Abstract
Cyclin-dependent kinase 5 (Cdk5) is essential for the proper development of the CNS, as is evident from the perinatal lethality of conventional Cdk5 knockout (Cdk5-/-) mice. Cdk5 is also implicated in numerous complex functions of the adult CNS such as synaptic transmission, synaptic plasticity, and neuronal signaling. To elucidate the molecular roles of Cdk5 in the adult CNS, we have abrogated neuronal expression of Cdk5 in perinatal mice by using a cre-loxP system. The Cdk5-loxP flanked mice were crossed with the cre-transgenic mice in which the cre expression is driven by the murine neurofilament-heavy chain promoter, resulting in generation of viable Cdk5 conditional knockout mice with the restricted deletion of the Cdk5 gene in specific neurons beginning around embryonic day 16.5. Twenty-five percent of the Cdk5 conditional knockout mice carrying the heterozygous cre allele had neuronal migration defects confined to brain areas where neuronal migration continues through the perinatal period. These results indicate that abrogation of Cdk5 expression in mature neurons results in a viable mouse model that offers further opportunities to investigate the molecular roles of Cdk5 in the adult CNS.
Cyclin-dependent kinase 5 (Cdk5) is a small serine/threonine kinase belonging to the cdk family of proline-directed kinases. Unlike other cdks, Cdk5 activity is detected mainly in postmitotic neurons (1). Association of Cdk5 with its neuron-specific regulatory subunit, either p35 or its isoform p39, is critical for its kinase activity (2–4). We have earlier reported in vivo roles of Cdk5 with conventional Cdk5-/- mice (5, 6). These mice exhibit embryonic lethality and disruption of cortical laminar structures due to defective neuronal migration (5, 6). Chromatolytic changes such as a ballooned cell soma with eccentric nuclei were also observed in the neurons of the Cdk5-/- mice (5). Perikaryal accumulation of phosphorylated murine neurofilament-heavy chain (pNFH) was seen in the cell soma of the motor neurons in the brainstem and spinal cord (5). Because of the embryonic lethality of Cdk5-/- mice, it was not possible to carry out further analysis of this neuronal pathology in the adult CNS.
Cdk5 is believed to phosphorylate numerous substrates in neurons and thereby regulate many cellular processes of the mature CNS such as phosphorylation of neuronal cytoskeletons (7–10), synaptic transmission (11, 12), and dopaminergic signaling (13). To determine the role of Cdk5 in the adult CNS, we generated a conditional knockout (KO) mouse by using a cre-loxP system in which the Cdk5 gene was disrupted in the CNS in a temporally and spatially regulated manner. To abrogate Cdk5 expression, we generated transgenic mice in which the Cdk5 gene was flanked by loxP motifs, and then crossed them with the previously described heterozygous murine (m) NFHcre mouse line 12, in which the cre recombinase is expressed only in certain neurons beginning around embryonic day 16.5 (E16.5) (14). Unlike Cdk5-/- mice, the Cdk5 conditional KO mice are viable and fertile. Abrogated Cdk5 expression in these mice was associated with neuronal migration defects in certain brain areas: in cerebral cortex where the defects were restricted to the later-generated cortical neurons and in the olfactory bulb and cerebellar cortex where neuronal migration continues through the perinatal period.
Materials and Methods
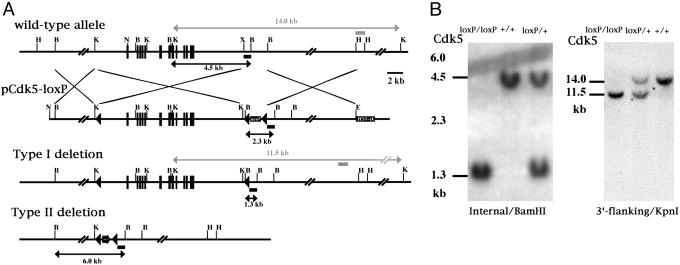
Generation of Cdk5-loxP Mice. To engineer the targeting vector for the Cdk5-loxP locus, three loxP motifs were introduced into a 18-kb fragment of the Cdk5 gene containing all of the exons (15). This targeting construct (pCdk5-loxP) also contained a neomycin-resistance gene flanked by loxP motifs 2.5 kb downstream of the last Cdk5 exon (Fig. 1A). Another loxP motif was located at 3.5 kb upstream from the Cdk5 translation starting site (Fig. 1 A). A detailed procedure for generating this construct will be provided on request. The pCdk5-loxP construct was linearized by NotI digestion and electroporated into R1 embryonic stem (ES) cells. The cells were subjected to drug selection as described (5). The homologous recombinant clones were identified by Southern blot analysis with the 3′-flanking probe (5). Three clones were selected for cre transient expression studies: 1 × 107 cells were transiently transfected with 30 μg of pIC-Cre plasmid (a gift from H. Gu, National Institutes of Health). ES cells were then expanded without drug selection and the colonies were picked, expanded, and stored at -130°C. We identified clones that had undergone a deletion of the neomycin resistance cassette from the targeted allele by the negative selection with G418 and confirmed them by Southern blot analysis. Two independent clones were injected into blastocysts for generation of the chimeric mice. The chimeric mice were bred with C57BL/6 mice to generate mice heterozygous for the floxed Cdk5 allele (flanked by two loxP motifs) termed fCdk5, which were further intercrossed to obtain mice homozygous for the fCdk5 (fCdk5/fCdk5).
Fig. 1.
Gene targeting and conditional deletion of the Cdk5 gene. (A) Restriction map of the wild-type allele, targeting vector, and strategy for cre-mediated deletion of the Cdk5 gene. Internal and 3′-flanking probes were used to confirm recombination events. Solid boxes indicate exons. PGK neomycin resistance gene (Neor) and thymidine kinase (TK) are positive and negative selection cassettes, respectively. Solid triangles represent loxP motifs. Restriction sites are as follows: B, BamHI; H, HindIII; K, KpnI; N, NotI; and X, XhoI. (B) Southern blot analysis of F1 pups derived from intercross of mice heterozygous for the floxed Cdk5 allele. (Left) Tail DNA digested with BamHI was hybridized with the internal probe: 4.5- and 1.3-kb bands represent the wild-type and floxed allele, respectively. (Right) Tail DNA digested with KpnI was hybridized with the 3′-flanking probe: 14.0- and 11.5-kb bands represent the wild-type and floxed allele, respectively.
Generation of Cdk5 Conditional KO Mice. fCdk5/fCdk5 mice were crossed with mNFHcre mouse line 12 (14) to generate mice heterozygous for both fCdk5 and mNFHcre (fCdk5/+; NFH-cre/+). The Cdk5 conditional KO mice (fCdk5/fCdk5; NFH-cre/+) were generated from crosses between mice heterozygous for both fCdk5 and mNFHcre and mice homozygous for the fCdk5 allele. This breeding strategy resulted in the generation of the conditional Cdk5 KO mice as well as fCdk5/fCdk5, fCdk5/+ and fCdk5/+; NFHcre/+ mice. All of these mice had a mixed genetic strain, C57BL/6, 129SV/J, and FVB/N. Mice were housed under a 12-h light/dark cycle. All care was given in compliance with National Institutes of Health guidelines on the use of laboratory and experimental animals.
Northern Blot and RT-PCR Analysis. Total RNA was isolated from brain, heart, lung, liver, kidney, spleen, and skeletal muscle of the Cdk5 conditional KO mice and the controls (fCdk5/fCdk5) by using an RNeasy mini kit (Qiagen, Valencia, CA). To determine the Cdk5 expression level in the brain, 20 μg of total RNA was used for Northern blot analysis as described (5). For RT-PCR, first-strand cDNA was synthesized with an oligo(dT)-adaptor primer by using an RNA PCR Ver2.1 kit (Takara Shuzo, Kyoto). To examine cre expression, 100 ng of cDNA from each tissue was denatured at 95°C for 15 min and then amplified for 35 cycles (30 sec at 94°C, 30 sec at 56°C, and 90 sec at 72°C) by using HotStar Taq DNA polymerase (Qiagen). The primer set corresponded to the following: cre-specific primer, CR5 5′-TGCCAGGATCAGGGTTAAAG-3′, and the adaptor primer attached to the PCR kit. GAPDH cDNA was amplified as described (16).
Histological Analysis. Animals were anesthetized with an i.p. injection of avertin (250 mg/kg of body weight, Fluka) and perfused transcardially with ice-cold 4% paraformaldehyde in PBS (pH 7.4). Brains were serially cut into 5- to 7-μm-thick paraffin sections or 15- to 20-μm-thick frozen sections. The sections were incubated overnight at 4°C with a primary antibody and processed with a Vectastain elite ABC kit or a Vector M.O.M. immunodetection kit (Vector Laboratories). Primary antibodies used in this study were as follows: polyclonal rabbit anti-Cdk5 antibody (C-8, 1:200, Santa Cruz Biotechnology), monoclonal anti-neuronal nuclei (NeuN) antibody (1:500 dilution, Chemicon), and monoclonal anti-calbindin-D-28K (1:2,000 dilution, Sigma). For immunofluorescence, mouse or rabbit primary antibodies were visualized with fluorescein- or Cy3-conjugated secondary antibodies (1:200 dilution, Jackson ImmunoResearch), respectively. All sections were examined by standard light and fluorescent microscopic techniques.
Results
Generation of fCdk5/fCdk5 and Cdk5 Conditional KO Mice. A Cdk5 targeting vector containing three loxP motifs in the same orientation was engineered (Fig. 1A). The first loxP motif was inserted at the XhoI site located 2 kb upstream of the first Cdk5 exon. This XhoI site was a few base pairs downstream of the KpnI site shown in Fig. 1A. A 2-kb fragment containing the neomycin-resistance gene flanked by two loxP motifs was inserted into the XhoI site located 3 kb downstream of the last exon of the Cdk5 gene. Finally, a thymidine kinase gene cassette was inserted into a HindIII site located 9 kb downstream of the last exon of the Cdk5 gene. After electroporation into the R1 ES cells and the subsequent selection of the ES cell colonies with G418 and ganciclovir, we obtained 18 homologous recombinants of 501 clones that were identified by Southern blot analysis. Three of the targeted ES cell clones were used for deletion of the neomycin cassette by a transient cre expression. ES cells from the selected targeted clone were electroporated with pIC-Cre plasmid. Cells were cultured to obtain robust colonies, and their genomic DNA was analyzed by Southern blot analysis to identify the types of cre-mediated recombination that had occurred at the three loxP motifs. The type-I and type-II cre-mediated recombinations resulted in deletion of the neomycin resistance gene cassette and the entire Cdk5 gene flanked by loxP motifs, respectively (Fig. 1A). The resultant clones with the type I deletion (targeted allele) were further confirmed by their sensitivity to G418. Two clones with the type I deletion were injected into C57BL/6 blastocysts to generate the chimera. Overt chimeras were bred with C57BL/6 females to generate mice heterozygous for the fCdk5 allele (fCdk5/+), which were further intercrossed to obtain fCdk5/fCdk5 mice and confirmed by Southern blot analysis (Fig. 1B). The fCdk5/fCdk5 mice were viable and fertile, and did not display any obvious phenotype. In mNFHcre transgenic mouse line 12, the cre recombinase was expressed only in neurons beginning around E16.5 (14). The fCdk5/fCdk5 mice were further crossed with fCdk5/+; NFHcre/+ mice, resulting in the generation of Cdk5 conditional KO (fCdk5/fCdk5; NFHcre/+) mice carrying only the heterozygous cre allele.
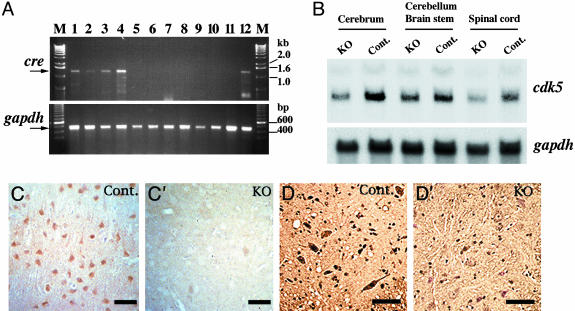
Postnatal Inactivation of the Cdk5 Gene in the CNS. The Cdk5 conditional KO mice were born normal and remained viable and fertile. They did not show any significant difference in body weight and in life span compared with the controls (data not shown). To determine the tissue specificity of cre expression, we analyzed the cre mRNA levels in the various tissues of the Cdk5 conditional KO mice by RT-PCR (Fig. 2A). This analysis revealed that cre expression was restricted to the CNS, confirming our earlier observation of the tissue-specific expression in the mNFHcre transgenic mouse line 12 (14). The highest level of cre expression was detected in the spinal cord of the Cdk5 conditional KO mice (Fig. 2 A). Northern blot analysis revealed 45–50% reduction of Cdk5 mRNA in the cerebrum and spinal cord of Cdk5 conditional KO mice compared with the controls (Fig. 2B). However, the reduction in Cdk5 mRNA levels was barely detected in the brainstem and cerebellum of Cdk5 conditional KO mice (Fig. 2B). Immunohistochemical analysis of the cerebral cortex and spinal cord revealed that most of the neurons lost Cdk5 immunoreactivity, whereas it was retained in glial cells (Fig. 2 C′ and D′).
Fig. 2.
Neuronal inactivation of the Cdk5 gene in Cdk5 conditional KO mice. (A) Expression levels of cre mRNA in the various tissues of Cdk5 conditional KO mice (lanes 1–10) and the controls (lanes 11 and 12) were analyzed by RT-PCR (Upper). The arrow indicates the amplified 1.2-kb fragment from cre mRNA. M, 1-kb ladder marker; lane 1, cerebrum; lane 2, cerebellum; lane 3, brainstem; lane 4, spinal cord; lane 5, heart; lane 6, skeletal muscle; lane 7, liver; lane 8, spleen; lane 9, kidney; lane 10, lung; lane 11, cerebrum from wild-type mouse; lane 12, cerebrum from mNFHcre mouse line 12. gapdh transcripts served as the internal controls and were seen in all of the tissues analyzed (Lower). (B) Northern blot analysis of brain RNA from the Cdk5 conditional KO mice and the controls (Cont.) at the age of 1 month. When the Cdk5 mRNA levels were normalized to the levels of gapdh mRNA, they were found to be reduced to 45–50% in the cerebrum and spinal cord of the Cdk5 conditional KO mice as compared to those of the controls. (C and D) Cdk5 protein levels were analyzed by immunohistochemistry with Cdk5 antibody (C-8) in the cerebral cortex (C and C′) and spinal cord (D and D′) of the Cdk5 conditional KO mice (C′ and D′) and the controls (C and D) at the age of 6 months. Note that Cdk5 immunoreactivities in the Cdk5 conditional KO mice were lost in the neurons but not in the glial cells.
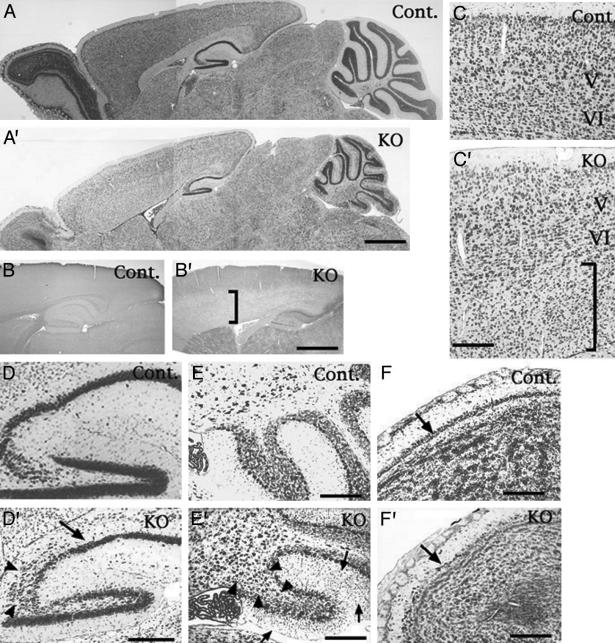
Neuronal Migration Defects in Cdk5 Conditional KO Mice. Because Cdk5-/- mice displayed extensive neuronal migration defects, we carried out a detailed histological analysis of the Cdk5 conditional KO mouse brain at 1 month of age. In the cerebral cortex of 25% (8 of 32) of Cdk5 conditional KO mice, we observed migration defects restricted to the later-generated cortical neurons (Fig. 3C′), whereas earlier-generated neurons in layer V and VI appeared to form normal laminar structure similar to the controls (Fig. 3C). Immunostaining for Cdk5 in the adjacent sections from Cdk5 conditional KO mice revealed a correlation between loss of Cdk5 and migration defects of later-generated cortical neurons in the cerebral cortex (Fig. 3 B′ and C′). In these mice, CA1 pyramidal neurons in the hippocampus appeared unaffected; however, a distinct disorganization of the CA3 region was observed (Fig. 3D′). In the cerebellum, many of the granule cells were arrested in the molecular layer (Fig. 3E′). In some cases, we also observed migration defects in the Purkinje cells in the cerebellum (Fig. 3E′). The mitral cell layer of the olfactory bulb was thinner and disorganized in the Cdk5 conditional KO mice (Fig. 3F′) compared to the controls (Fig. 3F). Additionally, the density of granule cells in the olfactory bulb appeared lower in the mutant mice than in the controls (Fig. 3 A, A′, F, and F′).
Fig. 3.
Neuronal migration defects in Cdk5 conditional KO mice. (A and A′) Nissl staining of parasagittal sections from 1-month-old control (Cont., fCdk5/fCdk5) and Cdk5 conditional KO mice (fCdk5/fCdk5; NFHcre/+). (B, B′, C, and C′) Immunostaining for Cdk5 (B and B′) and Nissl staining (C and C′) in the adjacent sections indicated the correlation between neuronal migration defects of cerebral cortical neurons (indicated by vertical black bar in C′) and loss of Cdk5-immunoreactivities (indicated by vertical black bar in B′). V and VI indicate layers V and VI in the cerebral cortex. D and D′ are higher-magnification images of A and A′, respectively, at hippocampal regions. Defective migration of hippocampal pyramidal neurons in the CA3 region (arrowhead) was observed in Cdk5 conditional KO mice. The arrow indicates the CA1 region where the pyramidal cell layer formed normally in the KO mice (D′). (E and E′) The migration defects (indicated by arrow in E′) of the granule cells from external granule cell layer to internal granule cells layer in the cerebellum. E and E′ were magnified from A and A′, respectively. Migration arrests of Purkinje cells (arrowheads in E′) were observed in 10% of the Cdk5 conditional KO mice. (F and F′) Nissl-stained sagittal sections of olfactory bulb from control (F) and Cdk5 conditional KO (F′) mice. Mitral cell layer in the KO mice (F′) appeared thinner than that in control mice (F). Arrows indicate the mitral cell layer. In addition, cells of the granule cell layer in the olfactory bulb seemed to be lower in Cdk5 conditional KO mice (A′ and F′) compared to the controls (A and F). (Scale bars: 1,000 μm in A′ and B′; 200 μm in C′, D′, E, E′, F, and F′.)
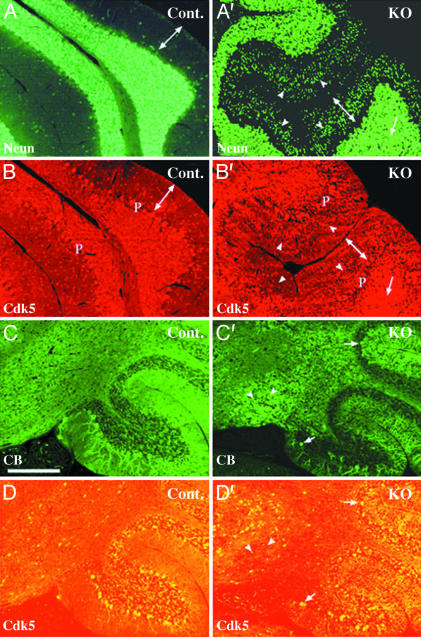
Previously, we demonstrated that the migration defects of cortical neurons in the cerebella from Cdk5+/+ ↔ Cdk5-/- chimeric mice were cell autonomous (17). Therefore, we studied the relation between loss of Cdk5 and migration defects of cortical neurons in the cerebella from the Cdk5 conditional KO mice by double immunostaining (Fig. 4). Few NeuN-immunopositive interneurons were located in the molecular layer of control cerebellum (Fig. 4A), whereas many NeuN-immunopositive granule cells were arrested in the molecular layer of the KO cerebellum (Fig. 4A′). Similar defects were noted in the Nissl-stained sections (Fig. 3E′). These accumulating granule cells in the molecular layer were negatively stained with Cdk5 antibody (Fig. 4B′), whereas granule cells in the granule cell layer were Cdk5-immunopositive in the control (Fig. 4B) and KO mice (Fig. 4B′). We carried out calbindin immunostaining to identify Purkinje cells specifically (Fig. 4 C and C′). In the Cdk5 conditional KO mice, ectopic Purkinje cells were negatively stained with Cdk5 antibody (Fig. 4D′), whereas Purkinje cells in the Purkinje cell layer were Cdk5-immunopositive in the control (Fig. 4D) and KO mice (Fig. 4D′). These results indicate that migration defects of cerebellar neurons in the Cdk5 conditional KO mice are cell autonomous.
Fig. 4.
Cell-autonomous migration defects of cerebellar cortical neurons in Cdk5 conditional KO mice. Immunostaining of the sagittal sections from 1-month-old control (Cont., fCdk5/fCdk5) (A, B, C, and D) and Cdk5 conditional KO (fCdk5/fCdk5; NFHCre/+) mice (A′ and B′, C′ and D′) are shown. Sections were double stained with antibodies against NeuN (A and A′) and Cdk5 (B and B′) or against calbindin (CB) (C and C′) and Cdk5 (D and D′). Neuronal cells except Purkinje cells (P) were positively stained with NeuN antibody (A and A′). Numerous NeuN-positive granule cells (indicated by arrowheads in A′) were accumulated in the molecular layers (indicated by a double-headed arrow) in the KO cerebellum. These granule cells in the molecular layer were negatively stained with Cdk5 antibody (arrowheads in B′), whereas granule cells in the granule cell layer were Cdk5-immunopositive (arrows in A′ and B′) in the KO mice. Ectopic Purkinje cells were negatively stained with Cdk5 antibody in the KO mice (arrowheads in C′ and D′), whereas Purkinje cells in the Purkinje cell layer were Cdk5-immunopositive (arrows in C′ and D′). A–D and A′–D′ are at the same magnification. (Scale bar in C: 200 μm.)
Discussion
Cdk5 is a unique kinase that phosphorylates multiple substrates in the CNS. Some of these substrates are involved in regulating cytoarchitecture of the CNS during development, and others are implicated in many physiological processes of the adult CNS such as synaptic plasticity, memory, learning, and behavior (18). Furthermore, recent evidence indicates that Cdk5 may be inappropriately activated in pathological conditions including drug addiction (19) and neurodegenerative diseases such as Alzheimer's disease (20) and Parkinson's disease (21). Because of these implications, Cdk5 is now considered as a therapeutic target molecule with a potential to prevent, ameliorate, and cure these aberrant disease processes. Currently, there is no animal model to examine the molecular roles of Cdk5 in the adult and aging CNS, thus hampering attempts to delineate its roles in brain functions and neurodegenerative diseases. We had earlier generated Cdk5-/- mice that displayed a striking CNS phenotype associated with mortality during the late embryonic stage and at birth (5). Additionally, we and others have also generated p35-/- and p39-/- mice that provided insights into the molecular roles of these Cdk5 activators (22–24). However, none of these mouse models provided the benefit of conditional deletion of Cdk5 gene in whole or specific areas of the adult CNS. We undertook the current investigation to generate such murine models. In the present report, we have demonstrated a general strategy for developing such a murine model, and also described the consequence of restricted inactivation of Cdk5 to neurons beginning around E16.5.
To circumvent the embryonic lethality associated with Cdk5-/- mice, we took advantage of a cre/loxP strategy to induce deletion of the Cdk5 gene in a temporally and spatially regulated manner. First, we generated mice homozygous for the Cdk5 allele flanked by loxP motifs (fCdk5/fCdk5) that were healthy and fertile without any obvious phenotype. Secondly, we generated a Cdk5 conditional KO mouse (fCdk5/fCdk5; NFHcre/+) in which Cdk5 expression was inactivated in certain population of neurons in the brain beginning around E16.5. This was achieved by crossing fCdk5/fCdk5 mice with the NFHcre mouse line 12 reported earlier (14). Unlike Cdk5-/- mice, the Cdk5 conditional KO mice carrying a heterozygous allele of NFHcre were viable and fertile, and displayed unique defects in neuronal migration.
RT-PCR analysis of fCdk5/fCdk5; NFHcre/+ mouse tissues indicated that the cre expression under the control of NFH promoter was restricted to the CNS. All other tissues failed to show any expression of cre recombinase, confirming our earlier studies on NFHcre mouse line 12 (14). Northern blot analysis of total RNA from the various brain regions revealed that Cdk5 mRNA was noticeably reduced in the cerebrum and spinal cord of Cdk5 conditional KO mice but not in the cerebellum and brainstem. As reported earlier, Cdk5 has been known to be expressed in both neurons and glial cells (25). Immunostaining of brain sections with Cdk5-specific antibody revealed a lack of Cdk5 immunoreactivity in the neurons but not in the glial cells of the Cdk5 conditional KO mice (Fig. 2). Therefore, the Cdk5 mRNA levels demonstrated by Northern blot analysis may include unaltered Cdk5 expression in the glial cells of these mice, complicating precise interpretation of the observed reduction in Cdk5 mRNA levels in the brain regions.
Despite the limited reduction in Cdk5 expression that is confined to specific areas of the brain, we observed neuronal migration defects in the mutant mice. Migration defects of the Cdk5 conditional KO mice were restricted to the later-generated neurons in the cerebral cortex and the pyramidal neurons in the CA3 region of the hippocampus. Many granule cells in the cerebellum of these mice were arrested in the molecular layer. About 10% of the mutant mice displayed migration defects of Purkinje cells in the cerebellum. These phenotypes of the cerebellum were reminiscent of our previous study with Cdk5+/+ ↔ Cdk5-/- chimeric mice that demonstrated the cell-autonomous migration defects of Cdk5-deficient neurons (17). A double-immunostaining study confirmed that the Cdk5-deficient neurons failed to migrate properly in the Cdk5 conditional KO mice (Fig. 4). The cell density of the granule cell layer in the olfactory bulbs was much lower in these mutant mice than in the controls. The migration of these interneurons takes place postnatally through the rostral migratory stream (26). These migration defects seemed to occur in the neurons that migrate after E16.5, coincident with the expression of cre recombinase under the control of the NFH promoter. It is puzzling to note that these mice do not develop an overt neurological phenotype associated with these defects. This subtle phenotype can be attributed to limited inactivation of Cdk5 in vital regions of the brain in the Cdk5 conditional KO mice. This limited inactivation of Cdk5 in turn is perhaps due to a lack of robust and uniform expression of cre recombinase in all neurons of different brain regions. Moreover, for these studies we have only used the mice heterozygous for the cre transgene because of the concerns about a potential complication due to the adverse effects of homozygosity for the cre transgene (data not shown). Another possibility that may complicate the robust and uniform cre expression in these mice is the mixed strains. Mice homozygous for the floxed Cdk5 allele (fCdk5/fCdk5) were generated in the C57BL/6 × 129SV/J strain and crossed to the NFHcre mouse line generated in the FVB/N strain, resulting in the generation of the Cdk5 conditional KO mice with a mixture of three strains. It is possible that rederiving these mice in a single strain may perhaps yield a different phenotype. On the other hand, generating additional conditional KO mice by crossing the fCdk5/fCdk5 mice with other transgenic mouse lines that express the cre recombinase in specific areas or cells of the brain will provide us with valuable mouse models to study the molecular roles of Cdk5 in brain development, function, and disease.
Acknowledgments
We thank Drs. Mary Jo Danton, Phil Graham, Mike Iadarola, and Eri Hirasawa for critical reading of the manuscript and Dr. Hua Gu for the gift of Cre and loxP plasmids.
Abbreviations: Cdk, cyclin-dependent kinase; E16.5, embryonic day 16.5; ES, embryonic stem; KO, knockout; NFH, neurofilament-heavy chain; mNFH, murine NFH.
References
- 1.Tsai, L. H., Takahashi, T., Caviness, V. S. & Harlow, E. (1993) Development (Cambridge, U.K.) 119, 1029-1040. [DOI] [PubMed] [Google Scholar]
- 2.Lew, J., Beaudette, K., Litwin, C. M. & Wang, J. H. (1992) J. Biol. Chem. 267, 13383-13390. [PubMed] [Google Scholar]
- 3.Tsai, L. H., Delalle, I., Caviness, V. S., Chae, T. & Harlow, E. (1994) Nature 371, 419-423. [DOI] [PubMed] [Google Scholar]
- 4.Tang, D., Yeung, J., Lee, K. Y., Matsushita, M., Matsui, H., Tomizawa, K., Hatase, O. & Wang, J. H. (1995) J. Biol. Chem. 270, 26897-26903. [DOI] [PubMed] [Google Scholar]
- 5.Ohshima, T., Ward, J. M., Huh, C. G., Longenecker, G., Veeranna, Pant, H. C., Brady, R. O., Martin, L. J. & Kulkarni, A. B. (1996) Proc. Natl. Acad. Sci. USA 93, 11173-11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilmore, E. C., Ohshima, T., Goffinet, A. M., Kulkarni, A. B. & Herrup, K. (1998) J. Neurosci. 18, 6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shetty, K. T., Link, W. T. & Pant, H. C. (1993) Proc. Natl. Acad. Sci. USA 90, 6844-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellmich, M. R., Pant, H. C., Wada, E. & Battey, J. F. (1992) Proc. Natl. Acad. Sci. USA 89, 10867-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, D., Leung, C. L. & Liem, R. K. (1996) J. Biol. Chem. 271, 14245-14251. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, S., Ishiguro, K., Omori, A., Takamatsu, M., Arioka, M., Imahori, K. & Uchida, T. (1993) FEBS Lett. 335, 171-175. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher, A. I., Shuang, R., Giovannucci, D. R., Zhang, L., Bittner, M. A. & Stuenkel, E. L. (1999) J. Biol. Chem. 274, 4027-4035. [DOI] [PubMed] [Google Scholar]
- 12.Tomizawa, K., Sunada, S., Lu, Y. F., Oda, Y., Kinuta, M., Ohshima, T., Saito, T., Wei, F. Y., Matsushita, M., Li, S. T., et al. (2003) J. Cell Biol. 163, 813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibb, J. A., Snyder, G. L., Nishi, A., Yan, Z., Meijer, L., Fienberg, A. A., Tsai, L. H., Kwon, Y. T., Girault, J. A., Czernik, A. J., et al. (1999) Nature 402, 669-671. [DOI] [PubMed] [Google Scholar]
- 14.Hirasawa, M., Cho, A., Sreenath, T., Sauer, B., Julien, J. P. & Kulkarni, A. B. (2001) Neurosci. Res. 40, 125-132. [DOI] [PubMed] [Google Scholar]
- 15.Ohshima, T., Nagle, J. W., Pant, H. C., Joshi, J. B., Kozak, C. A., Brady, R. O. & Kulkarni, A. B. (1995) Genomics 28, 585-588. [DOI] [PubMed] [Google Scholar]
- 16.Wullner, U., Isenmann, S., Gleichmann, M., Klockgether, T. & Bahr, M. (1998) Brain Res. Dev. Brain Res. 110, 1-6. [DOI] [PubMed] [Google Scholar]
- 17.Ohshima, T., Gilmore, E. C., Longenecker, G., Jacobowitz, D. M., Brady, R. O., Herrup, K. & Kulkarni, A. B. (1999) J. Neurosci. 19, 6017-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhavan, R. & Tsai, L. H. (2001) Nat. Rev. Mol. Cell Biol. 2, 749-759. [DOI] [PubMed] [Google Scholar]
- 19.Bibb, J. A., Chen, J., Taylor, J. R., Svenningsson, P., Nishi, A., Snyder, G. L., Yan, Z., Sagawa, Z. K., Ouimet, C. C., Nairn, A. C., et al. (2001) Nature 410, 376-380. [DOI] [PubMed] [Google Scholar]
- 20.Patrick, G. N., Zukerberg, L., Nikolic, M., de la Monte, S., Dikkes, P. & Tsai, L. H. (1999) Nature 402, 615-622. [DOI] [PubMed] [Google Scholar]
- 21.Smith, P. D., Crocker, S. J., Jackson-Lewis, V., Jordan-Sciutto, K. L., Hayley, S., Mount, M. P., O'Hare, M. J., Callaghan, S., Slack, R. S., Przedborski, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chae, T., Kwon, Y. T., Bronson, R., Dikkes, P., Li, E. & Tsai, L. H. (1997) Neuron 18, 29-42. [DOI] [PubMed] [Google Scholar]
- 23.Ohshima, T., Ogawa, M., Veeranna, Hirasawa, M., Longenecker, G., Ishiguro, K., Pant, H. C., Brady, R. O., Kulkarni, A. B. & Mikoshiba, K. (2001) Proc. Natl. Acad. Sci. USA 98, 2764-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko, J., Humbert, S., Bronson, R. T., Takahashi, S., Kulkarni, A. B., Li, E. & Tsai, L. H. (2001) J. Neurosci. 21, 6758-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, T., Veeranna, Ohshima, T., Rajan, P., Amin, N. D., Cho, A., Sreenath, T., Pant, H. C., Brady, R. O. & Kulkarni, A. B. (2001) J. Neurosci. 21, 550-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peretto, P., Merighi, A., Fasolo, A. & Bonfanti, L. (1999) Brain Res. Bull. 49, 221-243. [DOI] [PubMed] [Google Scholar]