Abstract
Circadian clocks are self-sustained cellular oscillators that synchronize oxidative and reductive cycles in anticipation of the solar cycle. We found that the clock transcription feedback loop produces cycles of nicotinamide adenine dinucleotide (NAD+) biosynthesis, adenosine triphosphate production, and mitochondrial respiration through modulation of mitochondrial protein acetylation to synchronize oxidative metabolic pathways with the 24-hour fasting and feeding cycle. Circadian control of the activity of the NAD+-dependent deacetylase sirtuin 3 (SIRT3) generated rhythms in the acetylation and activity of oxidative enzymes and respiration in isolated mitochondria, and NAD+ supplementation restored protein deacetylation and enhanced oxygen consumption in circadian mutant mice. Thus, circadian control of NAD+ bioavailability modulates mitochondrial oxidative function and organismal metabolism across the daily cycles of fasting and feeding.
The circadian clock is a molecular oscillator that enables light-sensitive organisms to coordinate nutrient storage and use in anticipation of daily periods of activity and rest. Circadian disruption in eubacteria and plants impairs metabolic efficiency and alters growth and reproduction (1, 2). Circadian and metabolic cycles are also closely coupled processes in animals, and at least 10% of all mRNAs exhibit daily oscillations in abundance in mammalian liver, including those from many genes involved in metabolic processes (3–9). Although mouse genetic studies have revealed that clock transcription factors modulate insulin secretion and maintain glucose homeostasis when animals are awake and feeding (10–13), whether the circadian system might also help to maintain energy homeostasis during fasting is less well understood.
Integration of circadian and metabolic cycles is suggested by studies showing that cellular redox status influences the activity of clock transcription factors (14). The abundance of the oxidoreductase factor nicotinamide adenine dinucleotide (NAD+) displays circadian rhythmicity due to direct clock transcriptional control of the rate-limiting enzyme in NAD+ biosynthesis, NAMPT (nicotinamide phosphoribosyltransferase) (15, 16). In turn, NAD+ salvage completes a short feedback loop by driving the activity of the deacetylase SIRT1, which regulates the activity of the CLOCK-and BMAL1-mediated forward limb of the clock (15, 16). In mitochondria, NAD+ also regulates SIRT3, a mitochondrial deacetylase important in fatty acid oxidation during fasting (17, 18). Thus, we hypothesized that the circadian clock may control mitochondrial function through rhythmic control of NAD+ biosynthesis and posttranslational protein deacetylation to facilitate lipid oxidation during fasting.
Circadian Clock Regulates Mitochondrial Oxidative Metabolism
We examined the role of the circadian clock in 24-hour oxidative cycles by determining whether fatty acid oxidation (FAO), measured by the rate of oxidation of [14C]oleate to [14C]CO2, displayed an endogenous circadian rhythm. We monitored FAO and NAD+ in liver homogenates every 4 hours over the course of 48 hours from fasted wild-type mice maintained in constant darkness (Fig. 1A and supplementary text). We observed ~24-hour oscillations of FAO (Fig. 1A, blue line), with peaks occurring near the end of the rest period [circadian time (CT) 8 to 12; CT 32 to 36] that coincided with the rhythms of total cellular NAD+ (Fig. 1A, dashed yellow line) and mitochondrial NAD+ (fig. S3A). Although an ultradian oscillation of NAD+ abundance occurred in ad libitum–fed mice (15), in fasted mice a single peak of NAD+ occurred during the rest period (Fig. 1A), suggesting that the 12-hour harmonic observed in the ad libitum state may be driven by behavioral feeding rhythms.
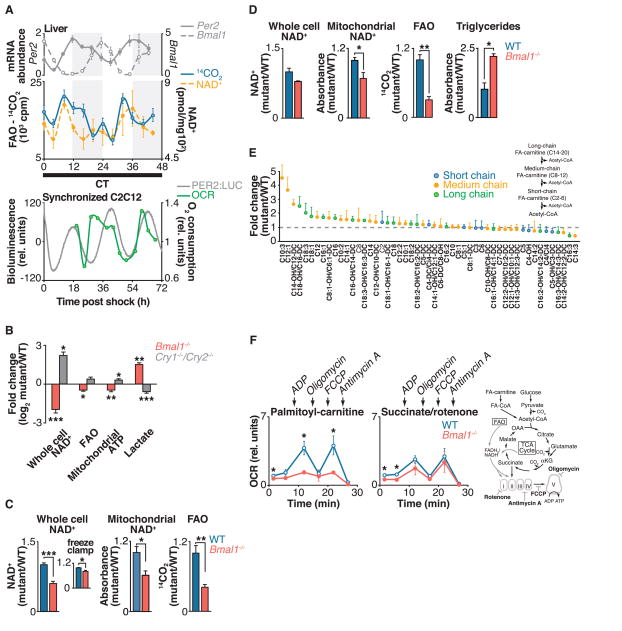
Fig. 1. Circadian clock control of mitochondrial oxidative function.
(A) Top: Liver FAO and NAD+ were measured every 4 hours for 48 hours in fasted wild-type mice kept in constant darkness. Shading indicates where light and dark periods would normally occur under 12:12 light-dark conditions. Relative mRNA expression of Per2 and Bmal1, as measured by qPCR, is shown above for reference to circadian phase (n = 4 per time point). Bottom: Relative oxygen consumption rate (OCR) in synchronized C2C12 cells, with PER2::LUC bioluminescence shown for reference to circadian phase (n = 1 per time point). (B) Relative amounts of NAD+, FAO, mitochondrial ATP (in galactose-containing media), and lactate (in glucose-containing media) in Bmal1−/− and Cry1−/−;Cry2−/− MEFs compared to wild-type (WT) controls (n = 6). (C and D) Relative whole-cell and mitochondrial NAD+, FAO, and triglycerides in liver from fasted Bmal1−/− liver compared to littermate controls at (C) ZT0 (n = 4 to 9) and (D) ZT12 (n = 3 or 4). Inset in (C): Relative NAD+ from freeze-clamped liver tissue (n = 5). (E) Acyl-carnitine profile in liver of Bmal1−/− mice compared to WT (n = 3). (F) OCR in mitochondria isolated from liver of fasted WT and Bmal1−/− mice in the presence of palmitoyl-carnitine or succinate plus rotenone treated sequentially with ADP, oligomycin, FCCP, and antimycin A at ZT0 (n = 3 or 4). Schematic shows FAO and TCA cycle pathways, and indicates both OCR substrates and metabolic inhibitors used to evaluate mitochondrial function in circadian mutants. *P < 0.05, **P < 0.01, ***P < 0.001 for Student’s two-tailed t test comparing single time points between WT and mutant averages. Data are represented as average ± SEM.
To determine whether the circadian clock regulates mitochondrial respiration cycles in addition to FAO, we monitored rhythms of oxygen consumption in live cell cultures of mouse C2C12 myoblasts, which when maintained as myotubes display robust PER2 rhythms after synchronization by exposure to a high concentration of serum (Fig. 1A). The C2C12 cycles of NAD+ accumulation and oxygen consumption were self-sustained across two complete circadian cycles and in phase with PER2 oscillation, indicating that they were not an acute response to serum exposure (Fig. 1A, green line, and fig. S4A). We further observed a 24-hour variation in glucose oxidation that was offset by 4 hours in phase from the rhythm of FAO (fig. S4B). Together, these data indicate the presence of intrinsic cycles of mitochondrial oxidative metabolism across the 24-hour day, independent of the feeding cycle.
To determine whether expression of the core molecular clock transcription factors has an impact on mitochondrial oxidative metabolism, we next analyzed respiration in mouse embryonic fibroblasts (MEFs) isolated from mice deficient in either the clock transcriptional activators (CLOCK and BMAL1) or repressors (CRY1 and CRY2) (Fig. 1B). MEFs lacking the clock activator gene Bmal1 displayed decreased FAO and NAD+ concentrations, whereas MEFs lacking the clock repressors Cry1 and Cry2 showed increased FAO and amounts of NAD+ (Fig. 1D and fig. S1), indicating opposing effects of the forward (CLOCK- and BMAL1-mediated) and reverse (CRY- and PER-mediated) limbs of the clock on both NAD+ biosynthesis and oxidative metabolism, independent of cellular nutrient availability.
Consistent with impaired mitochondrial energy production from FAO, MEFs lacking the clock activator Bmal1 produced ~30% less adenosine triphosphate (ATP) when maintained in medium containing galactose, which shifts ATP production from glycolysis to mitochondrial oxidative metabolism (19). In parallel, Bmal1−/− MEFs produced ~3 times as much lactate when maintained in glucose-containing medium (Fig. 1B and fig. S1), demonstrating a dependence in these cells on ATP production through glycolysis. Conversely, clock repressor mutant MEFs displayed increased mitochondrial ATP production by ~30% in galactose and decreased lactate production in glucose-containing medium (Fig. 1B and fig. S1), consistent with increased mitochondrial oxidative metabolism.
In support of altered glycolytic metabolism in Bmal1−/− mutants, we observed increased expression of key glycolytic genes at multiple time points, not only in MEFs but also in liver, including liver pyruvate kinase (L-Pk), a catalyst of the final ATP-generating step in glycolysis and an established clock gene target that oscillates during fasting (fig. S5A) (20), as well as phosphofructokinase 1 (Pfk1), a rate-limiting glycolytic enzyme (fig. S5, B and C). In contrast, we observed decreased expression of these genes in Cry1−/−;Cry2−/− MEFs (fig. S5B). The increased glycolysis in Bmal1 mutants is unlikely to be an adaptive response to decreased NAD+, because glycolytic gene expression was normal in mice with liver-specific Nampt ablation (fig. S5D), although these animals exhibited decreased NAD+, FAO, and mitochondrial oxygen consumption (Fig. 2C). Likewise, MEFs treated with n-[4-(1-benzoyl-4-piperidinyl)butyl]-3-(3-pyridinyl)-2E-propenamide (FK866), a specific inhibitor of nicotinamide phosphoribosyltransferase (NAMPT), displayed normal lactate production despite having low amounts of NAD+ (fig. S5E). Thus, circadian clock transcription factors appear to influence oxidative metabolism through modulation of lipid and glucose oxidation in addition to glycolysis.
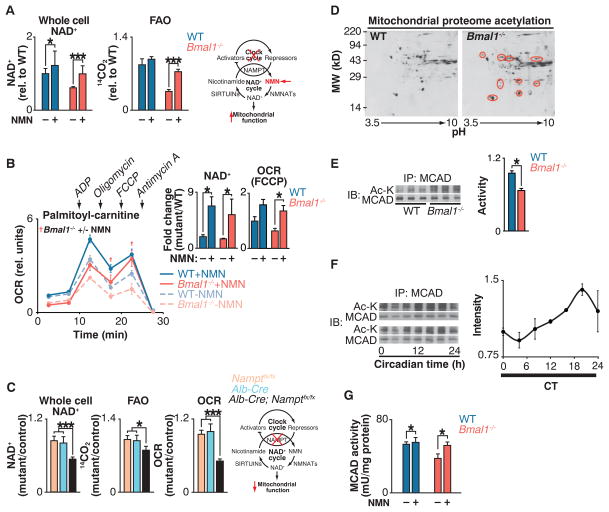
Fig. 2. Clock control of NAD+-dependent mitochondrial metabolism and protein acetylation.
(A) NAD+ and FAO in liver of WT and Bmal1−/− mice injected with either saline or NMN (250 mg/kg) 12 hours before killing at ZT0 (n = 6). Schematic shows interaction of clock with NAD+ salvage pathway, where the intermediate NMN bypasses the Nampt defect in circadian mutants. (B) OCR with palmitoyl-carnitine as substrate in mitochondria isolated from liver of fasted WT and Bmal1−/− mice that had been injected once a day for 10 days with either saline or NMN (500 mg/kg) before killing at ZT0 (n = 3). Inset: Whole-cell NAD+ after 10 days of NMN injections and quantification of uncoupled (FCCP) OCR. †P < 0.05 indicates Student’s two-tailed t test comparison between Bmal1−/− mitochondria with and without NMN treatment. (C) Relative NAD+, FAO, and palmitoyl-carnitine OCR in livers of liver-specific Nampt−/− animals (n = 3 to 6). (D) Two-dimensional gel electrophoresis of liver mitochondrial proteome from WT and Bmal1−/− mice at ZT0. Red circles represent acetylated proteins. (E) Relative MCAD acetylation and activity in WT and Bmal1−/− fasted liver mitochondrial extracts at ZT0 (n = 6). (F) Relative MCAD acetylation measured every 4 hours for 24 hours in fasted wild-type mice kept in constant darkness (n = 4). (G) Relative MCAD activity in liver of WT and Bmal1−/− mice injected with either saline or NMN (250 mg/kg) 12 hours before killing at ZT0 (n = 6). *P < 0.05, ***P < 0.001 for Student’s two-tailed t test comparing single time points between WT and mutant averages. Data are represented as average ± SEM.
We examined mitochondrial function in liver of mice lacking the clock activator gene Bmal1. Liver tissue of fasted mice at zeitgeber time (ZT) 0 and ZT12 [corresponding with the trough and peak of NAD+ in animals maintained on a 12:12 light-dark cycle (15)] showed reduced FAO and decreased amounts of NAD+ in whole cells and mitochondria (Fig. 1, C and D); Bmal1−/− MEFs. We used freeze-clamp isolation of liver to evaluate the potential for post mortem decline in amounts of NAD+. Freeze-clamping increased the absolute amount of liver NAD+ by ~17%, but we still observed a significant NAD+ deficit in Bmal1−/− liver relative to that in control liver (Fig. 1C, inset) (21). Amounts of hepatic FAO and NAD+ were decreased in liver-specific Bmal1−/− mutant mice, which suggested a tissue-specific role for the liver clock in regulation of NAD+ biosynthesis and mitochondrial oxidative function (fig. S2A). Consistent with defective FAO, hepatic triglyceride levels were increased by a factor of 2.2 in Bmal1−/− mutants at ZT12, corresponding with peak FAO in wild-type mice (Fig. 1D). Mass spectrometry analysis demonstrated an accumulation of long- and medium-chain fatty acyl-carnitines with normal amounts of the short-chain species in fasted Bmal1−/− liver, indicating a physiologic role for the clock in energy homeostasis during fasting (Fig. 1E).
Impaired Oxidative Capacity in Isolated Mitochondria After Circadian Disruption
To address the impact of the circadian clock on intrinsic mitochondrial biochemical pathways, we isolated intact mitochondria from liver of wild-type and Bmal1−/− mice at both ZT0 and ZT12 and monitored their oxygen consumption rate (OCR) in response to lipid and tricarboxylic acid (TCA) cycle intermediates (22). OCR measurements in isolated mitochondria were made in the presence of (i) adenosine diphosphate (ADP) to induce coupled respiration, (ii) the ATP synthase inhibitor oligomycin to measure OCR due to proton leak (uncoupled respiration), (iii) the drug FCCP [carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone] to uncouple electron transport from the proton gradient generated by ATP synthase and thereby measure maximal flux through the electron transport chain, and (iv) the complex III inhibitor antimycin A to completely halt electron flow and thereby measure nonrespiratory OCR (Fig. 1F, fig. S3, B and C, and supplementary text). Consistent with an intrinsic defect in mitochondrial FAO, Bmal1−/− mitochondria had reduced OCR in the presence of long-chain fatty acids (palmitoyl-carnitine) as substrate (Fig. 1F and fig. S3, B and C). This defect was apparent during both coupled (in the presence of ADP) and uncoupled (in the presence of FCCP) respiration, indicating that the mutant mitochondria possess defects in the delivery of electrons from oxidative pathways to the electron transport chain (ETC). In contrast, OCR was normal in circadian mutant mitochondria in response to succinate plus rotenone, which transfers electrons directly to complex II of the ETC, indicating that the ETC remains intact in the clock mutants (Fig. 1F). OCR in the presence of FCCP was also measured in circadian mutant mitochondria treated with other mitochondrial substrates, including medium-chain acyl-carnitine (octanoyl-carnitine), glutamate plus malate, and pyruvate plus malate (fig. S3, B and C). In addition to impaired OCR in circadian mutants in the presence of acyl-carnitines, we observed decreased OCR in the presence of pyruvate plus malate, but not in the presence of glutamate plus malate. Together, these data indicate defects at the level of the β-oxidation pathway in addition to pyruvate entry into the TCA cycle (Fig. 1F and fig. S3, B and C).
All OCR measurements were normalized to mitochondrial protein content in wild-type and mutant animals. Thus, the defects in OCR were not caused by differences in mitochondrial biogenesis. Further, Bmal1−/− mutant livers contained normal numbers of mitochondria and displayed normal expression of enzymes that function in mitochondrial biogenesis and FAO, including targets of peroxisome proliferator–activated receptor γ coactivator 1α (PGC-1α), a central regulator of mitochondrial biogenesis (fig. S6). Together with the experiments using isolated mitochondria, these data suggest intrinsic defects in liver mitochondrial oxidative pathways in circadian mutants.
Low NAD+ Contributes to Impaired Mitochondrial Function in Clock Mutants
To test whether NAD+ deficiency causes mitochondrial oxidative defects in circadian mutant mice, we sought to restore NAD+ levels in circadian mutant animals through administration of nicotinamide mononucleotide (NMN), a pro-drug that is the product of the NAMPT reaction (23, 24) (Fig. 2A). Injection of animals with NMN increased total cellular NAD+ in Bmal1−/− mouse liver to wild-type levels (Fig. 2A) and increased mitochondrial NAD+ in the mutants from 53% to 73% of that in wild-type livers (fig. S3D). NMN treatment increased FAO in Bmal1 mutant mice by a factor of ~2, from 41% to 84% of that in wild-type livers (Fig. 2A). Likewise, NMN significantly increased oxygen consumption during uncoupled (FCCP) respiration in isolated Bmal1−/− mitochondria to that of wild-type mitochondria in the presence of palmitoyl-carnitine (Fig. 2B and supplementary text), indicating that NMN supplementation restores intrinsic mitochondrial lipid oxidation in circadian mutants and that circadian mutants lack critical amounts of NAD+ that can be restored in vivo to improve mitochondrial oxidative capacity. Together with the observation that tissue-specific loss of Nampt in liver results in decreased NAD+, FAO, and OCR in the presence of palmitoyl-carnitine (Fig. 2C), these results indicate that the circadian clock controls mitochondrial oxidative metabolism through a posttranscriptional process involving NAD+ biosynthesis.
Clock Regulates Acetylation and Activity of Mitochondrial Oxidative Enzymes
NAD+ is an important cofactor in oxidative metabolism, and NAD+-dependent deacetylase activity affects protein acetylation in mitochondria (17, 25, 26). We therefore sought to identify differences in mitochondrial protein acetylation from livers of wild-type and Bmal1−/− mice. Proteins were separated by two-dimensional gel electrophoresis and subjected to immunoblotting using antibody to acetyl-lysine (Fig. 2D). Subsequent mass spectrometry revealed a cluster of proteins with increased acetylation in Bmal1−/− mitochondria, including several established SIRT3 targets [ornithine transcarbamylase (OTC), long-chain acyl dehydrogenase (LCAD), and 3-hydroxy-3-methylglutaryl–coenzyme A (CoA) synthase 2 (HMGCS2)] (17, 25, 27) as well as proteins that function in lipid metabolism [e.g., FAO enzymes such as LCAD, medium-chain acyl dehydrogenase (MCAD), electron-transferring flavoprotein (ETF), and enoyl-CoA hydratase short chain 2 (ECHS2)]. Bmal1−/− liver mitochondria also displayed decreased enzymatic activity of the medium-and long-chain acyl-CoA dehydrogenases MCAD and LCAD at saturating substrate concentrations (Fig. 2E and fig. S7A), which suggests that circadian control of protein acetylation affects function. Consistent with clock control of mitochondrial enzyme acetylation, we observed ~24-hour oscillation in acetylation of MCAD across the light-dark cycle, with a nadir during the light period and a zenith during the dark period (Fig. 2F). This rhythm is consistent with increased deacetylase activity and FAO during the light or fasting period (mice being nocturnal) because increased amounts of NAD+ increase deacetylase activity (24, 26, 28). Further, NMN supplementation restored MCAD activity in the Bmal1 mutants to that of wild-type animals (Fig. 2G). Together, these data are consistent with clock regulation of acetylation and activity of mitochondrial oxidative enzymes that is mediated through circadian control of NAD+.
Clock-Driven NAD+ Oscillations Regulate SIRT3 Activity
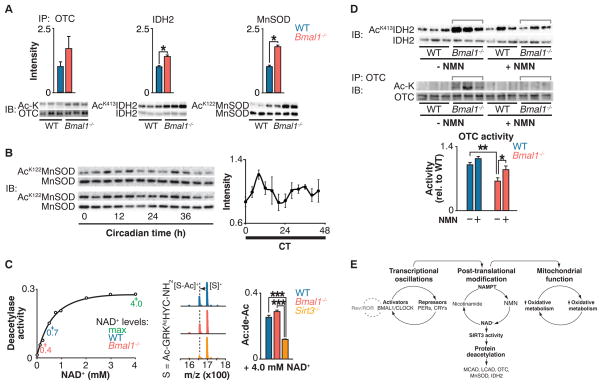
We observed increased acetylation of several targets of the NAD+-dependent deacetylase SIRT3, an important regulator of intrinsic mitochondrial function including FAO (29) (fig. S7C). To directly evaluate whether circadian disruption impaired SIRT3 activity, we analyzed the acetylation status of SIRT3 targets, including OTC, a urea-cycle protein identified in our proteomic screen (25); manganese superoxide dismutase (MnSOD) (30); and isocitrate dehydrogenase 2 (IDH2) (31, 32). Increased acetylation of OTC correlated with decreased enzymatic activity in circadian mutant liver, indicating that deficiency of SIRT3-mediated deacetylation of OTC reduced its activity (25) (Fig. 3A and fig. S7B). In Bmal1−/− mutant liver, we also observed increased acetylation at specific lysine residues within MnSOD (Lys122) (30) and IDH2 (Lys413) (31) that are established SIRT3 target sites (Fig. 3A). Further, MnSOD displayed ~24-hour rhythms of acetylation in wild-type mouse liver mitochondria (Fig. 3B), demonstrating that SIRT3 activity is under the control of the circadian clock. The abundance of SIRT3 remained unchanged in circadian mutant liver and across the circadian cycle in wild-type mice, which suggests that SIRT3 activity per se was altered, rather than the amount of the enzyme (fig. S8, A and B).
Fig. 3. Circadian regulation of SIRT3 activity.
(A) Immunoprecipitation of OTC followed by acetyl-lysine and anti-OTC blotting (left); IDH2 Lys413 and total IDH2 blotting (center); and MnSOD Lys122 and total MnSOD blotting (right) in liver from fasted WT and Bmal1−/− mice at ZT0. Quantitation of acetylated protein is normalized to the amount of the target protein. (B) MnSOD Lys122 acetylation and total MnSOD in liver measured every 4 hours for 48 hours in fasted wild-type mice kept in constant darkness. Quantification of acetyl-Lys122 MnSOD normalized to total MnSOD. (C) Left: In vitro deacetylase activity of recombinant SIRT3 with acetylated SIRT3-selective target peptide (S) in the presence of increasing concentrations of NAD+. Center and right: Representative shift in mass/charge ratio (m/z) and quantification of SIRT3 deacetylase activity (Ac:de-Ac) in liver from fasted WT, Bmal1−/−, and Sirt3−/− mice at ZT0 incubated with 4 mM NAD+ (n = 3). (D) Top: Western blots showing IDH2 Lys413 acetylation and IDH2 from livers of fasted WT and Bmal1−/− after injection with a single dose of saline or NMN 12 hours before ZT0 collection. Bottom: Relative OTC acetylation and OTC activity in livers of WT and Bmal1−/− mice that had been fasted after injection with NMN for 10 consecutive days. (E) Model of circadian regulation of NAD+-mediated mitochondrial oxidative function through SIRT3. *P < 0.05, **P < 0.01, ***P < 0.001 for Student’s two-tailed t test comparing single time points between WT and mutant averages. Data are represented as average ± SEM.
We examined the impact of NAD+ deficiency on SIRT3 activity with the use of self-assembled monolayer and matrix desorption ionization (SAMDI), a mass spectrometry–based assay in which acetylated hexapeptide substrates are assembled as a monolayer through thiolmaleimide binding to gold, avoiding the use of fluorescent labels (33). Using this assay, we established the NAD+ concentration dependence for deacetylation of a SIRT3-selective peptide substrate (Fig. 3C, left). Amounts of endogenous wild-type and Bmal1−/− liver NAD+ [0.7 mM and 0.4 mM, respectively, as measured by high-performance liquid chromatography (HPLC)] corresponded to changes in SIRT3 activity (Fig. 3C, left) and were consistent with the hypothesis that SIRT3 activity was impaired in the circadian mutants due to reduced amounts of NAD+. Further, supplementation of circadian mutant liver extracts with saturating amounts of NAD+ was sufficient to restore in vitro deacetylation of the SIRT3-selective peptide substrate, whereas SIRT3-deficient mice did not exhibit catalytic activity at the same concentration of NAD+ (Fig. 3C, right).
To determine whether changes in the ratio of NAD+ to its reduced form (NADH) contribute to the metabolic and acetylation changes observed in circadian mutants, we measured the amount of NAD+ and NADH in Bmal1−/− and control MEFs. Although amounts of both NAD+ and NADH were significantly decreased in circadian mutant MEFs, the NAD+/NADH ratio remained unchanged (fig. S9A). To determine whether a change in the amount of NADH was sufficient to alter SIRT3 function independent of the NAD+ concentration, we measured SIRT3 deacetylase activity in vitro with increasing doses of NADH (fig. S9B). SIRT3 deacetylase activity did not change in the presence of increasing NADH until concentrations were supraphysiological (fig. S9B). Together, these data indicate that SIRT3 activity is responsive to changes in NAD+ abundance independent of the NAD+/NADH ratio, and that decreased NAD+ biosynthesis itself may be sufficient to impair SIRT3 function.
Because nicotinamide is also able to inhibit sirtuin function (34), we measured amounts of nicotinamide in wild-type and Bmal1−/− liver tissue to determine whether altered nicotinamide concentrations contribute to decreased SIRT3 function in circadian mutants. However, we observed equal amounts of nicotinamide in wild-type and circadian mutants (fig. S9C), which suggests that changes in nicotinamide are unlikely to account for decreased SIRT3 activity in circadian mutants. Likewise, amounts of acetyl-CoA in circadian mutant and wild-type animals were also similar (fig. S9C), demonstrating that the increase in acetylation is unlikely to be caused by increased nonenzymatic acetylation (35).
Injection of Bmal1 in mutant mice with NMN normalized IDH2 and OTC acetylation to that of wild-type animals and also restored OTC activity (Fig. 3D), indicating that NAD+ supplementation restored endogenous SIRT3 activity. Note that amounts of SIRT3 after NMN supplementation were not different between wild-type and Bmal1−/− livers (fig. S8C). Consistent with NMN entry into the mitochondria (36), these data demonstrate that the clock regulates SIRT3 activity through control of concentrations of NAD+.
Although transcriptional profiling studies revealed rhythmicity of gene expression in liver of ad libitum–fed animals, mechanisms through which the circadian cycle affects physiologic processes have been less well defined. Our findings establish a link among circadian control of the NAD+ salvage pathway, mitochondrial protein acetylation, and bioenergetics. Mitochondria are central to energy homeostasis in eukaryotes, and our results indicate that oxidative capacity is coupled to the clock transcriptional cycle and varies according to circadian phase (Fig. 3E). Clocks also receive reciprocal input from metabolic signals, and metabolic cycles persist with 24-hour rhythmicity even in the absence of transcription (37). Use of NAD+ as a central node in coupling circadian and metabolic cycles adds flexibility by providing a rapid and reversible mechanism to augment mitochondrial oxidative function at the appropriate time in the light-dark cycle, and such cyclicity may be bidirectional.
Materials and Methods
Animals
All animal care and use procedures were in accordance with guidelines of the Institutional Animal Care and Use Committee. For the constant darkness (DD) experiments, male C57BL/6J mice at 12 weeks of age were purchased from the Jackson Laboratory and maintained for 2 weeks on a 12:12 light dark (LD) cycle at the Northwestern University Center for Comparative Medicine before placement in DD. Starting 30 hours after mice were placed in DD, groups of mice (n = 4 per group) were subjected to an 18-hour fast before tissue collection, which occurred every 4 hours over a period of 44 hours (i.e., two complete LD cycles). For acute NMN treatment, 3- to 4-month-old Bmal1−/− mutant and littermate control mice were intraperitoneally injected with either NMN (250 mg/kg) or saline 12 hours before tissue collection, and all mice were fasted 18 hours before killing. For longer-term NMN treatment, wild-type and Bmal1−/− mice were intraperitoneally injected at ZT12 for 10 consecutive days with either NMN (500 mg/kg) or saline, and tissues were collected at ZT0 the day after the final injection. Blood glucose levels were measured by Precision Xtra glucometer at the time of killing. For freeze-clamp studies, 5-month-old wild-type and Bmal1−/− mice were anesthetized with 100 μl of Nembutal before tissue collection. Livers were freeze-clamped in situ with forceps pre-cooled in liquid nitrogen and stored at −80°C (21).
Nucleotide Measurements
For measurement of whole-cell NAD+, frozen tissue or cells were extracted in 10% perchloric acid and neutralized in 0.75 M K2CO3 as described (15). NAD+ and ATP were measured by HPLC with Shimadzu LC-20A pump and UV-VIS detector with a Supelco LC-18-Tcolumn (15 cm × 4.6 cm). The HPLC was run at a flow rate of 1 ml/min with 100% buffer A (0.5 M KH2PO4, 0.5 M K2HPO4) from 0 to 5 min, a linear gradient to 95% buffer A/5% buffer B (100% methanol) from 5 to 6 min, 95% buffer A/5% buffer B from 6 to 11 min, a linear gradient to 85% buffer A/15% buffer B from 11 to 13 min, 85% buffer A/15% buffer B from 13 to 23 min, and a linear gradient to 100% buffer A from 23 to 30 min. NAD+ eluted as a sharp peak at 14 min and was normalized to tissue weight of frozen liver tissue or protein content of cultured cells. NAD+ and nicotinamide were further analyzed and confirmed with liquid chromatography–mass spectrometry (LC-MS/MS) by the Washington University Metabolomics Facility. Mitochondrial NAD+, whole-cell NAD+, and NADH were also measured using a cycling enzymatic assay (Bioassay Systems).
Acyl-carnitine and Acetyl-CoA Measurements
Snap-frozen liver sections were homogenized in 50% aqueous HPLC grade acetonitrile containing 0.3% formic acid, frozen in liquid nitrogen and stored at −80°C until further preparation for tandem mass spectrometry. After hepatic protein precipitation with methanol, supernatants were dried, esterified with hot, acidic methanol (for the acyl-carnitine extraction) and then analyzed by tandem mass spectrometry (Quattro Micro, Waters Corp., Milford, MA). Acyl-carnitines and acetyl-CoA were assayed by adapting described methods for analysis of amino acids in dried blood spots (38).
[14C]Oleate and [14C]Glucose Oxidation Assays
Whole liver tissue (200 mg) was excised and immediately placed in ice-cold buffer containing 250 mM sucrose, 1 mM EDTA, and 10 mM tris-Cl (pH 7.4). Tissue was homogenized using a Dounce homogenizer and incubated for 1 hour at 37°C in reaction buffer containing 100 mM sucrose, 10 mM tris-Cl (pH 7.4), 5 mM K2PO4, 80 mM KCl, 1 mM MgCl2, 2 mM L-carnitine, 0.1 mM malate, 2 mM ATP, 0.05 mM coenzyme A, 1 mM dithiothreitol (DTT), 0.2 mM EDTA, 0.3% bovine serum albumin (BSA), 0.5% fatty acid–free BSA, and 1 μCi (0.125 mM) [1-14C]oleic acid–BSA (Perkin Elmer). CO2 was released from the medium by addition of 35% perchloric acid and captured by Whatman filter paper saturated with 100% phenylethylamine. [14C]CO2 counts were determined by scintillation counting. MEFs and C2C12 cells were incubated overnight in Dulbecco’s modified Eagle medium (DMEM) containing either 0.5% fetal bovine serum (FBS) or no FBS, respectively, and 0.5% fatty acid–free BSA coupled to 100 μM oleic acid. [14C]Oleic acid (1.5 μCi) or U-[14C]glucose (2 μCi) was added to each well and cells were incubated for 2 hours at 37°C. [14C]CO2 was captured and measured as described above.
Mouse Embryonic Fibroblast (MEF) Isolation
Pregnant mice were killed at embryonic day 14 or 15, and embryos were dissected away from the uterus and placed in 1× phosphate-buffered saline (PBS). Heads and internal organs were removed and heads were subsequently used for genotyping. Blood was washed away from the remaining carcasses with 1× PBS, and tissue was minced in trypsin-EDTA, followed by incubation with stirring for 30 min. Trypsin was neutralized with heat-inactivated FBS, and cells were pelleted by centrifugation at 270g for 5 min. After centrifugation, cells were resuspended in DMEM containing 15% FBS and plated at 106 cells per 100-mm dish. Media was changed 24 hours after plating. Cells were maintained in DMEM containing 15% FBS.
Mitochondrial ATP Production
MEFs were plated in 10-cm dishes and grown to 80% confluence. Media was changed to DMEM containing galactose (4.5 g/liter) and incubated for 24 hours. ATP levels were measured using HPLC as described above and normalized to protein content.
Lactate Production
The rate of lactate production was determined using a Lactate Reagent Kit (Biovision). Bmal1−/−, Cry1−/−;Cry2−/−, and control MEFs were seeded onto a 6-well culture plate and incubated with fresh culture medium for 8 hours. An aliquot of medium was transferred to a 96-well plate and mixed with lactate reagent. Absorbance at 540 nm was measured using a V5 Spectromax spectrophotometer. Lactate values were normalized to cell number and divided by the length of incubation time. For FK866 treatments, MEFs were incubated with 100 μM FK866 for 24 hours before the media change.
C2C12 Synchronization
C2C12 myoblast cells were infected with Period 2-luciferase (Per2-luc) expressing lentivirus (gift of A. Liu, University of Memphis) (39) and maintained in DMEM that included 10% FBS and blastocidin (2.5 μg/ml) to select for stable Per2-luc integration. Two days before cell synchronization, confluent dishes were differentiated with 2% horse serum. Cells were then synchronized by serum shock every 4 hours for 44 hours with 50% horse serum as described (40). Twenty-four hours after the final shock, cells were collected and analyzed for NAD+, OCR, FAO, and glucose oxidation as described. In parallel, Per2-luc rhythms were simultaneously monitored in synchronized cells cultured in DMEM containing 0.1 mM luciferin using a LumiCycle apparatus (Actimetrics).
Liver Triglyceride Measurement
Frozen liver tissue was heated in alcoholic KOH at 60°C for 5 hours and precipitated with MgCl2. Samples were centrifuged for 30 min at 14,000 rpm and triglycerides were assayed with the Triglyceride Assay Kit (Stanbio).
Mitochondrial and Nuclear Isolation
For isolation of mitochondria for protein, fresh liver samples were dounce-homogenized in isolation buffer containing 0.25 M sucrose, 0.1 mM EDTA, 1 μM Trichostatin A (Sigma), and 20 mM nicotinamide, followed by centrifugation at 800g for 10 min at 4°C. Supernatants were centrifuged again at 8000g for 10 min at 4°C to pellet mitochondria. To isolate liver mitochondria for OCR experiments (see below), mitochondria were prepared as recommended by Seahorse Biosciences. Briefly, mitochondria were isolated by dounce homogenization in buffer containing 70 mM sucrose, 210 mM mannitol, 5 mM HEPES, 1 mM EGTA, and 0.5% (w/v) fatty acid–free BSA (pH 7.2). After homogenization, extracts were centrifuged at 800g for 10 min at 4°C. The lipid layer was aspirated, supernatants were centrifuged again at 8000g for 10 min at 4°C, and pellets were resuspended in 1× MAS buffer [70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1.0 mM EGTA, and 0.2% (w/v) fatty acid–free BSA (pH 7.2)]. For isolation of liver nuclei, fresh liver samples were first minced in hypotonic buffer containing 10 mM KCl, 1.5 mM MgCl2, 1 μM Trichostatin A, and 20 mM nicotinamide and incubated on ice for 20 min. Samples were centrifuged at 3000g for 3 min to pellet the nuclei.
Oxygen Consumption Rate (OCR) Measurements
OCR was measured in isolated liver mitochondria and C2C12 cells as described (22, 41). Mitochondria (isolated as described above) were plated on Seahorse Biosciences 24-well culture plates (50 μg of protein per well) and centrifuged for 20 min at 2000g at 4°C to adhere mitochondria to base of wells. Respiratory substrates (80 μM palmitoyl- or octanoyl-carnitine plus 0.5 mM malate, 10 mM glutamate plus 10 mM malate, 10 mM pyruvate plus 10 mM malate, or 10 mM succinate plus 2 μM rotenone) were diluted in 1× MAS buffer, added to mitochondria at 37°C, and incubated for 10 min. Plates were placed in Seahorse XF24 Bioanalyzer, and mitochondrial OCR was measured before and after sequential addition of 4 mM ADP, 10 μM Oligomycin, 10 μM FCCP, and 10 μM Antimycin A.
Mitochondrial Number
To estimate mitochondrial number, we extracted total DNA from frozen liver tissue by phenol-chloroform extraction; DNA was subjected to quantitative polymerase chain reaction (qPCR) using primers designed against Nd1 for mitochondrial DNA and Gapdh for nuclear DNA.
Protein Gel Electrophoresis and Immunoblotting
Protein from liver whole tissue, mitochondria, and nuclei was diluted in CelLytic MT Mammalian Tissue Lysis Reagent (Sigma) supplemented with protease inhibitors, 1 μM Trichostatin A, and 20 mM nicotinamide. Protein levels were quantified using DC Protein Assay (Biorad). Protein extracts were subject to SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes (GE Healthcare). We used primary antibodies to acetylated lysine (Cell Signaling), MCAD (Santa Cruz Biotechnology), SIRT3 (Cell Signaling and generated in J. Denu laboratory), OTC (Abcam), COX4 (Cell Signaling), PGC1 (Santa Cruz Biotechnology), Ac-122-MnSOD [generated in D. Gius laboratory (30)], MnSOD (Millipore), Ac-413-IDH2 (Genetel Laboratories), and IDH2 (Santa Cruz). Mitochondrial protein extracts were subjected to 2D gel electrophoresis and probed with antibody to acetyl-lysine (Cell Signaling) by Kendrick Laboratories in Madison, WI. The 10 most hyperacetylated proteins were identified using SameSpots software; spots were excised from the gel and subjected to mass spectrometry.
Immunoprecipitation
MCAD was immunoprecipitated from 1.5 mg of mitochondrial lysates using 2 μg of mouse antibody to MCAD (Abcam) and Protein A/G-sepharose beads (GE healthcare) in CelLytic MT Mammalian Tissue Lysis Reagent (Sigma) supplemented with protease inhibitors, 1 μM Trichostatin A, and 20 mM nicotinamide. MCAD was eluted by boiling in 1× SDS-PAGE loading buffer followed by immunoblotting for MCAD (Santa Cruz) and acetylated lysine (Cell Signaling). OTC immunoprecipitation and immunoblotting was performed as described (25). Briefly, mitochondrial lysate was incubated with antibody to OTC (Abcam; 1:500) at 4°C overnight. Protein A/G beads (Cell Signaling) were added and samples were incubated for another 3 hours. Beads were washed three times with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 50 mM NaF, 1.5 mM Na3VO4 containing protease inhibitors, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride). OTC was eluted by boiling in 1× SDS-PAGE loading buffer followed by immunoblotting for OTC (Abcam) and acetylated lysine [generated in J. Denu laboratory (25)].
Enzymatic Activity Assays
MCAD and LCAD activity was measured as previously described (17, 42). Briefly, mitochondrial extracts were diluted 1:10 in 50 mM Tris, pH 8.0, with 0.25% lubrol detergent to assist in solubilization. The samples were placed in a sonicating ice water bath for 30 s and then used directly in the anaerobic electron transfer flavoprotein (ETF) reduction assay. Activity was measured in 0.7 ml total volume of 50 mM Tris, pH 8.0, with 1 μM porcine ETF and 20 μM acyl-CoA substrate (either 2,6-dimethylheptanoyl-CoA or octanoyl-CoA for measurement of LCAD or MCAD activity, respectively). After addition of substrate, ETF fluorescence was followed for 1 min (excitation 340 nm, emission 490 nm). Activity measurements are expressed as milliunit (mU) per milligram of total mitochondrial protein, with 1 mU defined as the amount of enzyme required to fully reduce 1 nmol of ETF in 1 min. OTC enzymatic activity was measured as described (25). Briefly, 10 μl of diluted mitochondrial fraction was added to a solution containing ornithine and triethanolamine to a final volume of 675 μl. OTC reactions were started by adding 75 μl of 150 mM carbamoyl phosphate. Final concentrations in assay of each reagent were 5 mM ornithine, 15 mM carbamoyl phosphate, and 270 mM triethanolamine, pH 7.7. After 30 min of incubation at 37°C, reactions were stopped by adding 375 μl of phosphoric acid/sulfuric acid (3:1, v/v). Citrulline production was determined by adding 47 μl of 3% 2,3-butanedionemonoxime, boiling in the dark for 15 min, and reading absorbance at 490 nm. Relative OTC activities were normalized with OTC levels through Western blotting.
Sirtuin Activity Assays
Liver tissue was flash-frozen in Buffer D (20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA and protease inhibitors), thawed on ice, and homogenized in a Dounce homogenizer. Homogenates were centrifuged at 4000 rpm for 3 min, and supernatant was removed to a separate tube. The pellets were resuspended in Buffer D and centrifuged at 14,000 rpm for 10 min. The supernatant was combined with the first supernatant and flash frozen. Lysates were thawed on ice and spun twice at 14,000 rpm to clear the lysate. Specific sirtuin activity was then measured from liver extracts using the SAMDI mass spectrometry assay. Liver extracts were diluted in KDAC buffer (25 mM Tris, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2) to a final protein concentration of 1 mg/ml. TSA (Santa Cruz) was added to the diluted liver extracts to give a final concentration of 50 μM, and 5 μl of the resulting mixtures were distributed into separate wells of a 384-well plate. A protease inhibitor cocktail (1 μl, Roche) and NAD+ (1 μl, 4 mM final) were added to each reaction well. To initiate the reaction, 1 μl of the SIRT3-selective peptide substrate (Ac-GRKAcHYC-NH2) was added at a final concentration of 4 μM, and the reaction plate was incubated at 37°C for 4 hours. Small volumes of each reaction (2 μl) were then transferred onto the maleimide-presenting monolayers to allow immobilization of the peptide substrate and product. The monolayers were then rinsed with distilled water and ethanol, dried under nitrogen, and treated with matrix (2,4,6-trihydroxyacetophenone, 20 mg/ml in acetone). The monolayers were analyzed by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) MS to obtain a mass spectrum for each circular region, and activities were calculated from each spectrum as described (33).
Quantitative Real-Time PCR
Total RNA was extracted from frozen tissue with Tri Reagent (Molecular Research Center Inc.). cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Applied Bio-systems). Real-time PCR was performed and analyzed using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems). PCR conditions were: one cycle of 10 min at 95°C, then 35 cycles of 10 s at 95°C, 15 s at 60°C. Relative expression levels (normalized to Gapdh) were determined using the comparative CT method.
Primer sequences
| Primer | Sequence |
|---|---|
| Nd1 F | CCCATTCGCGTTATTCTT |
| Nd1 R | AAGTTGATCGTAACGGAAGC |
| Gapdh F | CAAGGAGTAAGAAACCCTGGACC |
| Gapdh R | CGAGTTGGGATAGGGCCTCT |
| L-Pk F | AATATCACCCAGGTCGTTGC |
| L-Pk R | AAGAAACCACCGTGTTCCAC |
| Ldha F | TCTCGGATGTTGTGAAGGTG |
| Ldha R | CTGCAGCTCCTTCTGGATTC |
| Pfk1 F | GCATCACCAACCTGTGTGTC |
| Pfk1 R | CGATGGTCAAGTGTGCGTAG |
| Pdk1 F | TGGCTATGAGAACGCTAGGC |
| Pdk1 R | TGTCTGTCCTGGTGATTTCG |
| Pgk F | ATCAAGGCTGCTGTTCCAAG |
Supplementary Material
Acknowledgments
We thank P. Schumacker, R. Allada, S. Imai, G. Barish, members of the Bass lab for discussion and comments on the manuscript; K. Lamia and P. Schumacker for Cry1−/−;Cry2−/− MEFs and Sirt3−/− mice, respectively; the Washington University Metabolomics Facility for nicotinamide measurements; the Northwestern University Skin Disease Research Center (NIH/NIAMS grant 5P30AR057216-05), and W. Song for technical assistance. Supported by National Institute of Diabetes and Digestive and Kidney Diseases grants F32 DK092034 (C.B.P.), F30 DK085936 (A.H.A.), and T32 DK007169 (B.M.); the Endocrine Society (D.J.B.); and NIH grants R01DK090242-03 (E.G.); P01DK58398 (C.B.N.); R01 AG038679 ( J.M.D.); R01 CA152601-01, R01 CA152799-01A1, R01 CA168292-01A1, and R01 CA16383801A1 (D.G.); 5P01HL071643-10 (N.S.C.); and P01AG011412-16, R01HL097817-01, and R01DK090625-01A1 ( J.B.). J.B. has financial interest in and serves as advisor to Reset Therapeutics, and has been a paid consultant for Merck, Janssen Pharmaceuticals, Vanda Pharmaceuticals, Gerson Lehrman Group, and Matsutani America. J.D. is a paid consultant for Sirtris, a GSK company. A provisional patent has been applied for by Northwestern University related to the role of the clock-NAD pathway in the mitochondrial control of cell growth, inflammation, and metabolism.
Footnotes
References and Notes
- 1.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 3.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. 10.1016/S0092-8674 (00)81199-X. [DOI] [PubMed] [Google Scholar]
- 4.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. 10.1016/S0092-8674 (02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 7.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 8.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLOS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: Replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 20.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies SP, Carling D, Munday MR, Hardie DG. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem. 1992;203:615–623. doi: 10.1111/j.1432-1033.1992.tb16591.x. [DOI] [PubMed] [Google Scholar]
- 22.Rogers GW, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLOS ONE. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai SI. A possibility of nutriceuticals as an anti-aging intervention: Activation of sirtuins by promoting mammalian NAD biosynthesis. Pharmacol Res. 2010;62:42–47. doi: 10.1016/j.phrs.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshino J, Mills KF, Yoon MJ, Imai SI. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantó C, et al. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belenky P, et al. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- 30.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurard-Levin ZA, Kilian KA, Kim J, Bähr K, Mrksich M. Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem Biol. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 35.Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213:513–522. doi: 10.1016/0005-2787. 7090058-4. [DOI] [PubMed] [Google Scholar]
- 36.Nikiforov A, Dölle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: From entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu JY, et al. ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J Clin Invest. 2004;113:434–440. doi: 10.1172/JCI19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLOS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, et al. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 2012;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholls DG, et al. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010;46:2511. doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goetzman ES. The regulation of acyl-CoA dehydrogenases in adipose tissue by rosiglitazone. Obesity. 2009;17:196–198. doi: 10.1038/oby.2008.467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.