Abstract
The apolipoprotein E epsilon 4 allele (ApoE-ε4) is the strongest known genetic risk factor for late onset Alzheimer’s disease. Expansion of the lateral ventricles occurs with normal aging, but dementia accelerates this process. Brain structure and function depend on ApoE genotype not just for AD patients but also in healthy elderly, and even in asymptomatic young individuals. Therefore, we hypothesized that the ApoE-ε4 allele is associated with altered patterns of longitudinal ventricular expansion, in dementia and normal aging. We tested this hypothesis in a large sample of elderly participants, using a Linear Discriminant Analysis-based approach. Carrying more ApoE-ε4 alleles was associated with faster ventricular expansion bilaterally and with regional patterns of lateral ventricle morphology at 1- and 2-year follow up, after controlling for sex, age, and dementia status. ApoE genotyping is considered critical in clinical trials of AD. These findings, combined with earlier investigations showing that ApoE is also directly implicated in other conditions, suggest that the selective enrollment of ApoE-ε4 carriers may empower clinical trials of other neurological disorders.
Keywords: ApoE, ventricular expansion, dementia, neuroimaging genetics, structural MRI
1. INTRODUCTION
Age- and disease-related expansion of the lateral ventricles reflects the accumulation of brain tissue loss throughout multiple brain regions. It co-occurs with gray and white matter degeneration globally and in nearby subcortical regions (Ferrarini et al., 2008; Qiu et al., 2009). In normal aging, ventricular enlargement rates accelerate around age sixty, and then continue at a stable pace even in those who survive into their late nineties (Jernigan et al., 2001; Walhovd et al., 2005). Dementia affects ventricular volumes (Carmichael et al., 2007a) and the rate of ventricular enlargement (Carmichael et al., 2007b), and this rate distinguishes Alzheimer’s disease (AD) patients from healthy elderly controls, even when cross-sectional ventricular volumes overlap between the two groups (Wang et al., 2002).
The apolipoprotein E epsilon 4 allele (ApoE-ε4) is the strongest known genetic risk factor for late onset Alzheimer’s disease (LOAD), and has unequivocally been linked to AD in over 100 studies (Bertram et al., 2007). Although there are ethnic and demographic differences in allele frequencies, around 25% of the world’s population carries at least one copy of the ε4 risk allele, and each copy of this risk allele is associated with an approximate threefold increase in the odds of developing AD (Bertram et al., 2007). The ApoE-ε4 allele increases not only the risk of developing LOAD but also the rate of dementia progression both in terms of cognitive decline (Stone et al., 2010) and temporal lobe atrophy (Lehtovirta et al., 1995; Filippini et al., 2009b; Hua et al., 2010).
A recent study examined how the ApoE-ε4 allele affects ventricular volume trajectory (Erten-Lyons et al., 2013). Greater ventricular volume enlargement over time was significantly associated with carrying the ApoE-ε4 allele (Erten-Lyons et al., 2013). However, this study had several limitations, including a small sample (N=71), and the use of a semi-automated methodological approach to measure ventricular volumes on MRI scans. Most importantly, the study included both healthy elderly and individuals with dementia, but the investigators did not control for dementia status when assessing how the ApoE-ε4 allele related to longitudinal ventricular expansion. As dementia affects the rate of ventricular enlargement (Wang et al., 2002; Carmichael et al., 2007b; Erten-Lyons et al., 2013), this study design did not reveal whether the ApoE-ε4 allele may independently influence ventricular volume trajectory, or whether the observed effects were mediated by dementia status, since people with AD are also more likely to carry the ApoE-ε4 allele.
Brain structure and function are related to ApoE genotype not just for AD patients but also in the healthy elderly, and even in asymptomatic young individuals who carry the ApoE-ε4 allele. For example, healthy young ApoE-ε4 carriers show different patterns of brain activity both in resting state and during memory encoding tasks, compared to non-carriers (Filippini et al., 2009a). Further, medial temporal lobe volumes are smaller in healthy middle-aged as well as elderly ApoE-ε4 carriers (Barboriak et al., 2000; den Heijer et al., 2002; Wishart et al., 2006) and hippocampal volumes are reduced in healthy young ApoE-ε4 carriers (O’Dwyer et al., 2012). Moreover, cognitively normal, late-middle-aged adults show whole-brain atrophy rates that are correlated with ApoE-ε4 gene dose (Chen et al., 2007). Healthy elderly ApoE-ε4 carriers also have localized abnormalities in ventricular shape and volume (Chou et al., 2008; Chou et al., 2010). Based on the above findings, we thus hypothesized that the ApoE-ε4 allele is associated with the overall trajectory of ventricular volume expansion both in dementia and in normal aging.
Here, we tested this primary hypothesis in a large sample of elderly participants (N=736 at baseline) from the Alzheimer’s Disease NeuroImaging Initiative (ADNI). To track the volume of the lateral ventricles across baseline, 1-year (N=622), and 2-year (N=479) follow-up scans, we used an innovative Linear Discriminant Analysis (LDA)-based approach previously developed by our group (Gutman et al., 2013). We also applied advanced surface-based anatomical modeling to examine associations between ApoE-ε4 loading and regional changes in the shape and volume of the lateral ventricles - after controlling for age, sex, and dementia status. This allowed us to create 3D maps of ventricular expansion associated with the ApoE-ε4 allele. We also conducted exploratory post-hoc analyses to examine the effects of ApoE genotype on ventricular expansion and expansion rate within each hemisphere and within each diagnostic group, to assess its possible impact on left-right asymmetry in expansion, and to test alternate models of allele effects.
2. METHODS
2.1 Subjects
Data used in this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to help researchers and clinicians develop new treatments and monitor their effectiveness, as well as decrease the time and cost of clinical trials.
All ADNI studies are conducted according to the Good Clinical Practice guidelines, the Declaration of Helsinki, and U.S. 21 CFR Part 50 (Protection of Human Subjects), and Part 56 (Institutional Review Boards). Written informed consent was obtained from all participants before protocol-specific procedures were performed. To avoid the known effects of population stratification on genetic analysis (Lander and Schork, 1994), we only included non-Hispanic Caucasian subjects identified by self-report and confirmed by multi-dimensional scaling (MDS) analysis (Stein et al., 2010). The ADNI cohort included 3 diagnostic groups: people with Alzheimer’s disease (AD), amnestic mild cognitive impairment (MCI), and healthy elderly (cognitively normal) participants (Aisen et al., 2010). About 14% of ADNI participants reported a paternal history of AD and 35% reported a maternal history of dementia (Andrawis et al., 2012) and the 2-year MCI to AD conversion rate was 37.7% (Tatsuoka et al., 2013). Our final analysis comprised 736 individuals (average age ± s.d. = 75.52 ± 6.79 years; 436 men/300 women) including 173 AD, 358 MCI, and 205 healthy participants at baseline (Table 1).
Table 1.
Demographic data by diagnostic and genotype groups
| CON | MCI | AD | Total | ||
|---|---|---|---|---|---|
| 0 APOE ε 4 allele | N0 | 151 (68 F) | 162 (56 F) | 60 (31 F) | 373 (155 F) |
| N12 | 130 (56 F) | 138 (49 F) | 45 (22 F) | 313 (127 F) | |
| N24 | 112 (52 F) | 98 (36 F) | 31 (16 F) | 241 (104 F) | |
| Age | 76.10 (± 5.16) | 76.27 (± 7.66) | 77.15 (± 8.53) | 76.34 (± 6.92) | |
| MMSE | 29.15 (± 0.94) | 27.24 (± 1.77) | 23.17 (± 2.07) | 27.36 (± 2.55) | |
| 1 APOE ε 4 allele | N0 | 50 (24 F) | 153 (54 F) | 79 (33 F) | 282 (111 F) |
| N12 | 46 (22 F) | 129 (45 F) | 62 (28 F) | 237 (95 F) | |
| N24 | 40 (20 F) | 94 (29 F) | 49 (21 F) | 183 (70 F) | |
| Age | 76.54 (± 4.52) | 74.85 (± 6.79) | 75.97 (± 6.52) | 75.46 (± 6.38) | |
| MMSE | 29.18 (± 1.02) | 27.01 (± 1.75) | 23.48 (± 2.02) | 26.40 (± 2.64) | |
| 2 APOE ε 4 alleles | N0 | 4 (2 F) | 43 (18 F) | 34 (14 F) | 81 (34 F) |
| N12 | 4 (2 F) | 39 (16 F) | 29 (11 F) | 72 (29 F) | |
| N24 | 4 (2 F) | 30 (11 F) | 21 (8 F) | 55 (21 F) | |
| Age | 73.75 (± 3.86) | 71.81 (± 5.98) | 71.91 (± 7.28) | 71.95 (± 6.43) | |
| MMSE | 29.00 (± 0.82) | 26.84 (± 1.98) | 23.44 (± 1.86) | 25.52 (± 2.62) | |
| Total | N0 | 205 (94 F) | 358 (128 F) | 173 (78 F) | 736 (300 F) |
| N12 | 180 (80 F) | 306 (110 F) | 136 (61 F) | 622 (251 F) | |
| N24 | 156 (74 F) | 222 (76 F) | 101 (45 F) | 479 (195 F) | |
| Age | 76.16 (± 4.99) | 75.13 (± 7.23) | 75.58 (± 7.61) | 75.52 (± 6.79) | |
| MMSE | 29.16 (± 0.95) | 27.09 (± 1.79) | 23.36 (± 2.00) | 26.79 (± 2.67) | |
N0, N12, and N24 indicate sample sizes at baseline, 12-month follow-up, and 24-month follow-up, respectively. Sample sizes are followed by the number of women in parentheses. Age and MMSE indicate baseline age and scores on the mini mental status examination. Means are followed by standard deviations in parentheses.
2.2 ApoE genotyping
ApoE genotyping was performed on DNA samples obtained from subjects’ blood, using an ApoE genotyping kit, as described in http://www.adni-info.org/Scientists/Pdfs/adniproceduresmanual12.pdf (also see http://www.adni-info.org for detailed information on blood sample collection, DNA preparation, and genotyping methods). The ApoE gene is polymorphic, with three major isoforms: ApoE2 (ε2 allele), ApoE3 (ε3 allele), and ApoE4 (ε4 allele). In all our analyses, ApoE genotype was coded as 0, 1, and 2 for the presence of 0, 1, and 2 ApoE-ε4 adverse alleles, respectively.
2.3 Image acquisition, correction, and pre-processing
Participants were scanned with a standardized MRI protocol developed for this cohort (Leow et al., 2006; Jack et al., 2008). Briefly, high-resolution structural brain MRI scans were acquired at 58 sites across North America, using 1.5 Tesla MRI scanners. There was no trend in the data whereby a great number of people with any one diagnosis were preferentially evaluated on any one scanner. In order to minimize variation due to technical nonuniformity in the data collected at different sites, a major effort was taken to establishing very specific standards in the MRI protocol and acquisition parameters. The ADNI MRI protocol was selected in a data-driven manner with considerable deliberation by a group of experts in the field (Jack et al., 2008). A sagittal 3D MP-RAGE sequence was used, and optimized for consistency across sites (TR/TE = 2400/1000 ms; flip angle = 8°; FOV = 24 cm; final reconstructed voxel resolution = 0.9375 × 0.9375 × 1.2 mm3) (Jack et al., 2008). In addition, phantom-based monitoring of the instruments themselves was conducted in order to monitor performance over time for each scanner involved in the ADNI project (Jack et al., 2008). Based on 622 phantom scans collected at 49 sites, the average standard deviations of scale factors across individual scanners were 0.04, 0.07, and 0.11% along the x, y, and z axes, respectively (Gunter et al., 2007). Furthermore, image quality control procedures and post-acquisition correction of various image artifacts were performed at a single site (Mayo clinic) to ensure the consistency of these preprocessing steps (Jack et al., 2008).
2.4 Segmentation of the lateral ventricles
For lateral ventricle segmentation, we analyzed baseline (N=736), one-year (N=622), and two-year (N=479) follow-up brain MRI scans from the ADNI cohort. Raw MRI scans were preprocessed to reduce signal inhomogeneity and linearly registered to a template (using 9 parameter registration). Prior methods for ventricular segmentation have used semi-automated, automated (Chou et al., 2008), and single-atlas or multi-atlas methods (Chou et al., 2009). Here we chose to segment the ventricles with our modified multi-atlas, LDA-based approach described previously (Gutman et al., 2013), which was further tested and expanded in two prior analyses (Madsen et al., 2012; Madsen et al., 2013, In Press). Our segmentation approach uses group-wise surface registration to existing templates in addition to surface-based template blending to yield more accurate results. The lateral ventricles were segmented in each subject using a validated method (Chou et al., 2008). Ventricular surfaces were then extracted from these segmentations and an inverse-consistent fluid registration with a mutual information fidelity term aligned a set of hand-labeled ventricular templates to each scan (Leow et al., 2007). The template surfaces were registered into homologous point-to-point correspondence as a group using medial-spherical registration (Gutman et al., 2012).
To construct a surface boundary of the new subject, a normalized similarity measure (mutual information) between each template image and the new image was computed around each vertex point of each deformed template surface. The position of each new boundary point was defined by the template with the best similarity score, and the final surface was then constrained to be a smooth approximation of this “winner-takes-all” construction. This approach is very similar to that of (Yushkevich et al., 2010), except the current approach is based on surface geometry rather than image voxels. This “patch-based” approach is advantageous compared to whole-template approaches typically used in multi-atlas segmentation, allowing more flexible segmentation, particularly for outliers. Segmentations were assessed visually for defects from multiple views. While visually inspecting ventricular surface segmentations, we removed 4 subjects (2 CON, 1 MCI, 1 AD) whose meshes deviated by several millimeters from the actual periventricular boundaries. Our final analysis included 736 ADNI subjects at baseline, 622 at 12-month follow-up, and 479 at 24-month follow-up (Table 1).
2.5 Volumetric analyses: statistical associations of ApoE genotype with ventricular volumes
In most analyses, the number of ApoE-ε4 alleles (0, 1, or 2) was used to predict variations in ventricular volumes and ventricular expansion using general linear models (GLMs) in SPSS 21.0. As each copy of the ε4 risk allele is associated with an approximate threefold increase in the odds of developing AD (Bertram et al., 2007), and ApoE-ε4 gene dose correlates with whole-brain atrophy rates (Chen et al., 2007), our primary modeling approach essentially assumed an additive model of allele effects. In some of the post-hoc analyses described in section 3.3.2, we tested other models of ε4 alleles effects. In the dominant model, ApoE-ε4 genotype was defined as carrying 0 ε4 allele versus carrying of 1 or 2 ε4 alleles; in the recessive model, it was defined as carrying of 0 or 1 ε4 allele versus being homozygous for ε4.
In the primary volumetric analyses (section 3.2), GLM outcome measures included ventricular volume at baseline (in cubic mm, N=736), difference between total ventricular volume at baseline and volume after one year (in cubic mm, N=622), and difference between total ventricular volume at baseline and volume after two years (in cubic mm, N=479). In the exploratory post-hoc volumetric analyses, we examined additional GLM outcome measures, including difference between ventricular volumes at baseline and volumes after one or 2 years, within each hemisphere (N=622 at 12-month and N=479 at 24-month follow-up, section 3.3.1), difference between ventricular volumes at baseline and volumes after one or 2 years divided by volume at baseline, within each hemisphere (i.e. “expansion rate”, N=622 at 12-month and N=479 at 24-month follow-up, section 3.3.1), and difference between left and right ventricular volumes and ventricular expansion at baseline, 12-month, and 24-month follow-up (i.e. “left-right asymmetry”, section 3.3.5).
Unless otherwise specified, all analyses controlled for age, sex, and diagnosis (i.e., baseline dementia status: healthy elderly control, MCI, or AD). As the volume images were already normalized for overall brain size by the 9-parameter affine alignment, additional volume normalization was unnecessary.
2.6 Surface-based analyses: mapping ApoE genotype effects across the ventricular surface
The methods for surface-based image analysis have been described in detail elsewhere (Thompson et al., 2004; Ballmaier et al., 2008; Coscia et al., 2009; Roussotte et al., 2012). The surface models constructed as described in Section 2.4 were also used for these analyses. Briefly, a 3D medial curve was computed along the long axis for the surface model of each structure and radial distance measures (distance from the medial core to the surface) were estimated and recorded at each corresponding surface point. These values were used to generate individual distance maps that allowed relationships between the number of ApoE-ε4 alleles (0, 1, or 2) and regional ventricular surface morphology to be assessed at high spatial resolution in 3D. As in the GLMs described above, age, sex, and dementia status (i.e., healthy elderly control, MCI, or AD) were included as nuisance covariates in the primary surface-based analyses (section 3.4.1). As also mentioned above, since the volume images were already normalized for overall brain size during the 9-parameter affine alignment, we did not control for total brain volume. Maps were corrected for multiple comparisons using a False Discovery Rate (FDR) of 5% (q-value < 0.05) according to the implementation by Storey and colleagues (Storey, 2002; Storey et al., 2004). In the post-hoc surface-based analyses (section 3.4.2), we generated individual distance maps to assess relationships between the number of ApoE-ε4 alleles (0, 1, or 2) and regional ventricular surface morphology within diagnostic groups, controlling for age and sex. Maps did not pass correction for multiple comparisons using a False Discovery Rate (FDR) of 5% (q-value < 0.05), so we presented uncorrected maps.
3. RESULTS
3.1 Analyses of demographic data
Table 1 indicates the sample sizes at baseline, 12-month follow-up, and 24-month follow-up in participants stratified by diagnosis, and substratified by ApoE-ε4 genotype groups. This table also shows the mean baseline ages and Mini-Mental State Examination (MMSE) scores (Folstein et al., 1975), and we examined whether the various subgroups of subjects differed in these measures. Diagnosis was not significantly related to age after controlling for sex and ApoE-ε4 genotype (Table S1). However, we found a significant linear relation between the number of ApoE-ε4 alleles carried and age in the entire sample (N=736), after controlling for sex and diagnosis (p<0.001, Table S1). The stratification by diagnosis revealed that carrying more ApoE-ε4 alleles was significantly associated with being younger, within the MCI (p=0.002) and AD groups (p=0.004), but not within the CON group (p=0.540, Table S1). ApoE-ε4 status was not significantly related to MMSE scores after controlling for sex and diagnosis (Table S1). However, we found a significant linear relation between diagnosis and MMSE scores in the entire sample, after controlling for sex and genotype (p<0.001, Table S1). The stratification by genotype showed that increasing levels of dementia were significantly associated with lower MMSE scores in all 3 genotype groups, which was expected given that MMSE scores are one of the diagnostic criteria for dementia, as described in http://www.adni-info.org/Scientists/Pdfs/adniproceduresmanual12.pdf.
3.2 Primary Volumetric analyses
The mean left, right, and total ventricular volumes for participants stratified by diagnosis, and substratified by ApoE-ε4 genotype groups are provided in Tables S2, S3, and S4, for baseline, 12-month, and 24-month follow-up, respectively. Diagnosis was not significantly related to total ventricular volumes at baseline (p=0.384) or at 12-month follow up (p=0.534), but at 24-month follow-up, increasing levels of dementia were significantly associated with larger ventricular volumes (p=0.032, F-ratio=3.479), consistent with the literature (Apostolova et al., 2012), after controlling for age, sex, and ApoE-ε4 status. ApoE-ε4 genotype was not significantly related to total ventricular volumes at any time point, after controlling for age, sex, and diagnosis (p=0.822 at baseline; p=0.757 at 1-year follow-up; and p=0.592 at 2-year follow-up).
To test our hypothesis that ApoE genotype affected the trajectory of ventricular volume expansion, we examined differences between ventricular volumes at baseline and volumes after one year (in cubic mm, N=622), and two years (in cubic mm, N=479), after controlling for sex, age, and dementia status. Total ventricular expansion relative to overall brain size showed a significant correlation with the number of ApoE-ε4 alleles carried. After one year, carrying more ε4 alleles was associated with greater overall ventricular expansion (p=0.011, F-ratio=4.546, Table 2). Effect sizes were even larger at 2-year follow-up, when we observed a more pronounced linear relation between the number of ApoE-ε4 alleles carried and total ventricular expansion, after controlling for age, sex, and diagnosis (p<0.001, F-ratio=10.252, Table 2).
Table 2.
Results of multiple regression analyses: associations between ApoE-ε4 genotype and ventricular expansion at 12-month and 24-month follow-up
| Additive model, 3 diagnostic groups1 | Dominant model, 3 diagnostic groups2 | Recessive model, 3 diagnostic groups3 | Additive model, CON & AD only4 | |
|---|---|---|---|---|
|
| ||||
| 12-month | N=622 | N=622 | N=622 | N=316 |
|
| ||||
| Total expansion (cubic mm) | 5,6F-ratio=4.546 | F-ratio=6.093 | F-ratio=5.795 | F-ratio=3.332 |
| p=0.011 | p=0.014 | p=0.016 | p=0.037 | |
|
| ||||
| Left expansion (cubic mm) | F-ratio=4.203 | F-ratio=6.785 | F-ratio=4.060 | F-ratio=2.411 |
| p=0.015 | p=0.009 | p=0.044 | p=0.091 | |
|
| ||||
| Right expansion (cubic mm) | F-ratio=4.622 | F-ratio=4.782 | F-ratio=7.200 | F-ratio=3.980 |
| p=0.010 | p=0.029 | p=0.007 | p=0.020 | |
|
| ||||
| Left expansion rate | F-ratio=8.115 | F-ratio=10.543 | F-ratio=10.596 | F-ratio=4.929 |
| p<0.001 | p=0.001 | p=0.001 | p=0.008 | |
|
| ||||
| Right expansion rate | F-ratio=9.514 | F-ratio=12.680 | F-ratio=12.063 | F-ratio=8.208 |
| p<0.001 | p<0.001 | p=0.001 | p<0.001 | |
|
| ||||
| 24-month | N=479 | N=479 | N=479 | N=257 |
|
| ||||
| Total expansion (cubic mm) | F-ratio=10.252 | F-ratio=19.495 | F-ratio=5.225 | F-ratio=5.431 |
| p<0.001 | p<0.001 | p=0.023 | p=0.005 | |
|
| ||||
| Left expansion (cubic mm) | F-ratio=9.923 | F-ratio=18.918 | F-ratio=4.971 | F-ratio=4.648 |
| p<0.001 | p<0.001 | p=0.026 | p=0.010 | |
|
| ||||
| Right expansion (cubic mm) | F-ratio=9.156 | F-ratio=17.366 | F-ratio=4.775 | F-ratio=5.403 |
| p<0.001 | p<0.001 | p=0.029 | p=0.005 | |
|
| ||||
| Left expansion rate | F-ratio=17.050 | F-ratio=27.995 | F-ratio=14.950 | F-ratio=7.596 |
| p<0.001 | p<0.001 | p<0.001 | p=0.001 | |
|
| ||||
| Right expansion rate | F-ratio=14.559 | F-ratio=27.400 | F-ratio=7.796 | F-ratio=6.641 |
| p<0.001 | p<0.001 | p=0.005 | p=0.002 | |
The number of ApoE-ε4 alleles (0, 1, or 2) was used to predict variations in ventricular expansion in the total sample of subjects, with age, sex, and diagnosis (CON, MCI, AD) regressed out.
ApoE-ε4 status (defined as carrying no ε4 allele, versus carrying at least 1) was used to predict variations in ventricular expansion in the total sample of subjects, with age, sex, and diagnosis (CON, MCI, AD) regressed out.
ApoE-ε4 status (defined as carrying 0 or 1 ε4 allele, versus being homozygous for ε4) was used to predict variations in ventricular expansion in the total sample of subjects, with age, sex, and diagnosis (CON, MCI, AD) regressed out.
The number of ApoE-ε4 alleles (0, 1, or 2) was used to predict variations in ventricular expansion in CON and AD subjects (MCI excluded), with age, sex, and diagnosis (CON, AD) regressed out.
In multiple regressions, the F-ratio is used to test the hypothesis that the slopes of the regression lines are 0. The F is large when the independent variable helps to explain the variation in the dependent variable, independently of the other explanatory variables that are regressed out. For instance, here we reject the hypothesis that the slope of the regression line is 0 (F-ratio=4.546, p=0.011), meaning that there is a significant linear relation between the number of APOE ε 4 alleles carried and total ventricular expansion at 12-month follow-up, independent of age, sex, and diagnosis.
Bold font indicates significant results (p<0.05), and regular font indicates results that did not reach statistical significance.
3.3 Post-hoc volumetric analyses
3.3.1 Ventricular expansion and expansion rate within each hemisphere
We conducted post-hoc analyses to determine if increasing numbers of ApoE-ε4 alleles related to the trajectory of ventricular volume expansion within each hemisphere. After one year, carrying more ε4 alleles was associated with greater expansion in the left (p=0.015, F-ratio=4.203) and right ventricle (p=0.010, F-ratio=4.622, Table 2), after controlling for age, sex, and diagnosis (N=622). As in the primary analyses of the effects of ApoE-ε4 genotype on total ventricular expansion, the effect sizes increased after 2 years. At 24-month follow-up, greater ApoE-ε4 loading was associated with greater expansion in the left (p<0.001, F-ratio=9.923) and right ventricle (p<0.001, F-ratio=9.156, Table 2), after controlling for age, sex, and dementia status (N=479).
We also sought to determine if ApoE-ε4 affected left and right ventricular expansion rates, defined as the difference between ventricular volume at baseline and volume after one or 2 years, divided by volume at baseline. After one year, carrying more ε4 alleles was associated with greater left (p<0.001, F-ratio=8.115) and right expansion rates (p<0.001, F-ratio=9.514, Table 2), with age, sex, and diagnosis regressed out (N=622). The effect sizes were even larger after 2 years, when greater ApoE-ε4 loading was associated with greater left (p<0.001, F-ratio=17.050) and right expansion rates (p<0.001, F-ratio=14.559, Table 2), after controlling for age, sex, and dementia status (N=479).
3.3.2 Alternate models of allele effects
Because some studies of hippocampal atrophy in healthy elderly reported no ApoE-ε4 gene dose effect (Lemaitre et al., 2005; Crivello et al., 2010), we also investigated dominant and recessive models of ε4 allele effects. First, we examined differences between ventricular volumes at baseline and volumes after one year (in cubic mm, N=622), and two years (in cubic mm, N=479), after controlling for sex, age, and dementia status. Total ventricular expansion relative to overall brain size showed a significant correlation with ApoE-ε4 status, in both the dominant (non-carriers of the ε4 allele versus carriers of 1 or 2 ε4 alleles) and recessive (carriers of 0 or 1 ε4 allele versus ε4 homozygotes) models of minor allele effects. After one year, ApoE-ε4 status was associated with greater overall ventricular expansion (dominant model: p=0.014, F-ratio=6.093, Table 2; recessive model: p=0.016, F-ratio=5.795, Table 2). At 2-year follow-up, we also observed a significant association between ApoE-ε4 status and total ventricular expansion, after controlling for age, sex, and diagnosis (dominant model: p<0.001, F-ratio=19.495, Table 2; recessive model: p=0.023, F-ratio=5.225, Table 2). As with the additive model, ApoE-ε4 status was also significantly associated with the trajectory of ventricular volume expansion within each hemisphere and with left and right ventricular expansion rates when using the dominant and recessive models of allele effects (Table 2).
3.3.3 Analyses restricted to controls and Alzheimer’s disease patients
MCI increases the risk of developing AD but some patients remain stable; therefore, MCI cannot simply be thought of an intermediate stage (Patel and Holland, 2012). Thus, we sought to determine if our findings remained significant when we restricted the analyses to the CON and AD groups (N=316 at 12-month and N=257 at 24-month follow-up). We found that carrying more ε4 alleles was still significantly associated with greater overall ventricular expansion, with greater expansion within each hemisphere, and with faster left and right ventricular expansion rates at both time points (Table 2). The only difference with the previous results including all 3 diagnostic groups was that, at 12-month follow-up, increasing numbers of ApoE-ε4 alleles showed only a trend-level association with the expansion of the left ventricle, when restricting analyses to the CON and AD groups (p=0.091, F-ratio=2.411, Table 2).
3.3.4 Genotype by diagnosis interaction and analyses within diagnostic groups
To determine whether the ApoE-ε4 allele affected ventricular expansion equally in healthy, MCI, and AD participants, we introduced a genotype by diagnosis interaction term in the regression models. We found no significant ApoE-ε4 by diagnosis interaction in the analyses of total ventricular expansion, or left and right expansion rates at 12-month follow-up. However, we detected a significant genotype by diagnosis interaction at 24-month follow-up (p=0.013, F-ratio=4.378 for total expansion; p=0.039, F-ratio=3.269 for left expansion rate; and p=0.005, F-ratio=5.280 for right expansion rate), suggesting that the effects of ApoE on ventricular expansion may be smaller in the AD than in the CON or MCI groups. We sought to further characterize the effects of ApoE in each diagnostic group by testing the association between ApoE-ε4 status and 2-year ventricular expansion separately within each group. Despite an important loss of statistical power, we found significant effects of ApoE on all 3 measures of ventricular expansion in every diagnostic group at 24-month follow-up (Table 3), though total expansion reached only a trend level of significance in the AD group (p=0.083, F-ratio=2.552, Table 3).
Table 3.
Results of multiple regression analyses: associations between ApoE-ε4 genotype and ventricular expansion at 24-month follow-up, within diagnostic groups
| CON group only1 | MCI group only2 | AD group only3 | |
|---|---|---|---|
|
| |||
| 24-month | N=156 | N=222 | N=101 |
|
| |||
| Total expansion (cubic mm) | 4,5F-ratio=3.291 | F-ratio=6.136 | F-ratio=2.552 |
| p=0.040 | p=0.003 | p=0.083 | |
|
| |||
| Left expansion rate | F-ratio=4.502 | F-ratio=10.573 | F-ratio=3.863 |
| p=0.013 | p<0.001 | p=0.024 | |
|
| |||
| Right expansion rate | F-ratio=3.843 | F-ratio=9.524 | F-ratio=3.733 |
| p=0.024 | p<0.001 | p=0.027 | |
The number of ApoE-ε4 alleles (0, 1, or 2) was used to predict variations in ventricular expansion in healthy elderly only, with age and sex regressed out.
The number of ApoE-ε4 alleles was used to predict variations in ventricular expansion in the MCI group only, with age and sex regressed out.
The number of ApoE-ε4 alleles was used to predict variations in ventricular expansion in AD patients only, with age and sex regressed out.
In multiple regressions, the F-ratio is used to test the hypothesis that the slopes of the regression lines are 0. The F is large when the independent variable helps to explain the variation in the dependent variable, independently of the other explanatory variables that are regressed out. For instance, here reject the hypothesis that the slope of the regression line is 0 (F-ratio=3.291, p=0.040), meaning that there is a significant linear relation between the number of ApoE-ε4 alleles carried and total ventricular expansion at 24-month follow-up within the control group, after controlling for age, sex, and diagnosis.
Bold font indicates significant results (p<0.05), and regular font indicates results that did not reach statistical significance.
3.3.5 Left-right asymmetry in ventricular volumes and ventricular expansion
Because previous reports suggested that both dementia (Madsen et al., 2010; Daianu et al., 2013) and ApoE genotype (Donix et al., 2013) could affect hemispheric asymmetry, we also investigated possible effects of diagnosis and ApoE-ε4 loading on left-right asymmetry in ventricular volumes and ventricular expansion. The mean difference between left and right ventricular volumes at baseline, 12-month, and 24-month follow-up for participants stratified by diagnosis, and substratified by ApoE-ε4 genotype groups are provided in Table S5. This table also shows the mean difference between left ventricular expansion and right ventricular expansion at 12-month and 24-month follow-up, for each subgroup of participants. We found that, overall, ApoE-ε4 genotype and diagnosis did not significantly affect left-right asymmetry in ventricular volumes and ventricular expansion (Table S6).
3.4 Surface-based analyses
3.4.1 Primary surface-based analyses
Consistent with the volumetric analyses, ApoE ε4 status was not significantly related to ventricular surface morphology at baseline. However, 3D surface statistics revealed significant (FDR-corrected at q<0.05) effects of the number ApoE-ε4 alleles on regional patterns of lateral ventricle morphology at 1- and 2-year follow-up, after controlling for sex, age, and dementia status. At 12-month follow-up, carrying more ApoE-ε4 alleles was associated with ventricular surface expansion, especially at the center (body) of the lateral ventricles. That is, higher ApoE-ε4 loading was associated with regions of expansion in the ventrolateral and dorsomedial ventricles bilaterally, while the frontal and temporal horns appeared less affected (N=622, Figure 1). At 24-month follow-up, carrying more ApoE-ε4 alleles was significantly associated with expansion of almost the entire ventricular surface. Only very small regions - notably the outer edges of the frontal and temporal horns - did not show a detectable association with ApoE ε4 status in our maps. This suggests that the association between higher ApoE-ε4 loading and expansion of the ventricular surfaces may “spread” from the body toward the edges of the structures over time (N=479, Figure 2).
Figure 1. Surface maps depicting relationships between number of ApoE-ε4 alleles (0, 1, or 2) and regional deformations of the lateral ventricular surface at 12 month follow-up (N=622).
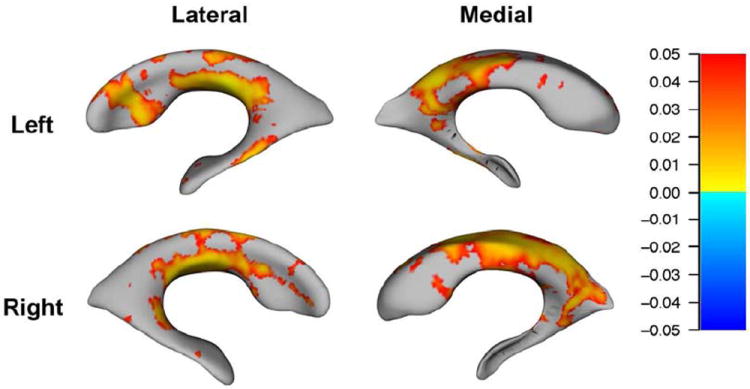
Red-to-yellow hues indicate regions where higher ApoE-ε4 loading is associated with expansion of the surfaces, after controlling for age, sex, and dementia status. For all statistical maps, the color bar encodes the FDR-corrected values (q<0.05) for the observed effects.
Figure 2. Surface maps depicting relationships between number of ApoE-ε4 alleles (0, 1, or 2) and regional deformations of the lateral ventricular surface at 24 month follow-up (N=479).
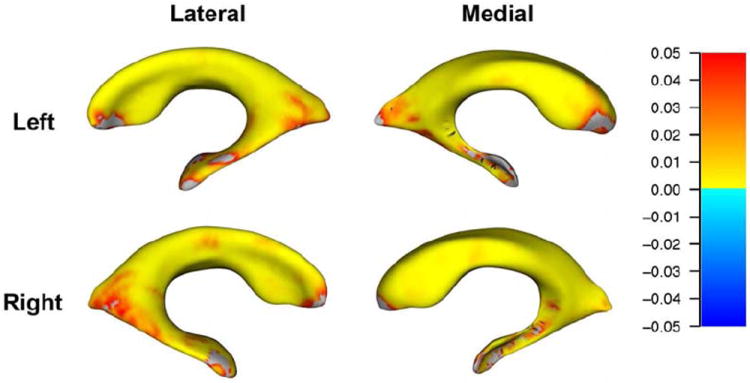
Red-to-yellow hues indicate regions where higher ApoE-ε4 loading is associated with expansion of the surfaces, after controlling for age, sex, and dementia status. For all statistical maps, the color bar encodes the FDR-corrected values (q<0.05) for the observed effects.
3.4.2 Post-hoc surface-based analyses
As with the volumetric analyses, we sought to further characterize the effects of ApoE in each diagnostic group by testing the association between genotype and ventricular surface morphology separately within each group. The uncorrected maps suggested associations between ApoE-ε4 status and ventricular morphology consistent with the results of the primary surface-based analyses across groups (Figure S1). Carrying more ApoE-ε4 alleles was associated with ventricular surface expansion, and this association was more widespread and more significant at 2-year than at 1-year follow-up, in all diagnostic groups (Figure S1). However, the effects of genotype on regional patterns of lateral ventricle morphology at 1- and 2-year follow-up after controlling for age and sex, did not pass FDR correction within any diagnostic group, probably as a result of insufficient statistical power, as sample size were much smaller than in the surface-based analyses across groups.
4. DISCUSSION
This study is the first to show that the ApoE-ε4 allele is directly associated with altered 3D profiles of longitudinal ventricular expansion rates and lateral ventricle surface morphology, both in dementia and in normal aging. Our 3D maps of ventricular surface morphology revealed that the effects of the ApoE-ε4 allele on longitudinal ventricular expansion were somewhat spatially distinct from the known effects of dementia progression. In contrast to prior studies showing that the frontal (Apostolova et al., 2012) and temporal horns (Thompson et al., 2004) were most sensitive to AD progression, we found that the association between higher ApoE-ε4 load and expansion of the ventricular surfaces started in the body of the lateral ventricles and extended toward the frontal and temporal horns over time.
Carrying more ApoE-ε4 alleles was associated with being younger, within the MCI and AD groups but not within the control group. Assuming that all participants enrolled in the ADNI study a similar amount of time after disease onset, this finding could reflect the fact that ApoE-ε4 carriers develop dementia at younger ages, consistent with the literature (Slooter et al., 1998; Thambisetty et al., 2013). Post-hoc analyses suggested that the association between ApoE-ε4 genotype and longitudinal ventricular expansion was significant using all possible models of allele effects. Previous studies focusing on medial temporal lobe atrophy reported no gene dose effects of ApoE-ε4 in the healthy elderly (Lemaitre et al., 2005; Crivello et al., 2010), and found significant effects of ApoE genotype and dementia on hemispheric asymmetry (Donix et al., 2013). In contrast, our results suggest that the ApoE-ε4 allele may affect longitudinal ventricular expansion in a dose-dependent manner, and that neither ApoE-ε4 genotype nor diagnosis affect left-right asymmetry in ventricular volumes and ventricular expansion.
We found a significant genotype by diagnosis interaction at 2-year follow-up, indicating that ApoE-ε4 status did not affect ventricular expansion equally in each diagnostic group. Within-group analyses of total ventricular expansion also suggested that at 24-month follow-up, the effects of ApoE genotype on ventricular expansion may be smaller in the AD than in the CON or MCI groups. It is likely that AD patients have more factors contributing to faster ventricular expansion than participants in the other 2 groups, including other dementia-related genetic factors, as well as additional developmental and lifestyle factors. As a result, ApoE genotype appears to account for a smaller portion of the variance in ventricular expansion in the AD group. Family history of dementia is related to genotype, but it may well be another factor influencing the trajectory of ventricular expansion differently in each diagnostic group, as emerging evidence suggests that ApoE-ε4 status and family history exert independent effects on brain atrophy (Andrawis et al., 2012).
Despite these group differences, the effects of ApoE-ε4 genotype on ventricular expansion became larger and more significant over time in the entire sample, after controlling for dementia status. In addition, the effects of ApoE-ε4 status after controlling for diagnosis, remained significant in studies restricting the analyses to the control and AD groups, even if the sample sizes were about half as large at both time points. Likewise, the effects of ApoE-ε4 on ventricular expansion remained significant within each diagnostic group in the volumetric studies of expansion rates. Furthermore, in all diagnostic groups, the uncorrected maps indicated associations between genotype and ventricular morphology consistent with the results across groups, even if the within-group surface based-analyses were not adequately powered to detect the effects of ApoE-ε4 status on lateral ventricle morphology after correction for multiple comparisons. Taken together, these findings strongly suggest that ApoE genotype is associated with ventricular expansion rate and surface morphology in both cognitively normal and demented participants.
Progressive ventricular expansion reflects atrophy of the surrounding gray and white matter, and there is growing evidence that the ApoE4 protein has direct neurotoxic effects that in turn can lead to secondary neuronal dysfunction and degeneration (Mahley and Huang, 2012b; Zlokovic, 2013). ApoE4 leads to the generation of neurotoxic fragments, which cause pathological mitochondrial dysfunction and cytoskeletal alterations. Protein instability and altered protein conformation (“domain interaction”) are responsible for these neurotoxic effects (Mahley and Huang, 2012b). As ventricular expansion reflects the accumulation of neuronal loss in neighboring tissues, future studies should examine whether ApoE-ε4 effects on regional brain volumes follow a similar pattern of spatial progression in the areas surrounding the lateral ventricles. This could be achieved by mapping the longitudinal effects of ApoE-ε4 loading on regional brain atrophy, to determine whether the association is first observed in regions adjacent to the callosal body and thalamus - located just above and below the body of the lateral ventricles, respectively. Future investigations could also examine the spatial progression of ApoE-ε4 effects on other areas in the brain, such as in the cortex, where thinner gray matter has been linked with ventricular expansion rates in the elderly (Madsen et al., 2013).
ApoE4 exerts its neurotoxic effects both via amyloid β-dependent and amyloid β-independent pathways (Zlokovic, 2013), consistent with the involvement of the ApoE-ε4 allele not only in AD but also in the etiology, pathogenesis, and prognosis of various other neurological disorders. Indeed, the ApoE-ε4 allele is associated with genetic predisposition to (Ely et al., 2007) and lower rates of recovery from delirium (Adamis et al., 2007). ApoE-ε4 also predicts poor neurological outcome after traumatic brain injury (Friedman et al., 1999) and cerebral hemorrhage (Alberts et al., 1995). Moreover, the same allele is a risk factor for hippocampal volume loss in alcoholics (Bleich et al., 2003; Wilhelm et al., 2008), suggesting that ApoE-ε4 may also promote neurodegeneration based on certain gene-environment interactions (Wilhelm et al., 2008).
ApoE genotyping is currently considered beneficial in clinical trials for Alzheimer’s disease and mild cognitive impairment, but the relevance of ApoE is not limited to AD and its prodromal states. ApoE participates in general cellular pathways designed to respond adaptively to many environmental, metabolic, and genetic stimuli (Mahley and Huang, 2012b). Our findings, combined with earlier investigations showing that ApoE-ε4 is a risk factor for other neurological conditions (Ely et al., 2007), is associated with reduced capacity for neuronal regeneration (Friedman et al., 1999) and modulates the neurotoxic effects of alcohol abuse (Bleich et al., 2003; Wilhelm et al., 2008), suggest that the selective enrollment of ApoE-ε4 carriers (or post-hoc stratification by ApoE status) may empower clinical trials of other disorders, such as alcoholism and traumatic brain injury.
ApoE genotype should also be an important factor when devising novel approaches to treat and prevent a large spectrum of neurological disorders. In fact, an emerging approach is to prevent neurodegeneration – as opposed to treating a specific brain disorder – by developing therapies to directly counteract ApoE4’s neurotoxic effects, before the onset of any sign of neuropathology (Mahley and Huang, 2012b). For example, as ApoE4’s detrimental effects result from its altered protein conformation (“domain interaction”), small-molecule-based therapeutics that induce proper ApoE4 folding and block domain interaction in ApoE4 have been shown to reverse many of its harmful effects, both in vitro and in vivo (Mahley and Huang, 2012a). Moreover, whole-transcriptome cerebral cortex gene expression data in unaffected ApoE-ε4 carriers recently identified a set of candidate core regulatory mediators, including SV2A, which encodes a well-described regulator of neuronal endocytosis (Rhinn et al., 2013). In human induced neuron cultures derived from human skin fibroblasts of ε4 carriers or non-carriers, a selective SV2A inhibitor was shown to suppress the accumulation of amyloid β species selectively in the ApoE-ε4 carrier cultures (Rhinn et al., 2013). It is still unclear whether such a therapeutic approach will be successful in human trials. Even so, this line of research represents an important shift from the current “reactive approach” to treating neurological disorders, which focuses on managing a specific condition after symptoms appear, to a more “proactive approach” based on early prevention and personalized treatments.
Supplementary Material
Acknowledgments
F.F.R. was supported, in part, by a postdoctoral fellowship from the A. P. Giannini Foundation. This work was additionally supported by National Institute of Health grants (R01 MH097268, R01 AG040060) to P.M.T. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Disclosure Statement
F.F.R. declares no actual or potential conflicts of interest.
B.A.G. declares no actual or potential conflicts of interest.
S.K.M. declares no actual or potential conflicts of interest.
J.B.C. declares no actual or potential conflicts of interest.
K.L.N. declares no actual or potential conflicts of interest.
P. M.T. declares no actual or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamis D, Treloar A, Martin FC, Gregson N, Hamilton G, Macdonald AJ. APOE and cytokines as biological markers for recovery of prevalent delirium in elderly medical inpatients. Int J Geriatr Psychiatry. 2007;22:688–694. doi: 10.1002/gps.1732. [DOI] [PubMed] [Google Scholar]
- Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, Walter S, Trojanowski JQ, Shaw LM, Beckett LA, Jack CR, Jr, Jagust W, Toga AW, Saykin AJ, Morris JC, Green RC, Weiner MW. Clinical Core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 2010;6:239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts MJ, Graffagnino C, McClenny C, DeLong D, Strittmatter W, Saunders AM, Roses AD. ApoE genotype and survival from intracerebral haemorrhage. Lancet. 1995;346:575. doi: 10.1016/s0140-6736(95)91411-0. [DOI] [PubMed] [Google Scholar]
- Andrawis JP, Hwang KS, Green AE, Kotlerman J, Elashoff D, Morra JH, Cummings JL, Toga AW, Thompson PM, Apostolova LG. Effects of ApoE4 and maternal history of dementia on hippocampal atrophy. Neurobiol Aging. 2012;33:856–866. doi: 10.1016/j.neurobiolaging.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Green AE, Babakchanian S, Hwang KS, Chou YY, Toga AW, Thompson PM. Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment (MCI), and Alzheimer Disease. Alzheimer Dis Assoc Disord. 2012;26:17–27. doi: 10.1097/WAD.0b013e3182163b62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmaier M, Narr KL, Toga AW, Elderkin-Thompson V, Thompson PM, Hamilton L, Haroon E, Pham D, Heinz A, Kumar A. Hippocampal morphology and distinguishing late-onset from early-onset elderly depression. Am J Psychiatry. 2008;165:229–237. doi: 10.1176/appi.ajp.2007.07030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboriak DP, Doraiswamy PM, Krishnan KR, Vidyarthi S, Sylvester J, Charles HC. Hippocampal sulcal cavities on MRI: relationship to age and apolipoprotein E genotype. Neurology. 2000;54:2150–2153. doi: 10.1212/wnl.54.11.2150. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bleich S, Wilhelm J, Graesel E, Degner D, Sperling W, Rossner V, Javaheripour K, Kornhuber J. Apolipoprotein E epsilon 4 is associated with hippocampal volume reduction in females with alcoholism. J Neural Transm. 2003;110:401–411. doi: 10.1007/s00702-002-0789-1. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Ventricular volume and dementia progression in the Cardiovascular Health Study. Neurobiol Aging. 2007a;28:389–397. doi: 10.1016/j.neurobiolaging.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Cerebral ventricular changes associated with transitions between normal cognitive function, mild cognitive impairment, and dementia. Alzheimer Dis Assoc Disord. 2007b;21:14–24. doi: 10.1097/WAD.0b013e318032d2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, Domb A, Osborne D, Fox N, Crum WR, Saunders AM, Hardy J. Correlations between apolipoprotein E epsilon4 gene dose and whole brain atrophy rates. Am J Psychiatry. 2007;164:916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- Chou YY, Lepore N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. Neuroimage. 2008;40:615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. Neuroimage. 2009;46:394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Lepore N, Saharan P, Madsen SK, Hua X, Jack CR, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. Ventricular maps in 804 ADNI subjects: correlations with CSF biomarkers and clinical decline. Neurobiol Aging. 2010;31:1386–1400. doi: 10.1016/j.neurobiolaging.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscia DM, Narr KL, Robinson DG, Hamilton LS, Sevy S, Burdick KE, Gunduz-Bruce H, McCormack J, Bilder RM, Szeszko PR. Volumetric and shape analysis of the thalamus in first-episode schizophrenia. Hum Brain Mapp. 2009;30:1236–1245. doi: 10.1002/hbm.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivello F, Lemaitre H, Dufouil C, Grassiot B, Delcroix N, Tzourio-Mazoyer N, Tzourio C, Mazoyer B. Effects of ApoE-epsilon4 allele load and age on the rates of grey matter and hippocampal volumes loss in a longitudinal cohort of 1186 healthy elderly persons. Neuroimage. 2010;53:1064–1069. doi: 10.1016/j.neuroimage.2009.12.116. [DOI] [PubMed] [Google Scholar]
- Daianu M, Jahanshad N, Nir TM, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Breakdown of brain connectivity between normal aging and Alzheimer’s disease: a structural k-core network analysis. Brain Connect. 2013;3:407–422. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Heijer T, Oudkerk M, Launer LJ, van Duijn CM, Hofman A, Breteler MM. Hippocampal, amygdalar, and global brain atrophy in different apolipoprotein E genotypes. Neurology. 2002;59:746–748. doi: 10.1212/wnl.59.5.746. [DOI] [PubMed] [Google Scholar]
- Donix M, Burggren AC, Scharf M, Marschner K, Suthana NA, Siddarth P, Krupa AK, Jones M, Martin-Harris L, Ercoli LM, Miller KJ, Werner A, von Kummer R, Sauer C, Small GW, Holthoff VA, Bookheimer SY. APOE associated hemispheric asymmetry of entorhinal cortical thickness in aging and Alzheimer’s disease. Psychiatry Res. 2013 doi: 10.1016/j.pscychresns.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely EW, Girard TD, Shintani AK, Jackson JC, Gordon SM, Thomason JW, Pun BT, Canonico AE, Light RW, Pandharipande P, Laskowitz DT. Apolipoprotein E4 polymorphism as a genetic predisposition to delirium in critically ill patients. Crit Care Med. 2007;35:112–117. doi: 10.1097/01.CCM.0000251925.18961.CA. [DOI] [PubMed] [Google Scholar]
- Erten-Lyons D, Dodge HH, Woltjer R, Silbert LC, Howieson DB, Kramer P, Kaye JA. Neuropathologic basis of age-associated brain atrophy. JAMA Neurol. 2013;70:616–622. doi: 10.1001/jamaneurol.2013.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini L, Palm WM, Olofsen H, van der Landen R, van Buchem MA, Reiber JH, Admiraal-Behloul F. Ventricular shape biomarkers for Alzheimer’s disease in clinical MR images. Magn Reson Med. 2008;59:260–267. doi: 10.1002/mrm.21471. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009a;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, Whitcher B, Matthews PM. Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer’s disease. Neuroimage. 2009b;44:724–728. doi: 10.1016/j.neuroimage.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda B, Groswasser Z. Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology. 1999;52:244–248. doi: 10.1212/wnl.52.2.244. [DOI] [PubMed] [Google Scholar]
- Gunter JL, Briston PJ, Felmlee JP, Ward CP, Schuff N, Weiner M, Levy J, Jack CR., J The ADNI Phantom and Analysis Algorithm: A New and Accurate Tool to Measure Scanner Performance. Proceedings of the International Society of Magnetic Resonance Imaging (ISMRM) 2007 [Google Scholar]
- Gutman BA, Wang Y, Thompson PM, Rajagopalan P, Toga AW. Shape matching with medial curves and 1-D group-wise registration. Biomedical Imaging (ISBI), 9th IEEE International Symposium.2012. [Google Scholar]
- Gutman BA, Hua X, Rajagopalan P, Chou YY, Wang Y, Yanovsky I, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage. 2013;70:386–401. doi: 10.1016/j.neuroimage.2012.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Hibar DP, Lee S, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Sex and age differences in atrophic rates: an ADNI study with n=1368 MRI scans. Neurobiol Aging. 2010;31:1463–1480. doi: 10.1016/j.neurobiolaging.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan TL, Archibald SL, Fennema-Notestine C, Gamst AC, Stout JC, Bonner J, Hesselink JR. Effects of age on tissues and regions of the cerebrum and cerebellum. Neurobiol Aging. 2001;22:581–594. doi: 10.1016/s0197-4580(01)00217-2. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Lehtovirta M, Laakso MP, Soininen H, Helisalmi S, Mannermaa A, Helkala EL, Partanen K, Ryynanen M, Vainio P, Hartikainen P, et al. Volumes of hippocampus, amygdala and frontal lobe in Alzheimer patients with different apolipoprotein E genotypes. Neuroscience. 1995;67:65–72. doi: 10.1016/0306-4522(95)00014-a. [DOI] [PubMed] [Google Scholar]
- Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alperovitch A, Mazoyer B. No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage. 2005;24:1205–1213. doi: 10.1016/j.neuroimage.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chiang MC, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26:822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen SK, Gutman BA, Joshi SH, Toga AW, Jack CRJ, Weiner MW, Thompson PM. Mapping dynamic changes in ventricular volume onto baseline cortical surface maps in normal aging, MCI, and Alzheimer’s disease. MICCAI MBIA Workshop; Japan: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen SK, Gutman BA, Joshi SH, Toga AW, Jack CRJ, Weiner MW, Thompson PM. Relating longitudinal ventricular expansion to cortical gray matter thinning in the elderly. Neurobiology of Aging – Special Issue on Novel Imaging Biomarkers for Alzheimer’s Disease and Related Disorders. In Press. [Google Scholar]
- Madsen SK, Ho AJ, Hua X, Saharan PS, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. 3D maps localize caudate nucleus atrophy in 400 Alzheimer’s disease, mild cognitive impairment, and healthy elderly subjects. Neurobiol Aging. 2010;31:1312–1325. doi: 10.1016/j.neurobiolaging.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Huang Y. Small-molecule structure correctors target abnormal protein structure and function: structure corrector rescue of apolipoprotein E4-associated neuropathology. J Med Chem. 2012a;55:8997–9008. doi: 10.1021/jm3008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012b;76:871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer L, Lamberton F, Matura S, Tanner C, Scheibe M, Miller J, Rujescu D, Prvulovic D, Hampel H. Reduced hippocampal volume in healthy young ApoE4 carriers: an MRI study. PLoS One. 2012;7:e48895. doi: 10.1371/journal.pone.0048895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BB, Holland NW. Mild cognitive impairment: hope for stability, plan for progression. Cleve Clin J Med. 2012;79:857–864. doi: 10.3949/ccjm.79a.11126. [DOI] [PubMed] [Google Scholar]
- Qiu A, Fennema-Notestine C, Dale AM, Miller MI. Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE epsilon4 effectors in Alzheimer’s disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- Roussotte F, Soderberg L, Warner T, Narr K, Lebel C, Behnke M, Davis-Eyler F, Sowell E. Adolescents with prenatal cocaine exposure show subtle alterations in striatal surface morphology and frontal cortical volumes. J Neurodev Disord. 2012;4:22. doi: 10.1186/1866-1955-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, Van Broeckhoven C, van Duijn CM. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- Stein JL, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. Neuroimage. 2010;51:542–554. doi: 10.1016/j.neuroimage.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DJ, Molony C, Suver C, Schadt EE, Potter WZ. ApoE genotyping as a progression-rate biomarker in phase II disease-modification trials for Alzheimer’s disease. Pharmacogenomics J. 2010;10:161–164. doi: 10.1038/tpj.2009.58. [DOI] [PubMed] [Google Scholar]
- Tatsuoka C, Tseng H, Jaeger J, Varadi F, Smith MA, Yamada T, Smyth KA, Lerner AJ. Modeling the heterogeneity in risk of progression to Alzheimer’s disease across cognitive profiles in mild cognitive impairment. Alzheimers Res Ther. 2013;5:14. doi: 10.1186/alzrt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, An Y, Tanaka T. Alzheimer’s disease risk genes and the age-at-onset phenotype. Neurobiol Aging. 2013;34:2696 e2691–2695. doi: 10.1016/j.neurobiolaging.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Reinvang I, Lundervold A, Dale AM, Eilertsen DE, Quinn BT, Salat D, Makris N, Fischl B. Effects of age on volumes of cortex, white matter and subcortical structures. Neurobiol Aging. 2005;26:1261–1270. doi: 10.1016/j.neurobiolaging.2005.05.020. discussion 1275-1268. [DOI] [PubMed] [Google Scholar]
- Wang D, Chalk JB, Rose SE, de Zubicaray G, Cowin G, Galloway GJ, Barnes D, Spooner D, Doddrell DM, Semple J. MR image-based measurement of rates of change in volumes of brain structures. Part II: application to a study of Alzheimer’s disease and normal aging. Magn Reson Imaging. 2002;20:41–48. doi: 10.1016/s0730-725x(02)00472-1. [DOI] [PubMed] [Google Scholar]
- Wilhelm J, Frieling H, von Ahsen N, Hillemacher T, Kornhuber J, Bleich S. Apolipoprotein E polymorphism, homocysteine serum levels and hippocampal volume in patients with alcoholism: an investigation of a gene-environment interaction. Pharmacogenomics J. 2008;8:117–121. doi: 10.1038/sj.tpj.6500453. [DOI] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, McAllister TW, Rabin LA, McDonald BC, Flashman LA, Roth RM, Mamourian AC, Tsongalis GJ, Rhodes CH. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53:1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol. 2013;70:440–444. doi: 10.1001/jamaneurol.2013.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


