Abstract
Significance: Reactive oxygen species (ROS) are generated by exogenous and environmental genotoxins, but also arise from mitochondria as byproducts of respiration in the body. ROS generate DNA damage of which pathological consequence, including cancer is well established. Research efforts are intense to understand the mechanism of DNA base excision repair, the primary mechanism to protect cells from genotoxicity caused by ROS. Recent Advances: In addition to the notion that oxidative DNA damage causes transformation of cells, recent studies have revealed how the mitochondrial deficiencies and ROS generation alter cell growth during the cancer transformation. Critical Issues: The emphasis of this review is to highlight the importance of the cellular response to oxidative DNA damage during carcinogenesis. Oxidative DNA damage, including 7,8-dihydro-8-oxoguanine, play an important role during the cellular transformation. It is also becoming apparent that the unusual activity and subcellular distribution of apurinic/apyrimidinic endonuclease 1, an essential DNA repair factor/redox sensor, affect cancer malignancy by increasing cellular resistance to oxidative stress and by positively influencing cell proliferation. Future Directions: Technological advancement in cancer cell biology and genetics has enabled us to monitor the detailed DNA repair activities in the microenvironment. Precise understanding of the intracellular activities of DNA repair proteins for oxidative DNA damage should provide help in understanding how mitochondria, ROS, DNA damage, and repair influence cancer transformation. Antioxid. Redox Signal. 20, 708–726.
Introduction
Shogyo Mujo: nothing can stay unchanged, a layman's term equal to the second law of thermodynamics. Genes cannot escape this truth, but can only delay the destruction of DNA through cellular mechanisms of genome integrity maintenance, that is, DNA repair (80). DNA repair pathways, which are enzymatically carried out in our body, have evolved to solve a number of stressful situations to survive as a single cell and as a multicellular organism. Decades of research have established several distinct DNA repair pathways: direct reversal, base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), damage bypassing, DNA double-strand breaks (DSBs) repair via nonhomologous end joining (DSBR via NHEJ), and DSBR via homologous recombination (DSBR via HR) (Fig. 1). The frameworks of the DNA repair pathways modeled in the 70s and 80s have been revised with subpathways as details emerged. Interactions among the repair pathways became apparent as studies identified particular repair proteins involved in multiple repair pathways. The convergence of DNA repair pathways demonstrates how organisms have evolved to respond and utilize available DNA repair proteins in a variety of environments. Despite the advances in the field, it is still far from clear how cells survive through many types of DNA damage in the genome, and elucidating each of these requires complicated modeling based on in vitro and in vivo experiments. However, these studies are necessary for designing future preventive medicine and therapeutic strategies for a plethora of diseases that rely on precise predictions of DNA damage responses. It has recently become apparent that reactive oxygen species (ROS) originated in the cells play a fundamental role in cellular transformation (139). This review discuses ROS generation from mitochondria first, as the cellular response to oxidative stress and DNA damage is closely linked to carcinogenesis. Then the primary DNA repair mechanism against oxidative DNA damage will be described in-depth, followed by discussion of how the components of the repair pathways may influence the cancer transformation.
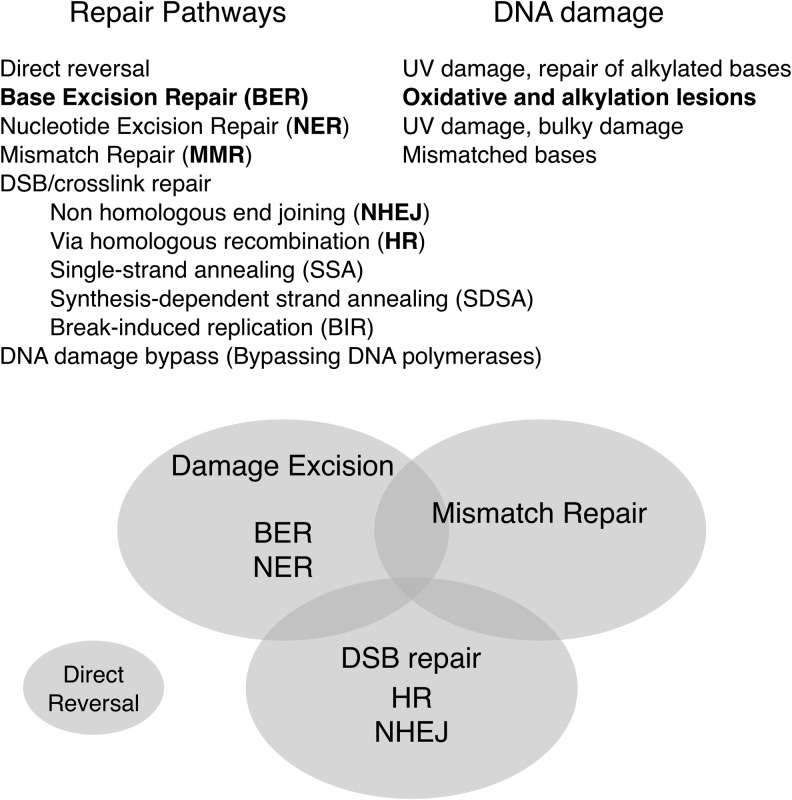
FIG. 1.
DNA repair pathways in mammals. The five major DNA repair pathways are direct reversal, DNA BER, NER, DNA MMR, and DNA DSBR. The Venn diagram illustrates relations of repair systems to one another. A few DNA repair enzymes function in multiple pathways, as denoted by the overlaps in the diagram. DNA damage can be bypassed during DNA replication by bypassing DNA polymerases. BER, base excision repair; NER, nucleotide excision repair; MMR, mismatch repair; DSBR, double-strand break repair.
ROS Generation in the Cell
ROS generation in mitochondria
Eukaryotic cells utilize oxidative phosphorylation (OXPHOS) in the mitochondria to generate more than 90% of the intracellular ATP (42). Although ROS can be generated by the enzymatic reactions of various intracellular oxidases, such as glucose oxidase and NADPH oxidase, mitochondria are the major source of ROS generation in the cells. The energy pooled by the proton (H+) gradient between the mitochondrial matrix and the space between inner and outer membrane is converted to ATP, while O2 is converted to H2O through electron transfer mediated by mitochondrial complex I, III, and IV (C-I, C-III, and C-IV, respectively) (101) (Fig. 2). The complexes ubiquinone (coenzyme Q10, CoQ10) and cytochrome b-c (cyto c) are the central molecules to transfer electrons (e−) from NADH (from C-I) or FAD (C-II) to O2. When Q10 and O2 directly contact, O2 is converted to more reactive O2−, the superoxide anion radical (29, 42). Rotenone, a plant extract commonly used as a pesticide, is a potent inhibitor of C-I and causes electron leakage from C-I and O2− formation (9). C-I is in the inner membrane of mitochondria and O2− from C-I is mainly generated inside the matrix. In contrast, C-III inhibitor antimycin A produces O2− outside of the inner membrane, and thus, O2− may be released from mitochondria (29). Subsequently, O2− can be converted to other reactive species, such as peroxynitrite (ONOO−) in the presence of nitric oxide, or cause lipid peroxidation (19, 26, 123). Helping to counteract the adverse effects of O2−, cells possess superoxide dismutases (SOD) that scavenges O2− and converts it into H2O2. H2O2 generated by the reactions of Cu/Zn-SOD (SOD1) and Mn-SOD (SOD2), present in the cytoplasm and mitochondria, respectively, can quickly convert O2− to H2O2, which is further catalyzed to H2O by catalase or glutathione peroxidases (93). Once O2− is converted to H2O2, however, a highly reactive hydroxyl radical OH• may be produced via Haber–Weiss reaction (19).
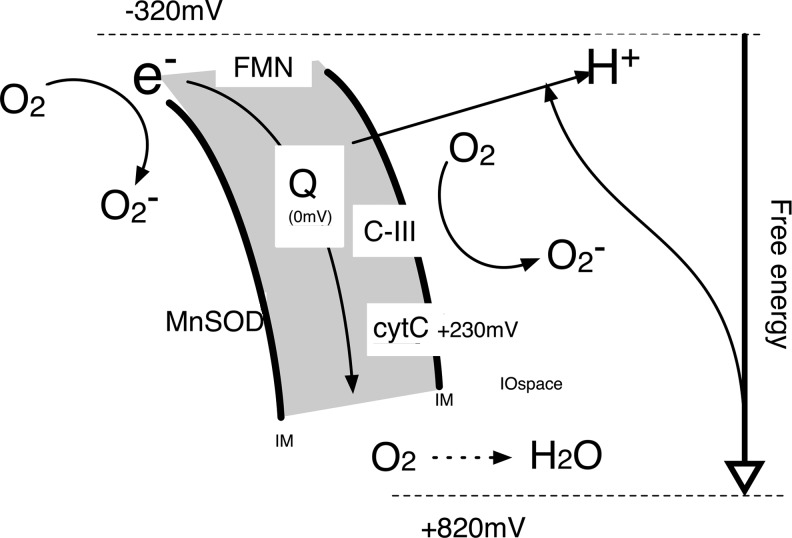
FIG. 2.
Generation of O2− in mitochondria. Electron transfer from Complex I and Complex II (at high redox potential) is carried by coenzyme Q to generate proton gradient between the inner membrane and inner and outer membrane space in mitochondria, before oxidized into H2O (at the lowest redox potential) (101). Immaturely trapped electrons generate O2−.
These ROS have profound influences on a number of cellular activities, including cell growth, inflammatory responses, and apoptosis (51). Although mitochondrial DNA has been shown to be the direct target, it has not been clear whether genomic DNA damage are caused by ROS directly derived from mitochondrial O2− generation. However, studies indeed showed that ROS generated in mitochondria cause oxidative DNA damage and mutations in nuclei (82, 104). A recent study also showed that rotenone treatment induced 7,8-dihydro-8-oxoguanine (8-oxoG) and γH2AX foci in the genomic DNA of undifferentiated Caco-2 cells, a human colorectal adenocarcinoma cell line (130). Considering that DNA damage increased within 2 h and significant apoptosis was not observed, it is likely that ROS generated by C-I interruption by rotenone was responsible for the genomic DNA damage directly or through lipid peroxidation. Whether or not ROS generated in mitochondria can travel into the nucleus and damage the genomic DNA is an important question that would help clarify the role of ROS generated in the mitochondria in genomic DNA damage and mutation.
Effect of ROS from mitochondria on cancer transformation
In addition to the direct effect of ROS on genomic DNA, ROS also trigger signal transduction cascades, including apoptosis activation (48). A high spike of ROS can also increase the vulnerability of mitochondrial membrane permeability transition, which may result in cell death unless the controlled autophagy (mitophagy) process takes place. Besides mitochondrial respiration, ROS are also generated from reactions catalyzed by oxidases, such as the glucose oxidases and NADPH oxidases, and importantly from macrophages that cause inflammation in the surrounding microenvironment.
Recent studies indicate the link between ROS generation and cell growth signaling influencing cell proliferation and stress response. Basal activity of c-Jun N-terminal kinase (JNK) is kept low due to its association with glutathione S-transferase pi (GSTp). Upon stress induction, including ROS, GSTp is oligomerized resulting in its dissociation from and activation of JNK (1). Therefore, ROS production at mitochondria may modulate cell proliferation signaling. Ohsawa et al. (99) discovered that in Drosophila, mitochondrial deficiency combined with a particular Ras oncogene mutation (RasV12) led to ROS generation, which triggered JNK activation and induction of IL-6 and Wnt homologue, paracrine factors that promoted cell growth of surrounding cells (nonautonomous cell growth). Interestingly, growth of the provocative cells that contained the Ras mutation and mitochondrial deficiency were not activated; rather, these cells were weak and eventually dissipated. The study is among many recent reports that highlight the importance of mitochondrial activity for the tumor microenvironment (107, 136).
Cancer cells are, in many cases, found in hypoxic conditions due to its unusual cell growth rate in undeveloped vasculature, and thus, maintain a lower mitochondrial respiration rate than normal tissues. This phenomenon is named the Warburg effect after Otto Warburg's pioneering observation. Importantly, the Warburg effect may lead to a lower rate of ROS generation due to their low OXPHOS reactions compared to normal cells. Indeed, using a high-sensitivity random mutation capture assay (15), Ericson et al. (41) recently found that the mutation rate in mitochondria was lower in cancer cells than in normal cells. This was a striking contrast from their previous report that cancer cells showed a higher rate of random mutations in the genomic DNA than normal cells (16). Therefore, the rates of mitochondrial DNA damage and mutations of transformed cells may be lower than previously studied. While many studies found mutations in mitochondrial DNA of cancer cells (53, 100, 115, 132), there are also studies that show no statistical difference in the mitochondrial mutation frequency between cancer and normal cells (88, 113). The observation reported by Ohsawa et al. implies that the growth initiating cells, that is, those with Ras and mitochondrial deficiency, are intrinsically weak and thus, may not be detected at the time patients are diagnosed with cancer. Importantly, these growth initiating cells may have completely different genetic traits and cellular characteristics from malignant cancer cells. To understand the influence of mitochondrial activity on cancer transformation, it may be necessary to involve the cancer microenvironment where stromal cells, including cancer associated fibroblasts, may facilitate the growth of cancer cells. While not showing a robust cell growth themselves, these stromal cells may provide the growth initiation through mitochondrial dysfunction or elevated OXPHOS and the associated increases of ROS generation. The mutation frequency of the mitochondrial DNA of the stromal cells may be completely different from the tumor cells whether benign, invasive or metastatic.
Damaged mitochondria are continuously removed and replaced with newly generated mitochondria. Cellular ability to maintain the mitochondrial integrity has a profound effect and is closely associated with the pathophysiology of cells, including cancer. Many kinds of DNA damage are generated by ROS attack (19, 43). In the following sections studies of DNA BER, the primary DNA repair mechanism for the oxidative DNA damage will be reviewed. Then the influence of the oxidative DNA damage and BER enzymes on cancer transformation and malignancy will be discussed.
DNA Base Excision Repair
An overview of BER with a simple scheme
BER was once regarded as the simplest DNA repair mechanism, next to direct reversal. The main reason for this perception was that separate biochemical analyses of each step of BER were possible. Thus, in vitro reconstituted BER assays and structural studies flourished with availability of recombinant proteins in the 90s (Fig. 3). A testament to the BER simplicity is that cDNAs of eukaryotic DNA glycosylases and the AP endonuclease can complement Escherichia coli mutants of the corresponding orthologous genes (108). Indeed, the human MPG (aka MAG) cDNA (N-methylpurine DNA glycosylase) was cloned based on a screening to complement an alkylation sensitive mutant of E. coli (27, 114). The studies were conceptual extensions of the pioneering study in this field that cloned the mammalian O6-methylguanine DNA methyltransferase (MGMT) gene, a DNA repair protein that complements an E. coli ada mutant via direct reversal of the alkylated guanine (128). This capability of interspecies complementation by some BER genes is a clear difference from other DNA repair pathways, such as NER in which the repair proteins need to interact with each other and with the transcription machinery (30). However, it is now clear that conservation between E. coli and the higher eukaryotes is limited, and that many mammalian specific factors and reactions are necessary for the whole BER pathway. In addition, the activity of BER to “edit” small base adducts and damage is utilized for nonrepair activity in the cells, for example, inducing hypermutability in immunoglobulin genes (74) and maintaining demethylated status of cytosine in the genome (32, 54, 63, 118). Conversely, it can also be argued that the high conservation of essential BER genes from prokaryotes to humans underscores the importance of BER for maintenance of genome integrity.
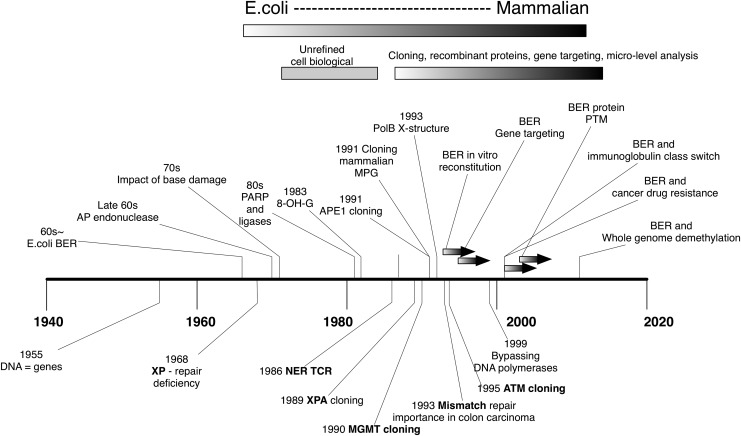
FIG. 3.
Advance of BER study. Above the time line shown are the major BER reports that impacted the study field. Selected DNA repair studies other than BER are shown below the timeline as a perspective. Impact of base damage: (81); PARP and DNA ligase: (34); 8-OH-G discovery: (73); APE1 cloning: (38, 108, 117); MPG cloning: (27, 114); BER reconstitution: (119, 120); BER ko mice: (52, 121, 146); PTM of BER: (14, 22, 47, 55, 56, 148); BER and hypermutability: (74); cancer resistance: (17); demethylation: (32, 33, 54). APE1, apurinic/apyrimidinic endonuclease 1; PARP, poly (ADP-ribose) polymerase; PTM, posttranslational modification; 8-OH-G, 8-hydroxyguanine; ko, knockout.
BER repairs most of the oxidized DNA damage. The basic components of BER are DNA glycosylases, AP-endonucleases, DNA polymerases, and DNA ligases (Fig. 4). The BER process can be considered as a series of DNA processing reactions each of which is carried out by a DNA repair enzyme. Briefly, the BER process from base adducts to repair completion are (i) base removal step, (ii) 3′-OH generation (3′-end cleaning) step, (iii) 5′-end cleaning step, (iv) DNA synthesis step, and (v) DNA nick-sealing step. DNA repair enzymes that process these steps, respectively are: (i) DNA glycosylases, (ii) AP-endonucleases, (iii) AP lyases commonly associated to many DNA glycosylases and DNA polymerases, (iv) DNA polymerases, and (v) DNA ligases.
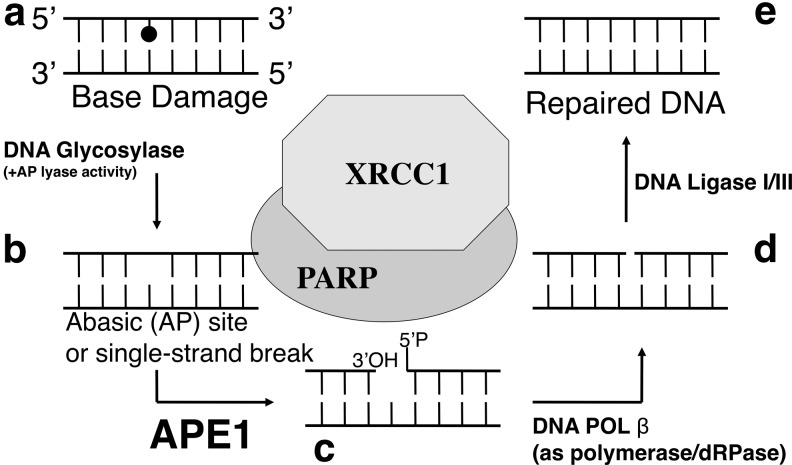
FIG. 4.
Basic steps of BER. (a) Abnormal base damage, including oxidative and alkylated bases (e.g., 8-oxoG, N3-methylG, and so on) are recognized and removed by DNA glycosylases. (b) Consequently, either AP sites or DNA SSBs are generated at the damaged sites. (c) APE1 generates 3′-OH termini for subsequent DNA repair synthesis step processed by DNA polymerases. (d) pol β is the primary BER DNA polymerase, which also removes 5′-ribose moieties to generate 5′-P termini at the gap. (e) The DNA nicks with 3′-OH and 5′-P ends (without gaps) will be ligated by DNA ligase I or III to complete the BER process. 8-oxoG, 7,8-dihydro-8-oxoguanine; SSB, single-strand break; pol β, polymerase beta.
An important consideration is that each reaction consequently leaves a new intermediate lesion in DNA, which is harmful if the entire repair reaction is not completed through nick sealing by the DNA ligases. Critically, the generation of single-strand break (SSBs) by DNA glycosylases or AP endonucleases (see below) is even more toxic than the oxidized bases before the reactions. Thus, the concept of BER coordination (“hand-off”) mechanism was proposed in the early 2000s (95, 142). Accordingly, a coordinated BER prevents the intermediate lesions from causing more harmful conditions, such as inducing apoptosis. This model was later supported by many studies thereafter (46, 60, 149).
Detailed mechanism of BER
The basic five steps described above have been through many major and minor revisions as BER researchers advanced the field. Notable additions and modifications include the following.
(i) There are multiple DNA glycosylases identified in mammals. DNA glycosylases that remove oxidized bases are 8-oxoguanine DNA glycosylase (OGG1), endonuclease III-like 1 (NTH1), endonuclease VIII-like (NEIL)1, NEIL2, NEIL3, and mutY homolog (MYH). This list excludes uracil DNA glycosylases (UNGs) and thymine DNA glycosylase (TDG) (31, 78, 102), which are necessary to remove U and T opposite G that arise from deamination of C and 5mC, respectively. Additional DNA glycosylases are described by Hegde et al. in detail (58, 59).
(ii) DNA glycosylases belong to either of two classes: those with their associated AP lyase activity and the others without the AP lyase activity. When damaged bases are removed by DNA glycosylases with an AP lyase activity, AP sites will be further processed through β-elimination (10) or sequential β- and δ-elimination reaction to become SSBs with 3′-phosphor-α, β-unsaturated aldehyde (3′-PUA), and 3′-phosphate, respectively (122, 153). Association of AP lyase reaction after the base removal has significant physiological consequences, as many of the base damage processed by DNA glycosylases become SSBs with “dirty ends.” Unlike 3′-OH termini generated by AP endonuclease, these SSBs should be regarded as intermediate DNA lesions more harmful than initial oxidative base damage, such as 8-oxoG. Besides the fact that DNA glycosylases produce 3′-PUA and 3′-phosphate, all these unusual end structures may be produced directly by DNA damaging reagents that directly cause SSBs.
(iii) The AP-endonuclease reaction, simply stated, is to produce the 3′-OH termini, an absolutely necessary priming structure for the subsequent DNA repair synthesis step carried out by DNA polymerases. apurinic/apyrimidinic endonuclease 1 (APE1), the mammalian AP endonuclease, has activities to generate the 3′-OH termini from AP sites, 3′-PUA, 3′-phosphoglycolate, and 3′-phosphate (66). This 3′-end cleaning process (3′-OH generation) can be replaced with sequential reactions of DNA glycosylases and polynucleotide kinase (PNK) (36, 141). The combined reaction on 3′-phosphate is more efficient than the reaction catalyzed by APE1 alone (141). However, it should be noted that this reaction only occurs when the DNA glycosylases carry out not only β-elimination but also subsequently δ-elimination to generate 3′-phosphate end, namely NEIL1, NEIL2. Also NEIL3 was recently shown to carry out β-δ-elimination reaction (83), although NEIL3-PNK repair has not been demonstrated in reconstituted biochemical assays. Topoisomerase Tyrosyl-DNA phosphodiesterase 1 (TDP1) solves a unique problem generated by stalled topoisomerase I reaction (25, 40). TopoI-DNA covalent linkage is accumulated in nonreplicative cells and causes SSBs accumulation and eventually to DSBs. TDP1 dissolves this particular DNA-topoI intermediate. In this repair event, gap-filling reaction by DNA polymerase is not necessary as there is no excision of damaged base or nuleotide (25, 40).
(iv) In E. coli, the 5′-end cleaning step that removes 5′-phosphor-ribose moiety as a result of incision by AP-endonuclease, was not an issue because the powerful nick translation activity of E. coli PolI had been already known. For the mammalian BER pathway, Matsumoto et al. (87) was the first to report that DNA polymerase beta (pol β) had an AP lyase activity. In this classic BER pathway also known as “one base filling repair” or “short patch (SP) repair,” pol β fills the one base-gap and DNA ligase I seals the nick. The 5′-end cleaning has to be processed by FEN1 when the 5′-moiety is resistant to AP lyase activity, such as oxidized AP site product (75). In this BER subpathway, the FEN1 reaction leaves a multiple bases-long gap in the DNA and thus, is called “long patch” (LP) repair. Thus, the importance of distinguishing the two modes of BER is that in the cells SP and LP BER may be separately necessary depending on the condition of intermediate lesions. Another situation where 5′-end cleaning is necessary is immature reaction of DNA ligase III at DNA nicks produced by APE1 reaction, which leaves 5′-adenylated lesion. Resolution of this particular lesion is carried out by Aprataxin (25, 105).
(v) Many mammalian DNA glycosylases and APE1, that is, BER enzymes that function before DNA repair synthesis step, possess polypeptide sequences that are not directly related to their corresponding DNA processing activities. Usually found at their N-termini, these polypeptides are about 60–150 amino acids-long, and not found in their prokaryotic orthologs. A number of studies have found that these unconserved regions provide interacting platforms among BER proteins for facilitating the reactions. Details of these domains in DNA glycosylases were reviewed previously (58, 59, 66).
(vi) Mammalian BER proteins undergo posttranslational modifications (PTMs), which modulates their activities. Most biochemical studies with reconstituted BER reactions in the 90s were based on recombinant BER proteins that were expressed and purified from E. coli. Based on these experiments, it was apparent that DNA glycosylases were relatively inefficient enzymes. Therefore, the possibility existed, that in cells BER proteins were post-translationally modified to increase repair efficiency. Although in vivo BER is far from understood, studies in the past decade or so identified multiple kinds of PTMs, including phosphorylation (61, 94), acetylation (14), ubiquitination, and SUMOylation (TDG) (6, 7, 22–24).
(vii) A new subpathway termed nucleotide incision repair (NIR) where APE1 can directly incise oxidized bases, and then the lesions are passed to DNA pol β has been reported (64, 65). In this situation, DNA glycosylases are not involved. The complete mechanism and impact on the cellular defense of NIR remain to be elucidated.
In addition to the aforementioned intricacies, poly (ADP-ribose) polymerase (PARP) and X-ray repair complementing defective repair in Chinese hamster cells 1 (XRCC1) deserve separate sections as these two molecules provide layers of complexities and opportunities for clinical applications with modulation of the mammalian BER pathway (109). PARP and XRCC1 do not participate in the direct DNA processing events, but form a scaffold that exists through almost the entire BER reaction.
In the early 1990s, PARP1, the major PARP protein, was found to act as an initial sensor molecule that recognizes SSBs in the genome. After binding to SSBs, PARP1 becomes active in catalyzing poly ADP-ribosylation reactions onto its target proteins (116). A number of proteins to be ADP-ribosylated have been identified, including DNA damage responding proteins DNA ligases, DNA polymerases, and histones (35, 98, 152). However, the main target of the ADP-ribosylation is PARP itself, and upon the polyADP-ribosylation PARP then decreases its affinity for the SSBs and thus, is released from the site, while the automodification results in interacting with DNA damage response proteins, including XRCC1, ATM, Ku70/86, and DNA-PKcs. It is believed that the initial binding by PARP1 has two functional roles for BER. One is to protect the toxic SSB from triggering cell death signal, and the other is to recruit BER proteins necessary in the later stages. An important interaction of PARP1 is established with XRCC1 at an early point of SSB repair. PARP1 homozygous knockout mice are viable, but PARP1−/− mouse embryonic fibroblasts showed hypersensitivity against many DNA damaging reagents, including ionizing radiation and ROS producing chemicals (37, 138), and double knockout (ko) of PARP1 and PARP2, a backup of PARP1, turned out to be lethal (91). These animal studies thus, confirmed PARP's critical activity in BER. Other ADP-ribosylase proteins of the PARP family that are clinically relevant, but less studied, with BER coordination function are reviewed by Poirier's group (109). Lethality of cancer cells, deficient in specific DNA repair, can be synthetically increased by inhibiting PARP. Two groups in 2005 independently observed efficient lethality of BRCA deficient cells with PARP inhibitors (20, 45), and use of PARP inhibitors in clinical trials are underway (85). More recently, PARP1 was shown to act at replication fork arrest caused by topoisomerase inhibitors to facilitate reversal of replication forks (106), and application of PARP inhibitors combined with topoisomerase I inhibitor has been discussed (13). Enzymatic activity of PARP has many biological consequences. In addition to BER enhancement, many pathophysiological conditions that overactivate PARP have been reported. In these conditions, NAD+, and thus, cellular ATP, is depleted due to PARP's ADP-ribosylation reaction, and the energy depletion induced necrosis. In the original study, where middle cerebral artery occlusion was induced in mice, PARP dependent increase of infarct volume in the brain was observed. Notably, the inflammation was significantly suppressed by pretreating the mice with PARP inhibitors. Together, PARP and XRCC1 interact with many BER proteins. Known interacting partners include DNA Lig III, APE1, and PNK. Among the XRCC1–BER protein interactions, Lig III and PNK interactions turned out to be critical for BER efficiency.
Oxidative Base Damage and Cancer
8-hydroxyguanine or 8-oxoG
Among many kinds of base damage that occur due to ROS and that increase cancer risks (59), the most studied base damage without a question is 8-oxoG. Although discovered as 8-hydroxyguanine (8-OH-G), the alternative term 8-oxoG is more frequently used because of its familiarity and tendency of this tautomer to mispair with adenine base (8-oxoG:A mispair). A detailed description of the terminology appears in a recent review (97) by Nishimura who led the pioneering studies to characterize this highly mutagenic base damage. 8-OxoG is not only the most studied oxidative base damage, but its effect on carcinogenesis is shown by many layers of evidence involving sophisticated chemistry, biochemistry, structural biology, genetics, and human subject studies. 8-OxoG are generated by singlet oxygen that is readily produced by UVA and methylene blue treatment of the cells. Its high rate of mispairing with A during DNA replication makes this base damage a potently mutagenic oxidative base. In mammals, the primary enzyme to remove 8-oxoG from the genome is OGG1. To remove 8-oxoG from the genome, OGG1 cleaves N-glycosylic bonds of the oxidized bases and leaves SSBs at the site (see below), which will then be processed by the later BER enzymes. Single nucleotide polymorphisms (SNP) in the human OGG1 have been reported. A recent meta analysis conducted by Wei et al. (140) found a significant association of the OGG1 S326C SNP and a high risk of lung cancer. However, OGG1−/− mice showed no or only marginal increase in pathogenesis and cancer frequency (4, 76, 112), although an accumulation of damage and increase of mutations were observed.
Two additional DNA repair enzymes important in the cellular defense against 8-oxoG are mutT homolog 1 (MTH1) and MYH. Cellular dGTP pool also contain significant amounts of 8-oxo-dGTP, which can be incorporated opposite to A in DNA, and in the next round of DNA replication the incorporated 8-oxoG would be fixed as a mutation (A:T to C:G transversion). MTH1 is a dGTPase, which specifically targets 8-oxo-dGTP, and it keeps the incorporation of 8-oxoG in the nascent strand to a minimum (86). MTH1 deficiency is known to accumulate 8-oxoG in the genomic DNA and increases cancer frequency in mice studies. MYH (E. coli MutY ortholog) is also critical in 8-oxoG repair. MutY was initially identified in E. coli as a mutator gene (96) and later shown to have a DNA glycosylase activity removing A from A:G mispair (5). Shortly after the mutagenic property of 8-oxoG became known, it was shown that MutY could also remove A from A:8-oxoG mispair (92). In a separate study, Tchou et al. (129) found that the relative activity of MutM (the OGG1 ortholog in E. coli) on 8-oxoG:C base pair was much higher than on a 8-oxoG:A pair. Thus, these studies revealed the well evolved defense against mutagenic 8-oxoG, whereby MutM repairs 8-oxoG in DNA generated de novo, but leaves misincorporated 8-oxoG opposite to A until MutY removes A (and C is filled in) to avoid mutation fixation in the next round of replication.
The mammalian MutY ortholog MYH is also a key repair enzyme to complete the 8-oxoG repair mechanism. The C:8-oxoG mispair generated de novo by ROS is not practically a mutation due to the ability of 8-oxoG to pair with C, despite its high 8-oxoG:A mispairing property. Therefore, mutation becomes more probable when A is incorporated in the nascent strand opposite 8-oxoG during DNA replication. Thus, accumulation of 8-oxoG may be tolerated at a surprisingly high capacity in the genome in the OGG1 ko background, until A is incorporated opposite to 8-oxoG. MYH needs to remove A that would fix the mutation. Hence, MYH would later be shown to be a bona fide mutator of which deficiency has been shown to increase colon cancer risk, and therefore, 8-oxoG generation definitely increases cancer risk (3). Moreover, MYH−/− mice were susceptible to small intestinal tumors when they were exposed to KBrO3 (111); double or triple deficiencies of OGG1, MYH, and MTH1 were predicted, and later shown to be more mutagenic (147).
Misincorporation of bases opposite 8-oxoG by bypassing DNA polymerase was also studied. In a cell biological study, NIH3T3 cells transfected with 8-oxoG carrying the c-Ha-ras gene increased its transforming activity significantly (71, 72); thus, underscoring the importance of 8-oxoG repair before mutation fixation. Interestingly, the detailed analysis revealed that not only G to T mutation, which is predicted by the misincorporation of A opposite to 8-oxoG, but also G to C mutation were frequent in the ras gene from transformed cells. This was most likely due to translesion DNA synthesis by pol η (eta) (68).
Still the question remains as to why OGG1 deficiency alone in mice did not show an apparent increase in tumorigenesis (4, 76), despite ample in vitro and cell biological studies showing accumulation of 8-oxoG, and genomic DNA mutations in the OGG1 deficient mice. Additionally, it is unclear why an SNP in OGG1 raises the risk of lung cancer even though ko mice did not show such an increase in cancer. It is likely that multiple factors, which vary tissue to tissue, influence the cancer risk in the situation where 8-oxoG repair is compromised (79). These include the activity of MMR proteins, extent of apoptosis (which may be enhanced when the amount of 8-oxoG is increased due to OGG1 deficiency), growth factor expression, inflammation, and immune responses. Although it will be a difficult task to delineate the development of cellular transformation and malignancy originated from 8-oxoG for each organ, as described above, 8-oxoG is clearly an initiator of carcinogenesis, and thus, improving its detection will be a key factor for cancer prevention.
Finally, a novel effect of 8-oxoG on cellular inflammation is worth noting. Boldogh et al. (18) reported that 8-oxoG, the free base generated by OGG1's DNA glycosylase activity from 8-oxoG containing DNA, triggered inflammatory response in human fibroblasts, HeLa cells, and BALB/c mice. Removed from DNA, the free 8-oxoG base is bound to OGG1, which then acts as a guanine exchange factor to facilitate the exchange of GDP to GTP of Ras family GTPases, which then activates proliferating signal transduction, including MEK1,2/ERK1,2 kinases (18). Moreover, the same group later implicated this signaling pathway to allergic lung inflammation (8). These studies highlight a role of 8-oxoG in ROS mediated inflammation and show a new development clearly different from the notion that 8-oxoG in DNA, not the free base, is the cause of cancer due to its mutagenicity.
Base damage repaired by NEIL2 and lung cancer
NEIL2 (E. coli endoVIII-like 2) is a DNA glycosylase first reported by Hazra et al. (57) in 2002 that showed DNA glycosylase activity at oxidative bases, including 5-hydroxyuracil (5-OHU) and at lesser extent of activity at 5,6-dihydrouracil and 5-hydroxycytosine. 5-OHU is produced by a sequential attacks of OH• and O2− at the 5,6 position of C (39). Recently, Dey et al. (39) reported a particular set of NEIL2 SNPs associated with lung carcinoma. From lung cancer patients' genomic DNA, R103Q and R257L mutations in NEIL2 were frequently observed. In a reconstituted BER assay, including PNKP, pol β, Lig IIIα, XRCC1, and the wild-type or above two mutant NEIL2, they observed significant decrease in the whole BER activity. Because in a lung cell line with NEIL2 down-regulation, they observed a six to sevenfold increase of HPRT mutation frequency. Taken together, NEIL2 may be critical in suppressing mutations arising from spontaneous oxidative DNA damage. While the relatively broad substrate specificity of NEIL2 makes it difficult to assess a particular kind of oxidative base damage to link to cancer risk, it is also independently reported that 5-OHU is highly mutagenic and thus, it is possible that cells carrying NEIL2 with the particular SNP accumulate 5-OHU, which increases cancer risk. Mutation spectrum of the mutant NEIL2-carrying cells may be informative in this regard.
Cancer Biology and APE1
Several unique characteristics make APE1 a primary subject of etiological studies relating BER to pathophysiology, including cancer and age-related diseases.
(i) To date there is no backup enzyme for its AP endonuclease activity identified in cell biological studies, although APE2 was reported to increase its 3′–5′ exonuclease activity in the presence of proliferating cell nuclear antigen in vitro (21). This is a surprising difference from other organisms, which possess multiple APE1 enzymes. It is expected that cellular level of APE1 should significantly influence the cellular sensitivity. Ways to downregulate or inhibit APE1 has been a focus for researchers who hope to improve therapeutic strategy of cancer by utilizing the vast knowledge of BER.
(ii) APE1's major gene regulatory function through redox activation of activator protein 1 (AP-1) factor may modulate cell growth signaling pathways significantly, as activation of cJun/cFos is a signature of cancer transformation.
(iii) Both of the above are consistent with multiple observations that APE1 is up-regulated during cancer progression. Chemo and radiation therapy resistance of malignant cancer cells have been associated with increased expression of APE1 in these tissues.
(iv) A particular SNP of APE1 at the 148th amino acid residue has been implicated to increase cancer risk when combined with an XRCC1 SNP (R194W) (62, 69). About 70% of human population carries Asp residue at the 148th amino acid residue, while the rest 30% carry Glu (23). The D to E amino acid substitution is a conservative change, and the site of SNP is on the surface of the protein almost opposite to its AP-endonuclease catalytic core in the tertiary structure. Therefore, the effect of D148E substitution is likely on APE1 interaction with XRCC1 or other partners.
APE1 is the sole mammalian AP endonuclease of which endonucleolytic activity has been extensively studied and its catalytic mechanism is understood very well. Still the current cancer etiology does not sufficiently explain the mechanism by which APE1 influences tumor development and adaptation to confer resistance to therapeutic treatments. In other words, although the mechanism of endonucleolytic DNA cleaving activity is thoroughly understood, the other APE1 functions are still not understood well. In addition, studies that link APE1 to disease pathology, including cancer are many but circumstantial and do not address how known APE1 activities can lead to cancer development. In addition to its multiple functions and interacting proteins, expression of APE1, its subcellular localization, and the status of PTMs change dynamically in a cellular response to stressful conditions, such as ROS increase. Reviewing studies concerning APE1 reactions associated with different cellular conditions should thus, help us establish a clearer model where APE1 influences pathological conditions. Of note, APE1's localization in nucleoli and interaction with nucleophosmin 1 (NPM1), as well as RNA processing capability are reviewed in the other articles in this Forum. PTM of APE1 was reviewed in a recent article (23). It will be briefly mentioned how multiple kinds of PTM affect APE1's localization and function. In addition, factors that interact with APE1 are listed in Tables 1 and 2 (adapted from the thebiogrid.org).
Table 1.
Proteins That APE1 Identified as Interacting Partners
| Interactor | Function | Experiment | Author |
|---|---|---|---|
| TCP1 | Chaparone | a | Vascotto (135) |
| PRPF19 | DNA repair, RNA splicing, ubiquitination | ||
| KRT8 | Cytoskeletal | ||
| RPSA | Ribosomal protein | ||
| WDR77 | RNA processing | ||
| TCEB1 | Transcription | a | Kristensen (78a) |
| TWF2 | Cytoskeletal associated | ||
| TXNRD1 | REDOX homeostasis | ||
| ANP32A | RNA processing, transcription | b | Fan (44) |
| SET | Nucleosome, RNA processing | ||
| MDM2 | Cell cycle, ubiquitination | b | Busso (22) |
| RNF4 | Transcription, ubiquitination | b | Hu (62a) |
| TP53 | Cell cycle, transcription, DNA damage response | b | Seemann (116a) |
| NPM1 | Nucleosome, DNA repair | b | Vascotto (135) |
| PRPF19 | DNA repair, RNA splicing, ubiquitination | ||
| WDR77 | RNA processing | ||
| PRDX6 | REDOX homeostasis | ||
| TRAF2 | NF-kappaB signaling | b | Merluzzi (91a) |
| STAT3 | JAK/STAT signaling | b | Gray (51c) |
| EP300 | Transcription | b | Sengupta (117a) |
| ASCL2 | Transcription | ||
| YBX1 | Transcription | ||
| SFPQ | DNA repair, RNA splicing, transcription | c | Havugimana (56a) |
| SNRPD1 | RNA processing | ||
| HIF1A | Hypoxia signaling | d | Carrero (25a) |
| XRCC5 | DNA repair | d | Chung (29b) |
| APP | Platelett and neuronal | d | Olah (100a) |
| EP300 | Transcription | d | Sengupta (117a) |
| XRCC1 | DNA repair | e | Vidal (136a) |
| POLR3D | Transcription | e | Ravasi (105a) |
| HOXC13 | Transcription | ||
| TCF21 | Transcription | ||
| UBE2I | Ubiquitination | e | Yan (148a) |
Affinity capture-MS.
Affinity capture-Western.
Cofractionation.
Reconstituted complex.
Two-hybrid (adapted from http://thebiogrid.org).
APE1, apurinic/apyrimidinic endonuclease 1.
Table 2.
Proteins That Identified APE1 as an Interacting Partner
| Interactor | Function | Experiments | Author |
|---|---|---|---|
| ARIH2 | Ubiquitination | a | Kristensen (78a) |
| CAPNS1 | Calpain proteolysis | ||
| CCDC124 | |||
| ESR1 | Estrogen signaling | a | Cheng (29a) |
| hnRNPK | Transcription, RNA processing | a | Kristensen (78a) |
| hnRNPUL1 | RNA processing | ||
| LGALS1 | Signal transduction | ||
| NAE1 | Neddylation | ||
| PABPC1 | RNA processing | ||
| PAPSS2 | Small molecule metabolism | ||
| PSMG1 | Proteosome | ||
| Rev | Virus replication | a | Naji (95a) |
| RIC8A | GTPase activity | a | Kristensen (78a) |
| SUMO1 | Sumolation | a | Grant (51b) |
| SUMO2 | Sumolation | a | Golebiowski (51a) |
| TFAP4 | DNA damage response, transcription | a | Ku (78b) |
| TXN | REDOX homeostasis, signal transduction | a | Kristensen (78a) |
| UBC | Ubiquitination | a | Meierhofer (89a) |
| UBC | Ubiquitination | a | Danielsen (35a) |
| UBC | Ubiquitination | a | Kim (74a) |
| XPOT | RNA transport | a | Kristensen (78a) |
| EP300 | Transcription | b | Sengupta (117a) |
| FEN1 | DNA repair | b | Dianova (39a) |
| MUTYH | DNA repair | b | Parker (101a) |
| RNF4 | Transcription, ubiquitination | b | Hu (62a) |
| TFAP4 | DNA damage response, transcription | b | Ku (78b) |
| TRAF2 | NF-kappaB signaling | b | Merluzzi (91a) |
| TXNRD1 | REDOX homeostasis | b | Seemann (116a) |
| UBC | Ubiquitination | b | Busso (22) |
| MDM2 | Cell cycle, ubiquitination | c | |
| TP53 | Cell cycle, transcription, DNA damage response | d | Jayaraman (68a) |
| HHV8GK18_gp81 | e | Shamay (117b) | |
| XRCC1 | DNA repair | e | Vidal (136a) |
| EP300 | Transcription | f | Sengupta (117a) |
| FEN1 | DNA repair | f | Dianova (39a) |
| hnRNPL | Transcription | f | Kuninger (78c) |
| MUTYH | DNA repair | f | Parker (101a) |
| PCNA | Cell cycle, DNA repair | f | Dianova (39a) |
| NUDT3 | Signal transduction | g | Vinayagam (136b) |
| TERF1 | Telomere | g | Lee (78d) |
| TERF2 | Telomere | ||
| TERF2IP | Telomere |
Affinity capture-MS.
Affinity capture-Western.
Biochemical activity.
Copurification.
Far-Western.
Reconstituted complex.
Two-hybrid (adapted from http://thebiogrid.org).
PCNA, proliferating cell nuclear antigen.
Multiple activities of APE1
In addition to the AP endonuclease function, which is the best characterized activity, reports have shown other cellular roles of APE1. As detailed reviews for the other functions are available in this Forum and elsewhere, they are briefly listed below with APE1's interacting partners.
Redox factor 1
In 1992 searching for a cellular factor that reduces specific Cys residues and thus, enhances the DNA binding activity of the c-Jun/c-Fos (AP-1), Curran's research group cloned the human redox factor 1 (Ref-1) cDNA, which was identical to the APE1 gene (145). APE1/Ref-1 physically interacts with AP-1 for their redox exchanging reaction. Later many groups reported similar roles of APE1 for enhancing other transcription factors, including p53, NFkB, HIF-1α, Pax8. The mechanism of the reduction is still not clear. It was reported that Cys65 in APE1 was the redox sensitive site and transfer the reduced state to AP-1 (44, 137), but there are some arguments against this scenario, including a mouse knock-in study and the fact that C65 is buried inside of the globular structure of APE1. However, Georgiadis et al. (49) proposed a conformational specific “gain of Ref-1 function” in which C65 plays the essential role.
nCaREi-dependent gene corepressor
In 1994 Okazaki et al. (99a) identified APE1 as a corepressor of human parathyroid receptor (PTH1) gene, which had been shown to be down-regulated in response to increase of intracellular Ca2+ concentration ([Ca2+]i). The promoter of PTH1 contained cis-elements named negative Ca2+ responsive elements (nCaRE), which was later shown to present upstream of other promoters, including the renin, APE1, and many other [Ca2+]i-responsive genes (89). Through investigating the nCaRE dependent regulation, it was discovered that APE1 interacts with AP-1, Ku70/86, hnRNP-L, YB-1, and HDAC1.
RNA processing by APE1
In 1995, APE1 was shown to possess RNaseH activity like its E. coli ortholog exonuclease III (12), while its significance in vivo was not clear. Much later in 2009, Barnes et al. (11) reported that purified APE1 showed endoribonuclease activity on c-myc mRNA. Whether APE1 specifically targets c-myc mRNA through its endoribonuclease activity or other RNA molecules are also processed by APE1 has not been clear. However, APE1 has activity to cleave AP-sites in RNA (135), and the N-terminal 33 amino acid residues in APE1 were necessary to bind to RNA (135). In the same report, NPM1 was shown to interact with this N-terminal APE1 segment, and NPM1 prevented APE1 from binding to RNA. Again, the latter report did not examine the inhibitory effect of NPM1 on the c-myc mRNA cleavage activity by APE1; thus, it is not clear whether NPM1 can modulate APE1‘s RNA processing in general.
Subcellular localization of APE1
Non-nuclear APE1 and clinical implication
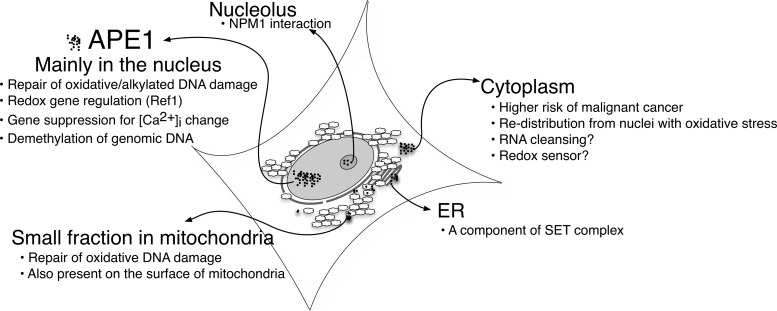
APE1 obviously needs to be imported into the nucleus for BER. The nCaRE-dependent gene regulation requires APE1 to bind to the consensus sequences and the redox-mediated activation (Ref-1) of AP-1 transcription factor assumes APE1 to be in the nuclei as well. A large number of studies indicate that considerable amount of APE1 is localized outside of the nucleus. These sites include cytoplasm, endoplasmic reticulum (ER), and mitochondria. Except for the mitochondrial APE1, biological significance of this localization is not clear. Soon the importance of APE1 in the above subcellular location will likely be revealed as details emerge. In particular, an advance in technology to identify more precise subcellular locations of APE1 and identifying interacting partners at each organelle will be important to understand clinical implication of these phenomena. Although it is far from a complete understanding, some studies that focused on the non-nuclear APE1 are worth mentioning (also illustrated in Fig. 5).
FIG. 5.
Subcellular distribution of APE1 and its influence on cell physiology. Details in the main text.
APE1 in mitochondria
AP endonuclease in mitochondria was first discovered in a biochemical study (131) before APE1, the major AP endonuclease in mammals, was cloned. A purified protein fraction from mouse mitochondria contained an AP endonuclease activity, which cross-reacted with an APE1 antibody, and its molecular weight appeared to be 65 kDa (APE1 is 36 kDa). Unlike APE1, the purified mitochondrial APE was not inhibited by adenine or NAD+, and not stimulated by Triton X-100, indicating the mitochondrial APE1 was distinguishable from APE1 and the observation was not due to a simple contamination of APE1 into the mitochondrial fraction. Although the identity of the 65 kDa APE is still unclear, it should be noted that APE1 may undergo a post-translational modification for translocation into mitochondria. Another possibility is that APE2 cross reacted with an APE1 antibody. APE2 has a calculated molecular mass of 57 kDa and was shown to be localized in mitochondria (133), and to cross react with APE1 antibody. The latter possibility should be tested, although AP-endonuclease activity in purified APE2 has been hardly detectable and therefore, it will be difficult to elucidate its resistance to adenine and NAD+.
Focusing on the localization of APE1, it was reported that APE1 was indeed translocalized into mitochondria (28). Purified from bovine liver, the mitochondrial APE1 activity showed about a three times higher kcat value than APE1 purified from nuclei. The mtAPE1 was truncated to be 33 kDa protein due to the cleavage process during mitochondrial translocalization. Interestingly, the optimum pH for the AP-endonuclease activity is at 9.2, which is significantly higher ompared with the nuclear APE1 (pH 7.5). The study also observed mitochondrial APE1 in immunocytochemistry although the amount was much smaller than the nuclear APE1.
As described in the previous section, mitochondrial respiration reaction OXPHOS is the main source of ROS generation in the cells. In addition, mitochondrial DNA is only loosely coated to form a nucleoid structure (50), unlike the chromatin structure in the genome. In 2008, two independent research groups reported rather detailed mtBER mechanisms and although there was a major difference at one BER reaction step, they were at most points in agreement and have been regarded as the framework model of mtBER. The mtBER detailed by Szczesny et al. (125, 127) consisted of mtAPE1 for 3′-OH generation, DNA pol γ with its associated AP lyase activity to generate 5′-phosphate and to carry out single nucleotide gap filling followed by the DNA ligation step by DNA Lig III. Oxidized AP sites (66, 75) are handled by APE1 too, but generate 5′-moiety, which is resistant to the AP lyase activity. The study found that in these cases 5′-exo/endonuclease (ExoG) carries out the 5′-end cleaning step. Liu et al. (84) in the same year proposed almost the same mtBER model independently, except for the difference of 5′-end processing, which in their study was carried out by FEN1. Both studies showed the presence of FEN1 in mitochondria, but other studies did not detect FEN1 in mitochondria, and its biological roles are not clear (2, 110). In any event, it is generally agreed that mtBER exists and is carried out in an almost identical manner as in the nuclear BER, using DNA glycosylases (UNG1, OGG1), APE1, ExoG or FEN1, pol γ, and DNA ligase III (2, 84, 125).
Many studies have identified mutations in mitochondrial DNA (www.mitomap.org/MITOMAP). However, studies of cancer associated mutations in mitochondrial DNA have not reached a consensus whether a particular type of mutations are represented in cancer cells. Moreover, two frequently discovered cancer associated mitochondrial DNA changes are of deletions (D310 and 4977), and thus, it is unlikely that mtBER affected the mutation frequency. Point mutations in mitochondrial DNA of cancer cells and other disease carrying cells, such as cardiomyopathy are also known and these mutations may arise from oxidative DNA damage and failure to repair with mtBER. Now that the mechanism of mtBER is known, the link between mtBER and diseases due to mitochondrial deficiency will hopefully become clearer.
APE1 in ER
During purification of mitochondrial protein, a considerable amount of APE1 was found in the ER (124). Also in an early study specifically in T-cells, it was reported that APE1 was in the ER as a component of the SET complex, and during apoptosis APE1 is translocated into nuclei with N-terminal 35 amino acid truncation, the site different from the one during mitochondrial translocalization. The truncated APE1 once in nuclei facilitates the apoptotic process by acting as a nonspecific exonuclease (44, 151). It should be noted that mitochondrial APE1, which is processed through a similar truncation (N-terminal 33 amino acid) did not show an increase in exonuclease activity, and therefore, APE1 must be associated with some other factors to facilitate exonuclease reaction. However, such a mechanism is still unknown. In any case, while multiple reports confirmed APE1‘s presence in ER, its biological or clinical implication of ER associated APE1 has not been elucidated yet. Since ER's main function is to facilitate proper folding and sorting of proteins to Golgi apparatus toward protein transportation, it may be reasonable to assume that APE1 in ER is a prerequisite for translocalization to known targets, such as nuclei and mitochondria. However, our unpublished results show almost equal amount of endogenous APE1 can exist in ER compared to nuclei, and so it is surprising that such a high ratio of APE1 is present in ER for the simple purpose of being sorted out. More dedicated study to identify its interacting partners and the fate of APE1 in ER should enlighten the rather curious observation.
APE1 in cytoplasm
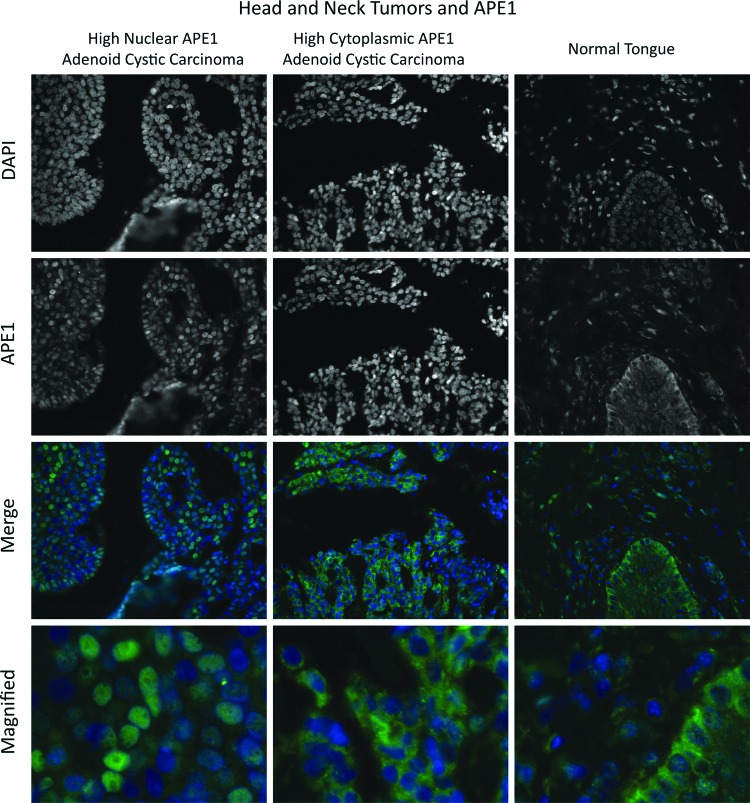
The presence of APE1 in the cytoplasm was first reported by Kakolyris et al. (70). The study discovered that significant amounts of APE1 were found outside of the nuclei in various human tissues. Remarkably, many of the tissues, including superficial cells of gastrointestinal tract and prostate glands, showed APE1 predominantly in the cytoplasm and not in the nucleus. The observation was unexpected and raised the possibility that in some circumstances APE1 needs to be sequestered from nuclei. As mentioned above, APE1 is also present in the ER (124), and thus, it is not clear how much of APE1 outside of the nucleus was in the ER or mitochondrial, or simply outside of any organelle. Nonetheless, the study by Kakolyris et al. raised the possibility that the cellular distribution of endogenous APE1 in vivo differs significantly from those observed in the cultured cell lines. Unsurprisingly, APE1 distribution was also examined in the past using cultured cell lines and transiently expressed APE1, which was tagged with artificial epitope, such as FLAG tag and EGFP (67, 126). These methodologies can increase the specificity for APE1 probes and thus, have advantages to reduce artificial signals due to cross-reactivities of antibodies to unrelated proteins. These studies unanimously observed that APE1 is almost exclusively in the nuclei. There are highly specific APE1 antibodies that show little cross-reactivity to unrelated proteins. Figure 6 shows two representative patterns of APE1 localization in head and neck cancer tissues examined by immunohistochemistry. The results point out the large variability of subcellular distributions of APE1 among individual tissues, corroborating the earlier observation by Kakolyris et al. and emphasize the importance of the studies linking unusual distribution to cancer malignancy (77, 143).
FIG. 6.
Immunohistochemical analysis of APE1 localization in the head and neck cancer tissues. Two different patterns of subcellular localization of APE1 (green) are apparent between two head and neck cancer tissues (left, predominantly in the nuclei; middle, in the cytoplasm) along with a normal tongue staining pattern. Nuclei are stained with DAPI. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
These staining profiles of histochemistry show a stark contrast from those observed in cultured cell lines, and raise interesting questions. One is why experimentally used cell lines show the nuclear APE1 almost exclusively, and another question is that if APE1 in the cytoplasm is common with tissue specimens, what role APE1 has in the cytoplasm. To answer these questions in cell biological studies, it is imperative to carefully define conditions where cultured cells induce cytoplasmic localization of APE1. Indeed, there are a few studies that identified such situations. Vascotto et al. (134) reported in 2011 that APE1 in glioblastoma cell line SF767 increased cytoplasmic APE1 in response to E3330, a redox-active compound that had been reported to inhibit APE1's Ref-1 activities. The mechanism is not completely clear yet, but it is likely that that E3330 changes the APE1 redox status and enhances relocalization of APE1 (134). To elucidate a relation between APE1 expression in the cells and malignancy of lung squamous cell carcinomas, Wu et al. reported that primary lung cancer cells are found to contain significant amount of cytoplasmic APE1 (143, 150). Certain lung cancer cell lines, including H441 were also identified as the cells rich in cytoplasmic APE1 (143). These findings are important not only in that the lung cancer malignancy was linked to cytoplasmic localization of APE1, but also in that cultured cell lines could be used to study the mechanism of and the physiological influence of cytoplasmic localization of APE1. The group also reported that E6 protein from human papilloma virus type 16 increased cytoplasmic APE1 (144). Although expression of E6 protein in nonsmall cell lung carcinoma may not be common, it would be interesting to test if E6 expression in other cell types, such as in squamous cell carcinoma of head and neck causes APE1 cytoplasmic localization.
How is APE1 enriched in the cytosol? In an independent study, Qu et al. (103) reported that a post-translational modification of APE1, S-nitrosation, occurred to APE1's two Cys residues, that is, C93 and C310. The group showed evidence that the S-nitrosation triggers nuclear export in a manner dependent on CRM1. Such nuclear export was abrogated by introducing point mutations at the two Cys residues. Therefore, these independent studies as a whole have proven that signal(s) to enhance cytoplasmic localization of APE1 exist in cells, and it may be relevant to malignancy of cancer cells. The mediating signals are not fully identified yet, but it is reasonably assumed to be triggered by reactive oxygen/nitrogen species as two studies indicated they are redox related mediators. There still needs investigation to identify a specific location where the S-nitrosation modification on APE1 may occur. If APE1 is modified by S-nitrosation before entering the nucleus, the modification may also inhibit nuclear localization.
It is not clear why increased cytoplasmic APE1 has a correlation with cancer malignancy. To answer this question, it is necessary to know the activity of APE1 in the cytoplasm. Also, as described above, most cultured cell lines do not show significant amount of cytoplasmic APE1. This is perplexing as almost all cell lines were established from cancer cells. One possibility is that APE1 in cytoplasm is very efficiently degraded, an idea consistent with the S-nitrosation study. It has been reported that APE1 is ubiquitinated by MDM2, the major negative regulator of p53, and later by UBR3 (90), a little characterized E3 ubiquitin ligases. Although in most cases ubiquitination induces highly efficient degradation pathway using 26S proteasome in the cells, monoubiquitinated APE1 was detected in both studies, suggesting that the distinct form of ubiquitinated APE1 is stable enough to exist in the cells. Although it is unclear whether the monoubiquitinated APE1 is an intermediate product for polyubiquitination toward the degradation, or reversed to the unmodified APE1 by deubiquitinase reactions, the post-translational modification can influence APE1's subcellular distribution significantly. While ubiquitin-APE1 fusion protein was found exclusively in the cytoplasm (22), APE1 ubiquitinated de novo was detected in the nuclei and in a chromatin bound complex (24). These results suggest that APE1 does not cross the nuclear membrane in its monoubiquitinated form. Therefore, it is likely that ubiquitination of cytosolic APE1 blocks its reentrance into the nucleus, and facilitates later polyubiquitination and degradation. The authors have recently found that Parkin, a ubiquitin E3 ligase, which cooperate with PTEN-induced kinase 1 (PINK1) for mitochondrial autophagy (mitophagy), degrades APE1 (manuscript in preparation). The efficiency of polyubiquitination and degradation of APE1 appears to be much more efficient than MDM2 and possibly UBR3. Considering that PARKIN and PINK1 function in the cytoplasm, it is possible that APE1 ubiquitination in the cytoplasm is carried out by PARKIN. Interestingly, PARKIN was recently reported to be a bona-fide tumor suppressor gene of which frequent LOH was found in at least glioblastoma, lung, and ovarian cancer. Therefore, cytoplasmic localization of APE1 may be a consequence of accumulation of APE1 and cellular inability to degrade APE1. Indeed, the amount of cytoplasmic APE1 was increased by treating cells with MG132, a potent 26S proteasome inhibitor.
Closing Remarks
ROS have influence on almost all kinds of cellular activities. Accidents happen: O2− generation during OXPHOS is unavoidable. Organisms have evolved to deal with it, and put the ROS generation into intra- and inter-cellular regulatory system that needs to coordinate the balanced cell growth that needs to involve autophagy and cell death. Since energy generation is inseparable from any kind of cellular activity, it is natural to use ROS as an essential signaling mediator in the cell. Genes are also continuously attacked by ROS resulting in mutations and contributing to variations of lives. However, DNA damage due to ROS formation causes diseases, including cancer. Understanding the role of ROS in unregulated cell growth, resistance to apoptosis, induction of nonautonomous cell outgrowth should be the work combining studies of cancer genetics and tumorigenesis involving stroma development. In the past decade reports established that abnormality of BER proteins in cancer cells. SNP and mutations in cancer cells are clear indications that 8-oxoG formation is involved in the cellular transformation.
APE1 is enigmatic not because of its DNA repair mechanism; the catalytic mechanism and biological significance of the AP-endonuclease activity is very well understood. It is the other functions of APE1 that still little is known, particularly in relation to cancer biology. However, recent studies described above are beginning to link the DNA repair and the other activities of APE1 toward understanding of APE1's influence on cancer malignancies. These studies form a front line to develop preventive measures, biomarkers, and therapeutic strategies based on BER proteins.
Abbreviations Used
- 5-OHU
5-hydroxyuracil
- 8-OH-G
8-hydroxyguanine
- 8-oxoG
7,8-dihydro-8-oxoguanine
- AP-1
activator protein 1
- APE1
apurinic/apyrimidinic endonuclease 1
- BER
base excision repair
- DSB
DNA double-strand break
- DSBR
DNA double-strand break repair
- ER
endoplasmic reticulum
- GSTp
glutathione S-transferase pi
- HDAC
histone deacetylase
- hnRNP
heterogenous nuclear ribonucleoprotein
- HR
homologous recombination
- JNK
c-Jun N-terminal kinase
- ko
knockout
- LP
long patch
- MGMT
O6-methylguanine DNA methyltransferase
- MMR
mismatch repair
- MTH
mutT homolog
- MYH
mutY homolog
- nCaRE
negative Ca2+ responsive element
- NEIL
endonuclease VIII-like
- NER
nucleotide excision repair
- NHEJ
nonhomologous end joining
- NIR
nucleotide incision repair
- NPM1
nucleophosmin 1
- NTH1
endonuclease III-like 1
- OGG1
8-oxoguanine DNA glycosylase
- OXPHOS
oxidative phosphorylation
- PARP
poly (ADP-ribose) polymerase
- PINK1
PTEN-induced kinase 1
- pol β
polymerase beta
- PTM
posttranslational modification
- Ref-1
redox factor 1
- ROS
reactive oxygen species
- SNP
single nucleotide polymorphism
- SOD
superoxide dismutase
- SP
short patch
- SSB
single-strand break
- TDG
thymine DNA glycosylase
- TDP1
tyrosyl-DNA phosphodiesterase 1
- UNG
uracil DNA glycosylase
- XRCC1
X-ray repair complementing defective repair in Chinese hamster cells 1
- YB-1
Y box binding protein 1
Acknowledgments
The authors would like to thank Dr. Susumu Nishimura for providing a number of critical suggestions, particularly for the section concerning 8-hydroxyguanine. This work was supported by NIH grants CA098664 and CA098664-06S1 (T.I.).
References
- 1.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR, Davis RJ, and Ronai Z. Regulation of JNK signaling by GSTp. EMBO J 18: 1321–1334, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbari M, Visnes T, Krokan HE, and Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 7: 605–616, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, and Cheadle JP. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet 30: 227–232, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Arai T, Kelly VP, Minowa O, Noda T, and Nishimura S. The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology 221: 179–186, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Au KG, Clark S, Miller JH, and Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci USA 86: 8877–8881, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, and Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979–982, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, and Shirakawa M. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J Mol Biol 359: 137–147, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Bacsi A, Aguilera-Aguirre L, Szczesny B, Radak Z, Hazra TK, Sur S, Ba X, and Boldogh I. Down-regulation of 8-oxoguanine DNA glycosylase 1 expression in the airway epithelium ameliorates allergic lung inflammation. DNA Repair (Amst) 12: 18–26, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacsi A, Woodberry M, Widger W, Papaconstantinou J, Mitra S, Peterson JW, and Boldogh I. Localization of superoxide anion production to mitochondrial electron transport chain in 3-NPA-treated cells. Mitochondrion 6: 235–244, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailly V. and Verly WG. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J 242: 565–572, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, and Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res 37: 3946–3958, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barzilay G, Walker LJ, Robson CN, and Hickson ID. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res 23: 1544–1550, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Jr, Lopes M, and Vindigni A. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20: 347–354, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhakat KK, Izumi T, Yang SH, Hazra TK, and Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J 22: 6299–6309, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bielas JH. and Loeb LA. Quantification of random genomic mutations. Nat Methods 2: 285–290, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Bielas JH, Loeb KR, Rubin BP, True LD, and Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA 103: 18238–18242, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobola MS, Blank A, Berger MS, Stevens BA, and Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res 7: 3510–3518, 2001 [PubMed] [Google Scholar]
- 18.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, and Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J Biol Chem 287: 20769–20773, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breen AP. and Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med 18: 1033–1077, 1995 [DOI] [PubMed] [Google Scholar]
- 20.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, and Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Burkovics P, Hajdu I, Szukacsov V, Unk I, and Haracska L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res 37: 4247–4255, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busso CS, Iwakuma T, and Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene 28: 1616–1625, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busso CS, Lake MW, and Izumi T. Posttranslational modification of mammalian AP endonuclease (APE1). Cell Mol Life Sci 67: 3609–3620, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busso CS, Wedgeworth CM, and Izumi T. Ubiquitination of human AP-endonuclease 1 (APE1) enhanced by T233E substitution and by CDK5. Nucleic Acids Res 39: 8017–8028, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631, 2008 [DOI] [PubMed] [Google Scholar]
- 25a.Carrero P, Okamoto K, Coumailleau P, O'Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol 20: 402–415, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casini A, Ceni E, Salzano R, Biondi P, Parola M, Galli A, Foschi M, Caligiuri A, Pinzani M, and Surrenti C. Neutrophil-derived superoxide anion induces lipid peroxidation and stimulates collagen synthesis in human hepatic stellate cells: role of nitric oxide. Hepatology 25: 361–367, 1997 [DOI] [PubMed] [Google Scholar]
- 27.Chakravarti D, Ibeanu GC, Tano K, and Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem 266: 15710–15715, 1991 [PubMed] [Google Scholar]
- 28.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, and Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res 34: 2067–2076, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, and Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem 278: 36027–36031, 2003 [DOI] [PubMed] [Google Scholar]
- 29a.Cheng PC, Chang HK, Chen SH. Quantitative nanoproteomics for protein complexes (QNanoPX) related to estrogen transcriptional action. Mol Cell Proteomics 9: 209–224, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29b.Chung U, Igarashi T, Nishishita T, Iwanari H, Iwamatsu A, Suwa A, Mimori T, Hata K, Ebisu S, Ogata E, Fujita T, Okazaki T. The interaction between Ku antigen and REF1 protein mediates negative gene regulation by extracellular calcium. J Biol Chem 271: 8593–8598, 1996 [DOI] [PubMed] [Google Scholar]
- 30.Cleaver JE, Lam ET, and Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet 10: 756–768, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Cortazar D, Kunz C, Saito Y, Steinacher R, and Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 6: 489–504, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, and Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470: 419–423, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, and Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creissen D. and Shall S. Regulation of DNA ligase activity by poly(ADP-ribose). Nature 296: 271–272, 1982 [DOI] [PubMed] [Google Scholar]
- 35.D'Amours D, Desnoyers S, D'Silva I, and Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342(Pt 2): 249–268, 1999 [PMC free article] [PubMed] [Google Scholar]
- 35a.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics 10: M110 003590, 2011. (DOI 10.1074/mcp.M110.003590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, and Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair (Amst) 5: 1439–1448, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, and de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA 94: 7303–7307, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demple B, Herman T, and Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA 88: 11450–11454, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey S, Maiti AK, Hegde ML, Hegde PM, Boldogh I, Sarkar PS, Abdel-Rahman SZ, Sarker AH, Hang B, Xie J, Tomkinson AE, Zhou M, Shen B, Wang G, Wu C, Yu D, Lin D, Cardenas V, and Hazra TK. Increased risk of lung cancer associated with a functionally impaired polymorphic variant of the human DNA glycosylase NEIL2. DNA Repair (Amst) 11: 570–578, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry 40: 12639–12644, 2001 [DOI] [PubMed] [Google Scholar]
- 40.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, and Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434: 108–113, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O'Sullivan J, Salk JJ, and Bielas JH. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet 8: e1002689, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernster L. and Dallner G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim Biophys Acta 1271: 195–204, 1995 [DOI] [PubMed] [Google Scholar]
- 43.Evans MD, Dizdaroglu M, and Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res 567: 1–61, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, and Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol 4: 145–153, 2003 [DOI] [PubMed] [Google Scholar]
- 45.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, and Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Fitzgerald ME. and Drohat AC. Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J Biol Chem 283: 32680–32690, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritz G. and Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene 18: 1033–1040, 1999 [DOI] [PubMed] [Google Scholar]
- 48.Fulda S, Galluzzi L, and Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 9: 447–464, 2010 [DOI] [PubMed] [Google Scholar]
- 49.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, and Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res 643: 54–63, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, and Quintanilla B. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol 5: a011080, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillies RJ, Verduzco D, and Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer 12: 487–493, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24, 2009 [DOI] [PubMed] [Google Scholar]
- 51b.Grant MM. Identification of SUMOylated proteins in neuroblastoma cells after treatment with hydrogen peroxide or ascorbate. BMB Rep 43: 720–725, 2010 [DOI] [PubMed] [Google Scholar]
- 51c.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene 24: 3110–3120, 2005 [DOI] [PubMed] [Google Scholar]
- 52.Gu H, Marth JD, Orban PC, Mossmann H, and Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265: 103–106, 1994 [DOI] [PubMed] [Google Scholar]
- 53.Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, and Califano JA. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res 8: 2260–2265, 2002 [PubMed] [Google Scholar]
- 54.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, and Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardeland U, Steinacher R, Jiricny J, and Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 21: 1456–1464, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, and Hottiger MO. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol Cell 10: 1213–1222, 2002 [DOI] [PubMed] [Google Scholar]
- 56a.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A. A census of human soluble protein complexes. Cell 150: 1068–1081, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, and Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem 277: 30417–30420, 2002 [DOI] [PubMed] [Google Scholar]
- 58.Hegde ML, Hazra TK, and Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell Mol Life Sci 67: 3573–3587, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hegde ML, Izumi T, and Mitra S. Oxidized base damage and single-strand break repair in Mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes. Prog Mol Biol Transl Sci 110: 123–153, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hill JW, Hazra TK, Izumi T, and Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res 29: 430–438, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu J, Imam SZ, Hashiguchi K, de Souza-Pinto NC, and Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res 33: 3271–3282, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, and Case LD. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis 22: 917–922, 2001 [DOI] [PubMed] [Google Scholar]
- 62a.Hu XV, Rodrigues TM, Tao H, Baker RK, Miraglia L, Orth AP, Lyons GE, Schultz PG, Wu X. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc Natl Acad Sci USA 107: 15087–15092, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue A, Shen L, Dai Q, He C, and Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21: 1670–1676, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ischenko AA. and Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415: 183–187, 2002 [DOI] [PubMed] [Google Scholar]
- 65.Ishchenko AA, Deprez E, Maksimenko A, Brochon JC, Tauc P, and Saparbaev MK. Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc Natl Acad Sci USA 103: 2564–2569, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, and Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology 193: 43–65, 2003 [DOI] [PubMed] [Google Scholar]
- 67.Jackson EB, Theriot CA, Chattopadhyay R, Mitra S, and Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1). Nucleic Acids Res 33: 3303–3312, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaloszynski P, Masutani C, Hanaoka F, Perez AB, and Nishimura S. 8-Hydroxyguanine in a mutational hotspot of the c-Ha-ras gene causes misreplication, ‘action-at-a-distance’ mutagenesis and inhibition of replication. Nucleic Acids Res 31: 6085–6095, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68a.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev 11: 558–570, 1997 [DOI] [PubMed] [Google Scholar]
- 69.Jiao L, Bondy ML, Hassan MM, Wolff RA, Evans DB, Abbruzzese JL, and Li D. Selected polymorphisms of DNA repair genes and risk of pancreatic cancer. Cancer Detect Prev 30: 284–291, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kakolyris S, Kaklamanis L, Giatromanolaki A, Koukourakis M, Hickson ID, Barzilay G, Turley H, Leek RD, Kanavaros P, Georgoulias V, Gatter KC, and Harris AL. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: a basis for its role in human disease. Histopathology 33: 561–569, 1998 [DOI] [PubMed] [Google Scholar]
- 71.Kamiya H, Miura K, Ishikawa H, Inoue H, Nishimura S, and Ohtsuka E. c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res 52: 3483–3485, 1992 [PubMed] [Google Scholar]
- 72.Kamiya H, Murata-Kamiya N, Koizume S, Inoue H, Nishimura S, and Ohtsuka E. 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene: effects of sequence contexts on mutation spectra. Carcinogenesis 16: 883–889, 1995 [DOI] [PubMed] [Google Scholar]
- 73.Kasai H. and Nishimura S. Hydroxylation of the C-8 position of deoxyguanosine by reducing agents in the presence of oxygen. Nucleic Acids Symp Ser 12: 165–167, 1983 [PubMed] [Google Scholar]
- 74.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, and Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J Immunol 188: 3559–3566, 2012 [DOI] [PubMed] [Google Scholar]
- 74a.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klungland A. and Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 16: 3341–3348, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, and Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA 96: 13300–13305, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, and Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Radiat Oncol Biol Phys 50: 27–36, 2001 [DOI] [PubMed] [Google Scholar]
- 78.Krokan HE, Standal R, and Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J 325(Pt 1): 1–16, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78a.Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nature Methods 9: 907–909, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78b.Ku WC, Chiu SK, Chen YJ, Huang HH, Wu WG, Chen YJ. Complementary quantitative proteomics reveals that transcription factor AP-4 mediates E-box-dependent complex formation for transcriptional repression of HDM2. Mol Cell Proteomics 8: 2034–2050, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78c.Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res 30: 823–829, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78d.Lee OH, Kim H, He Q, Baek HJ, Yang D, Chen LY, Liang J, Chae HK, Safari A, Liu D, Songyang Z. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol Cell Proteomics 10: M110 001628, 2011. ( 10.1074/mcp.M110.001628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liddiard K, Hills R, Burnett AK, Darley RL, and Tonks A. OGG1 is a novel prognostic indicator in acute myeloid leukaemia. Oncogene 29: 2005–2012, 2010 [DOI] [PubMed] [Google Scholar]
- 80.Lindahl T. Instability and decay of the primary structure of DNA. Nature 362: 709–715, 1993 [DOI] [PubMed] [Google Scholar]
- 81.Lindahl T. and Ljungquist S. Apurinic and apyrimidinic sites in DNA. Basic Life Sci 5A: 31–38, 1975 [DOI] [PubMed] [Google Scholar]
- 82.Liu D, Croteau DL, Souza-Pinto N, Pitta M, Tian J, Wu C, Jiang H, Mustafa K, Keijzers G, Bohr VA, and Mattson MP. Evidence that OGG1 glycosylase protects neurons against oxidative DNA damage and cell death under ischemic conditions. J Cereb Blood Flow Metab 31: 680–692, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu M, Bandaru V, Holmes A, Averill AM, Cannan W, and Wallace SS. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr Purif 84: 130–139, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, and Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 28: 4975–4987, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lord CJ. and Ashworth A. The DNA damage response and cancer therapy. Nature 481: 287–294, 2012 [DOI] [PubMed] [Google Scholar]
- 86.Maki H. and Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355: 273–275, 1992 [DOI] [PubMed] [Google Scholar]
- 87.Matsumoto Y. and Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science 269: 699–702, 1995 [DOI] [PubMed] [Google Scholar]
- 88.Maximo V, Lima J, Soares P, Botelho T, Gomes L, and Sobrinho-Simoes M. Mitochondrial D-Loop instability in thyroid tumours is not a marker of malignancy. Mitochondrion 5: 333–340, 2005 [DOI] [PubMed] [Google Scholar]
- 89.McHaffie GS. and Ralston SH. Origin of a negative calcium response element in an ALU-repeat: implications for regulation of gene expression by extracellular calcium. Bone 17: 11–14, 1995 [DOI] [PubMed] [Google Scholar]
- 89a.Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res 7: 4566–4576, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, Tasaki T, Kessler BM, Parsons JL, Kwon YT, and Dianov GL. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res 40: 701–711, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, and de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22: 2255–2263, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91a.Merluzzi S, D'Orlando O, Leonardi A, Vitale G, Pucillo C. TRAF2 and p38 are involved in B cells CD40-mediated APE/Ref-1 nuclear translocation: a novel pathway in B cell activation. Mol Immunol 45: 76–86, 2008 [DOI] [PubMed] [Google Scholar]
- 92.Michaels ML, Cruz C, Grollman AP, and Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA 89: 7022–7025, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Michiels C, Raes M, Toussaint O, and Remacle J. Importance of Se-glutathione peroxidase, catalase, and Cu/Zn-SOD for cell survival against oxidative stress. Free Radic Biol Med 17: 235–248, 1994 [DOI] [PubMed] [Google Scholar]
- 94.Mohan RD, Litchfield DW, Torchia J, and Tini M. Opposing regulatory roles of phosphorylation and acetylation in DNA mispair processing by thymine DNA glycosylase. Nucleic Acids Res 38: 1135–1148, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mol CD, Izumi T, Mitra S, and Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403: 451–456, 2000 [DOI] [PubMed] [Google Scholar]
- 95a.Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR, 3rd, Saphire AC, Gerace L. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol Cell Proteomics 11: M111 015313, 2012. ( 10.1074/mcp.M111.015313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nghiem Y, Cabrera M, Cupples CG, and Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G.C—T.A transversions. Proc Natl Acad Sci USA 85: 2709–2713, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nishimura S. 8-Hydroxyguanine: a base for discovery. DNA Repair (Amst) 10: 1078–1083, 2011 [PubMed] [Google Scholar]
- 98.Ohashi Y, Itaya A, Tanaka Y, Yoshihara K, Kamiya T, and Matsukage A. Poly(ADP-ribosyl)ation of DNA polymerase beta in vitro. Biochem Biophys Res Commun 140: 666–673, 1986 [DOI] [PubMed] [Google Scholar]
- 99.Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, and Igaki T. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature 490: 547–551, 2012 [DOI] [PubMed] [Google Scholar]
- 99a.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J Biol Chem 269: 27855–27862, 1994 [PubMed] [Google Scholar]
- 100.Okochi O, Hibi K, Uemura T, Inoue S, Takeda S, Kaneko T, and Nakao A. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res 8: 2875–2878, 2002 [PubMed] [Google Scholar]
- 100a.Olah J, Vincze O, Virok D, Simon D, Bozso Z, Tokesi N, Horvath I, Hlavanda E, Kovacs J, Magyar A, Szucs M, Orosz F, Penke B, Ovadi J. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J Biol Chem 286: 34088–34100, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Papa S, Martino PL, Capitanio G, Gaballo A, De Rasmo D, Signorile A, and Petruzzella V. The oxidative phosphorylation system in mammalian mitochondria. Adv Exp Med Biol 942: 3–37, 2012 [DOI] [PubMed] [Google Scholar]
- 101a.Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem 276: 5547–5555, 2001 [DOI] [PubMed] [Google Scholar]
- 102.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res 460: 165–181, 2000 [DOI] [PubMed] [Google Scholar]
- 103.Qu J, Liu GH, Huang B, and Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res 35: 2522–2532, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rasmussen AK, Chatterjee A, Rasmussen LJ, and Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res 31: 3909–3917, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rass U, Ahel I, and West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem 282: 9469–9474, 2007 [DOI] [PubMed] [Google Scholar]
- 105a.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, and Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423, 2012 [DOI] [PubMed] [Google Scholar]
- 107.Riemann A, Schneider B, Ihling A, Nowak M, Sauvant C, Thews O, and Gekle M. Acidic environment leads to ROS-induced MAPK signaling in cancer cells. PLoS One 6: e22445, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robson CN. and Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res 19: 5519–5523, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, and Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruhanen H, Ushakov K, and Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim Biophys Acta 1813: 2000–2007, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, Egashira A, Kura S, Yao T, Tsuneyoshi M, Maki H, Nakabeppu Y, and Tsuzuki T. MUTYH-null mice are susceptible to spontaneous and oxidative stress induced intestinal tumorigenesis. Cancer Res 67: 6599–6604, 2007 [DOI] [PubMed] [Google Scholar]
- 112.Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, and Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res 63: 902–905, 2003 [PubMed] [Google Scholar]
- 113.Salas A, Yao YG, Macaulay V, Vega A, Carracedo A, and Bandelt HJ. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med 2: e296, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Samson L, Derfler B, Boosalis M, and Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci USA 88: 9127–9131, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, and Sidransky D. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res 61: 7015–7019, 2001 [PubMed] [Google Scholar]
- 116.Satoh MS. and Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature 356: 356–358, 1992 [DOI] [PubMed] [Google Scholar]
- 116a.Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene 24: 3853–3863, 2005 [DOI] [PubMed] [Google Scholar]
- 117.Seki S, Akiyama K, Watanabe S, Hatsushika M, Ikeda S, and Tsutsui K. cDNA and deduced amino acid sequence of a mouse DNA repair enzyme (APEX nuclease) with significant homology to Escherichia coli exonuclease III. J Biol Chem 266: 20797–20802, 1991 [PubMed] [Google Scholar]
- 117a.Sengupta S, Mantha AK, Mitra S, Bhakat KK. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 30: 482–493, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117b.Shamay M, Liu J, Li R, Liao G, Shen L, Greenway M, Hu S, Zhu J, Xie Z, Ambinder RF, Qian J, Zhu H, Hayward SD. A protein array screen for Kaposi's sarcoma-associated herpesvirus LANA interactors links LANA to TIP60, PP2A activity, and telomere shortening. Journal of Virology 86: 5179–5191, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, and Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singhal RK. and Wilson SH. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem 268: 15906–15911, 1993 [PubMed] [Google Scholar]
- 120.Singhal RK, Prasad R, and Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem 270: 949–957, 1995 [DOI] [PubMed] [Google Scholar]
- 121.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, and Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 379: 183–186, 1996 [DOI] [PubMed] [Google Scholar]
- 122.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, and Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J 19: 3857–3869, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Szabo C, Ischiropoulos H, and Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 6: 662–680, 2007 [DOI] [PubMed] [Google Scholar]
- 124.Szczesny B. and Mitra S. Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech Ageing Dev 126: 1071–1078, 2005 [DOI] [PubMed] [Google Scholar]
- 125.Szczesny B, Tann AW, Longley MJ, Copeland WC, and Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem 283: 26349–26356, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takao M, Aburatani H, Kobayashi K, and Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res 26: 2917–2922, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, and Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J Biol Chem 286: 31975–31983, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tano K, Shiota S, Collier J, Foote RS, and Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci USA 87: 686–690, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, and Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA 88: 4690–4694, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thompson CM, Fedorov Y, Brown DD, Suh M, Proctor DM, Kuriakose L, Haws LC, and Harris MA. Assessment of Cr(VI)-induced cytotoxicity and genotoxicity using high content analysis. PLoS One 7: e42720, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tomkinson AE, Bonk RT, and Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem 263: 12532–12537, 1988 [PubMed] [Google Scholar]
- 132.Tong BC, Ha PK, Dhir K, Xing M, Westra WH, Sidransky D, and Califano JA. Mitochondrial DNA alterations in thyroid cancer. J Surg Oncol 82: 170–173, 2003 [DOI] [PubMed] [Google Scholar]
- 133.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, and Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res 29: 2349–2660, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vascotto C, Bisetto E, Li M, Zeef LA, D'Ambrosio C, Domenis R, Comelli M, Delneri D, Scaloni A, Altieri F, Mavelli I, Quadrifoglio F, Kelley MR, and Tell G. Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol Biol Cell 22: 3887–3901, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'Ambrosio C, Scaloni A, Quadrifoglio F, and Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol 29: 1834–1854, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Verschoor ML, Ungard R, Harbottle A, Jakupciak JP, Parr RL, and Singh G. Mitochondria and cancer: past, present, and future. Biomed Res Int 2013: 612369, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136a.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J 20: 6530–6539, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136b.Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, Assmus HE, Andrade-Navarro MA, Wanker EE. A directed protein interaction network for investigating intracellular signal transduction. Sci Signal 4: rs8, 2011 [DOI] [PubMed] [Google Scholar]
- 137.Walker LJ, Robson CN, Black E, Gillespie D, and Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol 13: 5370–5376, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, and Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 9: 509–520, 1995 [DOI] [PubMed] [Google Scholar]
- 139.Watson J. Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol 3: 120144, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wei W, He XF, Qin JB, Su J, Li SX, Liu Y, Zhang Y, and Wang W. Association between the OGG1 Ser326Cys and APEX1 Asp148Glu polymorphisms and lung cancer risk: a meta-analysis. Mol Biol Rep 39: 11249–11262, 2012 [DOI] [PubMed] [Google Scholar]
- 141.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, and Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell 15: 209–220, 2004 [DOI] [PubMed] [Google Scholar]
- 142.Wilson SH. and Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol 7: 176–178, 2000 [DOI] [PubMed] [Google Scholar]
- 143.Wu HH, Cheng YW, Chang JT, Wu TC, Liu WS, Chen CY, and Lee H. Subcellular localization of apurinic endonuclease 1 promotes lung tumor aggressiveness via NF-kappaB activation. Oncogene 29: 4330–4340, 2010 [DOI] [PubMed] [Google Scholar]
- 144.Wu HH, Chu YC, Wang L, Tsai LH, Lee MC, Chen CY, Shieh SH, Cheng YW, and Lee H. Cytoplasmic Ape1 expression elevated by p53 aberration may predict survival and relapse in resected non-small cell lung cancer. Ann Surg Oncol 2012[Epub ahead of print]; DOI: 10.1245/s10434-012-2431-2 [DOI] [PubMed] [Google Scholar]
- 145.Xanthoudakis S, Miao G, Wang F, Pan YC, and Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J 11: 3323–3335, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xanthoudakis S, Smeyne RJ, Wallace JD, and Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA 93: 8919–8923, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, and Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res 64: 3096–3102, 2004 [DOI] [PubMed] [Google Scholar]
- 148.Yacoub A, Kelley MR, and Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res 57: 5457–5459, 1997 [PubMed] [Google Scholar]
- 148a.Yan MD, Xu WJ, Lu LR, Sun LY, Liu XY, Zheng ZC. Ubiquitin conjugating enzyme Ubc9 is involved in protein degradation of redox factor-1 (Ref-1). Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 32: 63–68, 2000 [PubMed] [Google Scholar]
- 149.Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, and Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res 29: 743–752, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yoo DG, Song YJ, Cho EJ, Lee SK, Park JB, Yu JH, Lim SP, Kim JM, and Jeon BH. Alteration of APE1/ref-1 expression in non-small cell lung cancer: the implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung Cancer 60: 277–284, 2008 [DOI] [PubMed] [Google Scholar]
- 151.Yoshida A, Urasaki Y, Waltham M, Bergman AC, Pourquier P, Rothwell DG, Inuzuka M, Weinstein JN, Ueda T, Appella E, Hickson ID, and Y. Human apurinic/apyrimidinic endonuclease (Ape1) and its N-terminal truncated form (AN34) are involved in DNA fragmentation during apoptosis. J Biol Chem 278: 37768–37776, 2003 [DOI] [PubMed] [Google Scholar]
- 152.Yoshihara K, Itaya A, Tanaka Y, Ohashi Y, Ito K, Teraoka H, Tsukada K, Matsukage A, and Kamiya T. Inhibition of DNA polymerase alpha, DNA polymerase beta, terminal deoxynucleotidyl transferase, and DNA ligase II by poly(ADP-ribosyl)ation reaction in vitro. Biochem Biophys Res Commun 128: 61–67, 1985 [DOI] [PubMed] [Google Scholar]
- 153.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, and Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J 21: 789–800, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]