascar. Am Mus Novit 3669: 1–34. [Google Scholar]
- 51. Goodman SM, Maminirina CP, Bradman HM, Christidis L, Appleton B (2010a) Patterns of morphological and genetic variation in the endemic Malagasy bat Miniopterus gleni (Chiroptera: Miniopteridae), with the description of a new species, M. griffithsi . J Zool Syst Evol Res 48: 75–86. [Google Scholar]
- 52.Raxworthy CJ, Nussbaum RA (1997) Biogeographic patterns of reptiles in eastern Madagascar. In: Goodman SM, Patterson BP, editors. Natural change and human impact in Madagascar. Washington, D. C.: Smithsonian Institution Press. 124–140.
- 53. Carleton MD, Goodman SM (1998) New taxa of nesomyine rodents (Muroidea: Muridae) from Madagascar's northern highlands, with taxonomic comments on previously described forms. In: Fieldiana: Zoology, new series Goodman SM, editor. A floral and faunal inventory of the Réserve Spéciale d'Anjanaharibe-Sud, Madagascar: with reference to elevational variation, ed. S. M. Goodman. 90: 163–200. [Google Scholar]
- 54. Redondo RAF, Brina LPS, Silva RF, Ditchfield AD, Santos FR (2008) Molecular systematics of the genus Artibeus (Chiroptera: Phyllostomidae). Mol Phylo Evol 49: 44–58. [DOI] [PubMed] [Google Scholar]
- 55. Russell AL, Goodman SM, Florentino I, Yoder AD (2008) Population genetic analysis of Myzopoda (Chiroptera: Myzopodidae) in Madagascar. J Mam 89: 209–221. [Google Scholar]
- 56. Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13: 729–744. [DOI] [PubMed] [Google Scholar]
- 57.Kerth G (2006) Relatedness, life history, and social behaviour in the long-lived Bechstein's bat, Myotis bechsteinii. In: Zubaid A, McCracken GF, Kunz TH, editors. Functional and evolutionary ecology of bats. New York: Oxford University Press. 199–212.
- 58. Weyeneth N, Goodman SM, Appleton B, Wood R, Ruedi M (2011) Wings or winds: inferring bat migration in a stepping-stone archipelago. J Evol Biol 24: 1298–1306. [DOI] [PubMed] [Google Scholar]
- 59.Burney DA (1999) Rates, patterns, and processes of landscape transformation and extinction in Madagascar. In: MacPhee, RDE, editor. Extinction in near time. New York: Kluwer/Plenum. pp. 145–164.
- 60.Goodman SM, Jungers WL (In press) Windows into the extraordinary recent land animals and ecosystems of Madagascar. Chicago: The University of Chicago Press.
- 61. Goodman SM, Weyeneth N, Ibrahim Y, Saïd I, Ruedi M (2010b) A review of the bat fauna of the Comoro Archipelago. Acta Chiropterol 12: 117–141. [Google Scholar]
- 62.Monadjem A, Goodman SM, Stanley WT, Appleton B (in press) A cryptic new species of Miniopterus from south-eastern Africa based on molecular and morphological characters. Zootaxa. [DOI] [PubMed]
- 63.Simmons NB (2005) Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographical reference, 3rd edition. Baltimore: Johns Hopkins University Press. 312–529.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum Likelihood (ML) tree generated using RaXML Blackbox (see main text for citation) and derived from all available cyt -b sequences, corroborated with Bayesian analysis using MrBayes 3.2 (see main text for citation).
(DOCX)
Specimen data for individuals included in the study.
(DOCX)