Abstract
In the developing kidney, self-renewing progenitors respond to inductive signaling from the adjacent branching ureteric bud by undergoing mesenchyme-to-epithelium transition. Nascent nephrons subsequently undergo elongation, segmentation, and differentiation into a mature renal epithelium with diverse functions. Epigenetic mechanisms have been implicated in impacting cell fate decisions during nephrogenesis; however, the chromatin landscape of nephron progenitors and daughter differentiating cells are largely unknown. Here, we examined the spatiotemporal expression patterns of histone H3 methylation and histone methyltransferases in E15.5 mouse kidneys. Kidney sections were probed with antibodies against histone modifications, enzymes, and markers of progenitors and differentiation. The results revealed that: (1) nephron progenitor cells exhibit a broad histone methylation signature that comprises both “active” and “repressive” marks (H3K4me3/K9me3/K27me3/R2me2/R17me2); (2) nascent nephrons retain high H3K4me3 but show downregulation of H3K9/K27me3 and; (3) maturing epithelial tubules acquire high levels of H3K79me2/3. Consistent with respective histone marks, the H3K4 methyltransferase, Ash2l, is expressed in progenitors and nascent nephrons, whereas the H3K9/K27 methyltransferases, G9a/Ezh2, are more enriched in progenitors than nascent nephrons. We conclude that combinatorial histone signatures correlate with cell fate decisions during nephrogenesis.
Keywords: histone methylation, histone modifications, chromatin landscape, kidney development, nephrogenesis, nephron progenitors, differentiation
Introduction
Regulation of nucleosome structure and function via posttranslational histone modifications is a key epigenetic mechanism of gene regulation in development and disease.1,2 The nucleosome consists of 147 DNA base pairs wrapped around a core of 8 histones composed of 2 H3/H4 histone dimers flanked by 2 H2A/H2B dimers. Protruding from the histone globular domains are histone tails, which present a rich platform of discrete epitopes subject to chemical modifications, such as acetylation, methylation, and phosphorylation. It has been proposed that these combinatorial histone modifications form a so called “histone code or signature,” which can be recognized by specific chromatin modifiers and transcriptional regulators resulting in defined biological outcomes.3,4 Although this hypothesis is hotly debated, there is evidence that mutations in epigenetic modifiers and specific histone tail residues are coupled with different outcomes and disease states, suggesting that histone modifications regulate distinct developmental processes.5,6 Furthermore, genome-wide histone signatures (epigenetic states) in embryonic stem cells and cancer cells appear to correlate with transcriptional and functional outcomes.7-11 In contrast, little is known about the global epigenetic states of lineage-committed progenitors during organogenesis. For example, do progenitors exhibit a histone signature that sets them apart from the surrounding neighboring cells? And, do daughter cells retain or lose this signature? As we begin to answer some of these questions, we can begin to address epigenetic states of congenital and acquired diseases and whether original epigenetic states can be restored with therapy.
The metanephric kidney originates from the intermediate mesoderm via reciprocal interactions between two cell lineages, the metanephric mesenchyme (MM) and the ureteric bud (UB). A group of MM cells condenses around the UB branch tips to form a nephron progenitor niche (also called the cap mesenchyme), which lineage analysis has shown it gives rise to all cell types of the nephron from the glomerulus to the connecting tubule.12 Uninduced nephron progenitors express “stemness” genes such as Six2 but are also poised to undergo mesenchyme-to-epithelium transition in response to inductive Wnt signaling emanating from the adjacent UB.13-15 In this regard, Six2 interacts and cooperates with Lef/Tcf factors and β-catenin to initiate expression of nephrogenic genes such as Wnt4, Lhx1, Pax8, and Fgf8. We recently examined the chromatin landscape in MM cell lines using ChIP-Seq and reported that whereas H3K4me3 peaks predominantly decorate metabolic genes, H3K27me3 peaks are highly specific to developmental genes in each cell line. Moreover, we found a group of developmental genes that carries bivalent histone domains, which resolve during differentiation.16
In the present study, we examined the spatial pattern of histone H3 methylation and the developmental expression of the histone KMT in the mouse kidney. We hypothesized that nephron progenitors and differentiating daughter cells share common histone modifications yet express distinct global chromatin signatures.
Results
In embryonic stem/progenitor cells, cell renewal or differentiation correlates with net changes in histone modifications especially with methylation on H3K4, H3K9, and H3K27.17,18 Furthermore, in certain cancers there is a strong correlation between cellular invasive behavior and the global histone signature.19-21 To determine whether there is a correlation between progenitor cell renewal/differentiation and histone modifications during nephrogenesis, we used an in situ section immunofluorescence approach to define the distribution of histone H3 methyl-lysine derivatives during mouse nephrogenesis. The major compartments of the developing nephron and markers used for each compartment are shown in Figure 1.
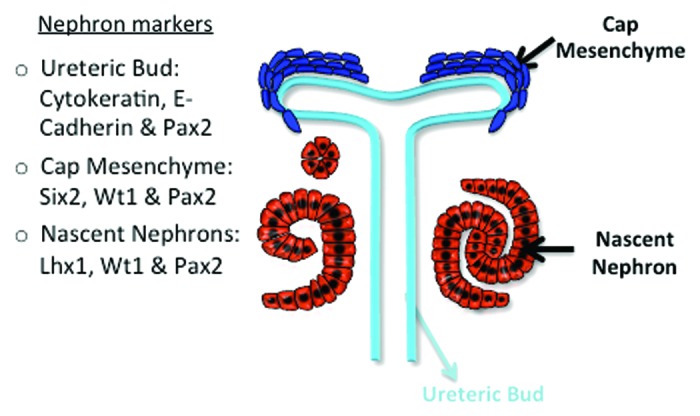
Figure 1. A schematic of a developing nephron and the markers used to detect the various compartments.
Immunolocalization of histone H3 methylation on lysines 4, 9, 27, and 79
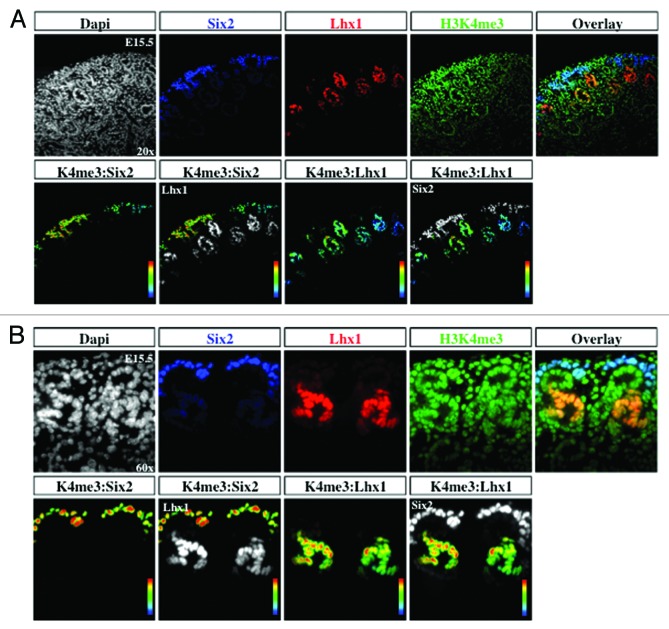
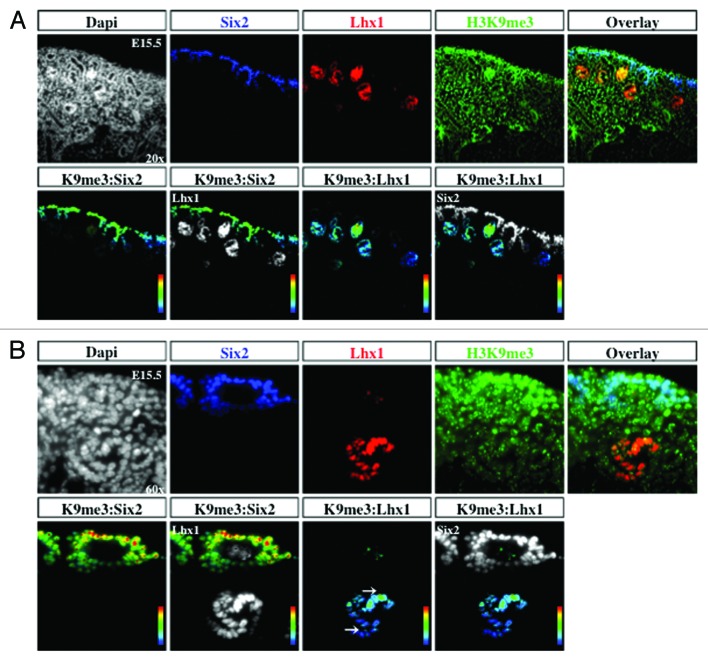
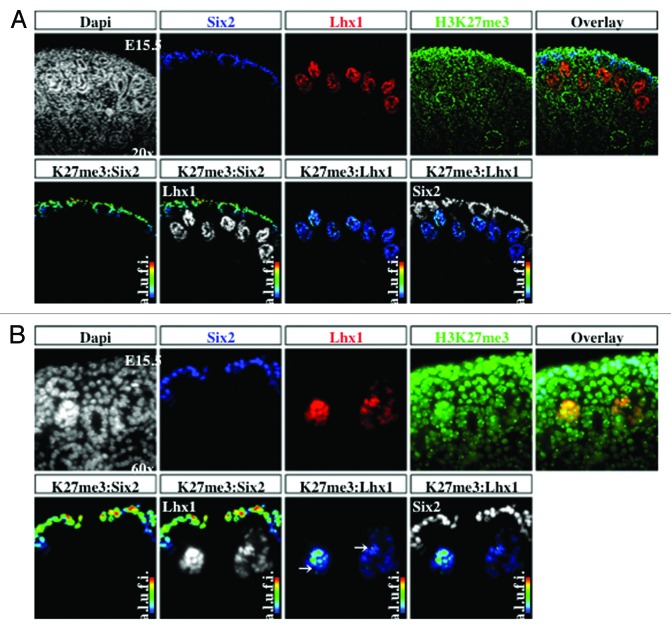
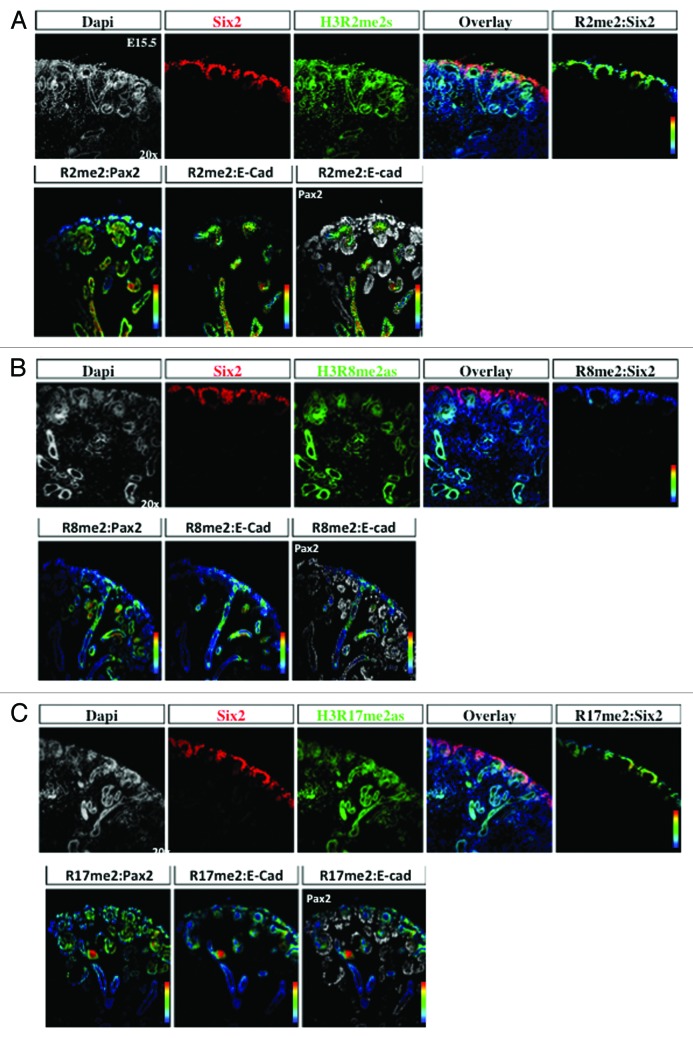
The transcription factors Six2 and Pax2 are markers of nephron progenitors in the cap mesenchyme. Pax2 also marks nascent nephrons and UB branches. The cell adhesion molecule, E-cadherin, is expressed in epithelial tubules. Lhx1 is a transcription factor expressed in nascent nephrons. We selected embryonic day 15.5 (E15.5) because nephrogenesis and branching morphogenesis are actively ongoing at this developmental stage. Genome-wide analyses performed in a multitude of cell lines and tissues including kidney cell lines have demonstrated a strong correlation between the presence of H3K4me3 peaks around the transcription start sites and gene activity.16,22,23 In contrast, H3K9me2/3 and H3K27me3 peaks decorate the promoters of repressed genes.9,24-26 We found a significant enrichment of H3K4me3 in Six2+/Pax2+ progenitors as well as in Lhx1+ nascent nephron cells (Figs. 2A, 2B, and 5A). In contrast, H3K9me3 and H3K27me3 marks are more abundant in Six2+ than Lhx1+ cells (Figs. 3, 4, 5B and 5C). Moreover, co-staining with E-cadherin antibodies revealed the presence of all 3 histone markers in the cortical epithelial tubules and collecting ducts (Fig. 5). Thus, whereas nephron progenitors are decorated with H3K4me3/K9me3/K27me3, nascent nephrons retain high H3K4me3 and downregulate H3K9me3/K27me3. The specificity of the H3K4me3, H3K9me3, and H3K27me3 antibodies was confirmed by pre-absorbing overnight with either an unmodified H3 peptide or one trimethylated on lysine 4, 9, or 27. Pre-absorption with the modified peptide significantly reduced immunostaining as compared with pre-absorption with an unmodified peptide (Fig. 6A–C).
Figure 2. Histone H3K4me3 is enriched in cap mesenchyme and nascent nephrons. The cap mesenchyme (nephron progenitors) marked by Six2 (blue) and nephron progenitors marked by Lhx1 (red) were co-stained with H3K4me3 (green) and analyzed at 20× (A) and 60× (B). RGB overlay and pseudocolor images demonstrate co-localization with H3K4me3. Arbitrary linear units of fluorescent intensity (a.l.u.f.i.).
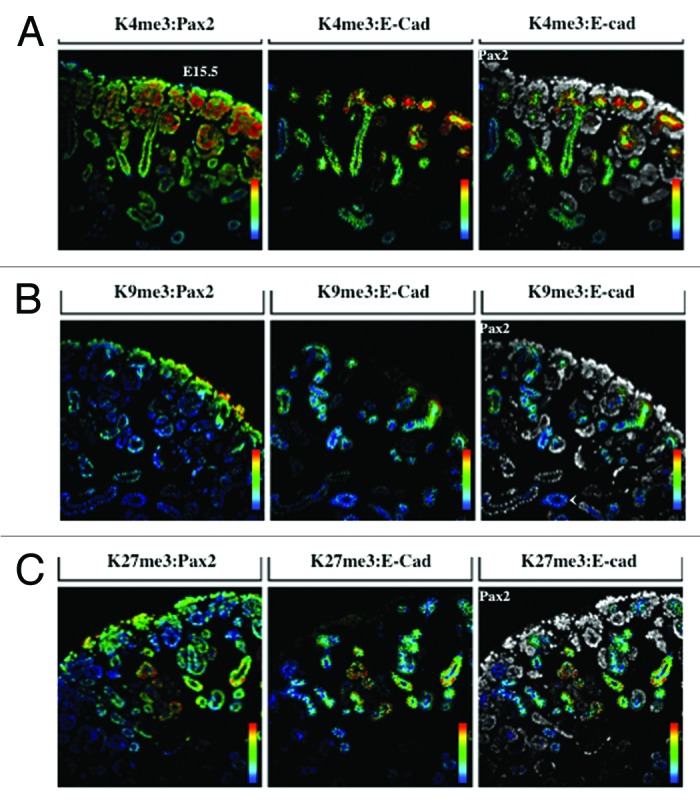
Figure 5. Distribution of H3K4me3, H3K9me3, and H3K27me3. The ureteric bud, cap mesenchyme, and nascent nephrons are marked by Pax2 (blue); ureteric bud structures and epithelial tubules are marked by E-cadherin (red); co-staining was performed with H3K4me3 (A), H3K9me3 (B) or H3K27me3 (C) (green) and analyzed at 20×.
Figure 3. H3K9me3 is enriched in cap mesenchyme. The cap mesenchyme marked by Six2 (blue) and nascent nephrons marked by Lhx1 (red) were co-stained with H3K9me3 (green) and analyzed at 20× (A) and 60× (B). RGB overlay and pseudocolor images demonstrate co-localization with H3K9me3.
Figure 4. H3K27me3 is more abundant in cap mesenchyme than nascent nephrons. The cap mesenchyme marked by Six2 (blue) and nascent nephrons marked by Lhx1 (red) were co-stained with H3K27me3 (green) and analyzed at 20× (A) and 60× (B). RGB overlay and pseudocolor images demonstrate co-localization with H3K27me3.
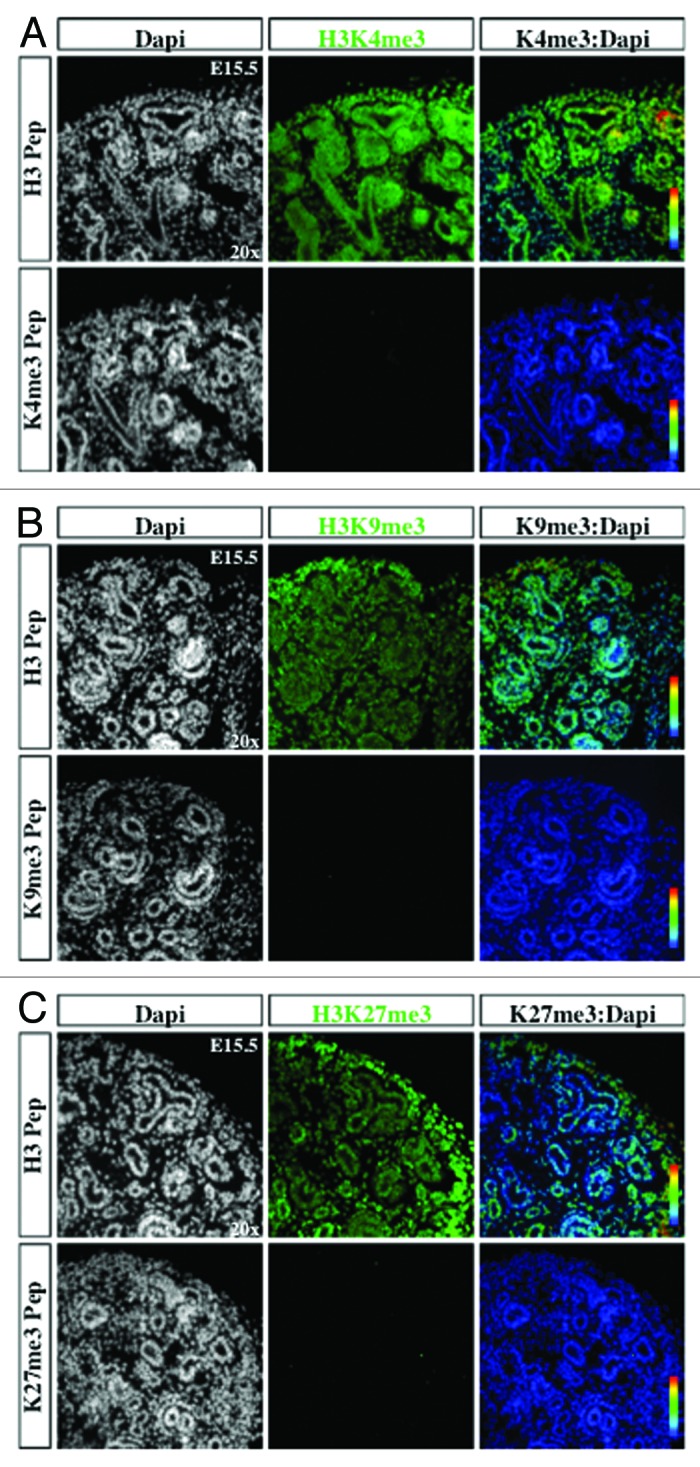
Figure 6. Antibody specificity. Kidney sections were stained with histone modification antibodies (green), either pre-absorbed with unmodified histone 3 N-terminal peptide (H3 Pep) or modified peptide with H3K4me3 (A), H3K9me3 (B) or H3K27me3 (C), and co-localized using pseudocolor (K4me3, K9me3, or K27me3:DAPI).
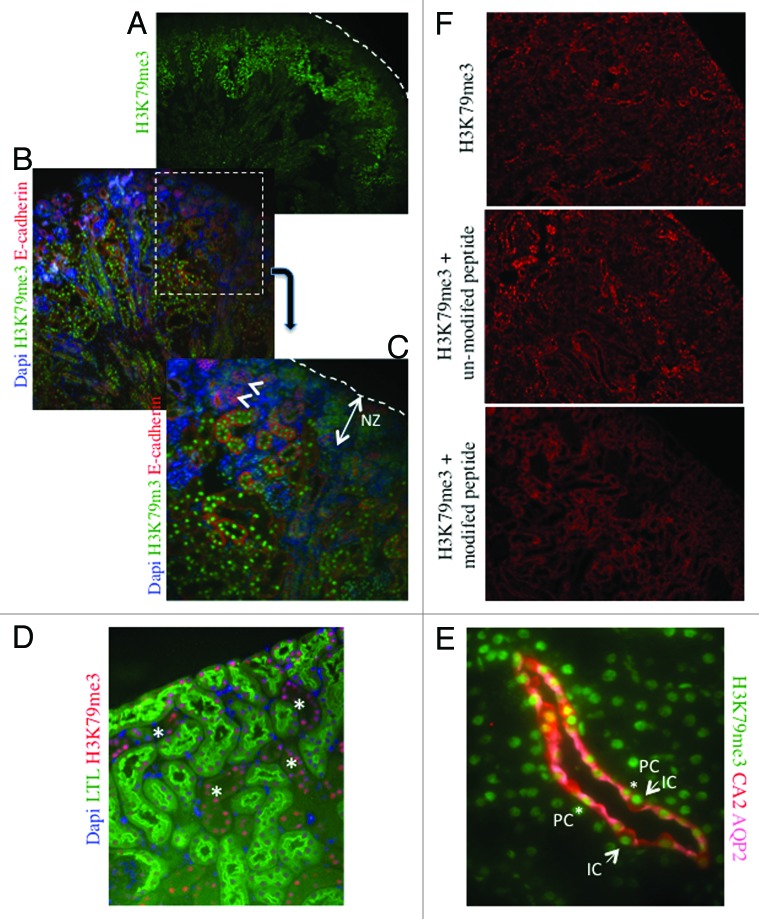
H3K79me3 is a histone mark that is abundant on exons of active genes.27,28 At E15.5, we find that H3K79me3 is more abundant in maturing inner cortical tubules than nascent nephrons or cap mesenchyme (Fig. 7A–C). In the adult kidney, H3K79me3 is abundant in nuclei of both proximal and distal nephron segments (Fig. 7D), and staining for principal and intercalated cell markers indicates that this mark is expressed in both cell types (Fig. 7E). The specificity of the H3K79me3 antibodies was confirmed by pre-absorbing overnight with either an unmodified H3 peptide or one tri-methylated on lysine 79. Pre-absorption with the modified peptide significantly reduced immunostaining as compared with pre-absorption with an unmodified peptide (Fig. 7F).
Figure 7. H3K79me3 is enriched in differentiating tubular epithelial cells at postnatal day 1. (A) Low power view (×4). Dotted line indicates the renal capsule. (B and C) Low (×10) and high (×40) power views of the renal cortex showing a progressive increase in H3K79me3 in differentiating E-cadherin+ tubules. The nephrogenic zone and nascent nephrons (arrowheads) are almost devoid of H3K79me3. (D) Low power view (×10) of the renal cortex in an adult kidney. LTL marks the proximal tubules. H3K79me3 is enriched in both proximal and non-proximal tubules. (E) Co-staining for H3K79me3, carbonic anhydrase-2 (CA2, a marker of intercalated cells), and aquaporin-2 (AQP-2, a marker of principal cells). Magnification ×60. (F) Low power view (×10) of the renal cortex in an adult kidney sections incubated with H3K79me3 antibody alone, H3K79me3 antibody + unmodified H3 N-terminal peptide, or H3K79me3 antibody + H3 K79me3 modified peptide. The specific nuclear staining is absent in the section incubated with the modified peptide.
Immunolocalization of histone methyltransferases Ash2l, Ezh2, Suz12, and G9a
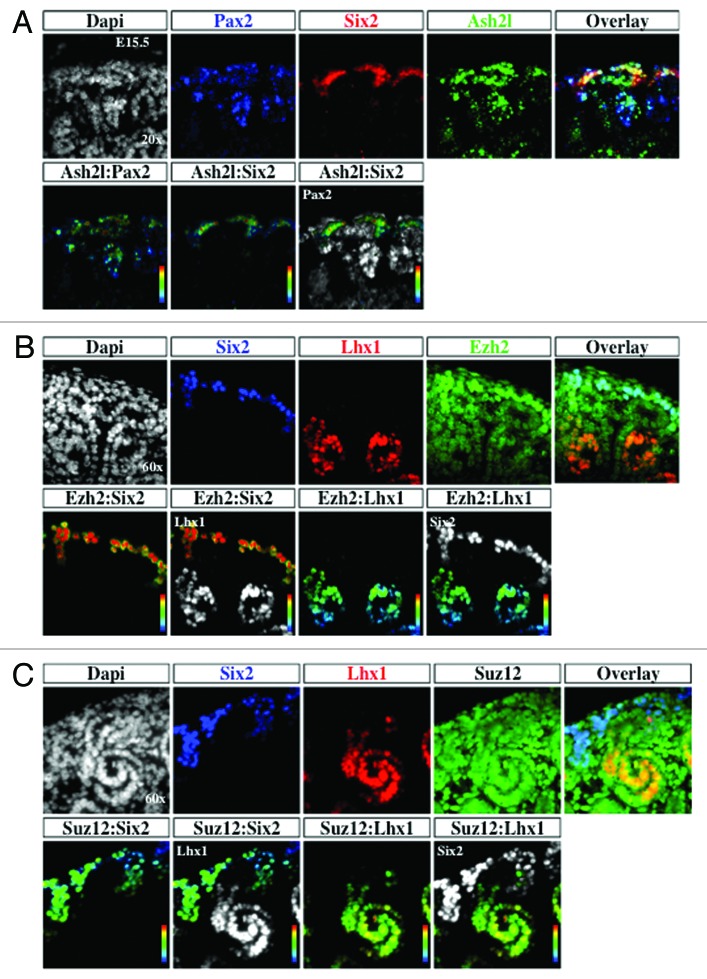
The enzymatic addition of methyl groups on H3K4 and H3K27 is catalyzed in part by Ash2l and Ezh2/Suz12, respectively. To determine the expression of these enzymes during nephrogenesis, E15.5 sections were co-stained with antibodies to nephrogenic markers and the enzymes. Ash2l and H3K4me3 share overlapping expression in the nephrogenic zone (Figs. 2 and 8). Ash2l is part of the Trithorax complex and is known to interact with PTIP, a Pax2 binding protein.29 Co-staining with Pax2 antibody demonstrated co-localization with Ash2l (Fig. 8A). Together, these results suggest that Ash2l may an effector of Pax2-dependent transcriptional regulation.
Figure 8. Distribution of histone modifiers Ash2l, Ezh2, and Suz12 in the developing kidney. The ureteric bud, cap mesenchyme, and nascent nephrons marked by Pax2 (blue) and cap mesenchyme marked by Six2 (red) were co-stained with the H3K4 KMT Ash2l (green) and analyzed at 60× (A). The cap mesenchyme marked by Six2 (blue) and nascent nephrons marked by Lhx1 (red) were co-stained with the H3K27 KMT Ezh2 or Suz12 (green) and analyzed at 60× (B and C).
Ezh2 is one of the core enzymes responsible for the catalytic activity of the PRC2 silencing complex and is a developmentally regulated enzyme.30 The localization of Ezh2 during nephrogenesis was similar to that of H3K27me3 with the greatest enrichment in the Six2+ cap mesenchyme (Fig. 8B). Immunostaining for Suz12, another member of the PRC2 complex, demonstrated diffuse expression in cap mesenchyme and nascent nephrons (Fig. 8C). One possible reason for enrichment of Ezh2 in the cap mesenchyme is the necessity of Ezh2 to maintain the identity of the Six2 population and to exert tight control over self-renewal and differentiation, as has been described in the skin.31
G9a is a histone lysine 9 KMT and catalyzes the formation of H3K9me2, a repressive chromatin mark.32 Staining for Pax2 and G9a demonstrated co-localization within the nephrogenic zone (Fig. 9A). Interestingly, there seemed to be a segment specific enrichment in G9a, with higher levels being seen in Pax2+ cap mesenchyme (white arrow) as well as the distal segment of the S-shaped body (white arrowhead) (Fig. 9A and B). In sections co-stained with Lhx1 and Wt1, markers of the distal and proximal portions of nascent nephrons respectively, we find a relative abundance of G9a in Lhx1+ cells (white arrowhead) as compared with the Wt1+ cells (white arrow) (Fig. 9C). This suggests that G9a may play a role in segmental nephron differentiation.
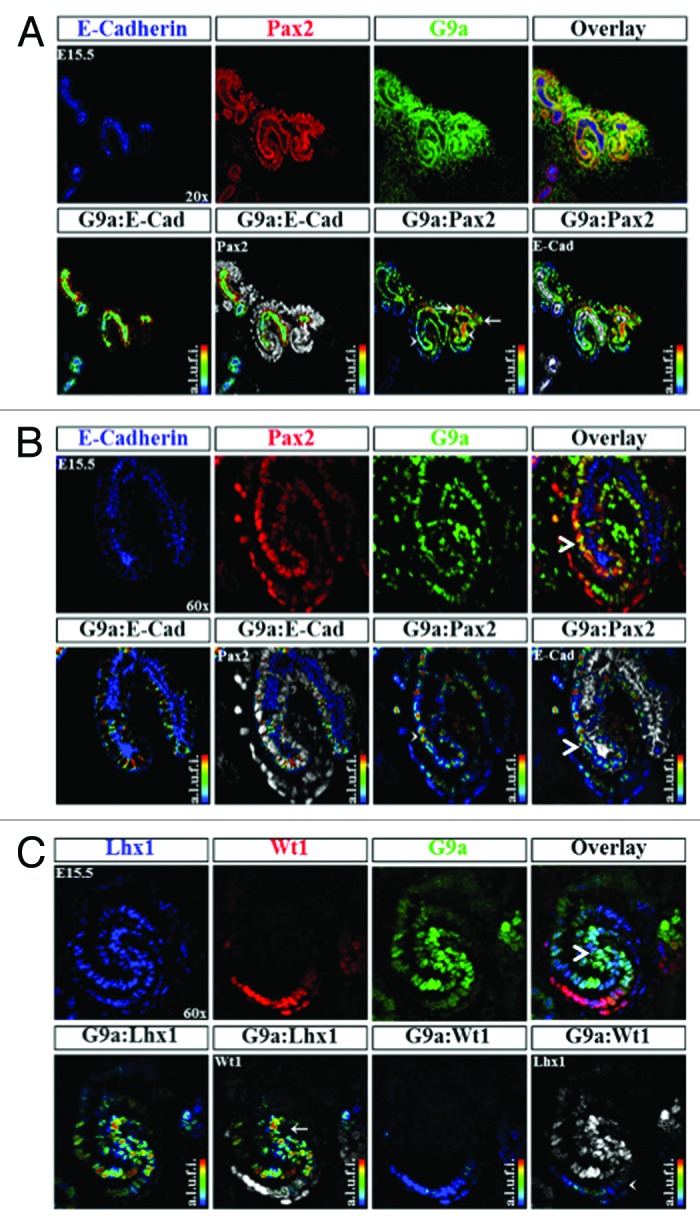
Figure 9. Distribution of H3K9 KMT, G9a, in the developing kidney. (A) G9a is expressed in epithelial (arrowhead) and mesenchymal components (arrows) ×20. (B) A higher power view (×60) showing G9a expression in an S-shaped body and its relative enrichment at the junction of the proximal and distal segments (arrowhead). (C) Co-staining of G9a with Lhx1 (a marker of nascent nephrons) and WT1 (a marker of podocytes) reveals G9a expression at the junction of proximal and distal segments of S-shaped body (arrow and arrowhead).
Histone Methylation of Arginine 2, 8, and 17 of Histone 3
Arginine residues on the H3 tails can be monomethylated, dimethylated symmetrically and dimethylated asymmetrically, and represent a less well-studied histone modification.17 H3R2me2s is a repressive histone modification,33,34 whereas and H3R8me2as may mediate either activation or repression.35-37 H3R17me2as is generally an activating mark.38,39 Interestingly, there was a differential enrichment of these three histone marks, since H3R2me2 and H3R17me2 are abundantly expressed in the cap mesenchyme and nascent nephrons, whereas H3R8me2 is more enriched in nascent nephrons (Fig. 10A–C). All three histone marks are expressed in the collecting ducts.
Figure 10. Differential distribution of histone H3 arginine methylation marks in the developing kidney. The cap mesenchyme, marked by Six2 and Pax2, and epithelial structures including UB branches, marked by Pax2 and E-cadherin, were co-stained with H3R2me2s (A), H3R8me2as (B), or H3R17me2as (C) and analyzed at 20×. RGB overlay and pseudocolor images demonstrate co-localization of H3R2me2 and H3R17me2 with Pax2 and Six2 in the cap mesenchyme, and H3R8me2 with Pax2 and E-cadherin in UB structures.
Developmental expression of histone modifications/modifiers
Developing organs are composed of cells undergoing proliferation, migration, and differentiation and therefore their overall histone modifications may differ from adult terminally differentiated organs. We therefore performed western blot analysis of acid-extracted histones for various histone marks. The results revealed that total histone H3K9ac, H3K9me3, H3K27me3 levels remain unchanged during kidney development (Fig. 11). In comparison, H3K79me3 levels increase in abundance postnatally correlating with tubular epithelial expression that we observed during nephron differentiation (Fig. 7). H3K4me3 abundance showed a reciprocal pattern to that of H3K79me3, higher in developing than mature kidneys (Fig. 11).
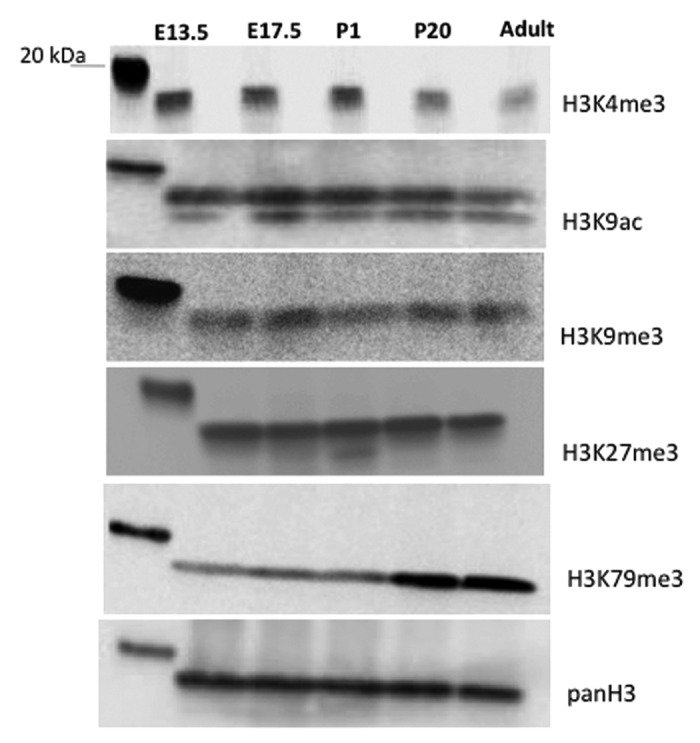
Figure 11. Global changes in histone modifications during kidney development. Western blot analysis of acid-extracted histones from embryonic and postnatal mouse kidneys.
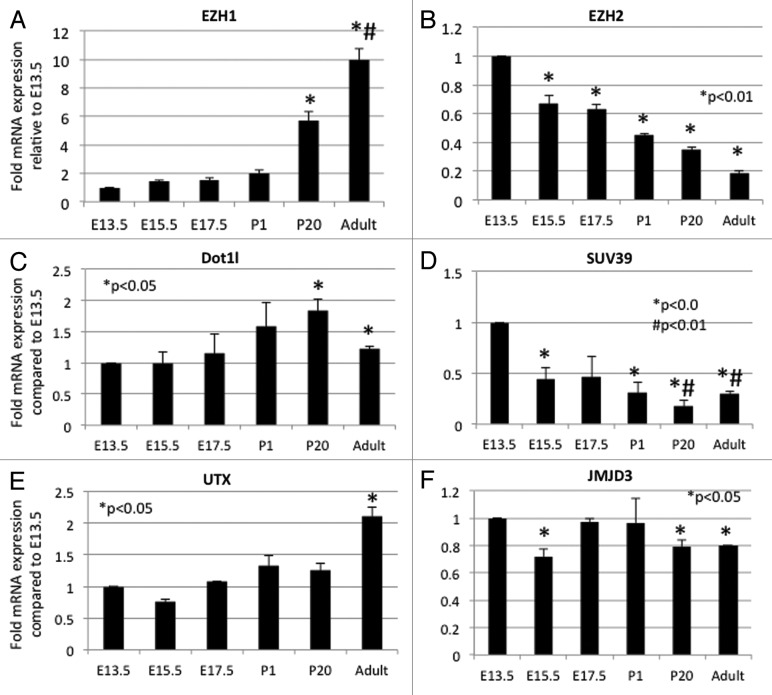
Because of difficulties in obtaining clean and reproducible signals by western blots using antibodies to histone modifiers such as Dot1l and Mll proteins, we performed quantitative RT-qPCR in order to determine whether the changes in histone modifications correlate temporally with corresponding developmental changes in histone modifiers (Fig. 12). The H3K27 KMTs, Ezh1 and Ezh2, exhibit reciprocal developmental expression patterns (Fig. 12A and B). This reciprocal pattern might explain the steady global levels of H3K27me3. Dot1l gene expression, the only known H3K79 KMT, increases postnatally which is consistent with the temporal changes in H3K79me3 (Fig. 12C). The histone KMT Suv39h mediates H3K9me3 formation, whereas Utx and Jmjd3 mediate demethylation of H3K27. We found that Suv39h expression is higher in embryonic than postnatal kidneys (Fig. 12D). The H3K27 KDM, Jmjd3, declines slightly post-weaning, while Utx gene expression increases postnatally (Fig. 12E and F). This may account for the maintained levels of H3K27me3 during kidney development.
Figure 12. Developmental expression of histone modifiers. Quantitative RT-PCR. (n = 3 animals per age group from a minimum of 2 different litters).
Cross referencing the spatial compartmentalization of the epigenetic modifiers in the Genito-Urinary Development Molecular Anatomy Project (GUDMAP)40 database revealed that the nephrogenic compartments express unique as well as shared histone modifiers and chromatin remodelers (Fig. 13A).
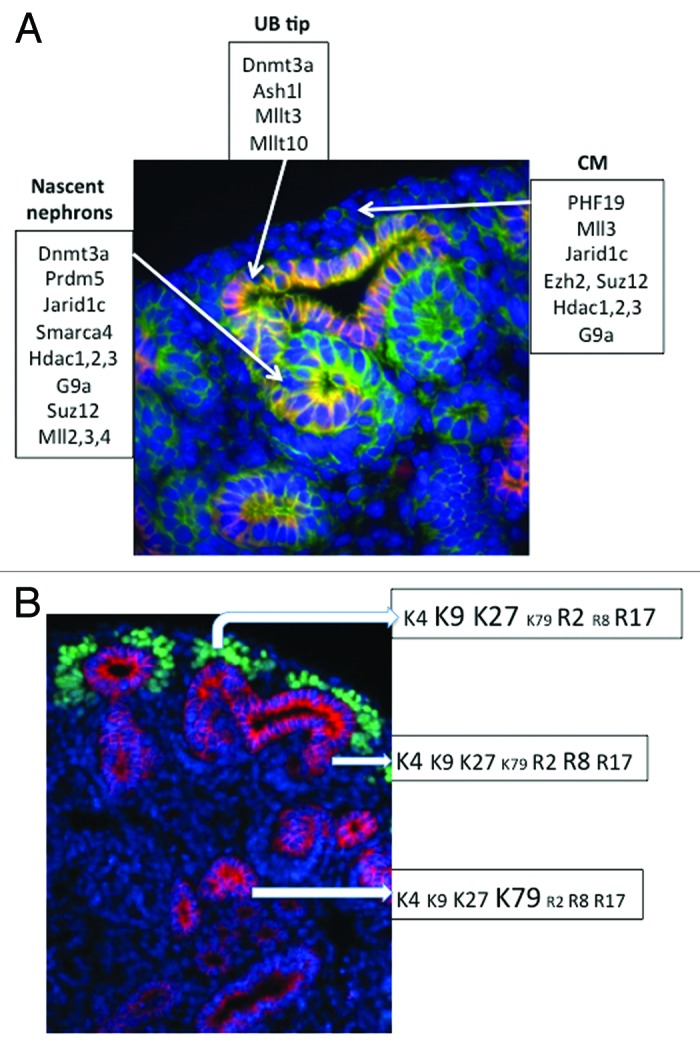
Figure 13. (A) Compartmentalization of histone modifiers in the developing nephron based on the GUDMAP40 database. (B) Histone signature of cap mesenchyme, nascent nephrons, and differentiated tubules.
Discussion
The present study examined the spatiotemporal distribution of histone modifications and modifiers during nephrogenesis. The results demonstrate that nephron progenitors and nascent nephrons share some chromatin marks (e.g., H3K4me3 and H3R17me2), whereas other histone marks (e.g., H3K9me3, H3K27me3, H3K79me3, H3R2me2, H3R8me2) tend to be more abundant in other compartments. These results suggest that nephrogenesis is accompanied by dynamic changes in histone modifications characterized by retention, gain or depletion of histone marks. The differential distribution of chromatin modifications imparts nephron compartments with global histone signatures. We propose that these combinatorial signatures may be important in maintaining the epigenetic states and in defining the functional identity of stage-specific nephron differentiation.
Histone H3K4 trimethylation is a mark that is enriched at the start site and 5′ end of actively transcribed genes.23 H3K4me3 is recognized by the basal transcription complex TFIID via the PHD finger of TAF3 and functions as a binding site for proteins that facilitate access by RNA Polymerase II to the DNA.41 There are at least 6 functionally non-redundant H3K4 lysine KMT in mammals, which carry essential and conserved functions such as Hox gene regulation. Recent studies in which KMT cofactors were disrupted in the mature heart and kidney have shown that H3K4me3 is important in specific aspects of adult terminally differentiated tissues, such as cardiac electrophysiology and renal concentrating mechanisms.42,43 Given its strong association with actively transcribed genes, the broad distribution of H3K4me3 and its KMT, Ash2l, in nephron progenitors and derivatives is not surprising. Our studies in clonal MM-like cells have demonstrated that H3K4me3 peaks decorate not only actively transcribed metabolic and housekeeping genes, but also promoters of developmental renal regulators such as Six2 and Osr1.16 The latter study also showed that H3K4me3 peaks in Six2 and Osr1 are depleted in differentiating cells concomitant with binding of H3K4 KDM. Conversely, gain of H3K4me3 and KMT occupancy correlates with expression of nascent nephron genes such as Pax8 and Lhx1.16 Thus, dynamic global and gene-specific changes in H3K4me3 occur during nephron lineage differentiation.
We find that H3K9me2 and H3K27me3, chromatin marks associated with transcriptional repression, are more abundant in Six2+ progenitors than Lhx1+ nascent nephrons. This correlates with differential expression of G9a and Ezh2, the respective KMTs, in these 2 compartments. Biron et al. have shown that H3K9me3 is highly abundant in mitotic cells lining the medial edges and luminal aspects of the neural tube in E9.5 embryos, and suggested that this histone mark may be a useful and novel marker to define embryonic regions with high proliferative activity.44 During folliculogenesis, primordial germ cells entrance into the genital ridge around E9.5–E11.5 is marked by accumulation of H3K9me3 and H3K27me3, whereas a more restricted peak in H3K4me3 occurs at E10.5.45 This suggests that follicle maturation is marked by a period of poised chromatin domains containing active and repressive histone marks as part of a differentiation gene expression program. In the present study, we observe that the Six2+ cells are enriched in active and repressive histone methylation marks (H3K4me3/K9me2/K27me3/H3R2me2) suggesting that differentiation programs are silent yet poised for later activation in response to external cues. In support of this, ChIP-Seq studies have shown that nephrogenic genes harbor bivalent H3K4me3/K27me3 domains in their promoters and that stimulation of Wnt-β-catenin signaling results in gain of H3K4me3 and/or depletion of H3K27me3.16 Genetic deletion of Ezh2 catalytic domain from nephron progenitors in mice impairs cell renewal and favors precocious differentiation, consistent with the role of H3K27me3 in maintenance of the undifferentiated state and lineage identity (McLaughlin, Kelly, Saifudeen, and El-Dahr, unpublished data).
Although some histone modifications exhibit spatial compartmentalization in the developing nephron, there was no discernible change in the temporal expression of these marks. One possible exception is H3K79me3 and its enzyme Dot1l, which showed significant stage and age-dependent increases during nephrogenesis. Chromatin analysis in yeast, fly, and mammalian systems indicates that H3K79me3 is a mark of active transcription.46 Developmental studies have shown that H3K79me3 is absent in the zygote and even in the blastocyst, suggesting that Dot1l functions post-early embryogenesis stages.47 Indeed, Dot1l-knockout mice die at E10.5 from cardiovascular defects and severe anemia.28 The function of Dot1l in the developing kidney is unknown but it has been suggested that Dot1l-H3K79 methylation plays a role in terminal differentiation of the collecting duct.48
Histone methylation occurs on lysine and arginine residues. Asymmetrically dimethylated H3R2me2a, catalyzed by Prmt6, lies adjacent to the active mark H3K4me3. It has been shown in yeast and human cells that H3R2me2a is enriched in heterochromatin in a mutually exclusive pattern with H3K4me3, and that methylation of H3R2 interferes with the binding of Ah2l/Mll and abrogates trimethylation of H3K4.33,34 H3R17me2s is catalyzed by CARM1 and promotes pluripotency of embryonic stem cells.17 In accord with these findings, our results show that H3R2me2 and H3R17me2 are abundant in nephron progenitors. In comparison, H3R8me2, a histone mark that is catalyzed by prmt5, is more concentrated in nascent nephrons than progenitors. H3R8me2 is associated with transcriptional repression of ribosomal genes and of tumor suppressors in cancer cells.36,49,50 The potential significance of histone arginine methylation in nephrogenesis remains to be determined, since relatively little is known about the functions of these chromatin marks.
The findings of the present study provide a general descriptive view of the histone landscape of the developing nephron but do not address other “epigenetic” marks such as DNA methylation and microRNA-based mechanisms. The data presented in this study are best interpreted within the context of functional data derived from existing and future ChIP-Seq data in nephron progenitors.14,16
In summary, the present study demonstrates differential spatial enrichment of specific histone marks and modifiers in nephrogenic compartments of the developing kidney. This is illustrated schematically in Figure 13B. The “repressive” marks H3-K9me3/K27me3/R2me2 are abundant in progenitor cells. The progenitor population is also rich in active marks, supporting the idea that some progenitors are “primed” for differentiation. Interestingly, H3K79 methylation is upregulated during the latter stages of epithelial differentiation. These differential spatiotemporal changes highlight the dynamic functional states of the epigenome during nephrogenesis. We propose that compartmentalization of histone signatures may be important in maintaining a healthy balance between self-renewal and differentiation. In the future, it will be interesting to elucidate the effects of embryonic stressors on the epigenetic landscape of nephron progenitors in vivo.
Materials and Methods
RNA isolation and QRT-PCR were performed as described.16 RT-PCR was performed on RNA extracted from CD-1 mouse kidneys (n = 3 mice/group, harvested from 2–4 litters per age group). One-step Brilliant quantitative RT-PCR master mix kit (Stratagene) was used to perform Taqman real-time PCR. Relative mRNA levels were normalized to GAPDH. The PCR conditions were as following: 50 °C 30 min, 1cycle; 95 °C 10 min, 1 cycle; 95 °C 15s, 56 °C 1 min, 72 °C 30s, 40 cycles. All QPCR probes were ordered from Applied Biosystems:
JMJD3 (mouse species): Mm01332672_g1
EZH1 (mouse species): Mm00468440_m1
EZH2 (mouse species): Mm00468464_m1
Dot1l (mouse species): Mm01173481_m1
Suv39 (mouse species): Mm01347696_g1
UTX (mouse species): Mm00801998_m1
GAPDH (mouse species): Mm99999915_g1
Statistical analysis was performed using one-way ANOVA. P < 0.05 was considered significant.
Histone extraction and western blot analysis
Histones were extracted from CD-1 mouse kidneys of 2–4 litters per age by the acid-extraction method. Homogenized tissue is lysed in Triton Extraction Buffer (TEB/PBS containing 0.5% Triton X-100 (v/v), 2 mM phenylmethylsulfonylfluoride (PMSF) and 0.02% (w/v) NaN3) at 0.1g/2.5ml TEB for tissues with gentle stirring on ice for 10 min. Following centrifugation at 2000 g for 10 min at 4 °C and removal of the supernatant, the pellet is washed in half volume of TEB and centrifuged again. Pellets were resuspended in 0.2N HCl overnight at 4 °C followed with centrifugation at 2000 g for 10 min. Supernatants containing histones were collected and stored at −80 °C. Histone protein concentration was assayed by Pierce BCA Protein Assay Kit (Thermo Scientific). Western blot was performed as previously described51 using the following primary antibodies: H3K4me3 (1:1000, Rabbit polyclonal, Abcam), H3K27me3 (1:1000, Rabbit polyclonal, Abcam), H3K9me3 (1:1000, Rabbit polyclonal, Abcam), H3K9-acetylation (1:1000, Rabbit polyclonal, Abcam), H3K79me3 (1:1000, Rabbit polyclonal, Abcam), pan H3 (1:1000, Rabbit polyclonal, Upstate Biotechnology).
Section immunofluorescence was performed as described51-53 using Tyramide Signal Amplification detection as per manufacturer recommendations (Perkin Elmer). Embryonic kidneys were fixed in 10% formalin at room temperature overnight then kept at 4 °C for up to 3 d until use. Paraffin-embedded kidneys were cut at 5 μm, placed on warn wet slides and allowed to dry overnight. Prior to use in immunohistochemistry, slides were placed in a 55 °C oven for 1 h to overnight. Sections were rehydrated by consecutive immersion in Xylene (2X, 10 min) and ethanol water dilutions (100%, 95%, 80%, 70%, 40%, 2 min each) followed by a quick rinse in tap water and 5 min in Tris-buffered saline (TBS). Antigen retrieval was performed by boiling slides for a total of 14 min in sodium citrate (10 mM, pH 6) using a conventional microwave, allowed to cool for 20 min and transferred to TBS. Blocking was performed for 1 h at room temperature or overnight at 4 °C using 10% normal donkey serum in TNB (Perkin Elmer), same as secondary host, supplemented with 20 μg/ml donkey anti-mouse FAB to block endogenous IgG (Jackson ImmunoResearch). Slides were washed in 10% hydrogen peroxide and then with TBS in 0.1% teween (TBSt) three times for 5 min with agitation, with final wash in TBS. Primary antibodies were diluted in TNB with 2% normal donkey serum (NDS) and incubated for 1 h at room temperature or overnight at 4 °C, and washed with TBSt 3 times for 5 min with agitation, with the final wash in TBS. Conventional fluorescent secondary antibodies (Donkey anti-rabbit, mouse or goat) (Life Technologies Corporation) conjugated to Alexa fluorophores (excitation 350 nm, 488 nm, 555 nm, or 747 nm, 1:300) diluted in TNB with 2% NDS were incubated for 1 h at room temperature. Hoerseradish peroxidase (HRP) conjugated secondary antibodies (Donkey anti-rabbit, mouse or goat) (Jakson ImmunoResearch) were used with Tyramide Signal Amplification detection (TSA) (Perkin Elmer). Slides were then washed in TBSt three times for five minutes with agitation, with the final wash being done in TBS. For TSA detection, TSA working solutions were prepared at a dilution 1:200 (TSA:Amplification Buffer) and sections incubated for 1–3 min. For downstream applications requiring comparison, TSA incubation time was the same ± 1–2 s. Following washing, the slides were incubated with DAPI (1:500, Life Technologies Corporation) for 10 min at room temperature, washed once with TBSt and once more with TBS and mounted using Vector Labs fluorescent media. Images were acquired with Olympus BX61 SDC fitted with a Hamamatsu CCD camera, as Z-stacks in 1-μm intervals using Intelligent Imaging Innovations SlideBook software, followed by constrained iterative deconvolution and Gaussian noise smoothing from system specific point spread functions. Primary antibodies used are as follows: E-Cadherin (BD Transduction, 610182, 1:200); Lhx1 (Developmental Hybridoma, 4F2, 1:100); Pax2 (Invitrogen, 71–6000, 1:200); Six2 (Proteintech, 11562–1AP, 1:100); WT1 (Abcam, ab15249, 1:200); H3K4me3 (Active Motif, 39160, 1:250); H3K9me3 (Abcam, ab8898, 1:200); H3K27me3 (Active Motif, 39535, 1:200); H3K79me2/3 (Abcam, ab3748, 1:150); H3R2me2s (Active Motif, 39703, 1:100), H3R8me2 (Active Motif, 39703, 1:200); H3R17me2as (Active Motif, 39709, 1:200); H3R17me2as (Abcam, ab8284, 1:200); ASH2L (Abcam, ab56099, 1:200); Ezh2 (Active Motif 39933, 1:250); SUZ12 (Active Motif, 39358, 1:100); Histone H3 Peptide: Unmodified 9Abcam, ab1425, 1:5–1:10); Histone H3 Peptide: H3K4me3 Modified (Abcam, ab1342, 1:5–1:10); Histone H3 Peptide: H3K9me3 Modified (Abcam, ab1773, 1:5–1:10); Histone H3 Peptide: H3K27me3 Modified (Abcam, ab1782, 1:5–1:10).
The specificity of H3K4me3, H3K9me2/3, H3K27me3, and H3K79me3 antibodies was tested by pre-incubating the primary antibodies with unmodified or modified histone peptides. Moreover, specificity of H3K79me2/3 antibodies was tested in kidneys section from conditional collecting duct Dot1l knockout mice (Hoxb7-cre::Dot1fl/fl) (not shown). Other antibodies were tested by omission of primary or secondary antibodies, or by using antibodies from different vendors (e.g., H3R17me2as). All immunohistochemistry experiments were performed on at least 4 different animals in triplicates.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH grants 1P50 DK096373 (Pediatric Center of Excellence in Nephrology), RO1-DK079886 and RO1-DK56264. We acknowledge the assistance of the Renal and Hypertension Center and The Louisiana Cancer Research Consortium Microscopy and FACS Cores. This work was performed as a part of PhD thesis (McLaughlin N).
Glossary
Abbreviations:
- MM
Metanephric mesenchyme
- UB
ureteric bud
- KMT
histone lysine methyltransferase
- KDM
histone lysine demethylase
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26793
References
- 1.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 2.Huang C, Sloan EA, Boerkoel CF. Chromatin remodeling and human disease. Curr Opin Genet Dev. 2003;13:246–52. doi: 10.1016/S0959-437X(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 5.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–90. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–6. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–70. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 11.Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–9. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mugford JW, Sipilä P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–9. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 14.Park JS, Ma W, O’Brien LL, Chung E, Guo JJ, Cheng JG, Valerius MT, McMahon JA, Wong WH, McMahon AP. Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev Cell. 2012;23:637–51. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–57. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin N, Yao X, Li Y, Saifudeen Z, El-Dahr SS. Histone signature of metanephric mesenchyme cell lines. Epigenetics. 2013;8:970–978. doi: 10.4161/epi.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–8. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- 20.Park YS, Jin MY, Kim YJ, Yook JH, Kim BS, Jang SJ. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol. 2008;15:1968–76. doi: 10.1245/s10434-008-9927-9. [DOI] [PubMed] [Google Scholar]
- 21.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 22.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 23.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–9. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 25.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–95. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–89. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–92. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Dev Biol. 2012;367:91–9. doi: 10.1016/j.ydbio.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–98. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–8. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–32. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–7. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 35.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–69. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majumder S, Alinari L, Roy S, Miller T, Datta J, Sif S, Baiocchi R, Jacob ST. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J Cell Biochem. 2010;109:553–63. doi: 10.1002/jcb.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384–94. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–23. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–6. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 40.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, et al. GUDMAP project GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–71. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 41.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–18. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121:2641–50. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Wang M, Cai Q, Brooks H, Dressler GR. Pax transactivation-domain interacting protein is required for urine concentration and osmotolerance in collecting duct epithelia. J Am Soc Nephrol. 2007;18:1458–65. doi: 10.1681/ASN.2006060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004;276:337–51. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 45.Pan Z, Zhang J, Li Q, Li Y, Shi F, Xie Z, Liu H. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J Genet Genomics. 2012;39:111–23. doi: 10.1016/j.jgg.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–39. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–58. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu H, Chen L, Zhou Q, Zhang X, Berger S, Bi J, Lewis DE, Xia Y, Zhang W. Aqp2-expressing cells give rise to renal intercalated cells. J Am Soc Nephrol. 2013;24:243–52. doi: 10.1681/ASN.2012080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tae S, Karkhanis V, Velasco K, Yaneva M, Erdjument-Bromage H, Tempst P, Sif S. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 2011;39:5424–38. doi: 10.1093/nar/gkr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen S, Bellew C, Yao X, Stefkova J, Dipp S, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J Biol Chem. 2011;286:32775–89. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saifudeen Z, Liu J, Dipp S, Yao X, Li Y, McLaughlin N, Aboudehen K, El-Dahr SS. A p53-Pax2 pathway in kidney development: implications for nephrogenesis. PLoS One. 2012;7:e44869. doi: 10.1371/journal.pone.0044869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aboudehen K, Hilliard S, Saifudeen Z, El-Dahr SS. Mechanisms of p53 activation and physiological relevance in the developing kidney. Am J Physiol Renal Physiol. 2012;302:F928–40. doi: 10.1152/ajprenal.00642.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]