SUMMARY
Tudor domain containing protein 3 (TDRD3) is a major methyl-arginine effector molecule that “reads” methyl-histone marks and facilitates gene transcription. However, the underlying mechanism by which TDRD3 functions as a transcriptional coactivator is unknown. We identified topoisomerase IIIB (TOP3B) as a component of the TDRD3 complex. TDRD3 serves as a molecular bridge between TOP3B and arginine-methylated histones. The TDRD3-TOP3B complex is recruited to the c-MYC gene promoter primarily by the H4R3me2a mark, and the complex promotes c-MYC gene expression. TOP3B relaxes negative supercoiled DNA and reduces transcription-generated R-loops in vitro. TDRD3 knockdown in cells increases R-loop formation at the c-MYC locus, and Tdrd3-null mice exhibit elevated R-loop formation at this locus in B cells. Tdrd3-null mice show significantly increased c-Myc/Igh translocation, a process driven by R-loop structures. By reducing negative supercoiling and resolving R-loop, TOP3B promotes transcription, protects against DNA damage and reduces the frequency of chromosomal translocations.
INTRODUCTION
Transcription levels are regulated by the recruitment of coactivators and corepressors that together orchestrate a cacophony of events with emergent order at enhancers, promoters, gene bodies and termination sites. Much of this regulation is mediated by enzymes that deposit post-translational modifications (PTMs) on histones and other proteins associated with chromatin. These modifications, such as acetylation and methylation, generate docking sites for effector molecules that “read” the PTM mark and help reinforce an active or repressed chromatin state (Badeaux and Shi, 2013). Protein arginine methyltransferases (PRMTs) are one such class of enzyme that regulates transcription, and the two primary transcriptional coactivators in this family are PRMT1 and CARM1 (coactivator associated arginine methyltransferase 1), which deposit the H4R3me2a and H3R17me2a marks respectively (Yang and Bedford, 2013). Both of these marks are recognized by the Tudor domain of TDRD3 (Yang et al., 2010), a protein that is enriched at the promoters of highly-transcribed genes and which can likely also associate with the C-terminal domain (CTD) of RNA Polymerase II (RNAP II) (Sims et al., 2011). TDRD3 has no enzymatic activity of its own, but here we show that it is tightly complexed with DNA topoisomerase IIIβ (TOP3B), an interaction that bestows, in part, coactivator activity on TDRD3. TOP3B is a member of the 1A subfamily of DNA topoisomerases, and as such, targets underwound or negatively supercoiled DNA (Wang, 2002). This subfamily of topoisomerases has been implicated in the resolution of R-loops (Wilson-Sali and Hsieh, 2002), which are nucleic acid structures formed by a RNA/DNA hybrid and the displacement of the DNA strand.
Transcription-mediated R-loop formation occurs when the nascent RNA transcript anneals back to the template DNA strand in the wake of RNAP II. R-loops form there owing to the presence of a negatively supercoiled region behind RNAP II and to the fact that RNA/DNA hybrids, particularly those formed by G-rich RNAs base-paired to C-rich DNA templates, are more stable than double stranded DNA (Roberts and Crothers, 1992). R-loops have been described at class-switch sequences in the IgH locus (Yu et al., 2003). There, R-loops are thought to help initiate double-strand DNA breaks and trigger class switch recombination through the recruitment of the AID cytidine deaminase. R-loops have also been detected at the 5’-end of human genes, particularly those transcribed from CpG island promoters, where they contribute to a protective pathway against DNA methylation (Ginno et al., 2012). Recent evidence also suggests that R-loop formation at the 3’-end of genes is common and mediates efficient transcription termination (Ginno et al., 2013; Skourti-Stathaki et al., 2011). Interestingly, the co-transcriptional formation of R-loops impedes the progression of elongating RNAP II (Belotserkovskii et al., 2010). Excessive R-loop formation is also linked to genomic instability (Aguilera and Garcia-Muse, 2012). This suggests that the potential benefits associated with R-loop formation must be carefully balanced with possible deleterious effects. At least three different surveillance mechanisms are thought to regulate R-loop formation: 1) RNase H enzymes, which degrade RNA in the context of RNA/DNA hybrids, 2) RNA/DNA helicases that specifically unwind these structures, and 3) topoisomerases that act to relax negative supercoiled regions and thereby prevent the persistence of R-loops (Aguilera and Garcia-Muse, 2012). However, how these redundant systems are targeted to R-loop prone genomic regions is unclear.
The interaction of TDRD3 with TOP3B provides a mechanism by which the topoisomerase activity can be recruited to regions of chromatin that are decorated with H4R3me2a, H3R17me2a marks and to methylated RNAP II at sites actively undergoing transcription. PRMTs (PRMT1 and CARM1) function as transcriptional coactivators in vivo by depositing methyl-marks that recruit the TDRD3-TOP3B protein complex to dampen R-loop formation.
RESULTS
TDRD3 and TOP3B are tightly associated
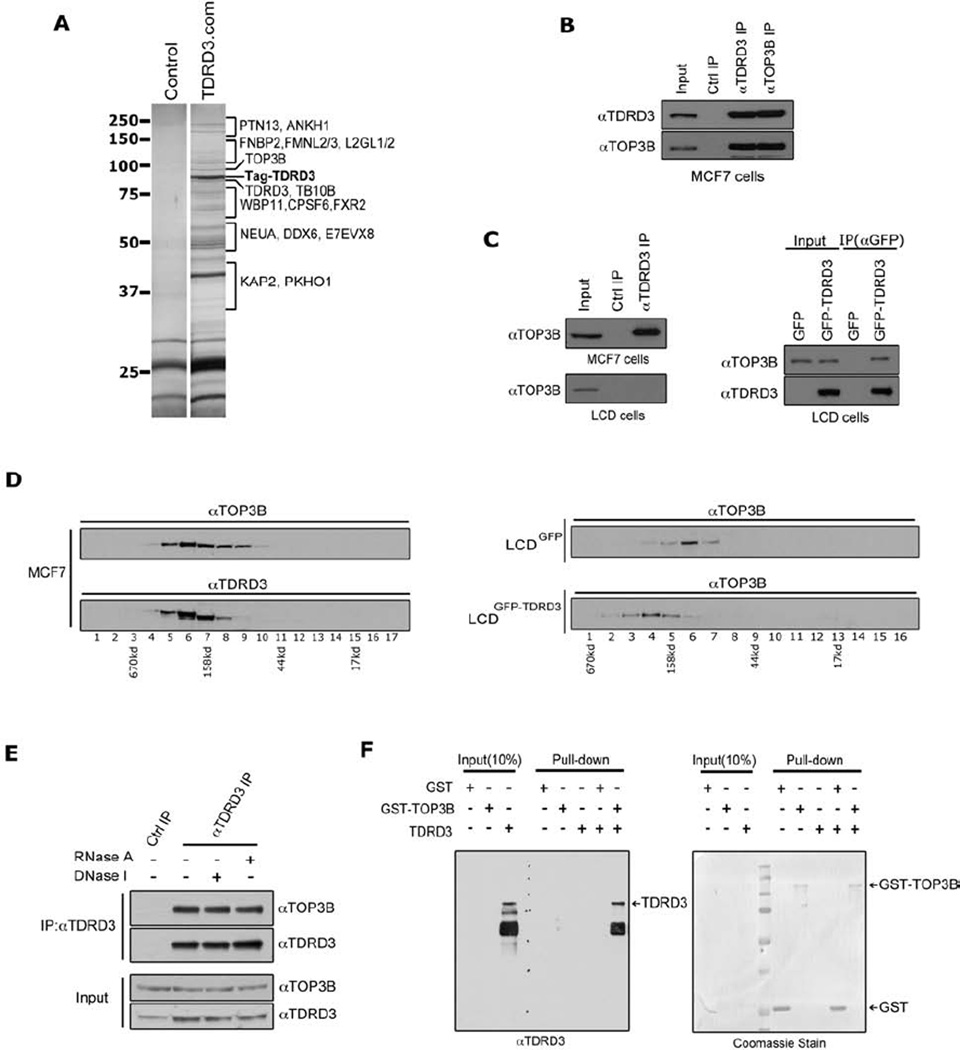
We identified TDRD3 as an effector molecule for methylarginine marks on histone tails (Yang et al., 2010). At that time, we speculated that TDRD3 would function as a scaffolding molecule that would recruit a protein complex to mediate the coactivator functions of PRMT1 and CARM1 methylation events. To address this possibility, we performed tandem affinity purification, followed by tryptic digestion and mass spectrometry to identify the TDRD3 protein complex (Figure 1A & Table S1). The TDRD3 complex primarily harbored proteins involved in mRNA metabolism and transcriptional regulation, but TOP3B was also repeatedly isolated in this complex, and it is involved in DNA topological conformation control (Seki et al., 1998). We performed experiments to confirm the protein-protein interaction between TDRD3 and TOP3B. Using antibodies to endogenous TOP3B and TDRD3, the proteins were reciprocally co-immunoprecipitated (co-IP) from MCF7 cells (Figure 1B). Sub-cellular fractionation reveals that TOP3B and TDRD3 are present in both the nuclear and cytoplasmic cellular compartments, and that these two proteins interact in the nuclear compartment (Figure S1). The lung adenocarcinoma cell line (LCD) was reported to have lost both alleles of TDRD3 (Imoto et al., 2006). Western analysis confirms that the TDRD3 antibody cannot co-IP TOP3B from this LCD cell line (Figure 1C, left panel), and the transient introduction of GFP-TDRD3 re-established the co-IP (Figure 1C, right panel). Using glycerol gradient fractionation, we found that TDRD3 and TOP3B co-fractionate (Figure 1D, left panels). We also stably introduced GFP-TDRD3, or GFP only, back into the LCD cell line. When glycerol gradient fractionation was performed with the LCDGFP and LCDGFP-TDRD3 lines, we saw that the presence of TDRD3 can shift the position of TOP3B (Figure 1D, right panels), indicating that TDRD3 complexes with TOP3B. The interaction between TDRD3 and TOP3B is not mediated by DNA or RNA, because neither RNase A nor DNase I treatment impacts the efficiency of the co-IPs (Figure 1E). Furthermore, recombinant TDRD3 and TOP3B interact directly with each other as demonstrated using a pull-down assay (Figure 1F). For this GST pull-down assay, TDRD3 was expressed as a GST fusion protein, cleaved off the GST using PreScission protease, and then mixed with either GST or GST-TOP3B. The TDRD3-TOP3B interaction is not impacted by the presence of methylated peptides that bind the Tudor domain (Figure S2). Thus, these different approaches provide independent lines of evidence to support a direct, strong and physiologically relevant interaction between TDRD3 and TOP3B.
Figure 1. TDRD3 interacts with TOP3B.
(A) TAP-tag purification of TDRD3 protein complex from HEK293 cells. HEK293 cells stably expressing the empty TAP-tag vector (control) or TAP-tag-TDRD3 (TDRD3.com) were harvested, and a standard TAP procedure was applied. The eluted protein complex was separated by SDS-PAGE and silver-stained. The indicated gel slices were processed for protein identification using mass spectrometry.
(B) TDRD3 and TOP3B co-immunoprecipitate. MCF7 cells were immunoprecipitated with rabbit control IgG, αTDRD3 antibody, and αTOP3B antibody. The eluted protein samples were detected with αTDRD3 and αTOP3B antibodies.
(C) TDRD3 antibody does not cross-react with TOP3B protein. TDRD3 protein complex was immunoprecipitated from either MCF7 cells or LCD cells (TDRD3 deletion of both alleles) and detected with αTOP3B antibody (left panels). LCD cells were transiently transfected with GFP vector or GFP-TDRD3 and immunoprecipitated with αGFP antibody. The eluted samples were detected with αTOP3B and αTDRD3 antibodies (right panels).
(D) TDRD3 complexes with TOP3B in cells. MCF7 cell lysates were separated by glycerol gradient. The collected samples were detected with αTOP3B and αTDRD3 antibodies (left panels). Cell lysates from GFP or GFP-TDRD3 stable expressing LCD cells were separated by glycerol gradient and the collected samples were detected with αTOP3B antibody (right panels). The collected fractions are numbered below, and the complex size is indicated, as determined by standards.
(E) The TDRD3-TOP3B interaction is DNA and RNA independent. Co-immunoprecipitation experiments were performed as in (B), except that cell lysates were treated with either DNase I or RNase A before immunoprecipitation.
(F) TDRD3 directly interacts with TOP3B in vitro. GST pull-down assays were performed using recombinant GST, GST-TOP3B and TDRD3 proteins. Both input samples and pull-down samples were detected with αTDRD3 antibody (left). The GST-tag recombinant proteins in the pull-down samples were visualized by coomassie staining (right).
See also Figure S1, S2 and Table S1.
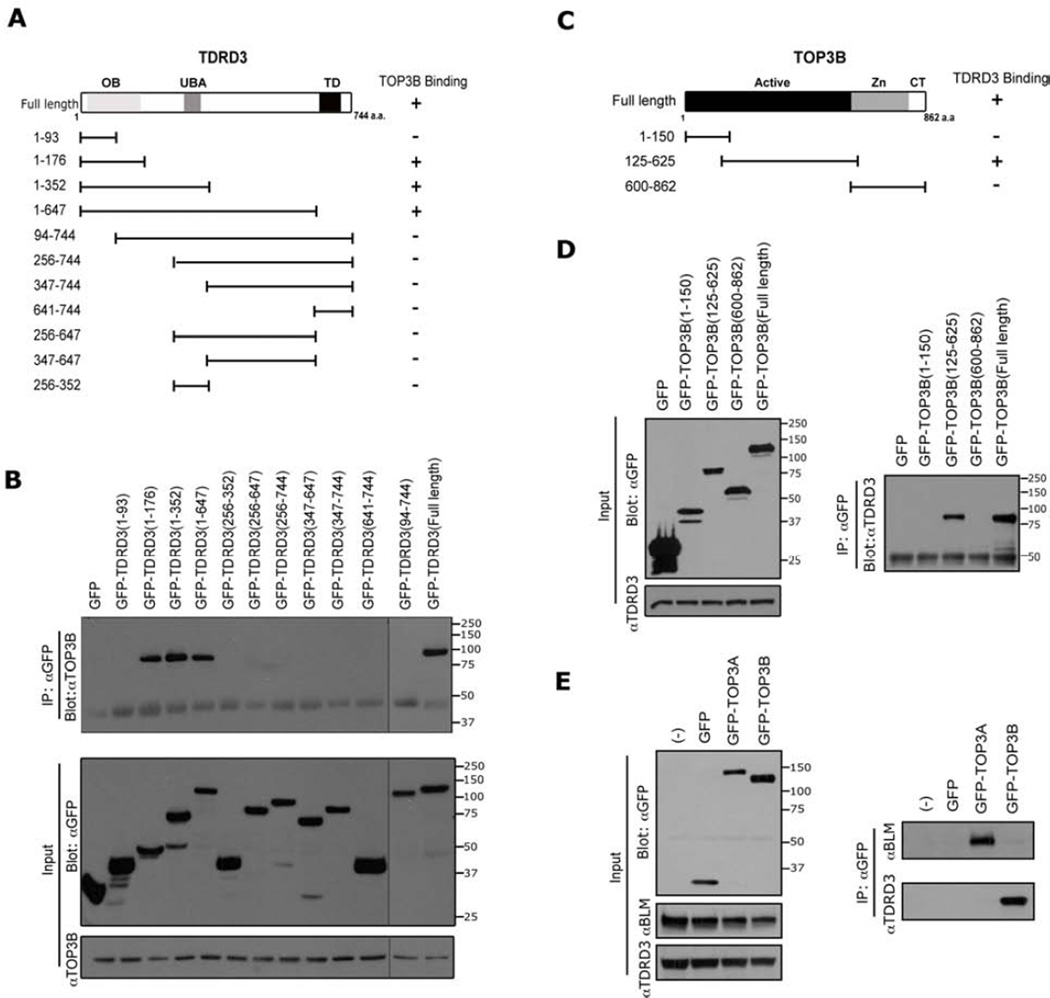
TOP3B interacts with the N-terminal region of TDRD3
We next mapped the interaction domains of TDRD3 and TOP3B. First, 11 different fragments spanning TDRD3 were transiently expressed as GFP fusion proteins in HeLa cells (Figure 2A & B). Immunoprecipitation of this GFP fusion series revealed that TOP3B interacts with three of these truncation constructs - the three that harbor an intact OB-fold (Figure 2B, top panel). An OB-fold refers to a motif that primarily binds single-stranded DNA or RNA (Theobald et al., 2003), although this motif has also been observed to function as a protein interacting interface (Jardetzky et al., 1994). It is unclear whether the OB-fold of TDRD3 can also bind RNA or DNA, and if these potential interactions could be impacted by TOP3B binding. Reciprocal mapping experiments were performed for TOP3B, and we found that the catalytically active cleavage/strand passage domain interacts with TDRD3 in transfected cells (Figure 2C & D). Interestingly, the RMI1 protein (RecQ-mediated genome instability) harbors an OB-fold that is required for the recruitment of TOP3A to the BLM protein complex (Raynard et al., 2008). TOP3A and TOP3B are both members of the 1A subfamily of DNA topoisomerases (Wang, 2002), and they share 40% identity over the catalytically active domains. Even with this high degree of shared identity, TOP3B is not found in the purified BLM- or RMI1-protein complexes, whereas TOP3A is (Xu et al., 2008) (Figure 2E, right panel). Conversely, we find that TDRD3 interacts with TOP3B, but not TOP3A (Figure 2E, right panel).
Figure 2. Mapping the interaction regions of TDRD3 with TOP3B.
(A) A series of GFP-fusion deletions of TDRD3 were generated. The locations of the OB-fold (OB), the ubiquitin-binding domain (UBA), and the Tudor domain (TD) are indicated. The graphic summary of the interactions observed in (B) is shown.
(B) A coIP assay was performed in HeLa cells transfected with the different TDRD3 GFP-fusion vectors. The cell lysates were immunoprecipitated with GFP antibody and eluted samples were blotted with αTOP3B. To observe all the samples, two membranes were spliced together. A gray line indicates the seam.
(C) Three GFP-fusion deletions of TOP3B constructs were generated. The locations of the catalytically active cleavage/strand passage domain (Active), the zinc-binding domain (Zn) and C-terminal domain (CT) are indicated. The graphic summary of the interactions observed in (D) is shown.
(D) A coIP assay was performed in HeLa cells transfected with the different TOP3B GFP-fusion vectors. Samples were prepared as described in (B) and blotted with αTDRD3 (right panel), and is shown (left panel).
(E) TDRD3 does not interact with TOP3A. HeLa cells were transiently transfected with GFP, GFP-TOP3A and GFP-TOP3B constructs. The cell lysates were immunoprecipitated with αGFP antibody and the eluted samples were assayed by Western blot analysis using αGFP antibody (left panel), and αTDRD3 and αBLM (right panel).
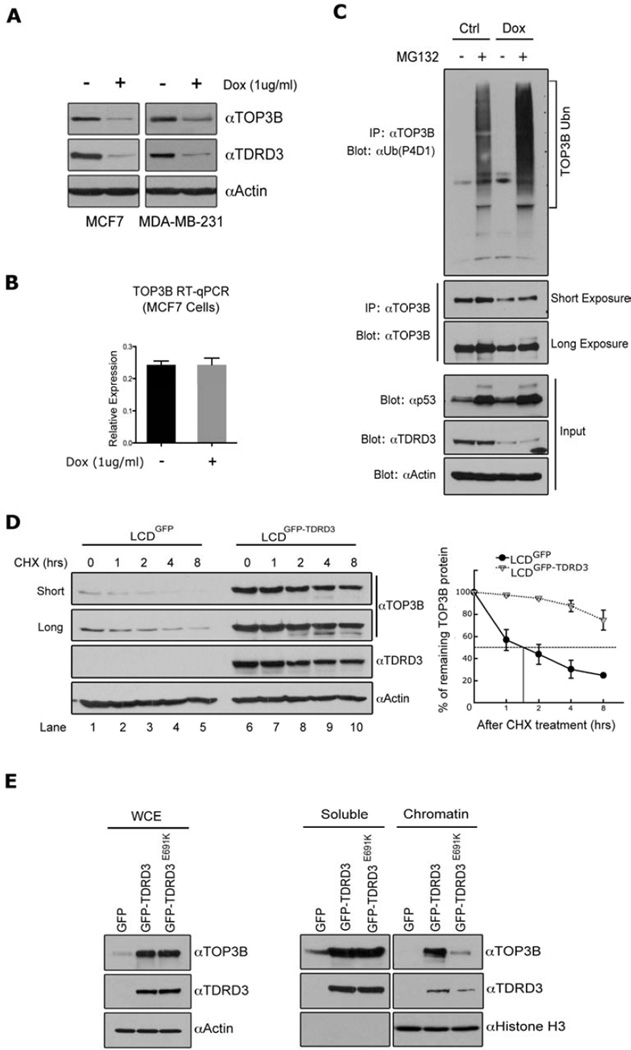
TDRD3 regulates TOP3B stability and recruitment to chromatin
To start evaluating the role of TDRD3 in cells, we generated two stable breast cancer cell lines (MCF7 and MDA-MB-231) carrying a tetracycline-inducible shRNA construct to knock-down endogenous TDRD3 levels. When these cells were treated with doxycycline (Dox), we observed an efficient reduction of TDRD3 levels together with an unexpected reduction in TOP3B levels (Figure 3A). A similar phenomenon was also seen with transient transfection of ezRNA targeting TDRD3 (Figure S3A). This suggests that TDRD3 either regulates the expression of TOP3B, or that an intact TDRD3-TOP3B complex is required to maintain TOP3B protein stability. We evaluated TOP3B RNA levels after TDRD3 knock-down, and detected no change in the expression of TOP3B (Figure 3B), suggesting that the TOP3B protein is stabilized by the TDRD3 interaction. Indeed, when MCF7 cells were treated with a proteasome inhibitor (MG132), we observe an increase in ubiquitination of TOP3B as detected with an anti-ubiquitin antibody (Figure 3C, upper panel – lanes 1 & 2). This laddering is exacerbated when TDRD3 is knocked-down in the presence of MG132 (Figure 3C, upper panel – lanes 3 & 4), even though there is less TOP3B. This experiment shows that under normal conditions, TOP3B is ubiquitylated (at low levels), and that the presence of TDRD3 protects it from enhanced ubiquitylation and subsequent targeting to the proteasome for degradation. To independently verify the requirement of TDRD3 for TOP3B protein stability, the LCDGFP and LCDGFP-TDRD3 lines were subjected to a time-course of cycloheximide (CHX) treatment to evaluate the stability of TOP3B in the absence and presence of TDRD3 (Figure 3D). Clearly, LCD cells transfected with GFP alone displayed very low levels of endogenous TOP3B, which had a very short half-life (50 minutes). By contrast, LCD cells that were rescued with GFP-TDRD3 display robust endogenous TOP3B stabilization.
Figure 3. TDRD3 stabilizes TOP3B and recruits it to chromatin.
(A) Reduction of TDRD3 levels destabilizes TOP3B. MCF7 and MDA-MB-231 cells were stably transfected with an inducible shRNA vector targeting TDRD3 mRNA. Both cells were treated with either vehicle or Doxycycline (Dox) (1ug/ml) for 6 days. The protein expressions of TOP3B and TDRD3 were detected by western blotting using αTOP3B and αTDRD3 antibodies. Actin served as a loading control.
(B) The mRNA levels of TOP3B in MCF7 cells treated under the same condition as in (A) were detected by RT-qPCR. Error bars represent standard deviation calculated from triplicate qPCR reactions.
(C) TOP3B is ubiquitinated in cells. Both control MCF7 cells and Dox-inducible TDRD3 shRNA expressing MCF7 cells were treated with vehicle or MG132 (10uM) for 16 hours. Cells were lysed in RIPA buffer and immunoprecipitated with αTOP3B antibody. The eluted samples were subjected to Western blot analysis using a αUbiquitin specific antibody (P4D1) and a αTOP3B antibody. The input samples were subjected to Western blot analysis using αp53 (positive control for MG132-treatment), αActin (loading control), and αTDRD3 antibodies.
(D) The re-expression of TDRD3 stabilizes the TOP3B protein. LCD cells stably expressing GFP or GFP-TDRD3 were treated with 50 ug/ml Cycloheximide (CHX) for the indicated time points. The cell lysates were detected by western blot using αTOP3B, αTDRD3 and αActin antibodies (left panel). TOP3B expression was quantified by densitometric analysis. Expression is represented as the percentage remaining relative to time zero. The half-life value was calculated using lines of best fit. Error bars represent standard deviation calculated from three independent Western blots (right panel).
(E) Wild-type Tudor domain, but not Tudor mutant TDRD3 recruits TOP3B to the chromatin. LCD cells stably expressing GFP, GFP-TDRD3 and GFP-TDRD3 (E691K) constructs were lysed. Total cell lysates (left panel), as well as soluble and chromatin fractions (right panel) were prepared and the expression levels of TOP3B and TDRD3 were detected by Western blot analysis. αHistone H3 blotting shows the quality of soluble/chromatin fractions and equal loadings.
See also Figure S3.
Apart from stabilizing TOP3B, TDRD3 could potentially recruit this topoisomerase to chromatin, because TOP3B interacts with the N-terminal portion of TDRD3 (Figure 2A & B) leaving the C-terminal Tudor domain free to interact with arginine methylated histones. To test this hypothesis, we generated a point mutation (E691K) in the Tudor domain of TDRD3, which blocks the ability of this domain to bind arginine methylated peptides (Figure S3B), but does not disrupt TDRD3‘s ability to stabilize TOP3B (Figure 3E, left panel). Next, total cellular proteins were separated into soluble and chromatin-associated fractions. A dramatic reduction in the recruitment of TOP3B to chromatin was observed when the Tudor domain of TDRD3 was non-functional (Figure 3E, right panel). This data supports the notion that TDRD3 functions as a scaffolding molecule that links TOP3B to chromatin through the ability of its Tudor domain to bind methylarginine motifs.
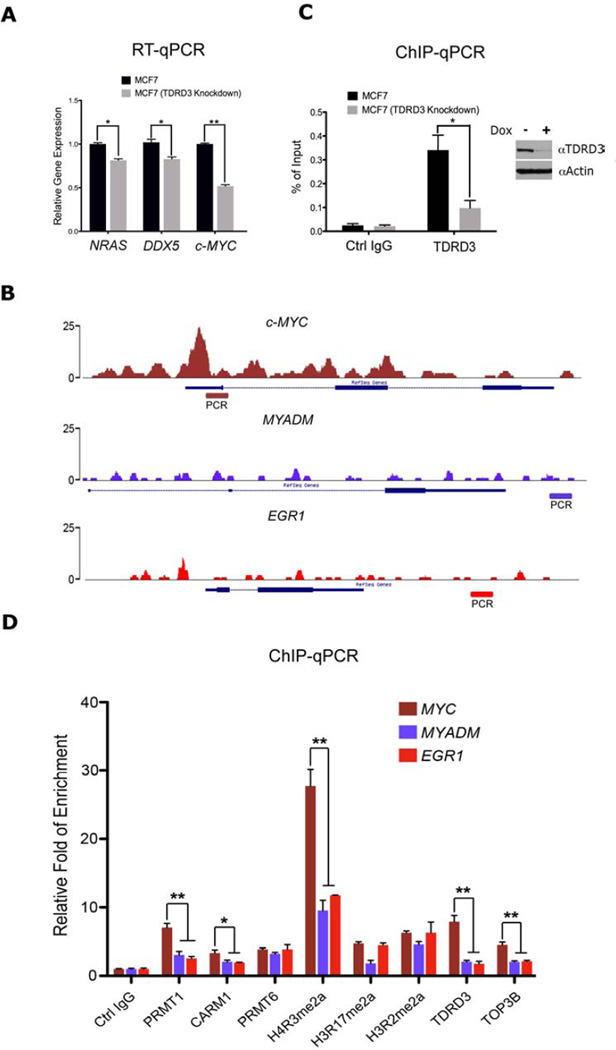
TDRD3 and TOP3B are recruited to the c-MYC locus to promote transcription
We previously reported that TDRD3 is recruited to the transcriptional start sites (TSSs) of NRAS, DDX5 and c-MYC (Yang et al., 2010). In parallel, the Reinberg group showed that an arginine methylation site on the CTD of RNAP II can bind TDRD3, and that this methylation site is important for the regulation of snoRNA expression (Sims et al., 2011). Quantitative measurements show that induced knock-down of TDRD3 results in significantly reduced expression of NRAS, DDX5 and c-MYC (Figure 4A), as well as snoRNA expression (Figure S4A & B). TDRD3 is not only enriched at the c-MYC TSS, but also in the body of the gene (Figure 4B, S4C & D). The MYADM and EGR1 genes do not display TDRD3 enrichment and are used as negative controls in later experiments. Using PCR primers that span exon 1 and part of the promoter of c-MYC, we confirm that TDRD3 is recruited to this region by ChIP-qPCR (Figure 4C), and as would be expected, knock-down of TDRD3 dramatically reduces the ChIP signal at this site. We next performed an in-depth analysis of PRMT pathways at this locus. We found that PRMT1 and CARM1 display significant enrichment at the c-MYC promoter region, in comparison to the control genes (MYADM and EGR1) (Figure 4D). In addition, the H4R3me2a mark, which is deposited by PRMT1, is the primary methyl-arginine mark at this region, and the effector complex (TDRD3-TOP3B) is also present. Similar observations were made at the NRAS and DDX5 loci, supporting the wide spread nature of this mode of regulation (Figure S4E & F). Thus, the complete PRMT•TDRD3•TOP3B axis is intact in the vicinity of the c-MYC, NRAS and DDX5 transcriptional start sites.
Figure 4. TDRD3-TOP3B complex regulates c-MYC gene expression.
(A) The mRNA levels of several TDRD3 target genes, in control and TDRD3 knockdown MCF7 cells, were detected by RT-qPCR. Error bars represent standard deviation calculated from triplicate qPCR reactions.
(B) Raw reads of the ChIP-seq tracings for three genes – c-MYC, MYADM, and EGR1 – demonstrating the enrichment of TDRD3 at c-MYC, but not negative controls. The regions that were subjected to qPCR in (C) and (D) are depicted.
(C) Chromatin immunoprecipitation experiment was performed in control and TDRD3 knockdown MCF7 cells using control IgG and αTDRD3 antibodies. The ChIP DNA was analyzed by qPCR with primers for the indicated region (B).
(D) Chromatin fractions from MCF7 cells were immunoprecipitated with αPRMT1, αCARM1, αPRMT6, H4R3me2a, H3R17me2a, H3R2me2a, αTDRD3, αTOP3B and control antibodies. The ChIP DNA was analyzed by qPCR with primers for the indicated region (B). * = p-value of 0.01 to 0.05, and ** = p-value of 0.001 to 0.01.
See also Figure S4.
The PRMT•TDRD3•TOP3B axis resolves R-loops at the c-MYC locus
To further study the underlying mechanisms of how TDRD3 regulates c-MYC gene transcription, we focused on its interaction partner – TOP3B. Both drosophila and mouse TOP3B have relaxation activity on negative supercoils (Seki et al., 1998; Wilson et al., 2000). To examine the topoisomerase activity of human TOP3B, we first incubated increasing amounts of purified TOP3B protein with negative supercoiled DNA substrates at 37 °C for 1 hour. Partial relaxation of supercoiled DNA was observed at a 1:12 DNA:enzyme ratio (Figure S5A & B). When conducted at a 1:60 DNA:enzyme ratio partial relaxation of substrate could be observed at 15 minutes and was complete by 60 minutes (Figure S5C). We next checked whether TDRD3 could regulate the activity of TOP3B by adding increasing amounts of TDRD3 protein to the reaction; however, substrate relaxation was not impacted (Figure S5D). Nor was TOP3B activity altered with the further addition of methylated peptides that can bind the Tudor domain of TDRD3 (Figure S5E–G). This indicates that TDRD3 does not affect TOP3B catalytic activity in vitro.
R-loop formation is dependent on DNA negative supercoiling (Aguilera and Garcia-Muse, 2012). The S9.6 monoclonal antibody recognizes RNA/DNA hybrids in a sequence independent manner (Boguslawski et al., 1986), and thus can be used to gauge the degree of R-loop formation in a DNA/RNA Immunoprecipitation (DRIP) experiment (Skourti-Stathaki et al., 2011). We applied this DRIP approach to assay whether TOP3B activity can reduce the levels of cotranscriptionally R-loops formation in vitro (Figure S5H). For this, we used an R-loop forming plasmid (pFC53), which contains a 1.2 kb DNA sequence from the murine Airn CpG island (CGI) (Ginno et al., 2012). In vitro transcription of this plasmid produced R-loops efficiently, as shown by agarose gel separation and quantified by in vitro DRIP-qPCR (Figure S5I – note smearing in lane 3). When TOP3B was added to the in vitro transcription reaction, the level of R-loop formation decreased ~8 fold (Figure S5J). These results indicate that TOP3B relaxes negative supercoiled DNA and reduces cotranscriptional R-loop formation in vitro.
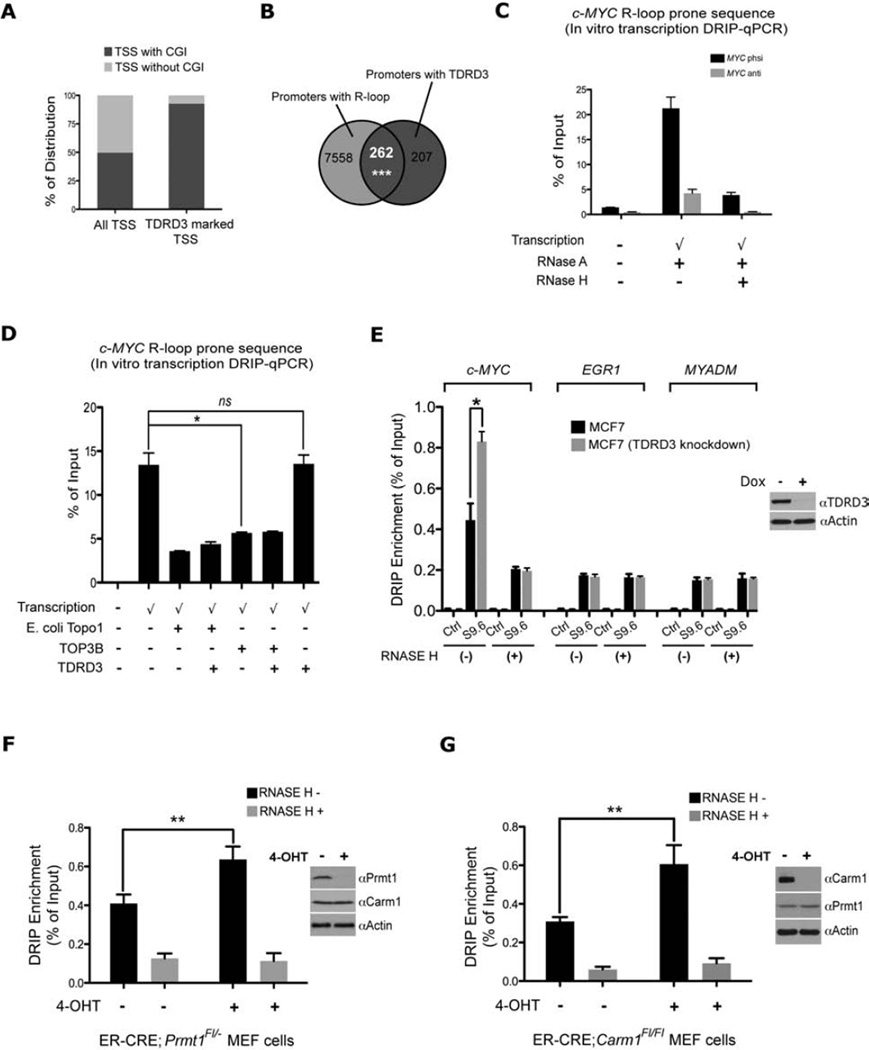
Previously we showed that more than 60% of TDRD3 peaks are at TSSs (Yang et al., 2010). An in-depth analysis of the TDRD3 occupied TSSs, revealed that the majority of these TSSs (92.6%) are CGI-containing TSSs, whereas genome-wide distribution of TSSs with CGI is comparable with non-CGI TSSs (49.7% vs 50.3%) (Figure 5A). CGI promoters are characterized by GC skew downstream of TSSs. R-loops form when the transcription machinery goes through skewed CGI promoters (Ginno et al., 2012). The fact that TDRD3 mainly peaks at CGI TSSs leads us to investigate whether TDRD3 promoter peaks overlap with R-loop forming promoters. Indeed, we found 55.9% of TDRD3 promoter peaks fall into predicted R-loop forming promoters (p<0.001) (Figure 5B). In cells, TDRD3 is likely to be the effector molecule that targets TOP3B activity to regions that are prone to R-loop formation. Importantly, the c-MYC locus is GC skewed over its CpG island and R-loops have been predicted to form there using an in silico approach (Wongsurawat et al., 2012). Furthermore, R-loops have been imaged at the in vitro transcribed c-MYC locus using transmission electron microscopy (Duquette et al., 2005). R-loop formation has been reported to prevent RNA pol II elongation and gene transcription (Belotserkovskii et al., 2010), and perhaps the ability of TDRD3 to regulate c-MYC gene expression is mediated by targeted R-loop reduction at the locus. To test this, we first confirmed that c-MYC undergoes efficient R-loop formation upon in vitro transcription (Figure 5C). Adding TOP3B to the in vitro transcription reaction decreases c-MYC R-loop levels, as efficiently as E. coli Topo1 (Figure 5D). The addition of TDRD3 does not affect TOP3B enzymatic activity (Figure 5D). To confirm c-MYC forms R-loop in vivo, we conducted a DRIP experiment on genomic DNA from control and TDRD3 knockdown MCF7 cells. Knockdown of TDRD3 in MCF7 cells leads to a significant increase in c-MYC R-loop formation, compared with control MCF7 cells (Figure 5E & S5K). Similar observations were made at the NRAS and DDX5 loci (Figure S5L & M). When we checked genomic regions that do not show TDRD3 occupancy (EGR1 & MYADM), this phenomenon was not observed. These data demonstrate that loss of TDRD3 expression leads to elevation of c-MYC, NRAS and DDX5 R-loop in vivo. Furthermore, overexpression of RNase H1 in cells negates the effects of TDRD3 loss on the DRIP signal and c-MYC expression (Figure S5N & O), but does not impact the recruitment of TDRD3 to chromatin (Figure S5P & Q).
Figure 5. TDRD3-TOP3B complex regulates co-transcriptional formed c-MYC R-loop.
(A) TDRD3 peaks mark CGI promoters. TDRD3 marked TSSs (579) were analyzed for CGI occupancy, showing 92.6% (536) are CGI associated. Human genome-wide analysis of 28875 TSSs reveals that roughly half of them contain CGIs. TDRD3 ChIP-seq data used for this comparison was previously published (Yang et al., 2010).
(B) TDRD3 peaks are enriched in R loop forming promoters. TDRD3 ChIP-seq data (Yang et al., 2010) was compared with 7820 predicted R-loop forming promoters (Ginno et al., 2012). The p value was determined by a permutation test: the overlap between TDRD3 and a random list of 7820 promoters (defined as −1000 to +1000 from TSS) was tested 10000 times to decide the p value.
(C) In vitro generation R-loops at the c-MYC gene. The c-MYC gene R-loop forming region was cloned and transcribed in the physiological direction of transcription (phsi) or anti-physiological orientation (Anti). Each transcribed sample was split equally and one half treated with RNase H. The in vitro transcribed samples were immunoprecipitated with S9.6 antibody and the DRIP DNA were analyzed by qPCR. An untranscribed control is included.
(D) TOP3B resolves c-MYC R-loop during in vitro transcription. c-MYC gene R-loop forming region was transcribed in the physiological direction in the presence or absence of E. coli Topo1, TOP3B and TDRD3. The DRIP DNA samples were analyzed by qPCR.
(E) Loss of TDRD3 increases c-MYC gene R-loop formation in MCF7 cells. DRIP experiment was performed on RNase H treated or not treated (− or +) genomic DNA from parental cells, and TDRD3 induce knockdown MCF7 cells. The DRIP DNA samples were analyzed by qPCR.
(F) Loss of PRMT1 increases c-MYC gene R-loop formation in MEF cells. Prmt1Fl/− ER-Cre MEFs were untreated or treated with 4-hydroxytamoxifen (OHT) for 6 days to remove Prmt1 (see inset). Genomic DNA from these cells were untreated or treated with RNase H. DRIP experiment was performed with S9.6 antibody and the DRIP DNA were analyzed by qPCR with primers for mouse c-Myc genes. ** = p-value of 0.001 to 0.01.
(G) Loss of CARM1 also increases c-Myc gene R-loop formation in MEF cells. Carm1Fl/Fl ER-Cre MEFs were untreated or treated with 4-hydroxytamoxifen (OHT) for 6 days to remove Carm1 (see inset). DRIP experiment was performed as described in (E). All DRIP-qPCR experiments were independently performed at least three times. Error bars represent standard deviation calculated from triplicate qPCR reactions of one representative experiment.
See also Figure S5.
Considering the fact that recruitment of TDRD3 to c-MYC gene is mediated by methylarginine histone marks deposited by PRMT1 and CARM1 (Figure 4D), we next assessed whether PRMT1 and CARM1 regulate c-MYC R-loop level. Using inducible knockout systems, we showed that loss of either PRMT1 or CARM1 increased R-loop formation at the c-MYC locus (Figure 5F & G). The loss of TDRD3 has a greater effect on R-loop formation (Figure 5E) than the loss of either PRMT on its own (Figure 5F & G), likely because these two PRMTs are known to function synergistically (Bedford and Clarke, 2009). These data suggest that loss of methylarginine effector TDRD3 (the reader), as well as the arginine methyltransferases (the writers), increase co-transcriptionally formed R-loops at the c-MYC locus.
TDRD3 deficient mice display decreased TOP3B stability and increased genomic instability
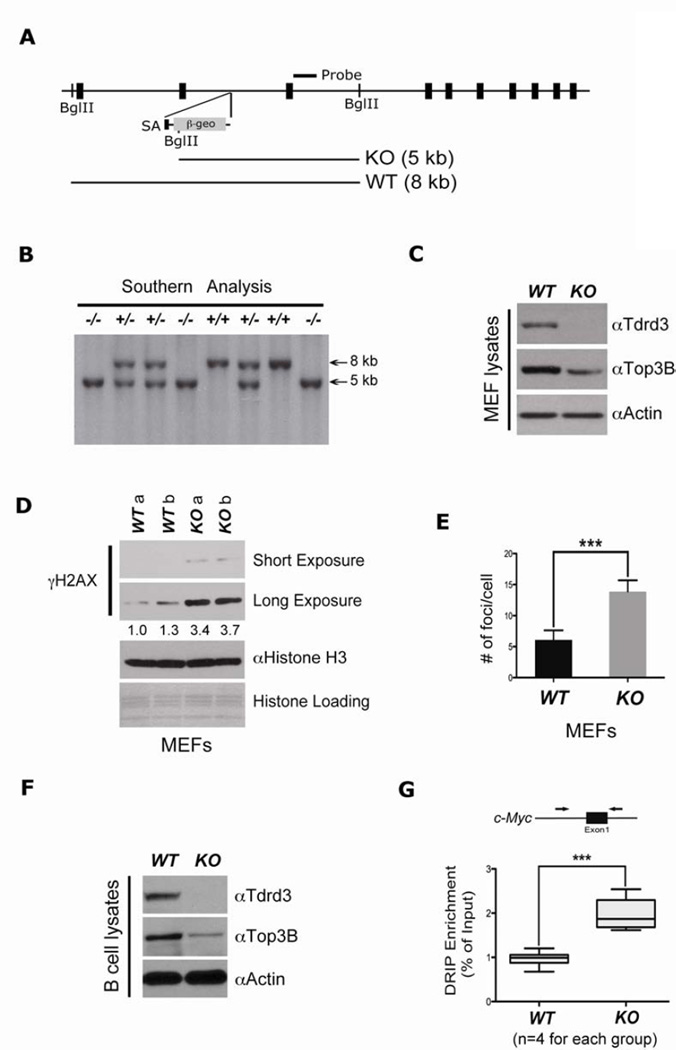
Tdrd3 knockout mice were generated from ES cells that harbored a gene-trap insertional mutation between exon 2 and 3 (Figure 6A). The gene-trap vector introduces a new Bgl II site, which results in a three-kilobase down-shift of the Southern blotting band for mice carrying the mutant allele (Figure 6B). Tdrd3−/− embryos are developmentally normal and the adults are viable and fertile. To facilitate additional studies of Tdrd3 cellular functions, we cultured mouse embryonic fibroblasts (MEFs) derived from E12.5 embryos, and confirmed the lack of Tdrd3 expression by Western blot analysis (Figure 6C). The Tdrd3−/− MEF line displayed reduced Top3B levels, consistent with previous findings (Figure 3A & D).
Figure 6. Tdrd3 knockout mice show genomic instability.
(A) The exon/intron structure of the Tdrd3 locus. Position of the external probe is shown. The trapping vector is integrated into the intronic region between exons 2 and 3.
(B) Southern blot analysis of tail DNA from a mouse litter, derived from a Tdrd3+/− intercross. The DNA was digested with BglII, resulting in a 5-kb mutant band and a 8-kb wild-type band.
(C) Primary MEFs were generated from E12.5 embryos. Western blot validated the loss of Tdrd3 expression and reduced expression of TOP3B in Tdrd3−/− MEFs.
(D) Elevated genomic instability in Tdrd3−/− MEFs compared with wild-type MEFs detected by γH2AX Western blot (right). MEFs derived from two different wild-type and knock-out E12.5 embryos were assessed. Band intensities were quantified and normalized relative to their respective histone H3 bands. Quantification was expressed as folds of the lowest value.
(E) Quantitation of genomic instability as a result of TDRD3 loss, as evidenced by primary MEFs γH2AX foci counting.
(F) Naïve B cells purified from wild-type and Tdrd3−/− mice spleen were examined for TDRD3 and TOP3B expression.
(G) Elevated c-Myc R-loop formation in Tdrd3−/− B cells. R-loop DRIP experiments were carried out with naïve B cells cultured in IL-4 and LPS for 72 hours. c-Myc R-loops were detected using mouse-specific primers as indicated in the methods section. Four mice per group were tested.
See also Figure S6.
Clearly, the loss of Tdrd3 will impair Top3B cellular functions, either by preventing its recruitment to chromatin, or by substantially destabilizing it, or both. Like Tdrd3−/− mice, Top3B−/− mice are viable, but exhibit a lifespan that is reduced by about one third (Kwan and Wang, 2001), and are more inclined to develop autoantibodies against nuclear antigens (Kwan et al., 2007). It was recently shown that Top3B−/− MEFs have increased DNA double-strand breaks as manifested by an increased γH2AX signal (Mohanty et al., 2008). Tdrd3−/− primary MEFs display a very similar phenotype, with increased H2AX phosphorylation and more γH2AX foci (Figure 6D & E). When Tdrd3−/− primary MEFs are challenged with a DNA damaging agent, they also display more H2AX phosphorylation than their wild-type counterparts (Figure S6A). It is possible that increased R-loop formation in Tdrd3−/− primary MEFs could account for the increase in γH2AX signal, because these single stranded DNA regions are more prone to breakage (Helmrich et al., 2011; Mischo et al., 2011).
Next we asked whether R-loop formation was elevated in cells isolated from Tdrd3−/− mice. We focused on B cells because of the known role for R-loop formation in class switch recombination (CSR) and the formation of c-Myc/Igh translocation in these cells (Duquette et al., 2005). We first confirmed that Tdrd3 is lost and Top3B levels are reduced in B cells isolated from Tdrd3−/− mice (Figure 6F), as we have observed in other mouse cells (Figure 6C) and in human cells (Figure 3A). We next evaluated the levels of R-loop formation at the c-Myc and Igh loci. For this experiment, Naïve B cells were cultured for three days in the presence of IL-4 and LPS. This stimulation induces proliferation, upregulation of c-Myc and Igh gene expression, and CSR from IgM to IgG1 (Stavnezer et al., 2008; Wierstra and Alves, 2008). The dramatically elevated transcription levels at these two loci are accompanied by R-loop formation (Yu et al., 2003). DRIP experiments revealed a doubling of R-loop formation at both the c-Myc locus (Figure 6G), and the Igh locus (Figure S6B), when Tdrd3 is lost.
Some gene-trap mutants display “leaky” expression of the endogenous gene, likely due to a small amount of splicing that can occur to bypass the trapping cassette (Swiercz et al., 2007). When we immunoprecipitated large amounts of TDRD3 from wild-type and knock-out B cells and subjected the samples to Western analysis, we did detect a small amount of endogenous Tdrd3 protein in knock-out cells (Figure S6C). We conclude that the insertional mutation results in a hypomorphic Tdrd3 allele, with 5–7% (as determined by densitometric quantitation) Tdrd3 expression levels in Tdrd3−/− cells.
TDRD3 null mice display increased translocation between the c-Myc and Igh loci
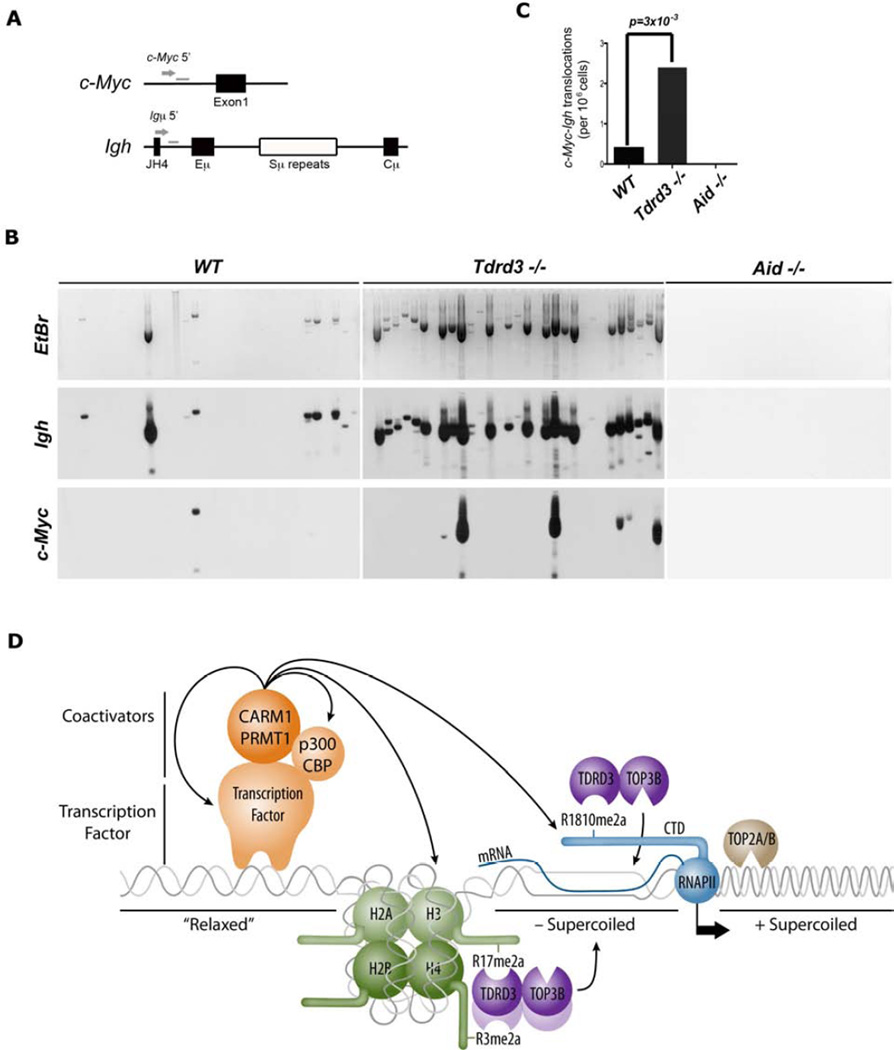
The observations that 1) there is an increased frequency of double-stranded DNA breaks in Tdrd3−/− MEFs (Figure 6D & E), 2) Tdrd3−/− B cells express reduced levels of Top3B (Figure 6F), and 3) R-loop formation is enhanced at both the c-Myc and Igh loci in these cells (Figure 6G & S6B) prompted us to pursue the hypothesis that the loss of Tdrd3 could cause an increase in the rate of c-Myc/Igh translocations. This is a process demonstrated to be driven by a combination of R-loop prone regions and AID-mediated DNA breaks (Ruiz et al., 2011). To examine the effect of Tdrd3-loss on these translocations, we assayed stimulated B cells for aberrant juxtaposition of the chromosomal regions that carry c-Myc and Igh, using a previously described PCR/Southern blotting approach (Ramiro et al., 2004) (Figure 7A & B). A six-fold increase in translocation was seen in Tdrd3−/− B cells compared to wild-type controls (Figure 7C). We also saw a dramatic increase in rearrangements of the Igh switch Mu region upon loss of Tdrd3 (Figure 7B). As expected, no translocations are observed in the absence of AID (Figure 7B). Interestingly, the loss of Tdrd3 does not impact CSR frequencies in these activated B cells (Figure S7A & B). Thus, Tdrd3 separates the CSR and chromosome translocation phenotypes that are associated with AID loss.
Figure 7. Increased c-Myc/Igh translocations in TDRD3 knockout mice.
(A) Schematic showing primers used for the PCR translocation amplification and the Southern blot probes.
(B) Naïve B cells from wild-type, Tdrd3−/− and Aid−/− mice were cultured in IL-4 and LPS for 72 hours and assayed for translocations by long-range PCR. Representative ethidium bromide gel and Southern blot with Igh and c-Myc probes are shown. Translocations were amplified from genomic DNA isolated from 105 cells per lane.
(C) Summary of translocation events detected (5×106 total cells assayed in 50 lanes) from six spleens of each genotype in three independent experiments. P value was determined by Fisher’s exact test.
(D) Model of how TDRD3-TOP3B complex regulates the formation of R-loop. TDRD3 functions as an effector molecule recognizing methyl-arginine histone marks and the methylated Pol II CTD recruiting TOP3B, which in turn helps resolve negative supercoiled DNA and prevent R-loop formation caused by active gene transcription.
See also Figure S7.
DISCUSSION
PRMTs promote transcription by directing the machinery for R-loop resolution
PRMT1 and CARM1 are the two major PRMTs that display transcriptional coactivator activity (Bedford and Clarke, 2009). They carry out their coactivator functions by not only methylating histone tails, but also RNA processing factors, transcriptional regulators, as well as transcription factors themselves (Lee and Stallcup, 2009). Because there are a large number of substrates for these two PRMTs, it is likely that multiple mechanisms exist, some of them possibly redundant, to increase transcription at specific loci. It has been shown that dysregulation of transcription and RNA processing pathways increase R-loop formation and genome instability in yeast (Aguilera and Garcia-Muse, 2012; Mischo et al., 2011; Wahba et al., 2011). Results presented in Figure 5F & G demonstrate that knock-down of PRMT1 and CARM1 results in elevated R-loop formation, as does the knock-down of TDRD3 (Figure 5D). The fact that TDRD3 is recruited to R-loop prone regions (Figure 5A) bolsters the idea that PRMT activity at these sites helps concentrate TDRD3-TOP3B where it is most needed, and broadly influences transcription efficiency in this manner. The involvement of a DNA topoisomerase, which actively works on underwound DNA in the wake of RNAP II, provides a fundamental novel mechanism by which PRMTs can coactivate gene transcription.
Topoisomerases and transcriptional regulation in mammalian cells
Direct involvement of DNA topoisomerases in transcription has been proposed (Liu and Wang, 1987). In this “twin-domain” model, advancing RNAP II generates positive supercoils in the DNA template ahead of it and negative supercoils behind it, and topoisomerase activity is required to relax the tension generated by this supercoiling and facilitate efficient transcription. This soon proved to be the case, with the identification of a purified transcriptional coactivator from HeLa cells as TOP1 (Kretzschmar et al., 1993). Subsequently, both TOP2A and TOP2B have been shown to regulate in vitro transcription on chromatin templates and nuclear receptor transcription in cells (Ju et al., 2006; Mondal and Parvin, 2001). Under normal conditions, TOP3A likely does not play a role in transcriptional regulation, but rather DNA repair (Raynard et al., 2008). However, the mislocalization of TOP3A, through a MLL translocation has been reported in an acute myeloid leukemia patient (Herbaux et al., 2012). This chimeric protein, which is an in-frame fusion of the complete TOP3A open reading frame, will likely redirect its activity to gene promoters to promote transcription. Thus, although TOP1, TOP2A and TOP2B have all been implicated as activators in transcriptional regulation, and TOP2B does co-localize with H3K4me2 marks (Tiwari et al., 2012), no mechanism for their recruitment to chromatin has been established. Here we show that TDRD3 performs this role for TOP3B. It is intriguing that TDRD3 also harbors a functional UBA domain, between its Tudor domain and OB fold (Linder et al., 2008). Thus, TDRD3-TOP3B could also be directed to specific loci where ubiquitin signals are enriched.
TDRD3 is implicated in c-Myc/Igh translocation but not class switch recombination
Although Aid activity is focused at the Ig loci, considerable “off-target” activity has been demonstrated (Chiarle et al., 2011; Klein et al., 2011), resulting in mutations and chromosome translocations that are frequently observed in lymphomas, including the oncogenic c-Myc/Igh translocation of Burkitt’s lymphoma. In B cells, this translocation requires Aid, which induces the DSB at the Igh and c-Myc loci, however DSB at c-Myc are less frequent and rate-limiting (Robbiani et al., 2008). We found here that loss of TDRD3 results in increased R-loop formation at c-Myc (Figure 6G), and we observed a significant increase in chromosome translocation to Igh (Figure 7B & C). The driving force behind the elevated c-Myc/Igh translocation frequency may be increased R-loop formation at the rate-limiting c-Myc locus.
R-loops have also been proposed to have a major role in Aid mediated CSR. During an immune response, B lymphocytes diversify the class of expressed antibody (IgM, IgG1, etc) by CSR of the IgH (Stavnezer et al., 2008). Recombination is triggered by Aid mediated DSB formation in transcribed non-coding, switch regions. Mammalian switch regions are composed of repetitive 1–12 kb elements with a G-rich non-template sequence that form stable R-loops in vitro and in vivo (Yu et al., 2003). A role for R-loops in CSR is supported by several lines of genetic evidence. Deletion of the G-rich content, inversion of R-loop forming non-template strand to the template strand, and decreasing the G content all lead to significant decreases in CSR (Chaudhuri et al., 2003; Shinkura et al., 2004). Furthermore, a non-switch region that forms R-loops can support CSR (Shinkura et al., 2004). Loss of Tdrd3 results in increased R-loop detection at the switch mu region (Figure S6B), however we do not see enhancement of CSR (Figure S7B). It is possible that R-loop contribution is already optimal, and further enhancement, due to TDRD3 loss, does not boost CSR. On the other hand TDRD3 may not target the switch γ1 switch region, but instead preferentially affect short-range recombination. During CSR intra-switch deletions and rearrangements at switch mu normally occur at a fairly high rate (Dudley et al., 2002). Following TDRD3 loss, we detect greater rearrangement of the switch mu region, which is indicative of enhanced intra-switch region recombination (Figure 7B).
A dual role for the TDRD3-TOP3B complex – one nuclear, the other cytoplasmic
Recently, two research groups identified the TDRD3-TOP3B as binding partners for fragile × mental retardation protein (FMRP) (Stoll et al., 2013; Xu et al., 2013). We also found FMRP and its homologs (FXR1 & 2) in the TDRD3 protein complex (Figure 1A), as has been previously reported (Linder et al., 2008). FMRP is a RNA-binding protein that associates with polyribosomes, and can function as a regulator of translation (Laggerbauer et al., 2001). Indeed, the TDRD3-TOP3B-FMRP complex is associated with the mRNA pool that is undergoing translation (Stoll et al., 2013). Importantly, TOP3B possesses RNA topoisomerase activity (Stoll et al., 2013; Xu et al., 2013). Although, it is not yet clear how this RNA-directed activity is implicated in FMRP function at the ribosomes, it has been proposed to reduce mRNA topological stress and thus promote translation (Xu et al., 2013). Thus, TDRD3-TOP3B containing complexes regulate both transcription and translation. Considering the emerging evidence that dysregulated R-loop formation contributes to neuronal disorders (Powell et al., 2013; Skourti-Stathaki et al., 2011), the elevation of R-loops observed with TDRD3 loss might also help explain the mechanisms underlying certain neurodevelopment disorders, where deletion of TOP3B is found (Stoll et al., 2013).
The PRMT•TDRD3•TOP3B axis model is depicted in Figure 7D. We envision that PRMT1/CARM1 are recruited to promoters by transcription factors. There, they methylate a number of different proteins, including histone H3, H4 and RNAPII. These methylation events are “read” by TDRD3, which in turn recruits TOP3B to heavily transcribed genes. The TDRD3-TOP3B complex will be concentrated between the transcriptional start site and the elongating RNAPII, the precise region that is underwound and prone to R-loop formation. TOP3B is thus delivered by the methylarginine effector molecule TDRD3 to the region of chromatin where it is most needed.
EXPERIMENTAL PROCEDURES
Antibodies
TDRD3 antibodies have been described, as have the antibodies for GFP, GST, PRMT1, CARM1, PRMT6, H4R3me2a, H3R17me2a and H3R2me2a antibodies (Yang et al., 2010). Mouse monoclonal TOP3B antibody (ab56445) and rabbit anti-BLM antibody (ab476) were purchased from Abcam. Mouse monoclonal DNA/RNA hybrid antibody (S9.6) has been described (Ginno et al., 2012). γH2AX (ser139) antibody (05–636) was purchased from Millipore. Mouse monoclonal Flag antibody (F3165) was purchased from Sigma.
Mice and Cell lines
Tdrd3 knockout mice were generated from a targeted mES clone RRK474 (BayGenomics). The Aid knockout mice have been described before (Ramiro et al., 2004). MCF7 cells, MCF7-tet-on-shCARM1, human VMRC-LCD cells, and GFP/GFP-TDRD3 stably expressing LCD cells have been described before (Yang et al., 2010). MCF7 tet-on-shTDRD3 cell line, tamoxifen induced Prmt1fl/− and Carm1fl/fl MEFs, and Tdrd3−/− MEFs are described in supplemental material.
DNA/RNA immunoprecipitation (DRIP)
The DRIP approach was modified from published studies (Ginno et al., 2012; Skourti-Stathaki et al., 2011), and described in supplemental material.
Supplementary Material
HIGHLIGHTS.
TDRD3 interacts with TOP3B to stabilize this methylarginine effector complex.
The Tudor domain of TDRD3 facilitates the recruitment of this complex to chromatin.
The TDRD3-TOP3B complex resolves R-loops at the c-MYC locus.
The absence of Tdrd3 results in increased c-Myc/Igh translocation in vivo.
ACKNOWLEDGEMENTS
We thank Maria Person for mass spectrometry analysis at the “Protein and Metabolite Analysis Facility” (UT Austin) supported by RP110782 (CPRIT). MT Bedford is supported by an NIH grant (DK062248); F Chedin is supported by an NIH grant (GM094299). MT Bedford is a cofounder of EpiCypher.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc Natl Acad Sci U S A. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf IF, Chedin F. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013 doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Herbaux C, Poulain S, Meyer C, Marschalek R, Renneville A, Fernandes J, Theisen O, Tricot S, Simon M, Duthilleul P, Daudignon A. TOP3A, a new partner gene fused to MLL in an adult patient with de novo acute myeloid leukaemia. Br J Haematol. 2012;157:128–131. doi: 10.1111/j.1365-2141.2011.08908.x. [DOI] [PubMed] [Google Scholar]
- Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Chi YI, Stauffacher C, Strominger JL, Wiley DC. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature. 1994;368:711–718. doi: 10.1038/368711a0. [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci U S A. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Greenwald RJ, Mohanty S, Sharpe AH, Shaw AC, Wang JC. Development of autoimmunity in mice lacking DNA topoisomerase 3beta. Proc Natl Acad Sci U S A. 2007;104:9242–9247. doi: 10.1073/pnas.0703587104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Wang JC. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc Natl Acad Sci U S A. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile × mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Plottner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17:3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S, Town T, Yagi T, Scheidig C, Kwan KY, Allore HG, Flavell RA, Shaw AC. Defective p53 engagement after the induction of DNA damage in cells deficient in topoisomerase 3beta. Proc Natl Acad Sci U S A. 2008;105:5063–5068. doi: 10.1073/pnas.0801235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- Powell WT, Coulson RL, Gonzales ML, Crary FK, Wong SS, Adams S, Ach RA, Tsang P, Yamada NA, Yasui DH, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent Holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet. 2011;7:e1002009. doi: 10.1371/journal.pgen.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Seki M, Onodera R, Katada T, Enomoto T. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIbeta) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. The Journal of biological chemistry. 1998;273:28553–28556. doi: 10.1074/jbc.273.44.28553. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G, Pietilainen OP, Linder B, Suvisaari J, Brosi C, Hennah W, Leppa V, Torniainen M, Ripatti S, Ala-Mello S, et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013 doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Burger L, Nikoletopoulou V, Deogracias R, Thakurela S, Wirbelauer C, Kaut J, Terranova R, Hoerner L, Mielke C, et al. Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci U S A. 2012;109:E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- Wilson TM, Chen AD, Hsieh T. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J Biol Chem. 2000;275:1533–1540. doi: 10.1074/jbc.275.3.1533. [DOI] [PubMed] [Google Scholar]
- Wilson-Sali T, Hsieh TS. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc Natl Acad Sci U S A. 2002;99:7974–7979. doi: 10.1073/pnas.122007999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsurawat T, Jenjaroenpun P, Kwoh CK, Kuznetsov V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012;40:e16. doi: 10.1093/nar/gkr1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shen W, Guo R, Xue Y, Peng W, Sima J, Yang J, Sharov A, Srikantan S, Fox D, 3rd, et al. Top3beta is an RNA topoisomerase that works with fragile × syndrome protein to promote synapse formation. Nat Neurosci. 2013 doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.