Abstract
Histone methyltransferases and demethylases reversibly modulate histone lysine methylation, which is considered a key epigenetic mark associated with gene regulation. Recently, aberrant regulation of gene expression by histone methylation modifiers has emerged as an important mechanism for tumorigenesis. However, it remains largely unknown how histone methyltransferases and demethylases co-regulate transcriptional profiles for cancer cell characteristics. Here, we show that in breast cancer cells, the histone H3 lysine 27 (H3K27) demethylase UTX (also known as KDM6A) positively regulates gene expression programs associated with cell proliferation and invasion. The majority of UTX-controlled genes, including a cohort of oncogenes and pro-metastatic genes, are co-regulated by the H3K4 methyltransferase mixed lineage leukemia 4 (MLL4, also called ALR, KMT2D, and MLL2). UTX interacted with a C-terminal region of MLL4. UTX knockdown resulted in significant decreases in the proliferation and invasiveness of breast cancer cells in vitro and in a mouse xenograft model. Such defective cellular characteristics of UTX-depleted cells were phenocopied by MLL4 knockdown cells. UTX-catalyzed demethylation of trimethylated H3K27 and MLL4-mediated trimethylation at H3K4 occurred inter-dependently at co-target genes of UTX and MLL4. Clinically, high levels of UTX or MLL4 were associated with poor prognosis in breast cancer patients. Taken together, these findings uncover that coordinated regulation of gene expression programs by a histone methyltransferase and a histone demethylase is coupled to the proliferation and invasion of breast cancer cells.
Introduction
Histone lysine methylation is central to epigenetic regulation of gene expression at the genome-wide level (1, 2). Among the genome-wide histone methylation marks that affect numerous genes are methylated histone H3 lysine 27 (H3K27) and H3K4. Methylated H3K27 is associated with gene repression, whereas methylated H3K4 is linked to gene activation or poised states (1, 3–6). Histone lysine methylation is present in mono-, di- or trimethylated states at specific lysine residues (7, 8) and is catalyzed by histone methyltransferases. For example, H3K4 methylation is generated by several specific H3K4 methyltransferases, such as mixed lineage leukemia (MLL) 1–5 and SET1A/B (3), while H3K27 methylation is catalyzed by EZH1- or EZH2-containing complexes. Histone lysine methylation can be reversed by histone lysine demethylases (KDMs) (9, 10). H3K4 methylation is removed by several H3K4 demethylases (e.g., LSD1/2 and JARID1a–d), while H3K27 methylation is demethylated by UTX (also known as KDM6A) and JMJD3 (also called KDM6B) (10).
Recent studies have shown that aberrations in histone lysine methylation may be clinically relevant to breast cancer, which is the most common malignancy in women. For example, it has been shown that global levels of trimethylated H3K27 (H3K27me3) are decreased in many breast tumors and that these altered levels correlate with poor prognosis (11). In addition, low H3K4me2 levels may be associated with poor patient survival (12).
It also has become evident that dysregulation of histone methylation modifiers may be important for breast cancer phenotypes. The H3K27 methyltransferase EZH2 is significantly overexpressed in breast tumors, and high levels of EZH2 have been shown to be associated with poor prognoses of breast cancer, including inflammatory breast cancer (13, 14). In addition, EZH2 expression in benign breast samples may be linked to high risk for breast cancer (15, 16). Mechanistically, EZH2 may promote cancer progression by repressing two key cellular senescence genes, p14ARF and p16INK4A (17), and may increase cancer cell invasion by transcriptionally repressing the metastasis suppressor RKIP (18). Recent studies have indicated that the oncogenic function of EZH2 may be antagonized by the H3K27 demethylase JMJD3, which de-represses p14ARF and p16INK4A genes (19). Interestingly, it has been shown that the histone H3K9 and H3K36 demethylase JMJD2C (alias GASC1) is a predictive marker in invasive breast cancer and is gene-amplified in breast cancer samples (20, 21).
The gene encoding the H3K27me3 demethylase UTX often undergoes somatic loss-of-function mutations in multiple cancer types, such as medulloblastoma, renal carcinoma, bladder cancer, leukemia, and prostate tumors [reviewed in (22)]. In addition, it has been shown that in normal fibroblast cells, UTX transcriptionally activates Rb pathway genes to suppress cell growth (23). Therefore, UTX has been thought to be a tumor suppressor in these types of tumors. In contrast, it has been reported that in breast tumors, UTX may be overexpressed (24) and is rarely mutated (25). To better understand the role of UTX in breast cancer, we sought to determine 1) the effect of UTX on breast cancer cell behaviors, such as cell proliferation and invasion; 2) which transcriptional programs are modulated by UTX; and 3) how UTX regulates transcriptional programs in breast cancer cells.
In the present study, we found that UTX contributes to cellular proliferation and invasiveness by activating oncogenic gene expression programs in breast cancer cells. Expression of most UTX-modulated genes was also regulated by the H3K4 methyltransferase MLL4 (also called ALR, KMT2D, and MLL2) of which C-terminal region interacts with UTX. UTX-depleted and MLL4-depleted cells had similar defects in the proliferation and invasion of breast cancer cells. Moreover, our results indicate that UTX and MLL4 upregulate gene expression by decreasing H3K27me3 levels and in parallel increasing H3K4me3 levels at their co-target genes in a coordinated manner. These findings suggest that UTX and MLL4 cooperatively regulate gene expression programs for cell proliferation and invasiveness in breast cancer cells.
Materials and Methods
Cell culture
The HMLE, MCF10A, MCF-7, MDA-MB-231, MDA-MB-468, BT-474, BT-549, SUM-159, Hs578T, T-47D, 293T, and HeLa cells were from American Type Culture Collection (ATCC; Manassas, VA, USA) and were maintained in low passage (< 15) for this study. ATCC validates cell lines by short tandem repeat profiles that are generated by simultaneous amplification of multiple short tandem repeat loci and amelogenin (for gender identification). The cell lines, with the exception of HMLE and MCF10A, were cultured in Dulbecco’s modified eagle’s medium (DMEM; Thermo Scientific, Waltham, MA, USA) containing 10% FBS and penicillin and streptomycin (Mediatech, Manassas, VA, USA) at 37°C in a humidified atmosphere of 5% CO2. HMLE and MCF10A cell lines were cultured in defined mammary epithelial growth media (CC-2571, MEGM Bullet Kit, Lonza, Inc., Basel, Switzerland) supplemented with bovine pituitary extracts and epidermal growth factor.
Plasmids, reagents, and antibodies
The cDNA construct encoding a C-terminus (Flag-MLL4-C) of MLL-4 (GenBank protein accession no. NP_003473.3) and its deletion constructs [i.e., Flag-MLL4-C (N), and Flag-MLL4-C (C)] were cloned into the expression vector pFLAG-CMV2. The shUTX6-resistant UTX and shUTX6-resistant UTX catalytic mutant (mUTX) were generated using pFLAG-CMV2-UTX (26) (Supplementary Table S1). The anti-UTX antibody was previously described (26). Anti-MLL4/MLL2-specific antibody (AP6183a) was from Abgent, SuZhou, China (or San Diego, CA, USA). Anti-Flag mouse monoclonal M2 (F3165) was from Sigma. Anti-RbBP5 (A300-109A) and anti-ASH2L (A300-107A) polyclonal antibodies were from Bethyl Laboratories, Montgomery, TX, USA. Anti-WDR5 polyclonal antibody (07-706) was from Millipore, Billerica, MA, USA.
Cell invasion and proliferation assays
For the cell invasion assay, the Boyden chamber assay with a modification was performed. Cells (1 × 105) in 200 μl of serum free media containing 0.1% bovine serum albumin (BSA) were seeded on the Matrigel-precoated membrane in the inserts. After 48 hours, cells that had invaded the Matrigel and migrated to the other side of the membrane were fixed and stained. Cell numbers were counted or the relative intensities of stained cells were quantified using ImageJ 1.46r. For cell proliferation assay, cells were seeded in triplicate at a density of 2 × 104 cells per well in a 12 well plate and were counted for 6 days at a 2-day interval. For all the assays, cells were infected with lentiviruses containing small hairpin (sh) UTX-6, and shUTX-8, shMLL4-15, shMLL4-18, or shLuciferase (shLuc) (Supplementary Table S1).
Soft agar assays
For soft agar assays, MDA-MB-231 cells (1 × 103) were infected with lentiviruses containing shUTX-6, shUTX-8 or shLuc, resuspended in 1 ml of 0.35% (w/v) agar with DMEM containing 10% FBS, and overlaid onto a 0.5% (w/v) agar solution in a well of a 6 well plate. Colonies were visible after an incubation of 10–14 days and were stained with crystal violet.
Mouse xenograft Experiments
Female athymic nu/nu mice were purchased from The University of Texas MD Anderson Cancer Center. The care and use of all mice were approved by the Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center. Luciferase-expressing MDA-MB-231 cells (2 × 106) were treated with Scrambled shRNA (shScrambled) or shUTX-6 for 72 hours. Each group of cells were injected into mice (n = 7) via the tail veins. The transplanted animals were in vivo monitored using an IVIS100 imaging system every two weeks. After bioluminescence images were captured, their intensities were quantitated using Living Image 3.2 acquisition and analysis software (Caliper Life Sciences, Hopkinton, MA). At the end of 8 weeks, lungs were collected from mouse and processed for hematoxylin and eosin staining.
Immunoprecipitation
pFlag-CMV2 plasmids harboring MLL4-C, MLL4-C (N), or MLL4-C (C) cDNAs were transiently transfected into human embryonic kidney 293T cells. Immunoprecipitation using anti-Flag resin was performed as previously described (26).
Gene expression microarray analysis
RNA was extracted from virus-infected MDA-MB-231 cells using micro RNeasy kit (Qiagen, Hilden, Germany), and labeled cRNAs were hybridized to an oligonucleotide microarray consisting of 54,675 gene probes (Affymetrix U133P Plus 2.0 GeneChip; Santa Clara, CA, USA). RMAexpress was used to compute gene expression summary values. The web-based Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov) (27) was used to analyze gene sets that were down- or upregulated (≥ 2 fold change) in the microarray.
RNA isolation, cDNA synthesis, and quantitative reverse transcription (RT)–PCR
Total RNAs were isolated using RNeasy kits (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions and amplified with iQ SYBR Green Supermix (Bio-Rad) using the CFX384 real-time PCR detection system (Bio-Rad). Expression levels were quantified using CFX Manager software (Bio-Rad) and normalized to 18s RNA. The relative mRNA levels represent the fold changes over the control.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the Millipore ChIP protocol with minor modifications as described previously (28). Chromatin immunoprecipitates for proteins and methyl marks were amplified by quantitative PCR, normalized to input, and calculated as percentages of inputs. Fold enrichment levels indicate the fold changes over the negative control immunoglobulin G (IgG). The PCR primer sequences are described in Supplementary Table S1.
Analysis of expression levels, survival and correlation
The overall survival and correlation analysis were performed using the dataset from The Cancer Genome Atlas (TCGA) database. For expression analysis, Oncomine and TCGA databases, which represents a unifying microarray database resource with an integrated data-searching tool (https://www.oncomine.org), was used.
Statistical analysis of Data
The statistical significance between two groups was analyzed by Student’s t-test. Survival rate and correlation analysis were carried out by the Logrank test and Pearson’s correlation test in Prism 5 software (GraphPad Software Inc., La Jolla, CA), respectively. Data are presented as the mean ± SEM (error bars). P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) denote statistically significant changes.
Results
UTX is critical for the proliferation and invasiveness of breast cancer cells
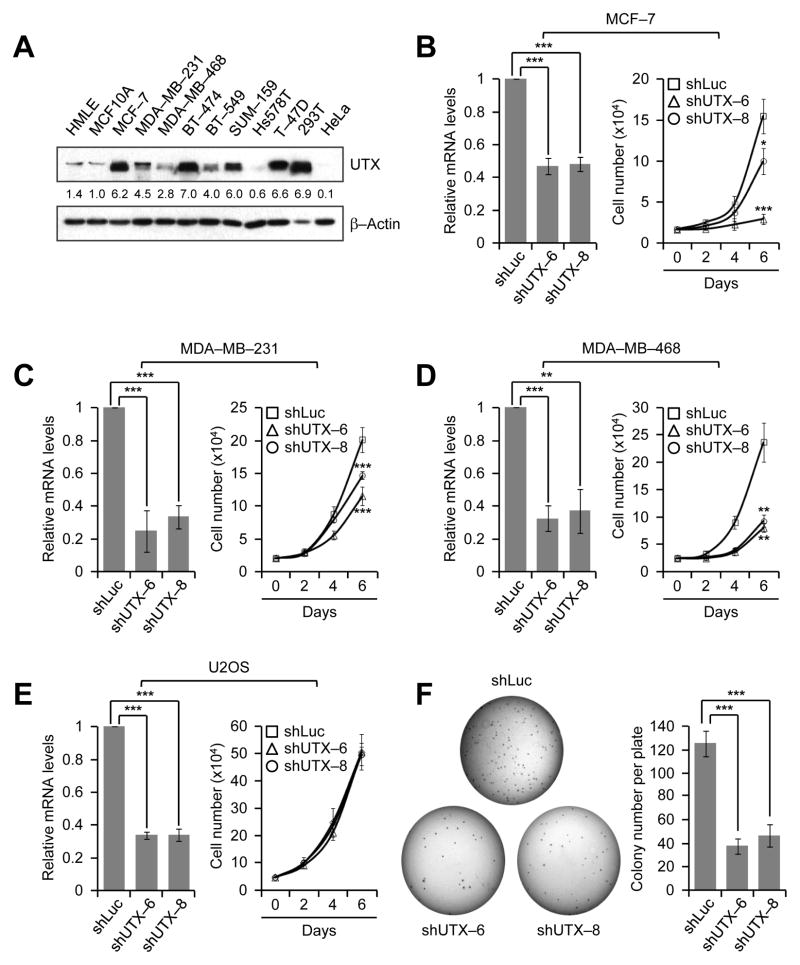
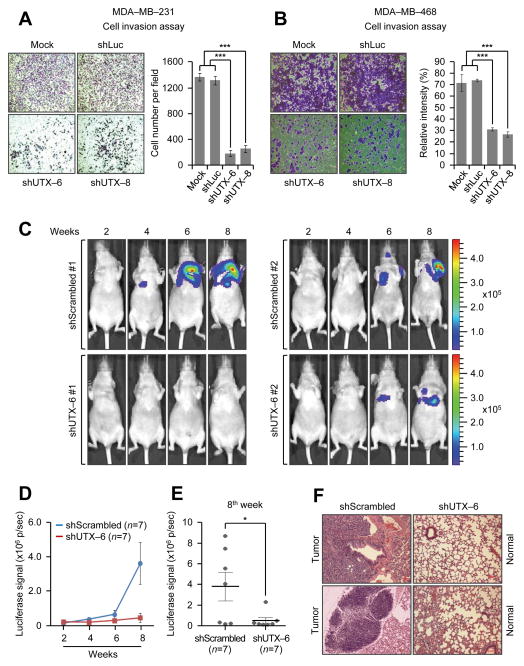
In an effort in determining the role of UTX in breast cancer, we first assessed UTX levels in two non-transformed mammary epithelial cell lines (MCF10A and HMLE) and several breast cancer cell lines. UTX protein levels were generally higher in breast cancer cell lines than in the normal cell lines (Fig. 1A). Next, to examine whether UTX is critical for breast cancer cell proliferation, we depleted UTX in the estrogen receptor (ER)-positive cell line MCF-7 and two triple-negative (ER/PR/HER2 negative) breast cancer cell lines MDA-MB-231 and MDA-MB-468 using small hairpin (sh) RNAs. UTX knockdown substantially reduced the proliferation of MCF-7, MDA-MB-231, and MDA-MB-468 cells regardless of ER status of breast cancer cell lines (Fig. 1 B–D). In contrast, the cell proliferation rate of the non-breast cancer cell line U2OS (a human osteosarcoma cell line) was not affected by UTX depletion (Fig. 1E). Previously, it has been shown that knockdown of UTX in normal fibroblasts increased cell proliferation (23). This previous report and our results suggest that UTX may regulate cell proliferation in a cell type-specific manner. Consistent with the inhibitory effect of UTX knockdown on the proliferation of MDA-MB-231 cells, UTX knockdown also decreased colony formation abilities of MDA-MB-231 cells in a soft agar assay (Fig. 1F). Furthermore, we determined the effect of UTX knockdown on the invasive abilities of two metastatic cell lines MDA-MB-231 and MDA-MB-468. UTX depletion remarkably inhibited the in vitro invasive abilities of both cell lines (Fig. 2A and B), although it did not have any significant effect on the migration capacities of MDA-MB-231 cells (Supplementary Fig. S1A). These results indicate that UTX is important for the proliferation, anchorage-independent growth, and invasion of breast cancer cells, such as MDA-MB-231 cells.
Figure 1. UTX knockdown attenuates the proliferation and colony formation abilities of breast cancer cells.
A, UTX levels in normal and breast cancer cell lines. UTX protein levels were examined by Western blot analysis. For comparison, the breast-unrelated cell lines HEK293T and HeLa were included. B–E, Effect of UTX knockdown on the proliferation of MCF-7 (B), MDA-MB-231 (C), MDA-MB-468 (D), and U2OS (E) cells. Cells were treated with control shRNA (shLuc) and two shRNAs against UTX. Knockdown efficiencies by shRNAs were examined by quantitative RT-PCR (Left panels). Cell numbers were counted at the indicated time points (Right panels). Data are presented as the mean ± SD (error bars) of at least three independent experiments (B, n = 4; C, n = 3; D, n = 3; E, n = 3). F, The inhibitory effect of UTX knockdown on colony formation of MDA-MB-231 cells in soft agar. Data are presented as the mean ± SEM (error bars) of five independent experiments.
Figure 2. UTX depletion suppresses in vitro and in vivo invasive abilities of breast cancer cells.
A and B, The effect of UTX knockdown on the invasiveness of MDA-MB-231 (A) and MDA-MB-468 cells (B). For invasion assay, six different microscopic fields (4 x magnification) from at least three independent experiments were examined: cells in the fields were counted (A, n ≥ 3) or relative intensities of the fields were measured (B, n ≥ 3). C–F, Effect of UTX knockdown on cellular invasiveness and tumor growth in a mouse xenograft model based on tail vein injection. Luciferase-expressing MDA-MB-231 (MDA-MB-231-Luc) cells were treated with viruses containing shScrambled (control shRNA) or shUTX-6. Then, shScrambled-treated or UTX-depleted cells were injected into mice via tail veins, and tumor colonization in the lung was monitored using an IVIS-100 imaging system. Representative bioluminescent images of mice harboring tumors are shown in (C). The luciferase signals were compared between the shScrambled group (n = 7) and the shUTX-6 group (n = 7) (D). In addition, the luciferase signals for both groups at the 8th week after tail vein injection were individually plotted (E). Metastatic lesions in lungs from mice at the 8th week were analyzed by hematoxylin and eosin staining (F). Data are presented as the mean ± SEM (error bars).
Cancer cell metastasis involves intravasation (entrance into blood systems), extravasation (exit from the blood vessels), and colonization in a distant organ (29). For these processes, the invasive and proliferative abilities of cancer cells are essential. To further assess the role of UTX in the invasion and tumor growth of breast cancer cells, we treated luciferase-expressing MDA-MB-231 (MDA-MB-231-Luc) cells with shScrambled (control)- or shUTX-6-containing viruses and subsequently injected these two groups of cells into mice via tail veins. In vivo imaging analysis demonstrated that mice (n = 7) injected with shScrambled-treated MDA-MB-231-Luc cells had stronger luciferase signals than those (n = 7) injected with shUTX-treated MDA-MB-231-Luc cells (Fig. 2C and D). In particular, luciferase signals between the shScrambled group and the shUTX-6 group were remarkably different at the 8th week (Fig. 2E). Tumors were confirmed by hematoxylin and eosin staining (Fig. 2F). Because the mouse xenograft models based on tail vein injection generally involve both cancer cell invasion during extravasation and tumor growth in a distant organ (primarily, lung), these results suggest that UTX is required for the in vivo invasiveness and tumor formation capacities of MDA-MB-231 cells.
UTX’s demethylation activity contributes to the proliferative and invasive abilities of breast cancer cells
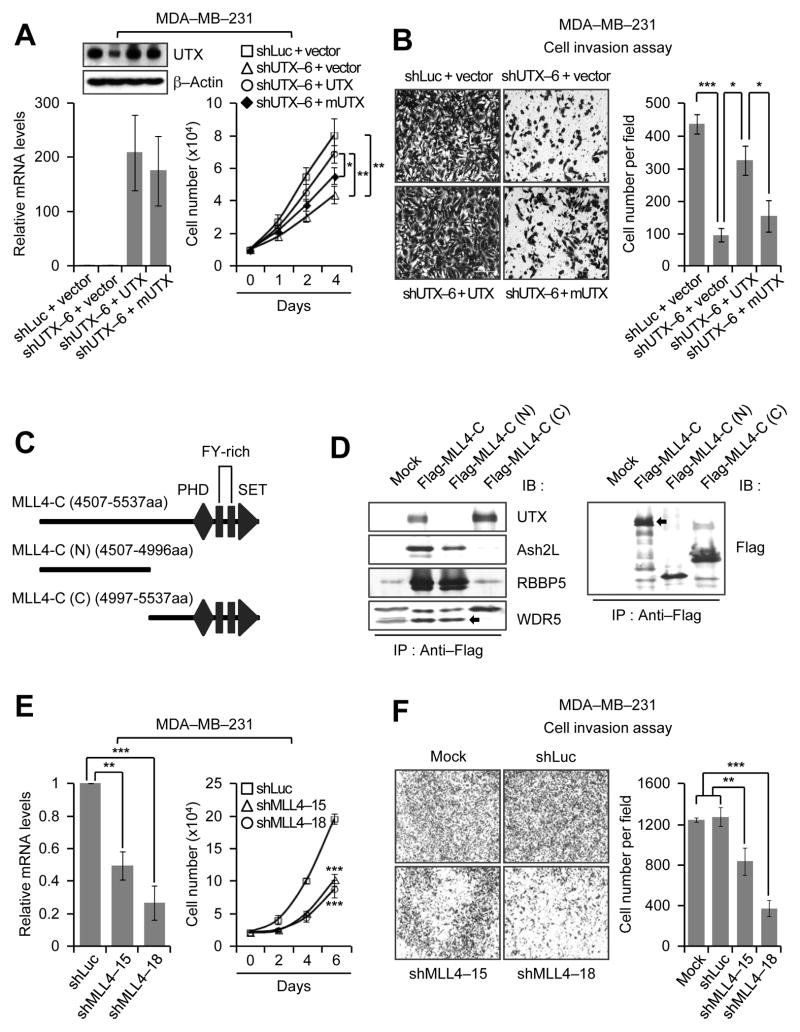
It has been reported that UTX’s demethylase activity is not required for T-box family member-dependent gene expression (30). To examine whether the demethylation activity of UTX is necessary for the phenotypic defects of breast cancer cells and whether defective phenotypes of UTX knockdown cells result from specific knockdown of UTX, we ectopically expressed GFP, shUTX6-resistant UTX, and shUTX6-resistant UTX catalytic mutant (mUTX) in UTX-depleted MDA-MB-231 cells (Fig. 3A, left panel; ). As shown in Fig. 3A and B, wild-type UTX significantly rescued defective proliferation and invasiveness of UTX-depleted MDA-MB-231 cells, whereas the catalytic mutant mUTX weakly or insignificantly restored such cellular phenotypes. These results indicate that UTX and UTX’s catalytic activity are needed for the proliferation and invasion of breast cancer cells, such as MDA-MB-231 cells.
Figure 3. UTX’s demethylase activity is important for cell proliferation and invasion, and MLL4 knockdown impedes these cellular properties.
A and B, The rescue experiments of defective proliferation and invasiveness of UTX-depleted cells. Wild type UTX and its catalytic mutant mUTX were ectopically expressed in UTX-depleted MDA-MB-231 cells. The mRNA and protein levels of UTX and mUTX were assessed by quantitative RT-PCR and Western blot analysis, respectively (A, left panels). For cell proliferation assays, cell numbers were counted at the indicated time points (A, right panel). For invasion assays, cells in the fields (10 x magnification) from three independent experiments were counted (B). The shLuc virus-treated cells that were transfected with the empty vector were used as controls. C, Schematic representation of a C-terminal region of MLL4. PHD, plant homeodomain; SET, the catalytic domain. D, Mapping of an MLL4’s C-terminal region responsible for the interaction with UTX. Flag-tagged MLL4-C, MLL4-C (N) and MLL4-C (C) were expressed in 293T cells. Whole cell extracts were immunoprecipitated with anti-Flag antibody, followed by Western blot analysis with anti-UTX, anti-ASH2L, anti-RBBP5 and anti-WDR5 antibodies. E, Negative effect of MLL4 knockdown on MDA-MB-231 cell proliferation. Knockdown efficiency was assessed by quantitative RT-PCR (left panel). Cells were counted at the indicated time points (right panel). F, Inhibitory effect of MLL4 knockdown on the invasiveness of MDA-MB-231 cells (4 x magnification). Data are presented as the mean ± SEM (error bars) of three independent experiments.
UTX interacts with a C-terminal region of MLL4, and MLL4 is required for the proliferation and invasiveness of breast cancer cells
The H3K27me3 demethylase UTX is present in multi-protein complexes with the H3K4 methyltransferase MLL4 (26, 31, 32). We previously showed that a C-terminus of MLL4 (MLL4-C; 4507–5537aa) is responsible for MLL4’s interaction with the core-subunits (e.g., ASH2L, RBBP5 and WDR5) in the MLL4 complex (28). To examine whether MLL4-C also mediates the association between UTX and MLL4, we transfected an expression plasmid encoding Flag-tagged MLL4-C into human embryonic kidney 293T cells and purified Flag-tagged MLL4-C using anti-Flag immunoprecipitation. Western blot analysis of immunoprecipitation eluates showed that MLL4-C physically interacted with UTX (Fig. 3C and D). To further map what region of MLL4-C is responsible for the association with UTX, we generated the N-terminal region of MLL4-C [MLL-C (N), 4507–4996aa] and the C-terminal region of MLL4-C [MLL-C (C), 4997–5537aa]. Immunoprecipitation experiments demonstrated that UTX associated with the C-terminal region of MLL4-C and that ASH2L, RBBP5 and WDR5 interacted with the N-terminal region of MLL4-C (Fig. 3C and D). These results not only confirm the association of UTX and MLL4, but also indicate that UTX interacts with MLL4 via the C-terminal region of MLL4.
Because MLL4 interacts with UTX, we sought to determine whether MLL4 has an effect on cell growth and invasion like UTX. We depleted MLL4 in MDA-MB-231 cells using shRNAs (Fig. 3E, left panel) and then performed cell proliferation and invasion assays. MLL4 depletion reduced the proliferation of MDA-MB-231 cells (Fig. 3E, right panel) and also impeded in vitro invasive abilities of MDA-MB-231 cells (Fig. 3F). Of interest, MLL4 knockdown did have a moderate effect on the migration of MDA-MB-231 cells (Supplementary Fig. S1B). These results indicate that similar to UTX, MLL4 is necessary for the proliferation and invasion of breast cancer cells.
Most UTX-regulated genes are co-modulated by MLL4
Because UTX-depleted and MLL4-depleted cells shared similar defects in cell proliferation and invasiveness and because UTX physically interacts with MLL4, we reasoned that the transcriptional co-activators, UTX and MLL4, may regulate a similar set of genes. For this, we assessed the effect of UTX or MLL4 knockdown on gene expression profiles in MDA-MB-231 cells. Specifically, we compared the mRNA expression levels between UTX (or MLL4)-depleted cells and control (shLuc-treated) cells using whole genome mRNA expression analysis.
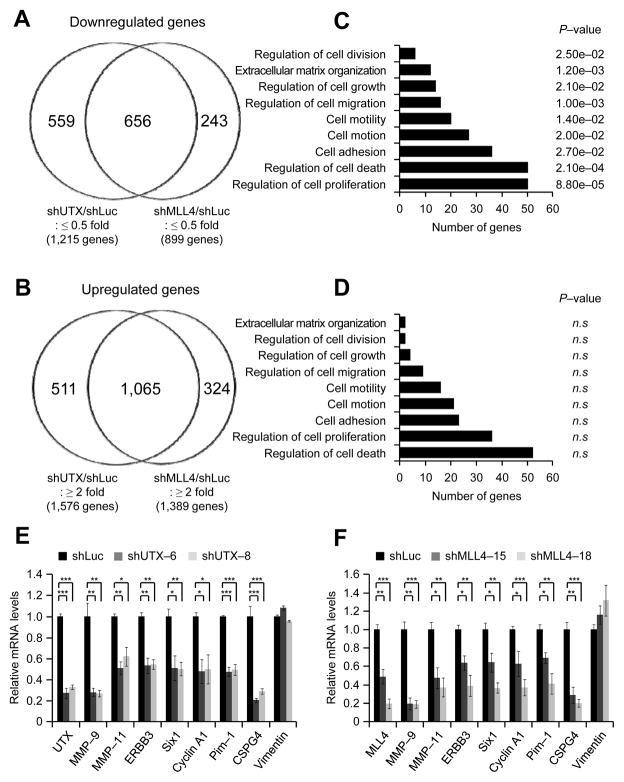
In particular, we analyzed genes that were at least 2-fold down- or upregulated by UTX or MLL4 knockdown in the microarray assays. Remarkably, 54% of shUTX-downregulated genes (656/1,215) overlapped with 73% of shMLL4-downreglated genes (656/899) (Fig. 4A), and 68% of shUTX-upregulated genes (1,065/1,576) were identical with 77% of shMLL4-upregulated genes (1,065/1,389) (Fig. 4B). These results indicate that most UTX-controlled genes are co-regulated by MLL4.
Figure 4. UTX and MLL4 positively co-regulate gene expression programs related to cell proliferation and invasion.
A and B, Venn diagrams for genes that are downregulated (A) or upregulated (B) by UTX and MLL4 knockdown. MDA-MB-231 cells were treated with viruses containing shLuciferase, shUTX-6 or shMLL4-18. The mRNA levels in UTX- or MLL4-depleted cells were measured by the expression microarray Affymetrix U133P and compared with those in the control cells. C and D, Gene ontology analysis of genes that are co-downregulated (C) or co-upregulated (D) by UTX and MLL4 knockdown. Gene groups that are related only to cell proliferation and invasiveness are shown. Gene ontology analysis was performed using the program DAVID. n.s, not significant. E and F, Quantitative RT-PCR analysis of several genes that are co-downregulated by UTX (E) and MLL4 (F) knockdown in MDA-MB-231 cells. Data are presented as the mean ± SEM (error bars) of at least three independent experiments. *, P<0.05; **, P<0.01; ***, P<0.001.
UTX and MLL4 co-regulate transcriptional programs that are associated with cell proliferation and invasiveness
As described above, knockdown of UTX or MLL4 inhibited cell proliferation and invasiveness. Therefore, we carried out gene ontology analysis of our whole genome mRNA expression data to assess whether UTX and MLL4 co-regulate gene sets that are associated with such knockdown phenotypes. Of interest, a cohort of cell proliferation-associated genes and pro-invasiveness genes were significantly downregulated by UTX and MLL4 depletion (Fig. 4C; See Supplementary Table S2 for each category of genes). In contrast, UTX and MLL4 knockdown did not have any significant effect on the upregulation of these categories of genes (Fig. 4D). These results indicate that UTX and MLL4 contribute to breast cancer cell proliferation and invasion by activating cell proliferation-associated genes and pro-invasiveness genes.
To confirm our whole genome mRNA expression analysis, we individually analyzed expression levels of several cell proliferation-associated genes and pro-invasiveness genes, including MMP-9, MMP-11, ERBB3, Six1, Cyclin A1, Pim-1, and CSPG4, using quantitative RT-PCR. MMP-9 and MMP-11 have been known to promote cell invasion (33). ERBB3, Six1, Cyclin A1, Pim-1, and CSPG4 are involved in tumorigenesis by promoting cell proliferation and invasion (33–37). Consistent with whole genome expression results, quantitative RT-PCR data demonstrated that in MDA-MB-231 cells, these genes were down-regulated by UTX or MLL4 knockdown (Fig. 4E and F). Similarly, UTX or MLL4 depletion also decreased expression levels of MMP-9, MMP-11, Six1, Cyclin A1, and CSPG4 in MDA-MB-468 cells (Supplementary Fig. S2A and B). Interestingly, although UTX was shown to regulate Rb pathway genes in mouse embryonic fibroblast cells (23), our quantitative RT-PCR and microarray results showed that Rb pathway genes (e.g., Rb and Rbbp4–6) in MDA-MB-231 cells were mostly unaffected by UTX knockdown (Supplementary Fig. S3; data not shown).
UTX-mediated demethylation of trimethylated H3K27 is linked to MLL4-catalyzed H3K4 trimethylation at co-target genes of UTX and MLL4
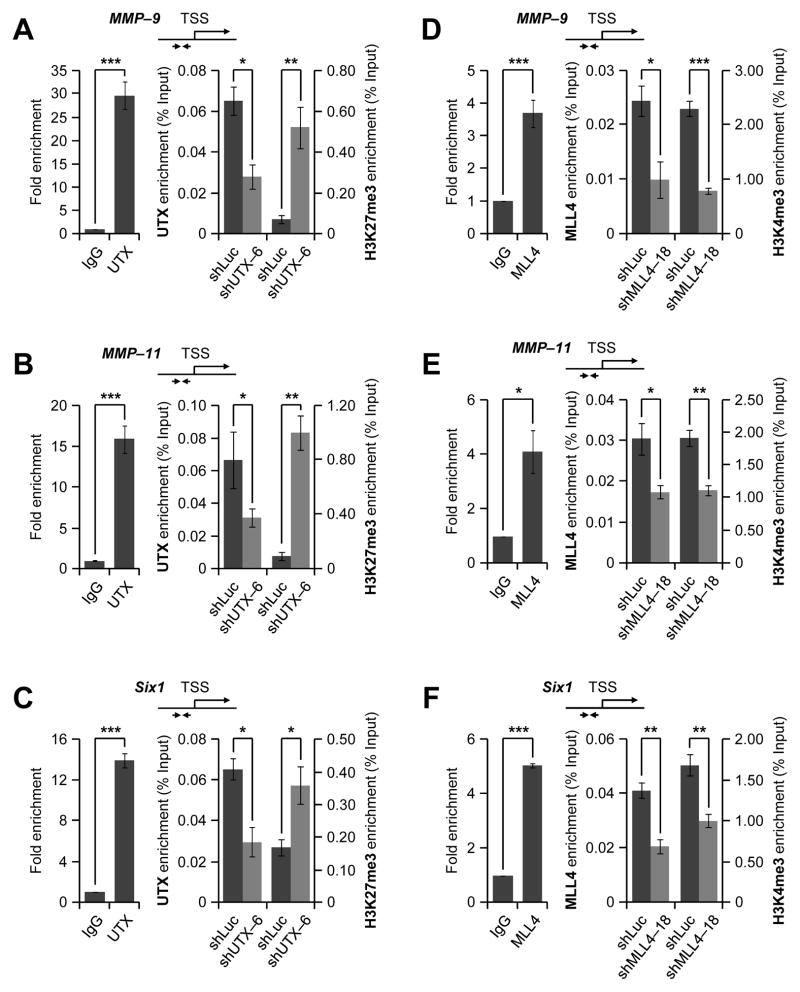
Co-target genes of UTX and MLL4 are likely down-regulated by UTX or MLL4 knockdown, because UTX and MLL4 generally act as transcriptional co-activators. Therefore, we examined whether UTX and MLL4 are co-recruited to genes (e.g., MMP-9, MMP-11, and Six1) that were downregulated by UTX and MLL4 knockdown. Quantitative ChIP results showed that UTX and MLL4 co-occupied the promoters of the MMP-9, MMP-11, and Six1 genes (Fig. 5A–F, left panels). In particular, MLL4 occupancy at these genes was confirmed using two different antibodies (Supplementary Fig. S4).
Figure 5. UTX and MLL4 are co-recruited to MMP-9, MMP-11, and Six1 genes.
A–C, The effect of UTX knockdown on chromatin levels of UTX and H3K27me3 at the proximal promoters of MMP-9 (A), MMP-11 (B), and Six1 (C) in MDA-MB-231 cells. Fold enrichments of UTX over the negative control IgG were measured by quantitative ChIP (left panels). In addition, chromatin levels of UTX and H3K27me3 were compared between shLuc-treated (control) cells and UTX-depleted cells using ChIP (right panels). D–F, The effect of MLL4 knockdown on chromatin levels of MLL4 and H3K4me3 at the proximal promoters of MMP-9 (D), MMP-11 (E), and Six1 (F) in MDA-MB-231 cells. Fold enrichments of MLL4 over IgG were measured by quantitative ChIP (left panels). In addition, chromatin levels of MLL4 and H3K4me3 were compared between shLuc-treated (control) cells and MLL4-depleted cells using ChIP (right panels). Data are presented as the mean ± SEM (error bars) of three independent experiments (n=3). *, P<0.05; **, P<0.01; ***, P<0.001.
UTX has been shown to remove methyl groups from the repressive mark H3K27me3 at the proximal promoters near the transcription start sites (26). To determine whether UTX demethylates H3K27me3 at the proximal promoters of the MMP-9, MMP-11, and Six1 genes, we examined the effect of UTX knockdown on H3K27me3 levels at the promoters of these genes. Quantitative ChIP results showed that UTX depletion increased H3K27me3 levels at the proximal promoters of the MMP-9, MMP-11, and Six1 genes (Fig. 5A–C, right panels). These results indicate that expression of the MMP-9, MMP-11, and Six1 genes is positively regulated by UTX-catalyzed demethylation of H3K27me3 at their proximal promoters.
We also assessed the effect of MLL4 knockdown on H3K4me3 levels at the proximal promoters of the MMP-9, MMP-11, and Six1 genes. Our ChIP assays showed that MLL4 depletion reduced H3K4me3 levels at the promoters of these genes. These results indicate that MLL4 may upregulate the MMP-9, MMP-11, and Six1 genes by enzymatically generating H3K4me3 at their proximal promoter regions (Fig. 5D–F, right panels).
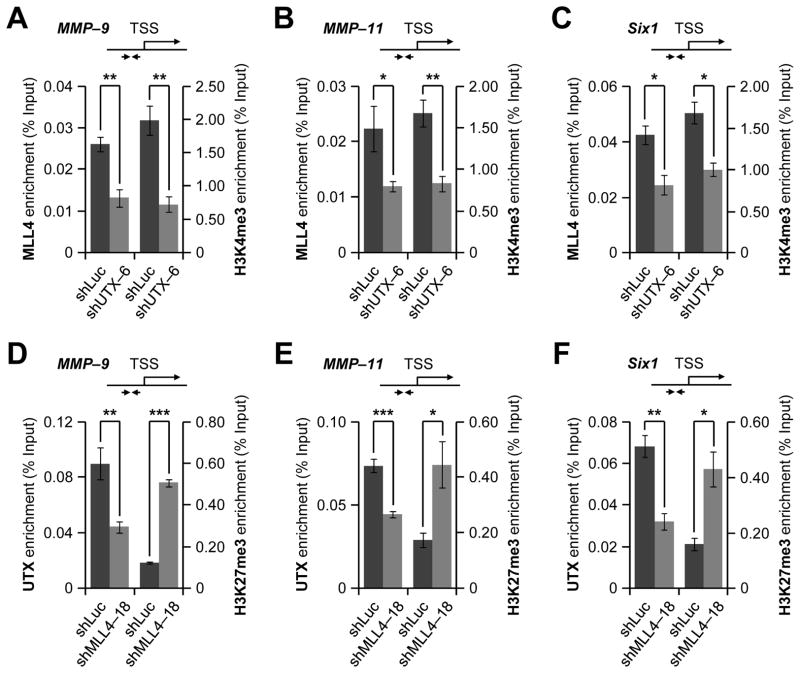
To determine whether UTX affects MLL4-catalyzed H3K4 methylation at the MMP-9, MMP-11, and Six1 genes, we examined the effect of UTX knockdown on H3K4me3 and MLL4 levels at the proximal promoters of the MMP-9, MMP-11, and Six1 genes. Our ChIP experiments demonstrated that UTX knockdown decreased MLL4 and H3K4me3 levels on these promoters (Fig. 6A–C). We also assessed whether MLL4 has any effect on UTX-catalyzed demethylation of H3K27me3 at the MMP-9, MMP-11, and Six1 genes. As shown in Fig. 6D–F, MLL4 knockdown decreased UTX levels while increasing H3K27me3 levels at these genes. Furthermore, we determined whether UTX (or MLL4) knockdown affects total cellular levels of H3K4me3, H3K27me3, MLL4 (UTX in case of MLL4 knockdown). UTX or MLL4 knockdown did not result in obvious changes in total cellular levels of H3K4me3 and H3K27me3 (Supplementary Fig. S5). Interestingly, MLL4 knockdown appeared to slightly down-regulate cellular UTX mRNA and protein levels (Supplementary Fig. S6A), although UTX knockdown did not change MLL4 mRNA levels (Supplementary Fig. S6B). Therefore, decreased levels of UTX at MMP-9, MMP-11, and Six1 genes by MLL4 knockdown may be at least in part due to transcriptional regulation of UTX expression by MLL4. In contrast, decreased chromatin levels of MLL4 by UTX depletion might result from reduced formation of the UTX-MLL4 complex due to diminished UTX levels, although we cannot exclude the possibility that UTX knockdown may affect MLL4’s protein stability. Nevertheless, these results indicate that UTX and MLL4 occupancy at their co-target genes may be inter-connected and that UTX-catalyzed H3K27me3 demethylation parallels MLL4-mediated H3K4 trimethylation in a gene-specific manner.
Figure 6. UTX-catalyzed H3K27me3 demethylation and MLL4-catalyzed H3K4 methylation occurs in a coordinate manner.
A–C, The effect of UTX knockdown on chromatin levels of MLL4 and H3K4me3 at the proximal promoters of MMP-9 (A), MMP-11 (B), and Six1 (C) in MDA-MB-231 cells. ChIP assays were carried out with anti-MLL4 and anti-H3K4me3 after UTX knockdown. D–F, The effect of MLL4 knockdown on chromatin levels of UTX and H3K27me3 at the proximal promoters of MMP-9 (D), MMP-11 (E), and Six1 (F) in MDA-MB-231 cells. ChIP assays were performed with anti-UTX and anti-H3K27me3 after MLL4 knockdown. Data are presented as the mean ± SEM (error bars) of three independent experiments. *, P<0.05; **, P<0.01; ***, P<0.001.
High mRNA levels of UTX and MLL4 are associated with poor prognosis in breast cancer patients
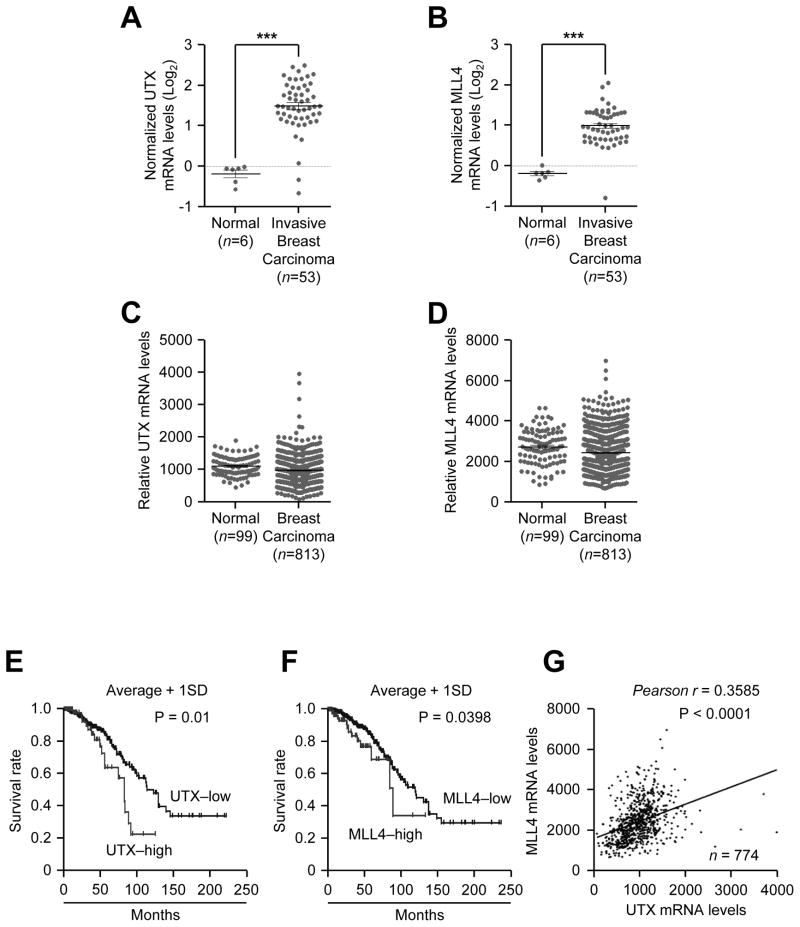
As mentioned earlier, UTX mRNA levels have been reported to be overexpressed in breast cancer samples (24). Thus, we determined whether both UTX and MLL4 mRNA levels are upregulated in breast tumors as compared to normal breast tissues using the following two publicly available databases: Oncomine and TCGA. Analysis of Oncomine database showed that although their average levels were not increased in breast cancer samples in most datasets (Data not shown), average UTX and MLL4 levels were higher in the invasive subtypes than in normal breast tissues in one study set (34) (Fig. 7A and B). In the TCGA database, there were no significant differences in average UTX and MLL4 levels between normal breast tissues and all breast tumor types combined, although their levels in some breast tumor samples were highly increased as compared to their normal average levels (Fig. 7C and D).
Figure 7. High mRNA levels of UTX and MLL4 correlate with poor survival in breast cancer patients.
A–D, Comparison of UTX (A and C) and MLL4 (B and D) mRNA levels between normal breast tissues and breast carcinoma. Oncomine (A and B) and The Cancer Genome Atlas (TCGA) (C and D) database were used. E and F, Survival analysis of breast cancer patients on the basis of UTX (E) and MLL4 (F) mRNA levels. The publicly available dataset from TCGA (774 human breast tumors) was analyzed. High UTX and high MLL4 denotes patients with mRNA levels that are greater than or equal to average plus one standard deviation (UTX-low, n = 683; UTX-high, n = 91; MLL4-low, n = 649; MLL4-high, n = 125). G, Correlation analysis between UTX and MLL4 mRNA levels in the TCGA dataset.
Next, we performed the Kaplan-Meier survival analysis to assess whether high levels of UTX and MLL4 in breast tumors are linked to the clinical outcomes of breast cancer patients. The survival analysis using the TCGA data with patient history demonstrated that high mRNA levels of UTX and MLL4 were linked to shorter survival in breast cancer patients (Fig. 7E and F). In addition, we found that UTX mRNA levels had a significant positive correlation with MLL4 mRNA levels in breast cancer patients (Fig. 7G). These results indicate that high UTX and MLL4 levels may contribute to poor prognosis in breast cancer patients.
Discussion
UTX plays an important role in regulating multiple cellular functions, including somatic cell reprogramming (35), germ cell development (35), embryo development in a sex-specific manner (36, 37), cardiac development (38), aging (39), and myogenesis (40). Results reported here showed that UTX knockdown suppressed cellular invasive and proliferative abilities of breast cancer cells, implicating a role for UTX in cell invasion and tumor growth. In line with this, intravenous mouse xenograft experiments in this study indicated that UTX may be required for in vivo invasion and tumor growth of breast cancer cells. Mechanistically, UTX activates gene expression programs associated with cell proliferation and invasiveness. Therefore, our findings reveal a novel UTX function in which UTX positively regulates breast cancer cell proliferation and invasiveness by transcriptionally activating the relevant gene expression programs.
Our results in the present study also showed that similar to UTX knockdown, MLL4 depletion inhibited the proliferative and invasive capacities of breast cancer cells. UTX and MLL4 co-regulated several gene expression programs, including those associated with cell proliferation and invasiveness. Our additional data showed a correlation of high UTX and MLL4 mRNA levels with poor survival of breast cancer patients, strengthening the clinical importance of UTX and MLL4 in breast cancer patients. Furthermore, our findings that UTX mRNA levels correlate with MLL4 mRNA levels in breast tumors support the physical and cell biological link between these two histone methylation modifiers. Together, our work uncovers that UTX and MLL4 regulate the proliferation and invasiveness of breast cancer cells in a cooperative manner.
Histone-modifying enzymes often cooperate for gene repression. The H3K4 demethylase LSD1 and the histone deacetylases HDAC 1 and 2 co-repress gene expression (41), and the H3K4 demethylase JARID1a and the H3K9 methyltransferase G9a cooperate for gene repression (42). Conversely, certain modifiers cooperatively activate their target genes. The H3K9 demethylase JMJD2B and the H3K4 methyltransferase MLL4 coordinately regulate ERα-activated transcription (43). In addition, our previous report showed that UTX and MLL4 were co-recruited to regulate the same target genes, such as the differentiation-specific HOX cluster genes, during stem cell differentiation (26). The notion that UTX-catalyzed H3K27 demethylation is linked to MLL4-mediated H3K4 methylation is further supported by our following results: 1) UTX knockdown not only increased the UTX substrate H3K27me3 but also decreased MLL4 and H3K4me3 levels; 2) MLL4 knockdown not only decreased UTX and the MLL4’s enzymatic product H3K4me3 but also increased H3K27me3 levels. Moreover, most of UTX-controlled genes were affected by MLL4 knockdown and vice versa. Therefore, it is likely that UTX and MLL4 regulate gene expression programs in a coordinated manner.
Similar to the UTX gene, the MLL4 gene also harbors somatic mutations and deletions in several types of cancers, including prostate cancer, medulloblastoma, and lymphoma [reviewed in (22)]. In contrast to these types of cancer, genomic alterations of UTX and MLL4 rarely occur in human breast tumors (25). Our results indicate that UTX and MLL4 may be positive contributors to breast tumor, because UTX or MLL4 knockdown impeded the invasiveness and proliferation of breast cancer cells. In support with our results, a recent report indicated that UTX is up-regulated in breast cancer and correlates with poor prognosis in breast cancer patients (24). In addition, UTX is overexpressed in renal cell carcinoma (44) and leukemia (45), and its knockdown in leukemia cell lines exhibits an anti-proliferation effect, implicating its role in the tumor-promoting function (45). Similarly, MLL4 has also been shown to be critical for the growth of HeLa cells (31).
These conflicting functions of UTX and MLL4 in different types of cancer might be explained by the following potential scenarios. First, tumor suppressive effect and oncogenic function of a histone modifier may not be mutually exclusive. EZH2 is overexpressed and plays an oncogenic role in several types of tumors (e.g., breast and prostate cancer), whereas it undergoes somatic loss-of-function mutations in malignant myeloid diseases [reviewed in (46)]. Similarly, it is possible that UTX and MLL4 act as either oncogenic factors or tumor suppressors in a tissue-dependent manner. In this regard, UTX and MLL4 might play oncogenic roles in breast cancer, but function as tumor suppressors in medulloblastoma, bladder carcinoma, leukemia and prostate tumors in which their loss-of-function mutation rates are significantly high. The second scenario is that UTX and MLL4 may function in a tumor stage-specific manner. It has been reported that UTX activates Rb-related tumor suppression pathways in normal human fibroblasts (23), whereas our results showed that UTX positively regulates oncogenic gene expression programs for cell growth and invasion in breast cancer cells. Therefore, it is tempting to speculate that UTX and MLL4 may act as tumor suppressors during the normal cellular stage or initial stages of breast cancer development and become a tumor-promoting protein in later stages and metastatic tumors. Intriguingly, this type of scenario has also been exemplified by TGF-beta, which behaves as an anti-proliferative protein in normal cells and at early oncogenesis but promotes tumor invasiveness in later stages (47). In addition, LSD1 is frequently overexpressed in ER-negative breast cancer (48) but inhibits breast cancer metastasis (49).
H3K27 methylation is regulated by the histone methyltransferases EZH1/2 and demethylases UTX and JMJD3. Our present study and others’ work support the notion that UTX likely contributes to the oncogenic pathogenesis in the breast (24, 44, 45). Interestingly, its biochemically opposing enzyme EZH2 appears to be an oncogenic protein in the same tissue (13, 14). These seemingly puzzling observations may be a result of the regulation of different sets of genes by UTX and EZH2. UTX may help cancer cells grow and invade by activating gene programs that may promote tumor growth and metastasis, as described herein. EZH2-containing polycomb repressive complex 2 transcriptionally suppresses p14ARF and p16INK4A, allowing cancer cells to resist various cellular senescence signals (17). Of note, it is also possible that EZH2 would promote breast tumorigenesis via an oncogene activation mechanism independent of its methylation activity, as EZH2 phosphorylation by AKT1 may contribute to castration-resistant prostate cancer by inhibiting EZH2’s methyltransferase activity and enabling EZH2 to act as a transcriptional co-activator of androgen receptor (50). Equally interesting, it is likely that UTX and JMJD3, albeit their same in vitro H3K27 demethylase activities, produce opposite biological outcomes by regulating different gene expression programs, because JMJD3 may have a tumor-suppressive function by activating p16INK4A and p14ARF (19).
In summary, our results provide evidence that a histone demethylase and a histone methyltransferase co-regulate gene expression profiles in a coordinated manner to contribute to the proliferation and invasiveness of breast cancer cells. Our findings might be relevant to the development of therapeutic agents, such as small molecule modulators of UTX or MLL4.
Supplementary Material
Acknowledgments
We are thankful to Dr. Mien-Chie Hung, Dr. Zhenbo Han, and Su Zhang for their help for bioluminescence imaging and to Luanne Jorewicz for manuscript editing. This work was supported by grants to M.G.L. from the NIH (R01 GM095659 and R01 CA157919) and Cancer Prevention and Research Institute of Texas (RP110183).
Footnotes
The authors disclose no potential conflicts of interest
References
- 1.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A. 1999;96:14967–72. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–39. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 9.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 10.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47:701–6. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–9. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 13.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Y, Huo L, Liu P, Sneige N, Sun X, Ueno NT, et al. Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer. 2011;117:5476–84. doi: 10.1002/cncr.26179. [DOI] [PubMed] [Google Scholar]
- 15.Kunju LP, Cookingham C, Toy KA, Chen W, Sabel MS, Kleer CG. EZH2 and ALDH-1 mark breast epithelium at risk for breast cancer development. Mod Pathol. 2011;24:786–93. doi: 10.1038/modpathol.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Erdmann C, Chinnaiyan AM, Merajver SD, Kleer CG. Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res. 2006;66:4095–9. doi: 10.1158/0008-5472.CAN-05-4300. [DOI] [PubMed] [Google Scholar]
- 17.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72:3091–104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 19.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berdel B, Nieminen K, Soini Y, Tengstrom M, Malinen M, Kosma VM, et al. Histone demethylase GASC1--a potential prognostic and predictive marker in invasive breast cancer. BMC Cancer. 2012;12:516. doi: 10.1186/1471-2407-12-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Liu S, Liu G, Dombkowski A, Abrams J, Martin-Trevino R, et al. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene. 2012;31:333–41. doi: 10.1038/onc.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, et al. The histone demethylase UTX enables RB-dependent cell fate control. Genes & development. 2010;24:327–32. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patani N, Jiang WG, Newbold RF, Mokbel K. Histone-modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res. 2011;31:4115–25. [PubMed] [Google Scholar]
- 25.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 27.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 28.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26:2749–62. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 30.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 35.Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 36.Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A. 2012;109:13004–9. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin C, Li J, Green CD, Yu X, Tang X, Han D, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–72. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaturvedi CP, Somasundaram B, Singh K, Carpenedo RL, Stanford WL, Dilworth FJ, et al. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc Natl Acad Sci U S A. 2012;109:18845–50. doi: 10.1073/pnas.1213951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:7541–6. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Y, Guo X, Wang Y, Qiu W, Chang Y, Zhang A, et al. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. BMC Cancer. 2012;12:470. doi: 10.1186/1471-2407-12-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Mercher T, Scholl C, Brumme K, Gilliland DG, Zhu N. A functional role for the histone demethylase UTX in normal and malignant hematopoietic cells. Experimental hematology. 2012;40:487–98. e3. doi: 10.1016/j.exphem.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–5. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 48.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–20. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 50.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.