Abstract
The analysis of genome-scale sequence data can be defined as the interrogation of a complete set of genetic instructions in a search for individual loci that produce or contribute to a pathological state. Bioinformatic analysis of sequence data requires sufficient discriminant power to find this needle in a haystack. Current approaches make choices about selectivity and specificity thresholds, and the quality, quantity and completeness of the data in these analyses. There are many software tools available for individual analytic component-tasks, including commercial and open source options. Three major types of techniques have been included in most published exome projects to date: frequency/population genetic analysis, inheritance state consistency, and predictions of deleteriousness. We discuss the required infrastructure and use of each technique during analysis of genomic sequence data for clinical and research applications. Future developments will alter the strategies and sequence of using these tools and are speculated on in the closing section.
Keywords: exome, Mendelian, next generation sequencing, bioinformatics, clinical sequencing
INTRODUCTION
Brief introduction to technology
Genome-scale sequencing (whole genome, exome, custom capture) is the result of the parallel production of a large number of short length sequences, or reads. Each short read must be mapped back to a genomic position. Methods for mapping short reads include the creation of contigs by assembly of overlapping sequences, gap-and/or-mismatch-tolerant alignment to a reference genome, or a combination of both. The short reads are compared to a reference sequence. Differences are detected, annotated and compiled into a list. The minimal working files for a genome-scale sequencing project include a structured, compressed file of aligned short reads plus a second file containing potential differences between the test subject and the given reference sequence.
We can define the problem of analyzing genome-wide data as part of a general class of problems of searching a large data space for a small number of candidate answers. Thus, there is a small signal size relative to the search space. As a result, the overall signal to noise ratio, even for low average noise levels, is problematic. Optimization of this process requires examination of the quality of the entire data set and recognition of the magnitude and locations of the types of noise present. Successful analysis involves being able to restrict the search space by the use of reduction “filters” that reject a very large amount of noise and still retain the small amount of true signal.
A second major challenge is characterization of the relationship between any given sequence variant and an organism-level phenotypic trait or traits. Common phenotypes can be studied using adequately-powered case-control studies and/or large pedigrees containing individuals with the trait. Definitive causation is more difficult to prove for rare phenotypes. In particular, determining causation is difficult when gene-phenotype relationships have not been previously established and when too few cases have been identified to allow epidemiological methods to reach statistical significance.
The total genome size for all humans is roughly 6.4x109 bases. The “signal” (the causative DNA variation) can be as small as a single base or as large as whole chromosome aneuploidy. Since large scale genomic alterations have had reliable karyotype and microarray tools available for decades, we restrict our discussion to the analysis of small scale genetic changes, on the order of a few exons or less, a range of 0.001Kb to 10Kb.
Targeted versus Non-Targeted Sequencing
Most current genome-scale sequencing utilizes one of two basic types of strategies: targeted and non-targeted. Both begin with a random fragmentation of a quantity of genomic DNA that contains multiple copies of the genome. This can be from samples like whole blood, mixed genome samples such as human saliva (containing both human and micro-organismal DNA), non-blood tissues, cultured tissues and even genomes amplified from a single cell. The purified DNA is sheared at random places to yield fragments that, if reassembled correctly, would allow overlapping fragment contigs to regenerate the original intact sequence of each chromosome. At this point, targeted strategies such as exome sequencing and non-targeted strategies such as whole genome sequencing diverge. In the targeted approach, a genomic DNA subset is selected by non-stringent hybridization to immobilized “bait” sequences. Non-hybridized fragments are then washed away. The baits can be customized to include any genomic subset of interest. Common examples include exomes and single chromosome regions. Non-targeted strategies do not select for a genomic subset; in ideal conditions the entire genome is included.
Sequencing
Once a library of fragments is generated, the individual fragments are sequenced, either by synthesis in parallel spatially separated microscopic clusters, polonies or other physical processes or by single molecule detection devices. The end result is a file of short reads that are each a small length (1 × 10-5) relative to the entire intact chromosome sequence. These short reads are typically stored in a FASTQ file format.
Alignment
All current modern and economically efficient techniques use alignment reconstruction, aligning individual reads to a pre-existing reference genomic sequence. An alternate technique, de novo assembly, has been explored on a research basis (Simpson and Durbin, 2012). Aligned short reads are stored in a standard Sequence Alignment/Map (SAM) file format, typically in compressed (BAM) form. An accompanying sorted BAM file index (BAI) file allows for rapid data access for processing and viewing.
Genotyping
Once the short reads are aligned to a reference genome, genotypes are called at each genomic position for which an adequate number of short reads have aligned or “piled up”. Various probabilistic models are used to determine the most likely genotype at positions where the short-reads contain a non-reference base. The most common approach uses a Bayesian algorithm conditioned on an estimated probability of variation at the given chromosomal position. Called variations are often stored in a standard Variant Call (VCF) file.
All the steps in sample preparation and sequencing can cause dropout of fragments or failure to generate fragments in some regions of the genome, in both random and systematic ways throughout the genome. Sources of systematic error include regions with high GC content (or other properties specific to the primary sequence) that interfere with the process of uniform and complete library generation/sequencing. Such errors degrade the quality of the sequence for the first exons in many genes. Amplification errors may lead to problems with allele drop out or allele skewing, which is reflected in a large difference in the expected 0.5 ratio of short reads between two different bases at a heterozygous position. Low amplification approaches to library generation can reduce this type of error, but are not currently available for most capture techniques like exome sequencing. They are in use for whole genome sequencing.
Annotation
The final step of genome-scale sequencing is annotation. Annotation is the process of combining information about individual variants with a registration of their position relative to known genes. Variants may need to be defined in the context of several potential transcripts. Other common annotations include an estimate of the variation's pathogenic potential (potential to disrupt protein function), the frequency of the variation in available populations, and the predicted consequences of the variation (deletion, insertion, missense, etc.). Annovar and SeattleSeq are examples of publically available annotation programs; several proprietary programs are also available (Wang et al., 2010) (http://gvs.gs.washington.edu/SeattleSeqAnnotation/). Different collections of gene transcripts such as Ensembl, UCSC Known Genes and Refseq are used or can be selected during annotation (Flicek et al., 2012; Hsu et al., 2006; Pruitt et al., 2009; Pruitt et al., 2012). Annotations are generally added to the VCF file used to store the called genotypes.
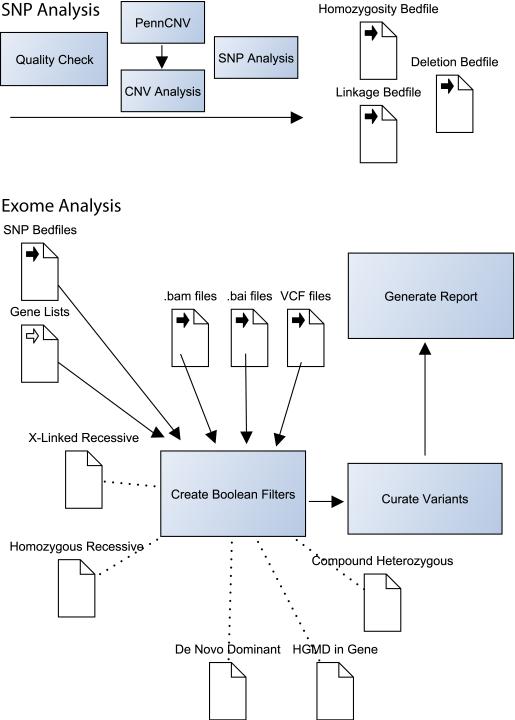
Figure 1 highlights some of the major components of the post-genotyping analytic strategy we use in the NIH Undiagnosed Diseases Program.
Figure 1. Selected Components of the NIH UDP Analysis Pipeline.
The NIH Undiagnosed Diseases Program analysis pipeline combines exome data with high-density SNP array data. We find that this is a cost-effective method for combining deep coverage of coding regions with a genome-spanning structural survey. SNP chips are checked for quality then analyzed for copy number variations (CNVs) with PennCNV (http://www.openbioinformatics.org/penncnv/). The list of CNVs is manually curated and combined with manual analysis for homozygosity and verification of parentage. If sufficient family members are available, Boolean searches and further manual curation are used to map recombination sites. CNVs, recombination sites and other regions of interest are defined in Browser Extensible Data (BED) file format for incorporation into later analysis. Subsequent exome analysis utilizes two primary programs: IGV and VarSifter (see text). The former is used to visualize pile-ups in the assembled BAM file and the second is used to incorporate BED file filters, allele frequency data, pathogenicity data and gene lists. VarSifter also allows the construction of arbitrary Boolean filters, providing fine control over searches for subsets of interest.
Scope and Characteristics of Genome-Scale Sequencing Results
Exome results from the current generation of capture kits target approximately 60 Mb of genomic regions. The average depth of coverage of the targeted regions is variable, but a >60x benchmark is commonly produced by commercial sequencing. Coverage of individual exons (or groups of closely packed exons) is highest in the center of the exon and tails off rapidly at the exon/intron boundaries. This pattern creates peri-exomic regions with low coverage that are generally not reliably interpretable for accurate genotype calls, especially for single exome interpretation. Thus, exome analyses require attention to the depth of coverage for each genotype call.
Whole genome sequencing typically utilizes a much lower average depth of coverage (10x or less). Reduced allele dropout and allele skew are present at the cost of a requirement for more starting material (fractionated DNA). Short read coverage is more uniform than the peaked short-read pile-ups associated with exome sequencing. As a result, edge detection techniques used to detect copy number variations may be employed. The cost for whole genome sequencing is currently 4X higher than equivalent exome sequencing. Costs for data storage and data processing are higher, approximately 10 to 20X for the same degree of accuracy.
“Research” vs “Clinical”
Genome-scale sequencing in humans may detect DNA sequence variations that are not of specific interest to the bioinformatician or to the person ordering the sequence, but are of potential medical importance nonetheless. Medical exomes performed in a clinical Laboratory, under the Clinical Laboratory Improvement Amendments (CLIA) act of 1988 require process controls to ensure reproducible, medical-grade results. Various standards for such testing have been proposed by the Center for Disease Control (Chen et al., 2009), the College of American Pathology, and the American College of Medical Genetics and Genomics (ACMG). The ACMG has published a list of genes/variants that it recommends be reported for exome sequencing performed for clinical diagnostics (Green et al., 2013). There is ongoing debate about whether variants found during research sequencing fall under similar or different standards; the ACMG recommendations were targeted to clinical sequencing.
ANALYTIC ASSUMPTIONS IMPLICIT TO GENOME-SCALE SEQUENCING
Ehrnfest model
The Ehrnfest model, as applied to genotype prediction, is based on a Bayesian calculation of the likelihood of obtaining the observed results at a specific location within a pileup of short reads. The likelihood calculation compares two models. In the first, a random, small sample has been taken of a large, hidden collection of mostly-reference alleles. It allows for a small number of non-reference alleles to be present due to sequencing error. In the second, a random, small sample has been taken from a large, hidden collection of alleles that are half reference and half variant. An excellent resource for understanding this process of inverse probability is described by Eigen (Eigen and Winkler, 1981) and popularized for general prediction by Sliver (Silver, 2012). The prior probability of variation at a given genomic site is, to a first approximation, the genome-wide average of the variation rate, taken over a sample of whole genomes. This number is about 1/1000 for an average human sample compared to the hg19 human genome reference. Using this basic model to calculate a genotype at any position therefore assumes a prior probability of ≈0.999 for a homozygous reference genotype at every site, and ≈0.001 prior probability for all other (variant) genotypes. This type of uniform estimate is problematic. Not all genomic sites mutate at the same rate. Any single genome-wide estimate will fail to capture the actual population history and structure that determines the allele frequency patterns across the genome for real populations. Better estimates of the prior probability of a non-reference allele can be obtained in several ways. SNP databases can be used to assess per-site variation where such information exists. Such databases are most useful when the population in the database closely matches the ancestry of the test subject. When parental sequences are available, genotypes can be called based on parental genotypes and Mendelian risks for inheritance. Parent-aware and population-aware genotypers are now available that significantly improve the Ehrnfest prior probability for called genotypes over the one-size-fits-all prior-probability genotypers. Such improvements are observed at many sites in the genome, but come at a cost of increased computation times and increased complexity of analysis-parameter choices (i.e. which population frequencies to use). Of note, the parent-aware approach is best suited to a global/naïve analysis and may make little or no difference for well-studied, small regions.
Reference sequences
Any single human reference sequence is an idealized construct. Compared with de novo assembly, the use of a reference sequence is more efficient for aligning short reads. But, the use of a reference sequence can be also cause misalignment in potentially important situations. The very notions that the total length is constant between any two parental chromosomes, or that the length of either is the same as that of a given reference sequence, are incorrect for many genomic regions. The idealized “reference sequence” for alignment is actually two references: the exact, haploid genome inherited from each parent. Strategies to approach this ideal are a focus of ongoing research and include limit dilution of source chromosomes, chromosome sorting prior to sequencing, and diploid alignment algorithms (Peters et al., 2012).
Genotype-Phenotype Relationships
Assessing the organism-level effect of any DNA sequence variation (or combination of variations) is challenging. The uncertainty associated with this determination is pervasive and touches on every aspect of the analysis of genome-scale data sets. In fact, unambiguous characterization of such genotype/phenotype relationships has only been obtained for a tiny fraction of the human genome. Extension to other genomic regions can only be made using inferential arguments. There are several ongoing efforts to improve this situation. Genome-wide variant databases have been established to collect phenotype association information about individual variants. Examples are the Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim; UNIT 9.13), HGMD (Stenson et al., 2003, CPBI UNIT 1.13), and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). In all such databases, the level of evidence for component genotype/phenotype relationships varies dramatically and cannot be used without interpretation. Many of the published observations used to populate existing databases are based on limited observations of the co-existence of a given phenotype and genotype, reflect assessments based on limited case populations, and refer to geographically and ethnically limited control groups.
Estimation of a single DNA variant's “pathogencity” or its potential to change protein function can be estimated to some extent by several methods. Current approaches include: amino acid residue conservation in sequence alignments from distant orthologous proteins or protein domains; prediction of amino acid substitution effects on protein structure; assessment of chemical/structural differences between native and substituted amino acids; population frequency of variants; large-scale association studies for common variants; and, segregation-based statistics if informative families are available.
STRATEGIC APPROACH TO DESIGNING THE SEQUENCING EXPERIMENT
Defining the Sequencing Targets
The ability to successfully analyze a genome-scale sequence is highly dependent on experimental design. An important initial decision is whether or not to use a targeted sequencing technology, and if so, whether to use exome targeting or a more specific experimental design. For instance, if the intent is to perform a cost-effective sequencing-battery for a limited number of known disease-genes, the sequencing protocol can be adjusted to produce reliable data. Additional Sanger sequencing may be required for regions that are not well covered by the exomic sequencing. Custom capture kits can be developed to focus more of the DNA library into sequencing the regions where accurate and complete genotype calling is desired. Analysis in such projects is similar to the analysis performed in Sanger-sequencing gene panels. Just as in Sanger sequencing, evaluation of the raw data is critical to a complete assessment. A determination of missing data (and potential false negatives) is critical to the gene-panel approach. It is especially critical when employing a stringent genotyping procedure that uses prior probabilities weighted toward reference alleles. The next-generation sequencing equivalent to the Sanger chromatogram is the BAM file. Individual alignment regions can be inspected for alignment, depth and context using tools like the Integrated Genome Viewer (IGV, see figure 2). To assess large groups of exons, many centers use small, home-grown computer programs to detect un-captured exons or shallow depth of coverage. Low depth-of-coverage, combined with even a small allele skewing bias during PCR amplification at the library construction stages of sequencing, can reduce the sensitivity of detection of non-reference alleles. In such cases, there is an insufficient depth of coverage to force the genotype calling Bayesian calculation to switch from a homozygous reference to a heterozygous call.
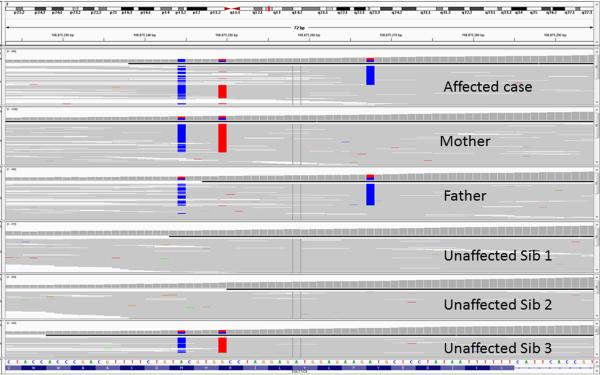
Figure 2. Integrated Genome Viewer Screenshot.
The Integrated Genome Viewer (IGV, http://www.broadinstitute.org/igv/) is a lightweight yet powerful tool for viewing short read pile ups. The example show includes pileups from six individuals: two parents, one affected child and three unaffected children. For convenience, a case was selected that shows two variants that are physically close to one another (and fit on the same screen). At the top of the display is a diagram of the chromosome being reviewed, with a small vertical red bar (between q12.1 and q13) highlighting the region being displayed below. The bulk of the display is taken up by six rows of pile-up data. Each row is an individual; each short read is a thin, gray horizontal line. Base positions that have been genotyped as non-reference are highlighted blue or red. In this case, the mother is heterozygous for two DNA variants. The father is heterozygous for one of the same variants and also for one different variant. The fact that each parent's pair of variants is cis-oriented is knowable because there are short reads with both variants, and short reads with neither variant. The affected sibling has DNA variations on both alleles, in contrast to any of the unaffected siblings.
Exhaustive scrutiny of individual genes becomes intractable as the number of genes rises to the 100's and 1000's present in an entire exome. A typical VCF file for one individual contains about 20,000 variants. In such cases, a series of “filtration” steps can be utilized to reduce the pool of variants for further more intense consideration. Experimental design can have a dramatic effect on the number of variants requiring extensive evaluation. Individual filtration steps are detailed below, but several specific examples are relevant to experimental design. Population-frequency-based variant exclusion removes variants above some fixed frequency in the population. Such filtration assumes that the population of the test subject matches that of the population used to derive the frequency estimate. Selecting test subjects from different populations will result in a higher rate of false positives—variants that appear to look rare will only be rare in the frequency-estimate population, not in the test subject's ancestral population.
Family versus Unrelated-Individual Study Design
Whenever possible, the authors recommend that every experimental design, beyond simple candidate gene searches for known variants, should include consideration of using small (nuclear) families rather than “single exomes” with no family data. As discussed below, parental data can be used to improve genotyping. In addition, segregation consistency constitutes a powerful variant filter. Even with the application of other filtering strategies, high quality exome sequencing may generate thousands of rare potentially deleterious candidate variants. Evaluation of such variants becomes especially complex when considering all the recessive alleles that can contribute to a compound heterozygous inheritance pattern. Family data that includes unaffected siblings can exclude identical combinations of variants inherited by unaffected siblings. Parents and siblings can be used to establish a variant as a de novo change in the affected individuals, including germline mosaic recurrences.
All uses of family data depend absolutely on correct assignment of affected and unaffected statuses. Such phenotyping may run counter to the structure and inertia of current medical practice. Many clinical encounters focus on the affected individual only. Finding resources to phenotype additional family members may be difficult. However, the advantages of segregation analysis are profound and worth making an effort to obtain. Phenotyping may also be complicated by age-of-onset penetrance factors present in some heritable conditions.
In our hands, an informal cost-benefit analysis suggests that certain family structures optimize signal-to-noise characteristics while maximizing practicality for real-world use. The nuclear family should include both parents, the proband and up to two additional siblings (if present). Clearly, such individuals are more likely to be available for pediatric cases, and less likely to be available for late-onset conditions in adults. Working with cohorts of such families is similarly difficult, but provides substantial power for not only finding genes, but supporting hypotheses regarding causation.
The use of multiple families with the same phenotype provides an opportunity to detect independent alleles at a common locus. It is important to note that adding unrelated individuals adds complexity and may increase the total noise of the data. Locus heterogeneity and phenocopy effects from separate diseases with a “final common pathway” or a common phenotype may occur. This is especially true for atypical cases in a cohort. Reliance on family-member reports and indirect references in medical records are should be avoided. An interesting study of these principles is the paper describing the discovery of the gene for Kabuki syndrome (Ng et al., 2010).
DNA Sources
The source of genomic DNA is important to consider briefly. True genomic DNA is not present in many blood cells due to genomic editing in antigen receptor and immunoglobulin loci. DNA from immortalized, and even primary, cell lines may be modified and rearranged to some extent. Alignments using these sources of DNA may produce problematic alignment errors in regions such as the one that includes the T cell receptor. Salivary derived (oral brushing) samples are frequently used. The total quantity of DNA in such samples is variable and may be contaminated with non-human DNA (bacterial, viral, and/or fungal). Foreign DNA sequences can cause sequence misalignment, although such misalignments are partially ameliorated by using “decoy” sequences in the reference sequence. Foreign DNA may be particularly deleterious in de novo assembly methods. Mitochondrial DNA may be enriched to provide the greater depth of coverage needed to assess heteroplasmy. This can be done by spiking a blood-derived DNA sample with additional mitochondrial DNA, bringing the percentage of mitochondrial DNA up to one to two percent of the total. The easiest source of pure mitochondrial DNA is a platelet pellet. However, as with any mitochondrial source, the platelets may or may not adequately represent the mitochondrial heteroplasmy present in the tissue(s) most affected by a given phenotype. Other sources of DNA can include tumor samples from tissues other than blood; these are often analyzed using a subtraction strategy to look for somatic differences. Very small samples may not produce sufficient DNA for sequencing. For whole genome approaches, 6 – 10 micrograms at 50 ng/μl or more concentration are typically required. Formalin-fixed paraffin-embedded (FFPE) samples and highly amplified whole genome DNA will be biased for uneven allele amplification. In general, targeted genome-scale strategies, especially when used for a specific candidate region or gene subset, will have smaller starting material requirements than non-targeted approaches used to survey the entire genome.
Consent
A full discussion of obtaining consent for human genome-scale sequencing is beyond the scope of this commentary. However, several important aspects of consent have become apparent as exome sequencing transitions from research applications to ever-more-common clinical use. Firstly, all of the usual principals of genetic testing consenting apply, including discussion of non-parentage, the possibility of ambiguous results and the possibility of revealing undisclosed family relationships. Secondly, people have varying opinions about what type of results they want to receive, including risk factors and unanticipated results. Adequate consent for clinical exome sequencing should strive to ascertain the individual's preferences in this regard. Finally, the issues surrounding genome-scale consenting are complex. Considerable time needs to be budgeted to adequately discuss consent-related issues.
Technology selection
Exome versus Genome
Most published clinical applications to date have used targeted approaches, and most of those have been exome sequencing. The likely reasons include cost, prioritization of improved signal-to-noise characteristics over uniformity of coverage, smaller starting material requirements and, perhaps most importantly, the presumption that the exome contains most of the important genetic information. Published basic-science projects have used a mixture of technologies. Non-targeted approaches have significant advantages, as discussed previously, and are becoming less costly. The decision to use one or the other technology in the future will be guided by experimental design and available resources.
Incorporation of Array Data
Genome-scale sequencing projects, particularly those using targeted approaches, may benefit from the inclusion of data from a DNA polymorphism array. In a clinical setting, karyotype, microarray and FISH may be used as a screening step preceding genome-scale sequencing. In both clinical and research contexts, array data can be used to determine genomic structural characteristics that are difficult to determine using sequence data alone. For instance, SNP chip data, in the context of a nuclear family, can be used to determine recombination segments (if multiple children are present), mosaicism, copy number variation, parentage and regions of homozygosity.
Selection of Targeted Sequencing Strategy
A variety of approaches are available for targeting specific regions of the genome. In the case of exome sequencing, the selection of any given commercially available capture kit was initially a price to performance question. As sequencing prices have declined, and capture kit technology has improved, the question has shifted to one of completeness. Current capture kits (potentially augmented with additional probes/baits when specific genes are going to be analyzed) have reached a plateau where there is no general preference to be considered. One very important point about new capture kits are that they now include sequences that were not produced by sequencing older kits, and thus a variant from an exome captured by a new kit may appear rare when a population sequenced by an older kit is used as the frequency standard. This is a common false positive that will only be perceived when enough of a cohort is sequenced using the same kit to obtain a true frequency at that position in the population ancestral to the individual whose exome is being sequenced.
INFRASTRUCTURE REQUIREMENTS FOR DATA HANDLING
Overview
Recall that genome scale sequencing involves several steps: DNA sequencing, alignment and/or assembly, genotyping, annotation and analysis of called genotypes. Starting with alignment and/or assembly, the resources needed to analyzed genome-scale data are substantial but decrease quickly. In concrete terms, most large projects (>50 exomes) use institution-scale, unix-based computers for alignment. At the other end of the process, many activities surrounding the analysis of called genotypes can be performed on a modestly powerful desktop workstation. The examples discussed below will focus on exome data. Whole-genome data increases most of the presented numbers by a factor of 20 or so. Anyone considering performing their own computational work with genome-scale data should familiarize themselves with the data available the Broad Institute Genome Analysis Toolkit Website (http://www.broadinstitute.org/gatk/). Whether or not their software is employed, the information they provide constitutes an invaluable resource for understanding the process. The field is changing so rapidly that a comprehensive list of software in is not worth committing to the printed page. An internet or Wikipedia search for “sequence alignment software” is a reasonable way to find a list of options. Choosing among the options is a complex matter that is largely beyond the scope of this commentary. The major considerations are price, performance for the particular experiment in hand, support options and computational resources (both physical and intellectual).
Alignment and/or Assembly (Alignment)
Typical output files from an Illumina HiSeq instrument (UNIT 18.2) for example, are in the gigabyte range, usually less that 10 Gb. Multiple output files may need to be incorporated into the alignment for a single individual depending on the sequencing strategy incorporated by the sequencing facility. For individual exomes, alignment of an exome can be performed on a standalone computer. NextGENe, one of the commercial products for aligning exomes on a workstation, specifies the following minimum system requirements: Windows 64 bit operating system with dual quad-core processors and 12 Gb of RAM. Some technologies, e.g. Ion Torrent, may require fewer resources. This type of machine is typical for currently available software. As noted, larger projects will benefit from a multiple-node unix environment.
Requirements for Remaining Analytic Steps
The subsequent analysis procedures have smaller hardware requirements. However, there are cases where the experimental design dictates that multiple data sets be analyzed simultaneously. In those cases, the requirements scale with the size of the dataset. The smallest data set to work with is a list of detected variants (genotypes) in VCF format. For one or a few exomes, this file can be handled on a typical desktop computer using familiar spreadsheet types of programs like Excel. For a single individual it will be on the order of 20,000 rows, with each row corresponding to a single called DNA variation/genotype. For a family of five, this approach will still be manageable for the resulting 100,000 entries. While the spreadsheet approach offers maximum flexibility, many users will opt for a freeware or commercial product with a more structured interface. Optimally, the files containing the aligned short reads will be available for inspection. These BAM files are typically on the order of 5 to 10 Gigabytes. As a rule of thumb with current technology, about 9 Gb of storage should be allocated for each sample. Genome wide data is roughly 20 times larger in storage requirements, needing about 100 to 120 Gb of storage per genome in a searchable format. These requirements increase linearly with the number of genomes/exomes per family or project. Unless VCF files are to be analyzed in isolation, consideration must be given to storage and processing facilities when planning a genome-scale analysis.
Software
VCF Genotype files can be opened directly with a text editor or spreadsheet program. Examples of commercial tools for manipulating such files include Cartagenia (Cartagenia, Inc. Cambridge, MA), Ingenuity (Ingenuity, Redwood City, CA), and Alamut (Interactive Biosoftware, Rouen, France). Freeware tools may be less integrated, but can be combined to provide many similar analyses. Examples include VAAST (Yandell et al., 2011) and exomizer (http://hem.bredband.net/magli143/exo/). BAM alignment files can be visualized using SamTools (Li et al., 2009) or the Integrated Genome Viewer (http://www.broadinstitute.org/igv/). GATK (McKenna et al., 2010) and Galaxy (Blankenberg et al., 2010; CPBI UNIT 10.5) provide rich sets of data analysis and manipulation tools. For complex analyses, custom computer programs (Perl and Python are popular languages) may be required. We have found that a graphical interface allowing for arbitrary Boolean searches is an exceptionally powerful analytic tool. We use an available but unsupported tool called VarSifter (Teer, 2011); we hope that this technology is incorporated into newer programs in the future.
Collaboration
Small scale experiments by users who are performing genome wide analysis on only a few individuals, or on a single cohort all with the same pathology, will benefit from collaboration with a center that has a larger database of exome/genome data. This allows recognition of variants that are shared (or not-shared) with large control populations. In addition, systematic sequencing and alignment errors are much easier to detect using large data sets compare with small ones.
GENERAL APPROACH TO DATA ANALYSIS
Classes of results
Genome-scale sequencing will produce several classes of results. Experimental design should include a strategy for dealing with each type of result.
Well-described variant, well-described gene, consistent phenotype.
Severe-appearing variant, well-described gene, consistent phenotype. These are variants that introduce termination codons or frameshifts or variants that alter amino acids known to be critical for protein function. The affected gene is known to be associated with the phenotype. The pattern of inheritance is biologically plausible given existing knowledge about dosage sensitivity and other factors.
Questionable variant, well-described gene, consistent phenotype. These variants recapitulate the “variant of unknown significance” (VUS) problem that is well known in molecular diagnostics. The variant itself provides incomplete evidence of causation, but may be studied further depending on the strength of the phenotype-gene association. Such variants likely have multiple lines of “soft” evidence for pathogenicity: high degree of conservation, significant change in amino acid structure, etc.
Severe-appearing variant, known gene, inconsistent phenotype. In this case the variant looks like it should cause disease, but the phenotype bears no resemblance to the phenotype associated with the known gene. These variant may represent incomplete penetrance, incorrect dogma about the gene, or evidence of genetic pleomorphism (multiple different phenotypes from the same gene).
Severe-appearing or questionable variant, gene in same pathway as well-described gene, phenotype consistent with known gene in pathway but not mutated gene. These may represent a new gene-phenotype causal relationship, but may also be difficult to prove experimentally. A subset of this situation is synergistic heterozygosity where two genes in the same pathway, both associated with recessive disease, carry one severe mutation each.
Severe-appearing or questionable variant, gene not associated with any phenotype. Such variants may represent new gene-phenotype discoveries or be unrelated to the phenotype. Biological hypotheses linking the gene to the phenotype may be present. This type of variant is common and presents a major challenge for establishing evidence of causation.
Severe-appearing mutations, known gene, consistent phenotype, inconsistent inheritance. In particular, these are “carrier” patterns, where a gene associated with autosomal recessive inheritance has only one detected variant. Depending on the strength of the gene-phenotype association. Some of these cases may be “rescued” from exclusion by finding a second mutation that was missed by sequencing. Examples include deletion of the second allele, allele-bias, failed capture of a mutated exon and deep-intronic mutations that affect splicing but were not captured in targeted sequencing.
Other. Other significant categories include false-positive variants, variants that can be excluded by conservative filtration or segregation analysis.
Incorporation of genome structure information from SNP chips
Hybridization experiments including array CGH and SNP chips, provide an orthogonal methodology (chemical thermodynamics versus numerical calculation) to determine a variant, either a single SNP, a deletion or the suspicion of some level of variance in the probe region of the oligonucleotide that causes an apparent deletion (failed hybridization). This information is genome-wide, highly accurate (in most cases) and can be used to complement and improve the analysis of genome -wide sequencing data. Linkage mapping utilizes chip data to identify chromosomal regions that have transmitted to offspring. Copy number variation information may include single copy deletions that can be matched with trans-oriented point mutations to form compound heterozygote pairs. With a close collaboration between the bioinformatics team and the sequencing facility, it is possible to use this data to alter the reference genome and improve the quality of the alignment processes even before a VCF file is produced. The detection of mosaicism and uniparental disomy requires some level of dynamic range beyond three diploid genotype states (0, 1 allele or 2 alleles). As a result, SNP data is preferable to exome data if those genetic mechanisms are suspected. The NIH Undiagnosed diseases program obtains SNP chip data for all cases it considers for genome-scale sequencing. Currently, adding a SNP chip to exome sequencing incurs approximately 30% additional costs. If the experimental goal is to obtain genomic structure and exome sequence, a SNP + exome is presently less expensive than a whole genome experiment. As whole genome costs decrease, this situation will need to be re-evaluated.
“Filtration”
Filtration is a general term for prioritizing the variants generated by genome-scale sequencing. The following section provides observations about the characteristics of common filtering components, where they have not been discussed earlier.
Pathogenicity
Filtration by predicted potential to alter protein function, as discussed in previous sections. Pathogenicity filtration must be used with caution, as it is error prone. In general, it should not be used as a static filter that is buried in an automated analytic pipeline. It is better used in an aid to an interactive process in the final stages of analysis.
Chromosome-Level Segregation
The inheritance state of any given chromosomal segment is determined by the crossover events that occurred during meiosis in the parents of the person being studied. In any series of children from the same parents, some chromosomal segments will be shared, and some will differ. These similarities and differences can be correlated with the affected status of each child. SNP chip (or whole genome) data can be used to identify and define crossover sites if sufficient family members are available. The smallest practical pedigree size for this procedure is two parents and two offspring. Crossover sites can be localized to within a few tens of kilobases, or well within the typical length of one genetic locus. Chromosomal regions demarcated by this mapping exercise can be included or excluded from further analysis depending on whether they segregate into the offspring in a manner consistent with a proposed genetic model. Excluded regions for a two-parent/two-offspring family ideally sum to the following percentages of the genome: inherited recessive (25%), dominant (50%) or X-linked (50%). In practice, the sum of the excluded regions is smaller because most genetically informative (gene containing regions) are not randomly distributed over the linkage map of the genome. This approach is becomes increasingly powerful with the addition of siblings. When there are two affected sibs, linkage bed files routinely exclude 70-80% of the genome from inherited recessive model analyses. Other means of including and excluding genomic regions include homozygosity mapping, which may be constructed using only proband DNA. Mosaicism and Uniparental disomy regions are also reliably defined by SNP chip data.
Whole genome data, in contrast to exome data, can potentially be used to for the same procedures described for SNP chips. Firstly, it covers a larger area than an exome data set and therefore allow for more even distribution of polymorphisms used in the analysis. Secondly, and importantly, it is also less susceptible to allele skewing and can be used to differentiate hemizygosity from homozygosity (based on read depth). The latter is problematic in exome analysis to the point where read depth cannot be used to reliably differentiate copy number differences. Targeted/exome experiments produce uneven depths of coverage and wide variances due to amplification during PCR reactions that frequently are taken to the saturation plateau on PCR curves. The result is that the distribution of read depths for single-copy (hemizygous) states and double-copy (dizygous) states may overlap.
Variant-Level Segregation
Construction of a VCF genotype file for a family allows each called genotype to be tested for segregation consistency (which is not always the same as the inheritance state). This can be done for homozygous recessive, X linked, and de novo dominant states by simple line by line Boolean sorting. Deletion plus point mutation recessive pairings can be identified by combining the VCF genotype file with a BED format file that defines the location of single copy deletions. Deletion files may be derived from array data or from whole-genome depth of coverage data. Future development of genotype callers that can reliably detect hemizygous alleles in whole-genome datasets will streamline this analysis.
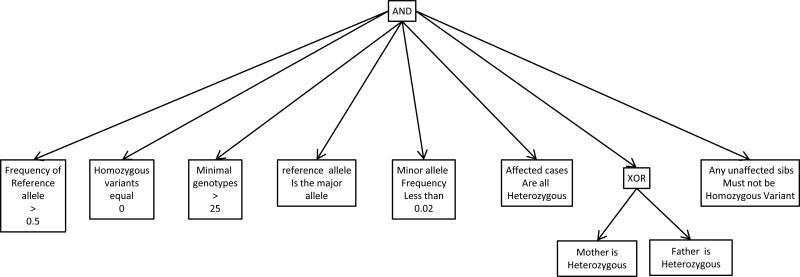
Compound heterozygous recessive inheritance is the most difficult analysis. There are usually more than five variants within any single locus and thus many potential combinations of variants. Any potential variant pair must furthermore be phased to verify trans-orientation. The authors approach this type of filtering by generating a list of potential “half-compound-heterozygotes”—variants that could be one of the two alleles in a compound heterozygous pairing. The Boolean search for such variants stipulates that a variant is present in one or the other parent but not both (the XOR Boolean operator), and that the variant is not homozygous in unaffected sibs (Figure 3). The resulting list of variants is sorted to group by genetic locus, generally using the annotated gene name. Pairs of alleles are manually assessed for pathogenic characteristics. Allele combinations that look promising are then further evaluated for segregation-consistent, trans-orientation. After testing that unaffected sibs do not have both alleles in the pair, the total number of compound het candidate pairs is typically reduced to a very small number, usually less than 10 for the entire genome and quite often only one or two loci. Of note, we generally apply a population low-frequency filter before, or at the same time as, Mendelian segregation filters. Typical cutoff values are between 1% and 5% maximum frequency of the variant allele. Genotyping programs may call multiple alleles at the same genomic location when a small insertion or deletion is present. Therefore, our first-pass analysis requires the major frequency allele to be greater than 50% to exclude variable length indels. These steps are discussed in detail in following sections.
Figure 3. Boolean Filter for finding compound-heterozygote “half hets”.
Boolean filtration can be used find variant subsets of interest within the called genotypes in a genome-scale sequencing data set. The schematic shown diagrams the criteria for all alleles to be one of two that can pair to fit a compound heterozygous recessive Mendelian model. After application of this filter, the resulting variant list is sorted by locus name. Variants of certain classes are prioritized, including those that result in stop, splice site, frame shift and non-synonymous amino acid changes. A normal number is about 300 to 900 total per exome. At any one locus there are at most a very small number of these types of variants, and typically there are only a very few loci with two or more. These must be inspected individually to see if there are two variants within loci that have more than one allele, to see if any pair are oppositely phased, one to each of the two parents. Pairs of variants that occur at the same loci, are of the type to change protein function, and are correctly phased (typically are no more than 0 to 5) constitute the compound heterozygous candidate variant pairs.
Population Variant Frequency
The appearance of both common and rare variants is normal in any genome-scale sequencing data set. The mutation pattern in any given individual is the result of multiple factors: segregation of variants from parental DNA; the history of the parents’ ancestral population(s); the DNA mutation rate associated with thermodynamic chemical limits on replication fidelity even after all molecular proof reading corrections are included; interphase DNA mutations; mutation effects on fitness/fecundity; and genetic drift. Common variants, except in exceptional circumstances, are assumed to have small effects on fitness. Rare variants may be neutral with respect to selection, but are close to extinction due to genetic drift or population history. Rare variants may also affect fitness, but remain in the population because they are newly introduced, incompletely penetrant or associated with disease only in a recessive state.
Population allele frequencies are commonly incorporated into genome-scale sequencing analysis in two ways: exclusion of frequent alleles and verification of Hardy-Weinberg Equilibrium consistency. The argument for excluding frequent alleles is that they are so common that they would result in the common occurrence of a homozygous phenotype. If the phenotype is known to be rare, such a situation would be inconsistent. The most important condition of this argument is that the phenotype is truly rare and not the extreme end of the normal distribution in a common phenotype. Exclusion of frequent alleles is likely reasonable in complex and severe phenotypes that are markedly different from normal and from any other known phenotype. A common, conservative threshold for this type of filter is an allele (or heterozygote) frequency of between 1 – 5 percent. Put in context, carrier rates for common deleterious alleles such as sickle cell anemia, hemochromatosis and cystic fibrosis are between 3% and 12%. Carrier rates may be higher in populations that are isolated or have a high coefficient of in breeding. Even so, the frequency of beta thalassemia in Sardinia was only 12.6% (Cao et al., 1978).
Early efforts in constructing polymorphism frequency filters utilized Entrez dbSNP (now NCBI dbSNP) (Ng et al., 2010; Teer et al., 2012). However, dbSNP was never intended as a uniform population record of allele frequencies, or as a list of verified, non-pathogenic variations. More recent versions of dbSNP (version 130 and later) have included large, annotated subsets (1000 genomes data, etc.) of data with good population-survey characteristics. Use of dbSNP data should include a careful selection of an appropriate subset. Direct use of a variety of databases is now possible. Popular examples include 1000 genomes (Abecasis et al., 2012) and the Exome Variant Server (http://evs.gs.washington.edu/EVS/). These databases are also beginning to have subpopulation-specific allele frequencies useful for exome filter functions (Tennessen et al., 2012). The ideal population frequency data would be gathered from carefully phenotyped individuals genotyped using identical DNA sources, laboratory techniques, and bioinformatics. This is a strong reason to recommend using a small number of common sequencing core laboratories and to carefully cross-validate each change in data-gathering protocols. Accumulated sequence data at any site may be used to estimate population allele frequencies. In fact, if a project is focusing on a poorly-studied population, the frequency data may be superior to larger public databases. Of note, only independent chromosomes should be counted toward the calculated frequencies; only “founder” data from each family should be included.
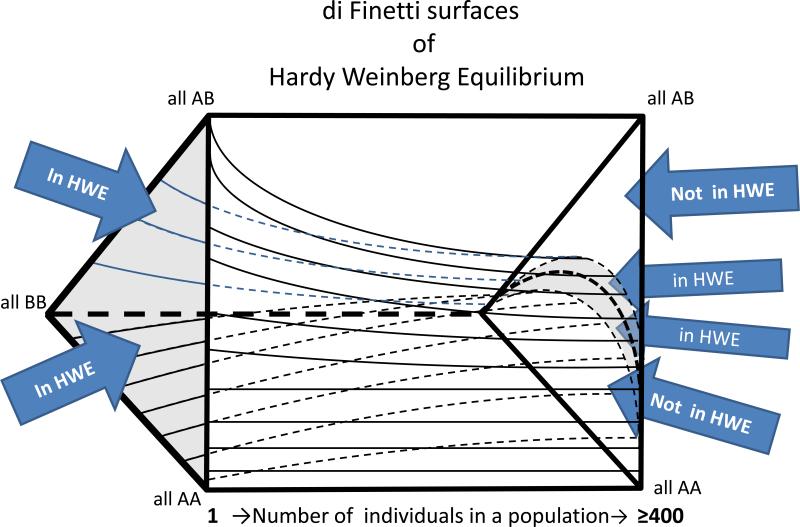
The second way frequency of an allele is used is to look at verification of Hardy-Weinberg Equilibrium consistency. One type of short-read misalignment is caused when two groups of nearly identical short read sequences align two different genomic regions. In this case, the pileups over each region will be a mixture of reads from both sites. This situation causes the apparent genotypes in those regions to appear as perfect heterozygous variants. Assuming that the same alignment error occurs with every person in the sequenced cohort, they will all appear to be heterozygous. Biological explanations for such a pattern are unlikely and include an extreme heterozygote advantage together with a large fetal wastage rate, or that all parents of every person sequenced were from two different homozygous and distinct subpopulations, with no homozygous pairings between two people from the same subpopulation. Therefore, such variants can be comfortably excluded as false positives. Even a sequenced cohort of a few hundred individuals will allow the exclusion of frequent polymorphisms and systematic errors. Such patterns can be used to filter out inconsistent phenotypes (Fuentes-Fajardo et al., 2011). The De Finetti diagram is a means of visualizing the range of possibilities of different proportions of population genotypes (Edwards, 2000). Furthermore, it provides a framework for designing a quantitative variant-exclusion criterion to use as a variant filter. The De Finetti surfaces change from a three state triangle to a very thin horizontal crescent moon shape around a central parabola, as the number of subjects genotyped from a large population in hardy Weinberg equilibrium is increased towards infinity (Figure 4). Given a sufficiently-sized collection of genome-scale sequencing data (300+ individuals works well in our hands), one can define a plus or minus 2-standard-deviation-surface for this crescent. The region outside this surface is large enough, for alleles with a minor allele frequency >5%, to allow exclusion of a given position based on the probability of alleles that are both common and out of Hardy Weinberg equilibrium due to an artifact like misalignment is greater than the likelihood that the candidate is real, common, and has one of the rare population genetic explanations for being far from HWE.
Figure 4. Di Finetti Diagram.
A de Finetti diagram is used to graph genotype frequencies in populations. It presumes two alleles, and can be used to plot genotype frequencies at which Hardy-Weinberg Equilibrium (HWE) is satisfied. The figure shows a rectangular prism with surfaces plotted in its interior. The vertices of the triangles on the ends of the prism correspond to genotypes as shown: AA, AB and BB. The length of the prism is a scale of individuals in the population from 1 (far left) to ≥ 400 (far right). The area between the upper and lower internal plot surfaces define the combinations of genotypes that are consistent with HWE given a particular population size. As the population size increases, an increasingly small proportion of all of the possible genotype combinations are in HWE. However, difference between the in-HWE and out-of-HWE regions changes increasingly gradually as the population size reaches hundreds of individuals. For this reason, a data set of 100's of individuals allows stringent criteria to be used in assessing whether a set of genotypes is out of HWE—potentially due to misalignment.
The calculation of a Chi-square statistic for each location in a VCF file can be done if there is a suitable population from the chosen sequencing center. The Exome Sequencing Project and the NCBI dbSNP ClinSeq™ subset have both frequency and Chi-square data for sufficient numbers of alleles to be a guide for HWE concerns during exome analysis. An ideal data base might include well-phenotyped individuals over the age of 70-90 years (who presumably have few genetic diseases), grouped by subpopulation type, and in a quantity somewhere near 50,000 to 100,000 individuals. Given recent advances, such a resource is feasible.
Quality scores (read, genotype) and coverage
Any candidate variant discovered during an analysis of a VCF file must be interrogated for the possibility of being a false positive due to genotyping or alignment error. Three characteristics of called genotypes are particularly useful for this determination: the depth of coverage, the phred-like short-read sequence quality scores and the MAQ (map quality) score. These should be used in concert with a direct inspection of the short read alignment (BAM file) using IGV or a similar tool. Marginally aligned sequences will have very low MAQ scores, meaning that the probability that the given short read was derived from the region it is nominally aligned to is not much greater than the probability that it was derived from another location in the genome. Some genotype callers use a threshold of 20 or 30 for MAQ and do not include any reads below that level. Leaving a substantial number of reads out of an alignment and genotyping process is risky in that it may violate assumptions of the Ehrenfest model and cause false-positive or false-negative calls. Manual examination of the .BAM files easily shows when this is occurring. Unlike the MAQ that applies to the entire read, the phred quality estimates are given to each base in every read, and are roughly correlated to the same score given in Sanger based capillary sequencing, approximately the 10*LOG of the odds that the base assigned in the read is the correct one (Ewing and Green, 1998; Ewing et al., 1998). The coverage is the total number of reads that contain a base at the given position. Some genotype software only counts the coverage of reads and phred scores above a given threshold. This “clipping” can cause aliasing bias in genotype calls, but greatly simplifies the Ehrenfest Bayesian calculations. Sensitivity and specificity are sacrificed, especially for poorly-covered and low-complexity regions of the genome. Using parent (pedigree)-aware, population-aware and genome-topology-aware genotype software may improve this aspect of genotyping.
Direct inspection of the pileup also allows assessment of the short reads and variants nearby the variant of interest. Typically a pile of overlapping reads will involve a region of around 200 bp with current technology. Regions with problematic alignments can be identified by the very large number (typically > 5) of non-homozygous positions over this short interval (considering that the genome-wide average of one heterozygous SNP per 1000bp predicts about 0.2 variants should be present). The newest generation of alignment and genotype callers (e.g. Stampy/Platypus) take this information into account and can use it to improve alignment and genotype calls (Rimmer A, 2012).
Analysis can be done without looking at the BAM files, and still incorporate partial information about quality score, and depth of coverage. By being aware of the nature of the Ehrenfest/Bayesian calling principle of genotyping, the analyst can recognize when a confidence score attached to a genotype is low relative to the depth of coverage. Another proxy for direct BAM file inspection is to note the close approximation (within 100 bp) of variants listed in a VCF genotype file. The genotype score/coverage ratio can be a be used to assess poor alignment as well (Wei et al., 2011). Calibration of this metric must be empirically adjusted to the type of quality score and to the conditions of the sequencing for an individual center, type of source DNA, type of capture, type of sequencing , type of alignment and finally type of genotype caller. This approach becomes increasingly powerful when a larger number (n>100) of exomes are able to be viewed simultaneously for their genotype, quality score and coverage in a sorted manner from lowest to highest. Such data can be used to assess variance across many exomes of the typical quality and coverage, and where the subject being analyzed fits into that distribution. Software has been developed to create a direct link between a variant file analysis interface that is pointed to a single variant, and to the same region in the BAM file, which the current authors find indispensable during analysis (Teer, 2011).
Predicted Pathogenicity
Pathogenicity prediction is the use of chemical, protein structure, ortholog conservation and other data to predict whether or not a given DNA variation is likely to affect protein function. There are numerous algorithms and tools that been designed to assess pathogenicity. Sift (Ng and Henikoff, 2006), Polyphen-2 (Adzhubei et al., 2010; UNIT 7.20), Grantham scores (Grantham, 1974), CDPred (Cherukuri, 2010) and Mutation Taster (Schwarz et al., 2010), phyloP (Cooper et al., 2005), PhastCons (Yang, 1995), and GERP (Davydov et al., 2010) are examples. The biggest unresolved issue for all models is the question of whether the training set of disease causing variants is a biased subset of causal mutations found by the very fact that these were the first and easiest diseases investigated. In that case these models will be a risk to exclude the very changes that do not look like traditional changes causing disease in the training set of variants. Additionally these algorithms have been trained on a set of presumed disease causing mutations from literature sources that were mostly reported in the era prior to genome-wide sequencing. All of the available procedures are, at best, estimates of average behavior and cannot predict or exclude the true effect of a variant at a single site. Such results must be considered judiciously during analysis. Numerical predictions can be included among per-variant annotations in genome-scale sequencing projects. The pathogenicity estimates can then be used to prioritize variants during analysis.
Special Genetic Cases De novo mutations and regions of homozygosity
New “de novo” mutations
New mutations are an important subset of genome-scale sequencing results. Their extraordinary rarity suggests that they should be considered with other potential candidates for disease causation. The Haldane/Bell estimate of 1.2 X 10-8 de novo mutations per locus per generation, based on hemophilia epidemiology in London England in the 1930-50s was confirmed in the whole genome sequencing of the Miller Syndrome Quartet (Muller, 1950; Roach et al., 2010). This rate predicts 1.2 mutations per average exome (60Mb) or 70-100 mutations per diploid genome. Using the Poisson distribution, the 95% confidence interval is between 0-4 de novo mutations per exome per generation. Since many genetic loci are recessive, many of the de novo mutations will only produce carrier states, even when truly deleterious. Some will result in dominant, penetrant phenotypes and a very few will complement a trans-inherited deleterious variant to produce a de novo recessive pair. When both parents are available, and especially when an unaffected sib has the same inheritance state for the specific region of the chromosome, de novo mutations are easy to detect.
One particular family structure is notably favorable for de novo analysis: a three generation pedigree with an unaffected grandparental generation and an affected parent with an affected child (or children).
In this case, a de novo mutation that is missing from the grandparents and present in all affected individuals is consistent with both the new appearance of a phenotype in the child of unaffected parents and the transmission of the phenotype in a dominant manner. Given unambiguous phenotyping in the grandparents, the parents, the affected child and one unaffected child, the probability that a de novo variant will randomly segregate in a manner consistent with disease association is less than 5%. This very favorable pedigree is the most statistically powerful small pedigree to allow discovery of a single gene causing disease. These pedigrees should be sought and attempts to study them should be offered.
Homozygosity Mapping
Homozygosity mapping has been repeatedly demonstrated to be a powerful technique for identifying disease genes. In genetic terms, cases of identity by descent (IBD) represent the entire linkage power of the consanguineous loop contained in the genotype of the single individual (Smith, 1953). However, there is also a substantial possibility that more than one deleterious change is present within the regions of homozygosity. Many severe rare phenotypes in a small consanguineous pedigree are not monogenetic. Separation of the effects of multiple mutated genes may not be straightforward.
Frequent pitfalls in exome experimental design
Strategy for recognizing true positives
The most common pitfall in experimental design is the presumption that a causal variant will be recognizable as such. Genome-scale sequencing not only generates many variants, but “false positives”. Here, false positives are any variants that appear to be likely to contribute to a phenotype, but in actuality do not. Improvement in this situation is unlikely to be possible for some time into the future, primarily due to the near impossibility of producing a false-positive-free data set. Even a data set that is 99.999% accurate will contain many hundreds of variants with some characteristics suggesting pathogenic potential. The use of careful and stringent filtering can reduce the number of false positives, but only at the cost of an increasing probability of excluding potential true positives. In practice, rapid determination of causation requires finding a known or severe variant in an already-well-established disease-causing gene. Establishing causation in other cases is a matter of current debate, but needs to include some combination of convincing functional studies and/or sufficient cases to use genetic or association statistics.
Inadequate starting material
Every choice that is made in the experimental design – e.g. using DNA from immortalized lymphocyte cell lines, using highly amplified DNA from dilute samples, using FFPI extracted DNA, - will potentially increase the false positive rate from hundreds to thousands of called genotypes in a VCF file. Given the effort required to analyze genome-scale sequence data, a high priority should be given to starting with large quantities of high quality (un-degraded) DNA whenever possible
Inadequate consideration of sequencing strategy
Filtration steps can be divided into those that remove variants based on a heuristic characteristic of the individual variant (frequency in population, predicted pathogenicity) and those that include or exclude regions of the genome. The latter strategy, when available, is often the most powerful. The most dramatic example would be a small candidate region or region of homozygosity. In such cases, genome-wide sequencing may not even be needed unless the region contains many genes/exons. The use of family/segregation data, with genomic-sequencing- or SNP-array-based recombination mapping becomes increasingly powerful with the addition of informative meioses. Further strategic decisions that will decrease false positives include using low amplification techniques in library construction and sequencing to the greatest depth affordable.
COMMENTARY
Expectations versus reality
Genome-scale sequencing is a powerful technology that is transforming genetics research and clinical practice. However, in our experience, nearly every new user of the technology experiences discomfiture when they are confronted with the fact that even the best analytic procedures fall short of establishing definitive genotype-phenotype causation. The situation is improved when there are many affected individuals to test for the presence of mutations in a candidate gene, or when an unambiguous diagnostic result is produced. For very rare phenotypes, the process of establishing causation is no different from other candidate generating procedures such as linkage analysis or homozygosity mapping. Exhaustive phenotyping, stepwise experiments in model organisms and searching for additional cases are the rule. Publishing trends are reflecting this fact. The requirements for publishing genotype-phenotype correlations were relaxed for a time when genome-scale sequencing first appeared. Since that time, there has been a general trend toward stricter requirements and more substantive evidence.
Future directions
The past few years have been a time of rapid improvement in genome-scale sequencing technology. Costs have decreased and supporting data sources such as variant databases have proliferated and improved. As discussed in this commentary, it is now possible to consider feasible, near-ideal characteristics for future sequencing resources. Paramount among these is an improvement in our understanding of the structure of major world populations. Some would argue that trends towards population admixture make this goal impossible. However, the needs of the sequencing community do not necessarily require static populations as much as the broadest possible survey of allele frequencies and a clearer picture of the ability of human homeostasis to tolerate variation at specific sites across the genome. For clinical applications, ethical and utility considerations will be forced upon the medical community a medical genome-scale sequencing undergoes rapid implementation. Whether such rapid deployment is wise, it is happening. As a result, we predict a continuation of the rapid changes of the past few years.
ACKNOWLEDGEMENTS
We would like to acknowledge William A Gahl, MD, PhD, who established the Undiagnosed Diseases Program and continues to support our work with enthusiasm. We need to acknowledge the team members of the Intramural UDP and the NIH Intramural Sequencing Center (NISC). We are especially grateful to the physicians who refer us study participants and the families who we work with at the NIH Clinical Center. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute (NIH, Bethesda, Maryland).
Works Cited
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous http://gvs.gs.washington.edu/SeattleSeqAnnotation/
- Anonymous . Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; National Center for Biotechnology Information, National Library of Medicine; Baltimore, MD: Bethesda, MD: [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Current protocols in molecular biology / edited by Frederick M. Ausubel ... [et al.] 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19:Unit 19 10 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A, Galanello R, Furbetta M, Muroni PP, Garbato L, Rosatelli C, Scalas MT, Addis M, Ruggeri R, Maccioni L, Melis MA. Thalassaemia types and their incidence in Sardinia. Journal of medical genetics. 1978;15:443–447. doi: 10.1136/jmg.15.6.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gagnon M, Shahangian S, Anderson NL, Howerton DA, Boone DJ. Good Laboratory Practices for Molecular Genetic Testing for Heritable Diseases and Conditions. Division of Laboratory Systems. National Center for Preparedness, Detection, and Control of Infectious Diseases, Coordinating Center for Infectious Diseases; 2009. [PubMed] [Google Scholar]
- Cherukuri PF. CDPred. 2010 [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome research. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AWF. Foundations of mathematical genetics. 2nd ed. Cambridge University Press; Cambridge, U.K. ; New York: 2000. [Google Scholar]
- Eigen M, Winkler R. Laws of the game : how the principles of nature govern chance. 1st American ed. Knopf : Distributed by Random House; New York: 1981. [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Harrow J, Herrero J, Hubbard TJ, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SM. Ensembl 2012. Nucleic acids research. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire A, Nussbaum RL, O'Daniel JM, Ormond KE, Rehm HL, Watson MSW, Williams MS, Biesecker LG. ACMG Recommendations for Reporting of Incidental Findings in Clinical Exome and Genome Sequencing. 2013 doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Our Load of Mutations. American Journal of Human Genetics. 1950;2:111–176. [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, Kermani BG, Sparks AB, Alferov O, Hong P, Alexeev A, Jiang Y, Dahl F, Tang YT, Haas J, Robasky K, Zaranek AW, Lee JH, Ball MP, Peterson JE, Perazich H, Yeung G, Liu J, Chen L, Kennemer MI, Pothuraju K, Konvicka K, Tsoupko-Sitnikov M, Pant KP, Ebert JC, Nilsen GB, Baccash J, Halpern AL, Church GM, Drmanac R. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein- coding gene set for the human and mouse genomes. Genome research. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic acids research. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A MI, Lunter G, McVean G. Platypus: An Integrated Variant Caller. 2012 [Google Scholar]
- Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease- causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Silver N. The signal and the noise : why so many predictions fail--but some don't. Penguin Press; New York: 2012. [Google Scholar]
- Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome research. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CAB. The Detection of Linkage in Human Genetics. J Roy Stat Soc B. 1953;15:153–192. [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Human mutation. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome- scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. 2011 http://research.nhgri.nih.gov/software/VarSifter/
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, Robinson W, Robinson S, Rosenberg SA, Samuels Y. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Huff C, Hu H, Singleton M, Moore B, Xing J, Jorde LB, Reese MG. A probabilistic disease-gene finder for personal genomes. Genome research. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. A space-time process model for the evolution of DNA sequences. Genetics. 1995;139:993–1005. doi: 10.1093/genetics/139.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]