This work uses genomic sequencing and microscopy to examine the differences in meiotic stability and fertility between synthetic and natural allopolyploids of Arabidopsis thaliana and A. arenosa. The authors find that the natural allopolyploid A. suecica and its parents have very similar genomic architecture and identify a single locus from A. suecica associated with increased meiotic stability.
Abstract
Whole-genome duplication resulting from polyploidy is ubiquitous in the evolutionary history of plant species. Yet, polyploids must overcome the meiotic challenge of pairing, recombining, and segregating more than two sets of chromosomes. Using genomic sequencing of synthetic and natural allopolyploids of Arabidopsis thaliana and Arabidopsis arenosa, we determined that dosage variation and chromosomal translocations consistent with homoeologous pairing were more frequent in the synthetic allopolyploids. To test the role of structural chromosomal differentiation versus genetic regulation of meiotic pairing, we performed sequenced-based, high-density genetic mapping in F2 hybrids between synthetic and natural lines. This F2 population displayed frequent dosage variation and deleterious homoeologous recombination. The genetic map derived from this population provided no indication of structural evolution of the genome of the natural allopolyploid Arabidopsis suecica, compared with its predicted parents. The F2 population displayed variation in meiotic regularity and pollen viability that correlated with a single quantitative trait locus, which we named BOY NAMED SUE, and whose beneficial allele was contributed by A. suecica. This demonstrates that an additive, gain-of-function allele contributes to meiotic stability and fertility in a recently established allopolyploid and provides an Arabidopsis system to decipher evolutionary and molecular mechanisms of meiotic regularity in polyploids.
INTRODUCTION
The establishment of new allopolyploid species, carrying two or more sets of chromosomes originating from different species, is a common event in plant evolution (Otto and Whitton, 2000; Doyle et al., 2008; Soltis et al., 2007). Yet, the challenges associated with the formation of allopolyploids are numerous (Comai, 2005), as indicated by the common observation that fertility and overall fitness are compromised (Moore, 2002). A key adaptation involves chromosome pairing and recombination during meiosis. Evidence for meiotic gene adaptation to autopolyploidy, involving more than two equivalent pairing partners, has been recently described (Hollister et al., 2012; Yant et al., 2013). In allopolyploids, pairing should be limited to two homologous partners (Comai, 2005; Cifuentes et al., 2010), as opposed to homoeologous partners, which originate from different parents. Strict homologous pairing ensures proper chromosome partitioning and reduces the risks of production of unbalanced gametes, which are often unviable. In paleopolyploid species, “diploid” species for which the latest known polyploidization event occurred in the distant past, chromosomes often appear to carry a mixture of segments from ancestral homoeologs (Doyle et al., 2008), suggesting that homoeologous exchanges occurred in their past but their rate has been reduced through the evolutionary cycle of diploidization (Paterson et al., 2010). The nature of the mechanisms that prevent homoeologous exchanges and maintain fitness in established allopolyploids and the timing of their establishment remain uncertain, but they constitute a key step in the establishment of polyploid species.
One hypothesis to explain meiotic regularity invokes the accumulation of structural chromosomal variation between the two parental genomes, which results in preferential pairing between homologs rather than homoeologs. Consistent with this proposal, rapid genomic changes have been observed in newly synthesized allopolyploids of various species, supporting the proposal that chromosomal remodeling is a major contributor to intragenomic separation. Many of the genomic changes observed in recent allopolyploids, such as in Tragopogon miscellus (Chester et al., 2012), Brassica napus (Gaeta et al., 2007; Szadkowski et al., 2010; Xiong et al., 2011), or cotton (Gossypium hirsutum; Flagel et al., 2012), are consistent with homoeologous exchanges. In other cases, the observed genomic changes may have a different origin, such as some of the changes reported in wheat (Triticum aestivum; Feldman et al., 1997; Han et al., 2005; Zhang et al., 2013). Furthermore, rapid changes did not ensue in newly synthesized allotetraploid cotton (Liu et al., 2001; Xiong et al., 2011), suggesting that different allopolyploids might respond differently to the challenge.
Another hypothesis involves the development of a molecular pairing editor that rejects chromosomal pairing between chromosome partners that cross a certain threshold of divergence (Jackson, 1982; Stebbins, 1984). For example, in allohexaploid wheat, the Ph1 locus reduces homoeologous pairing between the three diverged sets of chromosomes of this allopolyploid (Griffiths et al., 2006) and is thought to encode a cyclin-dependent kinase pseudogene array that affects the timing of chromosome condensation (Yousafzai et al., 2010). Similarly, the PrBn locus of the allopolyploid Brassica napus is thought to alter the rate of nonhomologous chromosome recombination, but its molecular function and mechanism of action remain to be determined (Jenczewski et al., 2003; Nicolas et al., 2009). Laboratory-made (synthetic) Brassica allopolyploids lacking PrBn display progressive dysgenesis correlated to homoeologous exchanges (Gaeta et al., 2007; Gaeta and Chris Pires, 2010; Xiong et al., 2011). Overall, these and other contributions demonstrate that control of meiotic pairing is necessary for stability of allopolyploid species (Moore, 2002; Comai, 2005; Chester et al., 2012). Little is known about such possible adaptation to polyploidy in other species.
In the genus Arabidopsis, a natural allopolyploid species, Arabidopsis suecica (Nat), originated from a single hybridization event that took place 20 to 300 thousand years ago (Jakobsson et al., 2006) between Arabidopsis thaliana (n = 5) and Arabidopsis arenosa (n = 8). A. suecica is self-fertile and fecund and is well established in various regions of Fennoscandinavia in Northern Europe (Schmickl and Koch, 2011). The cross that gave rise to A. suecica can easily be recreated in the laboratory, producing synthetic allopolyploids carrying the A. thaliana and A. arenosa genomes. Contrary to their natural counterpart, the synthetic allopolyploids exhibit poor fertility (Comai, 2000; Madlung et al., 2005). We asked whether this experimental system could be used to investigate the molecular mechanisms underlying meiotic stability in an allopolyploid context.
In this article, we investigated the genetic mechanisms underlying the adaptation to allopolyploidy in A. suecica. We produced synthetic allopolyploids carrying the same genomes as A. suecica and observed that, in addition to exhibiting lower pollen fertility, they also produced more frequent aneuploid progeny and other chromosomal abnormalities consistent with homoeologous recombination resulting from erroneous pairing at meiosis. Leveraging on this variability, we created an F2 population segregating for natural and synthetic allopolyploid alleles. We characterized the progeny in terms of fertility, meiotic stability, and chromosome dosage and correlated these data with genotype information obtained using next-generation sequencing. We found that the genomic architecture of A. suecica is overall highly similar to that of its parents but that presence of the A. suecica allele at a single locus was associated with higher pollen viability and meiotic stability.
RESULTS
Chromosome Dosage Analysis of Synthetic Arabidopsis Allopolyploids Indicates High Rates of Homoeologous Recombination
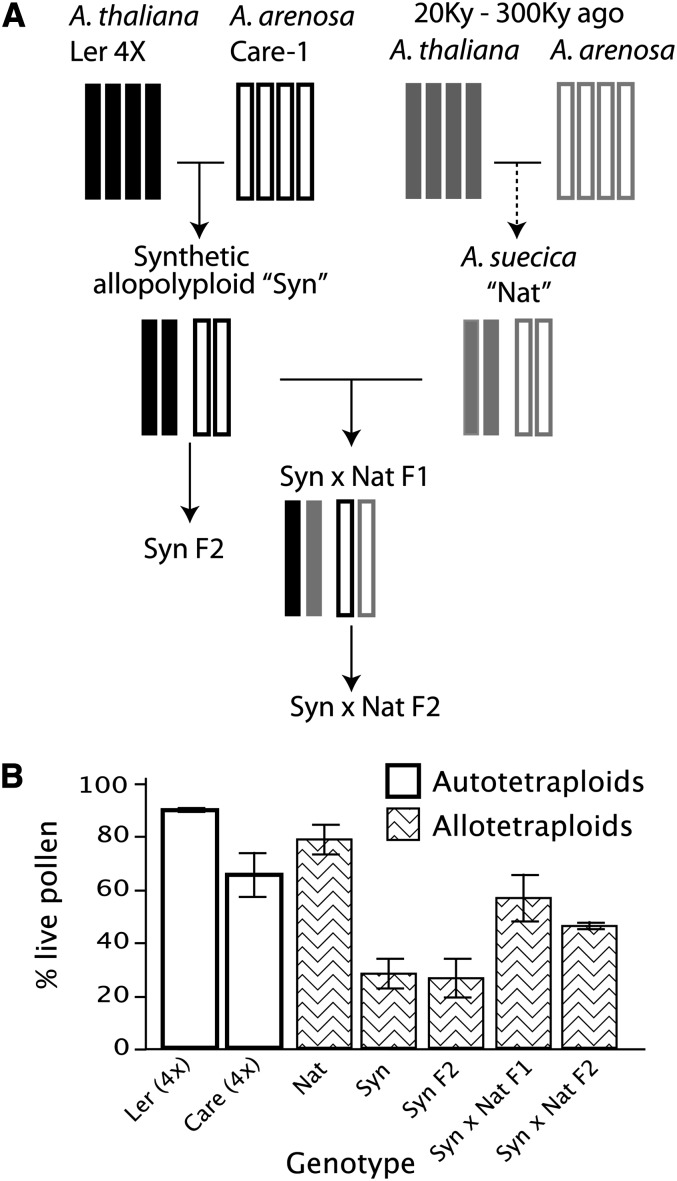
We created synthetic allopolyploids combining both the A. thaliana and A. arenosa genomes by crossing an A. thaliana tetraploid (accession Landsberg erecta-0 [Ler-0]) to the natural tetraploid A. arenosa (accession Care-1; Figure 1A). These newly synthesized allopolyploids (Syn F1s) carry the same genomic composition as the natural A. suecica but exhibited low pollen viability in comparison to their tetraploid parents and to the natural allopolyploids, A. suecica (Figure 1B). Next, we investigated meiotic regularity in these synthetic allopolyploids, as well as in a panel of naturally occurring A. suecica accessions. Specifically, progeny from nine different accessions of A. suecica and from synthetic allopolyploids originating from four independent tetraploid A. thaliana (4x-Ler) × tetraploid A. arenosa (Care-1) crosses were assessed for chromosomal imbalance. Each selfed F1 progeny was subjected to low-coverage next-generation sequencing for dosage analysis (see Methods for details). In short, sequencing reads were mapped to an in silico hybrid genome corresponding to the five chromosomes of the A. thaliana genome reference (TAIR9; Arabidopsis Genome Initiative, 2000) and the eight largest scaffolds of the A. lyrata v1.0 genome reference (Joint Genome Institute). Next, relative read coverage along the 13 chromosomes of the hybrid genome was plotted for each individual. Coverage values were normalized such that values for diploid segments oscillated around 2.0, while duplicated or deleted fragments oscillated around 3.0 or 1.0, respectively. In a “normal” allopolyploid dosage plot, all values are expected to cluster around 2.0 to indicate two copies of each homoeologous chromosome pairs.
Figure 1.
Pollen Viability in the Synthetic × Natural Allopolyploid Pedigree.
(A) Pedigree of allopolyploids used in this study. Each bar represents a genomic copy whose fill indicates the species of origin and whose contour indicates recent or prehistoric origin. The pedigree assumes that A. suecica was produced by hybridization of autotetraploids. Alternative modes, such as involving 2N gametes or a diploid hybrid that doubled its chromosomes, are possible but not shown.
(B) Mean pollen viability was measured for each plant, and the mean of the means was calculated for each genotype. Standard errors are indicated.
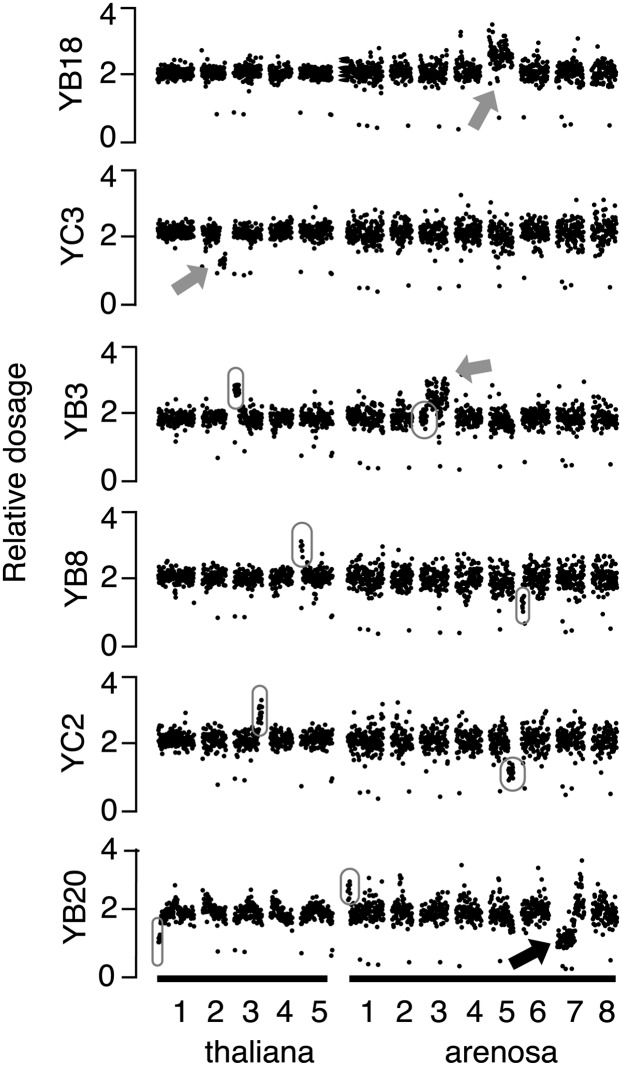
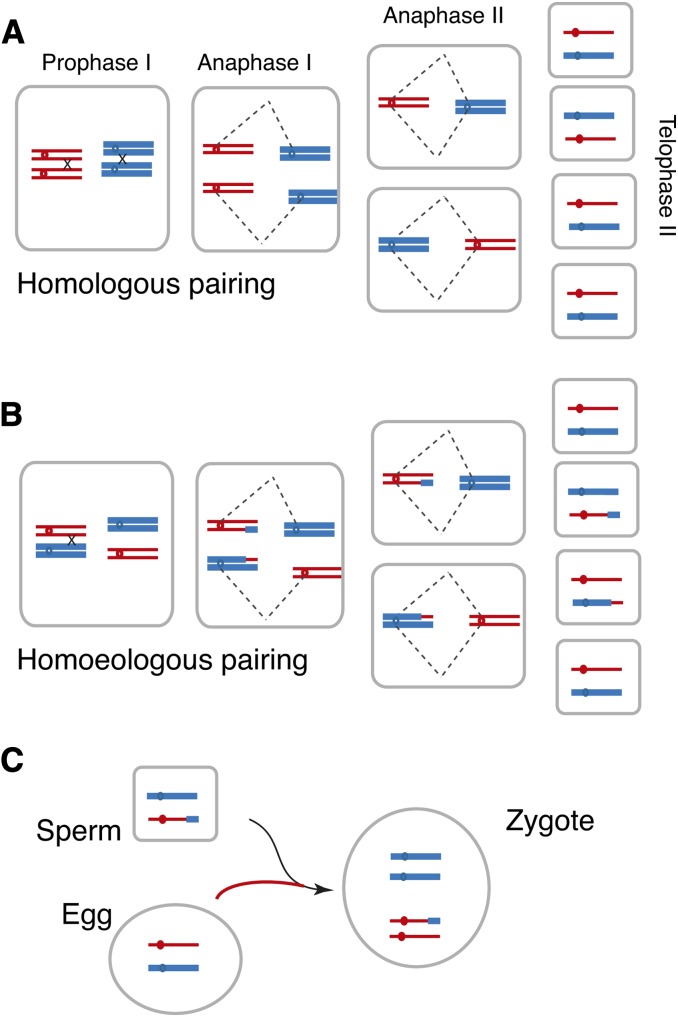
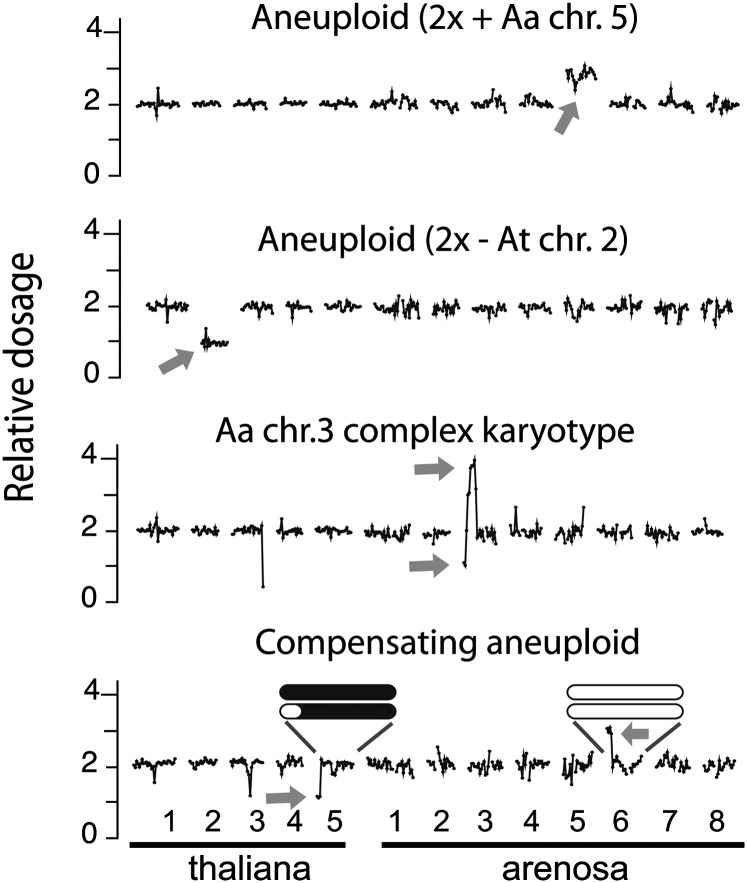
Aneuploidy is very common in polyploids (Ramsey and Schemske, 1998), including in A. thaliana (Henry et al., 2006), and the types and frequency of aneuploidy in a population can be diagnostic of meiotic irregularity and pairing preferences. Different types of irregularities were observed in our populations, such as whole chromosome aneuploidy (Figure 2, top), segmental aneuploidy (Figure 2, second plot), as well as more complex dosage configurations in which select portions of the genomes were present in more than one extra copy or exhibited both increased and decreased copy number within a single chromosome (Figure 2, bottom plot, black arrow). Finally, compensating dosage variants, in which increased dosage of a chromosome or chromosomal fragment from one genome is compensated for by the decreased dosage of the homoeologous fragment from the other genome, were observed as well (Figure 2, four bottom plots). As illustrated in Figure 3, the observation of compensating dosage variation between homoeologous fragments of the genomes is best explained as the result of homoeologous pairing and intergenomic exchange during meiosis in the F1 plants, as observed in recently formed allopolyploids of other species (Lim et al., 2004; Gaeta and Chris Pires, 2010; Chester et al., 2012).
Figure 2.
Evidence of Meiotic Irregularities and Homoeologous Exchange in the Synthetic Allopolyploid.
Examples of individual dosage plots. Sequencing reads were aligned to the reference genome and sorted into consecutive bins along the 13 chromosomes of the allopolyploid genome. For ease of visualization, relative read coverage is normalized such that dosage plots of fragments present in two copies fluctuated around 2.0. Changes up or down in relative coverage of consecutive bins indicates variation in dosage and correspond to the addition or deletion of a particular chromosome or chromosomal fragment, respectively. Relative coverage around the centromeric repeats consistently appears noisy most likely because of mis-mapping, quality of reference sequence, and changes in repeated elements. Gray arrows indicate aneuploidy of a whole or segment of a chromosome. The black arrow indicates an example of a complex dosage variation (part of a chromosome is present in one copy and another part is present in three copies). Compensating dosage variants, in which one chromosome fragment is missing and the corresponding homoeologous fragment from the other genome is present in three copies, are indicated by circles.
Figure 3.
Formation of a Homoeologous Exchange.
A hypothetical allopolyploid with four chromosomes is illustrated. Thin or thick lines depict chromosomes from one ancestral or the other ancestral genome. Hatched lines represent the spindle apparatus.
(A) Regular meiosis showing properly paired replicated chromosomes undergoing recombination and disjunction and the resulting meiotic products.
(B) Intergenomic pairing and recombination between homoeologous chromosomes leading to the formation of two unbalanced gametes.
(C) One unbalanced gamete fertilizes an egg leading to a zygote containing extra DNA from the thick-marked genome and missing the corresponding segment of the thin-marked genome. The outcome illustrated results from syntenic chromosomes. If the paired regions leading to exchanges are located on chromosomes that are structurally diverged (e.g., have inverted segments), the result can be catastrophic, leading to such aberrations as dicentric or acentric chromosomes, chromosomal breakage, and large deficiencies or duplication in the affected meiotic products. These, in turn, can lead to lethality in the gamete or in the zygote.
[See online article for color version of this figure.]
Individuals with irregular dosage profiles were found in the progeny of both synthetic and natural allopolyploids (Table 1). One synthetic allopolyploid (SynF) produced 5/17 complex dosage variants, with very similar dosage irregularities. This observation led us to believe that the parent (SynF) itself carried a similar dosage imbalance. Data from this individual were thus discarded from further analysis. While both the synthetic and the natural allopolyploids produced irregular dosage variants, the frequency of variants was higher in the progeny of the synthetic allopolyploids as a whole (90.1% versus 80%, χ-test P value = 0.045). Most importantly, compensating dosage variants were only found in the progeny of the synthetic allopolyploids (4/7 irregular dosage profiles versus 0/8 for the natural allopolyploids, Fisher exact P value = 0.026). These results confirm that the natural allopolyploid exhibits more regular meiosis than the synthetic allopolyploid, possibly via a reduction of homoeologous pairing.
Table 1. Chromosomal Imbalance in the Progeny of Natural and Synthetic Arabidopsis Allopolyploids.
| Allopolyploid Type | Accession (Sue)/Name (Syn) | Euploid | Aneuploidy/Dosage Variation | Homoeologous Rec. | Total | ||
|---|---|---|---|---|---|---|---|
| Whole Chr. | Chr. Fragment | Complex | |||||
| Sue | Sue-1 | 42 | 2 | 0 | 2 | 0 | 0 |
| Sue | Sue-2 | 5 | 0 | 0 | 0 | 0 | 5 |
| Sue | Sue-3 | 1 | 0 | 0 | 0 | 0 | 1 |
| Sue | Sue-4 | 2 | 0 | 0 | 0 | 0 | 2 |
| Sue | Sue-12 | 2 | 0 | 1 | 0 | 0 | 3 |
| Sue | Sue-13 | 6 | 1 | 0 | 0 | 0 | 7 |
| Sue | Sue-14 | 8 | 1 | 0 | 0 | 0 | 9 |
| Sue | Sue-16 | 5 | 0 | 0 | 0 | 0 | 5 |
| Sue | Sue-19 | 2 | 1 | 0 | 0 | 0 | 3 |
| Syn | Syn-A | 1 | 0 | 0 | 0 | 0 | 1 |
| Syn | Syn-B | 19 | 2 | 0 | 0 | 3 | 24 |
| Syn | Syn-C | 8 | 0 | 1 | 0 | 1 | 10 |
| Syn | Syn-Fa | 11 | 0 | 1 | 5 | 0 | 17 |
| Total Sue | 73 (90.1%) | 5 | 1 | 2 | 0 | 81 | |
| Total Syn | 28 (80.0%) | 2 | 1 | 0 | 4 | 35 | |
Chr., chromosome; Rec., recombination.
Data from the progeny of SynF were discarded from further analyses because of the likely presence of dosage imbalance in the parental plant itself (see Results for details).
Pollen Viability and Meiotic Regularity Segregate in the Progeny of a Cross between Newly Synthesized and Natural Allopolyploids
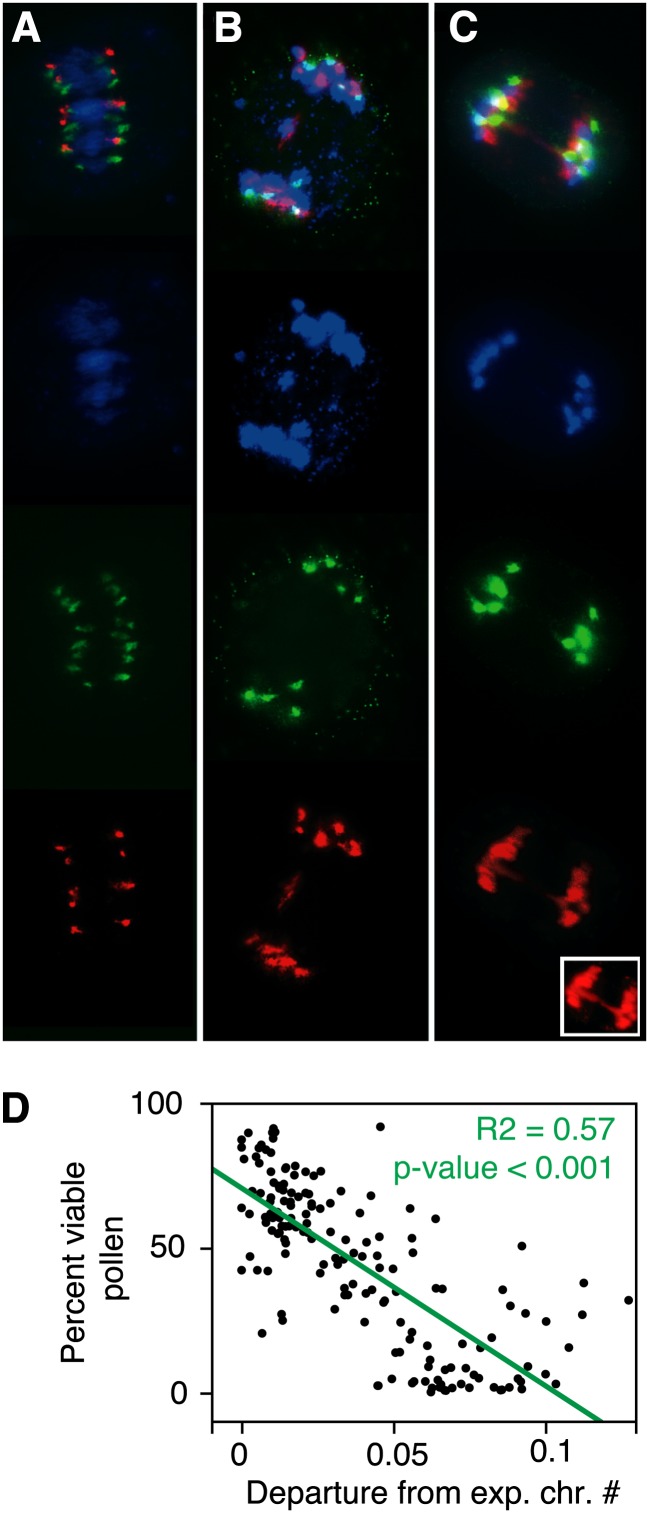
To investigate the genetic basis of the observed variability between the synthetic and the natural allopolyploid, we crossed Syn to Nat and produced a segregating F2 population (Figure 1A). Specifically, a single cross between a Syn (Ler × Care) plant and Sue-1 was performed to produce a family of Syn × Nat F1 plants. Three of those F1 plants were selfed and their progeny was planted to produce an F2 population. The hybrid Syn × Nat F1 plants exhibited intermediate pollen viability (Figure 1B). The Syn × Nat F2 progeny (from here on referred to as simply F2) displayed wide variation in pollen viability and other fertility-related traits, including transgressive segregation (Supplemental Figures 1 and 2). A total of 522 F2 plants were phenotyped for fertility-related traits, including pollen viability, pollen count, seed viability, and seed counts. Additionally, meiotic stability was assessed for each F2 individual by fluorescence in situ hybridization (FISH; Figure 4) using probes to the centromeric repeats of A. thaliana and A. arenosa (Comai et al., 2003). Chromosomes were counted in developing microspores and meiotic irregularities, such as lagging chromosomes or chromosomal bridges, were recorded (Figure 4). The frequency of meiotic irregularities was correlated with pollen viability (R2 = 0.57, P values < 0.001; Supplemental Figure 2), demonstrating that pollen viability can be used as a proxy for meiotic fidelity. The observed phenotypic variation in the F2 population also indicated that this population was well suited to investigate the mechanisms underlying meiotic adaptation to polyploidy.
Figure 4.
Meiotic Irregularities in the Syn × Nat F2 Plants.
(A) to (C) FISH observations of meiotic spreads of developing microspores from F2 individuals. From left to right: Overlay, 4′,6-diamidino-2-phenylindole staining in blue, At-cen labeled FISH in red, Aa-cen labeled FISH in green.
(A) Anaphase I, example of a regular meiosis.
(B) Anaphase I, one potential laggard from an arenosa chromosome is observed.
(C) Anaphase I, an arenosa chromosomal bridge is observed.
(D) Correlation between departure from expected chromosome number and the percentage of viable pollen in the F2 population. The regression P value and R2 value are indicated.
Alleles from the Adapted Allopolyploid Are Overrepresented in the F2 Population
We investigated whether the variation observed in the F2 was generated by structural remodeling of the A. suecica chromosomes that hinders homoeologous pairing or from a genetic regulation of meiosis. To distinguish between these hypotheses, the F2 individuals, which segregated for a maximum of four different alleles (Figure 1A) located on two different subgenomes (A. thaliana and A. arenosa), were genotyped via low-coverage sequencing (see Methods for details). In short, Illumina sequencing reads from the parental accessions (Ler-1, Care-1, and Sue-1) were used to identify robust polymorphisms between the two parental genomes (Ler versus A. suecica on the A. thaliana genome or Care versus A. suecica on the A. arenosa genome). Each F2 individual was then assessed for the presence of one or the other allele at each locus for which information was available and an overall recombinant genotype was obtained.
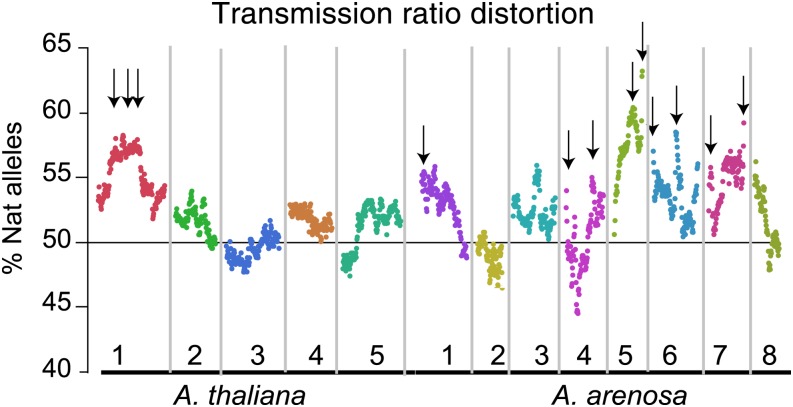
Data from all individuals were pooled to investigate the presence of chromosomal regions with biased allelic ratios. Overall, allelic ratios were biased for the presence of the natural allele from A. suecica on both parental genomes (52.7 ± 2.9 for the A. arenosa genome and 52.0 ± 2.4 for the A. thaliana genome). Twelve regions, all favoring the A. suecica allele, exhibited significant distortion (Figure 5) with the most distorted marker exhibiting 64% of A. suecica allele.
Figure 5.
Transmission Ratio Distortion in the F2.
The mean percentage of allele derived from natural A. suecica in the F2 population was calculated for each marker bin. Different colors represent different chromosomes. Arrows indicate significant distortion from the expected 50% of A. suecica natural allele.
[See online article for color version of this figure.]
Dosage Variation Is Common in the F2
We also used our genome-wide sequencing data to evaluate dosage variation in the F2 progeny. Aneuploidy for full or partial chromosomes or variation in dosage for smaller segments was detected in 57 of the 420 (13%) fully genotyped individuals (Table 2). The same general types of dosage variants were found in the F2 individuals as in the synthetic and natural allopolyploid F1 selfed progeny: Most aneuploid individuals were missing or carried an extra copy of a single chromosome or a piece of a chromosome (Figure 6, top two panels), but more complex cases of dosage variation (Figure 6, third panel) and compensating dosage variants (Figure 6, bottom panel) were observed as well (see Supplemental Data Set 1 for full karyotype plots of all individuals presenting dosage imbalances). The F2 population did not present multiple individuals with the same type of dosage variation suggesting that the parental Syn × Nat F1 plants themselves were not imbalanced and that the variation observed originated from irregular meiosis in the Syn × Nat F1.
Table 2. Karyotyping by Sequencing in the Syn × Nat Allopolyploid F2 Individuals.
| Type | n |
|---|---|
| Diploids | 363.0 |
| Diploids with sufficient coverage for genotyping and QTL mapping | 298.0 |
| Dosage variants | |
| Missing <1 chr. | 13.0 |
| Missing one chr. | 13.0 |
| One extra chr. | 11.0 |
| <1 Extra chr. | 1.0 |
| Compensating dosage variants | 7.0 |
| Complex dosage variants | 5.0 |
| Chr. Aa 3 or Aa 6 abnormal karyotypesa | 7.0 |
| Total | 420.0 |
| No. of dosage variants | 57.0 |
| Dosage variants (%) | 13.6 |
Chr., chromosome.
See Supplemental Data Set 1 and Figure 6.
Figure 6.
Dosage Variation in the Syn × Nat F2 Plants.
Examples of individual F2 dosage plots obtained through whole-genome sequencing. Sequencing reads were aligned to the reference genome and sorted into consecutive bins along the 13 chromosomes of the allopolyploid genome. For ease of visualization, relative read coverage is normalized such that fragments present in two copies fluctuated around 2.0. Changes up or down in relative coverage of consecutive bins (arrows) indicates variation in dosage and correspond to the addition or deletion of a particular chromosome or chromosomal fragment, respectively. Relative coverage around the centromeric repeats consistently appears noisy most likely because of mis-mapping, quality of reference sequence, and changes in repeat copy number.
Overall, 1.7% of the F2 individuals (7/420) exhibited dosage patterns consistent with homoeologous recombination events. This level is intermediate between what we observed in the A. suecica selfed population and the synthetic allopolyploid population (0 and 11.4%, respectively; see Table 1) and further confirms that A. suecica has acquired the ability to better promote homologous over homoeologous pairing. Correlating allelic bias and dosage variation for the regions that exhibited decreased dosage in the compensating dosage variants suggested that both the Sue and the Syn chromosomes were involved in homoeologous pairing in the context of the synthetic × natural hybrid cross. Indeed, of the chromosomal fragments exhibiting dosage variation consistent with homoeologous recombination, three were homozygous for the Syn allele and three were homozygous for the Sue allele at those positions and their flanking regions. The others were located in heterozygous regions and could not easily be used to draw conclusions as to the origin of the missing or additional chromosomal pieces.
Our measure of meiotic stability was consistent with incidence of dosage variation. For example, aneuploids carrying one extra chromosome exhibited higher mean chromosome counts in developing microspores than diploids, which exhibited higher mean chromosome counts than aneuploids missing one chromosome (Supplemental Figure 6). Finally, dosage variants as a group were not easily distinguishable from their diploid counterparts by any other phenotypic trait (t test P values all >0.01), confirming the need to detect them by genomic or cytological analysis.
Genomic Structure of the A. suecica Genome
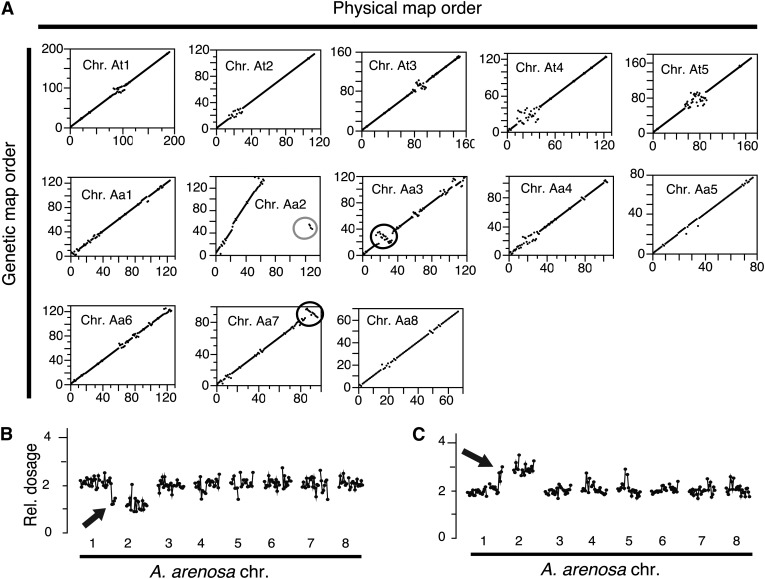
Using allelic calls in the F2 diploid individuals, we created a genetic map of the allopolyploid hybrid genome including 707 A. thaliana and 770 A. arenosa markers (Supplemental Table 2 and Supplemental Figure 7). Overall, the genomic organizations of the natural and synthetic allopolyploids were highly similar. Marker order was also similar to the published reference sequences of A. thaliana TAIR9 (Arabidopsis Genome Initiative, 2000) and A. lyrata v1.0 (Joint Genome Institute; http://www.jgi.doe.gov), which we used in lieu of an A. arenosa genomic reference sequence (Figure 7). Three major exceptions, all on the A. arenosa genome were identified (Figure 7A). Two were consistent with rearrangement between the A. lyrata and A. arenosa genomes. One of these was also confirmed by the karyotypes of several of our dosage variants (Figure 7B, inset) and was also detected in a population of Capsella rubella (Slotte et al., 2013). Taken together, these results suggest that it might originate from erroneous assembly of that particular segment of the A. lyrata genome (Slotte et al., 2013). Only the third rearrangement was consistent with an inversion between the two parental genomes (A. arenosa Care-1 and A. suecica Sue-1). Together, our data suggest that the A. suecica genome did not undergo major genomic rearrangements compared with those of present-day A. thaliana and A. arenosa.
Figure 7.
Comparison of the Genetic and Physical Maps.
(A) Marker bin order in the physical maps (published reference genomes of A. thaliana and A. lyrata) was compared with the marker order obtained from our F2 population. Two inversions (black circles) were identified. A few markers located on chromosome one of the A. lyrata genomic reference appeared reversed and translocated in the middle of chromosome two in our genetic map (gray circle).
(B) and (C) The translocation between chromosome one of the A. lyrata reference genome and chromosome 2 of A. arenosa is confirmed by the karyotypes of F2 individuals carrying altered number of copies of A. arenosa chromosomes 1 or 2.
A Quantitative Trait Locus Associated with Meiotic Stability
To identify loci associated with our fertility-related traits, we searched for quantitative trait loci (QTL) in the euploid F2 population (n = 298; see Methods). The traits analyzed were percentage of viable pollen, number of pollen grains per flower, percentage of viable seed, departure from expected chromosome number obtained from our FISH observations of male meiosis (Figure 4), and number of seed per silique (Supplemental Figure 1). We detected one locus on chromosome four of A. thaliana that correlated to the number of pollen grains per flower (Supplemental Figure 9), but no region was significantly correlated with either the percent of viable seed or the number of seed per silique.
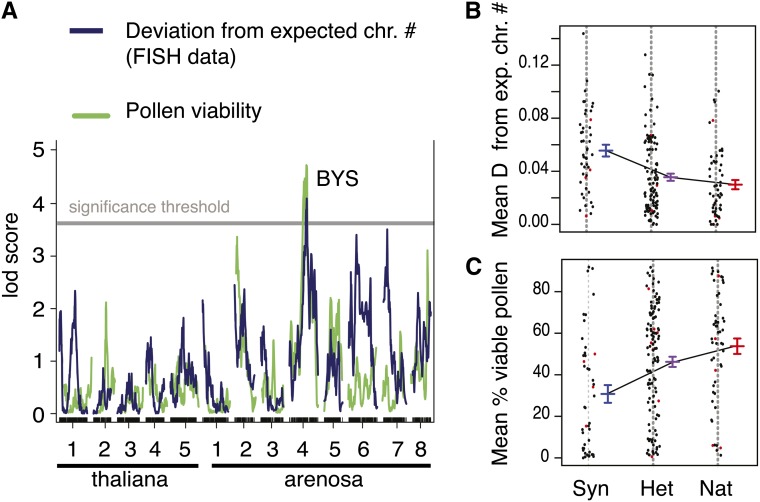
Most importantly, both pollen viability and our measure of meiotic regularity (mean deviation from expected chromosome number) were associated with a locus located on chromosome four of A. arenosa (position ∼16.9 Mb; Figure 8A). The genotype at this locus, which we named BOY NAMED SUE (BYS), explained 7.65% of the variation in pollen viability and 9.30% of the variation in our measure of meiotic regularity (Figure 8B). Neither pollen viability nor our measure of meiotic irregularity was correlated with overall seed number, suggesting that the effects were specific to the male gametophyte and not a more general loss of vigor or viability (Supplemental Figure 2).
Figure 8.
Identification of a QTL Associated with Pollen Viability and Meiotic Irregularity.
(A) Results of QTL detection using standard interval mapping in R/qtl (see Methods for a detailed description of the phenotypes and the QTL mapping parameters used). LOD scores are plotted along the five A. thaliana and the eight A. arenosa chromosomes. Significance thresholds of 3.74 (deviation from expected chromosome number) and 3.73 (pollen viability) were determined based on 1000 permutations and correspond to P values < 0.05. The locus corresponding to the peak exhibiting significant LOD scores for both phenotypes was named BYS.
(B) and (C) Individual (cloud) and mean pollen viability (blue) (B) and individual (cloud) and mean deviation from expected chromosome number (meiotic irregularity in green) (C) for F2 plants that were homozygous for Syn (synthetic allopolyploid allele, left), heterozygous (middle), or homozygous for Nat (natural allopolyploid allele, right) at the BYS locus (indicated on [A] and located approximately at 16.9 Mb on scaffold 4 of the A. lyrata genomic reference). Standard errors are represented. Imputed genotypes are plotted in red.
DISCUSSION
To investigate potential meiotic adaptations to polyploidy, we compared the natural, adapted allopolyploid A. suecica to a synthetic allopolyploid with the same genome composition. We observed higher pollen fertility in the natural polyploid (Figure 1B) but also observed that A. suecica produces fewer progeny with unbalanced chromosomes (Table 1, Figure 2). More importantly, we could not detect any signs of homoeologous recombination in the progeny of A. suecica, while the dosage irregularities observed in several of the synthetic progeny were consistent with homoeologous pairing and recombination (Figures 2 and 3). These results support the idea that A. suecica has evolved mechanisms for avoiding homoeologous pairing and are consistent with a recent study in wheat, documenting widespread aneuploidy in synthetic allohexaploids of wheat but no evidence for homoeologous exchanges (Zhang et al., 2013). The authors hypothesized that the presence of the functional allele of Ph1 in the tetraploid wheat parent used to produce the synthetic allohexaploid suppressed homoeologous pairing (Zhang et al., 2013).
The genomes of A. thaliana and A. arenosa are differentiated both by extensive sequence polymorphisms (5 to 8% in genes, higher in intergenic DNA) and large-scale structural variation. For example, the eight basal chromosomes found in A. arenosa and A. lyrata (Hu et al., 2011) have been reduced to 5 through multiple translocations and inversions. As a result, many expected products of homoeologous recombination include acentric or dicentric chromosomes, producing gametes that lack all copies of certain genomic loci, a situation that is likely to be lethal. Therefore, homoeologous recombination provides a potential explanation for the high pollen death phenotype observed in the synthetic allopolyploid (Figure 1B). Frequent lethality suggests that the incidence of homoeologous recombination is probably even higher than what we have observed. Importantly, the extensive rearrangements and considerable sequence divergence differentiating the two parental genomes (T and A) of A. suecica are not sufficient to prevent homoeologous recombination.
A. suecica Alleles Improve Viability
We observed a genome-wide trend for selection of A. suecica alleles over A. thaliana and A. arenosa alleles, with as many as 12 loci exhibiting suecica-biased transmission ratio distortion (Figure 5). The distorted loci were located mainly on the A. arenosa genome, with the exception of a small cluster of three loci on the A. thaliana chromosome 1 (Figure 5). Transmission rate distortion has been attributed to biased meiotic segregation, differential gametic success at fertilization, and postzygotic viability selection on certain genotypes (Wu et al., 2005). Consistent with the poor pollen and seed viability observed in the synthetic allopolyploid (Figure 1), we believe that both gametophytic and postzygotic selection for alleles and genomic regions contributed by the adapted allopolyploid A. suecica play a major role in determining biased inheritance. Often, genetic incompatibilities are determined by the interaction of two loci (Dobzhansky, 1936; Muller, 1942; Orr, 1995; Bomblies, 2010). We tested for association between the distorted loci, but we were not able to identify any specific pairs of loci exhibiting concordant distortion, suggesting that these loci were selected independently (data not shown). Distorted genomic regions have been frequently identified in populations of A. thaliana (Salomé et al., 2012), many also located on chromosome 1 (Salomé et al., 2012). It is thus possible that the factors underlying these regions affect viability in our system as well.
Dosage Imbalance Is Widespread
Dosage plots (as shown in Figures 2 and 6) can easily be generated from shotgun sequencing data and are informative about the types of meiotic irregularities that result in viable progeny. In our populations of F1 and F2 allopolyploids, we observed widespread aneuploidy and dosage imbalances, similar to those observed in newly synthesized allopolyploids of others species (Xiong et al., 2011; Chester et al., 2012; Zhang et al., 2013). Dosage imbalance was also present in the progeny of the natural A. suecica, indicating that, while it is clearly more fit than the synthetic allopolyploid, its meioses are often irregular. It is unclear whether this reduced fitness is compensated by the increased variation and adaptation potential generated by these dosage variants.
Interestingly, dosage variation preferentially involved altered dosage of A. arenosa chromosomes. Out of the 40 F2 individuals exhibiting dosage variation, 33 and 7 exhibited variation affecting A. arenosa– and A. thaliana–derived genomic DNA, respectively. This difference may be due to reduced negative selection against altered dosage of chromosomes that carry fewer genes (there are eight arenosa instead of five thaliana chromosomes) or could stem from more regular meiotic partitioning of one genome compared with the other. Alternatively, the two parental genomes in this allopolyploid may differ in overall epigenetic state leading to different overall expression levels and observed aneuploidy (Freeling et al., 2012).
In addition to being informative about the types of dosage variation found in our population, dosage plots can provide other insights. First, we were able to detect instances of homoeologous pairing that resulted in viable progeny (Figures 2 and 6). Notably, homoeologous recombination is recognizable because it results in an unbalanced genome (Figure 3). Balanced events would require both chromatids involved in recombination to enter the same meiotic product, an impossible event (Figure 3). Second, several of the individuals exhibited dosage imbalance in the F2 population consistent with the observed rearrangement between the A. lyrata and A. arenosa genomes (Figure 7B, inset), providing independent confirmation for these observations. Finally, while dosage variation is not necessarily easily detected phenotypically, it can drastically increase phenotypic variation in specific traits independently from allelic variation (Khush, 1973). It is thus imperative to discard individuals exhibiting dosage variation prior to QTL mapping.
What Is the Role of Structural Rearrangements in Polyploid Stability?
The accumulation of small and large scale structural variations between genomes is thought to contribute to meiotic regularity in polyploids. According to this hypothesis, genomic instability leading to structural variation in newly established polyploids has a stabilizing effect and could be required for efficient polyploidization. Evidence in support of this hypothesis is not found in all tested systems (see Introduction).
Previous reports used FISH to determine the location and number of nucleolus-organizer region (NOR) and 5S gene clusters in A. suecica and demonstrated the loss of a pair of NORs and 5S gene clusters in A. suecica compared with its presumed ancestors (Pontes et al., 2004). The authors hypothesize that the loss of the NOR can be explained by a double-stranded break, removing the NOR-containing tip of the A. thaliana chromosome 2. The missing 5S gene cluster is located in the pericentromeric region of the A. arenosa chromosome 8, a region rich in transposable elements. The loss of the gene cluster could thus be attributed to transposon-mediated rearrangements. While seemingly inconsistent with each other, these observations complement ours quite well. Indeed, this previous report focused on highly repetitive regions (readily visible by FISH), while our approach focused on single-copy regions onto which sequencing reads can be unambiguously assigned and for which coverage was obtained both on the A. suecica and the synthetic A. thaliana/A. arenosa genomes. Our markers therefore did not cover the specific regions targeted by Pontes but were otherwise well distributed along all 13 chromosomes of A. suecica (Supplemental Figure 7). Our genetic map was created de novo with no a priori information about linkage group or marker order and resulted in a map that is highly similar to those of A. thaliana and A. arenosa (Figure 7). Specifically, most regions exhibiting inconsistencies in marker order between A. suecica and its parental genomes were concentrated around the pericentromeric regions with the exceptions of two inversions and a few small-scale rearrangements (Figure 7). Taken together, these results do not support the hypothesis that structural differentiation between the parental genomes caused by chromosome inversions and translocations is sufficient to explain the observed reduction in homoeologous pairing in A. suecica and suggest that other mechanisms might be in place to increase fertility and meiotic regularity of natural Arabidopsis allopolyploids.
The BYS Locus Opens a New Door to Understanding the Evolution of Polyploidy Stability
Rather than widespread chromosome rearrangements, we instead document the genetic activity of an A. suecica allele at a single locus (BYS), whose favorable effect on both meiotic regularity and male fertility (Figure 8) is consistent with a gain-of-function innovation. These findings indicate that a major step in polyploidy adaptation can be mediated by a single gene change, which alone may address the challenge posed by polyploid meiosis. Unfortunately, no obvious candidate gene could be identified under the BYS QTL by perusing the syntenic regions of the A. thaliana and A. lyrata genomes. Additional loci may be present in the corresponding region of A. arenosa and A. suecica, for which no genomic reference genome is currently available.
The significance of the BYS QTL is relatively low compared with the threshold LOD score associated with our traits, despite a clear improvement in trait values for those F2 individuals carrying the A. suecica beneficial allele (Figure 8A versus Figures 8B and 8C). There are several possible explanations for this observation. First, nongenetic variance of our phenotypic assays is high. Aneuploidy is stochastic and rare. Pollen viability and deviation from expected chromosome numbers have a wide phenotypic range, even for the Syn × Nat F1 plants, which are not segregating for any allele (Supplemental Figure 1; Figures 8B and 8C). High variance decreases our power to detect QTL associated with these traits. Second, while meiotic stability appears to be affected by the BYS allele, it is not fully controlled by it. Yet, no other locus appears to be significant in our assay. It is possible that multiple other loci with limited impact affect the phenotype or that a genome-wide effect specific to the Sue or Syn genotype influences meiotic behavior.
Finally, we characterized this population for traits related to growth, organ development, shape, color, and others and found that, while the F2 displayed large variation in the corresponding phenotypes, we could only find associated QTL for a few of them, even if these traits have been previously shown to be genetically determined in other plant systems. Several of the plants analyzed in this F2 population exhibited stunted growth and dwarf phenotypes, suggesting karyotypic abnormalities, but did not carry any dosage imbalance according to our dosage assays. Together, these results lead us to believe that seed development and early sporophyte development might be affected by nongenetic factors. For example, aneuploidy in the endosperm or (epi)genetic shock associated with the hybridization event may increase stochastic phenotypes in the F2 plants independently of their sporophytic genotype. If this were the case, detection of the BYS locus despite these confounding factors suggests a stronger effect of BYS than documented in the QTL analysis.
Like the Ph1 gene of wheat, BYS is dominant (or additive), located on a single parental genome, and its active allele is found in the polyploid form of the organism. Since BYS originates from a natural allopolyploid, our preferred hypothesis is that it affects homoeologous pairing and multivalent formation. Because we lack direct evidence of a relationship between homoeologous recombination and the BYS QTL, its action may influence other mechanisms that directly or indirectly affect chromosome segregation, such as chromatid cohesion, centromere and spindle function, chromatin condensation, and recombination.
Our data suggest that BYS acts as a gain-of-function allele and its active form is present in the natural allopolyploid A. suecica but not in autotetraploid A. arenosa. It may be an allopolyploid-specific adaptation (i.e., beneficial only in allopolyploids and even detrimental in autotetraploids). Similar to the results presented here, new tetraploids of A. arenosa (diploids tetraploidized in the laboratory) exhibit more meiotic abnormalities than their natural autotetraploid counterparts (Hollister et al., 2012). Recent work comparing diploid and autotetraploid populations of A. arenosa has identified several genomic regions specifically associated with tetraploidy in this species (Hollister et al., 2012; Yant et al., 2013). The genes located within these regions are enriched in meiosis-related genes, confirming that rare variants or mutations that result in increased meiotic stability in a polyploid context have been selected upon tetraploidizaton in A. arenosa (Yant et al., 2013). BYS is not associated with tetraploidy in A. arenosa though, suggesting that the mechanisms of adaptation to auto- and allopolyploid meiosis may not be shared. Another interesting question is the timing of appearance of the A. suecica BYS allele. It is possible that it was required immediately after the initial hybridization event, in which case multiple accessions of A. suecica should carry the same function. Alternatively, it could have evolved gradually and different accessions of A. suecica may have evolved different mechanisms to reduce homoeologous pairing.
Meiosis is an ancient cellular function and its machinery is highly conserved and understanding the molecular basis for the action of the BYS locus will further our understanding of its intricacies. Whether BYS, the wheat Ph1 locus, and the Brassica PrBn locus share a common pathway or not, the existence of a locus associated with meiotic regularity in this highly tractable system should further our understanding of the mechanisms allowing the establishment of newly formed allopolyploids. Polyploid establishment and maintenance is a hurdle that many plant and animal species had to overcome at least once in their evolutionary path. Understanding the role of BYS and that of other loci with similar roles in other allopolyploid species will allow a broader understanding of the mechanisms underlying chromosome segregation and partitioning in successful polyploid species.
METHODS
Plant Material
All plants were grown on soil (Sunshine Professional Peat-Lite mix 4; SunGro Horticulture) in a growth room lit by fluorescent lamps (Model TL80; Phillips) at 22 ± 3°C with a 16-h:8-h light:dark photoperiod or in a greenhouse at similar temperatures and light regimes with supplemental light provided by sodium lamp illumination as required.
Tetraploid derivative of Arabidopsis thaliana ecotype Ler-0 (4x-Ler) was produced as previously described (Henry et al., 2005). The tetraploid Arabidopsis arenosa (Care-1) is a wild-collected accession corresponding to ABRC seed stock number CS3901. Crosses are always represented with the seed parent mentioned first and the pollen parent next. The synthetic allopolyploid (Syn) was obtained by crossing 4x-Ler as the seed parent to Care-1. The naturally occurring Arabidopsis suecica accessions were obtained as follows: Sue-1 was obtained from Fumiaki Katagiri (MIT, Cambridge, MA) (Hanfstingl et al., 1994). Sue-2 through Sue-6 were obtained from Craig Pikaard (Indiana University), and Sue-7 through Sue-19 were obtained from Magnus Nordborg (Gregor Mendel Institute, Vienna). Seed from Sue-1 through Sue-19 have been deposited at TAIR (accession numbers CS22505 to CS22517).
For the F1 selfed populations, Sue accessions and different synthetic (Syn) individuals were selfed and their progeny was grown in individual pots and assessed for dosage imbalances. The number of individuals for which sequence information was obtained are as follows: Sue-1 (n = 46), Sue-2 (n = 5), Sue-3 (n = 1), Sue-4 (n = 2), Sue-5 (n = 1), Sue-12 (n = 3), Sue-13 (n = 7), Sue-14 (n = 9), Sue-16 (n = 5), Sue-19 (n = 3), SynA (n = 1), SynB (n = 24), SynC (n = 10), and SynD (n = 17).
For the F2 population, the synthetic and natural allotetraploids were crossed to each other in order to produce the “Syn × Nat” F1 individuals. Three of these individuals were allowed to self-pollinate to produce the Syn × Nat F2 population (n = 522).
Trait Description
Each F2 plant was characterized for a variety of fertility-related traits. The distributions of values are illustrated in Supplemental Figure 1, and the pairwise regression analyses between trait values are shown in Supplemental Figure 2. For seed measurements, individual siliques were harvested into individual tubes and all the seeds from each fruit were counted using a dissecting microscope. Shriveled seeds that did not contain a visible embryo structure at least 20% the size of a wild-type seed were considered nonviable.
Pollen Viability
Mature flowers were harvested around 9 am, and at least three representative flowers from each plant were collected. Pollen fertility was estimated using 3% aceto-carmine stain. Random pollen samples from each flower were spread on a slide in a drop of 3% aceto-carmine stain. Viable (fertile) pollen stained dark red while nonviable pollen remained white or transparent. Fertile and sterile pollen were recorded as an average of three flowers from each of at least 10 microscopic fields under low power of a compound light microscope. Mean pollen viability was calculated for each of the following genotypes (number of plants observed): Ler-0 (22), Care-1 (16), Nat (16), Syn F1 (23), Syn F2 (11), Syn × Nat F1 (5), and Syn × Nat F2 (378). To compare genotypes, individual means were averaged and compared pairwise using Student’s t tests (Figure 1B).
Meiotic Regularity
To characterize meiotic stability, FISH of developing microspores was performed on each F2 individual, as well as the parental genotypes, as described previously (Henry et al., 2006). Briefly, fluorescent probes specific to the A. thaliana and A. arenosa centromeric repeats were used to distinguish between the two types of chromosomes, allowing us to record chromosome numbers in developing microspores at different stages of meiosis, as well as observations consistent with meiosis instability, such as chromosomal bridges or laggards (Figure 4). For each F2 individual, 10 to 15 meiotic figures were observed. The numbers of chromosomes detected in each developing microspore were averaged and used to derive the mean deviation from expected chromosome number (meanΔChr#).
Next-Generation Sequencing and Read Mapping
Genomic DNA was extracted using the Fast DNA-kit (MP Biomedicals) from all F1 and F2 individuals as well as the parental genotypes (Care-1, Sue-1, and Ler-0). For each individual, between 250 ng and 2.0 mg of DNA was further processed. Restriction enzyme sequence comparative analysis libraries were produced as previously described (Monson-Miller et al., 2012), using adaptors carrying 5-bp barcodes (Supplemental Table 1). Up to 96 F1 or F2 individuals were submitted in a single lane for 80-bp paired-end sequencing using Solexa Sequencing technology on an Illumina Genome Analyzer II or Illumina HiSeq-2000 (F1 individuals). Additional data describing the parental genotypes from an A. suecica library were generated on a full lane of a Genome Analyzer II flow cell, and Ler-0 and Care-1 were sequenced at half a lane each.
Sequencing reads were divided into individual pools according to their barcode, and the barcodes were removed from each read. Reads were further filtered for quality, the presence of ambiguous (N) bases, length, and adaptor/primer contamination using custom python scripts available online (http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools). The resulting read sequences were aligned to an in silico hybrid reference genome composed of the five chromosome of A. thaliana (TAIR 9.0 version) and the eight largest scaffolds of the A. lyrata reference genome (v1.0). Alignment was performed using the BWA aligner (version 0.6.0) and the default settings (Li and Durbin, 2009). Reads for which one single best match could be identified were retained as unique reads and further processed. Reads from A. arenosa (Care-1) and A. thaliana (Ler-1) were also aligned to the hybrid reference genome and the number of reads mapping onto the wrong genome was used to estimate the percentage of erroneous mapping on homoeologous regions (Supplemental Figure 3). Based on those percentages, reads mapping onto the A. thaliana chromosomes with more than one mismatch and reads mapping onto the A. lyrata scaffolds with more than four mismatches were discarded.
Karyotyping
For each individual, reads were pooled into nonoverlapping bins covering the whole reference genome using custom Python scripts, as described previously (Henry et al., 2010). For the F1 population, bins of 500 kb were used. For the F2 population, bins of 1 Mb were used. Coverage across the genome was derived by counting the number of reads that mapped to each bin. The mean percentage values across all F2/all F1 individuals was used as control, and relative coverage values were derived for each bin using the following formula: read coverage = (2 × reads in bin (sample) × total reads (control))/(total reads (sample) × reads in bin (control)). Using this formula, coverage values along chromosomes fluctuated around 2.0 for chromosome or chromosome fragments present in two copies and deviated up or down in individuals missing of carrying extra chromosomes or chromosomal pieces. The criteria used for identifying lesions were as follows: At least three consecutive bins had to exhibit relative coverage values closer to 1.0 (deletion) or 3.0 (insertion) than the expected 2.0 for an indel to be called. For various reasons, some regions of the genome exhibit much higher variations in coverage than others. Those regions could easily be mistaken for dosage variation if data from a single individual are considered. Therefore, for each instance of indel identified, coverage values for the same regions were examined in at least 10 other samples to rule out the possibility of false positives due to inherent variation for those regions. If another sample exhibited variation in the same region, that specific indel was discarded. In the case of the F1 populations, bins of 500 kb were used, and the smallest possible indel detectable was therefore 1.5 Mb. For the F2 population, bins of 1 Mb were used and the smallest possible indel was 3 Mb. A total of 15 and 57 individual with unbalanced chromosome dosage were identified for the F1 and F2 populations, respectively. Their karyotypes are presented in Supplemental Figure 4 for the Sue and Syn selfed individuals, Supplemental Figure 5 for the Syn × Nat F2 population, and Supplemental Data Set 1 for the Syn × Nat F2 individuals.
Marker Selection and Genotyping
The parental reads (Ler, Care, and Nat) were used to identify marker positions polymorphic between either Ler and Nat (on the A. thaliana chromosomes) or Care and Nat (mapped on the A. lyrata chromosomes) using custom python scripts as follows.
An mpileup table was created containing all base calls in the three parental genotypes, as well as the pooled F2 data, using the SAMtools package (Li and Durbin, 2009). The mpileup file was created using the python script “run-mpileup.py” (available at http://comailab.genomecenter.ucdavis.edu/index.php/Mpileup for download and documentation). To increase mapping accuracy and reduce file size, genome positions were filtered out according to the following criteria: (1) A. thaliana positions covered by any Care read and A. lyrata positions covered by any Ler reads were discarded; (2) positions had to be covered at least 10 times in the pooled F2 data and at least once in Nat; and (3) at least one basecall had to be different from the reference in the F2 alleles calls. This was performed using the python script “SynxNat1_Screen_mpup.py” (Supplemental Methods 1). This smaller mpileup-formatted file was parsed to obtain the percentages of each basecall for each position in each individual using “mpileup-parser.py” (see http://comailab.genomecenter.ucdavis.edu/index.php/Mpileup for download and documentation).
The final list of robust markers was generated by processing this output using the following criteria: (1) pooled F2 data coverage between 10 and 300; (2) coverage of at least five in both parental genotypes (Ler and Nat for A. thaliana positions and Care and Nat for A. arenosa positions); (3) the most common allele in the two parental genotypes represents at least 90% of their respective basecalls (not heterozygous positions) and are different from each other; and (4) the same two alleles are represented in the F2 pooled alleles calls, represent at least 20% of these calls each, and at least 95% combined. This step was performed using another custom python script (SynxNat2_Screen_Parsed_mpup.py; Supplemental Methods 2) and produced a set of 46,105 marker positions (see Supplemental Data Set 2 for the list of positions and the parental genotypes at those positions).
Next, the genotype of each F2 individual at these markers was determined and assigned to a parental allele. The following criteria were used: (1) if one base represented at least 90% of the basecalls and corresponded to one of the parental genotype, the genotype of that parent was assigned “S” for Sue or “Y” for the synthetic parent (Ler or Care); (2) if there are two alleles each representing at least 10% of the basecalls and those two alleles correspond to the two parental alleles, a heterozygous genotype was assigned “H”; and (3) in all other cases, no genotype was assigned “NA”. This was performed using script SynxNat3_CallAlleles.py (Supplemental Methods 3).
Because each F2 individual was sparsely covered, not all markers were covered in all F2 plants. More importantly, markers were usually only covered once or a few times, insufficient for distinguishing heterozygous from homozygous positions. To overcome this limitation and allow accurate scoring of genotypes for each F2 individual, markers were binned into nonoverlapping, consecutive bins of 30 markers. This generated 1533 marker bins. Individual F2 genotypes for each marker bin were calculated by summing the allele counts for each marker within each marker bin. The following criteria were applied to determine genotype: (1) a minimum of five allele calls had to be available; (2) a bin was annotated as homozygous if at least 95% of the allele calls corresponded to the same parental genotype; (3) bins were considered heterozygous if between 25 and 75% of the allele calls corresponded to one parental genotype and the remaining calls corresponded to the other parental genotype, otherwise the genotype was called “NA.” Possible genotypes at each marker bin were thus: Syn (Y) if it was homozygous for Ler or Care alleles, Sue (S) if it was homozygous for Sue alleles, Het (H) if it was heterozygous, and NA in all other cases. This step was performed using script SynxNat4_Create_MarkerBins.py (Supplemental Methods 4). A total of 1533 marker bins were generated from 424 sequenced F2 individuals (Supplemental Data Set 3). F2 individuals with fewer than 100 informative marker bins (12 lines) were discarded. Similarly, F2 individuals, for which <5% of the markers were Nat or Syn (61 lines) were discarded as potential contaminants, leaving 351 F2 lines.
To create a hybrid genetic map, marker bins were grouped using criteria that resulted in the expected number of linkage groups and subsequently ordered within each linkage group. All but three marker bins were assigned to one of these 13 linkage groups. Next, marker bin order was refined using the ripple function: The marker order that most closely matched the physical map was retained (Supplemental Figure 7).
Missing genotypes were imputed based on the genetic map obtained as follows: Walking along the chromosome, windows of marker bins were identified that started and ended with the same genotype included one additional instance of that genotype and contained a maximum of 15 marker bins. If a different genotype was found in more than one marker bin within that window, the genotypes for that window remained unchanged. Otherwise, all marker bins within the window were assigned the genotype identified at both ends of that window. At the beginning of chromosomes, if the first three assigned genotypes were consistent with each other, were <14 markers apart (total) and <14 markers from the beginning of the chromosome, marker bins were filled with that genotype. A similar approach was applied for the end of the chromosomes. This was performed using a custom python script (SynxNat5_Imputation.py; Supplemental Methods 5). In total, 149,422 NA genotypes were imputed, and 3829 outlier genotypes were corrected.
Next, the new genotypes were imported into R/qtl and 53 redundant markers, two individuals with identical genotypes and all individuals exhibiting dosage variation, were discarded, leaving 298 F2 individuals and 1477 markers.
QTL Mapping
The genotype data obtained in the previous step were imported into R/qtl (version 2.15.0 of R and version 1.23.16 of R/qtl) for the creation of a genetic map (Broman et al., 2003). The length of the genetic map was estimated at 443 centimorgans for the A. thaliana genome and 707 centimorgans for the A. arenosa genome (see Supplemental Table 2 and Supplemental Figure 7 for details). Pairwise recombination frequencies between all marker bins can be visualized in Supplemental Figure 8 and do not indicate substantial two-locus linkage disequilibrium as has been previously described in the progeny of some A. thaliana crosses (Törjék et al., 2006; Simon et al., 2008).
QTL detection was performed using the R/qtl package (Broman et al., 2003) using both the raw phenotypic values and the log transformed phenotypic values. The only difference in QTL detection between transformed and untransformed data analysis was the presence of one additional QTL using log transformed values for the trait “# seed per silique,” located at the very beginning of A. arenosa chromosome four. The more conservative results obtained using the nontransformed data are presented. Standard interval mapping was performed. LOD thresholds were calculated using 1000 permutations. All genotype and phenotype information can be found in Supplemental Data Set 4.
Accession Numbers
Next-generation sequence data generated in the context of this work can be found in the National Center for Biotechnology Information GenBank BioProject under ID PRJNA198011 and Sequence Read Archive accession number SRP029580.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of Phenotype Trait Values in the Syn × Nat F2 Population.
Supplemental Figure 2. Correlation between the Different Fertility-Related Traits in the Syn × Nat F2 Population.
Supplemental Figure 3. Mapping of Parental Reads to the Hybrid Reference Genome.
Supplemental Figure 4. Karyotype of the Individuals Exhibiting Dosage Variation in the Selfed Progeny of A. suecica Accessions.
Supplemental Figure 5. Karyotype of the Individuals Exhibiting Dosage Variation in the Selfed Progeny of Synthetic Allopolyploid (Syn) Individuals.
Supplemental Figure 6. FISH and Karyotyping Data Are Consistent with Each Other.
Supplemental Figure 7. Genetic Map of the Syn × Nat F2 Population.
Supplemental Figure 8. Pairwise Recombination Frequencies and LOD Scores between All Pairs of Marker Bins.
Supplemental Figure 9. Result of QTL Mapping for All Traits Analyzed.
Supplemental Table 1. List of Sequencing Barcodes Used for Library Demultiplexing.
Supplemental Table 2. Size of the Genetic Map.
Supplemental Data Set 1. Karyotype of the 57 Individuals Exhibiting Dosage Variation Identified in the Syn × Nat F2 Population
Supplemental Data Set 2. Marker List.
Supplemental Data Set 3. List of Marker Bins.
Supplemental Data Set 4. R/qtl Input File.
Supplementary Material
Acknowledgments
We thank the Biology Greenhouse (Biology Department, University of Washington) for material support and the UC Davis DNA Technologies Core for sequencing support. We thank Jennifer Monson-Miller, Brian Watson, Renee Weizbauer, and Meric C. Lieberman for technical and/or bioinformatics support. The A. lyrata genome sequence data were produced by the U.S. Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community. This work was supported by the National Science Foundation Project “Functional Genomics of Plant Polyploids” (DBI 0077774 and DBI 0733857 to L.C.) and the USDA National Research Initiative (2003-35300-13248 to B.P.D.).
AUTHOR CONTRIBUTIONS
B.P.D. and L.C. conceived and designed the crossing scheme and phenotyping part of this research. B.P.D. performed the crosses, generated the F2 population, and oversaw the phenotyping. B.C. performed the phenotypic analysis of the F2 plants. A.T. performed the FISH and pollen viability assays. I.M.H. conceived and led the genotyping-by-sequencing efforts of the F2 population. J.G. performed the sequencing and dosage analysis of the parental populations. I.M.H. and B.P.D. performed the QTL mapping analysis. I.M.H. wrote the article with contributions by L.C. and B.P.D. All authors read, reviewed, and approved the final article.
Glossary
- FISH
fluorescence in situ hybridization
- QTL
quantitative trait locus
- NOR
nucleolus-organizer region
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2010). Doomed lovers: Mechanisms of isolation and incompatibility in plants. Annu. Rev. Plant Biol. 61: 109–124 [DOI] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., Churchill G.A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Chester M., Gallagher J.P., Symonds V.V., Cruz da Silva A.V., Mavrodiev E.V., Leitch A.R., Soltis P.S., Soltis D.E. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M., Grandont L., Moore G., Chèvre A.M., Jenczewski E. (2010). Genetic regulation of meiosis in polyploid species: New insights into an old question. New Phytol. 186: 29–36 [DOI] [PubMed] [Google Scholar]
- Comai L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43: 387–399 [DOI] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A.P., Lysak M.A. (2003). FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 11: 217–226 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. (1936). Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Flagel L.E., Paterson A.H., Rapp R.A., Soltis D.E., Soltis P.S., Wendel J.F. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A.A., Vega J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F., Udall J.A. (2012). Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139 [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Chris Pires J. (2010). Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 186: 18–28 [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C., Iniguez-Luy F., Leon E., Osborn T.C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Han F., Fedak G., Guo W., Liu B. (2005). Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfstingl U., Berry A., Kellogg E.A., Costa J.T., III, Rüdiger W., Ausubel F.M. (1994). Haplotypic divergence coupled with lack of diversity at the Arabidopsis thaliana alcohol dehydrogenase locus: roles for both balancing and directional selection? Genetics 138: 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Comai L. (2006). Molecular karyotyping and aneuploidy detection in Arabidopsis thaliana using quantitative fluorescent polymerase chain reaction. Plant J. 48: 307–319 [DOI] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Miller E.S., Burkart-Waco D., Comai L. (2010). Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Young K., Watson B., Wu H., Comai L. (2005). Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Arnold B.J., Svedin E., Xue K.S., Dilkes B.P., Bomblies K. (2012). Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet. 8: e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T.T., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.C. (1982). Polyploidy and diploidy: New perspectives on chromosome pairing and its evolutionary implications. Am. J. Bot. 69: 1512–1523 [Google Scholar]
- Jakobsson M., Hagenblad J., Tavaré S., Säll T., Halldén C., Lind-Halldén C., Nordborg M. (2006). A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol. Biol. Evol. 23: 1217–1231 [DOI] [PubMed] [Google Scholar]
- Jenczewski E., Eber F., Grimaud A., Huet S., Lucas M.O., Monod H., Chèvre A.M. (2003). PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S. (1973). Cytogenetics of Aneuploids. (New York: Academic Press). [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.R., Matyasek R., Kovarik A., Leitch A.R. (2004). Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. Lond. 82: 599–606 [Google Scholar]
- Liu B., Brubaker C.L., Mergeai G., Cronn R.C., Wendel J.F. (2001). Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44: 321–330 [PubMed] [Google Scholar]
- Madlung A., Tyagi A.P., Watson B., Jiang H., Kagochi T., Doerge R.W., Martienssen R., Comai L. (2005). Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41: 221–230 [DOI] [PubMed] [Google Scholar]
- Monson-Miller J., Sanchez-Mendez D.C., Fass J., Henry I.M., Tai T.H., Comai L. (2012). Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing. BMC Genomics 13: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. (2002). Meiosis in allopolyploids — The importance of ‘Teflon’ chromosomes. Trends Genet. 18: 456–463 [DOI] [PubMed] [Google Scholar]
- Muller H.J. (1942). Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125 [Google Scholar]
- Nicolas S.D., Leflon M., Monod H., Eber F., Coriton O., Huteau V., Chèvre A.M., Jenczewski E. (2009). Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A. (1995). The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Freeling M., Tang H., Wang X. (2010). Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 61: 349–372 [DOI] [PubMed] [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S. (2004). Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 101: 18240–18245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J., Schemske D.W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501 [Google Scholar]
- Salomé P.A., Bomblies K., Fitz J., Laitinen R.A., Warthmann N., Yant L., Weigel D. (2012). The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb.) 108: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Koch M.A. (2011). Arabidopsis hybrid speciation processes. Proc. Natl. Acad. Sci. USA 108: 14192–14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Loudet O., Durand S., Bérard A., Brunel D., Sennesal F.X., Durand-Tardif M., Pelletier G., Camilleri C. (2008). Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835 [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Schemske D.W., Hancock J.F., Thompson J.N., Husband B.C., Judd W.S. (2007). Autopolyploidy in angiosperms: Have we grossly underestimated the number of species? Taxon 56: 13–30 [Google Scholar]
- Stebbins G.L. (1984). Chromosome pairing, hybrid sterility, and polyploidy: A reply to R. C. Jackson. Syst. Bot. 9: 119–121 [Google Scholar]
- Szadkowski E., et al. (2010). The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112 [DOI] [PubMed] [Google Scholar]
- Törjék O., Witucka-Wall H., Meyer R.C., von Korff M., Kusterer B., Rautengarten C., Altmann T. (2006). Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor. Appl. Genet. 113: 1551–1561 [DOI] [PubMed] [Google Scholar]
- Wu G., Hao L., Han Z., Gao S., Latham K.E., de Villena F.P., Sapienza C. (2005). Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division. Genetics 170: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R.T., Pires J.C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Hollister J.D., Wright K.M., Arnold B.J., Higgins J.D., Franklin F.C., Bomblies K. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23: 2151–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousafzai F.K., Al-Kaff N., Moore G. (2010). The molecular features of chromosome pairing at meiosis: The polyploid challenge using wheat as a reference. Funct. Integr. Genomics 10: 147–156 [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. Natl. Acad. Sci. USA 110: 3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.