Abstract
By immunoaffinity column chromatography, we have purified two RNA polymerase complexes, the transcriptase and replicase, from vesicular stomatitis virus-infected baby hamster kidney cells. The transcriptase is a multiprotein complex, containing the virus-encoded RNA polymerase L and P proteins, and two cellular proteins, translation elongation factor-1α and heat-shock protein 60. In addition, the complex contains a submolar amount of cellular mRNA cap guanylyltransferase. The replicase, on the other hand, is a complex containing the viral proteins, L, P, and the nucleocapsid (N), but lacking elongation factor-1α, heat-shock protein 60, and guanylyltransferase. The transcriptase complex synthesizes capped mRNAs and initiates transcription at the first gene (N) start site, whereas the replicase complex initiates RNA synthesis at the precise 3′ end of the genome RNA and synthesizes encapsidated replication products in the presence of the N–P complex. We propose that two RNA polymerase complexes that differ in their content of virally and host-encoded proteins are separately responsible for transcription and replication of vesicular stomatitis virus genome RNA.
A hallmark of all nonsegmented negative-strand (ns)RNA viruses (mononegavirales order) such as rabies, measles, Sendai, parainfluenza, Ebola, and many others is that mature virions contain a virally encoded RNA-dependent RNA polymerase (referred to as transcriptase), which transcribes the negative-sense genome RNA into discrete mRNAs on entry into the cell to initiate infection (1, 2). To replicate the genome RNA, the transcriptase is hypothesized to be modified by an unknown mechanism to form a replicase that synthesizes the full-length positive-strand genome RNA (3, 4); the replicase then synthesizes multiple copies of nsRNA using the positive RNA as template. During each step of the replication reaction, both plus- and minus-strand RNAs are concomitantly enwrapped by the newly synthesized nucleocapsid (N) protein (5, 6) to form the ribonucleoprotein (RNP) complex. The composition of the replicase and the process of replication remain an enigma.
We have been studying vesicular stomatitis virus (VSV) as a prototypic nonsegmented negative-strand RNA virus to probe the structure and function of transcriptase and the putative replicase to delineate the mechanism of transcription and replication of this class of viruses. VSV, like rabies virus, belongs to the rhabdovirus family and contains a single-strand genome RNA of negative polarity (≈11.2 kb long) tightly associated with the N protein and helically packed within a bullet-shaped shell that is surrounded by the host-cell plasma membrane (3, 4). Two membrane proteins, spike glycoprotein (G) and matrix (M) protein, are located outside and inside of the membrane, respectively. Two proteins are associated with the helical RNP, the RNA polymerase large (L) (241 kDa) and phosphoprotein (P) (29 kDa) (7), which together constitute the active transcriptase holoenzyme complex (3, 4). It is generally believed that the L protein is the multifunctional RNA polymerase that carries out such functions as RNA synthesis, capping, and polyadenylation of mRNAs and methylation of the capped mRNAs but manifests these activities when complexed with the P protein, which functions as a cofactor (4, 8). The RNP purified from the virions synthesizes in vitro a leader RNA (47 nucleotides) initiating from the 3′ end of the genome RNA (9, 10) followed sequentially by the synthesis of five 5′ capped and 3′ polyadenylated mRNAs (11) in the order 3′-N-P-M-G-L–5′ (12, 13). In a reconstituted in vitro transcription reaction containing an L and P fraction purified from the virions and transcriptionally inactive N-RNA template, Emerson (14) demonstrated that the transcriptase enters at a single site at the precise 3′ end of the genome RNA, synthesizes the leader RNA, and the same complex continues synthesis of the downstream genes sequentially. Thus, the synthesis of leader RNA is obligatory for transcription of subsequent genes. This widely accepted model is ascribed as the “single initiation stop-start model” of transcription (14). An obligate requirement of newly synthesized N protein for replication led to the proposition that an interaction of N protein with the nascent leader RNA leads to the modification of the transcriptase, which promotes read-through of the initiation and termination signals to copy the full-length genome RNA (15, 16). Subsequently, it was shown that the N protein remains complexed with the P protein in infected cells, thus implicating the involvement of N–P complex rather than the N protein alone in the replicative reaction (17, 18).
Several observations, however, raised questions as to the veracity of the proposed transcription model. Particularly puzzling is the synthesis of the leader RNA, which, unlike mRNAs, is uncapped and lacks a poly(A) tail (19) and thus appears to be a replication attempt by a putative replicase, because the leader RNA ultimately becomes part of the full-length plus-strand genome RNA. Moreover, if L and P indeed constitute the transcriptase, why is the leader RNA not capped, whereas the mRNAs are (8, 11)? Finally, based on the observation that a VSV mutant polR1 synthesizes in vitro molar excess of N transcripts over leader RNA, Chuang and Perrault (20) proposed a “two polymerase entry model,” one for leader RNA and the other for mRNA. Also recently, internal entry of the polymerase at the first gene start sequence was observed in the infected cells by Whelan and Wertz (21) by using recombinant VSV containing an additional small gene inserted at the leader-N gene junction and UV-mapping technique, implying two initiation events on the genome template. To date, however, there is no direct evidence that two distinct RNA polymerase entities are indeed present in the infected cells that carry out the two very different RNA synthetic reactions during the life cycle of the virus.
Some recent findings from our laboratory prompted us to further probe the mechanism of VSV RNA synthesis. First, while studying the structure and function of the L protein of VSV, we recently observed that two cellular proteins, specifically translation elongation factor (EF)-1αβγ (22) and the mRNA capping enzyme, guanylyltransferase (GT) (23), are tightly associated with the L protein when it is expressed in insect cells. These studies provided the first indication that the transcriptase may be a multiprotein complex containing cellular proteins, contrary to the notion that only two virally encoded proteins, L and P, constitute the transcriptase complex. Second, we demonstrated (24, 25) that the transcriptionally inactive P protein is fully competent to replicate a defective interfering particle RNA, which contains the replication promoters at both ends (26) in a three-plasmid-based (L, P, and N) reverse genetics system in vivo. Although the active transcriptase was not formed under this transfection condition, an N–Pmut complex, the required intermediate of replication, was efficiently made. These findings led us to propose that a putative tripartite complex comprising L–(N–P) of unknown composition may represent the replicase (25). In fact, such a complex with replicase activity was detected by coexpression of L, N, and P in Sf21 cells by recombinant baculoviruses that express the respective genes (27). Thus, the findings described above provided the initial indications that transcriptase and replicase of VSV may be two different entities and may be structurally distinct.
Here, we show directly that the transcriptase purified from VSV-infected baby hamster kidney (BHK) cells is indeed a multiprotein complex consisting of L, GT, EF-1α, and P. Surprisingly, we discovered an additional cellular protein identified as heat-shock protein (Hsp)60, bound to purified transcriptase complex in a molar amount. The transcriptase complex carried out synthesis of capped mRNAs in vitro but not the leader RNA. The replicase, on the other hand, is a complex of L, N, and P but devoid of such host proteins and initiates RNA synthesis at the 3′ end of the genome RNA and, in the presence of the N–P complex, carries out read-through replication products in vitro. These findings provide the basis for proposing a previously undescribed model of transcription and replication of VSV genome RNA.
Materials and Methods
Cell Cultures and Virus. A monolayer of BHK-21 cells was infected with VSV (Indiana serotype, Mudd–Summers strain) at a multiplicity of infection of 0.05, and the virus was purified as described (28).
Preparation of the Anti-EF-1α, Anti-P, and Anti-N Immunoaffinity Column. An anti-EF-1α monoclonal antibody (CBP-KK1) was obtained from Upstate Biotechnology (Lake Placid, NY). Five hundred micrograms of the monoclonal anti-EF-1α antibody was bound to cyanogen bromide-(CNBr) activated Sepharose CL-4B (Pharmacia) by the standard procedure of ligand coupling in 100 mM sodium bicarbonate/500 mM sodium chloride, pH 8.3 (29). The free groups of the CNBr-activated Sepharose were blocked by 100 mM Tris·HCL, pH 8.0, overnight, and then the slurry was washed by three cycles of alternating 100 mM Tris·HCL, pH 8.0, and 100 mM acetate buffer, pH 4.0. The slurry was then packed in a column and equilibrated in a buffer containing 50 mM sodium phosphate and 150 mM sodium chloride, pH 7.0.
Anti-P and -N polyclonal antibodies (raised against bacterially expressed proteins) were first purified by standard Protein A Sepharose immunoaffinity chromatography (29). The total IgG fraction was subsequently purified by P or N antigen affinity columns to obtain monospecific antibodies. Five hundred micrograms of monospecific anti-P or -N IgG bound to the cyanogen bromide-activated Sepharose CL-4B (Pharmacia) was used as described above.
Purification of Transcriptase and Replicase. BHK-21 cells were infected with VSVIND (Indiana serotype) at a multiplicity of 5 plaque-forming units per cell at 37°C. Sixty 150-mm plates were harvested 6 h postinfection and centrifuged at 800 × g for 10 min at 4°C; the pellet was resuspended and homogenized in the dounce buffer (10 mM Tris·HCl, pH 7.5/10 mM KCl/1.5 mM MgCl2/1 mM DTT) and centrifuged at 10,000 × g for 15 min at 4°C (30). The supernatant was then subjected to 100,000 × g centrifugation for 2 h at 4°C. The high-speed supernatant was adjusted to 40% saturation with ammonium sulfate [(NH4)2SO4] and centrifuged at 10,000 × g for 30 min at 4°C. The pellet was suspended in the dialysis buffer (50 mM sodium phosphate/150 mM sodium chloride, pH 7.0/1 mM DTT/10% glycerol) and dialyzed against the same buffer for 16 h. The dialyzed fraction was applied onto the anti-EF-1α immunoaffinity column (0.8 × 4 cm) and washed with buffer containing 50 mM sodium citrate/150 mM sodium chloride, pH 5.5 and 4.3, in a sequential manner. Finally the bound proteins were eluted by 100 mM glycine/150 mM sodium chloride, pH 2.3, and collected into microcentrifuge tubes containing 50 μl of 1.0 M Tris·Cl, pH 9.0. The eluted protein was then passed through an anti-P immunoaffinity column (0.8 × 4 cm) and washed and eluted in the same buffer as described above. Approximately 25 μg of purified transcriptase was obtained. For purification of the replicase, the flow-through of the anti-EF-1α immunoaffinity column was passed through an anti-P or -N immunoaffinity column, washed and eluted in the same buffers as described above. Approximately 20 μg of replicase was obtained.
PAGE and Western Blot Analysis. Proteins were subjected to 10% SDS/PAGE as described by Laemmli (31), and Western blot analysis was performed as described (32).
GT–GMP Complex Formation. Reaction mixtures containing 50 mM Tris·HCl (pH 8.0), 5 mM MgCl2, 1 mM DTT, 100 mM NaCl, and [α-32P]GTP (20 μCi, 3,000 Ci/mmol), and the enzyme were incubated for 1 h at 30°C and then analyzed in a 10% SDS/PAGE. The covalently labeled GT–GMP complex was visualized by autoradiography (23).
In Vitro Transcription/Replication of VSV mRNAs. Transcription reaction in vitro was carried out as described (28) in a standard reaction mixture containing the transcriptase and N-RNA template free of L and P proteins. To synthesize nonradioactive transcripts, 1 mM each NTP was used. For transcripts labeled with [α-32P]GTP (10 μCi, 3,000 Ci/mmol), the transcription reaction was carried out in the presence of 0.05 mM unlabeled GTP and 1 mM each ATP, CTP, and UTP. The RNA products were analyzed by 5% urea-PAGE and autoradiographed. To analyze the capping status of the transcripts, the synthesized unlabeled RNA products were capped with 1 unit vaccinia GT (Ambion, Austin, TX) and [α-32P]GTP (23). The capped transcripts were decapped with tobacco acid pyrophosphatase (25 units) at 37°C for 1 h, followed by incubation with HK phosphatase at 30°C for 1 h according to the manufacturer's protocol (Epicentre Technologies, Madison, WI). The dephosphorylated RNA products were then end-labeled by using [γ-32P]ATP and T4 polynucleotide kinase (10 units) at 37°C for 1 h (23) and visualized by autoradiography after a 5% urea-PAGE. In vitro replication reactions were carried out in the presence of [α-32P]GTP, and the RNAs were analyzed in a 0.8% formaldehyde–agarose gel as described (27). For analysis of the encapsidation status of the replication products, the reaction mixture after replication was treated with RNaseA (25 μg/ml) and incubated at 37°C for 30 min (33). The RNA products were analyzed by electrophoresis on a 0.8% formaldehyde-agarose gel.
Mass Spectrometry of Purified Proteins. The Coomassie bluestained band was cut from the gel, reduced, alkylated, and digested with trypsin. This tryptic digest was subsequently analyzed by capillary column HPLC-electrospray ionization-tandem mass spectrometry by using an ion trap mass spectrometer system, to sequence the resulting peptides according to Kinter and Sherman (34).
Results
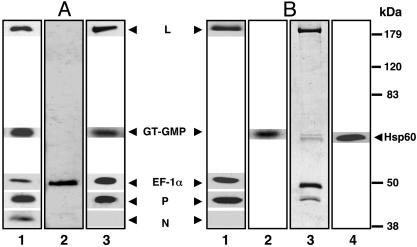
Purification of Transcriptase from Infected Cells. We have previously shown that a soluble postmicrosomal fraction prepared from VSV-infected cells containing the viral proteins L, P, and N can efficiently carry out mRNA synthesis in vitro when added to the N-RNA template (30). Thus, this fraction would serve as a rational source of the transcriptase and probably also the putative replicase. Coupled with our earlier studies that insect cell EF-1αβγ remains tightly bound to recombinant L protein (22), this prompted us to fractionate the 100,000 × g soluble fraction prepared 6 h after infection at 5 multiplicities of infection over a mammalian anti-EF-1α affinity column after an initial precipitation by 40% (NH4)2 SO4. Western blot analysis of the ammonium sulfate fraction after PAGE clearly established the presence of L, P, N, and EF-1α (Fig. 1A, lane 1) in the fraction. The presence of GT was confirmed by the formation of the GT–GMP32 complex, the required intermediate for the capping reaction (35), by incubating the fraction with [α-32P]GTP followed by SDS/PAGE and autoradiography (23). Subsequent anti-EF-1α affinity chromatography of the fraction followed by elution, SDS/PAGE, and Coomassie blue staining showed exclusively EF-1α, which represented the bulk of the protein in the fraction (Fig. 1 A, lane 2). However, Western blot analysis (Fig. 1 A, lane 3) clearly revealed the presence of L, EF-1α, P, and GT by the GT–GMP32 complex, but strikingly no N protein that was present in the flow-through fraction (see below). To remove the excess EF-1α and any EF-1α–GT complex, the fraction was passed over an anti-P column. The eluate from this column contained L, EF-1α, and P (Fig. 1B, lane 1) by Western blot analysis, and GT by the GT–GMP32 complex (lane 2), indicating that L and P proteins are complexed with EF-1α and GT. The flow-through fraction contained the excess EF-1α and GT but no L and P proteins (data not shown). By staining the gel with Coomassie blue, four distinct protein bands were discernible (Fig. 1B, lane 3). The GT–GMP complex comigrated with a minor band slightly slower than a major protein band, indicating that an additional host protein is also part of the transcriptase complex. Subsequent analysis of the major and minor proteins by tandem mass spectrometry (34) corresponded with the amino acid sequence of cellular Hsp60 and GT, respectively. This unexpected finding prompted us to confirm the identity of the major band by Western blot analyses, using the anti-Hsp60 antibody and, as shown in Fig. 1B, lane 4, the protein band indeed crossreacted with Hsp60. To further confirm that the host proteins are part of the transcriptase complex, the complex was treated with 0.8 M NaCl and fractionated over an anti-EF-1α column. All host proteins were quantitatively retained with the L protein and, as expected, the P protein was recovered in the flow-through fraction (Fig. 6, which is published as supporting information on the PNAS web site), indicating that the host proteins are tightly complexed with the L protein. From several batches of transcriptase purified by affinity chromatography described above and by comparing the staining intensities of the purified proteins and their respective molecular weights, an average molar ratio of 1:1:3(±1):3(±1) for L, Hsp60, EF-1α, and P was determined, with GT being reproducibly present in submolar amounts (≈0.2 mol) with respect to the L protein. Thus, based on the specificity of antibody binding, we were able to purify the VSV transcriptase complex, which contains two virally encoded proteins, L and P, two major cellular proteins, EF-1α and Hsp60, and a minor protein, GT.
Fig. 1.
Purification of transcriptase. (A) Western blot analysis of 40% (NH4)2SO4 fraction (500 ng) (lane 1), anti-EF-1α eluate (500 ng) (lane 3) was carried out by electrophoresis in a 10% SDS–polyacrylamide gel and transferred to a nitrocellulose membrane. The membranes were probed with antibodies against L, P, N, and EF-1α, and the protein bands were visualized by enhanced chemiluminescence. Covalent GT–GMP complex formation of GT was detected by incubation of reaction mixture with [α-32P]GTP at 30°C for 1 h and analyzed by 10% SDS/PAGE followed by autoradiography (lanes 1 and 3). Anti-EF-1α eluate (4 μg) (lane 2) was subjected to SDS/PAGE and visualized by staining with Coomassie blue dye. (B) Western blot analysis of anti-P eluate (500 ng) was done by using antibodies against L, P, N, and EF-1α (lane 1) and with Hsp60 antibody (lane 4). Covalent GT–GMP complex formation of GT was detected by incubation of reaction mixture with [α-32P]GTP at 30°C for 1 h and analyzed by 10% SDS/PAGE followed by autoradiography (lane 2). Anti-P eluate (4 μg) (lane 3) was subjected to SDS/PAGE and visualized by staining with Coomassie blue dye. Migration positions of the molecular mass markers are indicated.
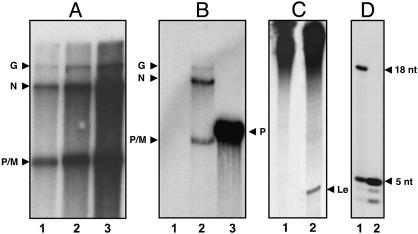
Properties of the Transcriptase Holoenzyme. The purified transcriptase efficiently synthesized mRNAs in vitro, and the yield of mRNA synthesis was proportional to the concentration of enzyme (Fig. 2A), indicating that the transcriptase is catalytically active. The L protein bound to host proteins after removal of the P protein by high salt, as expected, failed to transcribe (36) but was fully active when reconstituted with the P protein (Fig. 6). As demonstrated for recombinant L protein (23), the synthesized mRNAs by the transcriptase were fully capped (Fig. 2B) as shown (i) by the inability to label by [α-32P]GTP the 5′ ends of unlabeled mRNA by vaccinia GT (Fig. 2B, lane 1) and (ii) the ability to label the 5′ ends of mRNA after treatment by nucleotide pyrophosphatase, followed by phosphatase and labeling with polynucleotide kinase and [γ-32]ATP (lane 2). In a control experiment, uncapped P mRNA was fully capped by vaccinia virus GT (lane 3). Interestingly, analysis of the in vitro RNA products by 20% urea-PAGE revealed that the transcriptase did not synthesize the leader RNA (Fig. 2C, lane 1), whereas the RNP purified from the virus, as expected, clearly synthesized the leader RNA (Fig. 2C, lane 2). These results show that the transcriptase enters the genome RNA near the 3′ end but initiates RNA synthesis from the downstream genes. To confirm that the transcriptase indeed initiates at the N gene start site, we carried out RNA synthesis omitting UTP, which would synthesize a 32P-CMP-labeled pentanucleotide oligomer, AACAG of the 5′-terminal sequence of N-mRNA AACAG(U)AAUCA, but not the 18-nt leader RNA start sequence, [ACGAAGACAAACAAACCA(U)UA] if the initiation indeed occurred at that site (32). As shown in Fig. 2D, lane 2, a distinct labeled band was seen migrating similarly with the pentanucleotide synthesized by the virus (lane 1), whereas the purified virus synthesized both the 18-nt and the pentanucleotide (lane 1), as shown previously (37). Elution of the pentamer from the gel and its subsequent analysis, as described (37), confirmed the sequence as AACAG (data not shown). Two bands migrating faster than the pentamer (also seen in the RNP lane) represent AACA and AAC, the preterminated oligonucleotides. These results provided strong evidence that the transcriptase initiates exclusively from the N gene start site.
Fig. 2.
Characterization of transcriptase. (A) In vitro RNA synthesis was carried out by purified transcriptase fraction by using [α-32P]GTP as the labeled precursor as described in Materials and Methods. The RNA products were analyzed in 5% urea-PAGE. Lanes 1 (500 ng), 2 (1 μg), and 3 (2 μg). (B) Capping status of the transcriptase fraction from anti-P eluate was analyzed by capping the unlabeled RNA products with vaccinia GT and [α-32P]GTP (lane 1). The unlabeled RNA products were decapped with tobacco acid pyrophosphatase followed by dephosphorylation and end-labeling RNA products with [γ-32P]ATP and polynucleotide kinase (lane 2). As a control, the T7 transcript of VSV P was capped with vaccinia capping enzyme (lane 3). The transcripts were visualized by electrophoresis in 5% urea-PAGE followed by autoradiography. (C) Leader RNA synthesis. In vitro transcription reaction was carried out by using transcriptase fraction (500 ng) from anti-P eluate (lane 1) or RNP (300 μg) (lane 2), and the products were analyzed in 20% urea-PAGE. The position of the leader is marked as Le. (D) Oligonucleotide synthesis by the transcriptase in the absence of UTP. A standard in vitro transcription reaction was carried out in a 50-μl reaction containing 0.5 mM ATP, 150 μM CTP, 150 μM GTP, and 20 μCi of 6,000 Ci/mmol CTP, with virus (20 μg) (lane 1) or transcriptase (5 μg) (lane 2), and incubated at 30°C for 2 h. The reaction products were purified by phenol extraction followed by calf intestinal alkaline phosphatase treatment and analyzed in 20% urea-PAGE.
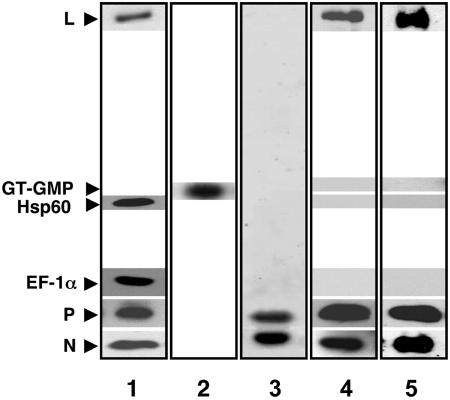
Purification of the Replicase. To purify the putative replicase, we then focused on the flow-through fraction of the anti-EF-1α column (Fig. 1, lane 2), which clearly removed the transcriptase but may contain the replicase. By Western blot analysis of the fraction (Fig. 3, lanes 1 and 2), it was apparent that a fraction of L, P, and all of the N protein was present in addition to GT, EF-1α, and Hsp60. It was then passed on an anti-P column, and the eluate was analyzed by staining with Coomassie blue (Fig. 3, lane 3) after PAGE. Only N and P proteins were discernible, indicating that the N–P complex constitutes the major proteins in the fraction. However, by Western blot analysis (Fig. 3, lane 4), the L protein was clearly seen, indicating that L is bound either to P or N-P and represents a minor component. Strikingly, the fraction did not contain any EF-1α, Hsp60, or GT (Fig. 3, lane 4). To eliminate the possibility that the complex recovered from the anti-P column may, in fact, be a mixture of L–P and N–P complexes, another aliquot of the anti-EF-1α flow-through fraction was passed through an anti-N column. As shown in Fig. 3, lane 5, the L protein was again quantitatively retained along with the N and P, because no L, P, or N proteins were detected in the flow-through fractions from both columns (not shown), indicating that L is complexed with N–P and not with L–P. Thus, the fraction contains two complexes, (i) L–(N–P), the putative replicase, and (ii) an N–P complex, which represents the bulk of the fraction.
Fig. 3.
Purification and characterization of replicase. Western blot analysis of anti-EF-1α flow-through fraction (40 μg) (lane 1), anti-P eluate fraction (500 ng) (lane 4), and anti-N eluate fraction (500 ng) (lane 5) was carried out by SDS/PAGE followed by immunoblot analysis with L, EF-1α, P, and N and Hsp60 specific antibodies. The bands were visualized by enhanced chemiluminescence. A GT–GMP assay (lane 2) was carried out (500 ng) as described in Fig. 1. Anti-P eluate (4 μg) was subjected to SDS/PAGE and visualized by staining with Coomassie blue dye (lane 3).
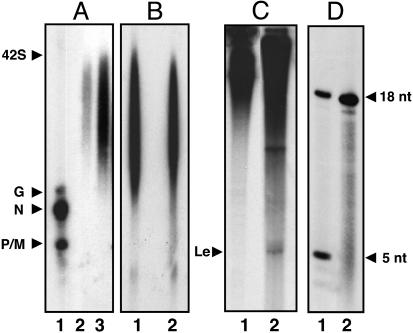
Properties of the Replicase. The putative replicase fraction was then tested for RNA synthesis in vitro by the addition of the N-RNA template. Strikingly, the RNA products larger than the G-mRNA and as large as genome-length RNA (42S) (Fig. 4A, lanes 2 and 3) were synthesized, indicating that the RNAs are the read-through replication products. Under the same electrophoresis condition, distinct mRNA bands, smaller than the read-through products, were synthesized by the purified transcriptase (Fig. 4A, lane 1). These results strongly suggested that the fraction contains replicase activity, which exclusively synthesizes read-through RNA products. Next, to determine whether the replication products are encapsidated during the reaction, we carried out RNase A digestion of the product RNAs. As shown in Fig. 4B, lane 2, RNA products isolated after treatment of RNase A, migrated similarly with the control RNAs (lane 1, no treatment), indicating that the read-through RNA products are indeed encapsidated, presumably by the N protein from the N–P complex, thus resistant to RNase A action. We then looked for the synthesis of leader RNA, if any, by analyzing the product RNAs in a 20% urea-PAGE. A distinct band, confirmed as leader RNA, migrating at the expected position, was discernible (Fig. 4C, lane 2), in contrast to purified transcriptase holoenzyme (Fig. 4C, lane 1). Finally, to confirm the start site of the replicase, we carried out the incomplete (–UTP) reaction as described for purified transcriptase (Fig. 4D). The replicase initiated exclusively a 32P-CMP-labeled 18-mer, confirmed as previously described (37), indicating that the initiation occurred at the 3′ end of the genome RNA (Fig. 4D, lane 2) (14). Thus, we have been able to purify two distinct complexes from VSV-infected cells, which carry out two separate and distinct RNA synthetic activities, i.e., transcription and replication.
Fig. 4.
Characterization of the replicase fraction. (A) Replication reaction was carried out in the presence of [α-32P]GTP as described in Materials and Methods by using transcriptase fraction (500 ng) (lane1) and replicase fraction [500 ng (lane 2) and 1 μg (lane 3)]. The products were analyzed by electrophoresis in 0.8% formaldehyde agarose gel. (B) Encapsidation of product RNAs. The replicase reaction products were treated with RNase A as detailed in Materials and Methods. The RNA products were analyzed by electrophoresis in 0.8% formaldehyde-agarose gel without (lane 1) and with RNase A (lane 2). (C) Leader RNA synthesis. An in vitro transcription reaction was carried out by using transcriptase fraction (500 ng) (lane 1) and replicase (5 μg) (lane 2), and the products were analyzed in 20% urea-PAGE. (D) Oligonucleotide synthesis by replicase in the absence of UTP. A standard in vitro transcription reaction was carried out in a 50-μl reaction containing 0.5 mM ATP, 150 μM CTP, 150 μM GTP, and 20 μCi of 6,000 Ci/mmol CTP with virus (20 μg) (lane 1) or replicase fraction (5 μg) (lane 2) and incubated at 30°C for 2 h. The reaction products were purified by phenol extraction followed by calf intestinal alkaline phosphatase. Reaction products were analyzed in 20% urea PAGE. Migration positions of 18 and 5 nucleotides are shown.
Discussion
One of the unique features of VSV and viruses of the mononegavirales order is that the genome RNA serves as template for both transcription (mRNA synthesis) and replication (genome RNA synthesis). Here, we show that these two RNA synthetic processes are carried out by two distinct complexes that contain the RNA polymerase L but differ in their content of other virally and host-encoded proteins, which we have purified from BHK-infected cells. We have previously shown that the recombinant L protein purified from insect cells (38) was tightly associated with cellular translation elongation factor EF-1α βγ and the mRNA capping enzyme (22, 23). However, the roles of these associated proteins were not apparent with regard to whether they are functional subunits or artifacts of insect cell protein expression. We thus sought to purify the transcriptase from VSV-infected mammalian cells, prepared 6 h after infection of BHK cells and purified by successive immunoaffinity chromatography using mammalian anti-EF-1α and anti-P antibody columns. The inclusion of the anti-EF-1α column in the purification step proved to be critical, because it allowed us to separate the transcriptase from the replicase, which was subsequently found to be devoid of EF-1α. This double immunoaffinity column chromatography resulted in the purification of the transcriptase to homogeneity and established the association of the mammalian homologue of EF-1α with the RNA polymerase L. Surprisingly, another major protein identified as cellular Hsp60, a molecular chaperone protein (39, 40), was found to be tightly bound to the transcriptase complex. The family of Hsps and molecular chaperones are abundant and ubiquitous and function constitutively in protein folding, protein translocation into membrane compartments, and the assembly and disassembly of oligomers (39, 40). The presence of a molar amount of Hsp60 in the transcriptase complex is unique for an RNA-dependent RNA polymerase, although the proteins of the heat shock family have been shown to be associated with the RNP as well as to stimulate RNA polymerase activity of several viruses of mononegavirales order (41–43), including VSV (New Jersey serotype) (42, 44) and influenza virus (45). We have also confirmed the association of Hsp60 in purified VSV (Indiana serotype) (Fig. 7, which is published as supporting information on the PNAS web site). The functional role, if any, of Hsp60, as well as EF-1α in L protein activity, will be of great interest for future work. Reproducibly, however, a submolar amount of GT remains tightly bound to the complex, suggesting that a small fraction of the complex associates with GT. If indeed GT has any role in the function of L protein, its requirement appears to be catalytic. The presence of 3 mol of P for each of L protein also confirms the previous finding obtained biochemically (46). It is important to stress that the observed molar ratio of the transcriptase complex, i.e., 1:1:3(±1):3(±1) with respect to L; Hsp60:EF-1α:P with a submolar amount of GT is an approximate value. The exact proportion of the polypeptides and whether the host proteins are indeed subunits will be established only on further biophysical analyses such as gel filtration, sedimentation analyses, and analytical centrifugation.
The transcriptase complex synthesized all mRNAs in vitro that are fully capped (Fig. 2 A and B) but strikingly failed to synthesize the leader RNA (Fig. 2C); and, after treatment with high salt (0.8 M NaCl), the L protein complex quantitatively retained the host proteins, but the P protein, as expected, was dissociated, indicating that the host proteins are indeed tightly bound to the L protein (Fig. 6). Finally omitting UTP from the reaction mixture, the transcriptase synthesized a pentanucleotide, AACAG, characteristic of the N gene start sequence but no 18 nt, representing the leader start sequence (Fig. 2D) (37). Thus, it became evident that the transcriptase complex is engaged specifically to synthesize mRNAs and initiates exclusively from the N gene start site.
We then purified the putative replicase fraction from the flow-through fraction obtained from the anti-EF-1α column by passing it through either an anti-P or -N column. The putative tripartite complex also contained an additional free N–P complex, because Coomassie blue staining of the gel detected only the N and P proteins (Fig. 3) but not the L protein. The latter can be seen only by Western blot analysis, similar to that observed for the transcriptase fraction (see Fig. 1, lanes 2 and 3), where EF-1α appeared as the major protein. Thus, further purification of the replicase fraction by affinity chromatography using an anti-L antibody or gel filtration would be needed to separate it from the N–P complex and to ascertain the molar ratio of the constituent polypeptides in the replicase complex. A striking finding is that the replicase fraction does not contain EF-1α, Hsp60, and GT, which differentiates it from the transcriptase complex, which is clearly associated with those host proteins. However, association of other cellular proteins with the replicase cannot be ruled out at the present time. The replicase fraction, when tested for RNA synthesis, produced read-through RNA transcripts of various lengths; also, the leader RNA and the replicated RNAs were encapsidated presumably by the N protein. Remarkably, the fraction synthesized only the 18 mer in a –UTP reaction, establishing that it initiates RNA synthesis precisely at the 3′ end of the genome RNA, suggesting that the promoter sites of replicase and transcriptase are separate, although they may overlap. Thus, we were able to purify a complex from the cytoplasmic supernatant fraction, which has all of the hallmarks of a replicase.
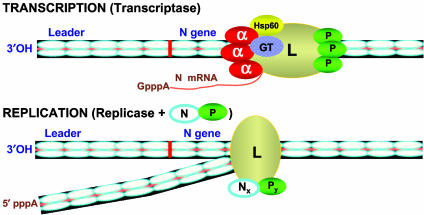
Based on our results, we propose that VSV RNA polymerase L exists in two distinct configurations in the infected cells (Fig. 5) that carry out two initiation events on the VSV genome RNA similar to that proposed previously (20). We envisage that when L protein is complexed with cellular EF-1α, Hsp60, and submolar quantity of GT, the resulting structure of the polymerase L allows it to interact with the phosphorylated P protein to form the transcriptase holoenzyme. The purified transcriptase initiates transcription precisely at the N gene start site, and sequentially synthesizes capped mRNAs (Fig. 5). At a specific time during infection, the L protein, either when free or while associated with host proteins, interacts with the N–P complex, resulting in the formation of a tripartite complex L–Nx–Py, the replicase of yet unknown composition. The replicase binds at a promoter site near the 3′ end of the genome RNA, which may overlap with the transcriptase promoter site and synthesizes the leader RNA in the absence of N–P complex. However, in the presence of the N–P complex, replication proceeds with the synthesis of read-through RNA concomitantly encapsidated with the N protein (Fig. 5). The antitermination at the leader-N gene junction as well as the downstream gene junctions is probably mediated by the N–P complex during its encapsidation of the leader RNA and interaction with the replicase.
Fig. 5.
A model for transcription and replication of VSV genome RNA.
The identification and characterization of the transcriptase and the replicase of VSV have led us to revisit the currently held “single initiation stop–start” mechanism of transcription of VSV genome RNA (14). This model was predicated on the experiment in which only two nucleotides, ATP and CTP, were used in the reconstitution reaction that initially formed the dinucleotide AC (leader sequence). Only after allowing the polymerase to copy the leader template, the AACA tetramer (N gene start sequence) was detected, consistent with single initiation at the 3′ end followed by sequential synthesis of downstream gene. Because there are a number of UG sequences present throughout the genome, the possibility exists that spurious AC initiations may have occurred at such sites obscuring the exact leader initiation site. Finally, our model postulates that virions also package the replicase in addition to the transcriptase. Because an extremely small quantity of the L (50 molecules) and P (500 molecules) per virion (7) is packaged within the purified virion, the purification may be difficult but is achievable and currently in progress. Further purification of the replicase fraction from the infected cells and elucidation of the role of host proteins in the transcriptase fraction will certainly lead to a better understanding of their structures and the mechanism of transcription and replication of VSV genome RNA.
Supplementary Material
Acknowledgments
We thank Philip Pellett for critically reading the manuscript and Santanu Bose for helpful discussion. We thank Dr. Michael Kinter, Director, Mass Spectrometry Core, Lerner Research Institute, Cleveland Clinic Foundation, for carrying out the tandem mass spectrometry. We thank Jacqueline A. Soos for secretarial assistance. This work was supported by a grant from the National Institutes of Health (AI26585, to A.K.B.).
Abbreviations: N, nucleocapsid; RNP, ribonucleoprotein; VSV, vesicular stomatitis virus; L, large; P, phosphoprotein; GT, guanylyltransferase; EF, elongation factor; BHK, baby hamster kidney; Hsp, heat-shock protein.
References
- 1.Baltimore, D., Huang, A. S. & Stampfer, M. (1970) Proc. Natl. Acad. Sci. USA 66, 572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyer, S. A. & Banerjee, A. K. (1975) Cell 4, 37–43. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, A. K. (1987) Micorbiol. Rev. 51, 66–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rose J. K. & Whitt, M. A. (2001) in Fields Virology, Knipe, D. M. & Howley, P. M., eds. (Lippincott Williams & Wilkins, Philadelphia), 4th Ed., pp. 1221–1244.
- 5.Blumberg, B. M., Leppert, M. & Kolakofsky, D. (1981) Cell 23, 837–845. [DOI] [PubMed] [Google Scholar]
- 6.Wertz, G. W., Davis, N. L. & Patton, J. (1987) in The Rhabdoviruses, Wagner, R. R., ed. (Plenum, New York.), pp 271–296.
- 7.Thomas, D., Newcomb, W. W., Brown, J. C., Wall, J. S., Hainfeld, J. F., Trus, B. L. & Steven, A. C. (1985) J. Virol. 54, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee, A. K. (1987b) Cell 48, 363–364. [DOI] [PubMed] [Google Scholar]
- 9.Colonno, R. J. & Banerjee, A. K. (1976) Cell 8, 197–204. [DOI] [PubMed] [Google Scholar]
- 10.Colonno, R. J. & Banerjee, A. K. (1977) Virology 77, 260–268. [DOI] [PubMed] [Google Scholar]
- 11.Abraham, G., Rhodes, D. P. & Banerjee, A. K. (1975) Cell 5, 51–58. [DOI] [PubMed] [Google Scholar]
- 12.Ball, L. A. & White, C. N. (1976) Proc. Natl. Acad, Sci. USA 73, 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham, G. & Banerjee, A. K. (1976) Virology 71, 230–241. [DOI] [PubMed] [Google Scholar]
- 14.Emerson, S. U. (1982) Cell 31, 635–642. [DOI] [PubMed] [Google Scholar]
- 15.Blumberg, B. M., Giorgi, C. & Kolakofsky, D. (1983) Cell 32, 559–567. [DOI] [PubMed] [Google Scholar]
- 16.Arnheiter, H., Davis, N. L., Wertz, G., Schubert, M. & Lazzarini, R. A. (1985) Cell 41, 259–267. [DOI] [PubMed] [Google Scholar]
- 17.Peluso, R. W. & Moyer, S. A. (1988) Virology 162, 369–376. [DOI] [PubMed] [Google Scholar]
- 18.La Ferla, F. M. & Peluso, R. W. (1989) J. Virol. 63, 3852–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonno, R. J. & Banerjee, A. K. (1978) Cell 15, 93–101. [DOI] [PubMed] [Google Scholar]
- 20.Chuang, J. L. & Perrault, J. (1997) J. Virol. 71, 1466–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whelan, S. P. & Wertz, G. W. (2002) Proc. Nat. Acad. Sci USA 99, 9178–9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das, T., Mathur, M., Gupta, A. K., Janssen, G. M. C. & Banerjee, A. K. (1998) Proc. Natl. Acad. Sci. USA 95, 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, A. K., Mathur, M. & Banerjee, A. K. (2002) Biochem. Biophys. Res. Commun. 293, 264–268. [DOI] [PubMed] [Google Scholar]
- 24.Pattnaik, A. K., Hwang, L., Li, T., Englund, N., Mathur, M, Das, T. & Banerjee, A. K. (1997) J. Virol. 71, 8167–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das, T., Pattnaik, A. K., Takacs, A. M., Li, T., Hwang, L. N. & Banerjee, A. K. (1997) Virology 238, 103–114. [DOI] [PubMed] [Google Scholar]
- 26.Pattnaik, A. K., Ball, L. A., LeGrone, A. W. & Wertz, G. W. (1992) Cell 69, 1011–1020. [DOI] [PubMed] [Google Scholar]
- 27.Gupta, A. K., Shaji, D. & Banerjee, A. K. (2003) J. Virol. 77, 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barik, S. & Banerjee, A. K. (1991) Virology 65, 1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow, E. & Lane, D. (1988) in Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 30.Masters, P. S. & Banerjee, A. K. (1986) Virology 154, 259–270. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 32.Towbin, H., Staehelin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das, T., Chakrabarti, B. K., Chattopadhyay, D. & Banerjee, A. K. (1999) Virology 259, 219–227. [DOI] [PubMed] [Google Scholar]
- 34.Kinter, M. & Sherman, N. E. (2000) in Protein Sequencing and Identification Using Tandem Mass Spectrometry (Wiley, New York), pp. 147–191.
- 35.Shuman, S. & Hurwitz, J. (1981) Proc. Natl. Acad. Sci. USA 78, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De, B. P. & Banerjee, A. K. (1985) Biochem. Biophys. Res. Commun. 126, 40–49. [DOI] [PubMed] [Google Scholar]
- 37.Chanda, P. K. & Banerjee, A. K. (1981) J. Virol. 39, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathur, M., Das, T. & Banerjee, A. K. (1996) J. Virol. 70, 2252–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morimoto, R. I., Tessieres, A. & Georgopoulos, C. (1994) in The Biology of Heat Shock Proteins and Molecular Chaperones (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 1–30.
- 40.Gething, M. J. & Sambrook, J. (1992) Nature 355, 33–95. [DOI] [PubMed] [Google Scholar]
- 41.Oglesbee, M. J., Liu, Z., Kenney, H. & Brooks, C. L. (1996) J. Gen. Virol. 77, 2125–2135. [DOI] [PubMed] [Google Scholar]
- 42.Sagara, J. & Kawai, A. (1992) Virology 190, 845–848. [DOI] [PubMed] [Google Scholar]
- 43.Parks, C. L., Lerch R. A., Walpita, P., Sidhu, M. & Udem, S. A. (1999) J. Virol. 73, 3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garry, R. F., Ulug, E. T. & Bose, H. R. (1983) Virology 129, 319–332. [DOI] [PubMed] [Google Scholar]
- 45.Momose, F., Naito, T., Yano, K., Sugimoto, S., Morikawa, Y. & Nagata, K. (2002) J. Biol. Chem., 277, 45306–45314. [DOI] [PubMed] [Google Scholar]
- 46.Gao, Y., Greenfield, N. J., Cleverley, D. Z. & Lenard, J. (1996) Biochemistry 35, 14569–14573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.