Abstract
Bacterial and viral mRNAs are often polycistronic. Akin to alternative splicing, alternative translation of polycistronic messages is a mechanism to generate protein diversity and regulate gene function. Although a few examples exist, the use of polycistronic messages in mammalian cells is not widely appreciated. Here we report an example of alternative translation as a means of regulating innate immune signaling. MAVS, a regulator of antiviral innate immunity, is expressed from a bicistronic mRNA encoding a second protein, miniMAVS. This truncated variant interferes with interferon production induced by full length MAVS, whereas both proteins positively regulate cell death. To identify other polycistronic messages, we carried out genome-wide ribosomal profiling and identified a class of antiviral truncated variants. This study therefore reveals the existence of a functionally important bicistronic antiviral mRNA, and suggests a widespread role for polycistronic mRNAs in the innate immune system.
Introduction
Signal transduction pathways are a critical part of the immune response to control the magnitude of inflammation. Regulation of such pathways maintains homeostasis in many cellular processes, and a common means of generating this regulation is through the diversification of protein form and function. From a single genetic locus this diversification can be achieved through alternative splicing and/or translation, resulting in the production of multiple proteins with distinct functions. This is a highly effective way of altering protein activities because it offers a mechanism for removing or adding functional domains.
In eukaryotes, protein diversification can be generated during mRNA processing and examples of this form of regulation include the gene RIG-I (Gack et al., 2008), a master regulator of antiviral innate immunity (Yoneyama et al., 2004). RIG-I is the founding member of the RIG-I like Receptor (RLR) family, which functions to detect viruses containing RNA (and in some instances DNA) genomes in the cytosol of infected cells (Nakhaei et al., 2009). Upon binding to viral RNA, RIG-I engages an adaptor protein called MAVS to induce the expression of antiviral factors such as type I interferon (IFN) and IFN stimulated genes (ISGs) (Sun et al., 2006). RIG-I encodes a full-length transcript controlling this pro-inflammatory response as well as a truncated splice variant that limits the signaling potential of its counterpart (Gack et al., 2008). The Toll-like Receptor (TLR) adaptor proteins MyD88 and TRAM offer additional examples of this phenomenon; both of these genes encode splice variants that can differentially regulate an inflammatory response (Burns et al., 2003; Palsson-McDermott et al., 2009). Thus, alternative splicing is an established means of generating protein diversity and controlling the activity of immune signaling pathways.
An alternative method to generate protein diversity is through the process of translation, whereby distinct proteins can be created from a single mRNA. Although the alternative translation of polycistronic messages is generally considered to be virus or prokaryote specific, recent genome-wide ribosomal profile studies suggest that polycistronic mRNAs may be more common in eukaryotes than previously appreciated (Guttman et al., 2013; Ingolia et al., 2011). For example, embryonic stem cells contain thousands of mRNAs that are predicted to have more than one translational start site (Ingolia et al., 2011). However, whether these newly annotated start sites actually produce protein products that are functional and stable remains an unanswered question. In fact, there are very few bona fide examples in mammalian cells of more than one protein being produced by a single mRNA (Burkart et al., 2012; Cocka and Bates, 2012; Descombes and Schibler, 1991; Shinohara et al., 2008; Yin et al., 2002), and no example of this type of gene regulation exists in the common signaling pathways of the innate immune system.
In this report, we identify two regulators of antiviral innate immunity that are translated from the same bicistronic message. The transcript encoding the RLR adaptor protein MAVS produces the well-characterized full length (FL) MAVS adaptor and a truncated variant we refer to as miniMAVS. These proteins are functionally distinct and uniquely regulate antiviral signal transduction. Moreover, genome-wide ribosomal profiling in human monocytes identified additional innate immune regulators that contain multiple translation start sites. This study highlights how protein diversification in the innate immune system can be achieved at the level of translation and suggests that eukaryotic polycistronic messages may have widespread roles in controlling immunity.
Results
Identification of miniMAVS, a truncated variant of Full-length (FL) MAVS
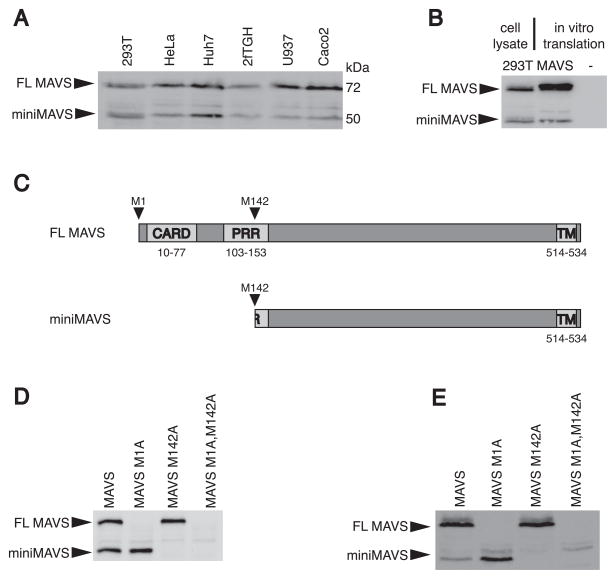
One of the original reports identifying the MAVS gene described the generation of a MAVS-specific antibody raised against a peptide consisting of amino acids 131-291 (Seth et al., 2005). This antibody detected two MAVS proteins with apparent molecular weights of 50 and 72 kilodaltons (kDa). It was speculated that the 50kDa variant represented a degradation product or processed version of the 72kDa full-length variant FL MAVS (Seth et al., 2005). To date, all antiviral activities of the MAVS gene have been attributed to FL MAVS (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). The origin and function of the smaller protein, miniMAVS, has yet to be defined. These two MAVS proteins can be detected in various human cell lines, indicating that the expression of both MAVS proteins is ubiquitous (Figure 1A). We considered the possibility that alternative mRNA splicing could explain the existence of a second MAVS variant. However, although several MAVS splice variants have been identified (Lad et al., 2008), none correspond to the correct size of miniMAVS (~50kDa) (data not shown). Additionally, both FL MAVS and miniMAVS were expressed from the MAVS coding region (CDS) by in vitro transcription and translation (Figure 1B). These data indicate that the two MAVS variants are generated from a single transcript and are therefore not generated by differential mRNA splicing.
Figure 1. miniMAVS is expressed from a second translational start site.
(A) Lysates from several different human cell lines were separated by SDS-PAGE and endogenous MAVS expression was detected with a MAVS antibody.
(B) in vitro transcription and translation of the MAVS CDS was compared with 293T cell lysates with an anti-MAVS antibody.
(C) Schematic of MAVS with predicted translation products FL MAVS and miniMAVS from the start sites corresponding to Met 1 and Met 142.
(D–E) Point mutations of translational start sites at Met 1 and Met 142 were made in the MAVS CDS and expressed in vitro (D) and in vivo (E) from MAVS deficient MEFs. The translation products were detected by immunoblot with a MAVS antibody.
See also Figure S1.
The presence of a methionine at amino acid 142 of the MAVS CDS suggested that miniMAVS expression is the result of translation initiation at an alternative start site (Figure 1C). Consistent with this hypothesis, initiation at this putative start codon (Met 142) would generate a protein corresponding to the molecular weight of miniMAVS (~50kDa) and share sequence homology with FL MAVS. To determine if Met 142 was required for the production of miniMAVS, we mutated the corresponding start sites by replacing the methionine with an alanine. Mutation of either the methionine at position 1 or the methionine at position 142 resulted in the respective loss of FL MAVS or miniMAVS expression in vitro (Figure 1D). Furthermore, stable expression of these mutant alleles in MAVS deficient mouse embryo fibroblasts (MEFs) had a similar expression pattern (Figure 1E). The putative start site corresponding to Met 142 of human MAVS is conserved among primates and other higher mammals (Figure S1A). In contrast, rodent MAVS sequences (e.g. ferret, guinea pig, mouse, rat and squirrel) do not contain a corresponding Met 142. Thus miniMAVS appears to have evolved later in evolution than the MAVS protein itself. These results suggest that the human MAVS transcript is bicistronic and that miniMAVS is the product of a unique open reading frame (ORF) downstream of the FL MAVS start site.
The MAVS mRNA is bicistronic and can produce two distinct proteins
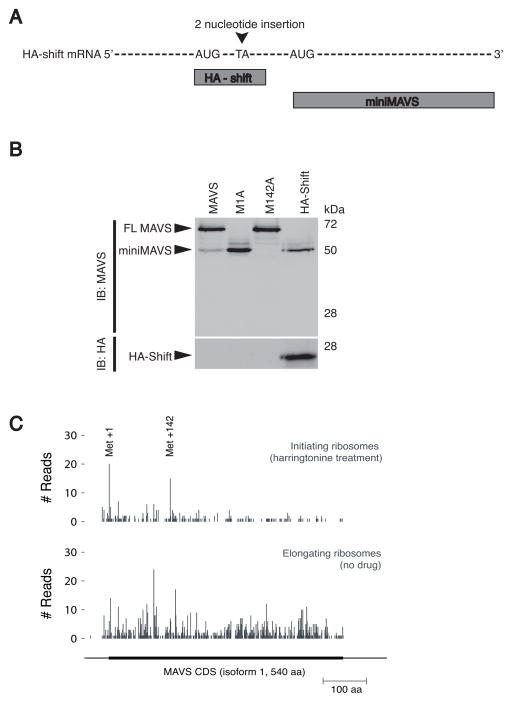
The methionine mutations described above suggest that miniMAVS expression is the result of alternative translation of a bicistronic MAVS transcript. If the MAVS transcript is truly bicistronic, then this mRNA should be capable of producing two distinct protein products that share no amino acid homology. To this end, a two-nucleotide insertion was introduced between the FL MAVS and miniMAVS start sites in a MAVS construct containing an amino-terminal HA epitope tag (Figure 2A). This insertion will shift the reading frame of HA-tagged FL MAVS, resulting in an altered amino acid sequence and a truncated protein called HA-shift. However, since the insertion is upstream of the miniMAVS start site, the reading frame and amino acid sequence of miniMAVS should not be affected. While the HA-shift protein could be detected by antibodies specific for the HA epitope tag, the shift in reading frame rendered the protein undetectable by the MAVS antibody (Figure 2B). Interestingly, this transcript still produced miniMAVS, as detected with the MAVS antibody (Figure 2B). The expression of these two distinct proteins from the same transcript demonstrates the bicistronic nature of the MAVS mRNA. Additionally, the frame-shift mutation rules out the possibility that miniMAVS is generated by post-translational proteolysis of FL MAVS.
Figure 2. MAVS is bicistronic and in vivo ribosome initiation is detected at the FL MAVS and miniMAVS start sites.
(A) Schematic of the HA-shift expression vector containing a frame shift mutation and the predicted translation products “HA-shift” and miniMAVS.
(B) Lysates from stable MEF lines expressing the MAVS and HA-shift constructs were separated by SDS-PAGE and protein expression was determined with MAVS and HA antibodies.
(C) Pattern of ribosome initiation (harringtonine treatment) and elongation on endogenous MAVS mRNA in 293T cells.
We next investigated the bicistronic nature of the endogenous MAVS mRNA inside of human cells. Ribosomal profiling of MAVS mRNA was performed in HEK293T cells. Ribosomal profiling is a strategy that utilizes deep sequencing of ribosome-protected mRNA fragments to investigate different aspects of translation (Ingolia et al., 2012; Ingolia et al., 2013; Ingolia et al., 2011). In conjunction with the drug harringtonine, which stalls ribosomes at initiation codons, this technique allows for the identification of functional translational start sites on endogenous mRNAs (Ingolia et al., 2012; Ingolia et al., 2013; Ingolia et al., 2011). In the absence of harringtonine, ribosomes were found throughout the open reading frame of MAVS, indicating active translation (Figure 2C). However, in the presence of harringtonine, ribosomes on the MAVS mRNA were predominately stalled at the two start sites we identified that correspond to methionine 1 and methionine 142 of MAVS (Figure 2C). Therefore, the same translational start sites that are required for FL MAVS and miniMAVS expression in vitro are sites of translation initiation on the endogenous MAVS mRNA in vivo. Taken together, these results establish that the MAVS mRNA is bicistronic and encodes for FL MAVS and miniMAVS by alternative translation of two distinct start sites.
Cis-acting elements that regulate the translation of miniMAVS
To further understand how the expression of FL MAVS and miniMAVS is regulated, we characterized cis-acting elements that control the expression of these variants. One mechanism by which downstream ORFs are expressed from a single transcript involves leaky ribosomal scanning through upstream start codons (Kozak, 2002; Somers et al., 2013). Typically, ribosomal scanning begins at the 5′ cap of a transcript and translation is initiated at the first optimal start site. Optimal translational start sites depend on the nucleotide context directly surrounding a start codon (Kozak, 1999). Leaky ribosomal scanning occurs when the start site is suboptimal and ribosomes fail to initiate translation (Kozak, 2002). Under these conditions, ribosomes will ‘leak’ through the initial start site, continue scanning along the mRNA, and initiate at a downstream start site. This mechanism predicts that the expression of downstream proteins is dependent on the translational context of upstream start sites.
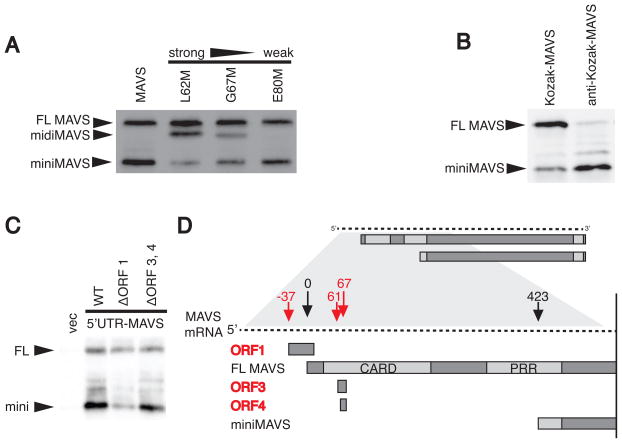
If miniMAVS expression requires leaky ribosomal scanning, then blocking ribosome scanning should decrease miniMAVS expression. To test this, we blocked ribosomal scanning by introducing a new start codon between the FL MAVS start codon and the miniMAVS start codon. Initiation at this new start codon would block scanning by translating a third protein, ‘midiMAVS’, thus preventing ribosomes from reaching the miniMAVS start site. Introduction of a new start codon in a position that has a naturally strong start context (L62M) suppressed miniMAVS expression (Figure 3A, lane 2). However, artificial start codons with weaker translational start contexts (G67M and E80M) were leaky, allowing ribosomes to proceed and more efficiently translate miniMAVS (Figure 3A, lanes 3 and 4). These results are consistent with the idea that miniMAVS expression relies on leaky ribosomal scanning from the FL MAVS start site to the miniMAVS start site.
Figure 3. Cis-regulatory elements of the MAVS transcript control miniMAVS expression.
(A) Translational start sites of varying strength were introduced at Leu62, Gly67, and Glu80 of the MAVS CDS to block ribosomal scanning between the FL MAVS and miniMAVS start sites. The constructs were expressed in vitro and the resulting MAVS products were detected by immunoblot with an anti-MAVS antibody.
(B) In vitro expression of two MAVS CDS constructs containing a strong (Kozak) or weak (anti-Kozak) translational context at the FL MAVS start site.
(C) Expression of the FL MAVS and miniMAVS in MAVS deficient MEFS transfected with expression constructs containing the endogenous 5′UTR of MAVS or constructs with mutated start sites for ORF1 or ORF3, 4.
See also Supplemental Table S1
(D) Schematic of the MAVS mRNA containing the endogenous 5′UTR and highlighting the 3 open reading frames (red) that are out-of-frame with FL MAVS and miniMAVS. Numbers indicate the distance (in nucleotides) each start site is from the FL MAVS start site.
Based on these data, the translational context of any upstream start site, including the FL MAVS start site, could affect the expression of miniMAVS. This possibility was addressed by placing an artificially strong (Kozak) and weak (anti-Kozak) translational context at the FL MAVS start site (Kozak, 2002). A strong translational context at the FL MAVS start site resulted in the high expression of FL MAVS compared to miniMAVS, whereas a weak translational context resulted in the lower expression of FL MAVS and high expression of miniMAVS (Figure 3B). These results establish the translational context surrounding the FL MAVS start site as a cis element that controls the expression of miniMAVS.
The above-described experiments all point to an important role for the endogenous 5′ untranslated region (UTR) of the MAVS transcript in controlling the expression of miniMAVS, as this region contains the natural translational context of the FL MAVS start site. To address this directly, a MAVS expression vector containing the endogenous 5′UTR was created. When MAVS-deficient MEFs were transiently transfected with this vector, both FL MAVS and miniMAVS were expressed (Figure 3C, lane 2) indicating that the endogenous context at the FL MAVS start site is sufficient for miniMAVS expression.
Examination of all the natural start codons present within the 5′UTR and coding region upstream of miniMAVS suggested a mechanism by which the FL MAVS start site is skipped en route to translating miniMAVS. Three additional start codons are present within this region including one in the 5′UTR (ORF1) coding for an out-of-frame upstream ORF (uORF) (Figure 3D). The translation of uORFs in 5′UTRs is emerging as a means by which translation of downstream ORFs can be regulated (Somers et al., 2013). For example, if initiation occurs at a uORF that overlaps with the start site of a canonical ORF, the translating ribosome will skip the start codon of the canonical ORF (Somers et al., 2013). After termination of uORF translation, the ribosome may resume scanning and re-initiate translation at downstream ORFs. ORF1 is an overlapping uORF, predicted to initiate the translation of a small peptide that overlaps with the coding region of FL MAVS, terminating past its start site (Figure 3D). We predicted that translation of ORF1 might allow ribosomes to bypass the FL MAVS start site, resume scanning, and re-initiate at the miniMAVS start site. To test this, the start site of ORF1 was mutated, as were the start sites for ORF3 and ORF4, which may create small peptides within the MAVS coding region (Figure 3D). The resulting constructs were then tested for the expression of FL MAVS and miniMAVS in MAVS deficient MEFs. Interestingly, mutating the start codon of ORF1 reduced the level of miniMAVS relative to FL MAVS, whereas mutating ORF3 and ORF4 had a minimal effect on miniMAVS expression (Figure 3C). These data suggest that ORF3 and ORF4 are likely bypassed by leaky scanning whereas translation of ORF1 allows ribosomes to skip the FL MAVS start site and facilitate the translation of miniMAVS, likely by re-initiation. However, because FL MAVS is expressed when ORF1 is present (Figure 3C), skipping of the FL MAVS start site cannot occur 100% of the time. We therefore speculated that leaky scanning might occur at the ORF1 start site, allowing for FL MAVS translation. Consistent with this hypothesis, the translational context at the ORF1 is suboptimal, suggesting a mechanism by which leaky scanning may occur (Supplemental Table 1). It should be noted however, that we cannot exclude the possibility that the mutation at the ORF1 start site influences the mRNA in additional ways (e.g. changes in secondary structure), which may contribute to altering the regulation of translation. Overall, our collective data reveal cis-regulatory elements in the 5′UTR of the MAVS transcript that explain the relative translation efficiency of FL MAVS and miniMAVS.
miniMAVS interferes with the signaling function of FL MAVS
Having established that the MAVS transcript encodes two proteins, we were interested in determining the role of each protein in the antiviral activities attributed to the MAVS gene. To study the respective signaling functions of FL MAVS and miniMAVS, we utilized the tools generated to characterize their expression. First, the start site mutations (Figure 1D, E) could be used to test each variant individually for their ability to activate a given cellular response. Second, changes in the translational context of the FL MAVS start site could be used to manipulate the expression ratio of FL MAVS to miniMAVS and determine how they function in conjunction. This latter point was of interest, as we noted a change in the ratio of FL MAVS to miniMAVS following viral infection (Figure S2A). Whereas FL MAVS became less abundant in infected cells over time, miniMAVS protein levels were not affected (Figure S2A). Thus, as the infection progressed, miniMAVS became the dominant MAVS variant in the cell.
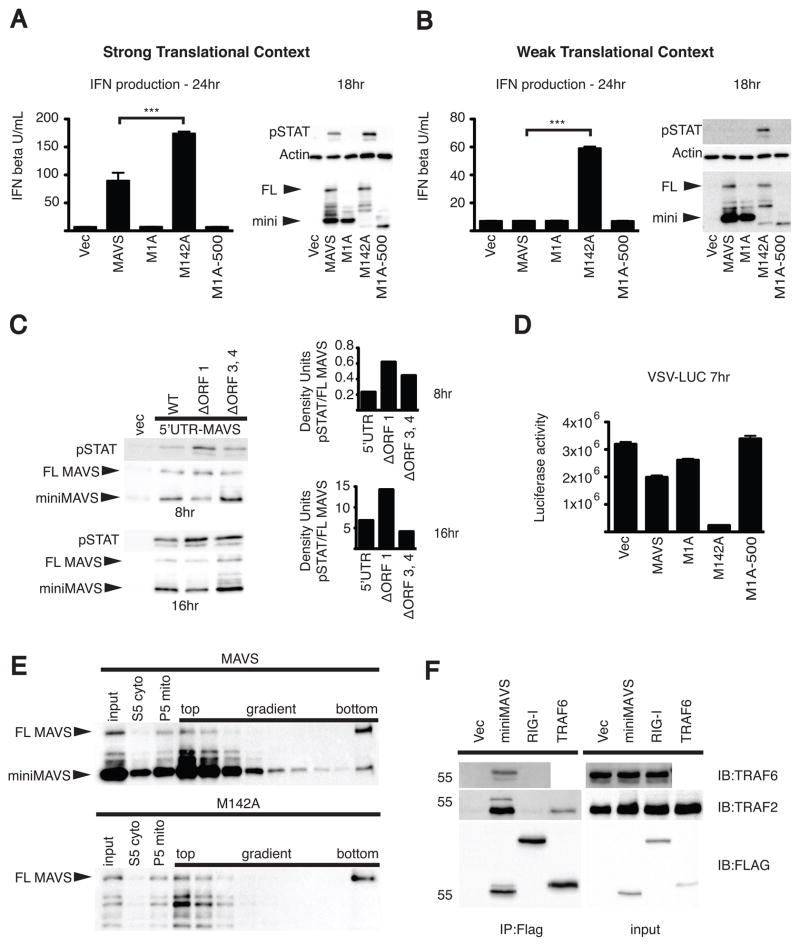
We began by overexpressing the MAVS start site mutants in 293T cells and measuring the resulting production of type I IFN. When only the FL MAVS variant (M142A) was expressed, robust production of type I IFN was observed (Figure 4A). Conversely, when only miniMAVS (M1A) was expressed there was no induction of type I IFN. In addition, a miniMAVS deletion mutant lacking the C-terminal localization signal (M1A-500) was not capable of inducing the production of IFN (Seth et al., 2005). This experiment suggests that FL MAVS is sufficient to positively regulate the production of IFN whereas miniMAVS is not sufficient to activate the pathway. However, when the two variants were expressed in conjunction (MAVS), there was a decrease in type I IFN production compared to FL MAVS expression alone (Figure 4A). To corroborate these findings, the phosphorylation of STAT1, an indicator of IFN signaling (Stark and Darnell, 2012), was monitored over the course of 24hrs following transfection. Compared to FL MAVS expression alone (M142A), cells expressing both MAVS variants (MAVS) contained lower levels of phosphorylated STAT1 over time (Figure 4A and Figure S2B). This difference in signaling activity between cells expressing FL MAVS alone and cells expressing both MAVS variants was not the result of differential expression of FL MAVS. Indeed, western analysis indicated comparable expression of FL MAVS when expressed alone (M142A) or when expressed in conjunction with miniMAVS (MAVS) (Figure 4A and Figure S2B). Taken together, these results suggest that miniMAVS antagonizes the signaling function of FL MAVS and inhibits IFN production.
Figure 4. FL MAVS-dependent IFN production is inhibited by miniMAVS.
(A–B) The MAVS dependent antiviral response was measured by IFN bioassay and STAT1 phosphorylation. The MAVS CDS and translational start point mutants were transfected into 293T cells using vectors with a strong translational context (A) as well as a weak translational context (B). FL MAVS and miniMAVS expression is shown by MAVS immunoblot.
(C) STAT1 phosphorylation at 8 and 16hrs following the transient expression of MAVS with the endogenous 5′UTR or uORF point mutants in 293T cells. The ratio of STAT1 phosphorylation to FL MAVS expression was quantified by densitometry. Densitometry is from a representative image of an experiment done in triplicate.
(D) 293T cells were transfected with the MAVS constructs from 4B for 24 hours and then infected with VSV encoding firefly luciferase. Luciferase activity was determined 7 hours following VSV infection.
(E) Crude mitochondria (P5) isolated from 293T cells transfected with MAVS or the M142A point mutant were separated by sucrose gradient ultracentrifugation. FL MAVS oligomers segregated to the bottom of the gradient (right) and were detected by SDS-PAGE followed by immunoblot.
(F) 293T cells were transfected with Flag-tagged miniMAVS, RIG-I or TRAF6 and Flag-immunoprecipitates were probed for endogenous TRAF2 and TRAF6.
*** p<0.001 by ANOVA with Tukey’s multiple comparison test. Error bars represent SD.
See also Figure S2.
To more directly test the hypothesis that miniMAVS can inhibit the production of IFN, we used an expression construct with a weak translational context at the FL MAVS start site. Due to leaky scanning, this weak translational context would increase the ratio of miniMAVS to FL MAVS in the cell, and we hypothesized that this increase in ratio would further inhibit the production of type I IFN. A weak translational context resulted in higher abundance of miniMAVS relative to FL MAVS when both variants were expressed (MAVS, Figure 4B), as compared to the experiments using MAVS with a strong translational context (Figure 4A). Remarkably, when both variants were expressed in conjunction (MAVS), the effect was a complete abrogation of IFN production and STAT1 phosphorylation (Figure 4B and Figure S2C). However, under the same conditions, when FL MAVS was expressed alone (M142A), a robust production of IFN and STAT1 activation was observed (Figure 4B and Figure S2C). Taken together, these data reveal miniMAVS as an inhibitor of FL MAVS signaling and that the ratio of FL MAVS to miniMAVS determines whether an antiviral response will occur.
To further test miniMAVS inhibition of IFN signaling, expression constructs that more closely mimicked the natural MAVS transcript were examined. We assessed the activation of IFN signaling following expression from constructs containing the endogenous 5′UTR and MAVS CDS. As described earlier, ORF1 in the 5′UTR of the transcript can regulate the expression of miniMAVS, and when mutated there is a decrease in miniMAVS expression (Figure 3C). Due to this, we hypothesized that mutating the ORF1 start site would increase IFN production. Consistent with this idea, when compared to the WT 5′UTR construct, expression of the uORF1 mutant resulted in increased STAT1 activation (Figure 4C). These data further establish that miniMAVS can interfere with the FL MAVS IFN response and identify uORF1 as a regulator of both the expression and function of miniMAVS.
Having established regulatory effects of FL MAVS and miniMAVS on antiviral signaling, we predicted that differential expression of the two proteins would also affect viral replication. While the expression of miniMAVS alone (M1A) had little effect on the replication of vesicular stomatitis virus (VSV), FL MAVS expression alone (M142A) dramatically reduced VSV replication (Figure 4D). Interestingly, expression of the two proteins in conjunction (MAVS, as in Figure 4B) was less effective at limiting viral replication as compared to expression of FL MAVS alone (Figure 4D). These data therefore establish that miniMAVS acts to restrict the signaling functions of FL MAVS, the physiological consequence of which is that FL MAVS is less able to create an antiviral cellular state.
During viral infections, large aggregates of FL MAVS form that recruit downstream enzymes to promote the expression of type I IFNs (Hou et al., 2011). It was therefore possible that miniMAVS restricts the signaling functions of FL MAVS by preventing the formation of these large protein aggregates. To address this possibility, FL MAVS was expressed alone or in conjunction with miniMAVS, and FL MAVS aggregates were detected following sucrose gradient ultracentrifugation. For these studies, the expression constructs containing the weak translational context from Figure 4B were used, as under these conditions, miniMAVS completely abrogated the production of IFN. When FL MAVS alone (M142A) was expressed, aggregates of FL MAVS could be detected at the bottom of the sucrose gradient (Figure 4E). This was expected because the expression of FL MAVS results in the production of IFN (Figure 4B), and it is thought that IFN signaling is a result of MAVS aggregation (Hou et al., 2011). Interestingly when both FL MAVS and miniMAVS were expressed in conjunction (MAVS), we also detected the formation of FL MAVS aggregates (Figure 4E). This was surprising, because under these conditions, miniMAVS completely blocks the production of IFN (Figure 4B). These data suggest that miniMAVS cannot block FL MAVS aggregate formation, even under conditions where the signaling functions of FL MAVS are completely prevented. Consistent with the idea that miniMAVS does not influence the aggregate-forming activity of its full length counterpart, we found that in response to Sendai virus infections, endogenous miniMAVS does not co-sediment with FL MAVS aggregates (Figure S2D). Thus, miniMAVS is neither a component of FL MAVS aggregates nor does it regulate their formation.
Aggregates of FL MAVS promote antiviral signaling by recruitment of the E3 ubiquitin ligases TRAF2 and TRAF6 (Liu et al., 2013). Since miniMAVS was not capable of blocking FL MAVS aggregation, we hypothesized that it may interfere with signal transduction by interacting with these downstream signaling proteins. To test this, we used a Flag-tagged miniMAVS expression vector and tested Flag-immunoprecipitates for the presence of endogenous TRAF2 and TRAF6. Both endogenous TRAF proteins interacted specifically with Flag-miniMAVS as compared to Flag-tagged RIG-I or a vector control (Figure 4F). Flag-tagged TRAF6 formed a modest complex with endogenous TRAF2. Additionally, when Flag-miniMAVS was co-expressed with HA-TRAF6 or HA-TIRAP, TRAF6 was detected in the Flag-immunoprecipitates whereas the Toll-like Receptor adaptor TIRAP was largely absent (Figure S2E). Taken together, these data indicate that miniMAVS forms a complex with TRAF proteins that are known to promote antiviral signaling and IFN production. A possible mechanism of miniMAVS function may therefore be proposed whereby two protein complexes exist that contain MAVS. One complex consists of FL MAVS aggregates and TRAF proteins, and is capable of activating type I IFN expression (Liu et al., 2013). The second complex consists of miniMAVS and the same TRAFs (Figure 4F and S2E), yet is incapable of activating type I IFN expression. The regulation of the functional competition between these two complexes remains an open area of inquiry.
miniMAVS positively regulates cell death
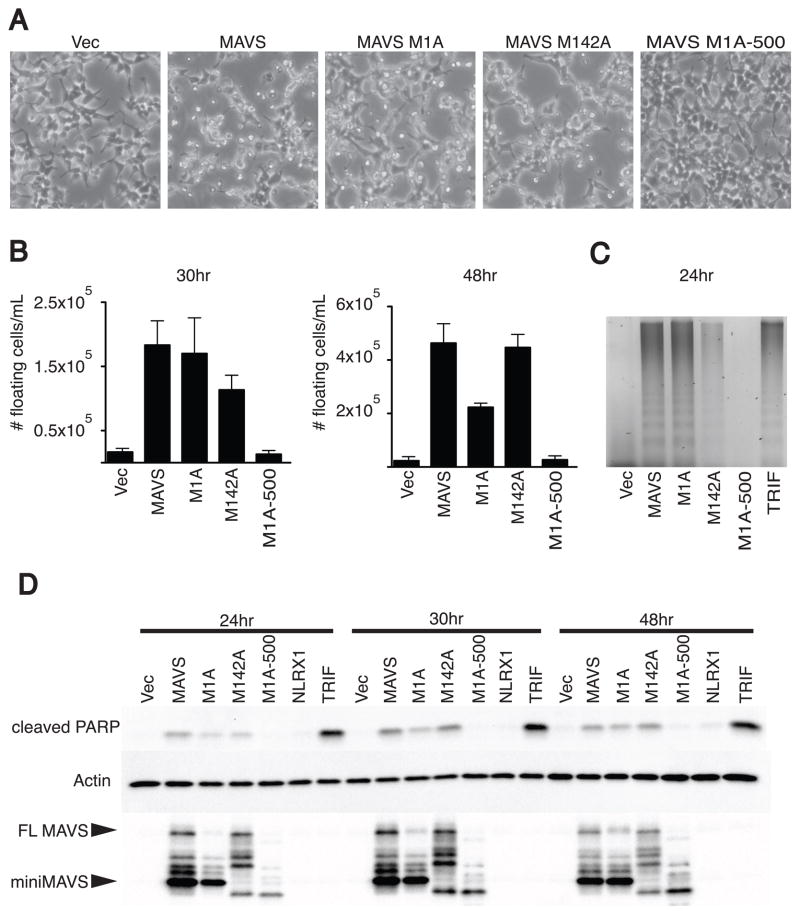
In addition to activating antiviral gene expression, MAVS can promote cell death upon overexpression or in response to certain viral infections (Lei et al., 2009). As with the IFN response, the role of miniMAVS in cell death is unknown. To test whether either MAVS variant is sufficient to activate cell death, we determined if overexpression of either variant was sufficient to kill transfected 293T cells. When both variants were overexpressed in conjunction (MAVS), there were visible signs of cell death compared to cells transfected with a vector control (Figure 5A). Interestingly, when miniMAVS (M1A) or FL MAVS (M142A) were expressed alone, we also observed signs of cell death. Quantification of the number of cells that detached from the tissue culture plate revealed that FL MAVS and miniMAVS induce comparable amounts of cell death at 30 hours following transfection (Figure 5B, left panel). However by 48 hours, cell death induced by FL MAVS exceeded that of miniMAVS (Figure 5B right panel). The increase in cell death induced by FL MAVS may be the result of secreted IFNs, which can positively influence cell death (Chawla-Sarkar et al., 2003). Interestingly, a miniMAVS deletion mutant lacking the C-terminal localization domain (M1A-500) did not show signs of cell death compared to the vector control (Figure 5A, B). Based on these data, we hypothesized that miniMAVS may function to positively regulate cell death in a localization-dependent, but IFN-independent manner.
Figure 5. miniMAVS is sufficient to induce cell death.
(A) Micrographs of 293T cells 48hrs following transfection with MAVS and start site point mutant expression vectors.
(B) Subsequent measurements were made at 30 and 48hrs following transfection to quantify the density of floating cells in the media. Transfection of MAVS and the start site mutants were done in triplicate. Error bars represent SD.
(C) Detection of fragmented genomic DNA was performed 24 hours following transfection of various MAVS constructs and TRIF and samples were separated on a 2% agarose gel.
(D) Cell lysates were collected at 24, 30, 48 hours post transfection of MAVS, the translational start point mutants, NLRX1, and TRIF. PARP cleavage and MAVS expression was determined by immunoblot following SDS-PAGE.
To further investigate the induction of cell death by FL MAVS and miniMAVS we assessed whether they can induce two hallmarks of this process. Programmed cell death, including apoptosis and necroptosis, is often characterized by the fragmentation of genomic DNA (Green and Reed, 1998). Both miniMAVS (M1A) and FL MAVS (M142A) induced the fragmentation of genomic DNA following expression in 293T cells (Figure 5C). In support of our visual observations of cell death, the miniMAVS mutant (M1A-500) lacking the localization signal was not capable of inducing DNA fragmentation. As a control, we monitored DNA fragmentation induced by a known regulator of cell death, the TLR adapter TRIF (Figure 5C) (Han et al., 2004; Ruckdeschel et al., 2004). Prior to the commitment toward cell death and DNA fragmentation, caspases become activated and subsequently cleave a variety of target substrates to carry out apoptosis (Green and Kroemer, 1998). PARP is one of the targets of these activated caspases, making detection of the cleaved product of PARP a reliable marker for cell death. To further investigate the induction of cell death by FL MAVS and miniMAVS, we assessed whether they can induce PARP cleavage individually or in conjunction. At several time points following the expression of both variants in conjunction (MAVS), the cleaved product of PARP was observed (Figure 5D). Again, TRIF was used as a positive control for cell death to monitor PARP cleavage. In agreement with our DNA fragmentation results, PARP cleavage was detected in cells individually expressing either miniMAVS (M1A) or FL MAVS (M142A) but not cells expressing the improperly localized miniMAVS mutant M1A-500. These data indicate that unlike their antagonizing activities towards IFN expression, FL MAVS or miniMAVS can both promote PARP cleavage and cell death. While the MAVS localization domain directs this adaptor to mitochondria and peroxisomes (Dixit et al., 2010; Horner et al., 2011), the central role of mitochondria in programmed cell death lead us to speculate that the death-inducing signal from MAVS probably emerges from this organelle (Green and Kroemer, 1998). When another mitochondrial protein (NLRX1) (Moore et al., 2008) was examined in the PARP cleavage assay, no PARP cleavage was observed (Figure 5D). Therefore, the observed cell death phenotype is specific to FL MAVS and miniMAVS, and is not a general response to ectopic expression of another mitochondrial membrane protein. Thus, in addition to their antagonistic actions in regulating IFN expression, FL MAVS and miniMAVS can each promote the cell death response.
Ribosomal profiling predicts a class of bicistronic mRNAs that encode regulators of innate immunity
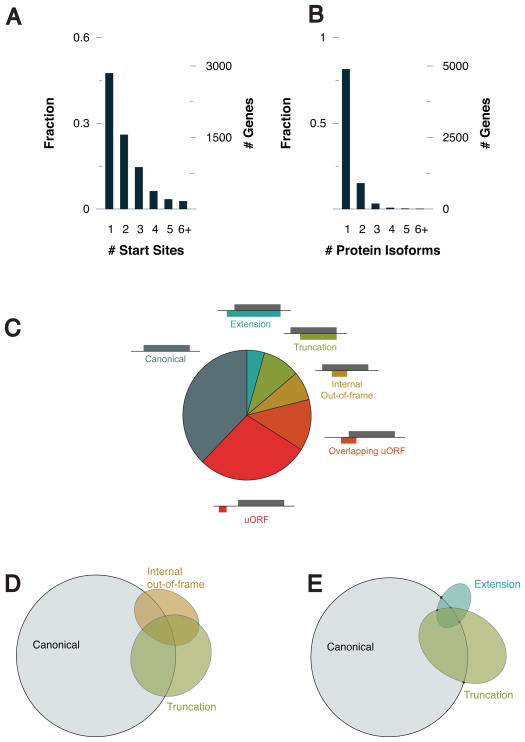
MAVS can now be added to a small list of eukaryotic genes known to produce bicistronic transcripts. However, based on previous ribosomal profiling studies in embryonic stem cells (Ingolia et al., 2011), there may be many more examples that exist but have yet to be identified. To determine if other regulators of antiviral innate immunity encode bicistronic transcripts, we carried out ribosomal profiling in U937 cells, a human monocyte cell line. In the presence of harringtonine we identified 14,336 start sites on 8893 transcripts (Supplemental Table 2). Many transcripts present in U937 cells had more than 1 start site (Figure 6A, B). These start sites consist of a number of different classes relative to the reading frame of the annotated CDS. These include start sites for the canonical CDS, uORFs, internal out-of-frame products, truncations, and extensions (Figure 6C). Because our work with MAVS has highlighted the importance of protein diversification via alternative translation, we focused our analysis on transcripts with start sites that resulted in variant protein isoforms such as truncations and extensions. These alternatively translated products are of particular interest, because they can either lose or gain a functional domain relative to the canonical CDS. MAVS is a clear example of this mode of regulation, since the truncation miniMAVS lacks the CARD domain present in the amino terminus of FL MAVS. In addition, our profiling data indicate that truncations are more prevalent than would be expected from random chance. Based on the use of triplet codons, a third of possible start codons would be in-frame with the canonical ORF and two thirds would be out-of-frame. If start site selection were random, we would expect a 1:2 ratio of truncations to internal out-of-frame ORFs. However we observed about a 4:3 ratio in the favor of truncations (Figure 6D). Additionally, truncations appear to be more frequent than extensions, suggesting these variants may have more biological significance (Figure 6E).
Figure 6. Ribosomal profiling of human monocytes identifies extension and truncation variants similar to miniMAVS.
(A) The fraction and number of genes that were detected to have one or more translational start site.
(B) The fraction and number of genes that have more than one translational start site resulting in either an extension or truncation.
(C) Classification of each of start site relative to the reading frame of the annotated CDS.
(D) Venn diagram showing the number of genes identified containing one or more canonical, truncation, or internal out-of-frame start site.
(E) Venn diagram showing the number of genes identified containing one or more canonical, truncation, or extension start site.
From the list of potential truncations, we chose several genes involved in antiviral immunity to further investigate. The patterns of ribosomal profiling indicate that like MAVS, IFIH1 (also known as MDA-5), MX2, IFITM2, and TRIM25 might also encode for truncated protein variants (Figure S3A-D). However, other genes related to antiviral immunity such as DDX58 (RIG-I) and TMEM173 (STING) were not identified as having truncations (Figure S3E, F). While additional work is needed to verify the abundance and function of these predicted protein variants, this analysis highlights the potential existence of a class of bicistronic regulators of antiviral innate immunity.
Discussion
Regulation of innate immune signal transduction is crucial for many aspects of cell and organ physiology during infection and homeostasis. Eukaryotes often regulate signaling pathways by producing proteins with diverse function through gene-extrinsic means (different genes encoding different regulators) or gene-intrinsic means (alternative splice variants encoding different regulators). The use of mRNA-intrinsic protein diversification strategies (multiple translation products from the same transcript) has traditionally been characterized for prokaryotes and viruses (Alberts, 2008; Powell, 2010). Our finding that MAVS encodes a bicistronic mRNA that produces proteins with different functions represents a novel means by which protein diversity can be generated in the innate immune system.
Several lines of evidence support our conclusion that the MAVS transcript is bicistronic. 1) The cDNA of MAVS can produce both FL MAVS and miniMAVS, and the molecular weight of miniMAVS does not correspond to that of any possible product of alternative splicing. 2) Profiling of ribosomes arrested at translational start sites within the endogenous MAVS mRNA revealed two start codons. These start codons are predicted to produce proteins of the size corresponding to FL MAVS and miniMAVS. When these start codons were mutated, the resulting transcripts lost the ability to produce the corresponding MAVS variant. 3) Shifting the reading frame of the MAVS coding sequence at a site between these two start sites resulted in the production of two distinct protein products (HA-shift and miniMAVS). Since FL MAVS is not produced under these conditions, the existence of miniMAVS cannot be explained by proteolytic cleavage of the full length protein. Collectively, the above observations can only be explained by the conclusion that FL MAVS and miniMAVS are produced from a bicistronic mRNA encoded by the MAVS gene.
Our genome-wide ribosome profiling analysis suggests that FL MAVS and miniMAVS are not the only regulators of innate immunity that are encoded by a bicistronic mRNA. Indeed, this analysis revealed the existence of hundreds of transcripts that are predicted to encode more than one protein product. This result is consistent with prior work in embryonic stem cells (Ingolia et al., 2011), which first suggested that polycistronic mRNAs are prevalent in mammalian cells. Our analysis in immune cells indicates that this feature of mammalian mRNAs is not unique to stem cells. Rather, we suggest that polycistronic mRNAs may be used to diversify protein function in differentiated cells as well. Interestingly, our analysis suggests the existence of additional bicistronic regulators of antiviral immunity. While much work needs to be done to verify the existence and function of any predicted product of alternative translation, the ability of ribosome profiling approaches to predict the existence of FL MAVS and miniMAVS strengthens confidence in these analyses.
Our functional studies of miniMAVS revealed that FL MAVS and miniMAVS antagonize one another and that the strength of antiviral gene expression induced by MAVS is the result of the collective actions of these two MAVS variants. We suggest that a competition exists within cells at the level of the MAVS proteins, and the relative abundance of each variant may determine the signaling potential of the RLR pathway. Mechanistically, this competition may be occurring at the level of interactions with downstream TRAF proteins. Evidence in support of this suggestion comes from our studies indicating that miniMAVS, like its full length counterpart (Hou et al., 2011; Liu et al., 2013), can form a complex with TRAF2 and TRAF6. It will be important to further characterize the means by which TRAFs can be recruited into functionally distinct protein complexes consisting of either FL MAVS and miniMAVS. Interestingly, in our attempts to understand the inhibitory role that miniMAVS plays during IFN production, we noted that results were less reliable when the variants were expressed in trans. Whether this indicates a requirement for expression in cis or rather a limitation of experimental design was not determined. It is interesting to speculate that the expression of regulatory variants from bicistronic transcripts may have evolved based on a requirement for expression in cis.
Due to the importance of MAVS in controlling viral infections (Kumar et al., 2006; Sun et al., 2006), one might have expected that the MAVS mRNA would have evolved to contain a highly efficient translational start site. However, our analysis of cis-acting sequences in the MAVS transcript revealed otherwise. The nucleotides surrounding the FL MAVS start codon do not fit the classic definition of a Kozak sequence (Kozak, 1986; Kozak, 1987), and FL MAVS translation appears to be limited by the actions of a uORF that overlaps with the start codon of FL MAVS. Therefore, uORF-mediated start codon skipping, a mode of regulation previously found to maintain the polycistronic nature of the mRNA encoding the transcription factor C/EBP (Calkhoven et al., 2000; Raught et al., 1996), may facilitate the expression of miniMAVS. We suggest that (in advanced mammalian species) the endogenous 5′UTR of the MAVS transcript evolved not to maximize the translation of FL MAVS, but rather to balance the production of FL MAVS with the production of its downstream miniMAVS partner.
Based on these data, we suggest that the gene encoding FL MAVS and miniMAVS is functionally analogous to a bacterial operon, in which multiple proteins can be produced from a single mRNA that collectively control a cellular activity. The fact that FL MAVS and miniMAVS participate in both the antiviral and cell death responses of human cells suggests that this mammalian operon may be particularly important in maintaining tissue homeostasis before, during and after infections. These discoveries provide a mandate to consider the functions of additional polycistronic regulators of innate immunity and indicate that even “well-characterized” genes have much to reveal in terms of their functions in health and disease.
Experimental Procedures
Cloning and generation of MAVS mutants
The MAVS CDS from allele BC044952 was a gift of ZJ Chen (UTSW). Variants were cloned into a pcDNA3 vector containing an N-terminal HA tag. Variants were cloned with (strong translational context) or without (weak translational context) the N-terminal tag. Both used the same C-terminal restriction site XhoI, primer: AAAAACTCGAGCTAGTGCAGACGCCGCCGGTACAGC. The strong translational context variants were inserted into the vector with KpnI, fwd primer: AAAAAGGTACCGCACCGTTTGCTGAAGACAAGACCTAT. The translational context at the HA start codon of this vector: AAGCTTACGATGG. The weak translational context variants were inserted with HindIII, which removed the HA tag, fwd primer: TTTTTAAGCTTATGCCGTTTGCTGAAGACAAGACCTAT. The translational context at the start codon of this vector: CCCAAGCTTATGC. For the Kozak and anti-Kozak constructs the following sequences were placed directly upstream of the FL MAVS start codon: GCCGCCACC and ATATATTTT. The sequence used to generate the 5′UTR MAVS constructs is listed in the Ensemble database under transcript ID number ENST00000428216. The HA-shift construct was made by inserting two nucleotides ‘TA’ at bp number 254 of the MAVS CDS with the fwd primer: GTGAGCTAGTTGATCTCGTACGGACGAAGTGGCCTCTGTC. Stable MAVS cell lines were generated with pMSCV2.2 IRES GFP in MAVS deficient MEFs.
MAVS expression, antibodies, type I IFN bioassay, and viral infections
MAVS in vitro expression was performed using a coupled transcription and translation rabbit reticulocyte lysate kit (Promega) with a T7 pcDNA3 expression vector. MAVS was expressed in 293T cells cultured in DMEM, 10% serum by Fugene 6 (Promega) mediated transfection of pcDNA3 expression constructs. The antibodies used for western blots were MAVS (Bethyl Labs A300-782A), pSTAT (BD 612132), PARP (BD 611038), HA (Roche 3F10), Flag (Biolegend 637301) TRAF2 Cell Signaling (C192), TRAF6 Abcam 33915. The type I IFN bioassay was performed as previously described (Dixit et al., 2010). Statistics were performed using PRISM (Graphpad). Cells were infected with 50 U/mL of SeV or an MOI = 1 for VSV firefly luciferase. 3XFlag-miniMAVS was immunoprecipitated with an M2-affinity gel from Sigma and eluted with a FLAG peptide.
Detection of DNA fragmentation
Fragmented genomic DNA was observed by agarose gel electrophoresis following phenol chlororform extraction (Matassov et al., 2004).
Detection of FL MAVS oligomers by sucrose gradient ultracentrifugation
Sucrose gradient ultra centrifugation was performed as previously described (Hou et al., 2011). Briefly, 5x10^5 293T cells were plated in 10cm dishes and transfected with MAVS expression vectors. Ten hours following transfection cells were lifted and lysed by dounce homogenization. A P5 crude mitochondrial pellet was obtained and solubilized in 1% DDM (Hou et al., 2011). Soluble mitochondria were then loaded onto a 30–60% sucrose gradient and centrifuged for two hours at 170,000g 4°C. Fractions were then removed from the gradient with the bottom fraction containing MAVS oligomers.
Ribosomal Profiling
U937 cells were left untreated or treated for 5 minutes with 2ug/mL of harringtonine then immediately lysed. Ribosomal profiling and analysis was then carried out as previously described (Ingolia et al., 2012).
Supplementary Material
Research Highlights.
The MAVS mRNA is bicistronic and codes for MAVS and a truncated variant, miniMAVS
Cis-regulatory elements in the 5′ UTR determine the ratio of MAVS variants produced
miniMAVS restricts MAVS-induced antiviral responses; both proteins induce cell death
Ribosome profiling reveals additional polycistronic mRNAs in the innate immune system
Acknowledgments
We would like to thank Priya Dadd and members of the Horng lab for experimental help. We would like to thank Steen Hansen for reagents and advice. Finally, we are indebted to the members of the Kagan lab for experimental help and invaluable advice. The NIH grants AI093589, AI072955, P30DK34854 and an unrestricted gift from Mead Johnson & Company support the work performed in the lab of J. Kagan. J.K. and A.G. are also supported by the Bill and Melinda Gates Foundation (OPP1066203). N.I. is supported by the Searle Scholars Program. Dr. Kagan holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts B. Molecular biology of the cell. 5. New York: Garland Science; 2008. [Google Scholar]
- Burkart C, Fan JB, Zhang DE. Two independent mechanisms promote expression of an N-terminal truncated USP18 isoform with higher DeISGylation activity in the nucleus. J Biol Chem. 2012;287:4883–4893. doi: 10.1074/jbc.M111.255570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, Borden EC. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- Cocka LJ, Bates P. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog. 2012;8:e1002931. doi: 10.1371/journal.ppat.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D, Kroemer G. The central executioners of apoptosis: caspases or mitochondria? Trends Cell Biol. 1998;8:267–271. doi: 10.1016/s0962-8924(98)01273-2. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Genome-wide annotation and quantitation of translation by ribosome profiling. Curr Protoc Mol Biol. 2013;Chapter 4(Unit 4):18. doi: 10.1002/0471142727.mb0418s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Yang G, Scott DA, Chao TH, da Correia JS, de la Torre JC, Li E. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol Immunol. 2008;45:2277–2287. doi: 10.1016/j.molimm.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Lei Y, Moore CB, Liesman RM, O’Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, Gong M, Golenbock D, Espevik T, O’Neill LA. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- Powell ML. Translational termination-reinitiation in RNA viruses. Biochem Soc Trans. 2010;38:1558–1564. doi: 10.1042/BST0381558. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, James A, Medina D, Sonenberg N, Rosen JM. Expression of a translationally regulated, dominant-negative CCAAT/enhancer-binding protein beta isoform and up-regulation of the eukaryotic translation initiation factor 2alpha are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res. 1996;56:4382–4386. [PubMed] [Google Scholar]
- Ruckdeschel K, Pfaffinger G, Haase R, Sing A, Weighardt H, Hacker G, Holzmann B, Heesemann J. Signaling of apoptosis through TLRs critically involves toll/IL-1 receptor domain-containing adapter inducing IFN-beta, but not MyD88, in bacteria-infected murine macrophages. J Immunol. 2004;173:3320–3328. doi: 10.4049/jimmunol.173.5.3320. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shinohara ML, Kim HJ, Kim JH, Garcia VA, Cantor H. Alternative translation of osteopontin generates intracellular and secreted isoforms that mediate distinct biological activities in dendritic cells. Proc Natl Acad Sci U S A. 2008;105:7235–7239. doi: 10.1073/pnas.0802301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J, Poyry T, Willis AE. A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol. 2013;45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.