Abstract
Background and Purpose
ANRIL has long been considered as the strongest candidate gene at the 9p21 locus, robustly associated with stroke and coronary artery disease (CAD). However, the underlying molecular mechanism remains unknown. The present study works to elucidate such a mechanism.
Methods
Utilizing eQTL analysis we identified potential genes whose expression may be influenced by genetic variation in ANRIL. To verify the identified gene(s), knockdown and over-expression of ANRIL was evaluated in HUVECs and HepG2 cells. Ischemic stroke and CAD risk was then evaluated in the gene(s) demonstrated to be mediated by ANRIL in 3 populations of Chinese Han ancestry; two ischemic stroke populations including the Central China cohort (903 cases and 873 controls) and the Northern China cohort (816 cases and 879 controls), and one CAD cohort consisting of 772 patients and 873 controls.
Results
eQTL analysis identified CARD8 among others, with knockdown of ANRIL expression decreasing CARD8 expression and over-expression of ANRIL increasing CARD8 expression. The minor T allele of a previously identified CARD8 variant (rs2043211) was found to be significantly associated with a protective effect of ischemic stroke under the recessive model in two independent stroke cohorts. No significant association was found between rs2043211 and CAD.
Conclusion
CARD8 is a downstream target gene regulated by ANRIL. SNP rs2043211 in CARD8 is significantly associated with ischemic stroke. ANRIL may increase the risk of ischemic stroke through regulation of the CARD8 pathway.
Keywords: ischemic stroke, coronary artery disease, case-control study, single nucleotide polymorphisms, CARD8
Introduction
Atherosclerosis is one of the most common causes of both ischemic stroke and coronary artery diseases (CAD).1 Stroke is a leading cause of morbidity and mortality in China and other parts of the world.2 Ischemic stroke accounts for about 87% of all strokes,3 and is caused by both genetic and environmental factors. To date, large-scale genome-wide association studies (GWAS) have identified several risk loci for ischemic stroke, including HDAC9, PITX2, ZFHX3, 9p21, PRKCH, and NINJ2.4 However, most loci have small effects and may explain a small proportion of the heritability of ischemic stroke. Nearly fifty risk loci for CAD have been identified by GWAS, but explain approximately 10% of the heritability of CAD only. 5 Therefore, more genetic factors for ischemic stroke and CAD remain to be discovered.
SNPs (single nucleotide polymorphisms) at the chromosome 9p21 locus were found to be associated with both ischemic stroke and CAD by GWAS.4, 5 The 9p21 locus contains an annotated non-coding RNA, termed ANRIL (antisense non coding RNA in the INK4 locus). ANRIL is considered as a prime candidate gene for atherosclerosis at the 9p21 locus.6 First, SNPs associated with ischemic stroke and CAD (rs10116277, rs7865618, rs564398, rs496892, rs7044859) within the 9p21 region are located within the ANRIL gene. 7 Second, ANRIL is expressed in cell types and tissues that are involved in atherosclerosis. Third, several studies investigated ANRIL’s association with 9p21 SNP genotypes and showed a correlation of ANRIL expression with atherosclerosis severity, even though the direction of the effects is still in dispute.6, 7 Moreover, the risk alleles of rs10811656 and rs10757278 disrupted a binding site for transcriptional factor STAT1 and STAT1 in turn regulated ANRIL expression.8 The STAT1 signaling pathway mediates responses to inflammation upon stimulation of the pro-inflammatory cytokine interferon gamma.9 These results supported the notion that ANRIL might play a role in the inflammatory response and atherosclerosis. The molecular mechanism by which ANRIL mediates atherosclerosis is unknown. However, as a long noncoding RNA, ANRIL may play its role in atherosclerotic processes by influencing the expression of other genes.
In this study, we identified CARD8 as a downstream gene of ANRIL, and assessed the association between CARD8 SNP rs2043211 and ischemic stroke or CAD in Chinese Han populations.
Materials and Methods
Analysis of Expression Quantitative Loci (eQTLs) for ANRIL SNPs
In order to identify potential downstream genes regulated by ANRIL, we analyzed ischemic stroke- and CAD-associated SNPs rs10116277, rs7865618, rs564398, rs496892, and rs7044859 in ANRIL.7 These SNPs were shown to influence the mRNA level of ANRIL.7 We performed eQTL analysis for these SNPs by searching the database at University of Michigan Center for Statistical Genetics (http://www.sph.umich.edu/csg/liang/imputation/). These studies identified several genes whose expression may be associated with SNPs evaluated in our study. We chose to evaluate CARD8 over the other identified genes because of its increased expression in atherosclerotic lesions.10
Cell Transfection and Quantitative Real-Time PCR (qRT-PCR) Analysis
Details of cell transfection and qRT-PCR were described in online SUPPLEMENTAL MATERIAL. The sequence of ANRIL siRNA was as follows: 5’- GGAATGAGGAGCACAGTGA -3’. Plasmid pcDNA3.1-ANRIL (NR_003529.3) was synthesized by GENEWIZ (Beijing, China). The sequences of primers used for qRT-PCR are listed in Supplement Table I.
Study Subjects
All study participants were selected from the GeneID database.11 Diagnostic criteria for ischemic stroke, CAD, and related factors were described in detail in online SUPPLEMENTAL MATERIAL. This study followed the principals outlined in the Declaration of Helsinki and has been approved by local institutional review boards on human subject research. Written informed consent was obtained from all participants.
Genotyping and Statistical Analysis
Details of isolation of genomic DNA, SNP genotyping, and statistical analysis were described in online SUPPLEMENTAL MATERIAL.
Results
ANRIL Regulates Expression of CARD8
Five 9p21 SNPs rs10116277, rs7865618, rs564398, rs496892, and rs7044859 are located within ANRIL and affect the expression level of ANRIL mRNA.7 By searching a public eQTL database (http://www.sph.umich.edu/csg/liang/imputation/), we identified 87 genes whose expression may be associated with one of the five 9p21 SNPs (online-only Supplement Table I). One of the 87 genes, CARD8, became a strong candidate gene downstream of ANRIL because it also showed differential expression in a preliminary microarray analysis comparing HepG2 cells treated with ANRIL siRNA to those transfected with control siRNA (data not shown).
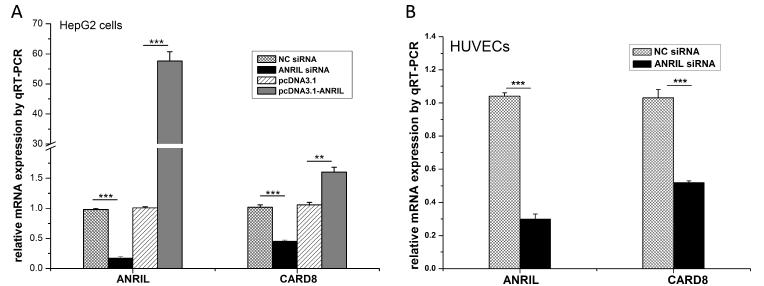
To verify that CARD8 is a downstream gene regulated by ANRIL, HepG2 cells were transfected with ANRIL specific siRNA to knock ANRIL expression down (NC siRNA as negative control) and used for qRT-PCR analysis. Compared to NC siRNA, ANRIL siRNA successfully reduced its own expression by about 83% (P<2.0×10−5) and the expression of CARD8 by about 55% (P<2.4×10−4) (Figure 1A). Similarly, HUVECs transfected with ANRIL specific siRNA showed significant reduction of ANRIL by 70% (P<1.5×10−5) and CARD8 by 48% (P<6.7×10×4) compared to cells with NC siRNA (Figure 1B). These data suggest that ANRIL regulates the expression of CARD8. Consistent with the siRNA studies, HepG2 cells transfected with pcDNA3.1-ANRIL for 48 hrs showed a 57-fold increase in ANRIL mRNA expression (P<5.84×10−5) and 1.6-fold increase in CARD8 mRNA expression (P<3.1×10−3) (Figure 1A). Due to a difficulty in transfection of the specific line of HUVECs under this study with plasmid DNA, we did not obtain any data on the effect of ANRIL overexpression on CARD8 in HUVECs.
Figure 1.
Effect of knockdown or over-expression of ANRIL on regulation of CARD8 by quantitative RT–PCR (qRT–PCR) analysis. The mRNA samples were prepared from transfected HUVECs or HepG2 cells. ACTIN was used as a control for normalization. (A) CARD8 gene was significantly down-regulated about two fold by ANRIL knockdown in HepG2 cells. Expression of ANRIL was decreased by 5.7-fold and CARD8 expression was reduced by 2-fold. Transfection with pcDNA3.1-ANRIL increased the expression of ANRIL by 57-fold and CARD8 expression by 1.6-fold. Each experiment was preformed three times in triplicate. (B) CARD8 gene was significantly down-regulated about two fold by ANRIL knockdown in HUVECs. ***: P<0.001; **: P <0.01; *: P<0.05.
Characteristics of Study Subjects
Two independent cohorts were used to assess whether CARD8 SNP rs2043211 is associated with ischemic stroke. The discovery cohort for the ischemic stroke study consisted of 903 cases and 873 controls enrolled from Hubei Province in Central China. The replication cohort for the ischemic stroke study consisted of 816 cases and 879 controls enrolled from hospitals in Northern China (Table 1). The case-control cohort for the CAD study consisted of 772 patients with CAD and 873 controls from Hubei Province in Central China (Table 1). Patients with ischemic stroke or CAD had a higher prevalence of conventional risk factors, including smoking, history of hypertension,diabetes and a lower level of HDL-C (Table 1).
Table 1.
Clinical and demographical characteristics of study populations.
| GenelD-Central |
GenelD-North |
GenelD-Central |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Ischemic Stroke |
Control | p value | Ischemic Stroke |
Control | p value | CAD | Control | p value |
| Subject numbers | 903 | 873 | N.A | 816 | 879 | N.A | 772 | 873 | N.A |
| Age (years) * | 68.3±14.0 | 45.2±15.1 | <1.00E-50 | 60.2±12.5 | 52.1±17.0 | <1.00E-50 | 66.1±12.5 | 45.2±15.1 | <1.00E-50 |
| Sex (male%) | 61.1 | 55.7 | 4.30E-03 | 57.7 | 55.9 | 4.71E-06 | 69.3 | 55.7 | 3.98E-15 |
| Smoking (%) | 18.6 | 6.6 | 5.98E-26 | 22.4 | 17.9 | <1.00E-50 | 26.8 | 6.6 | <1.00E-50 |
| BMI (kg/m2) | 24.3±0.3 | 23.4±2.0 | <1.00E-50 | 24.4±2.7 | 24.6±1.3 | 3.89E-04 | 24.1±1.5 | 23.4±2.0 | 1.08E-28 |
| Hypertension (%) | 80.1 | 13.9 | <1.00E-50 | 58.5 | 26.6 | <1.00E-50 | 69.9 | 13.9 | <1.00E-50 |
| DM | 24.4 | 3.6 | <1.00E-50 | 10.3 | 13.3 | <1.00E-50 | 25.3 | 3.6 | <1.00E-50 |
| Tch (mmol/1) | 4.58±1.01 | 4.36±0.46 | 3.53E-17 | 4.57±0.51 | 4.36±1.13 | 1.64E-12 | 4.40±0.92 | 4.36±0.46 | 0.15 |
| TG (mmol/1) | 1.49±0.97 | 1.50±0.50 | 0.79 | 1.75±0.78 | 1.54±1.12 | 9.80E-11 | 1.59±1.00 | 1.50±0.50 | 0.67 |
| HDL-c (mmol/1) | 1.10±0.27 | 1.19±0.14 | 1.09E-35 | 1.20±0.19 | 1.36±1.24 | 7.23E-08 | 1.12±0.27 | 1.19±0.14 | 9.51E-19 |
| LDL-c (mmol/1) | 2.77±0.80 | 2.60±0.39 | 3.14E-15 | 2.61±0.41 | 2.40±0.83 | 1.57E-19 | 2.62±0.76 | 2.60±0.39 | 0.36 |
The data are presented as the means ± standard deviation or percent;
Age for the case group is the age at diagnosis; age for the control group is the age at which the study subject was enrolled;
BMI, body mass index; DM, diabetes; Tch, total cholesterol; TG, triglyceride
HDL-C, high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol;
Statistical power analysis was performed for all three cohorts prior to each study. Each cohort had >90% of power to detect an association between rs2043211 and ischemic stroke or CAD with an OR of ≥1.20 at the nominal type I error rate of >0.05 and a minor allelic frequency of >0.43 for rs2043211 in the Chinese population (HapMap HCB data).10
Significant Genotypic Association between SNP rs2043211 and Ischemic Stroke in Two Independent Chinese Populations
The genotyping data for rs2043211 did not deviate from the Hardy-Weinberg equilibrium in the control group (P>0.05). In the discovery population for the ischemic stroke study, no significant allelic association was detected between rs2043211 and ischemic stroke (P-obs=0.077, P-adj=0.092, online-only Supplemental Table III). Similarly, no significant association between rs2043211 and ischemic stroke was detected in the replication cohort or the combined discovery/replication population (P>0.05, online-only Supplemental Table III).
Genotypic association analysis was then conducted because this type of study can provide genetic insights into the association under different inheritance models (additive, dominant or recessive). Interestingly, the minor allele T of SNP rs2043211 showed significant association with a protective effect of ischemic stroke under either a recessive model (P-obs=3.0×10−4) or an additive mode (P-obs=2.88×10−4) (Table 2). After multivariate logistic regression analysis by adjusting for covariates of the age, sex, BMI, smoking history, hypertension, diabetes mellitus and lipid concentrations, the genotypic association between rs2043211 and ischemic stroke remained significant only under the recessive model (P-adj=0.028, OR=0.68, Table 2).
Table 2.
Genotypic association analysis between rs2043211 and ischemic stroke.
| Cohorts | Model | Case(n) | Control(n) | P-obs | P-adj | OR(95%CI) |
|---|---|---|---|---|---|---|
| GenelD-Central IS (903/873) | ||||||
| Additive | 173/521/209 | 230/430/213 | 2.88E-04 | 0.082 | 0.83(0.67-1.02) | |
| Dominant | 694/209 | 660/213 | 0.535 | 0.520 | 1.12(0.80-1.56) | |
| Recessive | 173/730 | 230/643 | 3.00E-04 | 0.028 | 0.68(0.48-0.96) | |
| GenelD-North IS (816/879) | ||||||
| Additive | 174/464/178 | 235/404/240 | 4.26E-05 | 0.692 | 0.97(0.81-1.15) | |
| Dominant | 638/178 | 639/240 | 0.009 | 0.085 | 1.28(0.97-1.69) | |
| Recessive | 174/642 | 235/644 | 0.009 | 0.017 | 0.70(0.53-0.94) | |
| Combined IS (1719/1752) | ||||||
| entire cohort | Additive | 347/985/387 | 465/834/453 | 3.13E-08 | 0.324 | 0.94(0.83-1.06) |
| entire cohort | Dominant | 1332/387 | 1299/453 | 0.021 | 0.075 | 1.19(0.98-1.45) |
| entire cohort | Recessive | 347/1372 | 465/1287 | 9.78E-06 | 5.83E-04 | 0.70(0.57-0.86) |
| Female | Recessive | 127/561 | 203/563 | 2.57E-04 | 0.007 | 0.65(0.48-0.89) |
| Male | Recessive | 220/811 | 262/724 | 0.006 | 0.024 | 0.73(0.56-0.96) |
| Age<60 | Recessive | 104/442 | 360/965 | 2.17E-04 | 0.001 | 0.61(0.45-0.83) |
| Age≥60 | Recessive | 243/930 | 105/322 | 0.100 | 0.107 | 0.79(0.59-1.05) |
| Female and Age<60 | Recessive | 43/206 | 155/421 | 0.003 | 0.004 | 0.51(0.32-0.80) |
| Male and Age<60 | Recessive | 61/236 | 205/544 | 0.022 | 0.104 | 0.71(0.48-1.08) |
Model: Additive(TT/AT/AA), Dominant(TT+AT/AA),Recessive(TT/AT+AA); OR, odds ratio;
P-ohs, P value using 2 × 2 contingency table χ2 tests before adjustment for covariates;
P-adj, P value adjusted by multivariate logistic regression analysis for traditional risk factors, including age, sex, BMI, smoking history, hypertension, DM, TC, TG, HDL- and LDL-C)
To confirm the initial finding of genotypic association between rs2043211 and ischemic stroke in the discovery population, we validated the finding in an independent replication cohort. The results showed that rs2043211 was also significantly associated with a protective effect of ischemic stroke under either a recessive model (P-obs=9.00×10−3) or an additive mode (P-obs=4.26×10−5) (Table 2). The genotypic association between rs2043211 and ischemic stroke remained significant only under the recessive model in the replication population after multivariate logistic regression analysis (P-adj=0.017, OR=0.70, Table 2).
For the combined population of the discovery and replication cohorts, the P value for the genotypic association between rs2043211 and ischemic stroke under the recessive model became much more significant (P-obs=9.78×10−6, P-adj=5.83×10−4, OR=0.70, Table 2 and online-only Supplemental Figure I). These data suggest that SNP rs2043211 confers a protective effect of ischemic stroke under a recessive model of inheritance.
The genotypic association between rs2043211 and ischemic stroke under the recessive model was more significant in the female group (P-adj=0.007, OR=0.65) than in the male group (P-adj=0.024, OR=0.73, Table 2 and online-only Supplemental Figure I). The genotypic association was significant in the early-onset (<60 years) ischemic stroke group under the recessive model (P-adj=0.001, OR=0.61, Table 2 and online-only Supplemental Figure I). The genotypic association was stronger under the recessive model for the female early-onset ischemic stroke group (P-adj=0.004, OR=0.51, Table 2 and online-only Supplemental Figure I).
Lack of Significant Association between SNP rs2043211 and CAD
We also analysed SNP rs2043211 for its association with CAD. In a case-control study with 772 CAD cases and 873 controls, SNP rs2043211 did not show any significant association with CAD in the standard allelic association analysis (P-obs=0.235, P-adj=0.300) or in the genotypic association analysis under three different genetic models (all P>0.05). The association remained non-significant in either male or female CAD groups (online-only Supplemental Table IV).
Discussion
In the present study, we identified CARD8 as a downstream gene regulated by ANRIL, and demonstrated an association between the CARD8 SNP rs2043211 and ischemic stroke in two independent Chinese Han populations. CARD8 encodes a member of the caspase recruitment domain (CARD)-containing family, and is also known as TUCAN/CARDINAL. Previous population-based studies found that the functional SNP rs2043211 (p.C10X) located in exon 5 of CARD8 may be a genetic risk factor for chronic inflammatory diseases, such as inflammatory bowel disease (IBD) and rheumatoid arthritis (RA). Our finding that rs2043211 is associated with ischemic stroke could indicate a shared inflammatory response or pathway similar to IBD and/or RA.
SNP rs2043211 has been previously associated with other diseases. Roberts et al observed a significant genotypic association in New Zealand between rs2043211 and abdominal aortic aneurysm (P=0.047, OR=0.83),12 which is a disease that shared some similar pathological characteristics and risk factors with atherosclerosis, such as inflammation and angiogenesis. However, Paramel et al found that there was no significant association between rs2043211 and MI (FIA Cohort: P=0.10, OR=1.1; SCARF: P=0.66, OR=1.0).10 Garcia-Bermudez et al found that there was no evidence for the role of rs2043211 in the development of CV events in Spanish RA patients (P=0.67, OR=0.96). 13 Meanwhile, there were still some conflicting results in association studies between rs2043211 and some chronic inflammatory diseases. Roberts et al found that the minor allele T of rs2043211 conferred a potential protective effect against early disease onset of Crohn's disease in New Zealand Caucasians.14 But other studies reported that the minor allele T of rs2043211 was associated with increased severity of IBD(P=0.001, OR=1.5015 and RA16 as well as increased risk of Alzheimer’s disease in women (P=0.01, OR=2.39)17. Intriguingly, SNP rs2043211 was associated with ischemic stroke, but not with CAD in this study. These results are consistent with the findings for MI and CV events by Paramel et al10 and Garcia-Bermudez et al.13 Although the underlying molecular mechanism for the positive association with ischemic stroke and negative association with CAD by rs2043211 is not known, it is possible that the role of ANRIL-regulated CARD8 pathway may be limited to cerebral infarction but not to CAD.
CARD8 acts as an adaptor molecule that negatively regulates NF-κB activation, CASP1-dependent IL1β secretion, and apoptosis, and reduces the inflammatory response.18 SNP rs2043211 results in an A to T transversion that changes codon 10 into a stop codon in CARD8 mRNA (Cys10Stop). Previous studies showed that homozygotes for the stop codon allele T can reduce the expression of CARD8 and impair the NF-κB-inhibiting property of CARD8 in vitro.19 Moreover, the expression of CARD8 mRNA was significantly higher in atherosclerotic lesions from carotid artery plaque tissue in ischemic cerebrovascular patients compared to iliac arteries from brain dead transplant donors.10 But, more studies will be needed to investigate how CARD8 SNP rs2043211 influences the process of ischemic stroke.
These data suggest that ANRIL acts as an important modulator for expression of its downstream gene CARD8. This is consistent with the suggestion that ANRIL might modulate atherogenic diseases by cis- or trans-acting effects.6 Recently, Holdt et al suggested a novel role for Alu elements in epigenetic gene regulation by long ncRNAs.20 Importantly, trans-regulation was dependent on Alu motifs, which marked the promoters of ANRIL target genes and were mirrored in ANRIL RNA transcripts.20 As with other downstream target genes, the detailed mechanism by which ANRIL regulates CARD8 expression remains to be identified. We performed bioinformatic analysis of the promoter sequence of CARD8 and surprisingly found that the same core Alu motif was also present in the DNA promoter sequence of CARD8. Thus, the mechanism by which ANRIL regulates CARD8 expression may be that ANRIL binds to chromatin through interaction via the Alu motif.
There are some limitations in the present study. First, although the mRNA level of CARD8 is regulated by ANRIL, the precise underlying molecular mechanism is not clear. Second, the sample sizes are fixed and frequencies of covariates including age, gender, smoking history, hypertension, diabetes as well as lipid concentrations were significantly lower in controls than in cases. Nevertheless, the observed association remained significant after adjustment of covariates using multiple logistic regression analysis. Third, although rs2043211 is a functional SNP, it may still serve as a genetic marker, and further studies are needed to establish the cause-effect relationship. Fourth, in addition to CARD8, our preliminary microarray analysis identified another gene, DDX58, which also showed >2-fold differential expression in HepG2 cells treated with ANRIL siRNA compared to those transfected with control siRNA. DDX58, also referred to as RIG-1, is involved in host antivirus immunity by sensing viral RNAs and triggering innate antiviral responses.21 Because an antiviral response does not appear to be closely linked to atherosclerosis and stroke, DDX58 was not assessed for its association with stroke in this study. However, because DDX58 is regulated by ANRIL, it may still be interesting to investigate whether variants in DDX58 are associated with atherosclerosis in the future. Finally, previous studies showed that the 9p21 locus (the ANRIL locus) is associated with large-vessel atherosclerotic stroke, thus, it may be interesting to investigate whether CARD8 variant is associated with a specific stroke subtype in the future. Several schemes were reported to classify stroke subtypes, including the Trial of ORG 10172 in Acute Stroke Treatment (TOAST) classification system (TOAST), the Causative Classification System (CCS), or the phenotypic System A-S-C-O (A for atherosclerosis, S for small vessel disease, C for cardiac source, O for other cause).22 In the present study, many patients with specific subtypes were excluded, thus we were unable to perform association studies with specific stroke subtypes. In the future, we can collect more clinical information, including the data from the carotid study, classify the cases into different subtypes and assess the association between CARD8 SNP rs2043211 and individual stroke subtype.
Summary
In conclusion, here we show that ANRIL can regulate the expression level of CARD8 mRNA and a functional SNP in CARD8, rs2043211 located in exon 5 of CARD8 was significantly associated with ischemic stroke, but not with CAD in the Chinese Han population. Although the detailed mechanism of CARD8 in the pathogenesis of ischemic stroke remains unclear, this study indeed links the ANRIL regulated CARD8 pathway to the development of ischemic stroke.
Supplementary Material
Acknowledgements
We thank one of the reviewers for restructuring the Abstract and for many valuable suggestions to improve the manuscript.
Sources of Funding
This study was supported by grants from the National Basic Research Program of China (973 Program No. 2013CB0531101 and 2012CB517800), the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09), NIH R01 HL094498, the Program for New Century Excellent Talents in University of China (NCET-11-0181), the Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, the National Natural Science Foundation of China (No. 81270163 and 81070106), and a grant from the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FB16).
Footnotes
Disclosure
The research at Center for Human Genome Research on lipids and CAD is supported by a collaborative grant from Merck.
References
- 1.Duvall WL, Vorchheimer DA. Multi-bed vascular disease and atherothrombosis: scope of the problem. J Thromb Thrombolysis. 2004;17:51–61. doi: 10.1023/B:THRO.0000036029.56317.d1. [DOI] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2012 update: a report from the american heart association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 4.Amouyel P. From genes to stroke subtypes. Lancet Neurol. 2012;11:931–933. doi: 10.1016/S1474-4422(12)70235-1. [DOI] [PubMed] [Google Scholar]
- 5.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 7.Cunnington MS, Santibanez KM, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 snps associated with multiple disease phenotypes correlate with anril expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 dna variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaren JE, Ramji DP. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125–135. doi: 10.1016/j.cytogfr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Paramel GV, Folkersen L, Strawbridge RJ, Elmabsout AA, Sarndahl E, Lundman P, et al. Card8 gene encoding a protein of innate immunity is expressed in human atherosclerosis and associated with markers of inflammation. Clin Sci (Lond) 2013;125:401–407. doi: 10.1042/CS20120572. [DOI] [PubMed] [Google Scholar]
- 11.Shi L, Li C, Wang C, Xia Y, Wu G, Wang F, et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a chinese han population. Hum Genet. 2009;126:843–849. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- 12.Roberts RL, Van Rij AM, Phillips LV, Young S, McCormick SP, Merriman TR, et al. Interaction of the inflammasome genes card8 and nlrp3 in abdominal aortic aneurysms. Atherosclerosis. 2011;218:123–126. doi: 10.1016/j.atherosclerosis.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Bermudez M, Lopez-Mejias R, Gonzalez-Juanatey C, Corrales A, Castaneda S, Ortiz AM, et al. Card8 rs2043211 (p.c10x) polymorphism is not associated with disease susceptibility or cardiovascular events in spanish rheumatoid arthritis patients. DNA Cell Biol. 2013;32:28–33. doi: 10.1089/dna.2012.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts RL, Topless RK, Phipps-Green AJ, Gearry RB, Barclay ML, Merriman TR. Evidence of interaction of card8 rs2043211 with nalp3 rs35829419 in crohn's disease. Genes Immun. 2010;11:351–356. doi: 10.1038/gene.2010.11. [DOI] [PubMed] [Google Scholar]
- 15.Yang SK, Kim H, Hong M, Lim J, Choi E, Ye BD, et al. Association of card8 with inflammatory bowel disease in koreans. J Hum Genet. 2011;56:217–223. doi: 10.1038/jhg.2010.170. [DOI] [PubMed] [Google Scholar]
- 16.Kastbom A, Johansson M, Verma D, Soderkvist P, Rantapaa-Dahlqvist S. Card8 p.c10x polymorphism is associated with inflammatory activity in early rheumatoid arthritis. Ann Rheum Dis. 2010;69:723–726. doi: 10.1136/ard.2008.106989. [DOI] [PubMed] [Google Scholar]
- 17.Fontalba A, Gutierrez O, Llorca J, Mateo I, Berciano J, Fernandez-Luna JL, et al. Deficiency of card8 is associated with increased alzheimer's disease risk in women. Dement Geriatr Cogn Disord. 2008;26:247–250. doi: 10.1159/000160956. [DOI] [PubMed] [Google Scholar]
- 18.Razmara M, Srinivasula SM, Wang L, Poyet JL, Geddes BJ, DiStefano PS, et al. Card-8 protein, a new card family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 19.Bagnall RD, Roberts RG, Mirza MM, Torigoe T, Prescott NJ, Mathew CG. Novel isoforms of the card8 (tucan) gene evade a nonsense mutation. Eur J Hum Genet. 2008;16:619–625. doi: 10.1038/sj.ejhg.5201996. [DOI] [PubMed] [Google Scholar]
- 20.Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Alu elements in anril non-coding rna at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goubau D, Deddouche S, Reis ESC. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marnane M, Duggan CA, Sheehan OC, Merwick A, Hannon N, Curtin D, et al. Stroke subtype classification to mechanism-specific and undetermined categories by toast, a-s-c-o, and causative classification system: direct comparison in the north dublin population stroke study. Stroke. 2010;41:1579–1586. doi: 10.1161/STROKEAHA.109.575373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.