Abstract
The production of newly synthesized proteins is a key process of protein homeostasis that initiates the biosynthetic flux of proteins and thereby determines the composition, stability and functionality of the proteome. Protein synthesis is highly regulated on multiple levels to adapt the proteome to environmental and physiological challenges such as aging and proteotoxic conditions. Imbalances of protein folding conditions are sensed by the cell that then trigger a cascade of signaling pathways aiming to restore the protein folding equilibrium. One regulatory node to rebalance proteostasis upon stress is the control of protein synthesis itself. Translation is reduced as an immediate response to perturbations of the protein folding equilibrium that can be observed in the cytosol as well as in the organelles such as the endoplasmatic reticulum and mitochondria. As reduction of protein synthesis is linked to life span increase, the signaling pathways regu-lating protein synthesis might be putative targets for treatments of age-related diseases. Eukaryotic cells have evolved a complex system for protein synthesis regulation and this review will summarize cellular strategies to regulate mRNA translation upon stress and its impact on longevity.
Keywords: Aging, Chaperone, Life span, mRNA Translation, Proteostasis, Stress response, UPR.
INTRODUCTION
Ribosome biogenesis and mRNA translation are complex processes, which are tightly controlled in the nucleus and cytosol [1-4]. Protein synthesis is a highly energy-consuming process and is therefore linked to environmental conditions, such as nutrient availability and stress [3, 5, 6]. Stress leads to a shift of the energy distribution from production to survival mode, leading to a reduction of global protein synthesis and activation of stress response pathways [4, 7, 8]. Therefore, ribosome biogenesis and mRNA translation are important regulatory nodes for survival upon stress.
The eukaryotic ribosome consists of two subunits. The small 40S subunit is composed of 18S rRNA and up to 33 ribosomal proteins (r-proteins) whereas the large 60S subunit consists of 5S, 5.8S and 25-28S rRNAs and up to 49 r-proteins [9, 10]. Its biogenesis to a functional unit comprises many highly conserved and controlled steps including rRNA maturation, r-proteins synthesis and transports between nucleus and cytosol [9, 11]. More than 200 non-ribosomal factors, such as chaperones, are involved in RNA folding, cleavage, transport and stabilization [12, 13]. Thereby, the spatial separation of ribosome assembly and protein synthesis in nucleus and cytosol is one check point to control ribosome function by preventing premature translation through export control [14].
The mRNA translation process can be divided into four parts: initiation, elongation, termination and ribosome recycling, with a regulatory focus on translation initiation [15]. The eukaryotic cap-dependent translation initiation process includes mRNA initialization by initiation factor (eIF) complex eIF4F and poly(A)-binding protein PABP (for mRNA cyclization) as well as the recruitment of the 43S pre-initiation complex (PIC), which subsequently scans the mRNA for AUG start codon [15]. After AUG match, irreversible hydrolysis of GTP is performed and scanning is stopped, leading to eIF release and 60S recruitment to form the 80S ribosome for translation elongation [15]. Upon ribosomal exit, two ribosome-associated chaperones, namely ribosome-associated chaperone complex (RAC) in cooperation with Hsp70 family members and nascent polypeptide-associated complex (NAC), assist in the initial folding steps, protect the nascent chains, and guide them to the downstream cytosolic chaperone network for subsequent folding [16]. Together with protein degradation systems, a complex protein network is created, which is tightly regulated to maintain protein homeostasis (proteostasis).
Upon acute stress, such as heat shock and nutrient restriction, diverse stress responses are activated (Fig. 1). The cytosolic heat shock response (HSR) and unfolded protein responses (UPRs) in the endoplasmatic reticulum (ER) and mitochondria shift the cell from production to survival mode. This is achieved by a reduction of global protein synthesis as well as by a selective induction of chaperones and proteases [4, 17-20]. If stress conditions persist (e.g. chronic stress, aging), stress pathways often start to fail, leading to protein aggregation and cellular death, which is often described in age-related diseases like Alzheimer’s diseases, Parkinson and Huntington’s disease [21, 22]. Interestingly, deletion or inhibition of translation components, such as ribosomal proteins, initiation factors and regulatory kinases, were reported to increase life span [17, 23]. But how can alterations in the translational machinery have positive effects on longevity? In the following paragraphs, cellular pathways will be reviewed discussing mRNA translation adaptation in response to stress and their influence on life span.
Fig. (1).
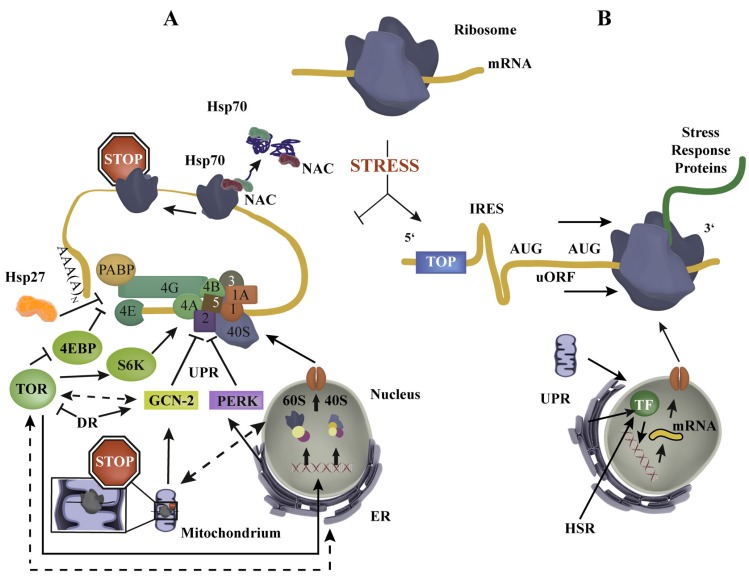
Protein synthesis regulation in stress – an overview. (A) Upon stress (e.g. dietary restriction, heat shock) global protein synthesis is reduced by a complex cross-compartmental network of stress response pathways (TOR, GCN-2, PERK), which interact with components of the eIF4F complex (eIF4A, eIF4E, eIF4G) and 43S (40S, eIF1, eIF1A, eIF2, eIF3, eIF5) and thereby disturb mRNA translation initiation [15]. Dotted lines indicate general pathway interconnections which can be activating and/or inhibiting. Chaperones also regulate mRNA translation either by ribosome dissociation, which causes ribosomal stalling (NAC, Hsp70) or by interaction with PABP/eIF4G (Hsp27). (B) Enhanced synthesis of stress response proteins, e.g. chaperones, degradation machineries, transcriptions factors (TF), is mediated by IRES, uORFs and TOP elements, enabling protein synthesis in conditions where global mRNA translation is reduced. Cytosolic, mitochondrial and ER stress responses recruit TFs (e.g. HSF-1, ATFS-1, XBP-1) to induce compartment-specific stress response protein production.
PROTEIN SYNTHESIS IN STRESSFUL CONDITIONS – KEEP THE MACHINERY RUNNING OR STEP ON THE BRAKE?
Protein folding already starts during mRNA translation and is tightly regulated to avoid early protein misfolding [24, 25]. One regulatory element is the mRNA itself. Optimal codons, whose matching tRNAs are high in number and guarantee an efficient amino acid transfer, are associated with conserved sites in proteins and are often found in aggregation-prone regions [26, 27]. In contrast to that, non-optimal codons cause a reduction of translational speed as its matching tRNAs are less abundant [24]. Thereby, clusters of non-optimal codons are associated with protein’s secondary structures, suggesting that codon usage serves as a fine-tuning element for translation elongation and co-translational folding [24, 28, 29]. The secondary structure of mRNAs can also cause translational pausing, as the ribosomal helicase activity has to unwind these structures to continue translation elongation [30]. In addition, positively charged residues are also able to slow down translation by interaction with ribosomal structures, which are linked to protein degradation by the ubiquitin proteasome system [31]. Although this regulatory system, in addition to ribosome-bound chaperones that will be discussed below, is very complex and adapted to the abundance and physico-chemical properties of proteins, it is not sufficient upon stress and is therefore supported by numerous stress response pathways discussed in this review.
Starving Cells – Responses to Dietary Restriction
If nutrient availability is restricted, energy has to be restored for cell survival. As mRNA translation is a highly energy-consuming process, protein synthesis is reduced in response to stress such as starvation [1]. Reduction in mRNA translation can be achieved in two ways, (1) by reduction of the number of ribosomes and (2) by inhibiting the mRNA translation process. The target of rapamycin (TOR) signaling pathway participates in both ribosome biogenesis and mRNA translation by its downstream targets S6 kinase (S6k) and eIF4E-binding protein (4EBP) [4, 32, 33] and is highly conserved from yeast to mammals [17, 34]. There are two different TOR complexes, TORC1 and TORC2, which differ in structure and function [35]. In mammals, mTORC2 is influenced by growth factors and regulates cell survival, metabolism and cytoskeletal organization [36, 37]. However, mTORC1 processes signals stemming from stress, energy levels and oxygen and modulates autophagy, cell growth and macromolecule biosynthesis [36]. As this section will focus on TORC1 it will be referred to hereafter as TOR. During an active TOR signaling, TOR binds eIF3 and promotes direct phosphorylation of S6k and 4EBP [38]. Phosphorylation leads to dissociation of 4EBP-eIF4E and S6K-eIF3-complexes, resulting in eIF4F complex formation and 43S complex recruitment [38, 39]. In addition, S6k phosphorylates numerous factors, which are involved in translation initiation, among others [38, 39]. Upon dietary restriction (DR), TOR is inactivated and causes enhanced autophagy, altered Insulin-like-signaling (ILS), a hypoxic response and a reduction of total mRNA translation [17, 36, 40]. Concerning the latter point, TOR participates in r-protein synthesis, rDNA transcription as well as mRNA translation initiation, which will be inhibited due to reduced TOR signaling upon stress (Fig. 1A) [32, 41, 42]. The importance of this signaling pathway and mRNA translation to stress response and longevity is demonstrated by the observations that a deletion of TOR pathway components or translation initiation factors lead to life span extension in yeast, nematodes, fruit flies and mice [4, 43-46]. For example, inhibition of nematode ifg-1 (orthologue of eIF4G), which is also down-regulated upon starvation, results in selective expression of stress response genes [44]. Furthermore, in mammalian cell culture experiments, it was reported that decreasing TOR activity is associated with enhanced Hsp70 translation [34]. These results demonstrate that, besides a reduction in chaperone load by mRNA translation decrease, dietary restriction is also able to activate stress responses, which may also contribute to an increase in life span.
Besides TOR signaling, a second pathway is involved in monitoring and responding to nutrient availability. In this general amino acid control pathway (GAAC), uncharged tRNAs accumulate due to nutrient restriction, which cause an activation of GCN-2 (general control nonderepressible 2) [47]. GCN-2 is a protein kinase that is activated by binding uncharged tRNAs and thus monitors nutrient restriction [48]. Active GCN-2 phosphorylates eIF2α (Fig. 1A) and that leads to mRNA translation inhibition as well as a selective expression of genes, encoding for proteins of the amino acid biogenesis pathways [47, 49]. An interconnection of this pathway and TOR signaling is indicated in yeast, where GCN-2 is activated by inhibition of TOR signaling [50]. Also in C. elegans, an interconnection between LET-363 (orthologue of TOR) and GCN-2 pathways is demonstrated as GCN-2 is able to regulate PHA-4 (orthologue of FoxA), a transcription factor downstream of LET-363 [49]. Similar to the TOR pathway, an important role of GCN-2 in life span regulation is indicated as a loss of GNC-2 function decreases life span upon amino acid limitation [49]. These results demonstrate a tight interconnection of TOR and GCN-2 pathways and indicate that DR can lead to activation of a complex system to regulate mRNA translation and longevity.
Regulation of mRNA Translation by Chaperones
To prevent unwanted intermolecular interactions, caused by the high abundance of macromolecules in the cytosol (300-400 mg/ml) [51], the emerging nascent chain is protected by two chaperone complexes that associate with the ribosome, NAC and RAC-Ssb in yeast or mRAC-Hsp70 in mammals [52].
NAC is required for efficient translational activity and assists in the folding of newly synthesized proteins, but can also dissociate from the ribosome to exhibit chaperone function independent of the ribosome [20, 51]. As deletion of NAC causes ubiquitination of longer and aggregation-prone proteins, NAC not only protects proteins from misfolding, but also from degradation [53]. In addition, NAC can act as a sensor of protein folding, transmitting information of the cytosolic protein folding conditions to the ribosomal translation machinery [20]. With increasing amounts of misfolded and aggregated proteins upon heat shock, NAC dissociates from the ribosome and acts as a chaperone by aiding in protein re-solubilization (Fig. 1A). Thereby, a dissociation of NAC from the ribosome leads to a reduction of mRNA translation rate [20]. The dissociation of NAC from the ribosome is reversible with cessation of the stress, once protein homeostasis is rebalanced. In contrast, upon chronic stress, caused for instance by the constitutive expression of aggregation-prone proteins or aging, NAC is not enabled to re-associate with the ribosome as protein folding conditions do not recover [20]. However, a number of questions regarding NACs role in the regulation of ribosomal activity or proteostasis remain open. First, it is not known yet how the translational attenuation by NAC is achieved. Possible nodes of translation regulation by NAC in response to stress are ribosome biogenesis and translation initiation, as a role for NAC in both processes was reported [54, 55]. Second, it is also not understood yet whether NAC directly interacts with substrates or whether it assists other chaperones in their remodeling functions [20]. Support for the latter stem from co-immunoprecipitation data of NAC using a whole cell lysate that identified chaperones from all major chaperone families as interaction partners of NAC, including Hsp70, Hsp90, sHsps and Hsp110 [20]. These results implicate a complex interaction network of NAC and its important role in cellular proteostasis [20].
The second ribosome-bound chaperone system is comprised of RAC and Ssb (Hsp70 family member) in yeast or mRAC and cytosolic Hsp70 in mammals [51]. About 70% of the nascent chains are thought to interact with the RAC-Ssb system in yeast, showing a strong enrichment of aggregation-prone, large, multi-domain and slow translated proteins [56]. In addition, RAC-Ssb was reported to participate in SRP (signal recognition particle)-independent transfer, degradation of nonstop mRNA and interaction with insoluble polyglutamine repeats in vivo [51, 56-59]. A similar chaperone titration strategy to regulate protein synthesis as shown for NAC [20] has recently been demonstrated for mammalian Hsp70 (Fig. 1A) [60, 61]. It was shown that acute proteotoxic stress causes translation attenuation and a stalling of the ribosomes in the first 50 – 65 codon region [60, 61], where the nascent chain is partially still inside the ribosomal tunnel and just starts to exit the ribosome [52, 60]. Cytosolic protein folding conditions could influence this stalling process by directly interfering with the folding of the nascent chain or by sequestration of Hsp70 as demonstrated before for NAC and RAC-Ssb [20, 60].
An additional player in the regulation of mRNA translation is the small heat shock protein Hsp27 that interacts with the translation initiation factor eIF4G and the PABP1 during heat shock [2, 62-64]. Upon heat shock, eIFG and PABP1 dissociate from PABP1-eIF4G complex to interact with Hsp27 and re-localize from the cytosol to the nucleus, suggesting that Hsp27 accompanies their transfer into the nucleus [2]. In earlier studies, it was shown that with increasing temperatures, eIF4G in complex with Hsp27 is localized in insoluble heat shock granules after dissociation from PABP1 [62]. During recovery, the Hsp27-eIF4G-PABP-complex is again detected in the cytosol, indicating that Hsp27 binding alone is not responsible for mRNA translation attenuation [2]. In addition, a link between mRNA nuclear exit in presence of eIF4G/PABP1 and the increased nuclear concentration of these proteins upon heat shock has been suggested [2]. With elevated temperatures, the export of eIF4G and/or PABP1 bound to mRNA might be prevented, indicating an uncoupling of mRNA nuclear export and translation, which could also explain the nuclear increase of eIF4G/PABP1 upon stress [2]. In summary, these data demonstrate that chaperones not only act as refolding machineries to ensure survival upon stress but can also sense stress and influence mRNA translation to reduce new protein influx and with that limit the additional chaperone burden.
ER Stress Response Affects Total Protein Synthesis
The ER is a membrane bound organelle, responsible for synthesis, maturation and modification of proteins of the secretory pathway. The ER has evolved a system that responds to imbalance of ER proteostasis – the UPRER, comprising the induction of ER chaperone expression, proteolytic pathways such as ER associated degradation (ERAD) and a global reduction in protein synthesis [18]. One important ER sensor chaperone is the ATPase and Hsp70 protein BiP, which is located in the ER lumen and activates three ER stress response pathways, the inositol-requiring element 1 (IRE-1), the PkR like ER kinase (PERK) and the activating transcription factor 6 (ATF6) in mammals [18]. Upon IRE-1 and ATF6 pathway activation, expression of diverse genes, involved in ER expansion, protein folding and degradation, are induced [18, 65, 66]. The serine-threonine kinase PERK, which is kept as a monomer by BiP, starts to homodimerize upon stress, which leads to self-activation by phosphorylation [18]. Activated PERK then phosphorylates eIF2α, causing inhibition of mRNA translation by 43S ternary complex stalling (Fig. 1A) [18, 67]. In addition, it was demonstrated that eIF2α phosphorylation by PERK also disturbs rRNA transcription in ribosome biogenesis and regulates ATF6 synthesis, indicating the integration of these pathways [68, 42]. As the UPRER progresses, the phosphatase GADD34 (growth arrest and DNA damage-inducible protein 34), whose translation is elevated upon global translational attenuation by eIF2α-P, starts to dephosphorylate eIF2α-P, which causes translational recovery [69]. In aging rodents, a decline of e.g. BiP and PERK is observed [70, 71]. This suggests that the proteostasis system declines in aging cells, which could exacerbate proteotoxic stress and may lead to neurodegenerative diseases. Besides the translational regulation through PERK in ER stress, there is also a crosstalk with other regulators of protein synthesis. The UPRER is triggered by hyperactive mTOR signaling, for instance, indicating that protein synthesis integrates many signaling pathways [72]. In addition, a connection between NAC and UPRER was suggested, as NAC deficient nematodes show an up-regulation of HSP-4 (orthologue of BiP) [73]. It was suggested that loss of NAC might cause an expanded target pool for the SPR-mediated translocation. Hence, proteins could be transported to the ER, which normally would not be translocated and might cause UPRER activation [73]. This again implicates the important role of NAC in cellular proteostasis as translocation coordinator as well as in global mRNA translation as a central regulatory node of diverse stress responses. For future studies, it will be interesting to investigate the impact of impaired NAC function during aging and upon elevated ER stress.
The Interconnection Between Mitochondrial and Cytosolic Protein Synthesis Upon Mitochondrial Stress
The mitochondrion plays an important role in ATP production and energy supply for numerous essential cellular processes. Although mitochondria harbor their own protein synthesis machinery, their proteome is complemented by nuclear encoded proteins and thus a complex chaperone and protease system has been developed, to maintain mitochondrial proteostasis [74]. Upon accumulation of unfolded or aggregated proteins, e.g. due to excessive ROS production, a stress response is induced, which is due to its similarities to the ER stress response, called the UPRmt. The UPRmt involves a retro-signalingfrom mitochondria to the nucleus, inducing the expression of nuclear encoded mitochondrial chaperones and proteases [74].
Besides the activation of additional chaperones, it was reported that mitochondrial stress also leads to cytosolic mRNA translation reduction, as it is observed for the kinase PERK and the phosphorylation of eIF2α as part of the UPRER. A putative pathway was identified in C. elegans, where mitochondrial dysfunction is sensed by a yet unknown mechanism that results in a GCN-2 mediated phosphorylation of eIF2α and thus to a global translational attenuation (Fig. 1A) [7]. It was hypothesized that reduced cytosolic translation would consequently lead to a reduction in mitochondrial protein import, which might down-regulate mitochondrial translation and thus aid in the regeneration processes to cope with mitochondrial proteotoxicity [7]. The importance of GCN-2 in mitochondrial dysfunction is demonstrated by the finding that GCN-2 is required for life span extension upon mitochondrial stress [7].
A direct correlation between cytosolic mRNA translation and mitochondrial dysfunction has been observed in a recent analysis of stress response profiles in long-lived yeast strains identifying the mitochondrial AAA protease AFG3 and r-proteins of the large ribosomal subunit (MRPL) [75]. AFG3 is a regulator of electron transport chain complexes and is also required for mito-ribosome maturation [75, 76]. It is thereby suggested, that loss of AFG3 causes failure of mito-ribosomal assembly, as the AFG3 target MRPL-32 cannot be cleaved for maturation. Failure in mitochondrial ribosome assembly may induce signals for cytosolic translation attenuation and thereby prevent an imbalance between nuclear and mitochondrial encoded proteins [75]. A putative role of the mitochondrial r-protein MRPL-32 in translation attenuation signaling pathway was suggested as MRPL-32Δ cells also show life span extension [75]. Mitochondrial ribosomal proteins in general seem to have an important influence on life span [17]. In mice and C. elegans, MRPS-5 was reported to be important for life span regulation [19]. Knockdown of mrps-5 leads to a reduction in cellular ATP levels, which is possibly caused by reduced mitochondrial mRNA translation and hence mitochondrial function. In addition, mrps-5 knockdown activates UPRmt, which seems to be specific as the UPRER and cytosolic heat shock response are not affected [19]. MRPS-5 expression decreases in muscles of aging mice [19]. As mrps-5 knockdown leads to life span increase, the age-related MRPS-5 decline may serve as early protein aggregation prevention strategy by reducing mRNA translation in aging cells. Age-related decrease of MRPS-5 protein synthesis can be rescued by nutrient restriction. In addition, mito-nuclear imbalance is also induced by the TOR inhibitor rapamycin via alteration in nDNA/mtDNA oxidative phosphorylation protein ratio and thus provides a connection to dietary restriction pathways [19]. Rapamycin also activates UPRmt, but not UPRER or cytosolic HSR, demonstrating the sensitivity of mito-nuclear balance and its effects on cross-compartmental stress responses. These results raise the question whether components of the mito-nuclear balance or the mitochondrial protein synthesis could serve as putative targets for age-related diseases.
Role of Ribosomal Proteins on Life Span
The reduction of mRNA translation, which is linked to life span increase, can be achieved by prevention of translation initiation or by reduction of the number of ribosomes. To identify the role of ribosomes on life span regulation, r-protein deletion and RNAi studies were performed in yeast and nematodes [4, 17, 33, 45, 77, 78]. In these analyses more than 10 r-proteins were identified, which correlated with life span extension. Life span regulating r-proteins that were identified in all screens and thus whose function in life span extension is conserved across species were RPL-6, RPL-9 and RPL-19 [17, 45, 79, 80]. A deletion of r-proteins could increase life span by two possible pathways: (1) alteration of mRNA translation (by reduction of the number of ribosomes or decreased translation) and (2) induction of stress response pathways. For example, it has been demonstrated that life span expansion by r-protein deletion relies in part on the transcription factor GCN-4 [45]. Furthermore, deletion of r-proteins was demonstrated to reduce sensitivity to ER stress, indicating cross-compartmental signaling of translational activity [79]. Recently it was shown that RPL-22 of the mammalian 60S subunit is able to regulate ribosome composition. A deletion of RPL-22 leads to the expression of its paralog and thereby restores translational activity [81]. These findings implicate the importance of the integrity of the ribosomal structure as conserved r-proteins influence life span and higher eukaryotes seem to have evolved a back-up system.
Keeping the Machinery Running – Enhanced mRNA Translation in Stress
Stress responses in the cytosol as well as UPRs in ER (UPRER) and mitochondria (UPRmt) include the up-regulation of stress response proteins, such as chaperones, transcription factors and components of the degradation machinery [18, 43, 65, 66, 74]. As reported earlier, general protein synthesis is down regulated upon stress, so how can these proteins efficiently be generated if translation machineries are repressed? One regulatory node is mRNA transcription, where specific transcription factors bind on elements in stress protein promoters (Fig. 1B). These factors are activated as part of diverse stress response pathways like UPRER, UPRmt and the cytosolic HSR and include the key stress transcription factors XBP-1 (X-box binding protein), ATFS-1(activating transcription factor associated with stress) and HSF-1 (heat shock factor), respectively [43, 65, 66, 74]. As these factors act on the transcriptional level, how can these stress proteins, e.g. chaperones, be efficiently translated in response to stress, where global mRNA translation is reduced?
Enhanced mRNA translation upon stress is achieved by mRNA structures in the 5’ untranslated region (5’ UTR) (Fig. 1B). IRES (internal ribosomal entry site) is one of them and allows for ribosome recruitment in a cap-independent manner [82]. About 10% of human mRNAs harbor these special mRNA structures [83], including the chaperone BiP (binding immunoglobulin protein), which enables translation in stressful conditions, when cap-dependent translation is inhibited [84]. IRES trans-acting factors (ITAFs) were reported to regulate the IRES-mediated translation where they may function as chaperones that remodel the IRES structure [83, 85]. Recently, it was reported that the insulin-receptor contains IRES structures in Drosophila and mice, whose signaling is also required for stress responses [86]. These results emphasize the importance of these conserved RNA structures as they regulate important receptors of conserved signaling pathways. Another possibility for enhanced mRNA translation in stress is the uORF strategy (upstream open reading frame), which is reported to occur in ~50% of total human mRNA [87]. The presence of uORFs interrupts cap-dependent translation initiation, but upon stress and eIF2a-P mediated translation reduction, uORF-regulated genes can be translated. One example is the translation of the human transcription factor ATF4 (activating transcription factor), which has two uORFs in its 5’UTR [88]. Besides that, many mRNAs, which encode proteins of the translation machinery, such as translation factors and ribosomal proteins, contain a terminal oligo pyrimidine tract (TOP) in their 5’UTR, which is important for enhanced protein synthesis in recovery periods [2, 64, 89]. The mechanism of TOP element regulation is still controversial, but it is suggested that TOR signaling and diverse trans-acting factors might play a role [2, 90-92] These strategies indicate how fine-tuned translation regulation in cells can become as cells can switch from global to specialized protein synthesis to ensure cell survival upon stress.
RIBOSTASIS, TISSUE-SPECIFIC SENSITIVITIES AND TRANS-TISSUE STRESS RESPONSES
Besides the many effects of genetic and environmental signals on translation machineries [4, 17-19], the RNA molecules themselves can be altered during aging due to oxidation by ROS [93, 94]. Lately, Ramaswami and co-workers introduced the concept of ribostasis, which was defined as the appropriate production and regulation of the cellular transcriptome, which has downstream effects on proteostasis [95]. Emphasizing the importance of ribostasis for the cellular and organismal survival, many factors like stress granules, which sequester mRNA, miRNAs or RNA-binding proteins, influence ribostasis and have an impact on downstream mRNA translation and proteostasis [93, 95]. Neurons were reported to be more sensitive to ribostasis changes as they are long-lived, have an increased risk of mRNP (messenger ribo-nucleoprotein particles) aggregates and, due to their inter-neuronal connectivity, exhibit a higher possibility to spread these alterations [95].
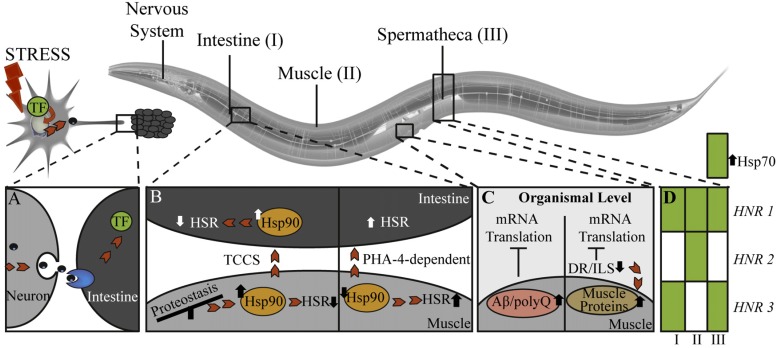
In addition, recent studies demonstrated tissue-selective as well as trans-tissue stress responses in C. elegans (Fig. 2) [20, 96-99]. In neurons, UPRER induction and XBP-1 activity lead to a secreted ER stress signal, which causes XBP-1 activation via IRE-1 in distal intestinal cells (Fig. 2A) [96]. In addition, an elevated Hsp90 expression in muscle cells due to metastable myosin proteins, also increased Hsp90 expression in intestinal cells, which is regulated by trans-cellular-chaperone signaling and PHA-4 (orthologue of FoxA) activity (Fig. 2B) [98]. As elevated Hsp90 causes repression of HSR, hsp-90 knockdown causes an organismal HSR which also involves PHA-4 activity (Fig. 2B) [98]. With respect to aging, accumulation of those proteostasis imbalances in muscles might induce Hsp90 expression in a distal tissue and hence an overall increased sensitivity to heat shock, promoting organismal aging. Interestingly, expression of stressor proteins, such as polyQ or Aβ proteins in muscle cells, lead to an organism-wide translational decrease (Fig. 2C) [20]. These findings again imply a widespread impact of a stress response in a single cell or tissue on global stress responses as well as longevity in multi-cellular organisms.
Fig. (2).
Tissue-specific and tissue-spanning stress responses in C. elegans. (A) Stress responses, e.g. UPRER, in neurons can generate signals that are transmitted to distal intestinal cells to induce similar stress responses there. This cell non-autonomous signaling could be achieved by exchange of small molecules (e.g. peptides, neurotransmitter, regulatory RNA) [96, 98]. (B) Trans-cellular-chaperone signaling (TCCS) allows cross-tissue Hsp90 adaptation, which causes also reduced HSR in distal tissues due to HSF-1 inactivation by Hsp90. PHA-4 (orthologue of FoxA) -mediated trans-cellular-signaling allows HSR activation upon Hsp90 knockdown. (C) Attenuation of global mRNA translation is caused by reduction of ILS signaling/dietary restriction and expression of proteotoxic proteins, e.g. Aβ and polyQ proteins. Upon ILS signaling reduction the expression of muscle components is maintained to allow locomotion upon starvation. (D) Knockdown of HSR negative regulators (HNR 1 to 3) causes induction of tissue-specific Hsp70 expression (filled squares) in intestine (I), muscles (II) and spermatheca (III).
Long-lived nematodes (DR or daf-2 mutants) show a global r-protein decline but also an increase in striated muscle proteins to maintain muscle integrity upon starvation (Fig. 2C) [99]. This organismal strategy allows locomotion upon nutrient restriction to find nutrient rich areas and demonstrates that stress signals can lead to fine-tuned cellular outputs. In line with that, a genome wide RNAi screen identified negative regulators of the heat shock response, which show tissue-specific induction of Hsp70 upon RNAi mediated knockdown (Fig. 2D) [97]. These results also suggest that tissues may have different signaling responses upon the same stress condition that allows them to adjust to the specific requirements of the tissue cell function [97]. At the same time, the organismal stress response network may also affect aging networks as aging cells and the subsequent decline in trans-cellular transmitted stress responses could increase stress sensitivities in other tissues, initiating an aging cascade. For future studies, it will be important to investigate the connection between these differences in stress response with respect to aging and age-related diseases. For example, are certain cell types more sensitive to aging due to weaker stress response patterns than other cells? Do aging cells “infect” other cells and thereby cause organismal aging? These and many more questions have to be answered in future to get an understanding of organismal aging and to advance treatment of age-related diseases.
CONCLUSION
Protein synthesis plays a central role in cellular and organismal adaptation to environmental conditions. Upon stress, protein synthesis is reduced, either by a decline of the number of ribosomes or mRNA translation inhibition, lowering chaperone burden by a reduction of protein influx [17, 100]. The different cross-compartmental stress responses that also affect protein synthesis, are connected [19, 50, 72, 79], setting up a multifaceted stress response network and allow tissue-specific, trans-cellular and cross-tissue responses [96-98]. Aging is associated with a decline in protein quality control and chaperone activity [18, 101, 102]. Aging cells or tissues, which cope with increasing protein misfolding and aggregation, might induce aging processes in distal tissues and cause global organismal aging. Interestingly, it was speculated whether aging is more a result of over-activated stress responses rather than an accumulation of damage [103]. The truth might lie in the middle and both, excessed responses and damage accumulation, might contribute to the aging process. TOR signaling was often reported to play a role in life span regulation as its down regulation affects mRNA translation and increases life span [4, 17, 32, 33]. As TOR inhibition by rapamycin increases life span in multicellular organisms, such as C. elegans and D. melanogaster and is conserved from yeast to mammals [17, 34], its putative impact on aging regulation in mammals was brought into question. An increase in life span and a decrease in age-related diseases upon rapamycin treatment were observed in mice [104, 105], but it is still controversial whether it is a direct consequence or caused by side effects of the rapamycin treatment. [104, 106]. Also in humans, a link between RPTOR (regulatory associated protein of mTOR) expression and longevity was shown [107], implicating that this longevity pathway is strongly conserved and might be a putative target for future therapies of age-related diseases.
ACKNOWLEDGEMENTS
JKM acknowledges funding by the DFG Excellence Cluster NeuroCure.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6(4):318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Ma S, Bhattacharjee R B, Bag J. Expression of poly(A)-binding protein is upregulated during recovery from heat shock in HeLa cells. FEBS J. 2009;276(2):552–570. doi: 10.1111/j.1742-4658.2008.06803.x. [DOI] [PubMed] [Google Scholar]
- 3.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25(48):6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 4.Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6(1):95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 5.Marion R M, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad Sci. USA. 2004;101(40):14315–22. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proud C G. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. FEBS. 2002;269(22):5338–49. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- 7.Baker B M, Nargund A M, Sun T, Haynes C M, Larsson N-G. Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2a Kinase GCN-2. PLoS Genet. 2012;8(6):e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morley JF. Regulation of Longevity in Caenorhabditis elegans by Heat Shock Factor and Molecular Chaperones. Mol. Biol. Cell. 2004;15(2):657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafontaine D L, Tollervey D. The function and synthesis of ribosomes. Nat. Rev. 2001;2(7):514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- 10.Zeidan Q, Wang Z, De Maio A, Hart G W. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell. 2010;21(12):1922–36. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann. Rev.Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 12.Kressler D, Hurt E, Bergler H, Baßler J. The power of AAA-ATPases on the road of pre-60S ribosome maturation — Molecular machines that strip pre-ribosomal particles. Biochimica et Biophysica Acta (BBA) - Mol. Cell Res. 2012;1823(1):92–100. doi: 10.1016/j.bbamcr.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 2010;189(1):69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23(5):242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y E, Hipp M S, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Ann. Rev. Biochem. 2013;82(1):323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 17.Mehta R, Chandler-Brown D, Ramos F J, Shamieh L S, Kaeberlein M. Regulation of mRNA translation as a conserved mechanism of longevity control. Adv. Exp. Med. Biol. 2010;694:14–29. doi: 10.1007/978-1-4419-7002-2_2. [DOI] [PubMed] [Google Scholar]
- 18.Brown M K, Naidoo N. The endoplasmic reticulum stress response in aging and age-related diseases. Front. Physiol. 2012;3:263. doi: 10.3389/fphys.2012.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houtkooper R H, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams R W, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497 (7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirstein-Miles J, Scior A, Deuerling E, Morimoto R I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32(10):1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross C A, Poirier M A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto M, Rockenstein E, Crews L, Masliah E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer's and Parkinson's diseases. Neuromol. Med. 2003;4(1-2):21–36. doi: 10.1385/NMM:4:1-2:21. [DOI] [PubMed] [Google Scholar]
- 23.Smith E D, Tsuchiya M, Fox L A, Dang N, Di H, Kerr E O, Johnston E D, Tchao B N, Pak D N, Welton K L, Promislow D E L, Thomas J H, Kaeberlein M, Kennedy B K. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Gen. Res. 2008;18(4):564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struc. Mol. Biol. 2013;20(2):237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell. 2013;49(3):411–21. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Zhou T, Tartaglia G G, Vendruscolo M, Wilke C O. Translationally optimal codons associate with aggregation-prone sites in proteins. Proteomics. 2010;10(23):4163–4171. doi: 10.1002/pmic.201000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond D A, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134(2):341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21(1):25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Chartier M, Gaudreault F, Najmanovich R. Large-scale analysis of conserved rare codon clusters suggests an involvement in co-translational molecular recognition events. Bioinorm.(Oxford, England) 2012;28(11):1438–1445. doi: 10.1093/bioinformatics/bts149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho C-P, Lin S-C, Chou M-Y, Hsu H-T, Chang K-Y. Regulation of Programmed Ribosomal Frameshifting by Co-Translational Refolding RNA Hairpins. PloS one. 2013;8(4):e62283. doi: 10.1371/journal.pone.0062283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384(1):73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber A, French S L, Tekotte H, Yerlikaya S, Stahl M, Perepelkina M P, Tyers M, Rougemont J, Beyer A L, Loewith R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30(15):3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaeberlein M. Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients. Science. 2005;310 (5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Conn C S, Han Y, Yeung V, Qian S B. PI3K-mTORC1 Attenuates Stress Response by Inhibiting Cap-independent Hsp70 Translation. J. Biol. Chem. 2011;286(8):6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall M N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 36.Laplante M, Sabatini D M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapierre L R, Hansen M. Lessons from C.elegans: signaling pathways for longevity. Trends Endocrinol. Metabol. 2012;23 (12):637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012;441(1):1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 39.Holz M K, Ballif B A, Gygi S P, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Kapahi P, Zid B. TOR pathway: linking nutrient sensing to life span. Neurobiol. ging. 2004;2004 (36):PE34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannan K M, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur G A, Pearson R B, Hannan R D. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell Biol. 2003;23 (23):8862–77. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DuRose J B, Scheuner D, Kaufman R J, Rothblum L I, Niwa M. Phosphorylation of eukaryotic translation initiation factor 2alpha coordinates rRNA transcription and translation inhibition during endoplasmic reticulum stress. Mol. Cell Biol. 2009;29(15):4295–307. doi: 10.1128/MCB.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shore D E, Ruvkun G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 2013;23(9):409–20. doi: 10.1016/j.tcb.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson B W, Hubbard A, Melov S, Lithgow G J, Kapahi P. Life Span Extension via eIF4G Inhibition Is Mediated by Posttranscriptional Remodeling of Stress Response Gene Expression in C.elegans. Cell Metabol. 2011;14(1):55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffen K K, MacKay V L, Kerr E O, Tsuchiya M, Di H, Fox L A, Dang N, Johnston E D, Oakes J A, Tchao B N, Pak D N, Fields S, Kennedy B K, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapahi P, Zid B M, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14(10):885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinnebusch A G. Translational regulation of GCN4 and the general amino acid control of yeast*. Ann. Rev. Microbiol. 2005;59(1):407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 48.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch A G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6(2):269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 49.Rousakis A, Vlassis A, Vlanti A, Patera S, Thireos G, Syntichaki P. The general control nonderepressible-2 kinase mediates stress response and longevity induced by target of rapamycin inactivation in Caenorhabditis elegans. Aging Cell. 2013;12(5):742–51. doi: 10.1111/acel.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherkasova V A, Hinnebusch A G. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17(7):859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 2012;37(7):274–283. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16(6):589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 53.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50(3):379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010;189(1):57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freire MA. Translation initiation factor (iso) 4E interacts with BTF3, the ß subunit of the nascent polypeptide-associated complex. Gene. 2005;345(2):271–277. doi: 10.1016/j.gene.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 56.Willmund F, del Alamo M, Pechmann S, Chen T, Albanèse V, Dammer E B, Peng J, Frydman J. The Cotranslational Function of Ribosome-Associated Hsp70 in Eukaryotic Protein Homeostasis. Cell. 2013;152(1-2):196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plath K, Rapoport T A. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 2000;151(1):167–78. doi: 10.1083/jcb.151.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiabudini M, Conz C, Reckmann F, Rospert S. Ribosome-Associated Complex and Ssb Are Required for Translational Repression Induced by Polylysine Segments within Nascent Chains. Mol. Cell. Biol. 2012;32(23):4769–4779. doi: 10.1128/MCB.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Meriin A B, Zaarur N, Romanova N V, Chernoff Y O, Costello C E, Sherman M Y. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23(2):451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu B, Han Y, Qian S-B. Cotranslational Response to Proteotoxic Stress by Elongation Pausing of Ribosomes. Mol. Cell. 2013;49(3):453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shalgi R, Hurt J A, Krykbaeva I, Taipale M, Lindquist S, Burge C B. Widespread Regulation of Translation by Elongation Pausing in Heat Shock. Mol. Cell. 2013;49(3):439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cuesta R, Laroia G, Schneider R J. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14(12):1460–1470. [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess H M, Gray N K. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly(A)-binding proteins. Comm. Integ. Biol. 2012;5(3):243–247. doi: 10.4161/cib.19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Datu A-K, Bag J. Enhanced translation of mRNAs encoding proteins involved in mRNA translation during recovery from heat shock. PloS One. 2013;8(5):e64171. doi: 10.1371/journal.pone.0064171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4(2):265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 66.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 67.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 68.Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, Wek RC. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell. 2011;22(22):4390–4405. doi: 10.1091/mbc.E11-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Majumder M, Huang C, Snider MD, Komar A A, Tanaka J, Kaufman R J, Krokowski D, Hatzoglou M. A Novel Feedback Loop Regulates the Response to Endoplasmic Reticulum Stress via the Cooperation of Cytoplasmic Splicing and mRNA Translation. Mol. Cell. Biol. 2012;32(5):992–1003. doi: 10.1128/MCB.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naidoo N, Ferber M, Master M, Zhu Y, Pack A I. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J. Neurosci. 2008;28(26):6539–48. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paz Gavilan M, Vela J, Castano A, Ramos B, del Rio J C, Vitorica J, Ruano D. Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol. Aging. 2006;27(7):973–82. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 72.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning B D, Hotamisligil G S. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol. Cell. 2008;29(5):541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arsenovic P T, Maldonado A T, Colleluori V D, Bloss T A, Aballay A. Depletion of the C.elegans NAC Engages the Unfolded Protein Resonse Resulting in Increased Chaperone Expression and Apoptosis. PLoS ONE. 2012; 7(9):e44038. doi: 10.1371/journal.pone.0044038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pellegrino M W, Nargund A M, Haynes C M. Signaling the mitochondrial unfolded protein response. Biochimica. et biophysica Acta. 2013;1833 (2):410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delaney JR, Ahmed U, Chou A, Sim S, Carr D, Murakami CJ, Schleit J, Sutphin G L, An E H, Castanza A, Fletcher M, Higgins S, Jelic M, Klum S, Muller B, Peng Z J, Rai D, Ros V, Singh M, Wende H V, Kennedy B K, Kaeberlein M. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell. 2013;12(1):156–166. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli E I, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123 (2):277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, Breitenbach M, Breitenbach-Koller L. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 2007;42(4):275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 78.Curran S P, Ruvkun G. Lifespan Regulation by Evolutionarily Conserved Genes Essential for Viability. PLoS Genet. 2007;3(4):e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steffen K K, McCormick M A, Pham K M, MacKay V L, Delaney J R, Murakami C J, Kaeberlein M, Kennedy B K. Ribosome Deficiency Protects Against ER Stress in Saccharomyces cerevisiae. Genetics. 2012;191(1):107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yanos M E, Bennett C F, Kaeberlein M. Genome-Wide RNAi Longevity Screens in Caenorhabditis elegans. Curr. Genomics. 2012;13(7):508–18. doi: 10.2174/138920212803251391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O'Leary M N, Schreiber K H, Zhang Y, Duc A C, Rao S, Hale J S, Academia E C, Shah S R, Morton J F, Holstein C A, Martin D B, Kaeberlein M, Ladiges W C, Fink P J, Mackay V L, Wiest D L, Kennedy B K. The ribosomal protein rpl22 controls ribosome composition by directly repressing expression of its own paralog, rpl22l1. PLoS Genet. 2013;9(8):e1003708. doi: 10.1371/journal.pgen.1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spriggs K A, Stoneley M, Bushell M, Willis A E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100(1):27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 83.Mitchell S A, Spriggs K A, Bushell M, Evans J R, Stoneley M, Le Quesne J P, Spriggs R V, Willis A E. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19(13):1556–71. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5' leader of a cellular mRNA. Nature. 1991;353(6339):90–4. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 85.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2008;28(1):40–9. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olson CM, Donovan MR, Spellberg MJ, Marr MT. 2nd The insulin receptor cellular IRES confers resistance to eIF4A inhibition. eLife. 2013;2:e00542. doi: 10.7554/eLife.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calvo S E, Pagliarini D J, Mootha V K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106(18):7507–12. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vattem K M, Wek R C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U S A. 2004;101(31):11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pichon X, Wilson L A, Stoneley M, Bastide A, King H A, Somers J, Willis A E. RNA binding protein/RNA element interactions and the control of translation. Curr. Prot. Pep. Sci. 2012;13(4):294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Loayza-Puch F, Drost J, Rooijers K, Lopes R, Elkon R, Agami R. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 2013;14(4):R32. doi: 10.1186/gb-2013-14-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thoreen C C, Chantranupong L, Keys H R, Wang T, Gray N S, Sabatini D M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Damgaard C K, Lykke-Andersen J. Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25(19):2057–68. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cookson M R. Aging--RNA in development and disease. Wiley interdisciplinary Rev RNA. 2012; 3(1):133–43. doi: 10.1002/wrna.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hofer T, Marzetti E, Xu J, Seo A Y, Gulec S, Knutson M D, Leeuwenburgh C, Dupont-Versteegden E E. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp. Gerontol. 2008;43(6):563–70. doi: 10.1016/j.exger.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramaswami M, Taylor JP, Parker R. Altered Ribostasis RNA-Protein Granules in Degenerative Disorders. Cell. 2013;154(4):727–36. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor R C, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–47. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guisbert E, Czyz D M, Richter K, McMullen P D, Morimoto R I. Identification of a tissue-selective heat shock response regulatory network. PLoS enet. 2013; 9(4):e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Oosten-Hawle P, Porter R S, Morimoto R I. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 2013;153(6):1366–78. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Depuydt G, Xie F, Petyuk V A, Shanmugam N, Smolders A, Dhondt I, Brewer H M, Camp D G, Smith R D, Braeckman B P. Reduced insulin/IGF-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol. Cell. Proteom. 2013 doi: 10.1074/mcp.M113.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaeberlein M. Longevity and aging. F1000prime Rep. 2013;5:5. doi: 10.12703/P5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ben-Zvi A, Miller E A, Morimoto R I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 2009;106 (35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haigis M C, Yankner B A. The Aging Stress Response. Mol. Cell. 2010;40(2):333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blagosklonny M V. Hormesis does not make sense except in the light of TOR-driven aging. Aging. 2011;3(11):1051–1062. doi: 10.18632/aging.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilkinson JE, Burmeister L, Brooks S V, Chan C C, Friedline S, Harrison D E, Hejtmancik J F, Nadon N, Strong R, Wood L K, Woodward M A, Miller RA. Rapamycin slows aging in mice. Aging Cell. 2012;11(4):675–82. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flynn J M, O'Leary M N, Zambataro C A, Academia E C, Presley M P, Garrett B J, Zykovich A, Mooney S D, Strong R, Rosen C J, Kapahi P, Nelson M D, Kennedy B K, Melov S. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12(5):851–62. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neff F, Flores-Dominguez D, Ryan D P, Horsch M, Schroder S, Adler T, Afonso L C, Aguilar-Pimentel J A, Becker L, Garrett L, Hans W, Hettich M M, Holtmeier R, Holter S M, Moreth K, Prehn C, Puk O, Racz I, Rathkolb B, Rozman J, Naton B, Ordemann R, Adamski J, Beckers J, Bekeredjian R, Busch D H, Ehninger G, Graw J, Hofler H, Klingenspor M, Klopstock T, Ollert M, Stypmann J, Wolf E, Wurst W, Zimmer A, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, Ehninger D. Rapamycin extends murine lifespan but has limited effects on aging. J. Clin. Invest. 2013;123(8):3272–91. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Passtoors WM, Beekman M. Gene expression analysis of mTOR pathway association with human longevity. . Aging Cell. 2013;12(1):24–31. doi: 10.1111/acel.12015. [DOI] [PubMed] [Google Scholar]