Abstract
Purpose
Tumor genotyping using cell free plasma DNA (cfDNA) has the potential to allow noninvasive assessment of tumor biology, yet many existing assays are cumbersome and vulnerable to false positive results. We sought to determine whether droplet digital PCR (ddPCR) of cfDNA would allow highly specific and quantitative assessment of tumor genotype.
Experimental Design
ddPCR assays for EGFR, KRAS, and BRAF mutations were developed using plasma collected from patients with advanced lung cancer or melanoma of a known tumor genotype. Sensitivity and specificity were determined using cancers with non-overlapping genotypes as positive and negative controls. Serial assessment of response and resistance was studied in EGFR-mutant lung cancer patients on a prospective trial of erlotinib.
Results
We identified a reference range for EGFR L858R and exon 19 deletions in specimens from KRAS-mutant lung cancer, allowing identification of candidate thresholds with high sensitivity and 100% specificity. Received operative characteristic (ROC) curve analysis of 4 assays demonstrated an area under the curve in the range of 0.80-0.94. Sensitivity improved in specimens with optimal cfDNA concentrations. Serial plasma genotyping of EGFR-mutant lung cancer on erlotinib demonstrated pretreatment detection of EGFR mutations, complete plasma response in most cases, and increasing levels of EGFR T790M emerging prior to objective progression.
Conclusions
Noninvasive genotyping of cfDNA using ddPCR demonstrates assay qualities that could allow effective translation into a clinical diagnostic. Serial quantification of plasma genotype allows noninvasive assessment of response and resistance, including detection of resistance mutations up to 16 weeks prior to radiographic progression.
Keywords: Cell-free DNA, droplet digital PCR, EGFR, lung cancer
Introduction
Tumor genotyping has proven to be an invaluable biomarker for identifying subsets of solid tumors with unique sensitivity to targeted therapies. Non-small cell lung cancer (NSCLC) harboring EGFR and ALK mutations and melanomas harboring BRAF mutations have been shown to be highly sensitive to targeted kinase inhibition (1-3). KRAS mutations have similarly been shown to have a negative predictive value in identifying cancers that will not respond to EGFR antibodies and EGFR kinase inhibitors (4, 5). With innumerable new genotypic biomarkers in development, the power of cancer genomics may become limited only by the availability of biopsy specimens for genotyping. Furthermore, the challenges of genotype-directed cancer care grow even greater when rebiopsy is needed to characterize and target specific resistance mechanisms.
Noninvasive techniques for tumor genotyping may be needed to fully realize the potential of genotype-directed cancer care. Early research suggested that circulating tumor cell (CTC) capture and analysis had potential as a noninvasive marker of tumor genotype (6), however clinical development of these technologies have been slow. Several studies have now suggested that highly sensitive genotyping assays can detect mutations in cell-free plasma DNA (cfDNA) from cancer patients, potentially reflecting the biology of a patient's cancer (7-10). Unfortunately, a challenge of highly sensitive genotyping assays is the detection of low prevalence mutant alleles of uncertain clinical significance. In a recent study, lung cancers positive for EGFR mutations only with a highly sensitive tumor genotyping assay did not demonstrate the expected durable benefit from EGFR kinase inhibitors, suggesting detection of false positives or mutations present in minor populations (11). The challenge of false positive results is even greater when studying plasma cfDNA: because cfDNA is mostly of germline origin from ruptured benign cells, tumor-derived mutations are inherently present at a low prevalence, lowering the signal-to-noise ratio of any detection assay.
Toward the goal of identifying an assay for noninvasive genotyping that has a high positive predictive value (PPV), is applicable to multiple genotype-defined solid tumors, and can be easily translated into clinical laboratories, we evaluated cfDNA genotyping using droplet digital PCR. By using a quantitative assay, we aimed to develop a biomarker both for accurate diagnosis of a targetable tumor genotype as well as for convenient monitoring of disease status.
Materials and Methods
Patient population
For our primary study population, we selected patients with advanced NSCLC undergoing routine tumor genotyping in our clinic. All patients consented to an IRB-approved protocol allowing collection and genomic analysis of blood specimens, limited to <50 mL of blood over any 3 month period. Patients were eligible for cfDNA analysis if they harbored a known EGFR or KRAS mutation in their NSCLC. Tumor genotyping of EGFR and KRAS was performed in a clinical, CLIA-approved laboratory. A second population of patients with advanced melanoma and a known BRAF genotype was also studied after consent to specimen collection on an IRB-approved protocol.
Plasma collection
For each eligible patient, plasma was collected during routine care either prior to first-line therapy or at a subsequent time when the cancer was progressing on therapy. Additional follow-up specimens were collected if possible during routine care. Each specimen was collected into one 10 mL EDTA-containing vacutainer and was spun into plasma within 4 hours of collection. Cell free DNA was extracted from 2 mL of plasma, and the final DNA eluent (~100 μL) was frozen at −80C until genotyping (Supplemental Materials and Methods). Mean isolated DNA mass per 1mL of plasma across all samples was 91.5 ng of DNA (interquartile range: 57-305 ng), quantified by PicoGreen as per manufacturer's recommendation.
Droplet Digital PCR
Droplet Digital PCR (ddPCR) is a digital PCR technology that takes advantage of recent developments in microfluidics and surfactant chemistries. Whereas conventional digital PCR involves a sometimes cumbersome process of diluting input DNA into individual wells for analysis (12, 13), ddPCR emulsifies input DNA into thousands of droplets that are PCR amplified and fluorescently labeled, and then read in an automated droplet flow cytometer (Figure 1) (14). Each droplet is individually assigned a positive or negative value based on the fluorescent intensity. The number of positive and negative droplets is read by a flow cytometer and is used to calculate the concentration of an allele. To minimize bias and to ensure the integrity of results, the laboratory was blinded to the tumor genotype when testing plasma specimens, but results were selectively unblinded for data analysis. A detailed protocol for each ddPCR assay is provided in the supplement. Each plasma sample was analyzed in triplicate with an increasing quantity of input DNA (e.g. 1 μL, 2 μL, and 4 μL) on a QX100 digital droplet reader (Supplemental Materials and Methods). Results were reported as copies of mutant allele per mL of plasma, as done by prior investigators (9, 10).
Figure 1.
Plasma genotyping using droplet digital PCR (ddPCR). Cell free DNA (cfDNA) is extracted from a plasma specimen and emulsified with oil into thousands of droplets, each containing approximately 0-1 molecules of target DNA. PCR is performed to endpoint in each droplet. These droplets are run through a flow cytometer, where droplets containing mutant and wildtype DNA emit different colored signals. The count of these signals allows quantification of allelic prevalence.
Results
Assay characteristics
We first developed two assays for EGFR L858R and exon 19 deletions; the latter assay was designed to detect loss of the wildtype signal and therefore could detect exon 19 deletions of variable sequence. Specifically, the assay is designed in such manner that a VIC-labeled “reference probe” sequence is shared by both the wildtype and the deletion mutants, while the FAM-labeled probe sequence spans the hotspots of the deletion and thus is only present in wildtype samples (13). An EGFR exon 19 wildtype sample will therefore show both FAM- and VIC-labeled droplets, while an EGFR exon 19 mutant will only have VIC-labeled droplets. To demonstrate the analytical sensitivity and specificity of each assay, each ddPCR cycling condition was optimized to yield the maximum fluorescent signal with minimal increase in background signal (Supplemental Figure 1). Using serial dilutions of mutant DNA, we found that ddPCR detects a mutation prevalence between 0.005% and 0.01% with a sensitivity of 5 to 50 mutant copies in a background of 10,000 wildtype copies (Supplemental Figure 2), depending on the mutation assayed. Experiments were repeated over three non-consecutive days. Both assays demonstrated linear quantification of allelic prevalence across a dynamic range spanning 4 orders of magnitude. From a technical standpoint, this suggests that ddPCR provides a reliable and quantitative measure of low prevalence EGFR mutant alleles within a plasma sample.
Maximizing positive predictive value
To optimize the specificity of our EGFR genotyping assays (and utility in guiding clinical decisions), we tested the incidence of false positive reads in a gold-standard negative population. To ensure selection of patients certain to be wildtype for EGFR, we studied patients with KRAS-mutant lung cancers. Large studies have found that EGFR and KRAS mutations are non-overlapping in NSCLC and represent distinct cancer populations (15, 16). Furthermore, KRAS-mutant lung cancers are recognized to be insensitive to treatment with EGFR kinase inhibitors (5, 17); while small subpopulations of cells within an individual KRAS-mutant lung cancer might hypothetically harbor mutations in EGFR, they evidently do not impact drug sensitivity for these cancers. Therefore any EGFR-mutant DNA found in the plasma of patients with KRAS-mutant NSCLC can be interpreted as biologically insignificant and representative of the “reference range” for our assay.
We first studied the EGFR L858R assay in 23 NSCLC patients, 12 with EGFR L858R and 11 with KRAS mutations in their cancers. Low levels of EGFR L858R were detected in 2 KRAS-mutant cases (18%) with a peak level of 0.9 copies/mL (Figure 2A). Using 1 copy/mL as our threshold for a positive result, 8 of 12 cases could be correctly identified as positive for EGFR L858R. We next studied the variable exon 19 deletion assay in a new cohort of 23 NSCLC patients, 9 with EGFR exon 19 deletions and 14 with KRAS mutations in their cancers. Low levels of EGFR exon 19 deletions were detected in 3 KRAS-mutant cases (21%) with a peak value of 5 copies/mL (Figure 2B). Using 6 copies/mL as our threshold for a positive result, 6 of 9 cases could be correctly identified as positive for EGFR exon 19 deletion. Lastly, we tested the reverse experiment in 31 NSCLC patients using a KRAS G12C assay that we developed as above. Of 17 patients with EGFR-mutant lung cancer, none had measurable mutant KRAS (Figure 2C). Using a threshold of 0.5 copies/mL, 10 of 14 KRAS G12C cases could be correctly identified as positive. For each assay, a receiver operating characteristic (ROC) curve was generated, with an area under the curve (AUC) in the range of 0.8-0.9 (Figure 2D-F).
Figure 2.
Detection of mutant alleles in gold standard positive and negative populations, using assays for EGFR L858R (A), EGFR exon 19 deletion (B), and KRAS G12C (C). Receiver operating characteristic (ROC) curves are also shown (D,E,F). By studying plasma from lung cancer patients with non-overlapping genotypes, a “reference range” for each assay can be identified. Dashed lines indicate one candidate threshold for positive with a very high specificity and acceptable sensitivity
To gauge the generalizability of this assay to other genotype-defined malignancies, we developed an assay for BRAF V600E in the fashion described above and tested plasma specimens from 13 melanoma patients. Using a threshold of 0.5 copies/mL for a positive result, 7 of 8 cases could be correctly identified as positive, and the ROC curve had a high AUC (Supplementary Figure 3), demonstrating potential value of ddPCR genotyping in a disease other than NSCLC.
Quality control to improve sensitivity
To better understand the false negative results, we measured human long interspersed element 1 (LINE-1) to assess the quantity and quality of cfDNA from the 32 EGFR- and KRAS-mutant lung cancer cases (true positives) studied in the above experiments. LINE-1 is an easily measured, genomically common retrotransposon that has been previously used to estimate total DNA in plasma (18)(Supplemental Figure 4). Median LINE-1 concentration was 186 ng/mL (interquartile range: 73-620 ng/mL) across the 32 specimens.
Detection of mutant alleles improved with increased levels of LINE-1 (Figure 3). Sensitivity was 81% in the 16 cases with LINE-1 levels higher than median, and 50% in the 16 cases with LINE-1 levels below median (p=0.07). However, three outlier cases with the highest levels of LINE-1 (greater than ~20,000 ng/mL) had no detectable levels of plasma genotype, likely indicating a high quantity of germline DNA obscuring detection of mutant cfDNA. These results suggest that LINE-1 levels may assist in identifying which plasma specimens are vulnerable to falsely negative genotyping result.
Figure 3.
Plasma DNA quantification to optimize sensitivity. Studying genotype concentration in gold standard positive cases, the false negative results all have either low or very high levels of LINE-1. Sensitivity is 81% above the median LINE-1 concentration of 168 ng/mL. Circles represents EGFR-mutant cases and squares represents KRAS-mutant cases.
Developing a disease monitoring biomarker
To assess the value of cfDNA genotype prevalence as a disease monitoring biomarker, we quantified the range of variability. Using the techniques described above, we generated a fifth genotyping assay to detect the EGFR T790M mutation. We generated human plasma DNA specimens that contained either 1,2,10, or 20 copies of EGFR T790M per 25 μL reaction, divided each into 32 individual specimens, and tested each of these for T790M prevalence by ddPCR. The assay exhibited a Poisson distribution between positives droplets and sample input with acceptable coefficient of variance in the range of 20-30% (Supplemental Figure 5), suggesting that changes exceeding this amount could represent a true change in tumor burden or biology.
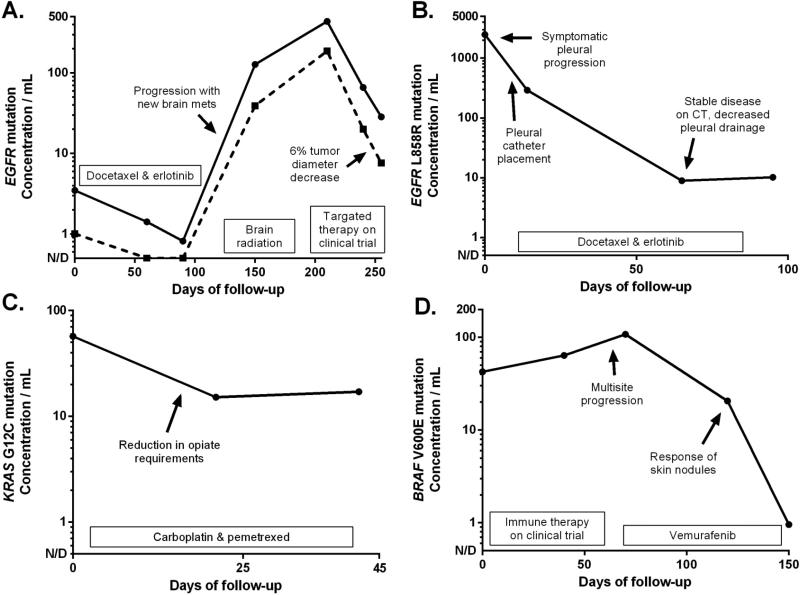
To gauge feasibility, we studied serial plasma specimens from patients with genotype-defined lung cancer or melanoma to determine whether changes in cfDNA were representative of tumor biology (Figure 4). In a patient with EGFR-mutant NSCLC receiving chemotherapy after failing erlotinib (Figure 4A), an increase in plasma L858R and T790M was seen with development of new brain metastases, followed by decreased plasma levels when treatment on a clinical trial was initiated. In a second case of EGFR-mutant NSCLC receiving chemotherapy (Figure 4B), plasma L858R decreased as the patient's pleural drainage resolved, though CT imaging of the non-measurable disease showed disease stability. In a patient with KRAS-mutant NSCLC and bone metastases (Figure 4C), chemotherapy caused a decrease in plasma G12C levels concordant with improved pain control and decreased opiate requirement. Lastly, a patient with BRAF-mutant melanoma had progression on experimental immune therapy followed by response to vemurafenib (Figure 4D), seen in the rise and fall of plasma V600E levels. This pilot experience suggests cfDNA genotyping has value for serial assessment of disease status, even in patients without objectively measurable disease on CT.
Figure 4.
Serial measurement of plasma genotype for disease monitoring. A wide dynamic range is seen in some cases (A, B). Decreases in plasma genotype can be seen both in cases of objective tumor shrinkage (A, D) and in cases of symptomatic response with no measurable disease (B, C). Concurrent EGFR L858R (A, solid line) and T790M (A, dashed line) mutations trend in parallel.
Monitoring for resistance mutations
To determine whether ddPCR could identify the development of resistance mutations after treatment with targeted therapy, we studied patients with advanced EGFR-mutant NSCLC treated on a prospective clinical trial of first-line erlotinib (NCT00997334), limiting our analysis to 13 patients that had serial plasma specimens collected until development of objective progression per the Response Evaluation Criteria In Solid Tumors (RECIST). In each of these patients, genotyping of archived tissue at diagnosis identified an EGFR exon 19 deletion without evidence of T790M. Four patients had no detectable pretreatment plasma genotype and were excluded, leaving 9 cases (69%) for analysis.
All 9 patients exhibited a plasma response to erlotinib, with 8 demonstrating a complete plasma response (Figure 5). In 6 of the patients, plasma levels of mutant EGFR were again detected at objective progression, with plasma progression detected 4-12 weeks prior to RECIST progression. In each of these patients, plasma T790M could also be identified at progression, generally at somewhat lower levels than the EGFR sensitizing mutation. Four of these patients had a tumor rebiopsy adequate for EGFR genotyping, and T790M was confirmed in each. The remaining 3 patients had no reemergence of plasma genotype at objective progression; notably, each of these patients had indolent asymptomatic progression in the chest only, such that they subsequently continued single-agent erlotinib off-protocol.
Figure 5.
Plasma levels of mutant EGFR in 9 patients receiving first-line erlotinib until objective disease progression (PD) by RECIST. In all patients, plasma levels of the EGFR sensitizing mutation (solid line) drop in response to treatment, with 8 patients (B-I) having a complete plasma response. In 6 patients, plasma genotype levels reemerge up to 4 months prior to PD, and a lower concentration of T790M (dashed line) is also detected. In 3 patients (G-I), plasma genotype was not detected at time of PD; all 3 had indolent progression in the chest only.
Discussion
We herein describe a new quantitative assay for plasma-based tumor genotyping which has been technically optimized for translation into clinical practice. By quantifying the prevalence of targetable genotypes in clinical plasma specimens, and through study of rigorous gold-standard negative cases harboring non-overlapping cancer genotypes, we have identified a reference range for EGFR and KRAS mutation detection using ddPCR. Using such a calculated threshold as the criteria for a positive results, as well as LINE-1 concentration to eliminate poor quality specimens, our data suggests this assay can have high sensitivity and specificity. These proposed thresholds require prospective validation.
Because many targetable genotypes are relatively uncommon, we have focused our assay development on maximizing specificity. Consider, for example, a plasma assay for detecting EGFR sensitizing mutations, present in 8.6% of 10,000 NSCLC patients from the large French experience (19). In this population, a plasma assay for EGFR mutations having with 80% sensitivity and 90% or 95% specificity would have a PPV of only 43% or 60%, respectively. For this reason, a clinical-grade assay will likely need to sacrifice sensitivity in order to optimize specificity. In the same population, an assay with 70% sensitivity and 99% or 100% specificity would result in a PPV of 87% or 100%, respectively. Furthermore, the need to maximize specificity is magnified when testing for rarer genotypes such as BRAF V600E in NSCLC, representing only 2% of patients (20). One valuable characteristic of a quantitative assay such as ddPCR is the flexibility to allow an alteration of the criterion for positive if the pretest probability changes (e.g. Asian lung cancer patients). This is in contrast to an allele-specific PCR assay, such as one which showed high concordance with tumor genotyping in a preliminary analysis of plasma from 241 Asian lung cancer patients (21); as such an assay is qualitative, it cannot easily be adjusted to a higher specificity criterion in populations with lower mutation prevalence.
This study is one of several that have investigated plasma genotyping as a way of noninvasively detecting the EGFR T790M resistance mutation in lung cancer patients treated with EGFR kinase inhibitors.(13, 22-24) Yet this is the first study to demonstrate, across multiple patients, that serial assessment of plasma genotype allows detection of resistance weeks (and sometimes months) prior to clinical development of resistance. Early detection of resistance has particular importance given the growing role of EGFR T790M as a biomarker for patients with EGFR-mutant lung cancer and acquired resistance. Firstly, acquired T790M has been associated with indolent growth and a favorable prognosis compared to T790M-negative acquired resistance (25). Secondly, third-generation EGFR kinase inhibitors with T790M-specific activity have recently been shown to have activity in patients with T790M-mediated acquired resistance (26-28). While pharmaceutical development of T790M-directed targeted therapies could be limited by the challenges of performing a repeat biopsy after resistance develops (29), our data suggests that early emergence of EGFR T790M can be identified noninvasively using ddPCR, and potentially used to guide subsequent treatment.
The quantitative nature of plasma genotyping with ddPCR also offers a mechanism for monitoring the prevalence of tumor clones harboring a specific genotype, potentially giving insight into the pharmacodynamics of a targeted therapy. In liquid malignancies like chronic myelogenous leukemia, rapidity of molecular response to kinase inhibitors has been established as an important biomarker of prognosis, and helps indicate which patients may need early salvage therapy (30). Diehl et al, also found that cfDNA levels have the potential to be used in colorectal cancer, much like CEA levels, to distinguish successful versus unsuccessful surgical resection.(31) Similarly, serial assessment of a plasma genotype may prove to be a valuable biomarker for genotype-defined solid tumors treated with targeted therapies, both as a clinical biomarker of favorable outcome and potentially as an early clinical trial endpoint. Indeed, this was even suggested in our small series – the one patient not exhibiting a complete plasma response to erlotinib had early progression – and will need to be studied in larger cohorts. In addition, response assessment using plasma genotype quantification could potentially allow trial accrual for those patients with genotype-defined solid tumors that are not objectively measurable using conventional response criteria.
While there is currently no standard unit for the reporting of plasma genotyping results, we have reported our results using copies per mL of plasma, as reported previously in the literature.(9, 10) Other studies have presented plasma genotyping results as the percent of reactions that are mutant.(7, 13) However, we worry that this relative concentration may be less precise, particularly at low concentrations – while 2 mutant copies / 2000 wildtype copies and 20 mutant copies / 20000 wildtype copies both can be calculated as 0.1% mutant, they are not equal, and the former is more likely to be a false positive. To facilitate comparisons, we have also provided our data recalculated using this alternate unit (see Data Supplement). As this is a dynamically changing field, we encourage other investigators to consider that there may also be other more precise strategies the presentation of plasma genotyping results.
In conclusion, we herein present a proof of concept demonstrating the clinical utility of cfDNA genotyping for detecting and monitoring EGFR sensitizing and drug resistance mutations for patients with non-small cell lung cancer. Droplet digital PCR is an attractive technology as its speed, cost, and ease of use is similar to other PCR-based assays, yet the sensitivity and quantitative nature of this assay offers broader clinical application. Prospective validation based upon this initial experience is needed, and is underway.
Supplementary Material
Statement of Translational Relevance.
A major limitation in the advancement of genotype-directed therapy in solid tumors is the challenge of tumor re-biopsy to characterize resistance to targeted therapies. This challenge is particularly important for EGFR-mutant lung cancer, where the T790M resistance mutation is a target of active pharmaceutical development. Here we demonstrate detection and monitoring of EGFR sensitizing and drug resistance mutations in cell-free plasma DNA from patients with EGFR-mutant lung cancer. In patients receiving first-line erlotinib, T790M-mediated acquired resistance could be detected up to 16 weeks prior to radiographic progression; in one patient, response of plasma EGFR T790M was seen with treatment on a subsequent clinical trial. These data suggests that noninvasive genotyping of cell-free plasma DNA has potential as a clinical biomarker for personalizing therapy of genotype-defined solid tumors.
Acknowledgments
Funding support:
This work was supported in part by the National Cancer Institute at the National Institutes of Health (R01-CA135257, P50-CA090578), by the Conquer Cancer Foundation of ASCO (Career Development Award), and by research funding from Genentech.
Footnotes
Disclosures:
GRO is a consultant/advisory board member for Astellas, Astra-Zeneca, AVEO, Boehringer Ingelheim, Genentech, and Novartis; and has received honoraria from Astra-Zeneca, Boehringer Ingelheim and Chugai. DMJ is a consultant/advisory board member for Foundation Medicine and Genentech; and has received honoraria from Chugai. PAJ is a consultant/advisory board member for Abbot, Astra-Zeneca, Boehringer Ingelheim, Chugai, Clovis, Genentech, Pfizer, and Sanofi. GRO, CPP, YK, and PAJ are inventors on a pending patent related to findings described in this manuscript. P.A. Jänne is a co-inventor on a patent held by the Dana-Farber Cancer Institute for the use of EGFR genotyping, and receives a share of post-market licensing revenue distributed by DFCI.
References
- 1.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic Lymphoma Kinase Inhibition in Non–Small-Cell Lung Cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 5.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater SA, Powers P, et al. Detection of tumor PIK3CA status in Metastatic Breast Cancer using Peripheral Blood. Clin Cancer Res. 2012;18:3462–9. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Zhang X-C, Chen Z-H, Yin X-L, Yang J-J, Xu C-R, et al. Relative Abundance of EGFR Mutations Predicts Benefit From Gefitinib Treatment for Advanced Non–Small-Cell Lung Cancer. J Clin Oncol. 2011;29:3316–21. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 12.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yung TKF, Chan KCA, Mok TSK, Tong J, To K-F, Lo YMD. Single-Molecule Detection of Epidermal Growth Factor Receptor Mutations in Plasma by Microfluidics Digital PCR in Non–Small Cell Lung Cancer Patients. Clin Cancer Res. 2009;15:2076–84. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 14.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardarella S, Ortiz TM, Joshi VA, Butaney M, Jackman DM, Kwiatkowski DJ, et al. The Introduction of Systematic Genomic Testing for Patients with Non–Small-Cell Lung Cancer. J Thorac Oncol. 2012;7:1767–74. doi: 10.1097/JTO.0b013e3182745bcb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119:356–62. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–70. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 19.Barlesi F, Blons H, Beau-Faller M, Rouquette I, Ouafik Lh, Mosser J, et al. Biomarkers (BM) France: Results of routine EGFR, HER2, KRAS, BRAF, PI3KCA mutations detection and EML4-ALK gene fusion assessment on the first 10,000 non-small cell lung cancer (NSCLC) patients (pts). ASCO Meeting Abstracts. 2013;31:8000. [Google Scholar]
- 20.Cardarella S, Ogino A, Nishino M, Butaney M, Shen J, Lydon C, et al. Clinical, Pathologic, and Biologic Features Associated with BRAF Mutations in Non–Small Cell Lung Cancer. Clin Cancer Res. 2013;19:4532–40. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok T, Wu YL, Lee JS, Yu C-J, Sriuranpong V, Wen W, et al. Detection of EGFR-activating mutations from plasma DNA as a potent predictor of survival outcomes in FASTACT 2: A randomized phase III study on intercalated combination of erlotinib (E) and chemotherapy (C). ASCO Meeting Abstracts. 2013;31:8021. [Google Scholar]
- 22.Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Sueoka-Aragane N, Iwanaga K, Sato A, Komiya K, Kobayashi N, et al. Application of a Highly Sensitive Detection System for Epidermal Growth Factor Receptor Mutations in Plasma DNA. J Thorac Oncol. 2012;7:1369–81. doi: 10.1097/JTO.0b013e31825f2821. 10.097/JTO.0b013e31825f2821. [DOI] [PubMed] [Google Scholar]
- 24.Sakai K, Horiike A, Irwin DL, Kudo K, Fujita Y, Tanimoto A, et al. Detection of epidermal growth factor receptor T790M mutation in plasma DNA from patients refractory to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer science. 2013;104:1198–204. doi: 10.1111/cas.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–22. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sequist LV, Soria J-C, Gadgeel SM, Wakelee HA, Camidge DR, Varga A, et al. First-in-human evaluation of CO-1686, an irreversible, selective, and potent tyrosine kinase inhibitor of EGFR T790M. ASCO Meeting Abstracts. 2013;31:2524. [Google Scholar]
- 28.Ranson M, Pao W, Kim D-W, Kim S-W, Ohe Y, Felip E, et al. AZD9291: an irreversible, potent and selective tyrosine kinase inhibitor (TKI) of activating (EGFRm+) and resistance (T790M) mutations in advanced NSCLC. J Thorac Oncol. 2013;8:MO21.12. [Google Scholar]
- 29.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski M, et al. Rebiopsy of Lung Cancer Patients with Acquired Resistance to EGFR Inhibitors and Enhanced Detection of the T790M Mutation Using a Locked Nucleic Acid-Based Assay. Clin Cancer Res. 2011;17:1169–80. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branford S, Kim DW, Soverini S, Haque A, Shou Y, Woodman RC, et al. Initial molecular response at 3 months may predict both response and event-free survival at 24 months in imatinib-resistant or -intolerant patients with Philadelphia chromosome-positive chronic myeloid leukemia in chronic phase treated with nilotinib. J Clin Oncol. 2012;30:4323–9. doi: 10.1200/JCO.2011.40.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.