Abstract
Previous studies suggest that the number of proteins containing covalently bound biotin is larger than previously thought. Here, we report the identity of some of these proteins. Using mass spectrometry we discovered 108 novel biotinylation sites in the human embryonic kidney HEK293 cell proteome; members of the heat shock protein (HSP) superfamily were overrepresented among the novel biotinylated proteins. About half of the biotinylated proteins also displayed various degrees of methionine oxidation, which is known to play an important role in the defense against reactive oxygen species; for biotinylated HSPs, the percent of methionine sulfoxidation approached 100%. Protein structure analysis suggests that methionine sulfoxides localize in close physical proximity to the biotinylated lysines on the protein surface. Mass spectrometric analysis revealed that between 1 and 5 of the methionine residues in the C-terminal KEEKDPGMGAMGGMGGGMGGGMF motif are oxidized in HSP60. The likelihood of methionine sulfoxidation is higher if one of the adjacent lysine residues is biotinylated. Knockdown of HSP60 caused a 60% increase in the level of reactive oxygen species in fibroblasts cultured in biotin-sufficient medium. When HEK293 cells were transferred from biotin-sufficient medium to biotin-free medium, the level of reactive oxygen species increased by >9 times compared with baseline controls and a time-response relationship was evident. High levels of methionine sulfoxidation coincided with cell cycle arrest in the G0/G1 and S phases in biotin-depleted cells. We conclude that biotinylation of lysines synergizes with sulfoxidation of methionines in heat-shock proteins such as HSP60 in the defense against reactive oxygen species.
Keywords: biotin, heat shock proteins, methionine oxidation, reactive oxygen species
INTRODUCTION
Methionine is sensitive to oxidation by reactive oxygen species (ROS), thereby creating methionine-S-sulfoxide and methionine-R-sulfoxide isomers in proteins [1, 2]. Methionine sulfoxide reductase (Msr) has catalytic activity to reduce methionine sulfoxides to methionine and, therefore, is a key enzyme in the repair of oxidatively damaged proteins [3, 4]. The oxidation of methionine and its subsequent reduction by Msr is considered a major pathway in the elimination of ROS. Msr deficiency accelerates aging [5], impairs cell signaling and protein function [6], produces a more aggressive phenotype in breast cancer [7], and prevent Parkinson’s-like symptoms [8]. Despite the biological importance of the methionine redox cycle, the sulfoxidation proteome is poorly characterized.
Here, we conducted a mass spectrometry screen of posttranslational modifications of proteins and discovered that many biotinylated proteins also contain methionine sulfoxides. This was a surprising observation, because to the best of our knowledge no links between biotin and protein oxidation have been described in the scientific literature. The only remote connection between biotin and redox biology is that the sulfur atom in the heterocyclic ring of biotin is a target for oxidation itself, creating biotin-l-sulfoxide and biotin-d-sulfoxide [9, 10]. The binding of biotin to lysine residues in proteins is catalyzed by holocarboxylase synthetase [11], which localizes in the cytoplasm, mitochondria, and nuclei in mammalian cells [12-15]. Previous studies suggest that the abundance of protein biotinylation marks depends on biotin supply in human cell cultures and in healthy adults [16, 17].
In this study we further characterized possible interactions between lysine biotinylation and methionine sulfoxidation. We focused primarily on HSP60 based on the following rationale. First, the role of HSP60 in ameliorating oxidative stress is unambiguous [18]. Second, HSP60 is particularly abundant in mitochondria, i.e., in the cellular compartment where many ROS are produced [19]. Third, our mass spectrometry analyses produced very robust biotinylation and sulfoxidation signals for HSP60. Specifically, we tested the hypothesis that biotinylation of HSP60 facilitates sulfoxidation of adjacent methionines, thereby decreasing oxidative stress.
MATERIALS AND METHODS
Cell cultures
Human embryonic kidney HEK293 cells and fetal lung IMR90 fibroblasts were maintained in MEM and RPMI-1640 media, respectively, following routine protocols [16, 20-22]. Cells were grown to ~70% confluence for subsequent analyses. In some experiments, cells were cultured in biotin-free media for up to three days [20]. Efficacy of treatment was assessed by measuring the abundance of biotinylated carboxylases using streptavidin blots [20, 22]. Biotinylated carboxylases are widely accepted as markers of cellular biotin status [10]. Cell viability was routinely monitored by using trypan blue exclusion and was typically >95%.
Purification of biotinylated proteins
A pellet of 30×106 HEK293 cells was lysed in 10 mL of Bug Buster lysis buffer (Novagen), containing 10 μL of protease inhibitor cocktail (Sigma) and 250 units Benzonase nuclease (Novagen). Samples were incubated on ice for 15 min. After centrifugation, the supernatant was filtered through a 22-μm membrane (Millipore) and run through a 2-mL monomeric avidin column according to the manufacturer’s instructions (Thermo Scientific; biotin-binding capacity = 1.2 mg biotinylated protein/mL settled resin). Biotinylated proteins were eluted with 4 mL of Regeneration Buffer (0.1 M glycine, pH 2.8), and samples were desalted and concentrated using 5 kDa Spin-X UF concentrators (Corning).
Analysis of protein modifications by mass spectrometry
Avidin-purified proteins were resolved using NuPAGE 4-12% Bis-Tris gels. Gels were stained with coomassie blue, and proteins were released from cut-out pieces and digested with trypsin as described previously [23]. Samples were loaded on the monolithic trap column in a Dionex U 3000 nano LC unit. The desalted peptides were separated and eluted using a C18 Pep Map column (75 μm inner diameter, 15 cm, 3 μm particle size) and an acetonitrile gradient (acetonitrile plus 0.1% formic acid) and introduced into the mass spectrometer using the nano spray source. The LCQ Fleet mass spectrometer (Thermo Electron Corporation) operates with the following parameters: nano spray voltage (2.0 kV), heated capillary temperature 200°C, full scan m/z range 400-2000. The LCQ was operated in data dependent mode with 4 MS/MS spectra for every full scan, 5 microscans averaged for full scans, and MS/MS scans in collision-induced dissociation mode. The acquired spectrum was compared to the human reference proteome for sequence identities using the MASCOT database analysis software [24].
Protein structures
Protein structures were modeled in silico to identify the location of biotinylated and sulfoxidated lysines and methionines, respectively. Briefly, proteins containing modified peptides were identified using the PDB database [25] and BLAST (version 2.2.26). Best matches were selected from top BLAST hits aligning to full-length peptides with ≥95% sequence identity and having E-values of less than 1.0×10−6. HSP60 was the only exception to this rule for which the best match was its microbial ortholog GroEL, which had 60% sequence identity. The secondary structures and solvent accessible areas surrounding biotinylation and sulfoxidation sites were determined using the DSSP program [26]. The relative solvent accessibility (RSA) was calculated as the percentage of the accessible surface area of residue X in the tripeptide G-X-G as described previously [27]. Residues with an RSA <20% and ≥ 20% were considered buried and exposed, respectively, to the solvent. All protein presentations were prepared using VMD version 1.9.1 [28]. Molecular surface representations were calculated using the MSMS 2.5.7 as plugin in VMD [29]. In graphical presentations, K and M residues were set at 100% opacity and the protein opacity was set at 75%. The average structural features at the biotinylation and oxidation sites were calculated using a custom perl script.
Cell cycle analysis
Cell cycle analyses were conducted in primary human fibroblast cultures. IMR90 fibroblasts were detached from culture flasks by treatment with trypsin for <10 min at room temperature, and cells were fixed in 70% ethanol (2.5×105 cells/mL) at 4°C overnight. Prior to analysis, cells were washed in phosphate-buffered saline (PBS) containing 2% bovine serum albumin and collected by centrifugation. Cell pellets were suspended in 0.5 mL of PBS containing DNase-free RNase (100 μg/mL) and incubated at 37°C for 0.5 h. Cells were then stained with propidium iodide (40 μg/mL final concentration) and analyzed using a BD FACSCalibur flow cytometer and the Cell Quest Pro software (BD Biosciences). The data are expressed as percent of cells in each phase of cell cycle.
ROS analysis
The membrane permeable 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) is a non-fluorescent probe that is hydrolyzed and oxidized to 2′,7′-dichlorofluorescein after entry into the cell; the level of the fluorescent 2′,7′-dichlorofluorescein is a measure for cellular levels of ROS [30]. Briefly, 2.5×105 IMR90 fibroblasts were incubated with 10 μM H2DCFDA (final concentration) in a volume of 5 mL of RPMI-1640 at 37°C for 1 h. Cultures were rinsed with phosphate-buffered saline, detached using trypsin, and subjected to flow cytometry analysis of 2′,7′-dichlorofluorescein (DCF). Results are expressed as percent change in ROS per 10,000 cells based on fluorescence ratios in treatment versus control.
HSP60 knockdown cells
HSP60 was knocked down by transfecting 2×106 HEK293 cells with 2 nM of HSP60 Trilencer-27 Human siRNA (OriGene) using Lipofectamine 2000 (Invitrogen). Controls were transfected with siRNA not targeting any human gene (Negative Control siRNA, Origene). Cells were collected 2 d after transfection and knockdown was confirmed by western blot analysis, using 3–8% Tris-Acetate gels (Invitrogen), anti-HSP60 Antibody (Abcam), IRDye® 800CW anti-rabbit IgG (LI-COR), and an Odyssey infrared imaging system [16]. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as control.
RESULTS
Co-localization of lysine biotinylation and methionine oxidation
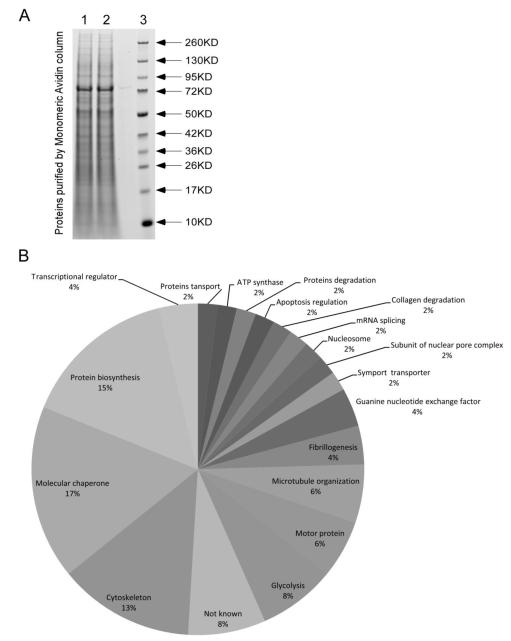
A large number of proteins eluted off the avidin column and were visualized using coomassie blue staining (Fig. 1A). LC/MS/MS analysis of gel pieces revealed the existence of 213 proteins, including 108 novel biotinylated proteins and 105 non-biotinylated proteins (Table 1). Epsilon amino groups in lysines were the only targets for covalent binding of biotin. Ninety of the 213 proteins that were detected by LC/MS/MS also carried methionine sulfoxidation marks. Fifty percent of the proteins that were biotinylated were also modified by methionine sulfoxidation, whereas the percentage was only about 34% (36 of 105 proteins) in biotin-free proteins, suggesting a link between protein lysine biotinylation and methionine sulfoxidation. In subsequent analyses we focused on proteins that were dually modified by lysine biotinylation and methionine sulfoxidation (Table 2). The majority of these dually modified proteins are members of the superfamily of HSPs (molecular chaperones), regulators of transcription and translation, proteins with functions in the cytoskeleton (tubulin and actin), and enzymes in glycolysis (Fig. 1B). HSP60 is unique among these proteins because the sulfoxidated methionines cluster in a unique C-terminal KEEKDPGMGAMGGMGGGMGGGMF motif. The motif is characterized by five methionine residues embedded in a glycine-rich sequence and the presence of two lysine residues that are targets for biotinylation (Table 3). Between one and five of the methione residues were oxidized in the HSP60 C-terminus, in the presence or absence of one lysine biotinylation mark.
Fig 1.
Biotinylated proteins in HEK293 whole cell extracts. (A) Proteins extracts were purified using monomeric avidin, resolved by SDS-PAGE,and and visualized using coomassie blue. Lanes 1 and 2 = independently prepared samples; lane 3 = markers. (B) Proteins containing both lysine biotinylation and methionine sulfoxidation marks were clustered by biological function.
Table 1.
Proteins purified by avidin and identified by LC/MS/MS analysis. Proteins are broken down by lysine biotinylation and methionine sulfoxidation marks.
| Count (% of all proteins) | Biotinylation | Total | ||
|---|---|---|---|---|
| − | + | |||
| Sulfoxidation | − | 69 (32.4%) | 54 (25.4%) | 123 (57.7%) |
| + | 36 (16.9%) | 54 (25.4%) | 90 (42.3%) | |
| Total | 105 (49.3%) | 108 (50.7%) | 213 (100%) | |
Table 2.
Proteins modified by both biotinylation of lysines and sulfoxidation of methionines.
| Name | Function | Amino acid sequence and modification sites1-3 |
|---|---|---|
| ACTA2 protein (Fragment) | Cytoskeleton | RKbDLYANNVLSGGTTMYPGIVDRMoQKE |
| ACTB Actin, cytoplasmic 1 | Cytoskeleton | KDLYANTVLSGGTTMYPGIADRMoQKbEITALAPSTMKI |
| ANKRD31 Ankyrin repeat domain-containing protein 31 |
Not known | RMQDPALMoIDGKbEKNMoHSARF |
| ARHGEF4 108 kDa protein | Guanine nucleotide exchanger |
GKbAVVLMoGKNTMMoR |
| ATP5B ATP synthase subunit beta, mitochondrial | ATP synthase | RFLSQPFQVAEVFTGHMoGK…RAAPTAVHPVRDYAAQ TSPSPKbAGAATGR |
| C18orf34 Isoform 3 of uncharacterized protein | Not known | MoKKbSIGILSPGVALGMAGSAMoSSKb |
| C2CD2L Isoform 2 of C2 domain-containing protein 2-like |
Proteins targeting | RELTLKbVLR…REILMoIASKGQGVNNVPKR |
| CCT2 T-complex protein 1 subunit beta | Molecular chaperone | KGMoDKILLSSGRDASLMVTNDGATILKb |
| CEP164 Isoform 1 of Centrosomal protein of 164 kDa |
Microtubule organization |
RGLVDTPPSALRGSQSVSLGSSVESGRQLGELMoLPSQG LKb |
| CLASP2 protein | Microtubule organization |
RVFSMoFLETLVDFIQVHKbDDLQDWLFVLLTQLLK |
| CLASP2 cDNA FLJ61103, highly similar to CLIP-associating protein 2 |
Microtubule organization |
KTYMoGLRNHFPGEAETLYNSLEPSYQKSLQTYLKb |
| COL11A1 alpha 1 type XI collagen isoform C preproprotein |
Fibrillogenesis | RGKbLLSYLDVEGNSINMVQMoTFLKb |
| COL27A1 Isoform 2 of Collagen alpha-1(XXVII) chain |
Fibrillogenesis | KGDLGPLGTPGEQGLIGQRGEPGLEGDSGPMoGPDGLKb |
| DYNC1H1 Cytoplasmic dynein 1 heavy chain 1 | Motor protein | KQMoSVKbEDLDKVEPAVIEAQNAVK |
| EEF1A2 Elongation factor 1-alpha 2 | Protein biosynthesis | KSVEMoHHEALSEALPGDNVGFNVKbNVSVK |
| EEF1A2 Eukaryotic translation elongation factor 1 alpha 2 variant (Fragment) |
Protein biosynthesis | KSGDAAIVDMoVPSKPMoCVESFSDYPPLGRFAVR…KE GNASGVSLLEALDTILPPTRPTDKbPLR |
| EIF2AK2 Interferon-induced, double-stranded RNA-activated protein kinase |
Protein biosynthesis | KTKCLFIQMEFCDKb…KRFGMoDFKEIELIGSGGFGQVFK |
| EIF4A3 Eukaryotic initiation factor 4A-III | Protein biosynthesis | REANFTVSSMoHGDMoPQKb |
| Elongation factor 1-alpha | Protein biosynthesis | KSVEMoHHEALSEALPGDNVGFNVKbNVSVK |
| ENO1 Isoform alpha-enolase of Alpha-enolase | Glycolysis | KAGYTDKVVIGMoDVAASEFFRSGKb |
| GCN1L1 Translational activator GCN1 | Protein biosynthesis | RKbNIVSLLLSMoLGHDEDNTR |
| General transcription factor IIH, polypeptide 1 | Transcriptional regulator |
KMoKbYAESVPHNMoTEK |
| HIST1H1C Histone H1.2 | Nucleosome | MoSETAPAAPAAAPPAEKb |
| HNRPU protein | mRNA splicing | KYNILGTNTIMoDKMMVAGFKb |
| HSP90AA1 Isoform 2 of Heat shock protein HSP 90-alpha |
Molecular chaperone | RTDTGEPMoGR…KEKEEKbESEDKPEIEDVGSDEEEEKb |
| HSP90AA2 Putative heat shock protein HSP 90- alpha A2 |
Molecular chaperone | KSGTKbAFMoEALQAGADISMIGQFGVSFYSAYLVAEKb |
| HSP90AB3P Putative heat shock protein HSP 90- beta-3 |
Molecular chaperone | KACMoEALQAEKLVVITKb |
| HSPA1B;HSPA1A Heat shock 70 kDa protein 1A/1B |
Molecular chaperone | KGETKbAFYPEEISSMoVLTKMoKb |
| HSPA1L cDNA FLJ56386, highly similar to Heat shock 70 kDa protein 1L |
Molecular chaperone | RNLDVSKbLK…KNALESYAFNMoK |
| HSPA8 Isoform 1 of Heat shock cognate 71 kDa protein |
Molecular chaperone | RLIGDAAKbNQVAMoNPTNTVFDAKbR |
| HSPD1 60 kDa heat shock protein, mitochondrial | Molecular chaperone | RALMoLQGVDLLADAVAVTMGPKGRTVIIEQSWGSPKb |
| HSPD1 Short heat shock protein 60 Hsp60s2 | Molecular chaperone | KIMQNSSEVGYDAMoVGDFMNMoVEKb |
| KIF1C Kinesin-like protein | Motor protein | KTPHLVNLNEDPLMoSECLLYHIKDGVTRVGQVDMoDIK b |
| KIF26A Kinesin-like protein | Motor protein | RAGPSVGAKAGRGTVMoGTKb |
| LDHAL6A L-lactate dehydrogenase A-like 6A | Glycolysis | KGETMoDLQHGSPFMoKbMPNIVSSKb |
| MECOM Isoform 2 of MDS1 and EVI1 complex locus protein EVI1 |
Transcriptional regulator |
RDLRSLPLKbMoEPQSPGEVK |
| MMP16 Isoform Long of Matrix metalloproteinase-16 |
Collagen degradation |
MoILLTFSTGR…KGTPRHILYCKb |
| NUP153 Nuclear pore complex protein Nup153 | Subunit of nuclear pore complex |
MoASGAGGVGGGGGGKb |
| OBSCN Isoform 3 of Obscurin | Muscle protein | KKLSSSSKbVCMoEATGCTR |
| PKM2 Isoform M2 of Pyruvate kinase isozymes M1/M2 |
Glycolysis | RLDIDSPPSKb…KFGVEQDVDMoVFASFIR |
| PKM2 Pyruvate kinase | Glycolysis | KDIQDLKbFGVEQDVDMoVFASFIR |
| POTEE Isoform 1 of POTE ankyrin domain family member E |
Actin-like protein | KQIEVVEKMoNSELSLSCKb |
| POTEJ POTE ankyrin domain family member J | Actin-like protein | KRTALTKbAVQCQEDECALMoLLEHGTDPNIPDEYGNTT LHYAIYNEDK |
| PSMC4 Isoform 1 of 26S protease regulatory subunit 6B |
Proteins degradation | KKEFLHAQEEVKbR…KVIMoATNRADTLDPALLRPGRL DR |
| RPLP0 60S acidic ribosomal protein P0 | Protein biosynthesis | RGKbAVVLMoGKNTMMoR |
| SATL1 Isoform 2 of Spermidine/spermine N(1)- acetyltransferase-like protein 1 |
Not known | KQPSMoSQAGMoRQSGTNLPDINQPGMoKb |
| SLC1A3 Excitatory amino acid transporter 1 | Symport transporter | KMGMoRAVVYYMoTTTIIAVVIGIIIVIIIHPGKbGTKb |
| SNORA4;EIF4A2;MIR1248;SNORA81;SNORA 63;SNORD2 26 kDa protein |
Protein synthesis | MoSGGSADYNREHGGPEGMoDPDGSNWNEIVDNFDDM NLKb |
| TP53BP2 Isoform 1 of Apoptosis-stimulating of p53 protein 2 |
Apoptosis regulation | KGVIYALWDYEPQNDDELPMoKbEGDCMoTIIHR |
| TUBB Tubulin, beta | Cytoskeleton | KTAVCDIPPRGLKbMoSATFIGNSTAIQELFK |
| TUBB2A Tubulin beta-2A chain | Cytoskeleton | RKbESESCDCLQGFQLTHSLGGGTGSGMoGTLLISKbIR |
| TUBB6 46 kDa protein | Cytoskeleton | RYLTVATVFRGPMSMoKbEVDEQMLAIQSK |
| UGGT1 Isoform 1 of UDP-glucose:glycoprotein glucosyltransferase 1 |
Protein glycosylation. |
KMAKbEGAAEALAAGADIAEFSVGGMoDFSLFK |
| ZBED4 Zinc finger BED domain-containing protein 4 |
Not known | RKbVEMoLFEETMoGIDTMLRSLKb |
One letter amino acid code is used.
Abbreviations: b, biotinylation; o, sulfoxidation.
Discontinuous sequences are denoted by “…” Proteins in which lysine biotinylation and methionine sulfoxidation marks were found in distinct peptide fragments are reported this way.
Table 3.
Permutations of lysine biotinylation and methionine sulfoxidations in the HSP60 C-terminus.
| Modifications in the HSP60 C-terminus1 | Oxidation count | Biotinylation count |
|---|---|---|
| KbDPGMOGAMOGGMOGGGMOGGGMoF | 1 | 0 |
| KbDPGMoGAMOGGMOGGGMOGGGMOF | 1 | 0 |
| KbDPGMOGAMoGGMOGGGMOGGGMoF | 2 | 1 |
| KbDPGMOGAMoGGMoGGGMoGGGMOF | 3 | 0 |
| KbDPGMoGAMoGGMOGGGMOGGGMoF | 3 | 0 |
| KbDPGMOGAMoGGMoGGGMoGGGMOF | 3 | 0 |
| KbDPGMoGAMoGGMOGGGMOGGGMoF | 3 | 0 |
| KbDPGMoGAMoGGMoGGGMoGGGMoF | 5 | 0 |
| KbDPGMoGAMoGGMoGGGMoGGGMoF | 5 | 0 |
| KbEEKbDPGMOGAMoGGMoGGGMOGGGMOF | 2 | 0 |
| KbEEKbDPGMoGAMoGGMOGGGMOGGGMOF | 2 | 0 |
| KbEEKbDPGMoGAMOGGMoGGGMoGGGMOF | 3 | 1 |
| KbEEKbDPGMoGAMOGGMoGGGMoGGGMoF | 4 | 0 |
| KbEEKbDPGMoGAMOGGMoGGGMoGGGMoF | 4 | 0 |
Abbreviations: b, lysine biotinylation; o, methionine sulfoxidation.
One might ask whether methionine sulfoxidation marks are artifacts due to sample preparation or storage. Previous studies suggest that recombinant proteins may be stored for up to 1.7 years with no methionine sulfoxidation detectable by LC/MS/MS [31]. This previous observation is consistent with the theory that methionine sulfoxidation marks occurred naturally as opposed to being in vitro artifacts in this study. Additional evidence in support of this theory is presented below.
Structural analyses
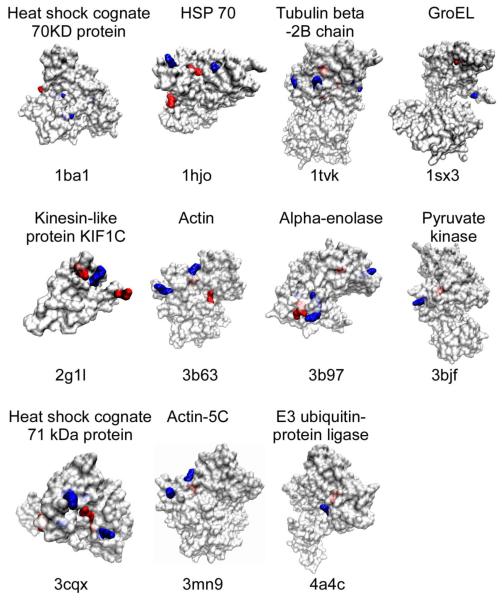
Forty biotinylated and sulfoxidated peptides were matched to PDB entries. Note that some peptides contained two biotinylation or sulfoxidation sites and that, therefore, the total number of modification sites was 44 for biotinylation and 46 for sulfoxidation. About 50% of biotinylation and sulfoxidation marks localized in α-helices, ~40% localized in loops, and ~10% localized in sheets (Table 4a). Solvent accessibility analysis suggests that the majority of biotinylated lysines (84%) are exposed to solvent, whereas the majority of sulfoxidated methionines (67%) are buried inside the protein (Table 4b). Note that the percentages were calculated by imposing novel biotinylation and sulfoxidation marks on existing PDB entries for unmodified proteins with the objective to illustrate the spatial distribution of these marks. These results can be visualized from the molecular surface representations showing the biotinylated and sulfoxidated positions in 11 proteins (Fig. 2). Note that GroEL is the bacterial ortholog of human HSP60, and that the peptide sequence is 60% identical to GroEL. The majority of biotinylated lysines are exposed on the protein surface, whereas a meaningful percentage of sulfoxidated methionines are buried inside proteins. Biotinylated lysine residues localize in close physical proximity to sulfoxidated methionines. The median distance between biotinylation and sulfoxidation sites was 12.1 Å based on the analysis of 55 sites in the 40 peptides (Table 4c).
Table 4.
Location of biotinylated lysines and oxidized methionines.
| a. Secondary structure: | |||
|---|---|---|---|
|
|
|||
| Biotinylated lysine | Oxidized methionine | ||
|
|
|||
| In helices | 54.6% (24/44) | 50.0% (23/46) | |
| In sheet | 13.6% (6/44) | 8.7% (4/46) | |
| In loop | 31.8% (14/44) | 41.3% (19/46) | |
|
|
|||
| b. Solvent accessibility: | |||
|---|---|---|---|
|
|
|||
| Biotinylated lysine | Oxidized methionine | ||
|
|
|||
| Buried | 15.9% (7/44) | 67.4% (31/46) | |
| Exposed | 84.1% (37/44) | 32.6% (15/46) | |
|
|
|||
| c. Distance between biotinylation and sulfoxidation sites: | ||
|---|---|---|
|
|
||
| Sites analyzed | 55 | |
| Minimum distance | 3.0 Å | |
| Maximum distance | 35.2 Å | |
| Average distance | 13.9 Å | |
| Median distance | 12.1 Å | |
|
|
||
Fig 2.
Localization of oxidized methionines and biotinylated lysines in proteins. Proteins are labeled with their name and corresponding Protein Databank (PDB) identifiers above and underneath the structure prediction, respectively. Oxidized methionines (M) and biotinylated lysines (K) are shown in red and blue, respectively. Bright colors denote residues that are exposed at the protein surface, whereas shaded colors denote buried residues.
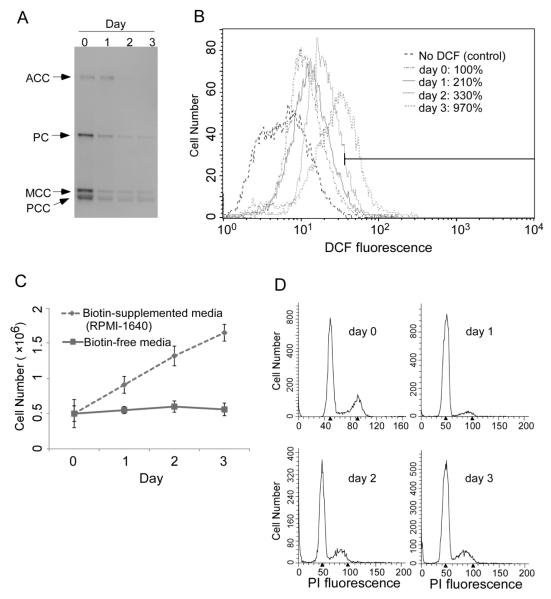
Biotinylation of lysines increased the likelihood of methionine sulfoxidation events. Fifty percent of the proteins that were biotinylated also carried sulfoxidation marks in HEK293 cells (54 of 108 proteins, Table 1). In contrast, 66% of the proteins that lacked a biotinylation mark also had no sulfoxidation marks (69 of 105 proteins). The theory that the removal of ROS is contingent on biotin-dependent creation of methionine sulfoxidation marks was further tested using biotin-depleted IMR-90 fibroblasts. Our rationale for studying fibroblasts was to demonstrate that interactions between biotinylation and sulfoxidation marks do not only exist in tranformed immortal cell lines such as HEK293, but also in primary cells with a finite life span such as IMR-90. When fibroblasts were cultured in biotin-free medium, the abundance of biotinylated proteins decreased to near background levels after three days of culture, as judged by the abundance of biotinylated carboxylases (Fig. 3A). The relative amount of ROS increased from 100% (day 0) to 210% (day 1), 330% (day 2), and 970% (day 3) in fibroblasts cultured in biotin-free media (Fig. 3B). When fibroblasts were stressed by biotin depletion, they responded with cessation of proliferation (Fig. 3C) due to an arrest in the G0/G1 and S phases of the cell cycle (Fig. 3D). Importantly, when the expression of HSP60 was knocked down by about 60% in HEK-293 cells (Fig. 4A), then the levels of endogenous ROS increased by 60% (Fig. 4B). Note that the experiment in HSP60 knockdown cells was conducted in regular RPMI-1640 which contains a pharmacological concentration of biotin (~ 1 μM). Currently, no antibody is available that is specific for biotinylated HSP60, i.e., we could not determine whether the abundance of HSP60 biotinylation marks depends on the concentration of biotin in cell culture media. Note, however, that we succeeded raising an antibody that is specific for the biotinylated species of another novel biotinylated protein, namely the transcription factor and tumor suppressor MBP-1. The abundance of biotinylated MBP-1 and transcriptional repressor activity of MBP-1 depended on the concentration of biotin in cell culture media (J. Zhou et al., in preparation).
Fig 3.
Biotin depletion causes an increase in oxidative stress and cell cycle arrest in IMR90 fibroblasts. Cells were transferred from biotin regular RPMI-1640 culture media (~1 μmol/l biotin) to biotin-depleted media and culture was continued for three days. (A) Depletion of biotinylated carboxylases at timed intervals after transfer of fibroblasts into biotin-free medium. ACC = acetyl-CoA carboxylases 1 and 2, PC = pyruvate carboxylase, MCC = 3-methylcrotonyl-CoA carboxylase, PCC = propionyl-CoA carboxylase. (B) Levels of ROS in fibroblasts at timed intervals after transfer of fibroblasts into biotin-free medium. No DCF control = IMR90 cells without H2DCFDA as stain for ROS in regular RPMI-1640. (C) Fibroblast counts at timed intervals after transfer into biotin-free medium. (D) Cell cycle analysis at timed intervals after transfer of fibroblasts into biotin-free medium.
Fig 4.
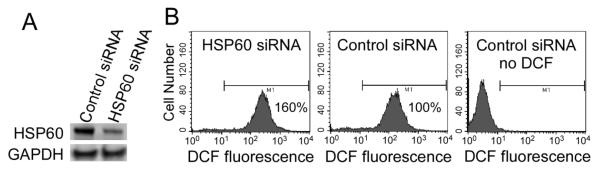
HSP60 knockdown causes ROS accumulation in HEK293 cells. (A) HSP60 abundance was assessed two days after knockdown with HSP60 Trilencer-27 Human siRNA; control cells were transfected with a siRNA not targeting any known gene in the human genome. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control. (B) Cellular ROS levels in HSP60 knockdown cells (left), knockdown controls (middle), and knockdown controls in the absence of fluorophore (right).
DISCUSSION
This study provides novel insights into biotin nutrition and redox biology at multiple levels. First, this is the first report to suggest that lysines in HSPs are biotinylated. Second, this is the first report of 108 novel biotinylated proteins using mass spectrometry analysis. Third, this is the first report to identify proteins containing methionine sulfoxidation marks in the absence of exogenous inducers of oxidative stress. Previous studies used superoxide and hydrogen peroxide [32, 33] and hypochlorous acid [34] to create methionine sulfoxidation marks, which may not represent the in vivo situation. Fourth, this is the first report to demonstrate the co-existence of lysine biotinylation and methionine sulfoxidation marks in endogenous proteins and an increase in ROS levels in response to biotin depletion. Fifth, this is the first report to suggest that proteins containing oxidized methionines may be enriched using avidin-based purification protocols. The availability of such a protocol is an important advancement, considering that the abundance of methionine sulfoxidation marks in proteins is low compared with the reduced normal protein [31]. Sixth, this is the first report directly implicating HSP60 with stress resistance.
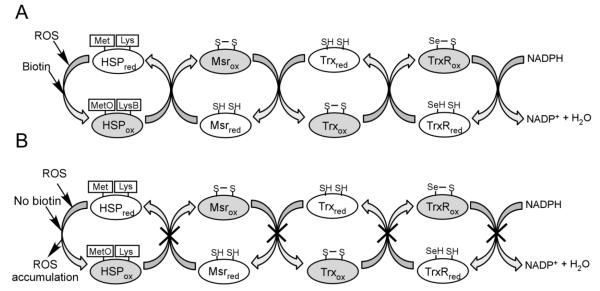
The KEEKDPGMGAMGGMGGGMGGGMF motif is highly conserved in different species [35], but its function is unknown. Our data suggest that the C-terminal motif in HSP60 might be a ROS acceptor. HSP60 is a high abundance protein which forms homooligomers composed of either 7 or 14 subunits in mitochondria [19]. Depending on the degree of polymerization, HSP60 oligomers have 35 or 70 methionine residues that could act as ROS scavengers, rendering the protein a high capacity ROS scavenger. We speculate that HSP60 is recycled through the action of Msr. The mechanism of methionine and Msr dependent elimination of ROS is firmly grounded in the literature [36, 37]. Here, we expand this model by proposing that biotinylation of lysines in heat shock proteins close proximity to sulfoxidation sites contributes toward the elimination of ROS through the methionine/Msr pathway (Fig. 5A and 5B).
Fig 5.
Model of the biotin-dependent methionine/Msr defense system in the presence (A) and absence (B) of biotin. Reduced and oxidized forms of proteins are denoted by using subscript red and ox. (A) ROS is intercepted by a methionine residue of heat shock protein (HSP), which is oxidized to MetO. Methionine sulfoxidation depends on lysine biotoinylation (LysB). HSP is recycled by methionine sulfoxide reductase (Msr). The oxidized Msr is reduced by thioredoxin (Trx), which is subsequently reduced by thioredoxin reductase (TrxR). Note that mammalian TrxR is a selenoprotein. TrxR is then reduced by NADPH. The net result is that ROS are scavenged in an NADPH-consuming process. (B) Biotin depletion causes an arrest in the methionine/Msr cycle, thereby leading to ROS accumulation.
This study is just the first step in advancing the discovery of novel biotinylation and sulfoxidation sites in proteins to a more mechanistic level. The following uncertainties will be addressed in future studies. (i) The theory that modified HSP60 is recycled by Msr is currently pure speculation. (ii) While it is clear that HSPs other than HSP60 are also target for biotinylation, it is currently uncertain whether they scavenge ROS. (iii) Evidence suggests that the protein biotin ligase holocarboxylase synthetase catalyzes the binding of biotin to HSPs (Xue et al., unpublished) but it is unknown whether the removal of biotin also is an enzyme-mediated process. (iv) The heterocyclic ring in the biotin molecule contains a sulfur atom that is known to undergo oxidation to produce biotin-d,l-sulfoxides and biotin sulfone in vivo [38, 39]. Sulfoxidation in the biotin molecule is an NADPH-dependent process [40], consistent with the diagram shown in Figure 5. It is currently unknown whether biotin sulfoxidation is an intermediate in the formation of methione sulfoxides in HSPs. We conclude that, despite a number of remaining uncertainties, evidence is emerging to suggest that biotin and methionine synergize in the protection against oxidative stress through HSPs, and that this possibility deserves further investigation.
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945 and DK077816.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Shechter Y, Burstein Y, Patchornik A. Selective oxidation of methionine residues in proteins. Biochemistry. 1975;14:4497–503. doi: 10.1021/bi00691a025. [DOI] [PubMed] [Google Scholar]
- [2].Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–8. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- [3].Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim Biophys Acta. 2009;1790:1471–7. doi: 10.1016/j.bbagen.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moskovitz J, Oien DB. Protein carbonyl and the methionine sulfoxide reductase system. Antioxid Redox Signal. 2010;12:405–15. doi: 10.1089/ars.2009.2809. [DOI] [PubMed] [Google Scholar]
- [5].Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–40. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- [6].Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal. 2003;5:577–82. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- [7].De Luca A, Sanna F, Sallese M, Ruggiero C, Grossi M, Sacchetta P, et al. Methionine sulfoxide reductase A down-regulation in human breast cancer cells results in a more aggressive phenotype. Proc Natl Acad Sci USA. 2010;107:18628–33. doi: 10.1073/pnas.1010171107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wassef R, Haenold R, Hansel A, Brot N, Heinemann SH, Hoshi T. Methionine sulfoxide reductase A and a dietary supplement S-methyl-L-cysteine prevent Parkinson’s-like symptoms. J Neurosci. 2007;27:12808–16. doi: 10.1523/JNEUROSCI.0322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McCormick DB, Wright LD. The metabolism of biotin and analogues. In: Florkin M, Stotz EH, editors. Metabolism of Vitamins and Trace Elements. Elsevier Publishing Company; Amsterdam, The Netherlands: 1971. pp. 81–110. [Google Scholar]
- [10].Zempleni J, Wijeratne SSK, Kuroishi T. Biotin. In: Erdman JW Jr., Macdonald I, Zeisel SH, editors. Present Knowledge in Nutrition. vol. 10th. International Life Sciences Institute; Washington, D.C.: 2012. pp. 587–609. [Google Scholar]
- [11].Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, et al. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat Genet. 1994;8:122–28. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- [12].Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- [14].Bailey LM, Wallace JC, Polyak SW. Holocarboxylase synthetase: correlation of protein localisation with biological function. Arch Biochem Biophys. 2010;496:45–52. doi: 10.1016/j.abb.2010.01.015. [DOI] [PubMed] [Google Scholar]
- [15].Bao B, Wijeratne SS, Rodriguez-Melendez R, Zempleni J. Human holocarboxylase synthetase with a start site at methionine-58 is the predominant nuclear variant of this protein and has catalytic activity. Biochem Biophys Res Commun. 2011;412:115–20. doi: 10.1016/j.bbrc.2011.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kaur Mall G, Chew YC, Zempleni J. Biotin requirements are lower in human Jurkat lymphoid cells but homeostatic mechanisms are similar to those of HepG2 liver cells. J Nutr. 2010;140:1086–92. doi: 10.3945/jn.110.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eng WK, Giraud D, Schlegel VL, Wang D, Lee BH, Zempleni J. Identification and assessment of markers of biotin status in healthy adults. British Journal of Nutrition. 2013:1–9. doi: 10.1017/S0007114512005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cabiscol E, Belli G, Tamarit J, Echave P, Herrero E, Ros J. Mitochondrial Hsp60, resistance to oxidative stress, and the labile iron pool are closely connected in Saccharomyces cerevisiae. J Biol Chem. 2002;277:44531–38. doi: 10.1074/jbc.M206525200. [DOI] [PubMed] [Google Scholar]
- [19].Cheng MY, Hartl FU, Horwich AL. The mitochondrial chaperonin hsp60 is required for its own assembly. Nature. 1990;348:455–8. doi: 10.1038/348455a0. [DOI] [PubMed] [Google Scholar]
- [20].Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- [21].Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J Nutr Biochem. 2010;21:310–16. doi: 10.1016/j.jnutbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuroishi T, Rios-Avila L, Pestinger V, Wijeratne SS, Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol Genet Metab. 2011;104:537–45. doi: 10.1016/j.ymgme.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature protocols. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- [24].Rodriguez-Rocha H, Garcia-Garcia A, Zavala-Flores L, Li S, Madayiputhiya N, Franco R. Glutaredoxin 1 protects dopaminergic cells by increased protein glutathionylation in experimental Parkinson’s disease. Antioxid Redox Signal. 2012;17:1676–93. doi: 10.1089/ars.2011.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D37. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- [27].Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- [28].Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [29].Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–20. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [30].Cathcart R, Schwiers E, Ames BN. Detection of picomole levels of hydroperoxides using a fluorescent dichlorofluorescein assay. Anal Biochem. 1983;134:111–6. doi: 10.1016/0003-2697(83)90270-1. [DOI] [PubMed] [Google Scholar]
- [31].Wehr NB, Levine RL. Wanted and wanting: Antibody against methionine sulfoxide. Free Radic Biol Med. 2012;53:1222–25. doi: 10.1016/j.freeradbiomed.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–40. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tarrago L, Kaya A, Weerapana E, Marino SM, Gladyshev VN. Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins and protect cells from oxidative protein unfolding. J Biol Chem. 287:24448–59. doi: 10.1074/jbc.M112.374520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dalle-Donne I, Rossi R, Giustarini D, Gagliano N, Di Simplicio P, Colombo R, et al. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free radical biology & medicine. 2002;32:927–37. doi: 10.1016/s0891-5849(02)00799-2. [DOI] [PubMed] [Google Scholar]
- [35].Brocchieri L, Karlin S. Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 2000;9:476–86. doi: 10.1110/ps.9.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–72. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stadtman ER. Cyclic oxidation and reduction of methionine residues of proteins in antioxidant defense and cellular regulation. Arch Biochem Biophys. 2004;423:2–5. doi: 10.1016/j.abb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- [38].Lee H-M, McCall NE, Wright LD, McCormick DB. Urinary excretion of biotin and metabolites in the rat. Proc Soc Exp Biol Med. 1973;142:642–44. doi: 10.3181/00379727-142-37085. [DOI] [PubMed] [Google Scholar]
- [39].Zempleni J, McCormick DB, Mock DM. Identification of biotin sulfone, bisnorbiotin methyl ketone, and tetranorbiotin-l- sulfoxide in human urine. Am J Clin Nutr. 1997;65:508–11. doi: 10.1093/ajcn/65.2.508. [DOI] [PubMed] [Google Scholar]
- [40].Lee YC, Joiner-Hayes MG, McCormick DB. Microsomal oxidation of a-thiocarboxylic acids to sulfoxides. Biochem Pharmacol. 1970;19:2825–32. doi: 10.1016/0006-2952(70)90021-3. [DOI] [PubMed] [Google Scholar]