Abstract
Purpose
Carcinogenesis is an adaptive process between nascent tumor cells and their microenvironment including the modification of inflammatory responses from anti-tumorigenic to pro-tumorigenic. Radiation exposure can stimulate inflammatory responses that inhibit or promote carcinogenesis. The purpose of this study is to determine the impact of radiation exposure on lung cancer progression in vivo and assess the relevance of this knowledge to human carcinogenesis.
Experimental Design
K-rasLA1 mice were irradiated with various doses and dose regimens and then monitored till death. Microarray analyses were performed using Illumina® BeadChips on whole lung tissue 70 days post-irradiation with a fractionated or acute dose of radiation and compared to age-matched unirradiated controls. Unique group classifiers were derived by comparative genomic analysis of three experimental cohorts. Survival analyses were performed using principal component analysis and k-means clustering on three lung adenocarcinoma, three breast adenocarcinoma, and two lung squamous carcinoma annotated microarray datasets.
Results
Radiation exposure accelerates lung cancer progression in the K-rasLA1 lung cancer mouse model with dose fractionation being more permissive for cancer progression. A non-random inflammatory signature associated with this progression was elicited from whole lung tissue containing only benign lesions and predicts human lung and breast cancer patient survival across multiple datasets. Immunohistochemical analyses suggest that tumor cells drive predictive signature.
Conclusions
These results demonstrate that radiation exposure can cooperate with benign lesions in a transgenic model of cancer by impacting inflammatory pathways, and that clinically relevant similarities exist between human lung and breast carcinogenesis.
Keywords: Radiation, Lung Cancer, Progression, Mouse models, Genomics
Introduction
Carcinogenesis is a complex multi-step process driven by the convergence of both intrinsic and extrinsic factors. DNA mutations that result in cellular dysregulation through the activation of an oncogene or the silencing of a tumor suppressor gene are classic examples of intrinsic factors. Extrinsic factors are those provided by physical and biochemical interactions between the emerging tumor and its microenvironment (1–4). Reciprocal interactions between these factors are dynamic throughout carcinogenesis and readily influence overall tumor responses (2, 4, 5). For processes, such as inflammation, this may result in pleiotropic or conflicting responses dependent on the initiated tissue type, mutational landscape of the tumor, and/or signal duration and intensity (2, 5, 6). The overall impact of modifying these factors, especially those regulating inflammation, may therefore fluctuate contextually throughout the carcinogenic process.
Radiation exposure is a known carcinogen whose effects may likewise be contradictory and contextual. The classical method in which radiation is thought to act as a carcinogen is intrinsic through the induction of DNA damage leading to an increased mutational load, chromosomal aberrations, and/or genomic instability (1, 3, 6). These effects are generally observed following the exposure to lower radiation doses as the exposure to higher doses is capable of inducing cell death (1, 3). Evidence is mounting, however, that radiation exposure can also modify extrinsic factors by disturbing cells in the microenvironment. These cells, in turn, affect the adjacent tumor cells in a cell non-autonomous fashion (1, 3, 7–9). Empirical evidence for this extrinsic role of radiation exposure on carcinogenesis has been largely derived from studies that rely on the use of normal and/or cancer cells in in vitro or transplantation assays (7–10). It is not clearly understood if initiated, non-transformed cells equally respond to these radiation-induced cues or if the transplantation procedure introduces extraneous damage that co-operates with radiation exposure.
Ionizing radiation is comprised of both electromagnetic (EM) and particulate radiation types, with the risk of exposure being higher for EM radiation types. The majority of radiation studies have examined the carcinogenic effect of EM radiation exposure and, as such, these effects are used as the baseline for determining the biological effectiveness of other radiation types (1). The therapeutic application of high-energy particle radiation and the mounting interest for deep space travel, however, is increasing the population exposed to high-energy particulate radiation types (11, 12). Extrapolation of the carcinogenic effects of EM radiation exposure to particulate radiation is confounded by differences in both their energy and methods of energy deposition. EM radiation types, such as X-rays and gamma rays, have lower energies and are more sparsely ionizing than particle radiation types. High-energy particulate radiation types densely ionize molecules along the particle trajectories, in addition to, indirectly ionizing molecules perpendicular to that track (1). It is currently not known how this method of energy deposition impacts the carcinogenic process.
Dose fractionation can induce a radio-protective effect and have a sparing effect in cells (1, 10, 11). Several studies have additionally suggested that dose fractionation may be more efficient at tumor induction in vivo and can affect the rate of radiation-induced transformation in vitro (13, 14). However, these studies were conducted using either EM or fast neutron particulate radiation, whose energy spectrum is lower than other charged particle types and that of high-energy neutron particles in space. Studies examining the effect of fractionation on high-energy charged particles or directly comparing acute and fractionated doses on promotion and progression in vivo are limited. Therefore, how dose fractionation impacts these stages of the carcinogenic process is not fully understood.
In this study, we examined the effect of radiation exposure on the later stages of the carcinogenic process using a lung cancer susceptible mouse model, K-rasLA1, in which lesions are spontaneously activated (15). Our results provide evidence that both EM and particulate radiation exposure is capable of accelerating lung cancer progression and that dose fractionation creates a more permissive environment for this progression. Comparative genomic analysis between whole lungs from unirradiated K-rasLA1 animals and those exposed to a fractionated or acute dose of high-energy particulate radiation revealed an expression signature that is capable of segregating K-rasLA1 animals irradiated with a fractionated dose from all others. This murine-derived “fractionated” gene classifier, which is driven by inflammatory networks, demonstrates relevance to human carcinogenesis as it retains the capacity to predict overall survival for human lung and breast cancer patients. Therefore, these results strongly support the concept that radiation exposure can enhance cancer progression through the disruption of inflammatory responses and identify an underlying biology related to inflammation with clinical relevance for both human lung and breast cancer.
Materials and Methods
Study Design
Irradiation studies were initiated to evaluate impact of radiation exposure on later stages of carcinogenesis in vivo. Experiments were initiated with a tolerable dose to wildtype mice, 1.0 Gy total dose, as there was no data on how radiation exposure would impact the carcinogenic process. Ensuing experiments were designed to assess the impact of radiation exposure at lower doses in addition to the impact of each dose fraction tested as an acute dose. Sample sizes for irradiation studies were established via power analysis of published survival data for K-rasLA1 mice. Cohort sizes were adjusted during the course of the study to accommodate the progression phenotype observed and were based on power analysis of background incidence of invasive adenocarcinoma in K-rasLA1 mice. Animals were randomly assigned to all experimental cohorts, irradiated as described below, and monitored until death or euthanization due to health concerns. Euthanized animals were censored in corresponding survival analyses and all histopathological analyses were blinded. For microarray studies, cohort sizes were determined based on power analysis of previous in-house expression data obtained from K-rasLA1 mice. Endpoint determination was guided by literature on studies examining the resolution of lung damage and overall radiation-induced effects in mice. Extraction and processing of all samples were blinded and then randomized across microarray chips. Samples for western and immunohistochemistry analyses were additionally extracted and processed blinded, as was the quantification of immunohistochemical results.
Mice
Animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees at the University of Texas Southwestern Medical Center at Dallas (UTSW) and Brookhaven National Laboratory (BNL) (Upton, NY). B6.129S2 K-rasLA1 mice were obtained from the National Cancer Institute Mouse Models of Human Cancers Consortium (Fredrick, MD) and 129S2 K-rasLA1 mice from Dr. Jonathan Kurie (University of Texas M.D. Anderson Cancer Center, Houston, TX). Animals were housed and bred in ventilated microisolator cages within a specific pathogen free (SPF) facility at UTSW. Heterozygote K-rasLA1 mice were crossed with wildtype animals of the same background.
Irradiation
Male and female K-rasLA1 mice, ages 5 to 15 weeks, were irradiated whole-body with either 250-kV X-rays or 1.0GeV/nucleon 56Fe- particles and then monitored twice daily in UTSW SPF facility until death or euthanasia due to health concerns. X-ray experiments were performed at UTSW with X-RAD 320 irradiator (Precision X-ray, Inc.) at an approximate dose rate of 0.14 Gy/min. For 56Fe- particle irradiation, animals were shipped to BNL, irradiated at the NASA Space Radiation Laboratory (NSRL) at dose rate of 0.2 Gy/min, and then returned to UTSW SPF facility following required quarantine period.
Lung Tumor Histology and Evaluation
At necropsy, the lungs were extracted, inflated via intra-tracheal infusion with 10% neutral buffered formalin (NBF), and then, after clamping the trachea, whole tissues were immersion-fixed overnight in 10% NBF. Liver, kidneys, spleens, and any tissues displaying gross abnormalities were also extracted and immersion-fixed overnight. Tissues were processed, paraffin-embedded, and 5μm sections were stained with hematoxylin and eosin for blinded histopathological evaluation. Hematological disorders required confirmation in one or more tissues. To quantify tumor size and number, three sections were cut per animal approximately 50 μm apart. Sections were hematoxylin and eosin stained and images of all discernable lesions were captured with Axiovision software v4.6.3 on Axioskop 2 plus microscope mounted with AxioCamHR color camera (Carl Zeiss Microscopy). Measurement of the surface area was performed with ImageJ software (16).
RNA Extraction
Age-matched 129S2 K-rasLA1 mice were sent to BNL and irradiated with an acute dose of 1.0 Gy 56Fe- particles, 5 daily doses of 0.2 Gy 56Fe- particles, or left unirradiated. 70 days post-irradiation, the lungs were extracted and flash frozen in liquid nitrogen. Frozen tissue was homogenized and RNA extracted with the Qiagen RNeasy Plus Kit (Qiagen) per manufacturer’s protocol.
Microarray and Survival Analyses
Mouse microarrays were performed using Illumina® MouseWG-6 v2.0 Expression BeadChips (Illumina). Samples were labeled and hybridized using Illumina® TotalPrep™ kit (Ambion) and then arrays were scanned using Illumina® Beadstation 500 BeadArray reader and data acquisitioned with BeadStudio (Illumina®). R 2.15.1 (http://www.R-project.org/) and tools in Bioconductor (http://www.bioconductor.org/) were used for all analyses unless otherwise stated. More detailed methodology, including flowchart and Sweave report, for microarray and survival analyses in Supplementary Methods.
Westerns
Age-matched 129S2 K-rasLA1 mice were irradiated, lungs extracted, and flash frozen 70 days post-irradiation as described above for RNA extraction. Frozen tissue was homogenized on ice in cold lysis buffer (50mM Tris-HCl, ph 7.5; 120mM NaCl; 1mM EDTA) supplemented with PhosStop phosphatase inhibitor and cOmplete protease inhibitor cocktail tablets (Roche). Immediately following homogenization, samples were mixed well with Triton™ X-100 (1% final concentration) (Sigma-Adrich) and incubated on ice for 20 min. Lysates were centrifuged at 13000rpm for 20 min and supernatants transferred to new tubes. Protein concentration was determined via Bradford assay (Bio-rad) and 15μg were separated on 4–15% Criterion™ TGX™ gels (Bio-rad). Gels were transferred onto nitrocellulose membranes (Bio-rad) with Trans-Blot® Turbo™ Transfer System (Bio-rad) and blocked with 5% milk. Blots were incubated with primary antibodies, diluted in 1X PBST, overnight at 4°C (p38 MAPK Cat# 9212, phospho-p38 MAPK Cat# 9211, c-JUN Cat# 9165, phospho-c-JUN Cat# 3270, phospho-STAT3 Y705 Cat# 9145, phospho-STAT3 S727 Cat# 9134 (Cell Signaling Technology); STAT3 Cat# 06-596 (Millipore). After three washes with 1x PBST, blots were incubated with secondary antibody (HRP-conjugated AffiniPure goat anti-mouse IgG or goat anti-rabbit IgG) diluted in 5% milk for one hour at room temperature and then washed again with 1X PBST. Blots were exposed with SuperSignal West Femto Chemiluminescent Subtrate diluted 1:5 in dH2O (Thermo Fisher Scientific). Images were acquired with GeneSnap software (Syngene) on G:BOX Chemi system (Syngene) and quantified using GeneTools software (Syngene).
Immunohistochemistry
Age-matched 129S2 K-rasLA1 mice were irradiated and, 70 days post-irradiation, lungs extracted and fixed with 10% NBF as described above for lung tumor evaluation. Tissues were processed, paraffin-embedded, and cut in to 5μm sections. Sections were de-paraffinized, rehydrated, and then antigens retrieved with citrate buffer (10mM sodium citrate, pH 6.0; 0.05% Tween 20) in pressure cooker. Endogenous peroxidase, biotin, and proteins were sequentially blocked with solutions of 3% hydrogen peroxide (Sigma-Adrich), Avidin/Biotin Blocking Kit (Vector Labs), and 10% bovine serum albumin (Vector Labs). Primary antibodies were diluted in 1x TBST with 5% BSA and then sections were incubated at overnight at 4°C (phospho-c-Jun Cat# 3270; phospho-STAT3 Y705 Cat# 9145). After three washes with 1x TBST, secondary antibody and ABC reagent were applied using VECTASTAIN® ABC kit (Vector Labs) following manufacturers’ protocol. Tissue sections were then incubated with ImmPACT DAB peroxidase substrate (Vector Labs), counterstained with methyl green, and then dried overnight prior to mounting coverslip. Images were captured with Axiovision software v4.6.3 on Axioskop 2 plus microscope mounted with AxioCamHR color camera (Carl Zeiss Microscopy) using Plan-APOCHROM 20X and 40X objectives. Quantification was performed on 20X fields of view using Image J software (16) by isolating the DAB and methyl green signals using the Colour Decovolution plugin and then quantifying each signal independently. Entire field were quantified for non-involved regions while only cells within tumor margins were quantified for hyperplasias and adenomas.
Statistics
Statistical analysis of histopathology was performed using two-tailed Fisher’s exact (95% CI) or chi-squared test when appropriate. For age-adjustment, all animals older than 500 days were censored prior to statistical analysis. Gender and background effects were evaluated for individual groups by univariate Cox regression analysis and between groups by either logistic regression or multivariate Cox regression analysis. Differences in survival were determined by log-rank test. Analysis of western blots was through one-way ANOVA with Tukey correction. Tumor burden and immunohistochemistry analysis was by two-way ANOVA with Tukey correction.
Results
Radiation exposure enhances lung cancer progression in K-rasLA1 mouse model
To determine the impact of radiation exposure on animals predisposed to the development of lung cancer, 5–15 week old K-rasLA1 mice were whole-body irradiated with varying doses of X-rays or high-energy 56Fe- particles. In this mouse model, pulmonary lesions are initiated through the spontaneous activation of a latent mutant K-ras G12D allele and predominantly result in the development of benign lung adenomas (15). Only 50% of K-rasLA1 mice will develop adenomas with atypia and, even fewer K-rasLA1 mice, 18%, will have lesions that will fully progress to invasive lung adenocarcinoma (Figures 1A – F). These adenocarcinomas are characterized by tumor cells with high nuclear-to-cytoplasmic ratios, definite nuclear pleomorphism, and have an undifferentiated appearance (Figures 1E and S1). Various adenocarcinomas also demonstrate metastatic potential (Figure S1). There is a marginally significant impact of the genetic background on the incidence of invasive carcinoma in this model as a higher incidence was observed in K-rasLA1 mice backcrossed to a C57Bl/6 background compared to those backcrossed to a 129S2 background (Table S1). A general trend for a gender effect was also observed with females being more susceptible to this transformation (Table S1).
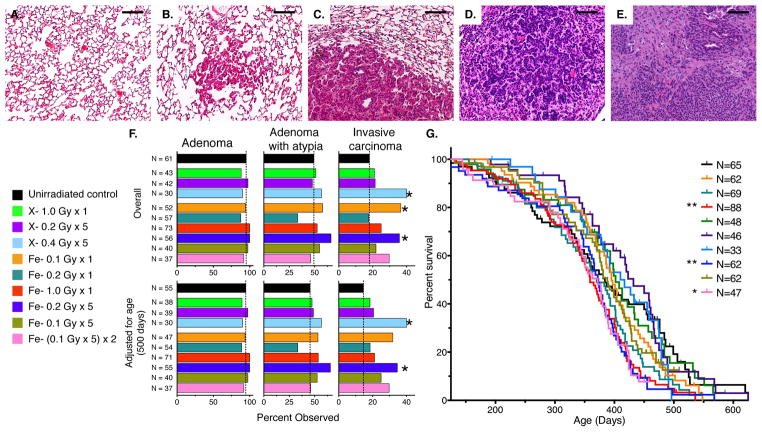
Figure 1. Radiation Exposure Enhances Lung Cancer Progression in K-rasLA1 Mice.
(A – E) Representative images of H&E-stained lungs in unirradiated K-rasLA1 mice demonstrating lung cancer progression in K-rasLA1 mouse model. Normal lung parenchyma (A), hyperplasia (B), adenoma compressing lung parenchyma (C), adenoma with atypia (D), and invasive adenocarcinoma (E).
(F) Quantification of overall and age-adjusted incidences of adenoma, adenoma with atypia, and invasive carcinoma in lungs of unirradiated or irradiated K-rasLA1 mice demonstrates increased incidence of invasive carcinoma following some radiation regimens.
(G) Kaplan-Meier survival plot of unirradiated or irradiate K-rasLA1 mice demonstrating that irradiation with only acute or fractionated dose of 1.0 Gy 56Fe- particles significantly decreases survival compared to unirradiated controls.
Scale bars denote 100 μm; Incidences in (F) evaluated by two-tailed Fisher’s exact test (95% CI); Survival differences were determined by log-rank test; *p < 0.05; **p < 0.01.
While radiation exposure at the doses used in these studies did not affect the overall incidence of adenomas or adenomas with atypia in this model, the overall incidence of invasive adenocarcinoma was substantially increased following some acute and fractionated irradiation regimens (Figure 1F). Logistic regression analysis to assess the impact of strain and sex on this increased incidence of invasive adenocarcinoma demonstrated an influence of strain as radiation exposure differentially increased the overall incidence in each mouse strain (OR = 4.00; 95% CI [0.95,16.92] versus OR = 1.90 95% CI [1.27, 2.85]) (Tables S1 – S3). Gender did not significantly impact this effect (Tables S2 – S3). Irradiation of K-rasLA1 mice with either an acute or fractionated dose of 1.0 Gy 56Fe- particles resulted in a significant decrease in survival, which was also influenced by strain (Figure 1G and Tables S4 – S5). This is in contrast to the survival of unirradiated K-rasLA1 mice, which is not affected by either strain or sex (HR = 0.97; 95% CI [0.53,1.75]; HR = 1.4; 95% CI [0.81,2.53], respectively). An age-adjustment to account for this radiation-induced decrease in survival did not impact the effect of radiation exposure on the incidence of invasive carcinoma (Figure 1F).
Approximately 25% of K-rasLA1 mice possess lung tumors that extend into the bronchial airways and these extensions were also more frequently observed in K-rasLA1 mice following irradiation with 56Fe- particles or a fractionated 2.0 Gy dose of X-rays (Figures S2A and Tables S1 – S2). The incidence of pneumonia is highly correlated with that of bronchial extensions in K-rasLA1 mice (p < 0.0001; chi-square test), but neither radiation exposure nor the increase in obstructed airways was found to impact the incidence of pneumonia (Figures S2B and Tables S1 – S2).
K-rasLA1 mice are also susceptible to thymic lymphoma (15) and radiation is known to impact the health of mice by inducing hematological malignancies including lymphoma (17). Examination of the incidence of lymphoma, leukemia, and myeloproliferative disorder revealed that the overall incidence of individual hematological malignancies was modestly suppressed in K-rasLA1 mice following various irradiation regimens (Figures S2C – F and Tables S1 – S2). The combined incidence of hematological malignancies, however, did not appear to be influenced by radiation exposure (Figure S2E) and thus may not directly contribute to the effect of radiation on survival or lung cancer progression in the K-rasLA1 mouse model. Interestingly, there was an inverse correlation found between the incidence of lymphoma and leukemia and invasive lung adenocarcinoma in these mice (p < 0.0001; chi-square test).
56Fe- charged particle irradiation increases the incidence of hepatocellular carcinoma in the CBA mouse strain, which has a relatively high background incidence, while X-ray irradiation has only a minimal effect (18). Although, K-rasLA1 mice are not susceptible to liver carcinogenesis, a similar trend was observed with the detection of small hyperplastic nodules or hepatomas in livers following exposure to 56Fe- particle radiation (Figure S2G).
Irradiated K-rasLA1 lungs have distinct expression profiles dependent on dose regimen
In order to evaluate how radiation exposure facilitates lung cancer progression in the K-rasLA1 mouse model, we proceeded to examine global gene expression changes post-irradiation. This was performed at the tissue level so that any influence of the microenvironment would also be included. To eliminate any strain effects, only K-rasLA1 mice backcrossed into a 129S2 background were utilized since unirradiated mice of this background are less susceptible for transformation (Table S1). Of particular interest for this analysis was the differential effect of 1.0 Gy 56Fe- particle irradiation on K-rasLA1 mice. Radiation exposure resulted in a significant decrease in survival of K-rasLA1 mice irrespective of dose regimen (Figure 1G), while only K-rasLA1 mice irradiated with a fractionated dose demonstrated an increased incidence of invasive lung adenocarcinoma (Figure 1F). Therefore, the effect of radiation on K-rasLA1 survival and lung cancer progression may be in part due to distinct biologies that are intertwined and collectively influence each phenotype.
Tumor cells and their microenvironment are known to change throughout the carcinogenic process (2, 4). To determine how radiation exposure impacted the carcinogenic process, with minimal confounding influences from the resultant cancer, total RNA was extracted from K-rasLA1 lungs seventy days post-irradiation with an acute or fractionated dose of 1.0 Gy 56Fe- particles and compared to age-matched unirradiated control K-rasLA1 lungs (Figure S4A). This allotted sufficient time for irradiated lungs to recover from any early radiation damage while short enough that tissues/tumors from all cohorts remained histologically identical (Figure 2). At this time point, no advanced adenomas or carcinomas were observed in either control or irradiated mice and inflammatory infiltrates, such as lymphocytes and macrophages, were negligible (Figure 2). Thus, any molecular changes observed would be predicted to be directly associated to changes in the evolving carcinogenic process.
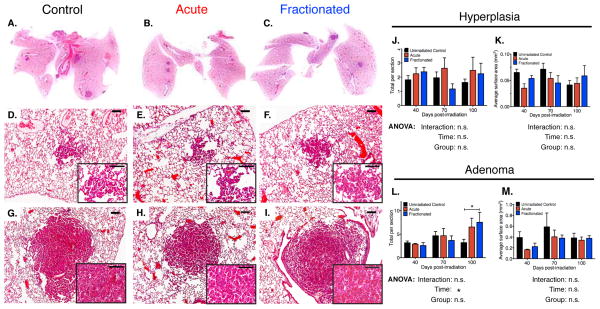
Figure 2. Tumor Burden and Grade in Irradiated K-rasLA1 Lungs Comparable to Unirradiated Controls Seventy Days Post-irradiation.
(A – C) Representative scans of hematoxylin and eosin stained K-rasLA1 lungs seventy days post-irradiation with an acute or fractionated dose of 1.0 Gy 56Fe- particles and age-matched unirradiated controls demonstrate low tumor burden.
(D – I) Representative images of seventy days lungs indicates lesions comprised of only benign lung hyperplasias (D – F) or adenomas (G – I). Inset higher magnification of lesion.
(J – M) Quantification of overall number and size of hyperplasias (J and K) and adenomas (L and M) in K-rasLA1 lungs from forty to one hundred days post-irradiation with an acute or fractionated dose of 1.0 Gy 56Fe- particles and age-matched unirradiated controls (N = 6 – 8 mice per group per timepoint); 3 sections per mouse quantified).
Scale bars denote 100 μm; Graphs show mean ± SEM; Tumor differences determined by two-way ANOVA with Tukey correction; *p < 0.05.
After background normalization and outlier exclusion, we applied a variation-based filter to eliminate any noise potentially introduced by tissue similarities and cellular heterogeneity (see supplementary methods and Figure S3). This resulted in 4580 unique probes representing 4311 genes available for analysis.
Although experimental groups could not be distinguished histologically, collective differences in gene expression were sufficient to segregate these groups by either hierarchical clustering (HCL) or principal component analysis (PCA) using 632 genes (Figures S3, S4B – C). The large number of genes required for this segregation further signifies the overwhelming complexity inherent to the genetic analysis of multi-cellular tissues that do not have overt histological differences. Interrogation of the underlying biology encompassed by this gene signature with Ingenuity Pathway Analysis (IPA) revealed that these genes could be ascribed to several networks associated with cancer, organismal injury and abnormalities, and infectious disease (Table S6).
Unique gene classifiers distinguish individual experimental cohorts
To determine how each group individually contributed to this global signature, we proceeded to identify classifiers that could discriminate each group from the others using only these 632 genes (Figures S3 and S4). Every group was compared to the other two groups separately (p < 0.05; t-test) generating gene lists that could distinguish either control mice from those irradiated with an acute dose, control mice from those irradiated with a fractionated dose, or mice irradiated with an acute dose versus a fractionated dose (199, 317, and 136 genes, respectively) (Figure S3). Overlapping genes between the two comparisons in which an individual group was tested defined the classifier for that particular group (Figures S3 and S4D – L). This culminated in classifiers consisting of 76 genes for control K-rasLA1 mice, 20 genes for K-rasLA1 mice irradiated with an acute dose, and 45 genes for K-rasLA1 mice irradiated with a fractionated dose. HCL, PCA and k-means clustering verified the capacity of these classifiers to isolate the associated experimental group from the others (Figures S4D – L).
“Fractionated” classifier capable of predicting lung cancer patient survival
Identification of a unique classifier for each experimental cohort indicated that these groups had already begun to diverge at the molecular level seventy days post-irradiation. The biological relevance of these classifiers, however, remained obscure due to the static window that which they were derived and dynamic nature of the carcinogenic process (2, 4). Increasingly, microarray profiling has been performed on primary human tumors at various stages and from different tissues, which are then consolidated into large databases replete with associated clinical and survival data (19, 20). This has enabled data mining for gene signatures correlated with patient survival in an attempt to refine clinical prognoses (19, 20). Since each of our experimental groups could be stratified based on the incidence of invasive lung adenocarcinoma and survival (Figures 1F – G), we hypothesized that our early stage classifiers would differentially predict patient survival if the captured gene expression was truly indicative of the resultant phenotypes. Classifiers were thus utilized to create prediction models with three independent human lung cancer microarray datasets, including two publicly available datasets (19, 20) and our University of Texas (UT) – Lung NCI SPORE dataset.
Consistent across all three datasets, only the “fractionated” classifier retained the capacity to predict overall patient survival when prediction models were built using either PCA or k-means clustering (Figures 3 and S5). Addition of the 45-gene “fractionated” classifier to clinical covariates improved survival prediction compared to using clinical criteria alone (p = 0.0001 (classifier + covariates) vs. p = 0.0028 (covariates alone); log-rank test; SPORE dataset). Interestingly, these analyses also demonstrated that the mutational status of K-ras is not correlated with survival and this correlation is not improved by the addition of the “fractionated” classifier (p = 0.16 vs p = 0.66; Cox analysis).
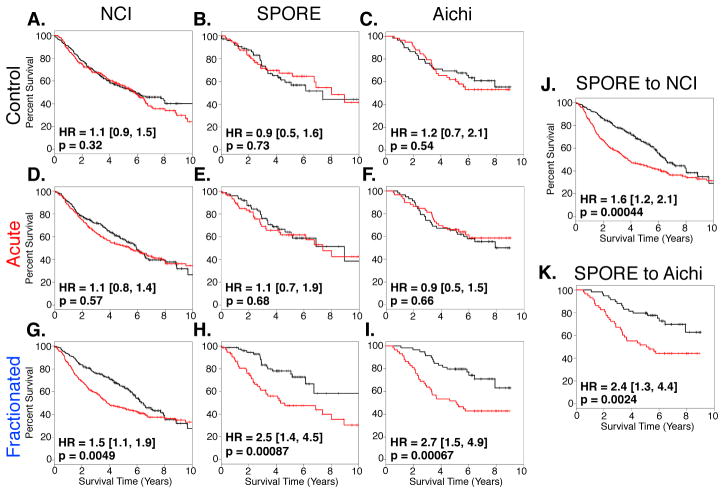
Figure 3. “Fractionated” Classifier Retains Capacity to Predict Overall Lung Cancer Patient Survival Across Multiple Human Lung Cancer Datasets.
(A – I) Prediction models constructed with PCA using gene classifiers that identify unirradiated K-rasLA1 control mice (A – C) or those irradiated with an acute (D – F) or fractionated dose of 1.0 Gy 56Fe- particles (G – I) and three independent human lung adenocarcinoma datasets. Results demonstrate that only “fractionated” classifier is capable of predicting patient survival and these results are independent of dataset specificity.
(J – K) Cross-validation of prediction model constructed with “fractionated” classifier. Prediction model was constructed with PCA using “fractionated” classifier and SPORE lung adenocarcinoma dataset and then model was tested in either NCI or Aichi adenocarcinoma dataset (J and K, respectively).
Red lines denote high-risk patients and black lines denote low-risk patients. Survival differences were determined by log-rank test. Hazard ratios and 95% confidence intervals are relative to high-risk patients.
To verify that our 45-gene classifier was non-random, bootstrap analysis (n = 1000) was performed using gene sets containing forty-five genes randomly selected from the 11051 genes in common between the three lung cancer datasets. Less than 1% of these random datasets performed comparably or better than our 45-gene “fractionated” classifier within the SPORE dataset. None of the random gene sets were capable of performing in all three datasets.
Inflammation-related signaling networks differentially activated across experimental groups
The capacity of the “fractionated” classifier to predict overall survival in patients with lung cancer demonstrated that these forty-five genes retained clinical relevance in humans. IPA annotations of this classifier indicated that these genes are highly associated with networks related to cancer, organismal injury and abnormalities, and infectious disease (Figures 4A – D and Table S6). To determine whether these networks were differentially expressed between our experimental groups, we analyzed the activation status of several integral or related network proteins in whole lungs from K-rasLA1 mice seventy days post-irradiation. Most notable was the coincident increase in the activation of the proto-oncogene JUN and the tumor suppressor mitogen-activated protein kinase 14 (MAPK14) p38 specifically in lung tissues from animals that were irradiated with an acute dose of radiation (Figures 4E and 4G). The transcription factor, signal transducer and activator of transcription 3 (STAT3), is constitutively activated in many solid tumors and promotes tumor immunosuppression (21). STAT3 activation is mediated by several growth factors and cytokines, including those induced by oncogenic Ras, environmental stress, and tissue damage (21, 22). Phosphorylation of tyrosine residue 705 was increased in the lungs of both irradiated groups compared to unirradiated control K-rasLA1 lungs. The phosphorylation of serine residue 727, however, was slightly increased only in K-rasLA1 lungs irradiated with fractionated dose (Figures 4E and 4H).
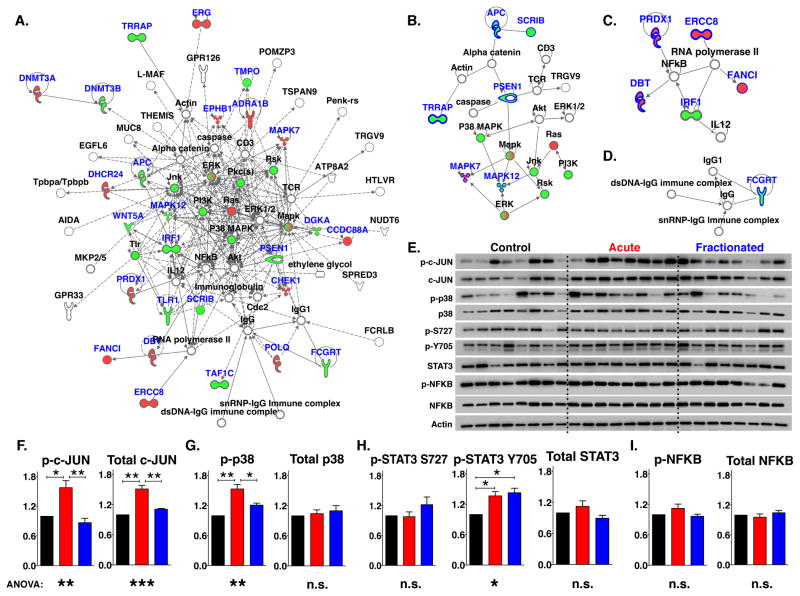
Figure 4. Disparate Activation of Inflammation-related Signaling Network Predicted by “Fractionated” Classifier.
(A) Top IPA network containing molecules interacting directly and indirectly with “fractionated” classifier genes.
(B – D) Direct networks within top network in (A).
(E) Western blot analysis of phosphorylated and total proteins directly or indirectly related to networks from “fractionated” classifier indicates differential activation between experimental groups.
(F – I) Quantification of phosphorylated and total proteins represented in (E). (N= 8 mice per group; 3 technical replicates analyzed; Black bars = Control; Red bars = Acute; Blue bars = Fractionated).
Genes contained within classifier are highlighted in blue. Red and green filled molecules indicate higher and lower expression of genes in animals in fractionated cohort compared to all others. Graphs show mean ± SEM; Western differences determined by one-way ANOVA with Tukey correction; *p < 0.05; **p < 0.01; ***p < 0.001.
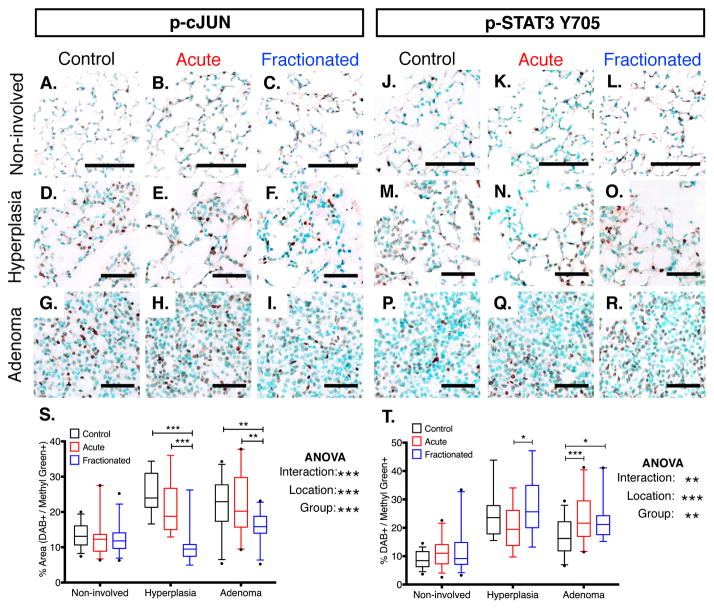
Activation of the canonical nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, which has been shown to be required for lung carcinogenesis (23, 24), was observed, but there was no difference between groups (Figures 4E and 4I). As the differential activation was detected in whole tissue extracts from K-rasLA1 lungs, immunohistochemical analysis of p-JUN and p-STAT3 Y705 was performed in order to determine which cells are responding in this fashion. Both activated signaling proteins were detected in the initiated lesions and in non-involved Type II cells and endothelial cells from the surrounding lung parenchyma (Figure 5). Cells contained within the margins of hyperplasias and adenomas, however, were those in which an activation differential was detected (Figures 5S – T). This apparent disparity in key inflammation-related networks between experimental groups suggested that radiation exposure affected lung cancer progression by altering the local inflammatory responses and this affect was dependent on the dose regimen.
Figure 5. Inflammation-related Signaling Networks Differentially Activated in Cells Associated with Pre-initiated Lesions.
(A – R) Representative images of cJUN (A – I) and STAT3 Y705 (J – R) phosphorylation within hyperplasias, adenomas, or non-involved K-rasLA1 lung seventy days post-irradiation with an acute or fractionated dose of 1.0 Gy 56Fe- particles and age-matched unirradiated controls.
(S – T) Quantification of p-cJUN and p-STAT3 Y705 IHC analysis represented in (A – I) and (J – R), respectively (5 – 7 animals per group region; Non-involved: N = 5 fields per animal; hyperplasias: N = 11 – 13 per protein analyzed; adenomas: N = 19 – 26; Black outlines = Control; Red outlines = Acute; Blue outlines = Fractionated).
For non-involved regions, scale bars denote 100 μm; For initiated lesions, scale bars denote 50 μm; Graphs indicate median and quartiles (95% CI); IHC differences determined by two-way ANOVA with Tukey correction; *p < 0.05; **p < 0.01; **p < 0.001.
Inflammation-based “fractionated” classifier relevant to breast cancer patient survival
Lung cancer is a heterogeneous disease consisting of several subtypes. Activation of mutant K-ras G12D in K-rasLA1 mice specifically results in the formation of adenocarcinoma, which is the most common form of lung cancer in humans (25). Inflammation has, however, been associated with several types of cancer, including breast and squamous cell carcinoma (5). As large-scale, annotated microarray datasets from primary human tumors are available for these cancer types, we proceeded to determine the global significance of our “fractionated” classifier by examining its relevance to these additional cancer types. Prediction models were constructed using our 45-gene “fractionated” classifier with several independent breast and squamous cell lung carcinoma datasets. While the expression of these genes alone was not capable of predicting the survival of patients with squamous cell lung cancer, breast cancer patients with a higher risk of mortality could be predicted in all three independent datasets (Figure 6 and S6). Therefore, identification of a unique inflammation-related classifier capable of segregating K-rasLA1 mice irradiated with a fractionated dose of radiation, which have an increased incidence of invasive carcinoma, has revealed a potential intimate relationship between the disruption of specific inflammation processes and the survival of human patients with both lung and breast adenocarcinoma.
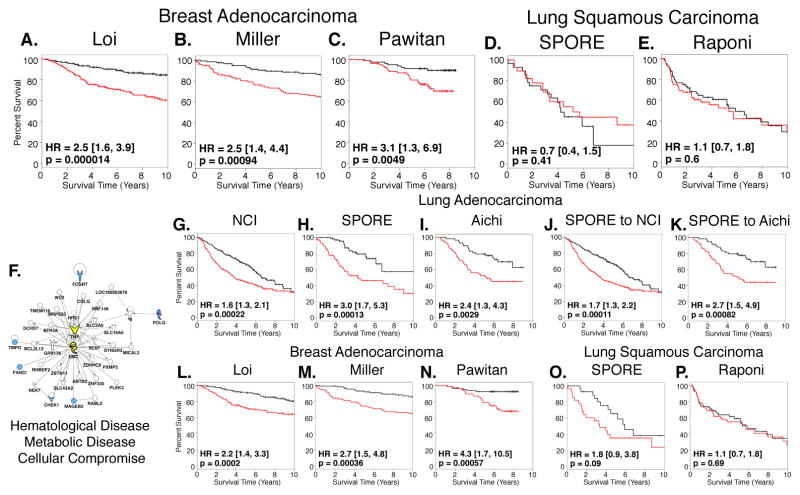
Figure 6. “Fractionated” Classifier Exposes Genetic Similarity Between Lung and Breast Cancer Patient Survival and May Be Condensed to 6 Genes.
(A – C) PCA prediction models were constructed with “fractionated” classifier and tested in three independent breast adenocarcinoma microarray datasets. Results indicate underlying biology encompassed by “fractionated” classifier is relevant for breast cancer survival.
(D – E) PCA prediction models constructed with “fractionated” classifier are not capable of predicting survival of patients with lung squamous cell carcinoma suggesting specificity for adenocarcinoma.
Univariate Cox regression analysis (p < 0.01) using SPORE dataset disclosed 6 genes highly correlated with survival in “fractionated” classifier.
(F) Only predicted network from Ingenuity Pathway Analysis of 6 genes suggests TNF signaling and its regulation by ubiquitylation is driving “fractionated” classifier.
(G – I) PCA prediction models constructed with condensed 6-gene “fractionated” classifier using three independent lung adenocarcinoma datasets demonstrates retention of predictive capacity of “fractionated” classifier for lung cancer.
(J – K) Cross-validation of prediction model constructed with 6-gene “fractionated” classifier and SPORE dataset and tested in NCI (E) and Aichi (F) lung adenocarcinoma datasets.
(L – N) PCA prediction models constructed with 6-gene classifier demonstrate predictive capacity for breast adenocarcinoma patient survival is retained.
(O – P) 6-gene PCA prediction models indicating that condensed “fractionated” classifier is not capable of predicting survival of patients with lung squamous cell carcinoma. 6 genes highlighted in blue in (A).
Red lines denote high-risk patients and black lines denote low-risk patients. Survival differences were determined by log-rank test. Hazard ratios and 95% confidence intervals are relative to high-risk patients.
Data reduction retains predictive capacity and implicates TNF as a potential driver
Although our 45-gene “fractionated” classifier demonstrated utility in multiple datasets derived from cancers of disparate tissues, the power of this classifier is reduced due to the complexity introduced by the overall number of genes. This is evident in the results from IPA network analysis as these genes can only be reduced to three non-overlapping networks with various associated functions (Figure 4A and Table S6). In order to minimize this complexity, we determined which genes within this classifier were most associated with human lung cancer survival. Univariate Cox regression analysis (p < 0.01) of the forty-five classifier genes using only one of the lung cancer microarray datasets, the University of Texas (UT) – Lung NCI SPORE dataset, resulted in six genes, Chek1, Fanci, Fcgrt, Mageb2, Polq, and Tmpo, that highly correlated with overall survival outcome (Figure 6F). Of these genes only Mageb2 was not contained within the first network predicted by IPA (Figure 4A). Condensing the “fractionated” classifier in this fashion demonstrates that this classifier is 3.5-fold enriched for genes that are highly correlated with overall survival (p < 0.01) when compared to the entire SPORE microarray dataset (13.3% vs. 3.8%; p < 0.01; univariate Cox). In fact, only 2.8% of the gene sets comprised of forty-five genes randomly selected from the SPORE dataset, have 6 genes or more associated with survival.
Univariate Cox analysis (p < 0.01) of the 11051 genes in common between the three lung cancer datasets reveals 576 genes significantly associated with survival in the SPORE dataset. The six genes most correlated with survival from these 576 are not predictive in all three datasets when tested as predictive signatures. Bootstrap analysis (n = 1000) using gene sets comprised of six genes randomly selected from the 576 survival-associated genes revealed that only five of these gene sets perform comparably or better in all three lung cancer datasets. One of these five “retrospective” gene sets was also predictive in all three breast cancer datasets at a level comparable to our “prospective” 6-gene signature. There are no overlapping genes between those derived from the 45-gene classifier and these 5 random gene sets.
IPA analysis of the condensed “fractionated” classifier further suggests that the complexity inherent to the 45-gene classifier was reduced, as only one network could be predicted. This network is associated with infectious disease, inflammatory response, respiratory disease, and immunological disease and is impacted by the cytokine tumor necrosis factor (TNF) (Figure 6F and Table S6). Validation that this data reduction did not alter the significance of the “fractionated” classifier for predicting overall human cancer patient survival was demonstrated by constructing prediction models using only these six genes and testing all lung and breast cancer datasets (Figures 6B – K and S7).
Discussion
These studies provide robust evidence that radiation exposure can enhance lung cancer progression in vivo. The breadth and scope of our experiments permitted evaluation of several endpoints and the influence of confounding variables such as genetic background and gender on these endpoints (Tables S1 – S5). Of particular interest is the observation that the incidence of invasive adenocarcinoma is increased following a fractionated dose of 56Fe- particles when compared to that following an equivalent acute dose or in unirradiated K-rasLA1 mice (Figure 1F and Table S3). When combined with the increased incidence observed following an acute lower dose of 56Fe- particles (Figure 1F and Table S3), this supports the controversial concept of an inverse dose-rate effect for carcinogenesis following high-energy radiation exposure (11).
Exploitation of the increased statistical power provided by the differential effect of dose fractionation on lung cancer progression enabled us to identify a genomic signature in whole lung that is driven by alterations to key inflammation-related pathways (Figures 5A, 6F, and Table S6). Inflammation has a significant impact on the carcinogenic process as both a tumor suppressor and promoter and radiation exposure can induce inflammatory responses that can elicit both these effects (5, 6, 26). For instance, tumor eradication is enhanced if radiation exposure results in anti-tumor immunity (6). Radiation-induced inflammation, in contrast, may also result in late normal tissue damage and/or secondary malignancies thereby restricting its therapeutic potential for cancer treatment (3, 11, 26). These responses are dose-dependent and mediated in part by the differential activation of interconnected signaling pathways by oxidative stress, growth factors, and cytokines (6, 26). Subtle variations in one or more of these pathways dictate the elicited response. Among these pathways are those that we found to be disproportionally perturbed in tissues with no overt histological differences (Figures S4 and 5E – I). Incorporation of the resultant phenotypes suggests that radiation exposure altered the normal inflammatory response in emerging tumors and that the inflammatory signature observed following a fractionated dose of radiation is permissive for tumor progression.
The contribution of inflammation on carcinogenic process has also been demonstrated in other tissues. Oncogenic K-RAS mutations are frequently observed in human pancreatic cancer (27) and inflammation cooperates with oncogenic KRAS in the transformation of adult pancreatic acinar cells (28, 29). Further, irradiated fibroblasts increase the invasive capacity of pancreatic cancer cells when co-cultured in vitro through the secretion of the hepatocyte growth factor, which has an anti-inflammatory and anti-fibrotic effect in vivo (8, 30).
The identification of a gene classifier that predicts overall survival for human patients with lung and breast cancer suggests that the inflammatory dysregulation represented by this classifier critically impacts carcinogenesis in both tissues. Data reduction by examining the genes that most correlate with survival within our “fractionated” classifier implies an essential role for TNF and the ubiquitylation system for regulating the observed phenotypes (Figure 6F). This is in concordance with the disrupted signaling networks observed in K-rasLA1 lungs as these are impacted by TNF stimulation (Figures 5 and 7A). Ubiquitylation of several key proteins within the TNF signaling network modulates the resultant cellular response and there are many E3 ubiquitin-protein ligases within the 632 genes that distinguish our experimental groups (Figure 3B – C and Table S6) (31). F-box protein 42, (Fbxo42) which regulates p53, is the only E3 ubiquitin-protein ligases unique to the “fractionated” classifier and is expressed at higher levels in animals that received the fractionated dose (32). The role of FBXO42 in the TNF response is currently not known.
Several other genes within our “fractionated” classifier provide further support for a role of an altered TNF response, including Toll-like receptor 1 and Wnt5a. Both genes are expressed at lower levels in the lungs of K-rasLA1 mice irradiated with a fractionated dose compared to the other two groups (Figure 5A). In addition, the tumor suppressor Scribbled planar cell polarity protein (Scrib) was also expressed at lower levels in K-rasLA1 lungs following a fractionated dose of radiation (Figures 5A – B). TNF stimulation induces cell death in cells expressing oncogenic KRAS or in which SCRIB expression is lost (33, 34). SCRIB deficiency in cells expressing oncogenic KRAS, however, subverts the TNF response to stimulate invasion (35). Finally, loss of SCRIB in mammary epithelia cooperates with c-Myc in the suppression of apoptosis and enhancing cellular transformation (36). How dose fractionation resulted in the downregulation of these genes requires further study.
Overall, we have demonstrated that radiation exposure may lead to several biological effects whose severity may be dependent on radiation dose and quality and radiation-induced inflammation appears to modulate these effects. Derivation of a genetic signature capable of predicting patient survival with lung or breast cancer suggests explicit similarities between these tissues in their inflammatory responses to emergent tumors. Therefore, evaluation of these responses prior to treatment, in particular those related to TNF, could potentially assist lung and breast cancer patient stratification.
Supplementary Material
Statement of Translational Relevance.
Radiation exposure is often used therapeutically as it readily induces DNA damage. However, radiation is a known carcinogen that is increasingly being utilized as a diagnostic tool and therefore may pose a potential risk to human health. In this study, we sought to determine how radiation exposure impacts cancer promotion and progression in vivo through the use of a mouse model of lung cancer and whether any gender or genetic background influences exist. Our results demonstrate that radiation exposure enhances lung cancer progression in K-rasLA1 mouse model of lung cancer. A non-random signature that is predictive of both human lung and breast cancer survival was elicited post-irradiation from murine lung tissue containing only benign lesions. This study provides evidence that the dysregulation of networks important for cancer progression and patient survival may be detectable in histologically identical tissues and that radiation exposure may cooperate with these networks for malignant transformation.
Acknowledgments
Financial Support: National Aeronautics and Space Administration (NSCOR NNJ05HD36GD), the National Cancer Institute (Lung SPORE P50CA70907, Cancer Center Support Grant 1P30CA142543, and CA-16672), and Department of Defense (W81XWH-07-1-0306).
We would like to acknowledge the assistance of the Genomics Shared Resource at the Harold C. Simmons Cancer Center, Drs. Yang Xie and Kevin Coombes for their guidance, and David Minna and Richard McDonnell for colony maintenance.
Footnotes
GEO Accession number: GSE42233
Conflicts of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Hall EJ, Giaccia AJ. Radiobiology for the radiologist. 7. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2012. [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Prise KM, O’Sullivan JM. Radiation-induced bystander signalling in cancer therapy. Nature reviews Cancer. 2009;9:351–60. doi: 10.1038/nrc2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends in genetics : TIG. 2009;25:30–8. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Multhoff G, Radons J. Radiation, inflammation, and immune responses in cancer. Frontiers in oncology. 2012;2:58. doi: 10.3389/fonc.2012.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen DH, Oketch-Rabah HA, Illa-Bochaca I, Geyer FC, Reis-Filho JS, Mao JH, et al. Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type. Cancer cell. 2011;19:640–51. doi: 10.1016/j.ccr.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohuchida K, Mizumoto K, Murakami M, Qian LW, Sato N, Nagai E, et al. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer research. 2004;64:3215–22. doi: 10.1158/0008-5472.can-03-2464. [DOI] [PubMed] [Google Scholar]
- 9.Gorski DH, Beckett MA, Jaskowiak NT, Calvin DP, Mauceri HJ, Salloum RM, et al. Blockage of the vascular endothelial growth factor stress response increases the antitumor effects of ionizing radiation. Cancer research. 1999;59:3374–8. [PubMed] [Google Scholar]
- 10.Qutob SS, Multani AS, Pathak S, McNamee JP, Bellier PV, Liu QY, et al. Fractionated X-radiation treatment can elicit an inducible-like radioprotective response that is not dependent on the intrinsic cellular X-radiation resistance/sensitivity. Radiation research. 2006;166:590–9. doi: 10.1667/RR0514.1. [DOI] [PubMed] [Google Scholar]
- 11.Newhauser WD, Durante M. Assessing the risk of second malignancies after modern radiotherapy. Nature reviews Cancer. 2011;11:438–48. doi: 10.1038/nrc3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nature reviews Cancer. 2008;8:465–72. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- 13.Ullrich RL. Tumor induction in BALB/c mice after fractionated or protracted exposures to fission-spectrum neutrons. Radiation research. 1984;97:587–97. [PubMed] [Google Scholar]
- 14.Ullrich RL. The rate of progression of radiation-transformed mammary epithelial cells is enhanced after low-dose-rate neutron irradiation. Radiation research. 1986;105:68–75. [PubMed] [Google Scholar]
- 15.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 16.Schneider CA, Rasband WS, Eliceriri KS. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan HS. Comparative susceptibility of the lymphoid tissues of strain C57 black mice to the induction of lymphoid tumors by irradiation. Journal of the National Cancer Institute. 1948;8:191–7. [PubMed] [Google Scholar]
- 18.Weil MM, Bedford JS, Bielefeldt-Ohmann H, Ray FA, Genik PC, Ehrhart EJ, et al. Incidence of acute myeloid leukemia and hepatocellular carcinoma in mice irradiated with 1 GeV/nucleon (56)Fe ions. Radiation research. 2009;172:213–9. doi: 10.1667/RR1648.1. [DOI] [PubMed] [Google Scholar]
- 19.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature medicine. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:2793–9. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kranenburg O, Gebbink MF, Voest EE. Stimulation of angiogenesis by Ras proteins. Biochimica et biophysica acta. 2004;1654:23–37. doi: 10.1016/j.bbcan.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–7. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer research. 2010;70:3537–46. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clinics in chest medicine. 2011;32:703–40. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao W, Robbins ME. Inflammation and chronic oxidative stress in radiation-induced late normal tissue injury: therapeutic implications. Current medicinal chemistry. 2009;16:130–43. doi: 10.2174/092986709787002790. [DOI] [PubMed] [Google Scholar]
- 27.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–54. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 28.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer cell. 2011;19:728–39. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Sakai K, Nakamura T, Matsumoto K. Hepatocyte growth factor twenty years on: Much more than a growth factor. Journal of gastroenterology and hepatology. 2011;26 (Suppl 1):188–202. doi: 10.1111/j.1440-1746.2010.06549.x. [DOI] [PubMed] [Google Scholar]
- 31.Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunological reviews. 2011;244:9–28. doi: 10.1111/j.1600-065X.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Shi L, Li W, Yu W, Liang J, Zhang H, et al. JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10195–200. doi: 10.1073/pnas.0901864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igaki T, Pastor-Pareja JC, Aonuma H, Miura M, Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Developmental cell. 2009;16:458–65. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreeger PK, Mandhana R, Alford SK, Haigis KM, Lauffenburger DA. RAS mutations affect tumor necrosis factor-induced apoptosis in colon carcinoma cells via ERK-modulatory negative and positive feedback circuits along with non-ERK pathway effects. Cancer research. 2009;69:8191–9. doi: 10.1158/0008-5472.CAN-09-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordero JB, Macagno JP, Stefanatos RK, Strathdee KE, Cagan RL, Vidal M. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Developmental cell. 2010;18:999–1011. doi: 10.1016/j.devcel.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–78. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.