Abstract
Background and Aims
Recently formed allopolyploid species represent excellent subjects for exploring early stages of polyploid evolution. The hexaploid Cardamine schulzii was regarded as one of the few nascent allopolyploid species formed within the past ∼150 years that presumably arose by autopolyploidization of a triploid hybrid, C. × insueta; however, the most recent investigations have shown that it is a trigenomic hybrid. The aims of this study were to explore the efficiency of progenitor-specific microsatellite markers in detecting the hybrid origins and genome composition of these two allopolyploids, to estimate the frequency of polyploid formation events, and to outline their evolutionary potential for long-term persistence and speciation.
Methods
Flow-cytometric ploidy-level screening and genotyping by progenitor-specific microsatellite markers (20 microsatellite loci) were carried out on samples focused on hybridizing populations at Urnerboden, Switzerland, but also including comparative material of the parental species from other sites in the Alps and more distant areas.
Key Results
It was confirmed that hybridization between the diploids C. amara and C. rivularis auct. gave rise to triploid C. × insueta, and it is inferred that this has occurred repeatedly. Evidence is provided that C. schulzii comprises three parental genomes and supports its origin from hybridization events between C. × insueta and the locally co-occurring hypotetraploid C. pratensis, leading to two cytotypes of C. schulzii: hypopentaploid and hypohexaploid. Each cytotype of C. schulzii is genetically uniform, suggesting their single origins.
Conclusions
Persistence of C. schulzii has presumably been achieved only by perennial growth and clonal reproduction. This contrasts with C. × insueta, in which multiple origins and occasional sexual reproduction have generated sufficient genetic variation for long-term survival and evolutionary success. This study illustrates a complex case of recurrent hybridization and polyploidization events, and highlights the role of triploids that promoted the origin of trigenomic hybrids.
Keywords: Allopolyploidy, Alps, Cardamine × insueta, Cardamine schulzii, Brassicaceae, hybridization, microsatellites, polyploid evolution, speciation, neoallopolyploidy
INTRODUCTION
The integrity of plant species is maintained by pre- and post-mating isolating mechanisms. They form barriers to interspecific gene flow, but these may be incomplete or may break down in certain circumstances. In fact, interspecific hybridization has been rather frequently reported in angiosperms, indicating that failure of isolating mechanisms is not rare (Arnold, 1997; Hegarty and Hiscock, 2005; Mallet, 2005; Rieseberg and Willis, 2007). Once hybridization occurs, it may have different outcomes, including hybrids that persist for a short time and reinforcement of barriers to gene exchange, the existence of highly variable hybrid swarms in contact zones, or new, stabilized hybrid (homoploid or allopolyploid) species (Urbanska and Landolt, 1998; Soltis and Soltis, 2009; Abbott et al., 2013). Indeed, hybridization and polyploidization have been recognized as major speciation forces, contributing significantly to plant diversity (Otto and Whitton, 2000; Soltis and Soltis, 2009; Soltis et al., 2009; Wood et al, 2009). It is particularly challenging to identify the processes responsible for the origin, establishment and persistence of a polyploid/hybrid and to discern its multiple formation, subsequent genome rearrangements, transcriptome dynamics and ecological selection, as well as the repeatability of these processes (e.g. Adams and Wendel, 2005; Gross and Rieseberg, 2005; Hegarty and Hiscock, 2005, 2008; Doyle et al., 2008; Soltis and Soltis, 2009). Such studies provide fascinating insights into the mechanisms of plant evolution and help us to understand plant species diversity.
A few allopolyploids inferred to have originated within the past ∼100–200 years have become models for the study of the early stages of polyploid evolution (e.g. Spartina anglica, Ainouche et al., 2004; Senecio cambrensis, Abbott and Lowe, 2004; Tragopogon mirus and T. miscellus, Soltis et al., 2004; Symonds et al., 2010). Although Cardamine schulzii has been listed among these few cases (Urbanska-Worytkiewicz, 1977a; Urbanska and Landolt, 1998; Soltis and Soltis, 2009), it has received significantly less attention and many aspects remain unexplored.
Cardamine (Brassicaceae) is a species-rich (∼200 species) genus of annual to perennial herbs distributed worldwide, characterized by high chromosome number variation and a high incidence of polyploidy (Lihová and Marhold, 2006; Carlsen et al., 2009). Both auto- and allopolyploidization have been fundamental for speciation in this genus, although the evolutionary histories of only a very few polyploids have been reconstructed in detail (e.g. Perný et al., 2005; Lihová et al., 2006, see also Lihová and Marhold, 2006). Several polyploid complexes composed of multiple closely related species have been recognized. The C. pratensis complex comprises ancestral diploids from southern Europe and diploid to polyploid species mainly from central and northern Europe (Franzke and Hurka, 2000; Lihová et al., 2003, 2004b). One of the latter is C. pratensis s. str. (further referred to as C. pratensis), a polymorphic species that displays high cytotype variation due to polyploidy (diploids to heptaploids) and dysploidy (Lövkvist, 1956; Mandáková et al., 2013). Diploid and tetraploid subalpine populations from the Alps have been segregated as C. rivularis auct. non Schur (e.g. Landolt, 1984; distinct from C. rivularis Schur, which is confined to the Southern Carpathians and Bulgarian mountains; see Franzke and Hurka, 2000). Cardamine amara is another polyploid complex comprising the widespread diploid C. amara subsp. amara and several more restricted diploid and tetraploid allopatric or parapatric taxa (Lihová et al., 2004a). In the Alps, the subspecies amara co-occurs with its autotetraploid derivative subspecies austriaca, although the latter usually grows at higher altitudes (Marhold, 1999).
The members of both polyploid complexes are outcrossing perennials capable of vegetative reproduction (by rhizome fragmentation, stolons or foliar vivipary), growing in mesic to wet habitats, quite often in sympatry or close vicinity, but on the fine scale separated by different habitat preferences (Lihová and Marhold, 2006). Reproductive barriers between the complexes are strong (Lövkvist, 1956), with very few hybrids reported (Lihová and Marhold, 2006). Two such hybrids, the triploid C. × insueta and hexaploid C. schulzii, were discovered at a high-mountain plateau at Urnerboden (Canton of Uri) in the Swiss Alps in the 1970s, and reported as cases of recent, human-influenced hybridization and allopolyploid speciation (Urbanska-Worytkiewicz and Landolt, 1972; Urbanska-Worytkiewicz, 1977a; Urbanska and Landolt, 1998). The triploid C. × insueta was inferred to have arisen from crosses between the sympatric diploids C. rivularis auct. (genome RR) and C. amara subsp. amara (genome AA), and was proposed to represent an RRA hybrid. Cardamine schulzii (RRRRAA) was thought to have originated by autopolyploidization of C. × insueta (Urbanska-Worytkiewicz, 1977a, 1980). Early molecular studies (Neuffer and Jahncke, 1997; Urbanska et al., 1997) confirmed the hybrid origin of both C. schulzii and C. × insueta. Their recent formation was inferred based on their occurrence in anthropogenic habitats, which were created at the beginning of the 20th century due to land use changes (Zimmerli, 1986). Cardamine × insueta grows mainly in fertilized hay meadows and C. schulzii is found in drainage ditches (Urbanska-Worytkiewicz, 1980; Urbanska and Landolt, 1998). Neither hybrid has been found outside of Urnerboden, although another triploid (RAA, without providing a hybrid name) was recorded from a neighbouring canton (Lej da Champfèr, Oberengadin, Canton of Graubünden; Urbanska-Worytkiewicz and Landolt, 1972), growing in sympatry with its proposed progenitors, the diploid C. rivularis auct. and the tetraploid C. amara subsp. austriaca, but not studied later.
Our recent cytogenetic and molecular study (Mandáková et al., 2013) has provided several lines of evidence that C. schulzii did not originate through the scenario previously postulated (Urbanska-Worytkiewicz, 1977a). Instead of an autopolyploid derivative of C. × insueta, we have inferred C. schulzii to be a trigenomic hybrid containing genomes derived from C. amara (A), C. rivularis auct. (R) and C. pratensis (P). Two cytotypes of C. schulzii were found, hypopentaploid (PPRRA) and hypohexaploid (PPPPRA). The presence of C. pratensis at the Urnerboden site had already been noted by Urbanska et al. (1997), but regarded as introduced there only very recently (in the 1990s) and not considered to be involved in hybrid formation at all. The allopolyploid genomes of C. × insueta and C. schulzii have appeared structurally stable (Mandáková et al., 2013), but the frequency of their formation, the amount of genetic variation and evolutionary potential have not been examined in detail so far.
Here we developed and employed progenitor-specific microsatellite markers to closely examine the polyploid origins and ongoing evolutionary processes in the hybridizing Cardamine populations in the Swiss Alps. Our aims were: (1) to prove the efficiency of using species-specific microsatellite markers in detecting hybridization events and parental contributions, and use these markers to verify the genome composition of the hybrids reported from the Urnerboden and Lej da Champfèr sites; (2) to test the hypotheses of single versus multiple hybrid formation by analysing the fine-scale genetic structure of the polyploids and their progenitors; and (3) to outline the evolutionary potential of the hybrids for long-term persistence and speciation by assessing their genetic variation and the extent of gene flow within and among the relevant taxa.
MATERIALS AND METHODS
Sampling
Our sampling focused on the populations at Urnerboden (Canton of Uri, Switzerland), delimiting 12 microlocalities there (Fig. 1) but also included comparative material for the parental species from other sites in the Alps and more distant areas (Appendix). Initial identification in the field was based on morphology and later revised by chromosome counting and ploidy level determination. At Urnerboden we identified and sampled C. amara subsp. amara, C. rivularis auct., C. pratensis and three hybrid types: triploid C. × insueta, hypopentaploids and hypohexaploids. For the hypohexaploids, the name C. schulzii (hereafter denoted as C. × schulzii; see Discussion for rationale) is fully applicable, since they fit the nomenclatural type and were found at the type locality (microlocality 3). Here we suggest a broad circumscription of C. × schulzii that also includes the hypopentaploids (found in neighbouring microlocalities 8 and 10 and having the same parentage; see Results). Cardamine amara, C. rivularis auct. and C. × insueta were abundant across the study area, whereas C. pratensis was rare and scattered, and the hypopentaploid and hypohexaploid C. × schulzii were present only locally. Based on their occurrence and frequency, we sampled 55 individuals of C. amara, 96 of C. rivularis auct., 15 of C. pratensis, 91 of C. × insueta, six of hypopentaploid C. × schulzii and 22 of hypohexaploid C. × schulzii (Fig. 1).
Fig. 1.
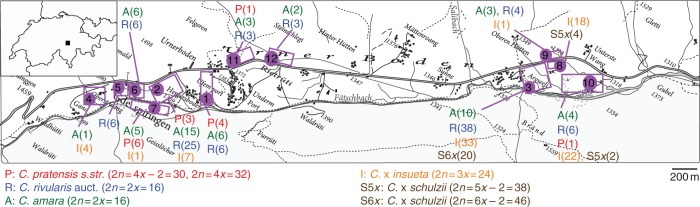
Map of the Urnerboden valley (Switzerland, Canton of Uri) showing the 12 microlocalities and the numbers of individuals sampled (in brackets).
At locations other than Urnerboden, we sampled 12 populations (77 individuals) of C. amara, eight populations of C. rivularis auct. (63 individuals) and four populations of C. pratensis (24 individuals). The hybrids from Lej da Champfèr (15 individuals) were also sampled (Appendix).
We collected fresh leaves in the field and dried them in silica gel. Dehydrated leaf tissue was used for both flow cytometry and DNA isolation. Flower buds were collected for chromosome number determination. Representative voucher specimens were deposited in the SAV herbarium (Bratislava, Slovakia). We also analysed herbarium specimens from ZT (ZT35743–35745, ZT35751–35756, Zürich, Switzerland) and OSBU (OSBU6917–6920, Osnabrück, Germany) herbaria that had been collected from the Urnerboden site at the time of hybrid discovery and included in subsequent studies (1970s–1980s), in order to compare their genetic composition and variation with those of the presently found individuals.
Chromosome counting and flow cytometric estimates of ploidy levels
The ploidy level was determined for each individual sampled from the Urnerboden and Lej da Champfèr sites and analysed for microsatellite variation. This was achieved by chromosome counting and/or by flow cytometric screening of nuclear DNA content. Mitotic chromosomes were counted from young anthers of flower buds fixed in an ethanol:acetic acid (3:1) mixture and stored in 70 % ethanol at –20 °C. Chromosome spreads were prepared following the protocol in Lysak and Mandáková (2013). The chromosomes were stained using 4′,6-diamidino-2-phenylindole (DAPI) and observed with an Olympus BX-61 epifluorescence microscope.
Relative nuclear DNA content was measured by flow cytometry using DAPI fluorochrome and following the protocol described in detail in Marhold et al. (2010). To relate the nuclear DNA content to the DNA ploidy levels, plants with known chromosome numbers were measured cytometrically to determine their nuclear DNA content values, and these were used as a reference.
For the other sampled populations, chromosome numbers or ploidy levels were either known from previous literature records or were assessed here by flow cytometry (see Appendix).
Microsatellite marker development
Microsatellite markers were developed from genomic sequence data (generated with the Genome Sequencer FLX system; Roche, Basel, Switzerland) obtained from two individuals sampled at Urnerboden, each representing one of the originally assumed diploid progenitor species, C. rivularis auct. and C. amara. Our strategy was to target progenitor-specific loci and track them in the hybrids. Species-specific microsatellite loci, expected to amplify in only one of the diploid species and its hybrid derivatives, were identified in silico and tested experimentally by PCR.
Sequencing libraries, sequencing of a half-picotiter plate for each sample with the Genome Sequencer FLX, and adapter trimming of the resulting data were performed at the Functional Genomics Center (Zürich, Switzerland). Identification of microsatellite repeats in the unassembled reads and high-throughput primer design were performed using msatcommander 0·8.2 (Faircloth, 2008). To ensure the ability of the microsatellites to discriminate between the two Cardamine genomes, the primers designed for each species were compared with the reads of the second species using the stand-alone version of BLAST (Altschul et al., 1990). The results of the BLAST searches were visualized using the PLAN web application (He et al., 2007). Potential species-specific markers were selected and their specificity was verified by cross-amplification tests. More details on marker development, cross-amplifications and marker characteristics, including the primer sequences, are presented in Supplementary Data ‘Marker Information’ and Table S1.
At the time of marker development, the involvement of C. pratensis in the hybrid origin was not expected, and thus screening for loci specific to C. pratensis was initially not done. However, due to its close relatedness to C. rivularis auct. (also confirmed by our initial PCR tests), C. rivularis auct. specificity extends well to C. pratensis: the C. rivularis auct.-specific loci amplified well also in C. pratensis, whereas C. amara-specific loci did not. The presence of specific alleles distinguishing between C. rivularis auct. and C. pratensis were tracked in the hybrids. In total, ten C. amara-specific microsatellite loci (hereafter denoted as Cama loci) and six C. rivularis auct./C. pratensis-specific loci (denoted as Criv loci) that showed successful amplification and unambiguous allele determination across multiple individuals were employed in the final analyses. In addition, four loci, originally considered as C. amara-specific, were found to amplify well also in C. rivularis auct. and C. pratensis (Supplementary Data ‘Marker Information’ and Table S1). Since our initial tests indicated that allele size differences may exist among the species, these four shared loci (denoted here as C-all loci) were also employed. Thus, variation at 20 microsatellite loci was finally examined.
Microsatellite amplification and genotyping
Total genomic DNA was isolated using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Microsatellite loci were amplified in five different multiplex reactions, with two to nine loci per reaction. The PCR was conducted in a reaction volume of 10 μL using a Multiplex PCR Kit (Qiagen). Primer concentrations ranged from 0·05 to 0·4 μm. Forward primers were fluorescently labelled using 6-FAM, NED, PET and VIC fluorescent dyes. The thermocycling conditions were 95 °C for 15 min, 25–30 cycles of 94 °C for 30 s, 55–58 °C for 90 s and 72 °C for 1 min, followed by the final extension step of 60 °C for 30 min. The PCR products were submitted for fragment analysis (using an ABI 3100 Avant capillary sequencer at the BITCET Consortium, Comenius University, Bratislava) with a GeneScan™ 500 LIZ® (Life Technologies) internal size standard. Allele sizes were recorded using DAx software (Van Mierlo Software Consultancy, The Netherlands). To check the consistency of amplification and allele determination, both within- and among-plate replicates were included.
Data analyses
Three datasets were assembled: (1) ten Cama loci × 266 individuals of C. amara and the hybrids; (2) six Criv loci × 333 individuals of C. rivularis auct., C. pratensis and the hybrids; and (3) four C-all loci × 464 individuals of all three progenitor species and the hybrids.
Due to the allele copy number ambiguity in these datasets, which encompassed both auto- and allopolyploids of different ploidy levels, determination of precise genotypes and allele frequencies was impossible. This limited the statistics and analyses available for data evaluation, although some advances in treating codominant microsatellite data in polyploid species have recently been achieved (discussed in Clark and Jasieniuk, 2011), and were utilized here. Here we employed two approaches to data treatment/coding to describe genetic patterns and compute genetic diversity and distances. The first corresponded to the multilocus genotype data (more accurately termed ‘multilocus allele phenotypes’, since individual alleles were recorded but with unknown copy numbers in polyploids; Sampson and Byrne, 2012). In the second, we coded each allele as a dominant marker, thus generating a binary presence/absence matrix (e.g. Vallejo-Marin and Lye, 2013). Conversion between these two data formats was done in R 2·15·1 (R Core Team, 2012) using the POLYSAT 1·3–0 package (Clark and Jasieniuk, 2011).
Genetic affinities between the progenitors and hybrids were explored by exact multilocus matches (with the clone assignment function in POLYSAT) and allele-sharing patterns. To examine genetic structure, both principal coordinates analysis (PCoA) and neighbour-joining clustering (NJ) were done in FAMD 1·25 (Schlüter and Harris, 2006), using two different genetic distance measures. The Bruvo distance (Bruvo et al., 2004) was calculated in POLYSAT from the multilocus allele phenotype data as a measure of genetic distance accounting for mutational distance between the alleles and allowing for allele copy number ambiguity, best suited for autopolyploids (in certain circumstances applicable to allopolyploids; Clark and Jasieniuk, 2011). The other distance was based on Jaccard's similarity coefficient, which does not take shared absence into account, and was calculated in FAMD from the binary presence/absence data matrix. For the Cama loci dataset, the Bruvo distance was an appropriate measure, since no allopolyploids were expected (considering the A subgenomes; see Results). The Criv and C-all loci datasets included individuals that could be regarded as allopolyploid, and because the progenitor species commonly displayed the same or very close alleles, for them the Bruvo distance was not appropriate (Clark and Jasieniuk, 2011), and the Jaccard coefficient was used.
Bayesian model-based clustering was employed, using STRUCTURE 2·3.3 (Pritchard et al., 2000), with an approach that enables the handling of dominant markers and genotype ambiguity in the codominant dataset (Falush et al., 2007). Due to the multiple ploidy levels present in our datasets, however, we opted for the dominant, presence/absence coding. For the hybrid individuals, only unique genotypes were kept in the datasets. This analysis was run to identify homogeneous genetic clusters and detect genetic admixture. Five to ten replicates were run for each K = 1–10 (user-defined number of clusters), with a burn-in length of 100 000 generations and data collection for an additional 1 000 000 generations, using the admixture model and uncorrelated allele frequencies. The analyses were performed on the Bioportal of the University of Oslo (www.bioportal.uio.no; Kumar et al., 2009). The STRUCTURE output data were parsed using STRUCTURE HARVESTER (Earl and vonHoldt, 2012) to determine the optimal K value, following the method of Evanno et al. (2005). Alignment of cluster assignments across replicate analyses was then conducted in CLUMPP 1·1·2b (Jakobsson and Rosenberg, 2007) and visualized using DISTRUCT 1·1 (Rosenberg, 2004).
The genetic diversity of each population/taxon was expressed as the total number of alleles observed over all loci (A), the average number of alleles per locus (A′), the average number of alleles per locus per individual (Ai′), the proportion of (partial) heterozygous individuals (Ho), the percentage of polymorphic loci [P(%)], the number of different multilocus allele phenotypes (Nphe) and the Shannon diversity index (a statistic based on genotype frequencies), computed manually or in POLYSAT.
RESULTS
Chromosome numbers and ploidy levels
Chromosome counting and flow cytometry showed that all C. rivularis auct. individuals sampled from Urnerboden were diploid (2n = 2x = 16). Cardamine pratensis was tetraploid (2n = 4x = 32) or hypotetraploid (2n = 4x – 2 = 30) at Urnerboden, the sole exception being a single hypopentaploid individual (determined as 2n = 38; however, this individual turned out to be a C. rivularis auct. × C. pratensis hybrid; see below). From the other localities, C. rivularis auct. included diploids and tetraploids, whereas C. pratensis was (hypo)tetraploid and (hypo)hexaploid. Cardamine amara from Urnerboden was diploid (a few tetraploids were also found but not analysed here), and from the other localities both diploid (i.e. subsp. amara, 2n = 2x = 16) and tetraploid populations (subsp. austriaca, 2n = 4x = 32) were recorded (Appendix).
Three ploidy levels were found among the hybrid individuals: most frequently triploids (2n = 3x = 24, C. × insueta from Urnerboden, hybrids from Lej da Champfèr), less frequently hypohexaploids (2n = 6x – 2 = 46, C. × schulzii, Urnerboden microlocality 3), and very few hypopentaploids (2n = 5x – 2 = 38, C. × schulzii, Urnerboden microlocalities 8 and 10). All the ploidy levels determined here or known from the literature are summarized in the Appendix.
Microsatellite data diversity, multilocus matches and patterns of allele sharing
Allelic variation. The dataset of the ten Cama loci encompassed 102 alleles in total (3–22 alleles/locus), that of the six Criv loci encompassed 74 alleles (6–21/locus) and that of the four shared C-all loci encompassed 49 alleles (8–18/locus). Three Criv loci (Criv15, Criv18 and Criv23) were found to have been duplicated, but were kept in the final analyses due to the scarcity of C. rivularis auct./C. pratensis-specific loci revealed during marker development (Supplementary Data ‘Marker Information’). All progenitor-specific loci amplified well in the hybrids from Urnerboden. Only one Criv locus (Criv34) was not amplified in the triploids from Lej da Champfèr, but this locus amplified in only a few individuals of the sympatric C. rivularis auct. Samples of C. rivularis auct. and C. pratensis from Urnerboden shared alleles at each of the Criv and C-all loci, but we also identified C. pratensis-specific (i.e. not present in C. rivularis auct.; 34 alleles) and C. rivularis auct.-specific alleles (eight alleles) (Table 1).
Table 1.
Patterns of allele sharing between Cardamine pratensis (P) and C. rivularis auct. (R) individuals from the Urnerboden locality, and occurrence of C. pratensis-specific and C. rivularis-specific alleles in the hybrids
| Locus | Number of alleles |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Present in P/R group1 | Shared by P and R2 | Specific to P3 | Specific to R4 | Specific to P3, present in C. insueta5 | Specific to P3, present in 5x/6x C. schulzii | Specific to R4, present in C. insueta | Specific to R4, present in 5x/6x C. schulzii | |
| Cama7 (C-all) | 12 | 11 | 1 | 4 | 0 | 0 | 0/0 | 0 | 0/0 |
| Cama19 (C-all) | 8 | 8 | 2 | 3 | 2 | 0 | 0/0 | 0 | 0/0 |
| Cama23 (C-all) | 18 | 13 | 3 | 3 | 3 | 0 | 0/1 | 0 | 0/0 |
| Cama25 (C-all) | 11 | 10 | 5 | 2 | 1 | 0 | 0/1 | 0 | 0/0 |
| Criv2 | 8 | 8 | 2 | 4 | 0 | 0 | 1/0 | 0 | 0/0 |
| Criv15 | 7 | 7 | 4 | 2 | 0 | 0 | 1/1 | 0 | 0/0 |
| Criv16 | 16 | 16 | 2 | 6 | 1 | 0 | 0/0 | 0 | 0/0 |
| Criv17 | 13 | 13 | 1 | 2 | 0 | 0 | 0/1 | 0 | 0/0 |
| Criv18 | 6 | 6 | 3 | 1 | 0 | 0 | 0/0 | 0 | 0/0 |
| Criv23 | 21 | 21 | 3 | 6 | 1 | 0 | 0/3 | 1 | 1/0 |
| Criv34 | 11 | 11 | 2 | 1 | 0 | 0 | 1/0 | 0 | 0/0 |
| Total | 131 | 124 | 28 | 34 | 8 | 0 | 3/7 | 1 | 1/0 |
1Alleles present in C. pratensis and C. rivularis auct. within and outside Urnerboden populations.
2Alleles shared between the two taxa in Urnerboden, irrespective of their presence or absence outside Urnerboden.
3Alleles present in C. pratensis from Urnerboden and absent from C. rivularis auct. from Urnerboden, irrespective of their presence or absence outside Urnerboden.
4Alleles present in C. rivularis auct. from Urnerboden and absent from C. pratensis from Urnerboden, irrespective of their presence or absence outside Urnerboden.
Multilocus matches and allele sharing at Cama loci. Multilocus matches at Cama loci were found for several individuals of C. amara within the same population. No exact match was observed between C. amara and hybrid individuals. Nevertheless, allele-sharing patterns revealed that each Cama allele observed in the Urnerboden hybrids was also found in the sympatric C. amara (Supplementary Data Table S3). The patterns at the Cama loci in the Lej da Champfèr triploid hybrid were more complex; several alleles present in the hybrid were not found in the sympatric C. amara, but only in some other populations, and two hybrid-specific alleles, not found elsewhere, were detected.
Multilocus matches and allele sharing at Criv loci. At the Criv loci, multilocus matches were observed within several populations of the progenitors, and also between C. × insueta and sympatric C. rivularis auct. Four exact matches between this triploid and its progenitor were detected, with 28 of 91 sampled C. × insueta individuals showing matches with one to five individuals of C. rivularis auct. The matches were not only among individuals from the same microlocality or very close microlocalities (e.g. 3, 8 and 10), but also between those from microlocalities spaced farther apart (e.g. 1 and 3). The rest of the C. × insueta individuals – those not matching the genotypes C. rivularis auct. exactly – possessed only the alleles found in sympatric C. rivularis auct. The Criv alleles recorded in C. × schulzii were also observed in sympatric C. rivularis auct. and/or C. pratensis, except for one allele in the hypohexaploids, and two in the hypopentaploids that were not found elsewhere. Cardamine rivularis auct.-specific (not present in C. pratensis) alleles were rare, and one such allele was recorded in both C. × insueta and hypopentaploid C. × schulzii (Table 1). Cardamine pratensis-specific (not present in C. rivularis auct.) alleles, on the other hand, were more abundant and were found also in C. × schulzii, with seven such alleles in the hypohexaploids and three in the hypopentaploids, but not in C. × insueta (Table 1; see also Supplementary Data Table S3). These numbers refer only to the C. rivularis auct. and C. pratensis samples from Urnerboden, due to the taxonomic uncertainty of the other sampled populations (see below). The triploid hybrid from Lej da Champfèr did not show an exact match with its progenitor, but all of its alleles were observed in sympatric C. rivularis auct.
Allele sharing at C-all loci. At the C-all loci, all alleles found in the Urnerboden hybrids were detected in the sympatric parental species. This, with the exception of one allele, also holds true for the Lej da Champfèr triploid hybrid. Allele-sharing patterns at the C-all loci clearly showed additivity for all four different types of hybrid (Table 2; Supplementary Data ‘Marker Information’ and Table S3). Even though allele sharing between the progenitors was common, progenitor-specific alleles could also be found and identified in the hybrids. Cardamine amara-specific and C. rivularis auct./C. pratensis-specific alleles were detected in C. × insueta, C. × schulzii and the Lej da Champfèr triploid. In addition, two C. pratensis-specific alleles were confirmed in the hypohexaploid C. × schulzii (Table 2).
Table 2.
Patterns of allele sharing and specificity at C-all loci
| Locus | Number of alleles |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total | Present/specific P/R group1 | Present/specific A | Shared by P/R and A | Present in hybrids, specific to P/R group2 | Present in hybrids, specific to A3 | Present in hybrids, equivocal origin4 | Present in C. schulzii, specific to P5 | |
| Cama7 (C-all) | 12 | 11/7 | 5/1 | 4 | 0 | 0 | 2 | 0 |
| Cama19 (C-all) | 8 | 8/3 | 5/0 | 5 | 0 | 0 | 1 | 0 |
| Cama23 (C-all) | 18 | 13/12 | 6/5 | 1 | 3 | 2 | 1 | 1 |
| Cama25 (C-all) | 11 | 10/5 | 6/1 | 5 | 5 | 1 | 1 | 1 |
| Total | 49 | 42/27 | 22/7 | 12 | 8 | 3 | 5 | 2 |
1Alleles present in/specific for C. pratensis and C. rivularis auct. within and outside Urnerboden populations.
2Alleles unequivocally originating from C. pratensis or C. rivularis auct.
3Alleles unequivocally originating from C. amara.
4Alleles not discriminating between the C. pratensis/C. rivularis auct. and C. amara.
5Alleles present in hypohexaploid C. × schulzii and unequivocally originating from C. pratensis from Urnerboden (none such were found in hypopentaploid C. × schulzii).
P, Cardamine pratensis, R, C. rivularis auct., A, C. amara.
Numbers of alleles in hybrids, indicating numbers of parental genomes. The numbers of alleles per locus and individual (excluding the duplicated loci) corresponded to the ploidy levels of the progenitor species and suggested the numbers of respective genomes in the hybrids to be one A (C. amara-derived) genome in C. × insueta and C. × schulzii, two R (C. rivularis auct.-derived) genomes in C. × insueta and two A genomes and one R genome in the triploid from Lej da Champfèr. The numbers of Criv alleles per locus in C. × schulzii individuals were lower than the expected number of P + R genomes (four and five in the hypopentaploids and hypohexaploids, respectively): a maximum of three and two alleles were observed in the hypopentaploids and hypohexaploids, respectively.
Genetic diversity of hybrids. Unequal sample sizes hampered statistical comparison of diversity parameters among populations and taxa, but some patterns are obvious. Hybrids harboured substantially less genetic variation than the progenitors (Supplementary Data Table S2). Cardamine × schulzii, including both cytotypes, was invariable at the Cama loci; at the Criv and C-all loci we found two multilocus allele phenotypes, with one displayed by all the hypohexaploids and the other by all the hypopentaploids. Cardamine × insueta possessed more genetic variation, with three multilocus allele phenotypes at the Cama loci, 16 at the Criv loci and six at the C-all loci (Nphe in Supplementary Data Table S2), but all diversity parameters were still below those of the parental species. The triploid from Lej da Champfèr showed two multilocus allele phenotypes at the Cama loci, two at the Criv loci and one at the C-all loci.
The microsatellite data retrieved from ZT herbarium specimens were, due to low DNA quality, only fragmentary. Nevertheless, all the alleles found in these old specimens were found also in the present-day populations at Urnerboden. In the OSBU herbarium specimens all loci could be amplified, and the multilocus phenotypes that were obtained matched the currently sampled individuals of C. × insueta and the hypohexaploid C. × schulzii.
Genetic structure at Cardamine amara-specific (Cama) loci
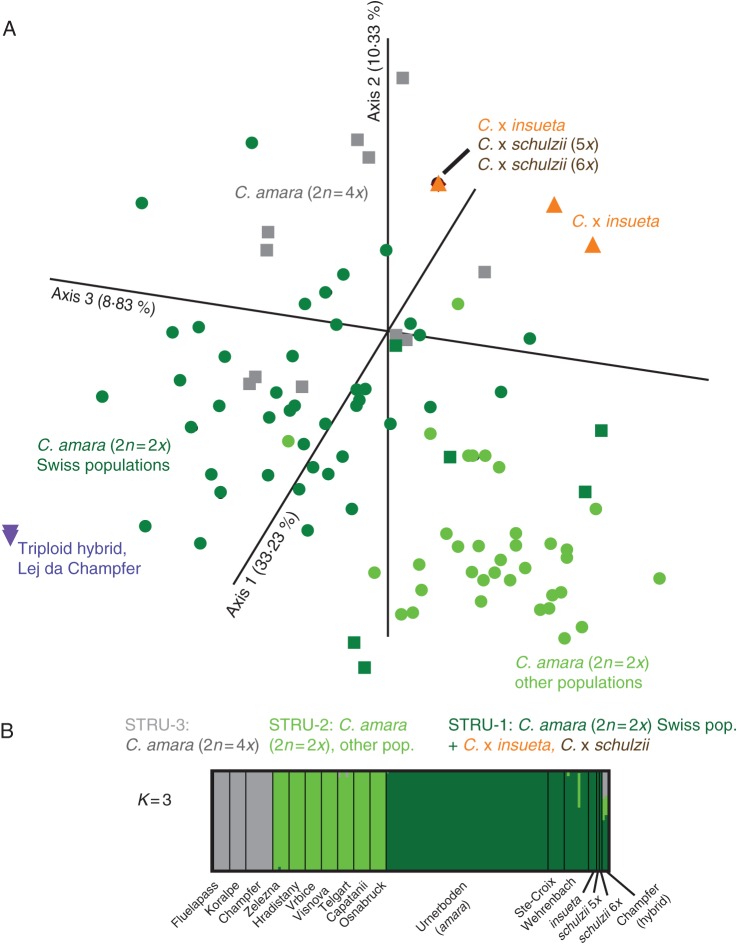
Both the PCoA plot (Fig. 2A) and the NJ tree (Supplementary Data Fig. S1) manifested shallow structure among the analysed accessions. The populations outside the Alps, however, appeared slightly differentiated from the Alpine ones (Urnerboden and two other Swiss populations). Extensive variation was found among the C. amara individuals from Urnerboden compared with the other sampled populations, but with no clustering related to the microlocalities. All Urnerboden hybrid individuals displayed only three multilocus allele phenotypes, which were placed close to the Alpine C. amara. One of these was shared by most C. × insueta (83/91 individuals from all microlocalities) and all of the C. × schulzii individuals, while the other two phenotypes characterized eight individuals of C. × insueta from microlocalities 3 and 8. The Lej da Champfèr triploid appeared in a somewhat distant position.
Fig. 2.
Genetic structure of 266 individuals based on ten Cardamine amara-specific (Cama) microsatellite loci. (A) Principal coordinates analysis based on the Bruvo coefficient. Percentage of variation represented by the axes is indicated. Colours and symbols indicate species/group affiliation (dark green circles stand for the C. amara accessions from Urnerboden, dark green squares for those from the other two Swiss populations). (B) Bayesian model-based clustering at K = 3. Vertical lines demarcate the populations, with the widths of the corresponding boxes proportional to the numbers of individuals analysed in the populations. The colouring indicates each individual's proportional cluster assignment. Population codes are presented in the Appendix.
Bayesian model-based clustering with optimal value K = 3 revealed the following genetic clusters (Fig. 2B): STRU-1, diploid C. amara from Urnerboden and two other Swiss localities, as well as all Urnerboden hybrids; STRU-2, diploid C. amara from the other studied localities; and STRU-3, tetraploid C. amara. Equivocal assignment was obtained for the Lej da Champfèr triploid hybrid.
Genetic structure at Cardamine rivularis auct./C. pratensis-specific (Criv) loci
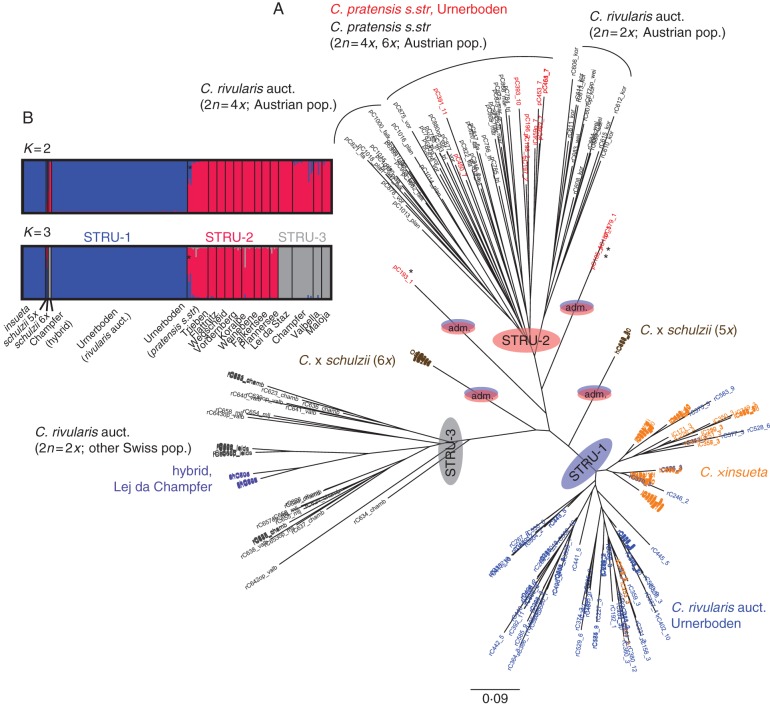
The NJ tree displayed three clearly divergent clusters and four isolated branches in intermediate positions (Fig. 3A). The three main clusters corresponded to: (1) C. rivularis auct. from Urnerboden, including all C. × insueta individuals; (2) C. pratensis from Urnerboden and the other sampled populations, along with C. rivularis auct. (diploids and tetraploids) from the Austrian localities; and (3) diploid C. rivularis auct. from the Swiss populations (other than Urnerboden), including also the Lej da Champfèr triploid hybrid. Grouping of individuals within the C. rivularis auct. + C. × insueta cluster did not follow the different Urnerboden microlocalities. Finally, the four intermediate branches consisted of hypohexaploid C. × schulzii (one branch), hypopentaploid C. × schulzii (one branch), and three C. pratensis individuals (two branches, including the individual with 38 chromosomes). These clustering patterns indicate the distinct origins of the four types of hybrid, as well as the intermediacy (additivity) of the hypohexaploid and hypopentaploid C. × schulzii containing both R and P genomes. These results also indicate genetic distinction between C. pratensis and C. rivularis auct. at Urnerboden, whereas the patterns of variation among the samples from other sites would argue against distinguishing between these two species as currently circumscribed. Indeed, our results revealed several genetic entities related to geography and ploidy levels, potentially representing cryptic species, which need to be explored and delimited in future (being beyond the scope of the present paper).
Fig. 3.
Clustering of 333 individuals based on six Cardamine rivularis auct./C. pratensis-specific (Criv) microsatellite loci. (A) Neighbour-joining tree based on the Jaccard coefficient. Colours indicate species/group affiliation. (B) Bayesian model-based clustering at K = 2 and K = 3. For explanation of graph see legend of Fig. 2. Asterisks highlight three individuals of C. pratensis from Urnerboden showing genetic admixture between the STRU-1 and STRU-2 clusters.
Bayesian model-based clustering suggested two consistent clustering solutions, K = 2 (highest ΔK value) and K = 3 (the second highest ΔK, also reaching the plateau of the likelihood distribution of K). At K = 2, C. rivularis auct. from Urnerboden and C. × insueta (NJ cluster 1 above) were separated from the rest of the individuals, whereas at K = 3 the three clusters corresponded to those in the NJ tree (Fig. 3B). In both cases, C. × schulzii (both cytotypes, but mainly hypohexaploid) and the three individuals of C. pratensis from two intermediate branches (marked with asterisks in Fig. 3) showed admixed ancestry drawn from two clusters containing Urnerboden individuals of C. pratensis and C. rivularis auct., respectively.
The PCoA plot was congruent with the results of the above analyses, supporting the three main groupings and the intermediacy of C. × schulzii (Supplementary Data Fig. S2).
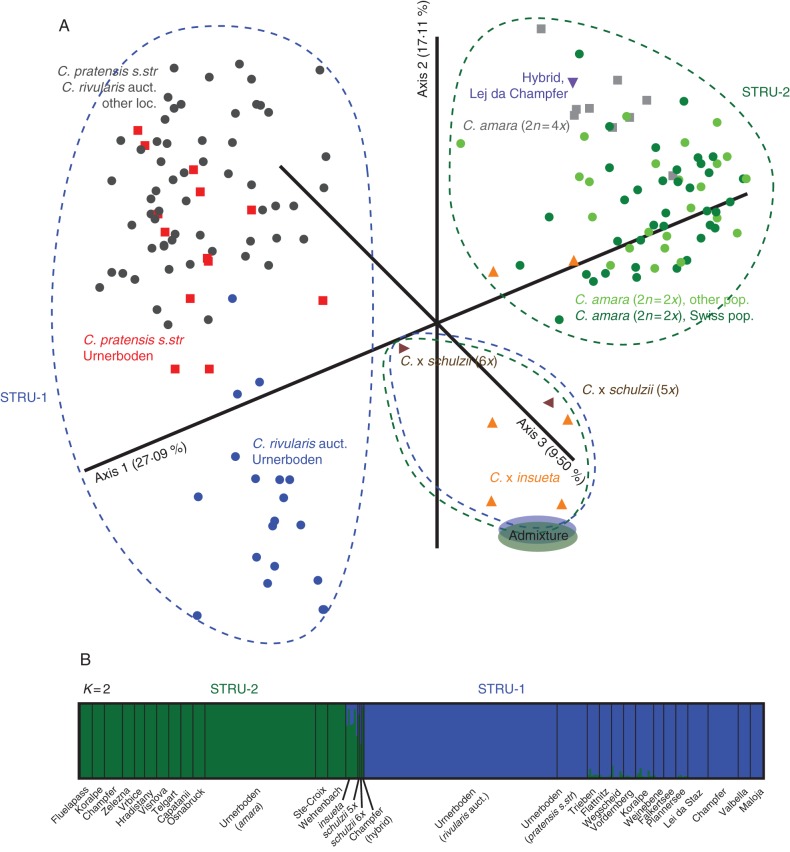
Genetic structure at shared (C-all) loci
The NJ tree recovered several mostly smaller clusters, which could be placed in three main groupings (Supplementary Data Fig. S3), as also recovered by PCoA (Fig. 4A). Cardamine amara was clearly separated from both C. rivularis auct. and C. pratensis along the first axis (Fig. 4A). The second axis, in turn, showed the distinctiveness of C. rivularis auct. from Urnerboden. The position of most Urnerboden hybrid individuals – hypohexaploid and hypopentaploid C. × schulzii and most C. × insueta (83 individuals representing four multilocus phenotypes) – was clearly intermediate. Eight individuals (from microlocalities 3 and 8) of C. × insueta, displaying two multilocus phenotypes, were at the variation margin of C. amara (Fig. 4A), although the NJ tree resolved them as a separate cluster in an intermediate position (Supplementary Data Fig. S3). The Lej da Champfèr triploid hybrid appeared close to tetraploid C. amara.
Fig. 4.
Genetic structure of 464 individuals based on four shared (C-all) microsatellite loci. (A) Principal coordinates analysis based on the Jaccard coefficient. Percentage of variation represented by the axes is indicated. Colours and symbols indicate species/group affiliation. (B) Bayesian model-based clustering at K = 2. For explanation of the graph see legend of Fig. 2.
Bayesian model-based clustering at optimal K = 2 differentiated between C. amara in one group and both C. rivularis auct. and C. pratensis in the other, with admixed ancestry detected in all three Urnerboden hybrids (Fig. 4B).
DISCUSSION
Newly arisen allopolyploid species provide an excellent resource for studying the processes of polyploid speciation and evolution at early stages. Cardamine × schulzii, discovered at a single site at Urnerboden, Switzerland, was one of the classic examples of neoallopolyploidy documented by cytological, experimental breeding and early molecular studies (summarized by Urbanska et al., 1997). Since that time, however, powerful cytogenetic and molecular methods that can provide deeper insights into reticulate and polyploid evolution have become available (Hegarty and Hiscock, 2005). Indeed, the multidisciplinary approach applied recently (Mandáková et al., 2013) rejected the postulated autoallopolyploid origin of C. × schulzii and showed that it is a trigenomic hybrid. Despite the close relatedness of the C. rivularis auct. and C. pratensis genomes, a few C. pratensis-specific genomic features proved the contribution of this species and made it possible to determine the ratio of P to R chromosomes and to infer the genome composition of the hypopentaploids as PPRRA and the hypohexaploids as PPPPRA (Mandáková et al., 2013). Here we follow these recent findings and, using high-resolution multilocus genotyping with species-specific markers, we provide additional evidence for the inferred hybridization scenarios and also gain insight into the genetic structure and variation of the parental species and their hybrid progeny.
Progenitor-specific microsatellite loci and alleles verify the hybrid origins and genome compositions of the allopolyploid Cardamine species at Urnerboden
All progenitor-specific microsatellite loci (Cama, Criv loci) discriminating between C. amara and C. pratensis/C. rivularis auct. amplified well in both C. × schulzii and C. × insueta, confirming their hybrid origins. Additionally, shared C-all loci displayed some species-specific alleles that showed additivity in the studied hybrids. The allelic variation also unequivocally indicates the involvement of C. pratensis in the origin of C. × schulzii, which supports the recent cytogenomic findings (Mandáková et al., 2013). Several loci present in both C. rivularis auct. and C. pratensis (Criv and C-all loci) displayed C. pratensis-specific alleles, and some of these were found in hypopentaploid and hypohexaploid C. × schulzii, but not in the triploid C. × insueta. Also, the ordination (PCoA) and clustering (NJ) methods congruently showed the intermediacy of C. × schulzii at Criv loci, strongly supported by the Bayesian model-based clustering that revealed genetic admixture within C. × schulzii, reflecting its ancestry in two genetic clusters, one comprising C. pratensis and the other C. rivularis auct. (both from Urnerboden).
Our finding of only one allele consistently recorded at each C. amara-specific (Cama) locus in all Urnerboden hybrid individuals supports the presence of only one A genome. Two R genomes were inferred for C. × insueta, in accordance with the expected RRA composition of this triploid. For C. × schulzii, however, due to allele sharing by C. pratensis and C. rivularis auct. and allele copy number ambiguity, the exact number of P and R genomes could not be inferred from the microsatellite data.
Importantly, analyses of specimens collected more than 20 − 30 years ago at Urnerboden (deposited in the ZT and OSBU herbaria) did not reveal genotypes or alleles distinct from those found in the currently sampled hybrid plants, and also proved the presence of C. pratensis-specific alleles in C. × schulzii. These findings, in accord with those by Mandáková et al. (2013), support our conclusion that the hybrids which we collected are the same as those reported and studied by Urbanska and co-authors.
Multilocus genotyping, which provides a high number of polymorphic and independent markers, has been commonly used to detect hybrid offspring and study hybrid zones. Nevertheless, considerable genotyping effort involving many loci may be needed, especially if divergence between parental species is low and/or later-generation and backcrossed individuals are also present (Vähä and Primmer, 2006). Species-specific markers can significantly facilitate such efforts. The present study illustrates the efficiency of using progenitor-specific microsatellite markers to detect hybridization as well as to infer parental contributions in hybrid progeny. A few other studies have employed a similar approach, for instance to examine interspecific gene flow in poplars (Khasa et al., 2005), recurrent formation in Tragopogon polyploids and gene flow among the independent polyploid lineages (Symonds et al., 2010), or to discriminate between two salmon species and recognize their hybrids (Perrier et al., 2011).
Scenarios proposed for the formation of Cardamine × schulzii
The exclusion of the autoallopolyploid origin of C. × schulzii from the triploid C. × insueta raised the question of the actual parentage of the hypohexaploid and hypopentaploid hybrids. The inferred genome composition, cpDNA haplotype sharing (implying that R genomes were maternally inherited) and gametes (polarized, unreduced and aneuploid) observed in C. × insueta suggested that the most plausible route for the formation of the hypohexaploid C. × schulzii (RPPPPA) is via hybridization between C. × insueta (assuming an RA egg cell with 16 chromosomes) and C. pratensis (unreduced PPPP pollen with 30 chromosomes) (Mandáková et al., 2013). Similarly, the hypopentaploid C. × schulzii (RRPPA) has probably originated via hybridization between C. × insueta (unreduced RRA egg cell) and C. pratensis (reduced PP pollen), one of the gametes being likely to be aneuploid (n − 1; Mandáková et al., 2013). The origin of both cytotypes of C. × schulzii via C. × insueta is strongly favoured by the microsatellite data, considering the fact that the same multilocus allelic phenotype at Cama loci was shared by all C. × schulzii and most C. × insueta individuals. In an alternative scenario, a C. pratensis × C. rivularis auct. hybrid might have arisen first, and then hybridized with C. amara. Such a hypopentaploid hybrid plant (C. pratensis × C. rivularis auct.) with 38 chromosomes has indeed been identified by the present microsatellite data. Genetic admixture was also indicated in two 30-chromosome plants of C. pratensis, which probably represent such hybrids as well. Nevertheless, the latter scenario of the origin of C. × schulzii is less likely because its multilocus allelic phenotypes at Criv loci differ from those of the C. pratensis × C. rivularis auct. hybrids, and also the abundance of C. × insueta contrasts with the scarcity of C. pratensis × C. rivularis auct. hybrids.
The observed allelic variation in both C. × insueta and C. × schulzii supports the inference of their recent origins, as originally proposed by Zimmerli (1986) on the basis of their occurrence in habitats created after land-use changes in the early 20th century. The level of allele sharing between the progenitors and the allopolyploids, with very few non-parental alleles detected in C. × schulzii, only at Criv loci, favours their recent origins. These seemingly C. × schulzii-specific alleles might represent alleles present in the progenitors but not sampled here (the sampling of C. rivularis auct. was extensive, but still some alleles may have escaped, whereas that of C. pratensis was indeed limited due to the species scarcity at the locality) or they may have evolved in the allopolyploid genomes. Human-induced habitat disturbance and generation of new niches have apparently played a significant role in the hybrid formation and establishment in Urnerboden (Urbanska and Landolt, 1998), as has been recognized also in other plant groups (Levin et al., 1996; see also Bleeker and Hurka, 2001; Lihová et al., 2007).
Multiple origins and interbreeding explain genetic variation in triploid Cardamine × insueta
It is now widely recognized that multiple and recurrent formations are common in allopolyploids (Soltis and Soltis, 1999). Recurrent hybridization and polyploidization events increase genetic variation and result in distinct hybrid genotypes, which can create additional diversity by interbreeding, differential homeologous recombination and segregation. Such processes can yield a highly variable polyploid species or even multiple cryptic species (Soltis and Soltis, 1999; Shimizu-Inatsugi et al., 2009; Symonds et al., 2010). In our study, both diploid progenitors displayed substantial variation at the studied microsatellite loci (despite their ability to reproduce clonally), enhancing the likelihood of detecting independent origins arising from genetically distinct parental individuals. Nevertheless, neither C. amara nor C. rivularis auct. showed geographically correlated genetic structure within the Urnerboden valley. Apparently, intraspecific gene flow is efficient within these outcrossing species, hindering distinction between multiple (in the sense of polytopic) and single origins of the studied hybrids. Moreover, even though discerning multiple hybrid origins is easier in recent polyploids, inferring the exact number of origins may be hampered by recombination and segregation of homeologous chromosomes, gene flow among the polyploids and backcrosses accumulating in later generations (Soltis and Soltis, 2009) – processes that may have obscured the true number of independent origins in this system.
Three multilocus genotypes were revealed in C. × insueta at Cama loci, as many as 16 at Criv loci, and six at C-all loci. Two of the three Cama genotypes in C. × insueta, however, are very similar to each other, differing at a single locus, which might be due to a recent mutation in the hybrid. Thus, only two of these multilocus genotypes clearly reflect distinct parental origins. Since C. × insueta contains only one A genome, these two genotypes observed at Cama loci could suggest at least two independent origins, i.e. involvement of two different male gametes. At Criv loci, much higher variation was found in C. × insueta, but recombination and segregation probably generated some of this variation. Although sexual reproduction is apparently limited in C. × insueta – as evidenced by meiotic irregularities, the prevalence of male-sterile plants with non-dehiscent anthers (Urbanska-Worytkiewicz, 1977b; Mandáková et al., 2013) and efficient vegetative propagation (J. Zozomová-Lihová et al., pers. obs.; Urbanska-Worytkiewicz, 1980) – viable gametes could be produced rarely and engage in sexual reproduction. Polarized meiosis (characterized by asymmetrical chromosome segregation), giving rise to two reduced euploid gamete types, RA and R, in addition to the unreduced RRA type, was previously reported and supported by experimental cross-pollinations (Urbanska-Worytkiewicz, 1977b). The four exact multilocus matches between C. × insueta and C. rivularis auct. could be regarded as evidence of independent origins, but some of these matches could also be due to recombination in the diploids or triploids (given the rather low number of investigated Criv loci and the high frequencies of most alleles). Thus, although we cannot infer the exact number of hybridization events that have resulted in C. × insueta, the relatively large genetic variation of C. × insueta is likely due to both multiple origins and interbreeding. Whether and to what extent backcrossing with C. rivularis auct. has shaped this variation remains unknown. Nevertheless, experimental backcrossing by Urbanska-Worytkiewicz (1977b) did yield diploid to pentaploid offspring, and although we did not detect individuals such as RRRA and RRRRA (raised experimentally by Urbanska-Worytkiewicz) in our study, some RR (C. rivularis auct.) and RRA (C. × insueta) individuals might be products of backcrossing.
Two hybridization events gave rise to two distinct cytotypes of Cardamine × schulzii
The present microsatellite data support two distinct hybridization events between C. × insueta and C. pratensis, giving rise to two cytotypes of C. × schulzii. These cytotypes were clearly genetically distinct, as seen from the alleles at Criv and C-all loci, and found in neighbouring microlocalities. Given the genetic uniformity within these two cytotypes, however, we can infer that each cytotype arose only once. Interestingly, the hypopentaploids have never previously been reported, meaning that they either escaped sampling or arose subsequently. The allelic composition of the old herbarium specimens matched either C. × insueta or hypohexaploid C. × schulzii.
Occurrence of another triploid hybrid with similar parentage but reciprocal genomic composition
We documented another hybridization event, from Lej da Champfèr, in accord with the earlier report by Urbanska-Worytkiewicz and Landolt (1972). We confirmed the genomic composition of this triploid, originally inferred as RAA, based on the number of progenitor-specific alleles retrieved from the triploid individuals. This triploid resembles C. × insueta morphologically (Urbanska-Worytkiewicz and Landolt, 1972), but is genetically distinct (as are its co-occurring progenitors from those at Urnerboden). It displayed a total of four different genotypes. This genetic diversity is somewhat surprising, given that it forms a single dense patch, for which a single origin and genetic uniformity would be expected. In addition, at Cama loci the hybrid contained several non-parental alleles, or alleles absent from the sympatric C. amara. Possibly these alleles are actually present in sympatric C. amara but were not sampled, or the population size and genetic variation of C. amara declined since the hybrid origin. The latter explanation is favoured by the genetic depauperation observed in C. amara and by its restricted occurrence on a disturbed lakeshore.
Reproductive behaviour and evolutionary implications of Cardamine allopolyploids
Diverse evolutionary fates of hybridization events, whether or not coupled with polyploidization, can be observed in natural populations. The progeny have a high chance of rapidly vanishing, due to minority cytotype disadvantage and small effective population size as well as likely increased sterility, lower fitness and the absence of suitable available niches. Even if some life history traits and reproductive strategies, such as the perennial habit, vegetative reproduction and self-compatibility, facilitate hybrid establishment, their populations remain genetically depauperate and thus vulnerable to extinction by environmental disturbances (reviewed by Arrigo and Barker, 2012). It has been suggested that recent polyploid species have higher extinction rates than their diploid relatives, and thus those that are extant represent rather rare successes (Arrigo and Barker, 2012). Indeed, recent allopolyploid lineage extinction has been observed in Senecio cambrensis (Abbott and Lowe, 2004).
Our findings on C. × schulzii, gathered several decades after its discovery, disagree to some extent with earlier observations. Previous studies, including field observations, experimental cultivation (Urbanska-Worytkiewicz, 1980) and analyses of pollen fertility (Urbanska et al., 1997), suggested that C. × schulzii had a balanced system of sexual and clonal reproduction. The genetic uniformity that we found in both cytotypes of C. × schulzii, however, indicates the presence of only vegetative propagation. This agrees with meiotic irregularities and pollen grains of different size and low fertility observed in these plants (Mandáková et al., 2013). Moreover, the peculiar trigenomic composition and the ratio of R/P to A genomes of 4:1 or 5:1 do not favour efficient sexual reproduction either.
Cardamine × schulzii is currently restricted to three adjacent microlocalities (3, 8 and 10), within or close to the locus classicus (locality 3; E. Landolt, ETH, Zurich, pers. comm.), and apparently is not spreading significantly. Thus, C. × schulzii remains a rare and locally persisting hybrid derivative without significant evolutionary potential, confined to a particular man-made niche, and is thus highly vulnerable to extinction. We refrain from treating it as a stabilized species as originally described (C. schulzii), and instead refer to these hybrid plants as C. × schulzii, as has been done in this paper. Sterile hybrids, however, can recover fertility or become widespread even if genetically depauperate (Ainouche et al., 2004; Vallejo-Marin, 2012; Vallejo-Marin and Lye, 2013), and long-term monitoring of C. × schulzii is therefore advisable.
The formation of hybrid swarms, another possible outcome of hybridization, depends upon weak reproductive barriers among the progenitors and hybrid derivatives, as well as sufficient gene flow between ploidy levels (e.g. Lihová et al., 2007). At Urnerboden, however, barriers to interspecific and interploidal gene flow appear to be rather strong, maintaining the integrity of the progenitor species.
The third possible, and evolutionarily most significant, outcome of hybridization is the formation of a new homoploid or allopolyploid species. Such a species is usually reproductively isolated from its progenitors, colonizes new habitats or further evolves by genome rearrangements and genetic, epigenetic and transcriptional changes (Hegarty and Hiscock, 2008; Arrigo and Barker, 2012). Our findings indicate that the C. × insueta studied here might be in transition from a temporary hybrid derivative to a stabilized allopolyploid species. This plant displays genetic variation, shows efficient clonal growth and high vigour, has successfully spread across multiple microlocalities in the Urnerboden valley (Fig. 1; see also Urbanska et al., 1997) and even promoted further hybridization by crossing with a non-parental species (C. pratensis), giving rise to a hybrid with a higher ploidy level, C. × schulzii. Although C. × insueta has reduced fertility (Mandáková et al., 2013), its multiple origins, coupled with occasional gene flow, can increase genetic variation and generate unique genotypes, thus ensuring long-term stability and evolutionary success.
In conclusion, this study describes a complex case of recurrent hybridization and polyploidization events resulting in multiple allopolyploids. This system and the findings that it has yielded call for further in-depth studies from various perspectives (evolution of duplicated genes, homeologue interactions, gene expression patterns, ecological divergence, etc.). We also suggest that revisiting earlier evolutionary hypotheses using more advanced molecular tools and approaches and larger population samples may significantly deepen our knowledge of the polyploid origins and evolution and can also bring completely new and exciting findings.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the curators of the herbaria in Zürich and Osnabrück for allowing us to study the voucher specimens from Urnerboden. We are grateful to E. Landolt for sharing his experience and knowledge of Urnerboden populations, E. Záveská for DNA isolations, P. Mikulíček for discussion of microsatellite data generation and analyses, colleagues (listed in the Appendix) who participated in the field sampling, and the Functional Genomics Center Zürich. This work was supported by the Czech Science Foundation (grant number P501/10/1014 to M.A.L.); the European Social Fund (grant numbers CZ.1·07/2·3.00/20·0189 and CZ.1·07/2·3.00/30·0037 to T.M. and CZ.1·07/2·3.00/30·0022 to S.Š.); the long-term research development project no. RVO 67985939 to K.K.; and by the Human Frontier Science Program to K.K.S.
APPENDIX
Cardamine species and populations sampled in the present study. Taxon name, population code (in bold), country code (AU, Austria; CH, Switzerland; CZ, Czech Republic; DE, Germany; RO, Romania; SK, Slovakia), administrative division (canton, Bundesland), locality, altitude and collector name (IK, I. Krásná; JŠ, J. Šibík; KM, K. Marhold; ML, M. Lysák; MP, M. Perný; SŠ, S. Španiel) are given. Chromosome numbers (2n =) and/or DNA ploidy levels assessed by flow cytometry (2n ∼) are specified, either determined in the present study (in bold, providing also the number of analysed individuals) or taken from the literature (summarized in the database available at http://www.cardamine.sav.sk (accessed 3 September 2013)).
Urnerboden site – CH, Uri, Urnerboden valley, 1318–1380 m, coll. ML et al.:
C. × insueta Urbanska (2n = 3x = 24, 32 ind.; 2n ∼ 3x, 60 ind.), C. × schulzii Urbanska (2n = 6x – 2 = 46, 21 ind.; 2n = 5x – 2 = 38, 6 ind.; 2n ∼ 6x, 5 ind.), C. rivularis auct. non Schur (2n = 2x = 16, 16 ind.; 2n ∼ 2x, 128 ind.), C. pratensis L. s. str. (2n = 4x = 32, 1 ind.; 2n = 4x – 2 = 30, 4 ind.; 2n ∼ 4x, 8 ind.), C. rivularis auct. non Schur × C. pratensis L. s. str. (2n = 5x – 2 = 38, 1 ind.), C. amara L. subsp. amara (2n = 2x = 16, 8 ind.; 2n ∼ 2x, 54 ind.).
Lej da Champfèr site – CH, Graubünden, Lej da Champfèr, 1800 m, coll. KM and JŠ:
C. amara subsp. austriaca Marhold (2n = 32, 2n ∼ 4x, 9 ind.), C. rivularis auct. non Schur (2n = 16, 2n ∼ 2x, 15 ind.), C. amara subsp. austriaca × C. rivularis auct. (2n = 24, 2n ∼ 3x, 15 ind.).
Comparative populations
Cardamine amara L. subsp. amara
Ste-Croix – CH, Vaud, Ste-Croix, 1209 m, coll. M. de la Harpe; Wehrenbach – CH, Zürich, Wehrenbach, coll. M. Helling; Železná – SK, Bratislava, Malé Karpaty Mts., Železná studnička, 270 m, coll. IK, 2n ∼ 2x; Vrbice – CZ, Podbořanská kotlina, Vrbice, 330 m, coll. IK, 2n ∼ 2x; Hradišťany – CZ, Plzeňská pahorkatina, Hradišťany, 400 m, coll. IK, 2n ∼ 2x; Višňová – CZ, Střední Povltaví, Višňová, 420 m, coll. IK, 2n ∼ 2x; Telgárt – SK, Telgárt, 890 m, coll. KM, ML, 2n ∼ 2x, 6 ind.; Capatanii – RO, Mt. Capatanii, Manastirei Pantrusa, 1000 m, coll. MP; Osnabrück – DE, Osnabrück, Sutthausen, 90 m, coll. K. Mummenhoff.
Cardamine amara subsp. austriaca Marhold
Flüelapass – CH, Graubünden, Flüelapass, 2380 m, coll. KM and JŠ, 2n = 32; Koralpe – AU, Styria, Wolfsberg, Mt. Koralpe, 1960 m, coll. KM and JŠ, 2n = 32.
Cardamine rivularis auct. non Schur
Valbella – CH, Graubünden, Valbella, lake Heidsee, 1480 m, coll. KM and JŠ, 2n = 16; Maloja – CH, Graubünden, Maloja, lake Sils, 1810 m, coll. KM and JŠ, 2n = 16; Lej da Staz – CH, Graubünden, St. Moritz, lake Lej da Staz, 1810 m, coll. KM and JŠ, 2n = 16; Weinebene – AU, Carinthia, Wolfsberg, from Weinebene to Grillitsch Hütte, 1720 m, coll. KM and JŠ, 2n = 16; Koralpe – AU, Styria, Wolfsberg, Mt. Koralpe, 1960 m, coll. KM and JŠ, 2n = 16; Falkertsee – AU, Carinthia, Nockberge, Falkertsee, 1940 m, coll. SŠ and MP, 2n = 32; Plannersee – AU, Styria, Niedere Tauern, Planneralm, Plannersee, 1780 m, coll. SŠ and MP, 2n = 32.
Cardamine pratensis L. s. str.
Trieben (P) – AU, Styria, Trieben, Hohentauern, 1065 m, coll. SŠ and MP, 2n ∼ 4x, 6 ind.; Flattnitz (P) – AU, Carinthia, Flattnitz, 1395 m, coll. SŠ and MP, 2n ∼ 4x, 6 ind.; Vordernberg (P) – AU, Styria, Vordernberg, 1080 m, coll. SŠ and MP, 2n ∼ 4x, 6 ind.; Wegscheid (P) – AU, Styria, Wegscheid, S of Mariazell, 910 m, coll. SŠ and MP, 2n ∼ 6x, 6 ind.
LITERATURE CITED
- Abbott RJ, Lowe AJ. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biological Journal of the Linnean Society. 2004;82:467–474. [Google Scholar]
- Abbott RJ, Albach D, Ansell S, et al. Hybridization and speciation. Journal of Evolutionary Biology. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ainouche ML, Baumel A, Salmon A. Spartina anglica C.E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biological Journal of the Linnean Society. 2004;82:475–484. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Natural hybridization and evolution. New York: Oxford University Press; 1997. [Google Scholar]
- Arrigo N, Barker MS. Rarely successful polyploids and their legacy in plant genomes. Current Opinion in Plant Biology. 2012;15:140–146. doi: 10.1016/j.pbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Bleeker K, Hurka H. Introgressive hybridization in Rorippa (Brassicaceae): gene flow and its consequences in natural and anthropogenic habitat. Molecular Ecology. 2001;10:2013–2022. doi: 10.1046/j.1365-294x.2001.01341.x. [DOI] [PubMed] [Google Scholar]
- Bruvo R, Michiels NK, D'Souza TG, Schulenburg H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molecular Ecology. 2004;13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- Carlsen T, Bleeker W, Hurka H, Elven R, Brochmann C. Biogeography and phylogeny of Cardamine (Brassicaceae) Annals of the Missouri Botanical Garden. 2009;96:215–236. [Google Scholar]
- Clark LV, Jasieniuk M. POLYSAT: an R package for polyploid microsatellite analysis. Molecular Ecology Resources. 2011;11:562–556. doi: 10.1111/j.1755-0998.2011.02985.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, et al. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics. 2008;42(21·1–21·19) doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Evanno GD, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Faircloth BC. MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Molecular Ecology Resources. 2008;8:92–94. doi: 10.1111/j.1471-8286.2007.01884.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, Hurka H. Molecular systematics and biogeography of the Cardamine pratensis complex (Brassicaceae) Plant Systematics and Evolution. 2000;224:213–234. [Google Scholar]
- Gross BL, Rieseberg LH. The ecological genetics of homoploid hybrid speciation. Journal of Heredity. 2005;96:241–252. doi: 10.1093/jhered/esi026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Dai X, Zhao X. PLAN: a web platform for automating high-throughput BLAST searches and for managing and mining results. BMC Bioinformatics. 2007;8:53. doi: 10.1186/1471-2105-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytologist. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Current Biology. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Khasa D, Pollefeys P, Navarro-Quezada A, Perinet P, Bousquet J. Species-specific microsatellite markers to monitor gene flow between exotic poplars and their natural relatives in eastern North America. Molecular Ecology Notes. 2005;5:920–923. [Google Scholar]
- Kumar S, Skjæveland A, Orr RJS, et al. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt E. Über die Artengruppe der Cardamine pratensis L. in der Schweiz. Dissertationes Botanicae. 1984;72:481–497. [Google Scholar]
- Levin DA, Francisco-Ortega J, Jansen RK. Hybridization and the extinction of rare plant species. Conservation Biology. 1996;10:10–16. [Google Scholar]
- Lihová J, Marhold K. Phylogenetic and diversity patterns in Cardamine (Brassicaceae) – a genus with conspicuous polyploid and reticulate evolution. In: Sharma AK, Sharma A, editors. Plant genome: biodiversity and evolution. Vol. 1C: Phanerogams (Angiosperms – Dicotyledons) Enfield, New Hampshire: Science Publishers; 2006. pp. 149–186. [Google Scholar]
- Lihová J, Tribsch A, Marhold K. The Cardamine pratensis (Brassicaceae) group in the Iberian Peninsula: taxonomy, polyploidy and distribution. Taxon. 2003;52:783–802. [Google Scholar]
- Lihová J, Marhold K, Tribsch A, Stuessy TF. Morphological and AFLP re-evaluation of tetraploid Cardamine amara (Brassicaceae) in the Mediterranean. Systematic Botany. 2004a;29:134–146. [Google Scholar]
- Lihová J, Tribsch A, Stuessy TF. Cardamine apennina: a new endemic diploid species of the C. pratensis group (Brassicaceae) from Italy. Plant Systematics and Evolution. 2004b;245:69–92. [Google Scholar]
- Lihová J, Shimizu KK, Marhold K. Allopolyploid origin of Cardamine asarifolia (Brassicaceae): incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Molecular Phylogenetics and Evolution. 2006;39:759–786. doi: 10.1016/j.ympev.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Lihová J, Kučera J, Perný M, Marhold K. Hybridization between two polyploid Cardamine (Brassicaceae) species in northwestern Spain: discordance between morphological and genetic variation patterns. Annals of Botany. 2007;99:1083–1096. doi: 10.1093/aob/mcm056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövkvist B. The Cardamine pratensis complex. Outline of its cytogenetics and taxonomy. Symbolae Botanicae Upsaliensis. 1956;14/2:1–131. [Google Scholar]
- Lysak MA, Mandáková T. Analysis of plant meiotic chromosomes by chromosome painting. Methods in Molecular Biology. 2013;990:13–24. doi: 10.1007/978-1-62703-333-6_2. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends in Ecology and Evolution. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Kovařík A, Zozomová-Lihová J, et al. The more the merrier: recent hybridization and polyploidy in Cardamine. Plant Cell. 2013;25:3280–3295. doi: 10.1105/tpc.113.114405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold K. Taxonomic evaluation of the tetraploid populations of Cardamine amara (Brassicaceae) from the Eastern Alps and adjacent areas. Botanica Helvetica. 1999;109:67–84. [Google Scholar]
- Marhold K, Kudoh H, Pak JH, Watanabe K, Španiel S, Lihová J. Cytotype diversity and genome size variation in Eastern Asian polyploid Cardamine (Brassicaceae) species. Annals of Botany. 2010;105:249–264. doi: 10.1093/aob/mcp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer B, Jahncke P. RAPD analyses of hybridization events in Cardamine (Brassicaceae) Folia Geobotanica Phytotaxonomica. 1997;32:57–67. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Perný M, Tribsch A, Stuessy TF, Marhold K. Allopolyploid origin of Cardamine silana (Brassicaceae) from Calabria (Southern Italy): karyological, morphological and molecular evidence. Botanical Journal of the Linnean Society. 2005;148:101–116. [Google Scholar]
- Perrier C, Grandjean F, Le Gentil J, Cherbonnel C, Evanno G. A species-specific microsatellite marker to discriminate European Atlantic salmon, brown trout, and their hybrids. Conservation Genetics Resources. 2011;3:131–133. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. http://www.R-project.org/ (accessed 3 September 2013) [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology. 2004;4:137–138. [Google Scholar]
- Sampson JF, Byrne M. Genetic diversity and multiple origins of polyploid Atriplex nummularia Lindl. (Chenopodiaceae) Biological Journal of the Linnean Society. 2012;105:218–230. [Google Scholar]
- Schlüter PM, Harris SA. Analysis of multilocus fingerprinting data sets containing missing data. Molecular Ecology Notes. 2006;6:569–572. [Google Scholar]
- Shimizu-Inatsugi R, Lihová J, Iwanaga H, et al. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Molecular Ecology. 2009;18:4024–4048. doi: 10.1111/j.1365-294X.2009.04329.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annual Review of Plant Biology. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biological Journal of the Linnean Society. 2004;82:485–501. [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Symonds VV, Soltis PS, Soltis DE. Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution. 2010;64:1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- Urbanska-Worytkiewicz K. An autoallohexaploid in Cardamine L., new to the Swiss flora. Berichte des Geobotanischen Institutes der ETH, Stiftung Rübel. 1977a;44:86–103. [Google Scholar]
- Urbanska-Worytkiewicz K. Reproduction in natural triploid hybrids (2n=24) between Cardamine rivularis Schur and C. amara L. Berichte des Geobotanischen Institutes der ETH, Stiftung Rübel. 1977b;44:42–85. [Google Scholar]
- Urbanska-Worytkiewicz K. Reproductive strategies in a hybridogenous population of Cardamine L. Acta Oecologica/Oecologia Plantarum. 1980;1:137–150. [Google Scholar]
- Urbanska-Worytkiewicz K, Landolt E. Natürliche Bastarde zwischen Cardamine amara L. und C. rivularis Schur aus den Schweizer Alpen. Berichte des Geobotanischen Institutes der ETH, Stiftung Rübel. 1972;41:88–100. [Google Scholar]
- Urbanska KM, Landolt E. Patterns and processes of man-influenced hybridisation in Cardamine L. In: van Raammsdonk LWD, den Nijs JCM, editors. Plant evolution in man-made habitats. Amsterdam: Hugo de Vries Laboratory, Institute for Systematics and Population Biology, University of Amsterdam; 1998. pp. 29–47. [Google Scholar]
- Urbanska KM, Hurka H, Landolt E, Neuffer B, Mummenhoff K. Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, central Switzerland: biosystematic and molecular evidence. Plant Systematics and Evolution. 1997;204:233–256. [Google Scholar]
- Vähä J-P, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marin M. Mimulus peregrinus (Phrymaceae): a new British allopolyploid species. PhytoKeys. 2012;14:1–14. doi: 10.3897/phytokeys.14.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marin M, Lye GC. Hybridisation and genetic diversity in introduced Mimulus (Phrymaceae) Heredity. 2013;110:111–122. doi: 10.1038/hdy.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploidy speciation in vascular plants. Proceedings of the National Academy of Sciences of the USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli S. Einfluss der Bewirtschaftung auf die Entstehung und Struktur der Cardamine-Populationen auf dem Urnerboden. Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Rübel, Zürich. 1986;87:141–154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.