SUMMARY
Inadequate adenosine-to-inosine editing of noncoding regions occurs in disease, often uncorrelated with ADAR levels, underscoring the need to study deaminase-independent control of editing. C. elegans have two ADAR proteins, ADR-2 and the theoretically catalytically inactive ADR-1. Using high-throughput RNA sequencing of wild-type and adr mutant worms, we expanded the repertoire of C. elegans edited transcripts over 5-fold and confirmed that ADR-2 is the only active deaminase in vivo. Despite lacking deaminase function, ADR-1 affects editing of over 60 adenosines within the 3′ UTRs of 16 different mRNAs. Furthermore, ADR-1 interacts directly with ADR-2 substrates, even in the absence of ADR-2; and mutations within its dsRNA binding domains abolished both binding and editing regulation. We conclude that ADR-1 acts as a major regulator of editing by binding ADR-2 substrates in vivo and raises the possibility that other dsRNA binding proteins, including the inactive human ADARs, regulate RNA editing by deaminase-independent mechanisms.
INTRODUCTION
RNA editing is a posttranscriptional process that introduces changes in RNA sequences and structures (Gott and Emeson, 2000). The most prevalent form of RNA editing in metazoa is the hydrolytic deamination of adenosine (A) to inosine (I) (Nishikura, 2010). Adenosine deaminases that act on RNA (ADARs) bind to double-stranded regions of RNA and catalyze this type of editing (Goodman et al., 2012; Savva et al., 2012). Although RNA editing was initially thought to be restricted to a few select mRNAs in the central nervous system, it is now clear that adenosine deamination is widespread, with current estimates of 400,000–1,000,000 A-to-I edits in the human transcriptome (Ramaswami et al., 2013).
Adenosine and inosine have different base-pairing properties; therefore, editing alters RNA structure. Furthermore, as inosine is recognized as guanosine by cellular machinery, RNA editing can modify splice sites, alter the amino acid encoded by a codon and redirect miRNAs and siRNAs to new targets (Hundley and Bass, 2010; Rosenthal and Seeburg, 2012). As the extent of RNA editing varies during development and between cell types (Wahlstedt et al., 2009), this type of modification dynamically regulates gene expression (Tan et al., 2009).
The molecular diversity generated by ADARs is most pronounced in the brain transcriptome (Blow et al., 2004; Paul and Bass, 1998). Consistent with this, deletion of ADARs in lower organisms, such as C. elegans and Drosophila, results in behavioral defects (Palladino et al., 2000; Tonkin et al., 2002), indicating that RNA editing is required for proper neuronal function. Furthermore, alterations in editing levels have been observed in a number of neuropathological diseases, including epilepsy, depression, amyotrophic lateral sclerosis, and brain tumors (Farajollahi and Maas, 2010; Tariq and Jantsch, 2012).
In both development and disease, ADAR expression levels do not directly correlate with the extent of editing (Maas et al., 2001; Wahlstedt et al., 2009), implying that other mechanisms exist to regulate ADAR-mediated RNA editing. Both alternative splicing (Lai et al., 1997; Rueter et al., 1999) and post-translational modification (Desterro et al., 2005) of ADARs generate less active variants of ADARs. Likewise, editing can be inhibited by sequestration of ADAR in the nucleolus (Sansam et al., 2003) or enhanced by proteins that promote nuclear localization of ADARs (Marcucci et al., 2011; Ohta et al., 2008). In addition to proteins that directly regulate ADARs, it has recently been demonstrated that both the local RNA structure (Daniel et al., 2012) and RNA binding protein (RBP) landscape of individual transcripts (Tariq et al., 2013) regulate ADAR activity. To date, none of these mechanisms have been linked to reduced RNA editing activity in disease (Orlandi et al., 2012). Furthermore, it is unlikely that regulators of specific transcripts will play a key role in the global hypoediting of transcripts observed in many human cancers and neurological diseases.
To identify mechanisms that could decrease global RNA editing levels, we focused on the role of catalytically inactive ADAR family members. The C. elegans genome encodes two proteins with the common ADAR family domain structure (ADR-1 and ADR-2). However, ADR-1 lacks several key amino acids required for deaminase activity. Worms lacking the adr-2 gene, have no detectable editing of the six known edited endogenous mRNAs (Tonkin et al., 2002), suggesting that ADR-2 is the catalytically active ADAR protein in worms. However, initial studies of worms lacking adr-1 revealed alterations in the editing efficiency of all six endogenous mRNAs examined (Tonkin et al., 2002). In addition, recent deep sequencing of C. elegans small RNAs identified over 30 small RNAs that are edited in vivo, and each have altered editing levels in worms lacking adr-1 (Warf et al., 2012). These prior observations suggest ADR-1 regulates editing. However, it is also possible that background mutations in the strains lacking adr-1 contribute to alterations in editing or that loss of adr-1 indirectly affects editing by ADR-2. To directly address these concerns, we developed a quantitative assay to measure in vivo editing levels of worms expressing adr-1 transgenes. About 40% of adenosines within three known edited mRNAs were affected by loss of adr-1. Furthermore, using a combination of high-throughput RNA sequencing of transgenic worms and probabilistic modeling we were able to identify 48 novel edited transcripts and demonstrate that loss of adr-1 affects editing of at least half of these newly identified ADAR targets. Using an RNA immunoprecipitation (RIP) assay, we demonstrate that ADR-1 directly binds to known editing targets in vivo, that disrupting this binding alters editing of the mRNAs, and that ADR-1 and ADR-2 co-occupy transcripts in vivo. In summary, we demonstrate that catalytically inactive ADR-1 acts as a global regulator of editing by binding to target mRNAs and modulating the accessibility of ADR-2 for target adenosines.
RESULTS
ADR-1 significantly alters RNA editing of multiple C. elegans mRNAs
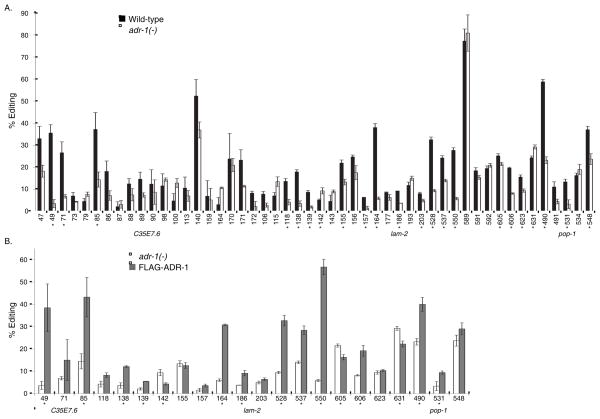
To determine the ability of ADR-1 to directly regulate RNA editing in vivo, we established a quantitative assay to measure changes in editing in worms lacking adr-1 and then tested if these changes were rescued by an ADR-1 transgene. First, we examined editing levels at 50 individual adenosines within three known edited mRNAs: C35E7.6, lam-2 and pop-1. These three mRNAs were chosen based on their diverse cellular functions and length of the double-stranded 3′ UTR, which range from 517 to 1423 nucleotides (nts). RNA was isolated from three independent biological replicates of wild-type and adr-1(−) adult worms. After reverse transcription, PCR amplification and Sanger sequencing, editing efficiency was quantitatively measured using the Bio-Edit program. Technical replicates of the editing assay suggest that editing at each site can be determined with <1% error (Figure S1A), which is consistent with published data on the accuracy of measuring editing efficiency by Sanger sequencing (Eggington et al., 2011). Of the 50 edited adenosines, we observed statistically significant differences in editing levels between wild-type and adr-1(−) worms at 22 individual sites (Figure 1A). The bulk of the statistically significant sites (91%) had decreased editing, ranging from 3–35%, in the absence of adr-1.
Figure 1. ADR-1 alters editing at specific adenosines in multiple mRNAs.
(A and B) Editing levels at individual nucleotides within the 3′ UTRs were measured for 3 biological replicates. Error bars represent standard error of the mean (SEM). Significant changes (p≤ 0.05) in editing levels between (A) wild-type and adr-1(−) or (B) adr-1(−) and FLAG-ADR-1 are marked with an asterisk.
To demonstrate that these sites are directly regulated by ADR-1, a 3X FLAG tagged genomic version of adr-1 was re-introduced to adr-1(−) worms by microinjection. Importantly, this transgenic worm rescues a known adr-1 dependent effect on neuronal protein expression (Hundley et al., 2008), indicating that the transgene expresses functional ADR-1 protein (Figure S1B). As the transgenic worms express FLAG-ADR-1 from an extrachromosomal array that is transmitted to progeny at a high frequency, but not 100%, a neuronal GFP marker was co-injected and flow cytometry was used to purify worms containing the ADR-1 transgene. In addition, to reduce effects of developmental timing on editing efficiency all worms were also sorted by size to obtain young adults. The quantitative editing assay showed that FLAG-ADR-1 significantly restored editing to 15 of the 22 editing sites altered in adr-1(−) worms (Figure 1B). It is important to note, that editing changes in the FLAG-ADR-1 worms are not a general phenomenon, as editing sites that are not affected by loss of adr-1 are not altered by the transgene (Figure S1C). The 15 ADR-1 regulated sites include both adenosines that have increased and decreased editing in the absence of adr-1. Together, these data indicate that ADR-1 alters editing of multiple transcripts, but the effects vary depending upon the individual adenosines examined.
ADR-1 binds directly to ADR-2 target mRNAs in vivo
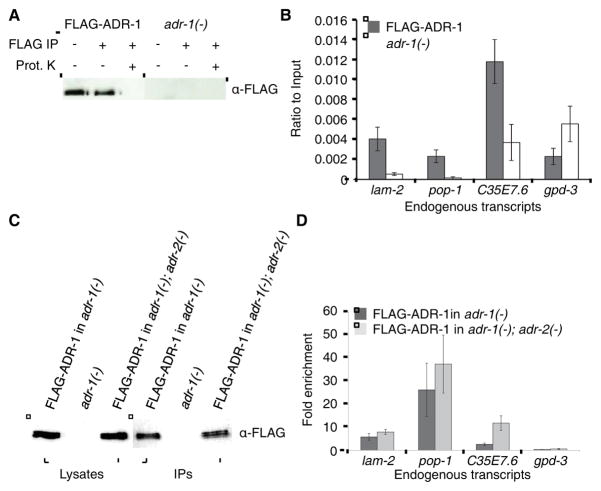
As the effects of adr-1 on editing are site specific, we hypothesized that ADR-1 is capable of regulating editing by utilizing two dsRNA binding domains (dsRBDs) to bind to potential editing substrates and alter accessibility of ADR-2 to particular nucleotides. To determine if ADR-1 could bind ADR-2 editing targets in vivo, we developed an RNA-immunoprecipitation (RIP) assay for ADR-1. As a previously generated polyclonal antibody to ADR-1 was incapable of immunoprecipitating ADR-1 efficiently, the 3x FLAG-tagged ADR-1 transgenic worm was utilized. To measure specific binding of ADR-1 to target mRNAs in vivo, we compared immunoprecipitates (IPs) from FLAG-ADR-1 and adr-1(−) worms that were subjected to UV irradiation (Fig 2A). The IP samples were treated with Proteinase K to degrade FLAG-ADR-1 and release ADR-1 associated RNAs into the supernatant. RNA was extracted from the supernatant, reverse transcribed and quantified using real-time PCR (qRT-PCR). Primers that amplify the three mRNAs tested in Figure 1, produced 3–15 fold more cDNA in the FLAG-ADR-1 IPs compared to adr-1(−) IPs (Fig 2B). In contrast, an mRNA that lacks dsRNA, gpd-3, is not enriched, indicating that, in vivo, ADR-1 specifically binds to these double-stranded ADR-2 target mRNAs.
Figure 2. ADR-1 binds ADR-2 substrates in vivo.
(A) Lysates from the indicated worm lines were subjected to FLAG IP and treatment with Proteinase K (Prot. K). A portion of the untreated lysate (IP−, Prot. K−), IP (IP+, Prot. K−) and beads after Prot. K treatment (IP+, Prot. K−) were subjected to immunoblotting for the FLAG epitope.
(B) cDNA levels for the indicated endogenous mRNAs were measured using qRT-PCR. Values from the IP samples of FLAG-ADR-1 in adr-1(−) and the negative control adr-1(−) were divided by their respective input levels. Error bars represent SEM for three biological replicates.
(C) Lysates from the indicated worm lines were subjected to immunoprecipitation with magnetic FLAG resin. A portion of the input lysate and IPs were subjected to immunoblotting for the FLAG epitope.
(D) cDNA levels for the indicated endogenous mRNAs were measured using qRT-PCR. Ratios of the cDNAs present in the IP samples of the indicated strains were divided by their respective input levels and normalized to the negative control adr-1(−) to give a fold enrichment. Error bars represent SEM for three biological replicates.
As these three mRNAs have both adenosines that are inhibited and enhanced by ADR-1, these data support the hypothesis that ADR-1 modulates editing via a direct interaction with dsRNA. However, in order to regulate editing, ADR-1 needs to bind to the dsRNA before it is edited. To test this possibility, we performed the RIP assay in cells expressing FLAG-ADR-1, but lacking adr-2 and RNA editing. FLAG-ADR-1 was expressed and immunoprecipitated to a similar level in the presence and absence of adr-2 (Fig 2C). Compared to the adr-1(−) worms, all three ADAR target mRNAs were enriched to a similar extent in the FLAG-ADR-1 IPs in the presence and absence of adr-2 (Fig 2D), indicating that binding of ADR-1 to known edited mRNAs is independent of ADR-2. Furthermore, as these mRNAs have no detectable editing in adr-2(−) worms, we conclude that ADR-1 binds unedited mRNAs in the cell.
ADR-1 alters RNA editing via binding to dsRNA in vivo
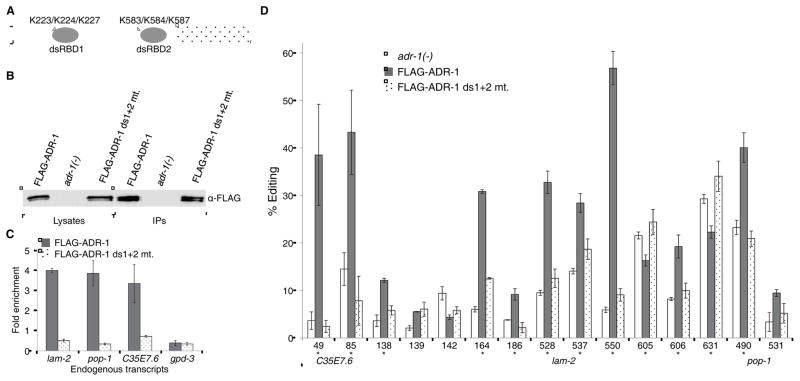
Our results indicate that ADR-1 binds to mRNAs that are targets for editing by ADR-2 in vivo. To determine if this binding is required for the ability of ADR-1 to alter editing in vivo, we created mutations in the dsRBDs of ADR-1 and examined the effects on endogenous RNA editing. A patch of lysine (K) residues, referred to as the KKxxK motif (x=any amino acid), is required for dsRNA binding proteins to bind dsRNA (Ramos et al., 2000; Ryter and Schultz, 1998). Mutation of the lysines to glutamate (E) and alanine (A) disrupts binding of human ADARs to dsRNA (Valente and Nishikura, 2007). To disrupt ADR-1 dsRNA binding, the KKxxK motif was mutated to EAxxA within both dsRBDs (referred to as the ds1+2 mutant) (Fig 3A). Similar to the aforementioned wild-type ADR-1, the ds1+2 mutant was 3XFLAG tagged and reintroduced in the adr-1(−) background. The FLAG-ADR-1 ds1+2 mutant protein is expressed in the transgenic worms to about the same level as transgenic wild-type FLAG-ADR-1 (Fig 3B). To test whether these mutations disrupt ADR-1 binding to dsRNA, the RIP assay was performed with the ds1+2 mutant. In contrast to wild-type ADR-1, the ds1+2 mutant IPs were not enriched for the ADR-2 editing targets (Fig 3C). Thus, the ds1+2 mutant has defects in mRNA binding in vivo.
Figure 3. Mutation of the KKxxK Motif within the dsRBDs of ADR-1 abolishes dsRNA binding and editing regulation.
(A) Schematic of ADR-1 protein with dsRBDs (grey ovals) and deaminase domain (patterned rectangle). Lysine (K) residues mutated are indicated above each dsRBD.
(B) FLAG Immunoblotting of lysates and IPs of the indicated strains.
(C) Ratio of the cDNA present in the IP samples divided by the input cDNA levels for the indicated strains were divided by the IP:input ratio of the adr-1(−) worms. Error bars represent SEM for three biological replicates.
(D) Calculated percent editing in the indicated strains for the endogenous mRNAs of C35E7.6, lam-2 and pop-1. Error bars represent SEM of 3 biological replicates. Significant changes (p≤ 0.05) in editing levels between FLAG-ADR-1 and FLAG-ADR-1 ds1+2 mutant are marked with an asterisk.
To determine if ADR-1 binding to target mRNAs influences editing efficiency, we compared in vivo editing levels of the FLAG-ADR-1 worms to the FLAG-ADR-1 ds1+2 mutant at the 15 sites that were identified as significantly regulated by ADR-1 (Figure 1B). As ADR-1 primarily promotes editing within these target mRNAs, most of the sites exhibit decreased editing in the absence of adr-1, with the exception of nt 631 of lam-2, which has increased editing in adr-1(−) worms (Figure 1A). The ADR-1 ds1+2 mutant failed to significantly restore editing to 11 of these 15 sites, including nt 631 of lam-2 (Fig 3D). Thus, ADR-1 binding to target mRNAs is required both for its ability to promote and inhibit editing of known edited mRNAs in vivo.
Binding of dsRNA by ADR-1 regulates editing across the transcriptome
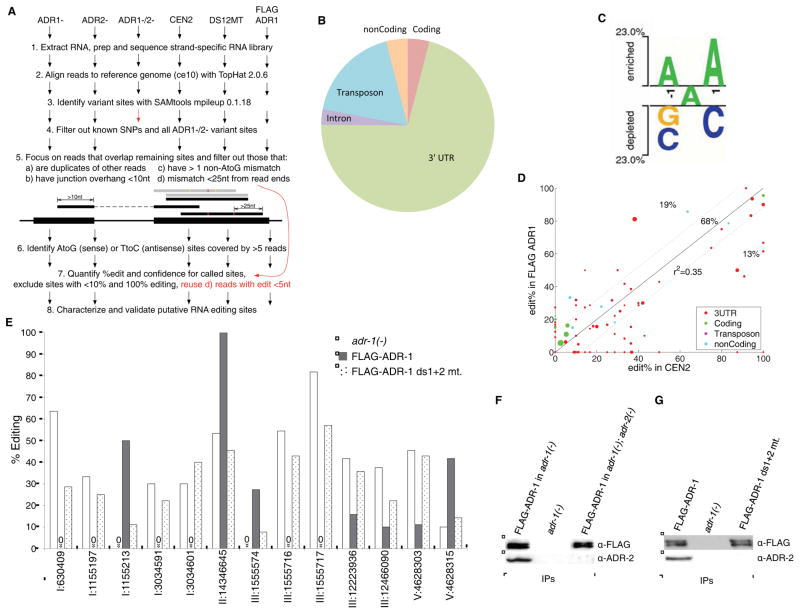
Our data indicates that ADR-1 binding to target mRNAs alters editing of specific adenosines in vivo. To understand the impact of ADR-1 across the transcriptome, we conducted strand-specific RNA-sequencing (RNA-Seq) of RNA from wild-type (N2), adr-1(−), adr-2(−), FLAG-ADR-1 and FLAG-ADR-1 ds1+2 mutant adult worms and compared the nucleotide changes amongst the strains and the published C. elegans genomic sequence (WS220,ce10) (Fig 4A). To distinguish true RNA editing events from single nucleotide polymorphisms (SNPs), we removed annotated SNPs using Illumina’s iGenomes collection. Unannotated single-nucleotide variants (SNVs) were further addressed by performing RNA-Seq on RNA from adr-1(−);adr-2(−) worms and identifying all SNVs between the adr-1(−);adr-2(−) RNA (which lacks all A-to-I editing) and the C. elegans genome. These 118,651 SNVs were subtracted from all other RNA-seq datasets. A Bayesian “inverse probability model” was then adapted (Li et al., 2008) to identify high-confidence A-to-I editing sites from the RNA-seq data, where a confidence value based on the number of reads is associated with each predicted site. Empirically, we found that a confidence threshold of 0.995 produced the largest number of predicted sites in all strains: 59 sites in N2, 141 sites in adr-1(−), 71 sites in FLAG-ADR-1, 102 sites in FLAG-ADR-1 ds1+2 mutant, while identifying the lowest number of edits in the adr-2(−) strain (6 sites) that we presumed represented false positives (Table S1).
Figure 4. Impact of dsRNA binding by ADR-1 on the editing transcriptome.
(A) Bioinformatics strategy depicting the major steps for processing RNA-seq data into A-to-I sites for each strain.
(B) Distribution of identified RNA editing sites within annotated transcriptome regions.
(C) Nucleotide preferences for the 270 candidate editing sites were calculated compared to a randomized control. Enriched and depleted nucleotides are shown above and below the axis, respectively. The level of conservation is represented by letter height. Logos were generated using a t-test with P<.005 and no Bonferroni correction.
(D) Scatter plots of percent editing of quantified sites that overlap in the wildtype (CEN2) and FLAG-ADR-1 datasets. The r2 fit to the y=x line (black diagonal). The margin (dotted line) between no-change and differentially-edited sites equals 12 units of change in the edit % (one standard deviation).
(E) Editing levels for 13 sites from the RNA-seq data where editing levels between adr-1(−) and FLAG-ADR-1 and between FLAG-ADR-1 and FLAG-ADR-1 ds1+2 mutant were greater than 12% (Table S3). Adenosines that had no observed editing are marked with a zero above the x-axis.
(F and G) Immunoblotting analysis of FLAG IPs from the indicated strains. IPs were performed as previously stated except worms were not subjected to UV-crosslinking and only light salt washes were employed.
Of the 270 unique high confidence editing sites that were identified, but not present in adr-2(−) worms (Table S1), 250 sites are novel editing events that occur within 48 different transcripts; the remaining 20 high confidence sites were located within the previously identified ADAR targets C35E7.6, lam-2 and rncs-1 (Morse et al., 2002; Morse and Bass, 1999). The majority (71%) of these candidate-editing events occur within non-coding regions of the genome (Fig 4B). Strikingly, the vast majority of editing events occurred in 3′ UTRs, consistent with the hypothesis that A-to-I editing controls gene expression by altering regulatory motifs in these regions. Interestingly, regions of the genome that encode for transposons were the second most highly identified (18%) category of editing events. In addition, we did identify 11 potential editing sites in coding regions of 8 different mRNAs. As editing events in the coding region of C. elegans mRNAs had not previously been identified, this suggests that similar to mammalian and Drosophila ADARs, C. elegans ADARs may also perform site selective editing in vivo.
Although ADARs target dsRNA of any sequence, the extent of editing at a particular site depends on the neighboring nucleotides (Wahlstedt and Ohman, 2011). Using the Two Sample Logo software (Vacic et al., 2006), the 270 candidate editing sites had an over-representation of A both immediately 5′ and 3′ to the edited adenosine, whereas both G and C are under-represented at the positions 5′ to the edited adenosine and C is under-represented 3′ to the edited adenosine (Fig 4C). Both in vitro biochemical studies and transcriptome-wide RNA-Seq data indicates that human ADARs have a similar 5′ preference. However human ADARs tend to favor a G at the 3′ position to the edited adenosine (Lehmann and Bass, 2000; Riedmann et al., 2008). It is important to note, that because of overlapping specificities of mammalian ADARs, human transcriptome-wide datasets apply to editing by both human ADAR1 and ADAR2. However, as C. elegans ADR-2 is responsible for deamination of all of the RNA-Seq sites, our data provides the first in vivo nucleotide preferences of a single ADAR acting primarily at noncoding regions.
To validate the potential editing sites, Sanger sequencing editing assays were performed for 9 novel edited transcripts (Figure S2A). Importantly, 50 of the 53 predicted sites were verified by Sanger sequencing, suggesting the false discovery rate of the pipeline is approximately 5.7%. In addition to the 50 editing sites identified from the RNA-Seq analysis, Sanger sequencing of these 9 novel transcripts revealed 179 additional editing sites (Table S2), indicating that our probabilistic model is capable of identifying highly edited transcripts.
To determine if ADR-1 affected editing in the transcriptome, the editing efficiency of the 270 high confidence editing sites was quantified using a novel Bayesian model. To ensure accurate quantification, we processed all the RNA-Seq reads through the bioinformatics pipeline described above (Fig 4A), with one exception: read filter 5d was relaxed from requiring an edit site to be 25 nt from each end down to a less-stringent 5 nt and required a minimum of 5 reads for a site in a given strain. With these criteria, we were able to quantify editing of over 100 sites for each of the four strains, with any two strains having an overlap of between 72–105 editing sites (Figure S2B–E). Pairwise comparison of the editing sites identified from the four RNA-Seq data sets indicated that editing efficiency is most consistent between the wild type and FLAG-ADR-1 strains (Fig 4D, Figure S2F–H). This is consistent with the Sanger sequencing data of known editing sites and provides further evidence that the FLAG-ADR-1 transgene is capable of restoring editing to the adr-1(−) strain at most sites. As over-two thirds of the wild-type and FLAG-ADR-1 sites fell within one standard deviation (12%) of the regression line on the scatter plot, we used this threshold to categorize our newly identified sites into ADR-1 and non-ADR-1 regulated (Table S3). As multiple RNA-Seq studies have shown that determination of editing levels increases with read coverage (Bahn et al., 2012; Lee et al., 2013), it is important to note that similar results (>80% overlap) were obtained when we utilized read density to estimate the error of editing at each site (Table S3), suggesting that the editing percent thresholds for ADR-1 regulated and non-regulated sites are accurate. Comparison of editing levels at the 81 sites common between wild-type and adr-1(−) RNA-Seq datasets revealed that over half (56%) of the edited adenosines have altered editing levels in the absence of adr-1 (Table S3). Interestingly, 44 of these 45 sites are located within the 3′ UTRs of 13 novel edited transcripts that we identified. This data is consistent with our quantitative Sanger sequencing analysis of the known ADAR targets 3′ UTRs (Fig 1A). In addition, at 38 of these ADR-1 regulated sites we were able to quantify editing levels for both the FLAG ADR-1 and FLAG-ADR-1 ds1+2 RNA-Seq datasets. Editing levels at 13 sites located within the 3′ UTRs of 8 newly identified ADAR target mRNAs were dependent upon dsRNA binding by ADR-1 (Fig 4E). Together these transcriptome-wide studies indicate that ADR-1 regulates editing of specific adenosines within the 3′ UTRs of the majority of C. elegans edited mRNAs and dsRNA binding is required for this function.
ADR-1 and ADR-2 co-occupy transcripts in vivo
At present it is unclear how ADR-1 binding to mRNAs affects editing by ADR-2. It is possible that ADR-1 and ADR-2 heterodimerize in the cell to edit certain transcripts, whereas others are edited by ADR-2 alone. Alternatively, it is possible that ADR-1 and ADR-2 interact on the same transcripts, but regulate editing in an adenosine-specific manner. To gain insight into these possibilities, we examined the wild-type and FLAG-ADR-1 RNA-Seq datasets to determine whether editing at ADR-1 regulated adenosines occurred on the same reads as edited adenosines that are not affected by loss of adr-1. For most of the novel transcripts edited in the 3′ UTR (9/12), editing was observed at both adenosines affected by adr-1 and non-regulated sites, within the same 75 nt read (Table S3).
To provide further evidence that ADR-1 and ADR-2 associate on common targets in vivo, we immunoprecipitated FLAG-ADR-1 and tested for the presence of ADR-2 with an ADR-2 specific antibody (Fig 4F). ADR-2 was present in IPs from FLAG-ADR-1 worms, but not FLAG-ADR-1 ds1+2 mutant or adr-1(−) worms (Fig 4G). Consistent with an RNA-dependent interaction of ADR-1 and ADR-2, IPs of wild-type ADR-1 treated with RNase also resulted in reduced ADR-2 co-immunoprecipitation (Figure S2J,K). Together, these data suggest that ADR-1 and ADR-2 interact on transcripts in vivo, but are not likely to heterodimerize independent of target mRNAs.
DISCUSSION
In this study, we have demonstrated that C. elegans ADR-1 utilizes its dsRNA binding function to regulate A-to-I editing levels in vivo. Using a high-throughput RNA sequencing approach coupled to probabilistic modeling, we were able to expand the number of known ADAR target mRNAs five-fold, as well as provide the first transcriptome-wide evidence that ADR-1 is a catalytically inactive member of the ADAR family. Furthermore, using both our extensive Sanger sequencing analysis of ADAR targets and quantification of transcriptome-wide RNA-Seq data, we demonstrate that ADR-1 regulates editing efficiency of specific adenosines within most ADAR target 3′ UTRs.
We propose that ADR-1 regulates editing by binding to target mRNAs and altering accessibility of ADR-2 for specific adenosines. Multiple recent studies support the idea that the RNA binding protein (RBP) landscape of ADAR target mRNAs affects editing levels (Bhogal et al., 2011; Garncarz et al., 2013; Tariq et al., 2013). However, in most of these studies, RNA binding by the regulators was not shown to be required for A-to-I regulatory activity and these regulators were all single-stranded RBPs that altered editing of specific coding editing events. In contrast, we demonstrate that ADR-1 binds to several target mRNAs via its dsRBDs, and that this binding is required for regulation of editing. This dsRNA binding activity would allow ADR-1 to interact with nearly all the same targets as ADR-2, thus allowing it to serve a more global role in regulating editing within long double-stranded regions. As dsRBDs are the second most abundant RNA recognition motif (Stefl et al., 2010), it is unlikely that this regulatory role is limited to C. elegans ADR-1. Consistent with this, 20% of our newly discovered edited transcripts overlap with recently identified targets of another dsRNA binding protein (dsRBP), C. elegans Staufen (LeGendre et al., 2013) (Table S1).
Our Sanger sequencing and transcriptome-wide analyses suggest that the regulatory role of ADR-1 is specific to certain adenosines (Fig 1A, Table S3). Although dsRBPs are generally presumed to lack sequence specificity (Tian et al., 2004), recent structural data suggests ADARs recognize specific nucleotides within dsRNA (Stefl et al., 2010). Our RIP assay indicates that ADR-1 binds to lam-2 and pop-1 mRNAs to a similar extent in the presence and absence of adr-2 (Fig 2D). Thus, at least for certain edited mRNAs, ADR-1 does not compete with ADR-2 for binding sites in vivo. Consistent with this, the majority of the ADR-1 regulated sites identified in both the RNA-Seq datasets and Sanger analysis have enhanced editing in the presence of adr-1 (Fig 1A, Table S3), suggesting that ADR-1 functions primarily to promote ADR-2 editing, not compete with ADR-2 for target adenosines. As editing is not required for ADR-1 to bind these mRNAs, we postulate that, in vivo, ADR-1 first binds to target mRNAs and then either alters binding of ADR-2 to specific regions and/or regulates the catalytic activity of ADR-2 (See Graphical abstract). Interestingly, it was recently demonstrated that human ADAR1 binding to mRNAs creates binding sites for another RBP, HuR, which results in increased RNA stability of HuR-ADAR1 bound transcripts (Wang et al., 2013). Similar to human ADAR1-HuR, we detected an in vivo interaction between wild-type ADR-2 and ADR-1, but not the ADR-1 ds1+2 mutant, which is consistent with ADR-1 and ADR-2 interacting on target mRNA. Interestingly, it has previously been suggested that human ADAR homodimerization on dsRNA is required for efficient editing in vitro (Jaikaran et al., 2002). Although our evidence indicates that ADR-1 utilizes dsRNA binding to regulate editing, it is possible that this regulatory function is due to effects of ADR-1 on expression of other RBPs, which in turn alter ADR-2 accessibility to target mRNAs. Future work aimed at both identifying ADR-1 and ADR-2 binding sites on mRNAs in vivo and determining the impact of ADR-1 on ADR-2 editing activity in vitro will be needed to determine if there is a correlation between binding site specificity and regulation of specific sites. In summary, our results indicate that ADR-1 utilizes dsRNA binding to regulate A-to-I editing across the C. elegans transcriptome. These studies not only suggest a potential biological function for the catalytically inactive ADARs present in humans, but also unveil a potential mechanism for other dsRBPs to regulate RNA editing levels.
EXPERIMENTAL PROCEDURES
Maintenance of worm strains and Transgenics
Worm strains were maintained by growth on NGM plates seeded with Escherichia coli OP50. A detailed description of the transgenic strains is given in the Extended Experimental Procedures.
RNA Isolation and Editing Assays
Total RNA was isolated using Trizol (Invitrogen). RNA was further treated with Turbo DNase (Ambion) and then isolated using the RNA Easy Extraction kit (Qiagen). Editing assays were performed using Thermoscript (Invitrogen) for reverse transcription and PFX Platinum DNA Polymerase (Invitrogen) for PCR amplification with gene-specific primers (Table S4). PCR products were gel purified and subjected to Sanger sequencing. For all editing assays, negative controls were conducted without Thermoscript RT to ensure that all DNA subjected to sequencing resulted from cDNA amplification.
Strand-specific RNA sequencing
Strand specific mRNA sequencing libraries were prepared as described previously (Parkhomchuk et al., 2009). Libraries were normalized to 2nM and sequenced for SE76 cycles on either HiSeq2000 (adr-1(−);adr-2(−)) or Illumina GAII (all other strains).
Bioinformatics Pipeline
To achieve accurate identification of editing sites, we combined filters from existing pipelines (Chen, 2013; Lee et al., 2013; Levanon et al., 2004; Ramaswami et al., 2012) in a strand-specific manner. Accurate quantification was performed by extending the existing Bayesian method for genomic variant calling used in the 1000 Genomes project (Li et al., 2008) with a custom-designed prior on the editing % (Figure S2I). In addition to leveraging established considerations with regards to read sequencing and alignment errors (Kleinman and Majewski, 2012; Lin et al., 2012; Pickrell et al., 2012) our approach benefits greatly from using the adr-1(−);adr-2(−) strain as a powerful filter for unannotated variants. Detailed steps of the pipeline and Bayesian method for variant calling are described in the Extended Experimental Procedures.
RNA Immunoprecipitation (RIP) Assay
After washing with IP Buffer (50mM HEPES, pH 7.4; 70mM K-Acetate, 5mM Mg-Acetate, .05% NP-40 and 10% glycerol), worms were subjected to 3J/cm2 of UV radiation using the Spectrolinker (Spectronics Corp.) and stored at −80°C. To obtain cell lysates, frozen worms were ground with a mortar and pestle on dry ice. After thawing, the lysate was centrifuged and protein concentration was measured with Bradford reagent (Sigma). Five micrograms of extract was added to anti-Flag magnetic beads (Sigma) that were washed with wash buffer (WB: 0.5M NaCl, 160mM Tris-HCl pH 7.5). After incubation for 1 hour at 4°C, the beads were washed with ice-cold WB, resuspended in low salt WB (0.11M NaCl), 1μl RNasin (Promega) and 0.5μl of 20mg/ml proteinase K (Sigma) and incubated at 42°C for 15 minutes to degrade protein and release bound RNA. Protein samples were subjected to SDS-PAGE and western blotting with a FLAG antibody (Sigma). RNA samples were isolated as described above. Following DNase treatment, qRT-PCR for known editing targets was performed as previously described (Hundley et al., 2008).
Flow Cytometry
Flow cytometry was conducted at the IUB Flow Cytometry Facility by a dedicated technician using the COPAS Select (Union Biometrica) large particle sorter. Parameters were adjusted to select adult worms and expressing GFP for transgenic lines.
Supplementary Material
HIGHLIGHTS.
Identification of >400 novel A-to-I editing sites, primarily within noncoding regions.
ADR-1 regulates editing of specific adenosines within 3′ UTRs of diverse transcripts.
ADR-1 regulates RNA editing by directly binding to ADR-2 target mRNAs.
ADR-1 and ADR-2 do not form heterodimers, but co-occupy transcripts in vivo.
Acknowledgments
This work was supported by funds to H.A.H from the Ralph W. and Grace M. Showalter Foundation and start-up from Indiana University School of Medicine, an NIH predoctoral training grant to M.C.W. (T32 GM007757), NSF graduate research fellowship to B.K., and partially supported by grants from the National Institute of Health to G.W.Y. (R01 HG004659 and R01 NS075449). G.W.Y. is also supported as an Alfred P. Sloan Research Fellow. We thank Christiane Hassel (IUB-Flow Cytometry) for assistance.
Footnotes
Supplemental information includes Extended Experimental Procedures, two figures and four tables and can be found with this article online at xxx. RNA-Sequencing data has been deposited in the GEO database (accession #GSE51556).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal B, Jepson JE, Savva YA, Pepper AS, Reenan RA, Jongens TA. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Characterization and comparison of human nuclear and cytosolic editomes. Proc Natl Acad Sci U S A. 2013;110:E2741–2747. doi: 10.1073/pnas.1218884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C, Veno MT, Ekdahl Y, Kjems J, Ohman M. A distant cis acting intronic element induces site-selective RNA editing. Nucleic Acids Res. 2012;40:9876–9886. doi: 10.1093/nar/gks691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Jaffray E, Hay RT, O’Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RA, Macbeth MR, Beal PA. ADAR proteins: structure and catalytic mechanism. Curr Top Microbiol Immunol. 2012;353:1–33. doi: 10.1007/82_2011_144. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran DC, Collins CH, MacMillan AM. Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J Biol Chem. 2002;277:37624–37629. doi: 10.1074/jbc.M204126200. [DOI] [PubMed] [Google Scholar]
- Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ang JK, Xiao X. Analysis and design of RNA sequencing experiments for identifying RNA editing and other single-nucleotide variants. RNA. 2013;19:725–732. doi: 10.1261/rna.037903.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre JB, Campbell ZT, Kroll-Conner P, Anderson P, Kimble J, Wickens M. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem. 2013;288:2532–2545. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Piskol R, Tan MH, Li JB. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210419. author reply 1302. [DOI] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, Bisso A, Keegan LP, Del Sal G, O’Connell MA. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci U S A. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc Natl Acad Sci U S A. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Fujiwara M, Ohshima Y, Ishihara T. ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans. Genetics. 2008;180:785–796. doi: 10.1534/genetics.108.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Barbon A, Barlati S. Activity regulation of adenosine deaminases acting on RNA (ADARs) Mol Neurobiol. 2012;45:61–75. doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012 doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal JJ, Seeburg PH. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron. 2012;74:432–439. doi: 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BZ, Huang H, Lam R, Soong TW. Dynamic regulation of RNA editing of ion channels and receptors in the mammalian nervous system. Mol Brain. 2009;2:13. doi: 10.1186/1756-6606-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq A, Garncarz W, Handl C, Balik A, Pusch O, Jantsch MF. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–2593. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq A, Jantsch MF. Transcript diversification in the nervous system: a to I RNA editing in CNS function and disease development. Front Neurosci. 2012;6:99. doi: 10.3389/fnins.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Ohman M. Site-selective versus promiscuous A-to-I editing. Wiley Interdiscip Rev RNA. 2011;2:761–771. doi: 10.1002/wrna.89. [DOI] [PubMed] [Google Scholar]
- Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG. ADAR Regulates RNA Editing, Transcript Stability, and Gene Expression. Cell Rep. 2013;5:849–860. doi: 10.1016/j.celrep.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Shepherd BA, Johnson WE, Bass BL. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012;22:1488–1498. doi: 10.1101/gr.134841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.