Abstract
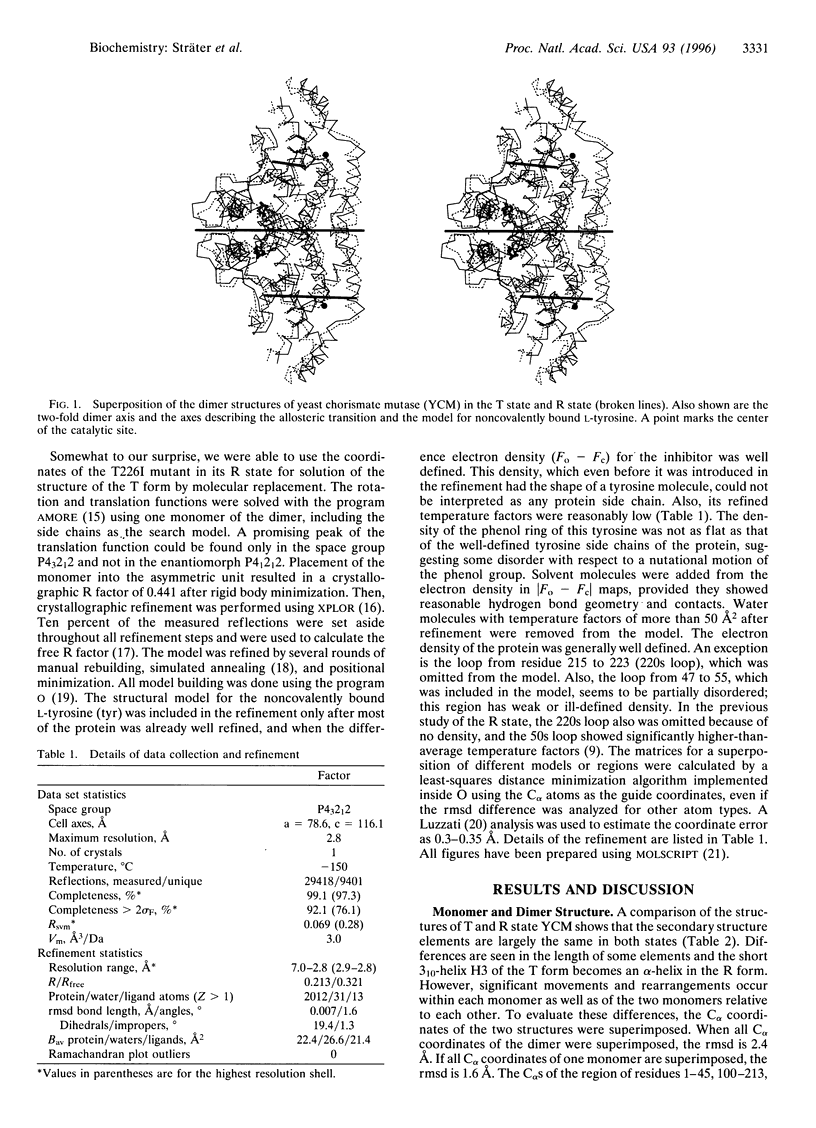
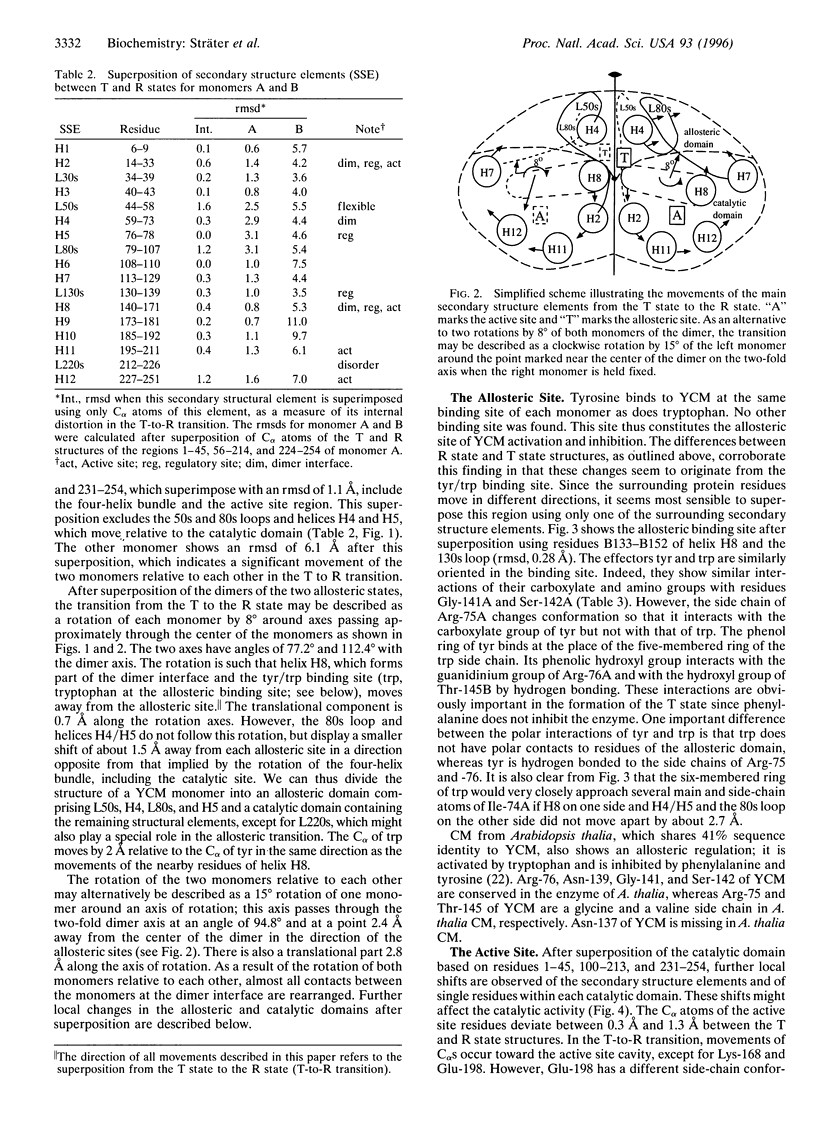
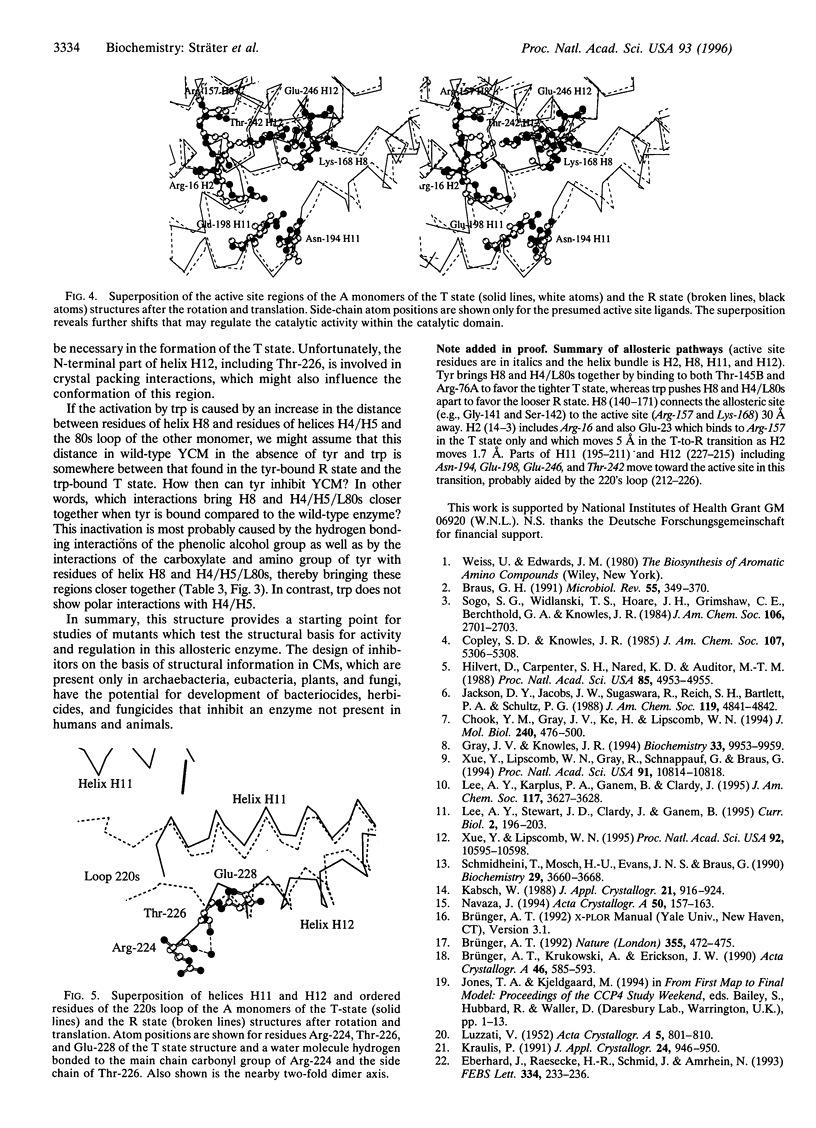
The crystal structure of the tyrosine-bound T state of allosteric yeast Saccharomyces cerevisiae chorismate mutase was solved by molecular replacement at a resolution of 2.8 angstroms using a monomer of the R-state structure as the search model. The allosteric inhibitor tyrosine was found to bind in the T state at the same binding site as the allosteric activator tryptophan binds in the R state, thus defining one regulatory binding site for each monomer. Activation by tryptophan is caused by the larger steric size of its side chain, thereby pushing apart the allosteric domain of one monomer and helix H8 of the catalytic domain of the other monomer. Inhibition is caused by polar contacts of tyrosine with Arg-75 and Arg-76 of one monomer and with Gly-141, Ser-142, and Thr-145 of the other monomer, thereby bringing the allosteric and catalytic domains closer together. The allosteric transition includes an 8 degree rotation of each of the two catalytic domains relative to the allosteric domains of each monomer (domain closure). Alternatively, this transition can be described as a 15 degree rotation of the catalytic domains of the dimer relative to each other.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braus G. H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev. 1991 Sep;55(3):349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Krukowski A., Erickson J. W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990 Jul 1;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Gray J. V., Ke H., Lipscomb W. N. The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction. J Mol Biol. 1994 Jul 29;240(5):476–500. doi: 10.1006/jmbi.1994.1462. [DOI] [PubMed] [Google Scholar]
- Eberhard J., Raesecke H. R., Schmid J., Amrhein N. Cloning and expression in yeast of a higher plant chorismate mutase. Molecular cloning, sequencing of the cDNA and characterization of the Arabidopsis thaliana enzyme expressed in yeast. FEBS Lett. 1993 Nov 15;334(2):233–236. doi: 10.1016/0014-5793(93)81718-f. [DOI] [PubMed] [Google Scholar]
- Gray J. V., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: FTIR studies and the mechanism of action of the enzyme. Biochemistry. 1994 Aug 23;33(33):9953–9959. doi: 10.1021/bi00199a018. [DOI] [PubMed] [Google Scholar]
- Hilvert D., Carpenter S. H., Nared K. D., Auditor M. T. Catalysis of concerted reactions by antibodies: the Claisen rearrangement. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4953–4955. doi: 10.1073/pnas.85.14.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. Y., Stewart J. D., Clardy J., Ganem B. New insight into the catalytic mechanism of chorismate mutases from structural studies. Chem Biol. 1995 Apr;2(4):195–203. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Schmidheini T., Mösch H. U., Evans J. N., Braus G. Yeast allosteric chorismate mutase is locked in the activated state by a single amino acid substitution. Biochemistry. 1990 Apr 17;29(15):3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- Xue Y., Lipscomb W. N., Graf R., Schnappauf G., Braus G. The crystal structure of allosteric chorismate mutase at 2.2-A resolution. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10814–10818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Lipscomb W. N. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]



