This work demonstrates that WRKY57 functions in the crosstalk between jasmonic acid (JA)– and auxin-mediated signaling during JA-induced leaf senescence. The findings provide a mechanistic insight into how JA-induced leaf senescence is antagonized by auxin via WRKY57.
Abstract
Leaf senescence is regulated by diverse developmental and environmental factors. Exogenous jasmonic acid (JA) can induce leaf senescence, whereas auxin suppresses this physiological process. Crosstalk between JA and auxin signaling has been well studied, but not during JA-induced leaf senescence. Here, we found that upon methyl jasmonate treatment, Arabidopsis thaliana wrky57 mutants produced typical leaf senescence symptoms, such as yellowing leaves, low chlorophyll content, and high cell death rates. Further investigation suggested that senescence-associated genes were upregulated in the wrky57 mutants. Chromatin immunoprecipitation experiments revealed that WRKY57 directly binds to the promoters of SENESCENCE4 and SENESCENCE-ASSOCIATED GENE12 and represses their transcription. In vivo and in vitro experiments suggested that WRKY57 interacts with JASMONATE ZIM-DOMAIN4/8 (JAZ4/8) and the AUX/IAA protein IAA29, repressors of the JA and auxin signaling pathways, respectively. Consistent with the opposing functions of JA and auxin in JA-induced leaf senescence, JAZ4/8 and IAA29 also displayed opposite functions in JA-induced leaf senescence and competitively interacted with WRKY57. Our results suggested that the JA-induced leaf senescence process can be antagonized by auxin via WRKY57. Moreover, WRKY57 protein levels were downregulated by JA but upregulated by auxin. Therefore, as a repressor in JA-induced leaf senescence, WRKY57 is a common component of the JA- and auxin-mediated signaling pathways.
INTRODUCTION
Leaf senescence, as a postmitotic senescence program, occurs not only with aging, but also in stressed or detached leaves (Gan, 2003). When environmental stresses and/or endogenous signals are perceived by plants, they are subsequently transmitted, resulting in changes in gene expression and/or physiological activities (Gan and Amasino, 1997) and finally bringing on senescence phenotypes. Plant growth regulators, such as abscisic acid (ABA), ethylene (ET), jasmonic acid (JA), and salicylic acid (SA), have been confirmed to promote leaf senescence in complex interconnecting pathways (Grbic and Bleecker, 1995; Morris et al., 2000; He et al., 2002).
JA functions in the induction of leaf senescence in many plant species (Ueda and Kato, 1980; He et al., 2002) by regulating the expression of various senescence-associated genes (Buchanan-Wollaston et al., 2005; Jung et al., 2007). For instance, the treatment of barley (Hordeum vulgare) leaves with exogenous JA or methyl jasmonate (MeJA) resulted in typical senescent phenotypes, such as the degradation of chlorophyll and reduced levels of Rubisco (Parthier, 1990). He et al. (2002) confirmed that exogenous JA promotes typical senescence in both attached and detached Arabidopsis thaliana leaves but fails to induce precocious leaf senescence in the JA-insensitive mutant coi1. Molecular, genetic, and physiological analyses showed that Rubisco activase plays an important role in JA-induced leaf senescence (Shan et al., 2011). Although several studies on leaf senescence induced by JA have been reported, the underlying molecular mechanism for JA-induced leaf senescence is not well understood.
Recent investigations have defined CORONATINE-INSENSITIVE1 (COI1), an F-box protein, as a key component of the receptor complex of the JA signaling pathway (Xie et al., 1998; Yan et al., 2009). JASMONATE ZIM-DOMAIN (JAZ) proteins are ubiquitinated via SCFCOI1 in response to JA (Thines et al., 2007). The JAZ family proteins function as repressors of the JA signaling pathway, and a recent structural and pharmacological study showed that COI1 and JAZ form a coreceptor complex (Sheard et al., 2010). JAZ proteins have been shown to directly bind to MYC2 and its close homologs, MYC3 and MYC4, to block their function (Chini et al., 2007; Cheng et al., 2009; Niu et al., 2011; Fernández-Calvo et al., 2011). JAZ proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect JA-regulated stamen development (Song et al., 2011) and with WD-Repeat/bHLH/MYB complexes to regulate JA-mediated anthocyanin accumulation and trichome initiation (Qi et al., 2011) in Arabidopsis.
Auxin plays a role in the suppression of leaf senescence (Lim et al., 2003). Classical studies have correlated auxin levels with senescence (Addicott, 1982; Sexton and Roberts, 1982; Nooden and Leopold, 1998). In bean (Phaseolus vulgaris) leaves, a gradient of auxin levels was detected between the leaf blade and the stalk (Shoji et al., 1951). Auxin levels decline with leaf age, and senescence occurs when auxin levels in the leaf and stalk are approximately equal (Shoji et al., 1951). These findings suggest that decreased auxin levels may signal the onset of senescence or enhance it. Moreover, auxin represses the transcription of some genes whose expression is correlated with senescence (Noh and Amasino, 1999; Hong et al., 2000; Tucker et al., 2002). In addition to the relationship of auxin level with leaf senescence, the auxin signaling pathway also functions in this process. AUXIN RESPONSE FACTOR2 (ARF2), a repressor of auxin signaling, functions in the auxin-mediated regulation of Arabidopsis leaf longevity by suppressing the expression of senescence-associated genes (Ellis et al., 2005; Lim et al., 2010). A study on neutron irradiation–induced senescence showed that the expression of some ARFs and Aux/IAAs was significantly changed during the senescence process (Fortunati et al., 2010). The ARF7 and ARF19 genes are also induced in senescing leaves (Lin and Wu, 2004). In barley, a novel senescence-induced gene, ARF1, plays an important role during senescence-associated transport processes (Ay et al., 2008). Although auxin levels and auxin signaling are related to leaf senescence, the underlying regulatory mechanism is still unclear.
One of the universal signaling pathways involved in the response to external stimuli is WRKY family protein regulation. The WRKY transcription factor family comprises 74 members in Arabidopsis (Eulgem et al., 2000; Dong et al., 2003; Eulgem and Somssich, 2007), and many members of the family have been shown to be involved in various biotic and abiotic stress responses (Eulgem and Somssich, 2007; Rushton et al., 2010; Parinita et al., 2011; Chen et al., 2012). Expression profiling indicates that many WRKY factors are strongly upregulated by senescence and that WRKY factors are the second largest group of transcription factors in the senescence transcriptome (Guo et al., 2004). Recently, functional analysis showed that several WRKY proteins play important roles in senescence-associated regulatory pathways. The WRKY53 protein plays a regulatory role in early events of leaf senescence (Hinderhofer and Zentgraf, 2001), and the study of protein–protein interactions of WRKY53 with ESR/ESP also shows that they mediate negative crosstalk between pathogen resistance and senescence influenced by the JA/SA equilibrium (Miao and Zentgraf, 2007). WRKY22 is one of the target genes of WRKY53 (Miao et al., 2004) and functions in dark-induced senescence (Zhou et al., 2011). WRKY6 positively influences senescence by specifically activating, through direct W-box interactions, the senescence-induced receptor-like kinase (SIRK) gene (Robatzek and Somssich, 2002). WRKY70 is located in the node of convergence for JA-mediated and SA-mediated signals (Li et al., 2004) and acts as a negative regulator of senescence (Ülker et al., 2007). WRKY30, interacting with WRKY53, participates in the senescence regulatory network with other WRKYs (Besseau et al., 2012).
In this study, we found that WRKY57 functions as a negative regulator in JA-induced leaf senescence in Arabidopsis. Further investigation indicated that WRKY57 directly represses the expression of SENESCENCE4 (SEN4) and SENESCENCE-ASSOCIATED GENE12 (SAG12). Moreover, JAZ and auxin/indole-3-acetic acid (IAA) proteins competitively interact with the zinc-finger domain of WRKY57, which accounts for the antagonism of auxin in JA-induced leaf senescence. Our work suggests that WRKY57 functions as a regulatory node in the JA-induced senescence signaling pathways mediated by auxin and JA. Because WRKYs have been suggested to integrate various hormonal and environmental signals in plants (Li et al., 2004; Miao and Zentgraf, 2007; Eulgem and Somssich, 2007; Rushton et al., 2010; Parinita et al., 2011; Chen et al., 2012), this study provides a mechanistic understanding of how the crosstalk between the JA and auxin signaling pathways in JA-induced leaf senescence is fine-tuned by WRKY57.
RESULTS
Loss of WRKY57 Function Accelerates JA-Induced Leaf Senescence
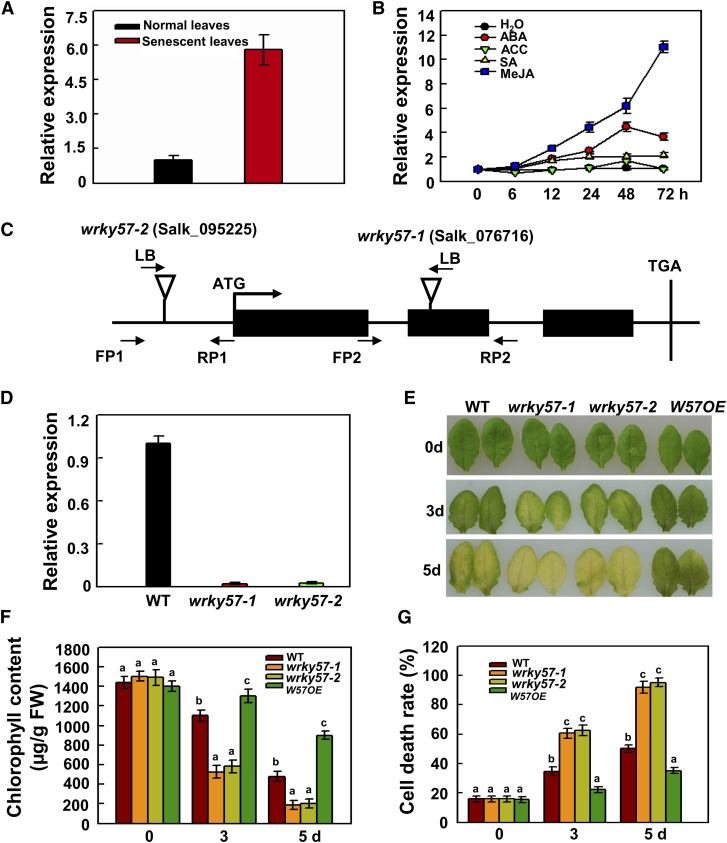
Our recent research showed that WRKY57 is induced by drought and its activated expression confers drought tolerance in Arabidopsis (Jiang et al., 2012). A microarray database for Arabidopsis indicated that WRKY57 is specifically expressed in senescent leaves (Winter et al., 2007), and we further confirmed this result (Figure 1A). In general, phytohormones (ABA, JA, SA, and ET) can facilitate leaf senescence in Arabidopsis (Vojislava and Anthony, 1995; Fan et al., 1997; Karl et al., 2000; Schippers et al., 2007; Kohki et al., 2009; Shan et al., 2011). To determine whether WRKY57 transcription is associated with phytohormones, we checked the transcript levels of WRKY57 in response to exogenous phytohormone treatments, revealing that JA significantly induces WRKY57 (Figure 1B). Given that JA induces leaf senescence, we speculated that WRKY57 may function in the JA-induced leaf senescence process.
Figure 1.
Loss of WRKY57 Function Accelerates JA-Induced Leaf Senescence.
(A) WRKY57 transcript levels in normal and senescent leaves. RNA samples were prepared from 4-week-old plants (normal green leaves) and 8-week-old plants (senescent leaves displaying obvious senescence phenotypes), respectively.
(B) WRKY57 transcript levels upon hormones treatments. Total RNA was extracted from plants harvested at 0, 6, 12, 24, 48, and 72 h after each treatment. ACC, 1-aminocyclopropane-1-carboxylic acid (the precursor of ET).
(C) T-DNA insertion position in two wrky57 mutants (Salk_076716 and Salk_095225).
(D) Real-time RT-PCR analysis of WRKY57 transcript levels in the wild-type and wrky57 mutants. In (A), (B), and (D), relative expression was normalized using the internal control ACTIN2.
(E) wrky57 leaves showed a more severe JA-induced senescence phenotype than the wild type and overexpression line upon 100 µM MeJA treatment.
(F) Chlorophyll content in detached full-grown rosette leaves. FW, fresh weight.
(G) Cell death. In (F) and (G), values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
To confirm our speculation, detached leaves from two wrky57 mutants (Figures 1C and 1D) and the wild type were used for JA-induced leaf senescence assays. After MeJA treatment, wrky57-1 and wrky57-2 leaves showed more serious yellowing, which is a typical characteristic of leaf senescence, when compared with wild-type and transgenic lines leaves (Figure 1E). It has been demonstrated that leaf senescence often accelerates the degradation of chlorophyll and cell death (Takamiya et al., 2000; Hörtensteiner and Feller, 2002; Lin and Wu, 2004). Measurement of chlorophyll content showed that chlorophyll was lost more quickly in the leaves of the wrky57-1 and wrky57-2 mutants than in the wild type and in a WRKY57 overexpression line under MeJA treatment (Figure 1F). Correspondingly, cell death rates were significantly higher in both wrky57 mutants than in the wild type and the overexpression line under MeJA treatment (Figure 1G). Further results also were obtained with the treatment of SA, ABA, and 1-aminocyclopropane-1-carboxylic acid (the precursor of ET) and implied that mutation of WRKY57 accelerated leaf senescence only when the senescence was induced by JA (Supplemental Figure 1). These results suggested that WRKY57 plays a negative role in JA-induced leaf senescence.
WRKY57 Directly Suppresses Expression of SEN4 and SAG12 under JA Treatment
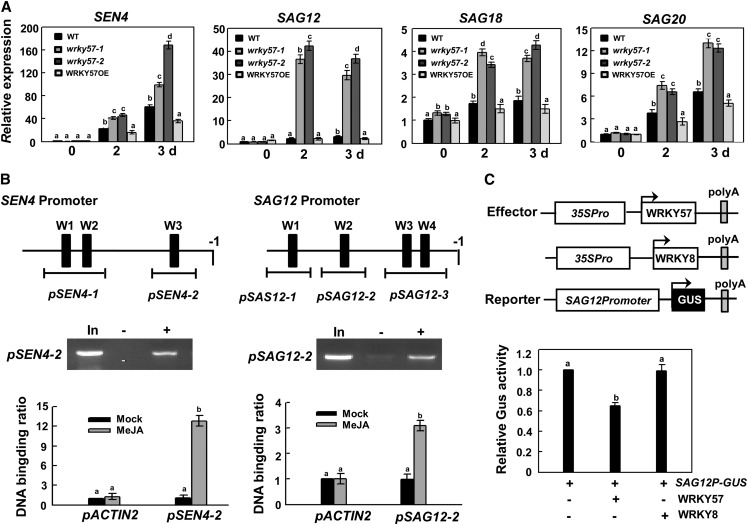
Many genes that are upregulated during the senescence process have been defined as senescence-associated genes. The transcript levels of four senescence-associated genes (SEN4, SAG12, SAG18, and SAG20) were determined in the wild type, the wrky57 mutants, and the WRKY57 overexpression line. After treatment with MeJA, their transcript levels were upregulated significantly in the wrky57 mutants compared with the wild type and the overexpression line (Figure 2A). These results suggested that WRKY57 negatively regulates the transcript levels of these senescence-associated genes, which then promotes the progression of leaf senescence.
Figure 2.
WRKY57 Directly Suppresses the Expression of Senescence-Associated Genes.
(A) Relative mRNA levels of SEN4, SAG12, SAG18, and SAG20. Three-week-old plants were detached from their roots for MeJA treatment. Values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
(B) The promoter structure of the SEN4 and SAG12 genes and fragments used in the ChIP assay. W1, W2, etc. denote each W-box, numbered from left to right with sequence sites relative to the start code (top). Lines indicate the sequences detected by ChIP assays. RT-PCR results showed that WRKY57 binds to the promoters of SEN4 and SAG12 (middle). In, PCR product from the chromatin DNA; −, PCR product from ChIP with preimmune serum (as a negative control); +, PCR product from ChIP with the antibody against Myc. Real-time RT-PCR data from ChIP assay with antibody against Myc with the ACTIN2 promoter (pACTIN2) as a negative control (bottom). Values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
(C) Schematic of the ProSAG12:GUS reporter and WRKY57 and GFP effectors (top). Transcription repression of the SAG12 promoter reporter by WRKY57 was assessed by GUS activity. GFP was used as a control. Values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
In (A) to (C), relative expression was normalized using the internal control ACTIN2.
It is well known that WRKY transcription factors function by binding typical cis-element W-boxes of target gene promoters. Several W-box motifs were found in the putative promoter regions of the four senescence-associated genes (Figure 2B; Supplemental Figure 2). To determine whether these genes are direct targets of WRKY57, chromatin immunoprecipitation (ChIP) experiments were conducted. The results from ChIP showed that WRKY57 binds to the promoters of the SEN4 and SAG12 genes via the W box sequence (pSEN4-2 and pSAG12-2, respectively) (Figure 2B). This suggested that WRKY57 directly regulates the transcription of SEN4 and SAG12.
To further confirm the negative regulatory function of WRKY57, we performed transient expression assays. A reporter plasmid was constructed by fusing a 2.7-kb promoter sequence of SAG12 with the β-glucuronidase (GUS) reporter gene. The effector plasmid had a WRKY57 or WRKY8 gene driven by the cauliflower mosaic virus (CaMV) 35S promoter. As shown in Figure 2C, coexpression of the WRKY57 gene resulted in significantly lower GUS activity than coexpression of the WRKY8 gene. This supported the hypothesis that WRKY57 is a negative regulator of senescence-associated genes.
WRKY57 Physically Interacts with JAZ4, JAZ8, and IAA29
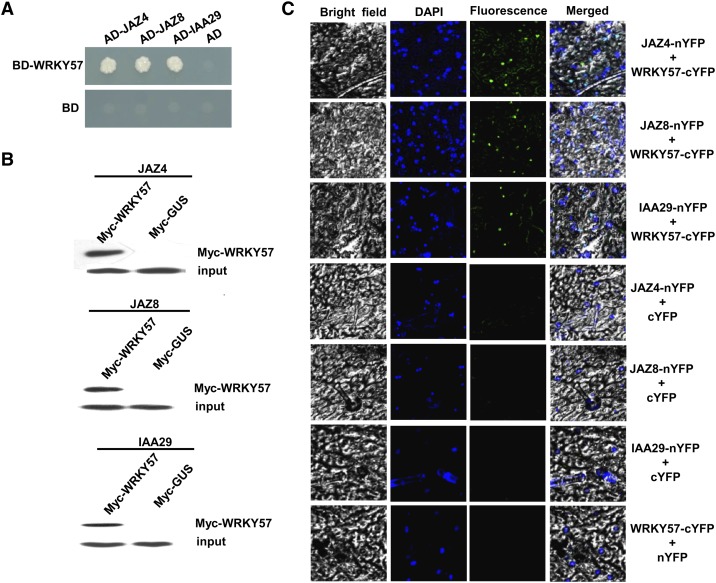
Increasing evidence suggests that WRKY proteins function by forming protein complexes with other interactors (Xu et al., 2006; Miao and Zentgraf, 2007; Hou et al., 2010; Shang et al., 2010; Hu et al., 2013a). To search for potential partners of the WRKY57 protein, we employed the yeast two-hybrid system. WRKY57 was fused with the BD domain of the pGBK-T7 vector as bait. Yeast cells harboring the bait were transformed with a cDNA library containing inserts for prey proteins fused to GAL4-AD. In total, 47 colonies were positive for the expression of the His3 and LacZ reporter genes. Among these candidate interactors, JAZ4, JAZ8, and IAA29 were frequently represented. To confirm their interaction in yeast, their open reading frame sequences were fused with the AD domain of the pGAD-T7 vector and used for further interaction experiments with WRKY57. As shown in Figure 3A, WRKY57 strongly interacted with JAZ4, JAZ8, and IAA29. We further tested their interaction in vitro by protein pull-down assays. Myc-WRKY57 and HA-JAZ4/JAZ8/IAA29 were coexpressed in yeast cells. The protein complexes were incubated with anti-HA and A/G-agarose beads and then separated on SDS-PAGE for immunoblotting with anti-Myc antibody. As a result, the JAZ4, JAZ8, and IAA29 proteins could be pulled down by WRKY57 (Figure 3B). To determine whether these interactions also occur in plant cells, we then employed the bimolecular fluorescence complementation (BiFC) system. Full-length JAZ4, JAZ8, or IAA29 cDNAs were fused to the N-terminal region of the yellow fluorescent protein (YFP). Agrobacterial cells harboring the corresponding interaction pair were infiltrated into tobacco (Nicotiana benthamiana) leaves. In parallel, empty vectors in combination with each fusion construct were coinfiltrated into tobacco leaves. After 2 d of incubation, YFP signals were observed with fluorescence microscopy. The samples coinfiltrated with an interaction pair showed YFP fluorescence in the cell nuclei, whereas all control samples failed to give any YFP signal (Figure 3C). These results indicated that WRKY57 and its partners colocalize and interact in plant cell nuclei.
Figure 3.
WRKY57 Physically Interacts with JAZ4, JAZ8, and IAA29.
(A) Yeast-two-hybrid assays. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu/Trp/His/Ade and containing 5 mM 3-aminotriazole. The Gal4 DNA binding domain was fused with WRKY57 (shown as BD-WRKY57) and the Gal4 activation domain was fused with JAZs or IAA29 (shown as AD-JAZs and AD-IAA29). The Gal4 DNA binding domain expressed by pGBKT7 (shown as BD) was used as a negative control.
(B) Coimmunoprecipitation assays. HA-fused JAZs or IAA29 were immunoprecipitated using anti-HA antibody, and co-immunoprecipitated Myc-WRKY57 was then detected using anti-Myc antibody. Protein input for HA-JAZs and HA-IAA29 in immunoprecipitated complexes were also detected and are shown.
(C) BiFC assays. Fluorescence was observed in the nuclear compartments of N. benthamiana leaf epidermal cells that resulted from complementation of the C-terminal part of YFP fused with WRKY57 (WRKY57-cYFP) and the N-terminal part of YFP fused with JAZs or IAA29 (JAZs-nYFP and IAA29-nYFP). No signals were observed from the negative controls. DAPI, 4′,6-diamidino-2-phenylindole.
JAZ4 and JAZ8 Negatively Regulate JA-Induced Leaf Senescence
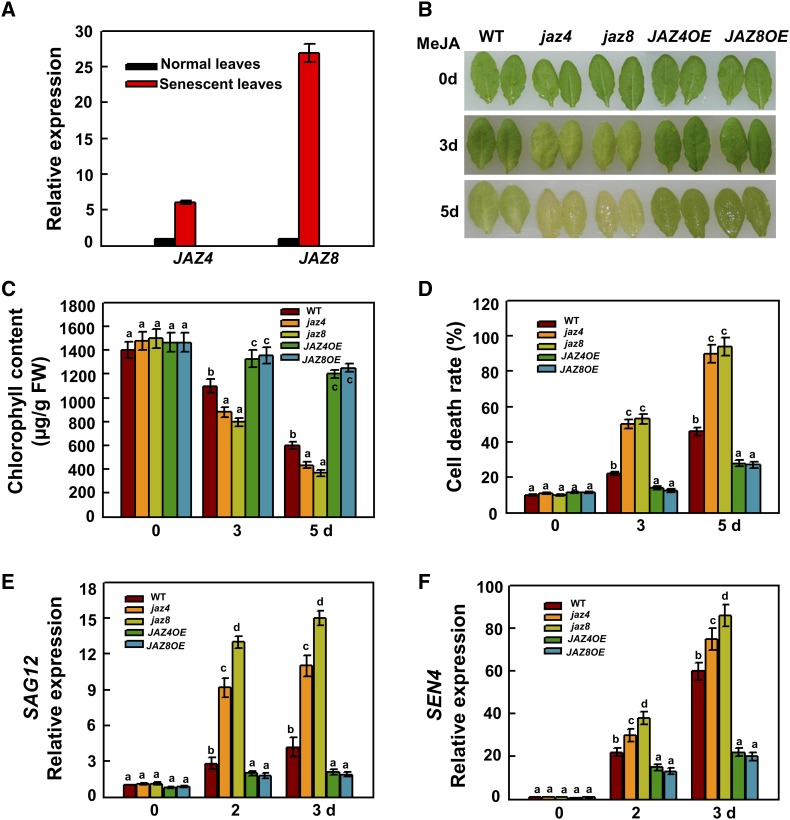
As the receptor of JA, COI1 is crucial for JA-induced senescence since its loss-of-function mutant is insensitive to JA-induced senescence (He et al., 2002; Shan et al., 2011). JAZ proteins are key negative regulators of JA signaling pathways and are incorporated into 26S the proteasome for degradation upon JA signaling (Pauwels and Goossens, 2011). Since JA directly promotes leaf senescence, JAZs should play opposite roles in this process. We first determined the transcript levels of JAZ4 and JAZ8 in senescent leaves. Consistent with WRKY57, the transcript levels of JAZ4 and JAZ8 were higher in senescent leaves than in normal leaves (Figure 4A). To investigate their potential functions in JA-induced senescence, we obtained two T-DNA insertion mutants and two overexpression lines (Supplemental Figure 3) for JAZ4 and JAZ8 and examined their phenotypes under MeJA treatment. The jaz4 and jaz8 mutants displayed more severe senescence phenotypes than the wild type (Figure 4B). By contrast, JAZ4 and JAZ8 overexpression plants displayed delayed senescence phenotypes (Figure 4B). Chlorophyll contents and cell death rates were also in agreement with the phenotypes (Figures 4C and 4D). We then determined the transcript levels of SEN4 and SAG12, demonstrating that SEN4 and SAG12 were upregulated in the jaz4 and jaz8 mutants, but downregulated in overexpression plants (Figures 4E and 4F). These results suggested that, as repressors of JA signaling pathways, JAZ4 and JAZ8 suppress JA-induced leaf senescence.
Figure 4.
JAZ4 and JAZ8 Negatively Regulate JA-Induced Leaf Senescence.
(A) Relative mRNA levels of JAZ4 and JAZ8 in normal and senescent leaves. Values are mean ± se (n = 5 experiments).
(B) jaz4 and jaz8 leaves showed more severe JA-induced senescence phenotypes than the wild type and overexpression lines upon 100 µM MeJA treatment.
(C) Chlorophyll content in detached full-grown rosette leaves. FW, fresh weight.
(D) Cell death.
(E) and (F) The expression levels of SAG12 and SEN4 in the wild type, mutants, and overexpression lines.
In (C) to (F), values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05). In (A), (E), and (F), relative expression was normalized using the internal control ACTIN2.
IAA29 Positively Regulates JA-Induced Leaf Senescence
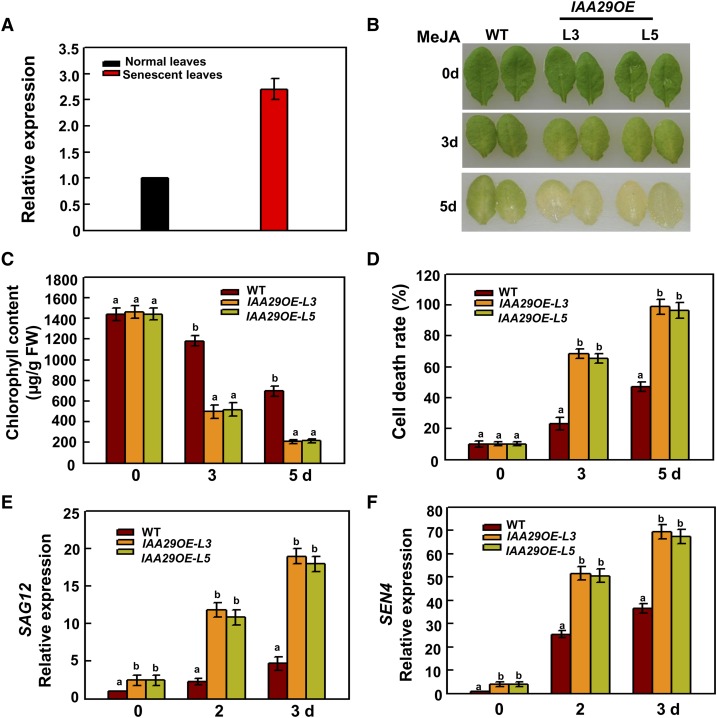
Given that WRKY57 and IAA29 interact, we thought it likely that IAA29 functions in senescence. To confirm our speculation, we first determined the transcript levels of IAA29, which were higher in senescent leaves than in normal leaves (Figure 5A). To explore the functions of IAA29 in JA-induced senescence, a T-DNA insertion mutant of IAA29 was used for a senescence assay (Supplemental Figures 4A and 4B). Under MeJA treatment conditions, the iaa29 mutant displayed no significant senescence phenotypes, similar to the wild type. To investigate whether elevated IAA29 levels could affect JA-induced senescence, IAA29 overexpression plants were generated (Supplemental Figure 4C) and used for senescence assay. As expected, leaf senescence symptoms appeared earlier in IAA29 overexpression lines compared with wild-type plants (Figure 5B). The senescence phenotypes were also in agreement with the reduced chlorophyll content and increased cell death rate in IAA29 overexpression plants (Figures 5C and 5D). Correspondingly, the transcript levels of SEN4 and SAG12 were higher in IAA29 overexpression plants than in the wild type (Figures 5E and 5F). These results suggested that IAA29 positively regulates JA-induced leaf senescence.
Figure 5.
IAA29 Positively Regulates JA-Induced Leaf Senescence.
(A) Relative mRNA levels of IAA29 in normal and senescent leaves. Values are mean ± se (n = 5 experiments).
(B) Leaves of IAA29 overexpression lines showed more severe senescence phenotypes than the wild type upon 100 µM MeJA treatment.
(C) Chlorophyll content in detached full-grown rosette leaves. FW, fresh weight.
(D) Cell death.
(E) and (F) Expression levels of SAG12 and SEN4 in the wild type and IAA29 overexpression lines.
In (C) to (F), values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05). In (A), (E), and (F), relative expression was normalized using the internal control ACTIN2.
WRKY57 Is Required for Auxin Antagonization of JA-Induced Leaf Senescence
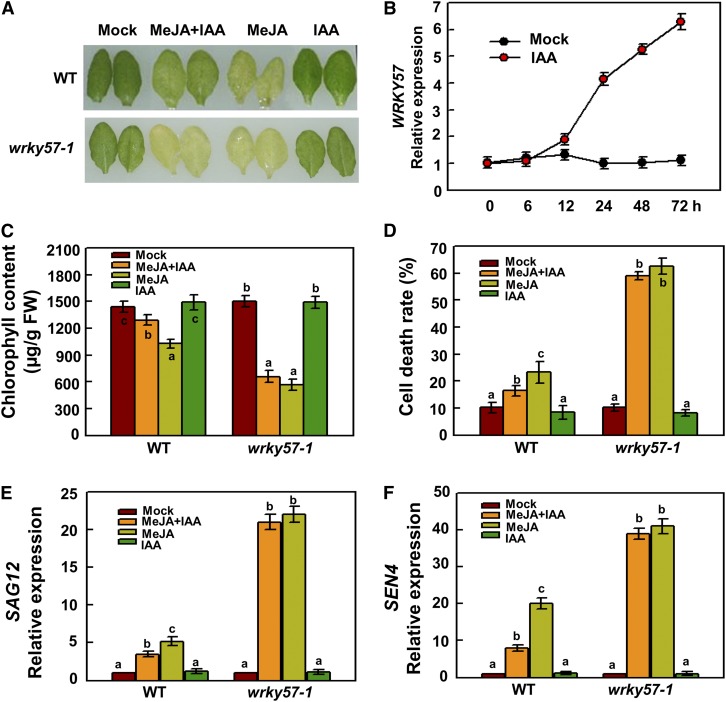
Previous studies have suggested that auxin can suppress senescence; however, the underlying mechanism is unclear. Considering that the function of auxin is opposite to that of JA in senescence, we questioned whether auxin could antagonize the function of exogenous JA in induced leaf senescence. Thus, detached wild-type leaves were treated with water (mock), MeJA, IAA, and MeJA+IAA. After 5 d, no senescence symptoms were observed for leaves in water or IAA; however, the leaves in MeJA showed obvious senescence symptoms whereas the leaves in MeJA+IAA showed slight senescence symptoms (Figure 6A). The chlorophyll content and cell death rate results supported these phenotypes (Figures 6C and 6D). Correspondingly, the transcript levels of SEN4 and SAG12 were significantly downregulated in leaves treated with MeJA+IAA compared with those treated with MeJA (Figures 6E and 6F). These results indicated that auxin can partially suppress JA-induced senescence.
Figure 6.
WRKY57 Is Required for Auxin Antagonization of JA-Induced Leaf Senescence.
(A) The JA-induced senescence phenotype was alleviated by treatment with 30 µM IAA in the wild type, but not in wrky57-1.
(B) WRKY57 transcript levels upon 10 µM IAA treatment.
(C) Chlorophyll content in detached full-grown rosette leaves. FW, fresh weight.
(D) Cell death.
(E) and (F) The expression levels of SAG12 and SEN4 in the wild type and wrky57-1 upon different treatments.
In (B) to (F), values are mean ± se (n = 5 experiments), and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05). In (B), (E), and (F), relative expression was normalized using the internal control ACTIN2.
Although WRKY57 negatively regulates JA-induced senescence, we did not know whether WRKY57 was also involved in auxin-suppressed senescence. We first examined the transcript levels of WRKY57 in response to auxin treatment and found that WRKY57 was dramatically induced by auxin treatment (Figure 6B), implying that WRKY57 may be involved in auxin signaling pathways. Subsequently, we determined whether WRKY57 was necessary for auxin suppression of senescence. When the wrky57-1 mutant was used for senescence assay, no difference was observed between leaves treated with MeJA or MeJA+IAA (Figure 6A). We also found that the transcript levels of SEN4 and SAG12 in the wrky57-1 mutant were not affected by treatment with MeJA+IAA compared with MeJA (Figures 6C and 6D). Chlorophyll content and cell death rate results further supported these phenotypes (Figures 6E and 6F). Taken together, our results suggested that auxin can antagonize JA-induced senescence and that WRKY57 is required for this antagonism.
JAZ and IAA Competitively Interact with WRKY57
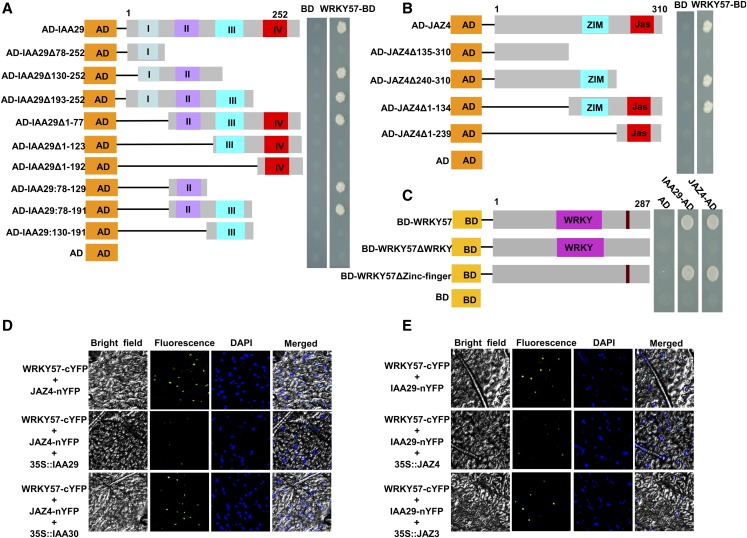
Given that WRKY57 physically interacts with JAZ (JAZ4 and JAZ8) and IAA (IAA29) and that the function of the JAZ4/8 proteins is opposite to that of IAA29, we questioned whether there is a competitive relationship between JAZ and IAA for WRKY57. To investigate which region of IAA29 is required for interaction with WRKY57, we fused 10 truncated IAA29 variants to the AD domain of the pGADT7 vector. The interaction between these derivatives and the WRKY57 protein (as determined using a yeast two-hybrid assay) revealed that domain II was specifically responsible for the interaction (Figure 7A). Similarly, five truncated JAZ4 variants fused to the AD domain of the pGADT7 vector were generated. The results revealed that the ZIM domain was responsible for interaction with AWRKY57 (Figure 7B). The pGBKT7 vectors containing a WRKY domain mutant or a zinc-finger domain mutant were used to analyze which domain of WRKY57 is responsible for interaction with the JAZ or IAA proteins. The results showed that the zinc-finger domain of WRKY57 was necessary for interaction with both the JAZ and IAA proteins (Figure 7C). Therefore, it is likely that the JAZ protein can interfere with the interaction between the IAA and WRKY57 proteins and vice versa.
Figure 7.
JAZ4 and IAA29 Competitively Interact with WRKY57.
(A) The Domain II of IAA29 is necessary for interaction with WRKY57. Left: Diagram of full-length and truncated IAA29 constructs with specific deletions. Right: Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The empty pGADT7 prey vector was used as a negative control.
(B) The ZIM domain of JAZ4 is required for interaction with WRKY57. Left: Diagram of full-length and truncated JAZ4 constructs with specific deletions. Right: Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The empty pGADT7 prey vector was used as a negative control.
(C) The zinc-finger domain of WRKY57 is necessary for interaction with JAZs and IAA29. Left: Diagram of full-length and truncated WRKY57 constructs with specific deletions. Right: Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The empty pGBKT7 prey vector was used as a negative control. In (A) to (C), AD and BD refer to the pGADT7 and pGBKT7 vector, respectively.
(D) and (E) Competition between JAZ4 and IAA29 for interaction with WRKY57. DAPI, 4′,6-diamidino-2-phenylindole.
To confirm our hypothesis, we employed a BiFC system enabling us to monitor changes in interaction between proteins in the presence or absence of a third protein. In the control, fused JAZ4-nYFP was coexpressed with fused WRKY57-cYFP in N. benthamiana leaves, and the YFP signal was very strong in the nucleus (Figure 7D). However, when a third protein, IAA29, was coexpressed with the fused JAZ4-nYFP and WRKY57-cYFP, the YFP signal became weaker than in the control (Figure 7D). When a different protein, IAA30, which cannot interact with WRKY57 or JAZ4, was coexpressed with JAZ4-nYFP and WRKY57-cYFP, the YFP signal was the same strength as in the control. Similarly, when JAZ4 was coexpressed with fused IAA29-nYFP and WRKY57-cYFP, the YFP signal became weaker than when IAA29-nYFP and WRKY57-cYFP were coexpressed alone (Figure 7E). By contrast, JAZ3, which cannot interact with WRKY57 or IAA29, did not affect the YFP signal when IAA29-nYFP and WRKY57-cYFP were coexpressed (Figure 7E). Taken together, our results suggested that JAZ4 and IAA29 competitively interact with WRKY57.
WRKY57 Proteins Are Induced by Auxin but Degraded by JA via the 26S Proteasome Pathway
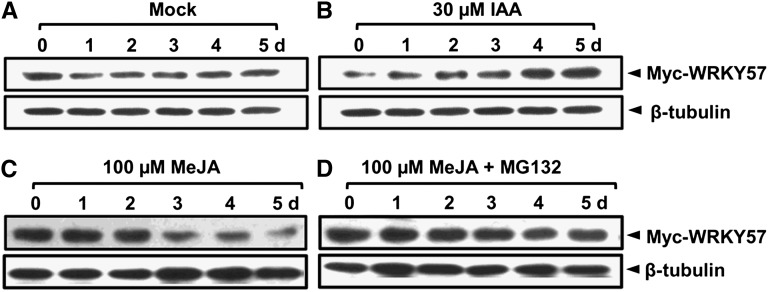
Although WRKY57 transcripts are induced by JA, WRKY57 functions as a negative regulator in JA-induced senescence. However, because mRNA levels sometimes do not reflect protein levels, it was necessary to determine the protein levels of WRKY57. For this purpose, WRKY57 cDNA fused with a Myc tag and driven by a 2.5-kb WRKY57 promoter was transformed into the wrky57-1 mutant. The expression of the Myc tag was used to monitor the protein levels of WRKY57 (by protein gel blot analysis). As shown in Figure 8A, WRKY57 protein levels in ProWRKY57: Myc-WRKY57 were not induced by water (mock) (Figure 8A). We then checked the response of WRKY57 to auxin at both the mRNA and protein levels in ProWRKY57:Myc-WRKY57, revealing that either mRNA or protein levels of WRKY57 were induced by auxin treatment (Figure 8B; Supplemental Figure 5). We also analyzed the responses of WRKY57 to MeJA. Similarly to the wild type, WRKY57 transcript levels in ProWRKY57:Myc-WRKY57 were upregulated after MeJA treatment (Supplemental Figure 5). By contrast, WRKY57 protein levels decreased with the treatment of MeJA (Figure 8C). The degradation of WRKY57 was clearly attenuated by the 26S proteasome inhibitor MG132 (Figure 8D), suggesting that the 26S proteasome pathway is required for WRKY57 protein degradation by JA. Taken together, these results indicated that at the protein level, WRKY57 is regulated by JA and auxin in an antagonistic manner. We also determined the protein levels of WRKY57 in coi1 mutants, and the results demonstrated that the protein degradation is partially dependent on COI1 (Supplemental Figure 6C) and suggested that other pathways might be involved in the turnover of WRKY57 upon JA treatments.
Figure 8.
Protein Blotting Analysis of WRKY57 Protein Levels.
Four-week-old ProWRKY57:Myc-WRKY57 transgenic plants were used for protein gel blotting analysis upon treatment with water (mock) (A), IAA (B), MeJA (C), or MeJA+MG132 (D) in the indicated time periods. The immunoblot was detected with β-tubulin antibody as a protein-loading control.
DISCUSSION
Although remarkable advances have been made in the last decade in understanding the mechanisms of JA signal transduction in model plants such as Arabidopsis, the specific mechanisms by which JA induces leaf senescence are not well understood. Moreover, few studies have focused on the crosstalk between the different hormones mediating senescence. Auxin modulates JA signaling, which is essential for development in plants. However, the molecular details of this phytohormone interaction also remain largely unknown. A great deal of indirect evidence has confirmed that plant WRKY proteins are involved in plant senescence, but information about the biological roles of specific WRKY proteins in JA-induced senescence is limited. Here, we reveal that auxin can antagonize JA-induced leaf senescence and that WRKY57 is a key modulator mediating the crosstalk between JA and auxin in JA-induced leaf senescence.
WRKY57 Negatively Regulates JA-Induced Leaf Senescence
Leaf senescence is the terminal stage of leaf development and is regulated by both intrinsic and environmental signals, with phytohormones and growth regulators, such as cytokinin, ABA, SA, JA, ET, brassinosteroids, and nitric oxide, acting in a precisely coordinated manner (Gepstein, 2004). As a class of specific transcription factors, WRKY genes are involved in diverse aspects of plant growth and development as well as biotic and abiotic stresses. In Arabidopsis, four members (WRKY30, WRKY53, WRKY54, and WRKY70) of the WRKY group IIIb proteins are involved in the regulation of plant senescence associated with SA (Besseau et al., 2012). WRKY54 and WRKY70 function as negative senescence regulators and WRKY53 as a positive regulator. It is possible that they regulate the progression of leaf senescence via interaction with WRKY30. Our previous study showed that WRKY22 mediates dark-induced leaf senescence (Zhou et al., 2011). In addition, WRKY6 can directly activate transcription of SIRK, which encodes a senescence-induced receptor-like protein kinase, to positively influence leaf senescence (Robatzek and Somssich, 2002). Arabidopsis WRKY57, which belongs to the WRKY group II proteins, has been found to regulate drought tolerance responses (Jiang et al., 2012). Our investigation suggested that WRKY57 transcripts were abundant in senescent leaves and induced by exogenous MeJA application. Although under normal conditions wrky57 mutants displayed no senescence symptoms compared with the wild type, accelerated leaf senescence was observed when they were subjected to MeJA treatment. After MeJA treatment, the transcript levels of WRKY57 in the coi1 mutant were significantly lower than that in the wild type, suggesting that the transcription of WRKY57 induced by JA is partially dependent on COI1 function (Supplemental Figure 6A). When wrky57coi1 double mutants were used for the JA-induced leaf senescence assay, the double mutants showed similar phenotype to coi1 single mutants (Supplemental Figure 6B). These results imply that the function of WRKY57 is dependent on COI1 function in JA-induced leaf senescence. Among the upregulated senescence-associated genes in wrky57 mutants, SEN4 and SAG12 were found to contain several WRKY-specific W-box cis-elements in their promoter regions. Our ChIP experiment indicated that WRKY57 can bind to the promoters of SEN4 and SAG12, suggesting that they are direct targets of WRKY57. The combination of their upregulation in wrky57 mutants and the downregulation of GUS activity in transient assays suggest that WRKY57 is a negative regulator of SEN4 and SAG12. Several WRKY proteins have been shown to act as both positive and negative transcriptional regulators. For example, WRKY6 acts as a negative regulator of its own expression and the expression of WRKY42 and as a positive regulator of SIRK (Robatzek and Somssich, 2002). Our recent research suggested that WRKY8 regulates ABI4 positively and ACS6 negatively when plants are subjected to virus infection (Chen et al., 2013). We confirmed that WRKY57 can directly activate RD29A transcription under drought conditions (Jiang et al., 2012). Here, we established that WRKY57 directly represses the transcription of SEN4 and SAG12 in leaf senescence. However, it is still unclear how the same protein displays completely opposite transcription activities upon binding to different promoters.
WRKY57 Mediates Crosstalk between the Auxin and JA Signaling Pathways in Leaf Senescence
Recently, the functions of JAZs as repressors of JA signaling pathway were well characterized (Chini et al., 2007; Cheng et al., 2009; Fernández-Calvo et al., 2011; Niu et al., 2011). Our latest research revealed that JA regulates plant tolerance to freezing via interaction between JAZ proteins and ICE1 (Hu et al., 2013b). Obviously, JAZs transduce the JA signal by interacting with different transcription factors. Our results showed that WRKY57 expression is responsive to JA treatment and that WRKY57 physically interacts with JAZ proteins. Similar to the bHLH subgroup IIId factors and MYC2, which interact with JAZs and function as negative regulators in plant immunity (Song et al., 2013; Zhai et al., 2013), WRKY57 is a negative regulator in JA-induced leaf senescence. However, it is unclear how JAZ proteins affect the function of WRKY57 proteins. It is worth noting that the ZIM domain of JAZ proteins is responsible for interaction with WRKY57, whereas the Jas domain interacts with MYB and bHLH transcription factors. It is unknown whether JAZs have different effects on their interactors.
A feature of plant development and adaptability to environmental stress is that major regulatory genes function as orchestrators of multiple hormone signaling pathways. As an important growth regulator, auxin interacts with other signaling pathways and alters the stability of IAA proteins to modulate the course of plant development and promote plant survival under adverse conditions (González-Lamothe et al., 2012). The combination of reduced auxin level in senescent leaves (Shoji et al., 1951) and delayed leaf senescence resulting from exogenous auxin application (Noh and Amasino, 1999) indicates that auxin acts as a suppressor of leaf senescence. Recently, ARF2, an auxin response factor, was confirmed to play a major role in regulating auxin-mediated leaf longevity (Lim et al., 2010). In our senescence assays, exogenous application of auxin caused a delay in MeJA-induced leaf senescence (Figure 6A), indicating that auxin can antagonize the functions of JA in inducing leaf senescence. However, it is still unclear which genes mediate the crosstalk between auxin and JA in leaf senescence. When the wrky57-1 mutant was used for senescence assays, exogenous auxin application could not delay JA-mediated leaf senescence, suggesting that WRKY57 is crucial for auxin antagonization of JA-induced leaf senescence. Similar to JAZs acting as repressors of the JA signaling pathway, IAA proteins function as repressors of the auxin signaling pathway. Domains III and IV of IAA proteins interact with ARF proteins (Guilfoyle et al., 1998; Liscum and Reed, 2002). By contrast, domain II of IAA29 is crucial for interaction with WRKY57. Consistent with the opposing functions of auxin and JA in leaf senescence, IAA29 positively regulates leaf senescence whereas JAZ4 and JAZ8 act as negative regulators. Interestingly, both JAZ4/8 and IAA29 physically interact with the zinc-finger domain of WRKY57. As shown in our competition experiments, elevated JAZ4 expression represses the interaction of IAA29 with WRKY57 and vice versa. It is likely that the opposite functions of JAZ4/8 and IAA29 are caused by their competitive binding to WRKY57. In addition, we revealed that WRKY57 protein levels are downregulated by JA but upregulated by auxin, demonstrating that WRKY57 protein levels are influenced antagonistically by auxin and JA. Taking our results together, we drew the conclusion that WRKY57 mediates the crosstalk between the auxin and JA signaling pathways in JA-induced leaf senescence.
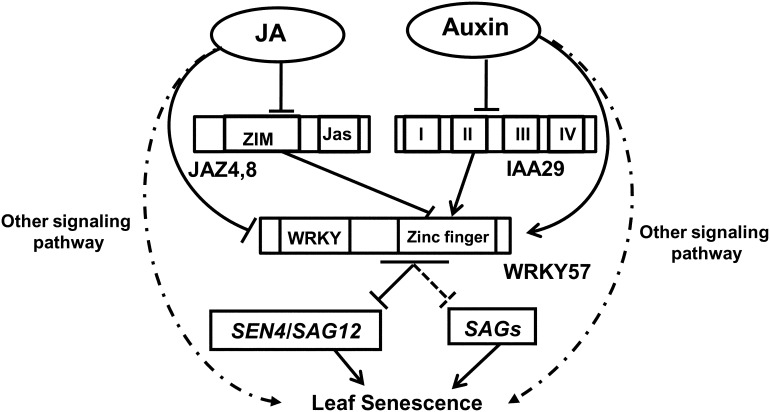
Finally, a working model of the role of WRKY57 in leaf senescence is shown in Figure 9. The exogenous JA-induced leaf senescence process can be antagonized by exogenous auxin. JAZ4/8 and IAA29, as the key negative regulators in JA and auxin signaling, respectively, competitively interact with the zinc-finger domain of WRKY57, which functions as a negative regulator during JA-induced leaf senescence. At the protein level, JA mediates the degradation of WRKY57, whereas auxin stabilizes WRKY57. WRKY57 directly binds to the promoters of SEN4 and SAG12 and represses their transcription. WRKY57 functions as a node of convergence for JA- and auxin-mediated signaling pathways during the JA-induced leaf senescence process.
Figure 9.
The Working Model of WRKY57 Functions in JA-Induced Leaf Senescence.
Auxin and JA antagonistically regulate leaf senescence through WRKY57. JAZ4/8 and IAA29 function as negative and positive regulators, respectively, in JA-induced leaf senescence, and they competitively interact with the zinc-finger domain of WRKY57. At the protein level, WRKY57 is induced by auxin but repressed by JA. WRKY57 directly binds to the promoters of SEN4 and SAG12 and then represses their transcription. The solid lines indicate direct interaction or regulation. The dashed lines indicate indirect regulation.
METHODS
Materials and Plant Growth Conditions
Arabidopsis thaliana (ecotype Columbia) seeds were surface-sterilized (20% [v/v] bleach for 15 min) before being sown on half-strength Murashige and Skoog medium and kept at 4°C for 3 d. One-week-old plants were transferred to soil. The plants were then kept in a growth cabinet at 22°C under long-day conditions (a 16-h-light [100 μE m−2 s−1]/8-h-dark cycle). Wild-type and mutant plants used in this study were in the Columbia-0 genetic background. The mutants used in this study are listed as follows: wrky57 (Salk_076716 and Salk_095225), jaz4 (Salk_141628), jaz8 (CS849856), and iaa29 (Salk_091933). To generate the overexpression transgenic plants, the full-length cDNAs of WRKY57, JAZ4, JAZ8, and IAA29 were cloned into the binary vector pOCA30 in the sense orientation behind the CaMV 35S promoter (Jiang et al., 2012).
Expression Analysis
Four-week-old normal leaves and 8-week-old senescent leaves were harvested for total RNA extraction. Hormone treatments were performed on mature plants. Four-week-old wild-type plants were detached from their roots and placed into deionized water supplemented with 100 µM MeJA, 30 µM IAA, 2 mM SA, 100 µM ABA, or 2 mM 1-aminocyclopropane-1-carboxylic acid. The plants were collected at designated time intervals and then used for total RNA extraction.
For real-time RT-PCR analysis, total RNA was extracted using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Fermentas) according to the manufacturer’s instructions. Total RNA (∼2 μg) was reverse-transcribed in a 20-μL reaction mixture using Superscript II (Invitrogen). After the reaction, 1-μL aliquots were used as a template for PCR amplification and SYBR green was used to monitor the kinetics of PCR products in real-time PCR. As an internal control, the ACT2 transcript was used to quantify the relative transcript levels of each target gene in each tissue type. Three biological replicates were conducted. The primers used for real-time RT-PCR are listed in Supplemental Table 1.
GUS activity was measured as previously described (Yoo et al., 2007), and the data were presented as the averages of five biological replicates.
For protein gel blotting, the aerial parts were cut from 4-week-old plants grown in soil, placed into solution containing 100 µM MeJA, 100 µM MeJA+MG132, 30 µM IAA, or water for various time periods and then harvested for protein extraction. Protein gel blotting analysis was performed as described previously (Xu et al., 2006). Crude antisera of anti-Myc were used at dilutions of 1:5000. For relative protein expression level assay, the protein gel blotting analysis was repeated three times.
Leaf Senescence Assays
Leaves from 4-week-old plants were used for the leaf senescence assay. Leaves at the same developmental stage were placed into dishes filled with distilled water supplemented with water (mock), 100 µM MeJA, and/or 30 µM IAA, allowing good water contact with leaf pedicels. The plates were then kept in weak light (20 μE m−2 s−1) at 22°C.
Chlorophyll was extracted with 80% acetone from detached leaves. Chlorophyll content was determined at 663 and 645 nm according to Lichtenthaler (1987). Cell death rate was detected by Evans blue staining as described previously (Guo and Crawford, 2005).
ChIP Assays
ChIP assay was performed essentially according to previously described protocols (Mukhopadhyay et al., 2008; Shang et al., 2010). Two-week-old ProWRKY57:Myc-WRKY57/wrky57-1 seedlings were immersed in cross-linking buffer under vacuum for 10 min followed by additional 10-min incubation with 0.1 M Gly. Seedlings were ground in liquid nitrogen and resuspended in nuclei isolation buffer. Nuclei were then collected by centrifugation at 11,000g for 20 min at 4°C, resuspended in nuclei lysis buffer, and sonicated to 200- to 1000-bp fragments. After centrifugation at 13,800g for 10 min at 4°C, the supernatants were incubated in ChIP dilution buffer containing 60 mL of protein A/G agarose beads (Santa Cruz Biotechnology) for 1 h at 48°C with gentle rotation to preclear the diluted sonicated chromatin and then centrifuged at 3800g for 2 min at 4°C. The supernatants were recovered and incubated with the antibody against Myc (with the preimmune serum instead of the antibody as a negative control) overnight at 4°C. Protein A/G agarose (60 mL) was added into the mixture for a further incubation for 2 h at 4°C and centrifuged at 3800g for 2 min at 4°C to collect the agarose beads and the chromatin. The agarose beads were washed for 10 min each time (except for TE buffer wash for 5 min each time) with gentle rotation at 4°C with 1 mL of each of the following buffers and centrifuged at 3800g for 2 min at 4°C: two times with low salt wash buffer, two times with high salt wash buffer, two times with LiCl wash buffer, and three times with TE buffer. The immunocomplexes were eluted from the agarose beads with 500 mL of the elution buffer (two times for 250 mL each) by incubating at room temperature for 30 min (two times for 15 min each) with gentle rotation. Cross-links were reversed by incubation at 65°C overnight followed by proteinase K treatment for 2 h at 45°C, phenol/chloroform/isoamyl alcohol extraction, and ethanol precipitation. Pellets were washed with 70% (v/v) ethanol and resuspended in double distilled water. The TTGACC/T motifs are W-box sequences recognized by WRKY transcription factors. The primers used for PCR amplification of different promoters are listed in Supplemental Table 1. PCR amplification was performed using 36 cycles at 60°C for all promoter fragments. Aliquots of the PCR reactions were resolved by electrophoresis on 2% agarose gels. The results presented here represent at least three independent experiments.
To quantitatively determine the WRKY57–DNA (target promoters) binding, real-time RT-PCR analysis was performed according to the procedure described previously (Mukhopadhyay et al., 2008) with the ACTIN2 3′-untranslated region sequence as an endogenous control. The relative quantity value is presented as the DNA binding ratio (differential site occupancy). The same primers used for the above-mentioned PCR analysis were used for real-time RT-PCR. A fragment of the ACTIN2 promoter was used as a negative control.
Yeast Two-Hybrid Screening and Confirmation
The full-length WRKY57 cDNA was cloned into the bait vector pGBKT7 and then transformed into the yeast strain Y2HGold (Clontech). Two-hybrid screening was performed via the mating protocol described in Clontech’s Matchmaker TM Gold Yeast Two-Hybrid user manual. To confirm protein–protein interactions, the full-length JAZ4, JAZ8, and IAA29 coding sequences (CDSs) were cloned into the prey vector pGADT7. The primers used for yeast two-hybrid are listed in Supplemental Table 2.
BiFC and Coimmunoprecipitation Assays
The cDNA sequences of enhanced YFP fragments, 173 amino acids located in the N terminus (nYFP), and 64 amino acids located in the C terminus (cYFP), were PCR amplified and cloned into the XbaI-XhoI and BamHI-XhoI sites of pFGC5941 to generate pFGC-nYFP and pFGC-cYFP, respectively (Chen and Chen, 2002; Hu et al., 2013a). The full-length WRKY57 CDS was inserted into pFGC-cYFP to generate a C-terminal in-frame fusion with cYFP, while JAZ4, JAZ8, and IAA29 CDSs were introduced into pFGC-nYFP to form N-terminal in-frame fusions with nYFP. The resulting plasmids were introduced into Agrobacterium tumefaciens (strain EHA105), and infiltration of Nicotiana benthamiana was performed as described previously (Hu et al., 2013a). Infected tissues were analyzed 48 h after infiltration. YFP and 4′,6-diamidino-2-phenylindole fluorescence were observed under a confocal laser scanning microscope (Olympus). The primers used for BiFC are listed in Supplemental Table 2.
For coimmunoprecipitation assays, WRKY57, JAZ4, JAZ8, and IAA29 were individually cloned into tagging plasmids behind the Myc or HA tag sequence in the sense orientation behind the CaMV 35S promoter. Myc-fused WRKY57 and HA-fused JAZ4, JAZ8, and IAA29 were then transiently coexpressed in the yeast strain Y2HGold. Coimmunoprecipitation assays were performed using yeast protein extracts. Briefly, HA-fused JAZ4, JAZ8, and IAA29 were immunoprecipitated using an anti-HA antibody diluted 5000-fold in a 20 mM Tris-HCl, pH 7.6, solution supplemented with 150 mM NaCl, 0.1% Tween 20, and 5% skim milk powder, and the coimmunoprecipitated proteins were then detected using an anti-Myc antibody (Sigma-Aldrich).
Accession Numbers
Nucleotide sequence data for the genes described in this article are available from the Arabidopsis Genome Initiative database under the following accession numbers: WRKY57 (AT1G69310), SEN4 (AT4G30270), SAG12 (AT5G45890), SAG18 (AT1G71190), SAG20 (AT3G10985), JAZ4 (AT1G48500), JAZ8 (AT1G30135), IAA29 (AT4G32280), and ACTIN2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Loss of WRKY57 Function Accelerated Leaf Senescence Only Induced by Exogenous MeJA.
Supplemental Figure 2. W-Box Information of SEN4, SAG12, SAG18, and SAG20 Genes Promoter Sequences in Detail.
Supplemental Figure 3. Mutant and Overexpression Line Screening of JAZ4 and JAZ8.
Supplemental Figure 4. Mutant and Overexpression Line Screening of IAA29.
Supplemental Figure 5. Expression Levels of WRKY57 in ProWRKY57:Myc-WRKY57 Transgenic Plants under IAA and MeJA Treatments.
Supplemental Figure 6. The Function of WRKY57 in JA-Induced Senescence Is Dependent on COI1.
Supplemental Table 1. qRT-PCR Primer Sets Used in This Study.
Supplemental Table 2. Primer Sets Used in Yeast Two-Hybrid and BiFC Assay.
Supplementary Material
Acknowledgments
We thank Zhixiang Chen (Purdue University, West Lafayette, IN) for Arabidopsis wrky57-1 mutant seeds and the ABRC at the Ohio State University for the wrky57-2, jaz4, jaz8, and iaa29 mutants. This work was supported by the National Natural Science Foundation of China (31300252 and 90817003).
AUTHOR CONTRIBUTIONS
Y.J. and G.L. found that WRKY57 functions as a negative regulator in JA-induced leaf senescence. Y.J. discovered that JAZ4/8 and IAA29 competitively interacted with WRKY57 in vivo and vitro and confirmed that WRKY57 was degraded by JA but induced by auxin. S.Y. screened mutants, constructed plasmids, and produced overexpression plants in this study. Y.J., G.L., and D.Y. designed the research and wrote the article. All authors read and approved the final article.
Glossary
- ABA
abscisic acid
- ET
ethylene
- JA
jasmonic acid
- SA
salicylic acid
- MeJA
methyl jasmonate
- ChIP
chromatin immunoprecipitation
- CaMV
cauliflower mosaic virus
- BiFC
bimolecular fluorescence complementation
- IAA
indole-3-acetic acid
- CDS
coding sequence
Footnotes
Online version contains Web-only data.
References
- Addicott, F.T., ed (1982). Abscission. (Berkeley, CA: University of California Press). [Google Scholar]
- Ay N., Clauss K., Barth O., Humbeck K. (2008). Identification and characterization of novel senescence-associated genes from barley (Hordeum vulgare) primary leaves. Plant Biol. (Stuttg.) 10 (suppl. 1): 121–135 [DOI] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E.T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen L.G., Zhang L.P., Li D.B., Wang F., Yu D.Q. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S.S., Xiao L.T., Soo H.M., Cheng Z.W., Xie D.X., Peng J.R. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Dong J.X., Chen C.H., Chen Z.X. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Ellis C.M., Nagpal P., Young J.C., Hagen G., Guilfoyle T.J., Reed J.W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Fan L., Zheng S.Q., Wang X.M. (1997). Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunati A., Tassone P., Damasso M., Migliaccio F. (2010). Neutron irradiation affects the expression of genes involved in the response to auxin, senescence and oxidative stress in Arabidopsis. Plant Signal. Behav. 5: 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S. (2003). Mitotic and postmitotic senescence in plants. Sci. SAGE KE 2003: RE7. [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1997). Making sense of senescence (Molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S. (2004). Leaf senescence—Not just a ‘wear and tear’ phenomenon. Genome Biol. 5: 212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R., Oirdi M.E., Brisson N., Bouarab K. (2012). The conjugated auxin indole-3-acetic acid-asparitic acid promotes plant disease development. 24: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V., Bleecker A.B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8: 595–602 [Google Scholar]
- Guilfoyle T., Hagen G., Ulmasov T., Murfett J. (1998). How does auxin turn on genes? Plant Physiol. 118: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.Q., Crawford N.M. (2005). Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cai Z., Gan S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27: 521–549 [Google Scholar]
- He Y., Fukushige H., Hildebrand D.F., Gan S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer K., Zentgraf U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Feller U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Hong S.B., Sexton R., Tucker M.L. (2000). Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol. 123: 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Lee L.Y.C., Xia K.F., Yan Y.Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hu Y.R., Chen L.G., Wang H.P., Zhang L.P., Wang F., Yu D.Q. (2013a). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Hu Y.R., Jiang L.Q., Wang F., Yu D.Q. (August 9, 2013b). Jasmonate regulates the ICE-CBF/DREB1 cascade and freezing tolerance in Arabidopsis. Plant Cell http://dx.doi.org/10.1105/tpc.113.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.J., Liang G., Yu D.Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5: 1375–1388 [DOI] [PubMed] [Google Scholar]
- Jung C., Lyou S.H., Yeu S., Kim M.A., Rhee S., Kim M., Lee J.S., Choi Y.D., Cheong J.J. (2007). Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep. 26: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Karl M., Soheila A.H.M., Tania P., Fred J., Alex M.M., John P.C., Vicky B. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Kohki Y., Yusuke J., Yuji K., Miyako K., Chiara C., Ralph P., Yoshinori C., Ken S. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids-pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Lim P.O., Lee I.C., Kim J., Kim H.J., Ryu J.S., Woo H.R., Nam H.G. (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 61: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Woo H.R., Nam H.G. (2003). Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 8: 272–278 [DOI] [PubMed] [Google Scholar]
- Lin J.F., Wu S.H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Liscum E., Reed J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49: 387–400 [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K., MacKerness S.A., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A.J.M., Tissenbaum H.A. (2008). Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y.J., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y.S., Amasino R.M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Nooden, L.D., and Leopold, A.C. (1998). Senescence and Aging in Plants. (San Diego, CA: Academic Press). [Google Scholar]
- Parinita A., Reddy M.P., Jitendra C. (2011). WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 38: 3883–3896 [DOI] [PubMed] [Google Scholar]
- Parthier B. (1990). Jasmonates: Hormonal regulators or stress factors in leaf senescence. J. Plant Growth Regul. 9: 445–454 [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T.C., Song S.S., Ren Q.C., Wu D.W., Huang H., Chen Y., Fan M., Peng W., Ren C.M., Xie D.X. (2011). The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Schippers, J., Jing, H., Hille, J., and Dijkwel, P. (2007). Developmental and hormonal control of leaf senescence. In Senescence Processes in Plants, S. Gan, ed (Oxford, UK: Blackwell Publishing), pp. 145–170. [Google Scholar]
- Sexton R., Roberts J.A. (1982). Cell biology of abscission. Annu. Rev. Plant Physiol. 33: 133–162 [Google Scholar]
- Shan X.Y., Wang J.X., Chua L.L., Jiang D., Peng W., Xie D.X. (2011). The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji K., Addicott F.T., Swets W.A. (1951). Auxin in relation to leaf blade abscission. Plant Physiol. 26: 189–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.S., Qi T.C., Fan M., Zhang X., Gao H., Huang H., Wu D.W., Guo H.W., Xie D.X. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9: e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.S., Qi T.C., Huang H., Ren Q.C., Wu D.W., Chang C.Q., Peng W., Liu Y.L., Peng J.R., Xie D.X. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamiya K.I., Tsuchiya T., Ohta H. (2000). Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci. 5: 426–431 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tucker M.L., Whitelaw C.A., Lyssenko N.N., Nath P. (2002). Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiol. 130: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J., Kato J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthum L.). Plant Physiol. 66: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Shahid Mukhtar M., Somssich I.E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Vojislava G., Anthony B.B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8: 595–602 [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu X.P., Chen C.H., Fan B.F., Chen Z.X. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D.X. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhai Q.Z., Yan L.H., Tan D.T., Chen R., Sun J.Q., Gao L.Y., Dong M.Q., Wang Y.C., Li C.Y. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang Y.J., Yu D.Q. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.