Abstract
Phytochrome-interacting factors (PIFs) are members of the Arabidopsis thaliana basic helix-loop-helix family of transcriptional regulators that interact specifically with the active Pfr conformer of phytochrome (phy) photoreceptors. PIFs are central regulators of photomorphogenic development that act to promote stem growth, and this activity is reversed upon interaction with phy in response to light. Recently, significant progress has been made in defining the transcriptional networks directly regulated by PIFs, as well as the convergence of other signaling pathways on the PIFs to modulate growth. Here, we summarize and highlight these findings in the context of PIFs acting as integrators of light and other signals. We discuss progress in our understanding of the transcriptional and posttranslational regulation of PIFs that illustrates the integration of light with hormonal pathways and the circadian clock, and we review seedling hypocotyl growth as a paradigm of PIFs acting at the interface of these signals. Based on these advances, PIFs are emerging as required factors for growth, acting as central components of a regulatory node that integrates multiple internal and external signals to optimize plant development.
INTRODUCTION: INTEGRATION OF ENVIRONMENTAL AND DEVELOPMENTAL SIGNALS THROUGH THE PIFs
It is not surprising that almost every aspect of plant physiology, growth, and development is influenced by light—the energy supply that fuels plant growth and development and also provides valuable seasonal and local environmental information. The ability to monitor and appropriately adjust to prevailing light conditions is mediated by photosensory photoreceptors, which perceive and transduce incoming light signals to the transcriptional network that implements downstream facets of morphogenesis (or photomorphogenesis) (reviewed in Jiao et al., 2007). In turn, light-regulated processes are highly coordinated with other environmental (temperature, presence of neighbors/competitors, biotic stress) and internal (circadian clock and hormones) signals. Plant survival and fitness require integration of all this developmental and external information to ensure that the responses occur at the appropriate time and place.
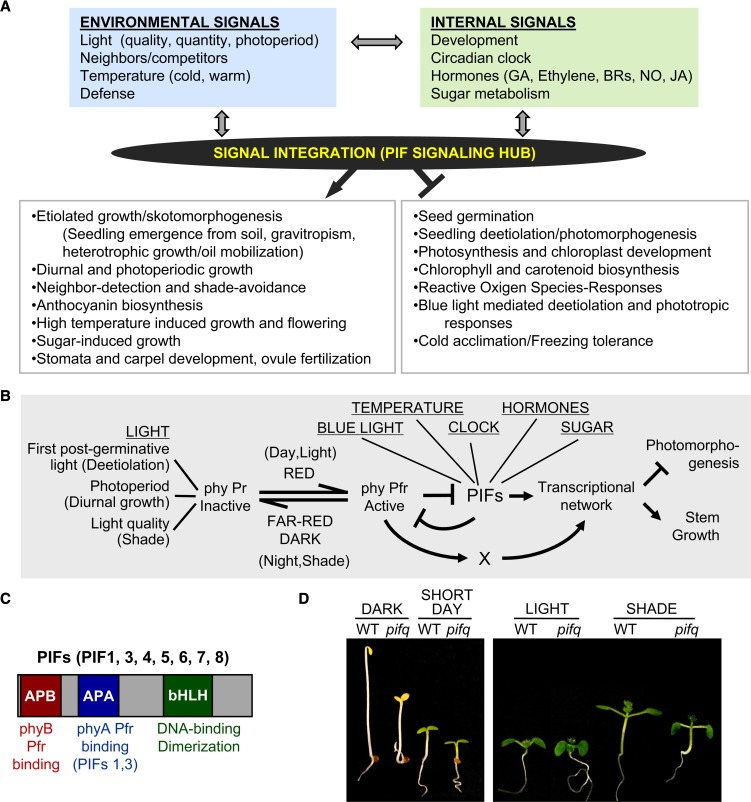
In this review, we focus on a small subset of basic helix-loop-helix (bHLH) transcription factors called phytochrome-interacting factors (PIFs) that are emerging as systems integrators in plant development due to their ability to integrate environmental and internal signals that drive pleiotropic facets of downstream morphogenesis (Figure 1A). PIFs were first described as central players in transducing light signals perceived by the red (R)/far-red (FR) light sensing phytochrome (phy) photoreceptors (phyA to phyE in Arabidopsis thaliana) during the seedling deetiolation developmental transition (reviewed in Castillon et al., 2007; Bae and Choi, 2008; Leivar and Quail, 2011). In the absence of phy activation (such as in seedlings germinated in the subterranean darkness), the phy Pr inactive conformer resides in the cytosol and allows the nuclear accumulation of PIFs, which implement an etiolated program of growth (skotomorphogenesis) characterized by long hypocotyls, a closed hook, and small appressed, nonphotosynthetic cotyledons, facilitating growth to seek for light at the soil surface (Figures 1A and 1B). Upon light exposure, phy Pr is converted into the biologically active Pfr conformer, which translocates into the nucleus and physically interacts with the PIFs, resulting in a reduction in the abundance of the transcription factors in the cell that triggers a rapid and massive transcriptional reprogramming that initiates photomorphogenic development (deetiolation), characterized by short hypocotyls, open hooks, and green expanded cotyledons (Figures 1A and 1B) (Leivar and Quail, 2011). In addition to this central phy/PIF pathway that also regulates several other facets of photomorphogenesis (Figures 1A and 1B), there are now extensive examples of other signaling pathways converging on the PIFs to regulate an increasing number of downstream processes (Figures 1A and 1B), including developmental processes like stomatal index, carpel formation, and ovule fertilization (Pagnussat et al., 2005; Casson et al., 2009; Reymond et al., 2012). Some of these pathways are triggered by internal signals, like the circadian clock, hormones, and developmental- and sugar-derived signals, whereas others are triggered by environmental cues like temperature (cold and warm), blue light, or defense responses (Figure 1A).
Figure 1.
Systems Integration of Environmental and Internal Signals through the PIF Transcription Factors.
(A) Summary of the biological functions of the PIFs as integrators of environmental and internal signals to implement multiple facets of downstream morphogenesis throughout the plant life cycle, acting as positive (→) or negative (−|) regulators.
(B) Simplified schematic illustration of PIF function in transducing light signals downstream of phy photoreceptors and in integrating information from other internal and environmental pathways to provide a coordinated transcriptional response that implements morphogenesis.
(C) Structural and functional domains of the PIF subfamily of bHLH transcription factors.
(D) PIFs are required to promote growth in etiolated seedlings, under diurnal SD conditions, and in response to shade. Visible phenotypes of wild-type and pifq mutant seedlings grown in darkness or in SDs for 3 d (left panel) or for 7 d in light (high R/FR) or simulated shade (low R/FR, 2 d in light + 5 d in shade) (right panel). (Right panel modified from Leivar et al. [2012b], Figure 1B.)
Several excellent reviews have covered the role of PIFs in regulating phy-signaling and transcriptional networks (Leivar and Quail, 2011; Jeong and Choi, 2013) or have focused on their participation in specific processes, such as seed germination, shade avoidance, thermal-induced responses, or crosstalk with specific hormonal pathways or the circadian clock (Franklin, 2008; Lau and Deng, 2010; Wang et al., 2012; Casal, 2013; de Wit et al., 2013; Kinmonth-Schultz et al., 2013; Proveniers and van Zanten, 2013; Shin et al., 2013; Wigge, 2013; Yamashino, 2013). Here, we will discuss evidence that emphasizes the role of PIFs as integrators of diverse signals to modulate plant growth and development to deliver the emerging view that PIFs function as required regulatory factors for growth. We have chosen to focus mainly on the integration of hormonal signals and the circadian clock. However, there is also evidence that PIFs integrate other pathways that are only briefly mentioned in this review, such as temperature (Koini et al., 2009; Stavang et al., 2009; Kumar et al., 2012) and sugar metabolism (Liu et al., 2011; Lilley et al., 2012; Sairanen et al., 2012). First, we summarize current knowledge of the PIF–phy interplay regulating morphogenesis. Next, we discuss progress made in defining the transcriptional networks directly regulated by the PIFs and how different light signals modulate common and specific transcriptional circuits in a highly dynamic fashion. We then provide an overview of functional profiling of PIF targets and discuss progress in our understanding of the transcriptional and posttranslational regulation of PIFs that illustrates the integration of light with hormonal pathways and the circadian clock. Finally, we review seedling hypocotyl growth as a paradigm of PIFs acting at the interface of these signals to regulate growth in accord with the environment.
PIF FUNCTION IN PHY-MEDIATED GROWTH RESPONSES TO LIGHT
phy Regulation of PIF Abundance and Activity, and Negative Feedback Modulation of phyB Levels
Most of the members of the PIF subfamily contain the conserved bHLH domain that provides dimerization (HLH domain) and DNA binding capacity (basic domain) (Toledo-Ortiz et al., 2003; Al-Sady et al., 2008; Shen et al., 2008; Bu et al., 2011a), a conserved motif called the APB (for active phyB binding) required for the conformer-specific binding to the R-light-activated Pfr form of phyB (Khanna et al., 2004) and a functional nuclear localization signal (Ni et al., 1998; Huq and Quail, 2002; Bauer et al., 2004; Huq et al., 2004; Oh et al., 2004; Leivar et al., 2008a) (Figure 1C). To date, 7 of the 15 PIF subfamily members have been shown to interact in vitro with phyB in a R/FR photoreversible fashion, including the founding member PIF3 (AT1G09530), PIF1 (also referred as PIF3-LIKE5 [PIL5], AT2G20180), PIF4 (AT2G43010), PIF5 (PIL6, AT3G59060), PIF6 (PIL2, AT3G62090), PIF7 (AT5G61270), and PIF8 (UNE10, AT4G00050) (Ni et al., 1999; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Oh et al., 2004; Leivar et al., 2008a; Leivar and Quail, 2011). The other subfamily members either do not bind phyB or this interaction has not been tested (Leivar and Quail, 2011); therefore, they are not the focus of this review. Some of these phyB–PIF interactions (e.g., for PIF1, PIF3, and PIF7) have been validated in vivo using dual fluorescent imaging or coimmunoprecipitation assays (Bauer et al., 2004; Leivar et al., 2008a; Shen et al., 2008; Clack et al., 2009; Kidokoro et al., 2009; Park et al., 2012; Ni et al., 2013). In addition to binding phyB, two members (PIF1 and PIF3) have also been shown to bind phyA in a Pfr conformer-specific manner, through a less conserved domain called the APA (for active phyA binding) (Figure 1C) (Huq et al., 2004; Oh et al., 2004; Al-Sady et al., 2006; Shen et al., 2008).
Current evidence shows that the direct physical binding of PIFs with the photoactive, nuclear-localized phy Pfr conformer leads to a reduction in PIF abundance and/or activity in the cell (reviewed in Leivar and Quail, 2011; Jeong and Choi, 2013) (Figure 1B). In dark-grown seedlings (i.e., etiolated), PIF proteins accumulate, whereas upon exposure to light (deetiolation response), photoactivated phy Pfr translocates into the nucleus and induces a rapid decline in the abundance of PIFs (particularly the PIF quartet [PIFq] members PIF1, PIF3, PIF4, and PIF5), with half-lives of 5 to 20 min (Bauer et al., 2004; Monte et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006; Nozue et al., 2007; Lorrain et al., 2008). This pathway involves direct interaction of the PIFs with phyA and/or phyB in nuclear speckles (Bauer et al., 2004; Al-Sady et al., 2006; Shen et al., 2008), which results in rapid phosphorylation, ubiquitylation, and degradation via the ubiquitin-proteasome system (Park et al., 2004; Al-Sady et al., 2006; Shen et al., 2007, 2008; Lorrain et al., 2008). Whereas both phyA and phyB dominate the rapid light-induced degradation of the PIFs (Bauer et al., 2004; Al-Sady et al., 2006; Shen et al., 2007, 2008), phyB appears to have a prominent function under prolonged light conditions (Leivar et al., 2012a; Soy et al., 2012).
Importantly, phy-induced PIF degradation is R/FR reversible (Al-Sady et al., 2006; Shen et al., 2007, 2008) and highly dynamic in the cell, and this interplay between phy activation and PIF levels also operates in other light environments where there are fluctuations in the Pr and Pfr levels, such as under diurnal light-dark cycles or under vegetational shade environments (Figure 1B). Under short-day (SD) photoperiods (8 h light + 16 h dark), photoactivated phy imposes a decrease in PIF1, PIF3, PIF4, and PIF5 proteins during the day, whereas the progressive decline in the Pfr levels during the night due to dark reversion allows reaccumulation of these PIFs, which peak toward the end of the night (Monte et al., 2004; Shen et al., 2005; Nozue et al., 2007; Soy et al., 2012; Yamashino et al., 2013). In the case of PIF3, these oscillations in protein levels are imposed by the action of phyA and phyB at dawn, which induce rapid degradation of PIF3 in response to light (dark-to-light transition) and by the action of phyB to maintain low levels of PIF3 at dusk and during early night (Soy et al., 2012). Phy regulation of PIF protein abundance is also observed in environments enriched in FR light, such as under vegetational shade, where there is a reduction in the R/FR ratio that imposes a shift in the phy photoequilibrium toward the Pr-inactive form, which results in rapid increases in PIF3, PIF4, and PIF5 protein levels (Lorrain et al., 2008; Leivar et al., 2012b).
Although the rapid phy-induced phosphorylation and proteolytic degradation of PIFq members is central to this pathway, a reduction of PIF levels in the cell may not be the only consequence of the phy–PIF interaction. In the case of PIF7, the interaction with phyB-Pfr does not lead to a significant rapid reduction of PIF7 protein during dark–light transitions (Leivar et al., 2008a; Kidokoro et al., 2009). Instead, the phy induces the accumulation of a relatively stable phosphorylated form of PIF7 under constant light (high R/FR), and this form is rapidly dephosphorylated in response to phy inactivation under simulated shade (low R/FR) conditions (Li et al., 2012). This shade-induced dephosphorylation increases binding of PIF7 to its target genes to activate their transcription (Li et al., 2012). It is currently unknown if phy-induced phosphorylation of light-stable forms regulates the DNA binding or transcriptional activity of other PIFs, but the presence of low mobility forms of PIF4 under certain diurnal conditions is consistent with this possibility (Foreman et al., 2011; Yamashino et al., 2013). In the case of PIF1 and PIF3, another report showed that the N-terminal domain of phyB is able to block PIF3 binding to DNA in chromatin immunoprecipitation (ChIP) assays under continuous R conditions (Rc) and that the full-length phyB-Pfr can block the DNA binding capacity of PIF3 and PIF1 in vitro (Park et al., 2012). This observation is in apparent contradiction to initial evidence showing in vitro formation of a ternary complex phyB-PIF3-DNA (Martínez-García et al., 2000); therefore, further analysis is required to assess the biological relevance of these two scenarios.
The interaction between photoactive Pfr with the PIFs not only induces rapid degradation of the PIFs, but also a concomitant, reciprocal, and relatively slower reduction of phyB photoreceptor levels under prolonged light conditions (Figure 1B) (Monte et al., 2004; Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a). PIFs facilitate this feedback process by enhancing phyB binding to COP1, an E3-ligase that promotes proteasome-mediated proteolytic degradation of the nuclear pool of photoactive phyB (Jang et al., 2010). PIF-induced phyB degradation appears to require direct interaction of photoactivated phyB with the APB motif of the PIFs (Khanna et al., 2007; Al-Sady et al., 2008) and, where examined (PIF3), the concomitant phyB-induced phosphorylation of PIFs (Ni et al., 2013). The additional observation that the rate of phyB degradation appears to be linked to the intrinsic rate of light-induced degradation of PIF3 has led to the proposal of a phyB-PIF3 codegradation mechanism (Ni et al., 2013), consistent with the formation of a negative feedback loop between phyB and PIFs that operates under prolonged light conditions. Whereas the initial and rapid phyB-induced degradation of PIFs is necessary to initiate seedling deetiolation, the slow, concomitant and reverse PIF-induced phyB degradation under prolonged light conditions has been proposed to attenuate the light signal and reduce the sensitivity of the seedlings to prevent an overresponse. Interestingly, PIF-induced phyB degradation does not operate under prolonged shade environments (Leivar et al., 2012a), consistent with the absence of interaction between PIFs and the Pr-inactive form of phy or under SD photoperiods (Soy et al., 2012), where the short light period is insufficient to promote significant slow PIF3-induced phyB degradation. Adding another layer of complexity, it has been reported that the interaction with PIFs mediates the nuclear import of phyB into isolated nuclei of the algae Acetabularia acetabulum (Pfeiffer et al., 2012). Consistent with this observation, the authors showed that phyB is not translocated into the nucleus of Arabidopsis pifq mutants during early irradiation of dark-grown seedlings. The authors hypothesized that the PIF-mediated nuclear import of phyB might be relevant only during early deetiolation and therefore would not operate under prolonged light conditions.
Antagonistic Interplay between phy and PIFs in Regulating Seedling Growth Responses during Deetiolation, Diurnal Growth, and Shade Avoidance
Current evidence establishes that PIF1, PIF3, PIF4, PIF5, and PIF7 act with varying degrees of overlapping redundancy to repress phy-imposed seedling photomorphogenesis during deetiolation, under diurnal growth conditions, and in response to shade (Figures 1B and 1D). A similar antagonistic functional interplay between phy and PIFs operates during seed germination, but this appears to be a PIF1-specific function (Oh et al., 2004; Penfield et al., 2005).
In etiolated seedlings, the pivotal observation that a quadruple mutant pif1 pif3 pif4 pif5 (or pifq) lacking PIFq members shows a partial constitutively photomorphogenic (cop) phenotype in strict dark conditions (Figure 1D) (Leivar et al., 2008b) established that these PIFs have the intrinsic capacity of promoting skotomorphogenesis by repressing photomorphogenesis in the absence of phy photoactivation. Whereas dark-grown wild-type seedlings display normal skotomorphogenic development, etiolated pifq mutants resemble a wild-type seedling grown in the light, including short agravitropic hypocotyls and expanded cotyledons that contain developed chloroplast (Leivar et al., 2009; Shin et al., 2009; Kim et al., 2011). These data suggest that removal of these PIFs (by phy-induced degradation in the light in the wild type or genetically in the dark in pifq) underlies the seedling deetiolation response. This view is further supported by the reported suppression of the phyB hyposensitive phenotype by the pifq mutation in R light in phyB pifq mutants (Leivar et al., 2012a), suggesting that higher levels of PIF3 and possibly other PIFq member proteins underlie the elongated phenotype of phyB mutants. Importantly, whereas monogenic pif1, pif3, pif4, and pif5 mutants show minor or absent cop-like phenotypes in darkness, additive to synergistic effects are observed in higher order mutant combinations that culminate in the prominent cop-like phenotype of pifq (Leivar et al., 2008b, 2012b; Shin et al., 2009; Stephenson et al., 2009). These studies show that PIF1 has a prominent role in promoting etiolated growth but that PIF3, PIF4, and PIF5 act together with PIF1 in a partially redundant manner. Altogether, current data establish a model whereby in the dark, PIF1, PIF3, PIF4, and PIF5 accumulate and collectively promote etiolated growth of the seedling, whereas upon light exposure, photoactivated phy reverses this PIF activity by targeting these PIFs to proteolytic degradation, thereby initiating the seedling deetiolation (Figure 1B).
Although PIFs are kept at very low levels or in an inactive form in the light, they conserve the capacity to promote growth under Rc conditions. Indeed, single pif3, pif4, pif5, and pif7 mutant seedlings show a prominent hypersensitive phenotype (i.e., short hypocotyls) under Rc, which contrasts with the absence of a robust phenotype of these mutants in the dark (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004; Monte et al., 2004; Oh et al., 2004; Shen et al., 2005; Khanna et al., 2007; Alabadí et al., 2008; Leivar et al., 2008a). Rc amplification of the short hypocotyl phenotype of these mutants compared with darkness can be explained in part by the redundancy of PIF function in the dark and by the reduced negative feedback modulation of phyB levels in these mutants in the light that results in higher photoreceptor levels (see above). In contrast with Rc, under constant FR light conditions (FRc), PIF4 and PIF5 repress deetiolation downstream of phyA, but the mechanism does not involve phyA-induced degradation of PIF4/5 nor a negative feedback modulation of phyA levels (Lorrain et al., 2009).
Deetiolated seedlings exposed to environments with reduced levels of photoactive phy (Pfr), such as under diurnal and shade conditions, experience an increase in PIF levels and/or activity that results in changes in the growth pattern (Figure 1D). Under FR-enriched shade environments, the inactivation of the phy pool results in the initiation of an adaptative growth response generally referred as shade avoidance syndrome (SAS) (Figure 1D). A comprehensive view of this response as well as the central role of the PIFs in this process has been covered in excellent reviews (Franklin, 2008; Casal, 2013). Briefly, a pivotal work (Lorrain et al., 2008) established that PIF4 and PIF5 collectively promote shade-induced hypocotyl and petiole growth. Recent work has shown that PIF1 and PIF3 also contribute modestly to promote shade-induced morphological and molecular responses (Leivar et al., 2012a, 2012b; Sellaro et al., 2012), whereas PIF7 appears to play a more dominant role in addition to PIFq members (Li et al., 2012). Together, the data support a model whereby in the light (high R/FR), photoactivated phy represses hypocotyl and petiole growth by targeting PIF1, PIF3, PIF4, and PIF5 to proteolytic degradation and by phosphorylating PIF7, whereas shade environments (low R/FR) induce a rapid increase in PIF3, PIF4, PIF5, and presumably PIF1, and a rapid dephosphorylation and activation of PIF7, which collectively promote shade-induced growth (Figure 1B). Consistent with this model, the constitutively shade-avoiding phenotype of the phyB mutant in light (high R/FR) is suppressed to various degrees by mutations in pif3 (Soy et al., 2012), pif7 (Li et al., 2012), pif4 and pif4pif5 (de Lucas et al., 2008; Lorrain et al., 2008), and pifq (Leivar et al., 2012a).
In addition to low R/FR, a reduction in PAR is also characteristic of shade environments under dense canopies. Recent studies under high R/FR conditions show that PIFq members also promote hypocotyl elongation in response to a reduction in PAR (Hornitschek et al., 2012; Leivar et al., 2012a), suggesting that PIFs exert integration of low R/FR and low PAR signals. Low PAR conditions involve a reduction in both R and blue light, so future examination is required to determine whether this role of PIFs in promoting growth in low PAR is due to a reduction in phyB signaling and/or a reduction in signaling via the blue light–sensing cryptochrome (cry) receptor. A reduction in phyB signaling presumably would result in increased PIF levels in high R/FR, as has been shown for PIF3 in phyB mutants (Leivar et al., 2012a) and for PIF1 under low PAR conditions (Chen et al., 2013). In turn, in response to blue light attenuation, PIF4 and PIF5 have been proposed to promote growth downstream of cry through an unknown mechanism (Keller et al., 2011). Intriguingly, although PIFq members have been shown to inhibit seedling photomorphogenic development in blue light (Kang and Ni, 2006; Castillon et al., 2009; Kunihiro et al., 2010; Bu et al., 2011a), this effect appears to be mediated by phy, at least in the case of PIF1 and possibly PIF3 (Castillon et al., 2009; Bu et al., 2011a). By contrast, PIF4 and PIF5 have been shown to repress phototropism downstream of the blue light sensor phototropin (Sun et al., 2013).
Taken together, the interplay between phy activation and PIF levels is highly dynamic and operates under different light environments. An integrated model can be proposed whereby in environments with low or absent levels of active phy, such as in etiolated seedlings, in shade, or at night, PIFs accumulate and have the intrinsic capacity to induce growth (Figure 1B). In response to light, the rapid phy-induced phosphorylation and degradation of the PIFs reduces PIF activity in the cell that leads to growth suppression, whereas the reciprocal and slow PIF-induced reduction of phyB levels tends to attenuate the initial signal. The antagonistic actions of phy and PIFs appear to operate under different light environments sensed by the photoreceptor, suggesting that PIFs exert integration of diverse light signals downstream of the phy.
Deciphering PIF-Regulated Transcriptional Networks during Deetiolation, Diurnal Growth, and Shade Avoidance
Several transcriptomic analyses using single and/or multiple pif mutants, coupled with ChIP-chip and/or ChIP-seq data for individual PIFs, are beginning to provide a genome-wide atlas of genes that are potential direct targets of transcriptional regulation by the PIFs. An initial comprehensive study was performed for PIF1/PIL5 during seed germination (Oh et al., 2009), in which through microarray and ChIP-chip analysis, authors identified a set of 166 genes as PIF1-regulated direct target genes, which mediate downstream facets of the PIF1 repression of seed germination.
Deetiolation
Microarray studies using dark-grown single mutants for PIF3 (Monte et al., 2004; Leivar et al., 2009; Sentandreu et al., 2011) or PIF1 (Moon et al., 2008), and double mutants for PIF4 and PIF5 (Lorrain et al., 2009), showed relatively modest contributions of individual PIFs in regulating gene expression genome wide, consistent with the absence of robust morphological phenotypes of these mutants in the dark due to genetic redundancy (Leivar et al., 2008b, 2012b; Shin et al., 2009). In sharp contrast, the transcriptomic profile of pifq mutants in the dark largely resemble that of wild-type seedlings grown in the light (Leivar et al., 2009; Shin et al., 2009). In a comprehensive microarray study aimed at identifying both rapid and sustained gene expression responses during deetiolation, Leivar et al. (2009) identified a subset of genes whose expression is strongly associated with the rapid light-induced proteolytic removal of PIFq proteins. Together, these data established that PIFq members function as constitutive transcriptional regulators in the dark and that the transcriptional response elicited by light-induced PIF proteolysis is a major component of the seedling deetiolation response.
New sequencing technologies are providing a new dimension to the definition of PIF-regulated transcriptional networks in etiolated seedlings. RNA-seq studies allow the identification of PIFq-regulated genes with full genome coverage, such as the identification of 2025 genes that are misregulated in the pifq mutant in the dark (Zhang et al., 2013), nearly twice the number of genes identified by microarray analysis under equivalent growth conditions (Leivar et al., 2009). ChIP-seq technology has been used to define the genomic occupancy of PIF3 (Zhang et al., 2013) and PIF4 (Oh et al., 2012) in etiolated seedlings and of PIF5 in shade (Hornitschek et al., 2012) (see below). These studies have defined high confidence binding sites in the genome for each of these PIFs and the associated genes, including 828 genes for PIF3, 4363 genes for PIF4, and 1218 genes for PIF5. The defined binding sites for these PIFs are located predominantly in promoter regions of target genes and are strongly enriched in the DNA motif G-box (CACGTG) and the E-box variant (CACATG and CATGTG) that has been called the PBE-box (for PIF binding E-box), suggesting that these PIFs have similar sequence recognition requirements in terms of DNA binding (Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013). In vitro assays have shown specific binding to the G-box for all the PIFs tested (PIF1, PIF3, PIF4, PIF5, and PIF7) (Martínez-García et al., 2000; Huq and Quail, 2002; Leivar et al., 2008a; Moon et al., 2008; Hornitschek et al., 2009) and to the PBE-box for PIF1, PIF3, and PIF4 (Kim et al., 2008; Hornitschek et al., 2012; Zhang et al., 2013).
By merging the list of PIF3 promoter-bound genes with the list of PIFq-regulated genes in etiolated seedlings, Zhang et al. (2013) identified 128 genes as potential direct targets of transcriptional regulation by PIF3. Expression analysis of a subset of these genes in triple pif-mutant combinations compared with pifq show different degrees of regulation by all PIFq members (from marginal to robust), resulting in a mosaic of differential transcriptional responsiveness of individual genes to the different PIFs and of differential regulatory activity of individual PIFs toward the different genes (Zhang et al., 2013). Because a majority of these genes are also bound by PIF4 and/or PIF5 (Zhang et al., 2013), the data suggest that the collective transcriptional activity of PIFq members may be exerted via shared occupancy of binding sites in target promoters.
Another important insight from these studies is that the majority of PIFq-regulated PIF3- and PIF4-bound genes (84 and 58%, respectively) correspond to PIF-induced genes (Oh et al., 2012; Zhang et al., 2013), thus indicating that PIFs appear to function predominantly as transcriptional activators in etiolated seedlings. Consistently, PIF3- and PIF4-bound genes are strongly enriched among the PIFq induced genes in the dark that are rapidly repressed by light like PIL1 (Leivar et al., 2009; Oh et al., 2012; Zhang et al., 2013), which shows a half-repression time of 5 min during the dark to light transition that parallels light-induced PIF proteolysis (Figure 2A; Leivar et al., 2009). By contrast, PIF3-bound genes are poorly represented among the PIFq repressed genes in the dark that are rapidly induced by light (Leivar et al., 2009; Zhang et al., 2013), suggesting that the vast majority of these genes are not direct targets of transcriptional regulation by PIF3.
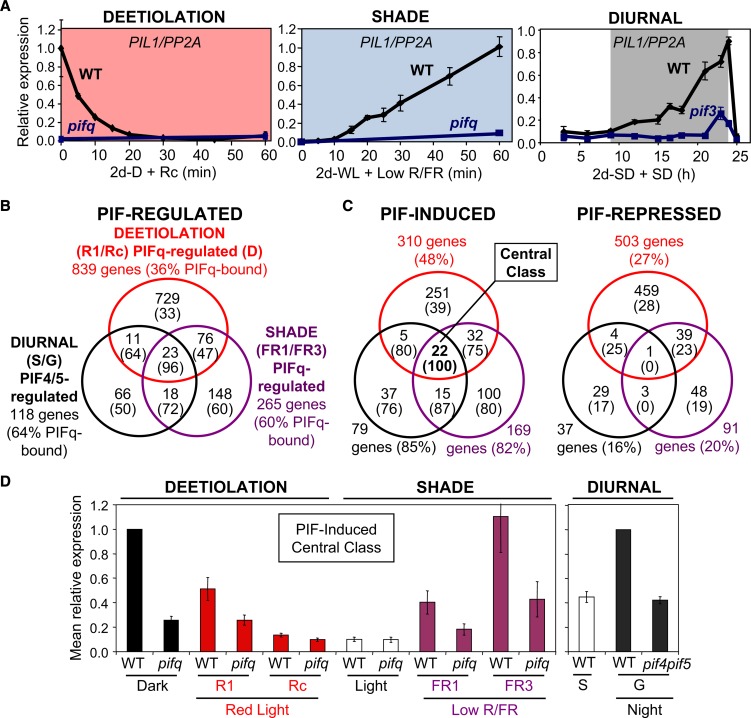
Figure 2.
Integration of Phytochrome-Regulated Transcriptional Networks through the PIF Transcription Factors.
(A) PIL1 expression is reciprocally regulated by light and PIFs during deetiolation, shade, and diurnal conditions. The wild type and pif mutants were grown in the dark for 2 d and then transferred to Rc (left panel), in white light (WL; high R/FR) for 2 d and then transferred to simulated shade (low R/FR) (middle panel), or in SD for 2 d and then kept in SD for an additional day (right panel).
(B) and (C) Comparative transcriptomic analysis of PIF-regulated genes during deetiolation, shade, and diurnal conditions. The percentage of genes that are bound by PIFq members is shown in parentheses. PIFq-bound genes were defined by combining the list of published PIF1-, PIF3-, PIF4-, and PIF5-bound genes (Oh et al., 2009, 2012; Hornitschek et al., 2012; Zhang et al., 2013), which results in a combined list of 5073 PIFq-bound genes that are targeted by one or more PIF, representing ∼15% of the Arabidopsis genome. A list of these genes is presented in Supplemental Data Set 1.
(B) Comparison of the following sets of light/growth-responsive PIF-dependent genes: (1) During deetiolation, the 839 genes that respond rapidly to R light (1 h, R1) or in a sustained manner (2d, Rc) in wild-type seedlings compared with darkness and that show PIFq regulation in the dark (Leivar et al., 2009). (2) In response to shade, the 265 genes that respond rapidly (1 h, FR1) or slowly (3 h, FR3) to low R/FR in wild-type seedlings that show moderately to robustly (>1.5-fold) PIFq dependency (Leivar et al., 2012b). (3) In diurnal conditions, the 118 genes whose expression correlates with the stationary (S) or growth (G) phases that show moderate to robust (>1.5-fold) PIF4/5 dependency (Nozue et al., 2011).
(C) The PIF-regulated genes in (B) were divided in PIF-induced and PIF-repressed genes. Most of the PIF-induced genes are light (R1, Rc) repressed, shade induced, and/or growth induced, whereas the PIF-repressed set shows the opposite light-responsive pattern. Genes showing complex patterns were removed from this analysis.
(D) Mean fold change in expression relative to wild type–dark (left graph) or wild type–G (right graph) of the 22 PIF-induced central class genes defined in (C), showing reciprocal regulation by light and PIFs during deetiolation, shade, and diurnal conditions.
([A] is modified from Leivar et al. [2009], Figure 6; Leivar et al. [2012b], Figure 4; and Soy et al. [2012], Figure 3A.]
Shade Avoidance
Exposure of Arabidopsis seedlings to simulated shade conditions also results in rapid transcriptional reprogramming, exemplified by PIL1, which shows shade induction that is detectable within 15 min (Figure 2A; Leivar et al., 2012b). This rapid response requires the participation to varying degrees of several PIFs, including PIFq members and PIF7, which bind to the G-box elements in the PIL1 promoter to activate its transcription (Hornitschek et al., 2009, 2012; Leivar et al., 2012b; Li et al., 2012).
Leivar et al. (2012b) addressed the collective role of PIFq members in shade-induced gene expression genome wide and established that a significant part of the rapid transcriptional response to shade is elicited by the collective action of the PIFq members, which mainly act as transcriptional activators. However, RNA-seq analysis of wild-type and monogenic pif7 mutant seedlings showed that ∼76% (109 genes) of the genes rapidly responding to shade require PIF7 (Li et al., 2012), suggesting that PIF7 is a prominent player in promoting rapid shade-induced transcriptional reprogramming. These two studies show significant overlap between the genes identified as PIF7 regulated (Li et al., 2012) and PIFq regulated (Leivar et al., 2012b) in response to shade, and several of the shade-responsive PIF7-dependent genes (Li et al., 2012) were also defined as PIF5-bound genes in shade (e.g., PIL1, ATHB2, HFR1, YUC8, and IAA29; Hornitschek et al., 2009, 2012). These data thus suggest that PIF7 and PIF5 (and presumably other PIFq members) share a significant number of target genes in regulating shade-induced gene expression. The identification of PIF7-bound genes genome wide and the analysis of higher order pif mutants, including pif7, should provide better understanding of the contribution of each PIF to the collective transcriptional changes elicited by shade environments.
Using ChIP-seq to investigate the genomic occupancy of PIF5 in shade conditions, Hornitschek et al. (2012) identified 1218 genes associated with PIF5 binding sites. Intriguingly, although protein binding microarrays showed only preferential binding of PIF5 to the G-box motif in vitro, analysis of the in vivo PIF5 binding sites in the genome shows strong enrichment in both G-and PBE-boxes, similar to PIF3 and PIF4. In a parallel microarray analysis of the wild type and pif4 pif5 mutants, the authors identified genes regulated by PIF4 and/or PIF5 in low PAR and/or in response to low R/FR conditions. A subset of these genes, also identified as PIF5-bound genes, were considered as potential direct targets of transcriptional regulation by PIF5 under these different shade environments.
Diurnal Growth
Under diurnal SD conditions, the expression of growth-related genes such as PIL1 (Figure 2A) peaks toward the end of the night, and this expression pattern is implemented at least by PIF1, PIF3, PIF4, and PIF5 (Nomoto et al., 2012; Soy et al., 2012, 2014). Nozue et al. (2011) undertook microarray analysis of pif4 pif5 double mutants under a modified SD schedule after entrainment in SD. The experiment aimed at identifying genes associated with the stationary phase (S genes) of growth at subjective early night and with the growth phase (G genes) at subjective dawn. This study identified 120 genes misregulated in pif4 pif5 mutants in the G or S phases of growth. Interestingly, the majority of these genes (67.5%) were upregulated in the G phase and are PIF-induced genes, whereas the remaining genes (32.5%) were upregulated in the S phase and are PIF-repressed. These data suggest that PIF4/5 act predominantly to induce growth under diurnal conditions by activating the expression of genes associated with growth.
Integration of Diverse Light Signals into a Partially Common PIF-Regulated Transcriptional Network
Comparison of the lists of PIF-regulated genes during deetiolation, shade, and diurnal conditions shows certain overlap, which suggest that these genes are under a reciprocal regulation by light and PIFs under different phy-regulated light environments. A prime example of this reciprocal regulation is provided by PIL1 (Figure 2A). In order to explore this concept genome wide, Leivar et al. (2012b) performed a meta-analysis to identify the rapid (within 1 h) R light and shade-responsive PIFq-dependent transcriptome and identified a subset of 14 genes with a reciprocal pattern of rapid regulation by phy and PIFs in response to light and shade signals. This subset is strongly enriched in PIF3-, PIF4-, and PIF5-bound genes (Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013), with 71% being bound by the three PIFs, indicating that these genes are under dynamic constant regulation by the antagonistic actions of phy and PIFs during deetiolation and shade, similar to PIL1. The data thus suggest that different light signals perceived by phy are transmitted through the PIFs to common sectors of the downstream transcriptional network.
To identify additional genes downstream of the PIFs that might mediate common aspects of growth under different light environments, here, we extended this analysis by performing a new comparative transcriptomic analysis including three experimental conditions (deetiolation, shade, and diurnal) (Figure 2B). By merging the three gene sets, we identified genes that respond to light/growth in a PIF-dependent manner in one or more experimental conditions. For each subset of genes, we calculated the percentage of genes that are bound by PIFq members. Finally, we performed separate analysis for PIF-induced and PIF-repressed genes (Figure 2C). Although this comparative transcriptomic analysis suffers from the shortcoming that it compares different genotypes, experimental designs, and data analysis, the data nevertheless provide several interesting insights. First, a significant number of genes are present in the overlapping sectors, which suggests that these genes are under reciprocal regulation by light and PIFs under more than one of the tested light environments. The number of overlapping genes is probably an underestimate, since the diurnal experiment only includes the pif4 pif5 double mutant (Nozue et al., 2011), so it misses the effect of PIF1 and PIF3 (Soy et al., 2012, 2014). Second, the 23 genes in the central sector of the Venn diagram (Figure 2B) are under continuous reciprocal regulation by light and PIFs under the three experimental conditions (deetiolation, diurnal, and shade). Third, although all these light-responsive PIFq-dependent gene sets show enrichment in PIFq-bound genes, this enrichment is generally more robust in the overlapping sectors compared with the nonoverlapping sectors, especially in the central sector, showing 96% of PIFq-bound genes. This enrichment in PIFq-bound genes is especially striking in the PIF-induced subset compared with the PIF-repressed one (Figure 2C), with an impressive 100% of PIFq-bound genes among the 22 genes belonging to the PIF-induced central class. These 22 genes include different transcriptional regulators (e.g., HFR1, ATHB2, ATHB52, IAA19, IAA29, and BEE1), hormone-related genes (e.g., SAUR23, SAUR25, and ST2A), or cell wall–modifying enzymes (e.g., XTR7) and, similar to PIL1 (Figure 2A), show transcriptional activation by the PIFs and reversal of this activity by light-induced PIF proteolysis (Figure 2D). Together, these data suggest that light signals associated with different light environments are transmitted by the phy/PIF system to common cellular components to implement growth.
Another interesting insight from this comparative analysis is that a large segment of the PIF-downstream transcriptional network appears to be specific to a particular light environment, suggesting that these genes might implement specific aspects of growth during deetiolation, diurnal, or shade conditions. This is particularly evident in the deetiolation response, where a large number of genes appear to be expressed only under this condition (Figure 2B). This is consistent with the seedling deetiolation switch being an irreversible developmental transition that involves an essential change in the plant growth habit from heterotrophic to autotrophic. What determines the specificity of the PIF regulation of a particular gene in a given light environment is unknown but might involve crosstalk with development-, hormone-, and clock-derived signals.
Functional Profiling of PIF-Target Genes
Broad functional categorization of the PIF-induced gene subset during deetiolation, shade, and diurnal growth conditions shows a similar pattern of enrichment in transcription-, cell wall–, and hormone-related genes, consistent with the PIFs functioning in promoting pleiotropic aspects of seedling growth under different light environments (Leivar et al., 2009, 2012b; Shin et al., 2009; Nozue et al., 2011; Hornitschek et al., 2012; Li et al., 2012; Oh et al., 2012; Jeong and Choi, 2013; Zhang et al., 2013). In contrast with this pattern, the PIFq-repressed subset during deetiolation is strongly enriched in photosynthesis/chloroplast-related genes (Leivar et al., 2009; Shin et al., 2009; Oh et al., 2012), consistent with the PIFs functioning in repressing chloroplast development and photosynthetic competency in etiolated seedlings (Leivar et al., 2009; Stephenson et al., 2009). Although these studies are providing a genomic view on how the light signal is diversified through the PIFs to specific sectors of the transcriptional network, our understanding of how the hundreds of PIF-regulated genes implement specific facets of downstream photomorphogenesis is limited.
A growing number of reports have addressed the morphogenic function of genes that are potential or confirmed direct targets of transcriptional regulation by the PIFs. Supplemental Table 1 provides a summary of a selection of genes defined as PIFq bound and PIFq regulated under one or more of the experimental conditions used in the transcriptomic comparison (Figure 2B) and/or that have an established or a potential regulatory role as PIF-downstream effectors of phy-regulated seedling morphogenesis. Although the functional interaction of these genes with the PIFs has only been tested genetically in few cases (i.e., HFR1, IAA29, IAA19, PRE1, YUCCA8, PAR1, GAI, SOM, SAUR19, and GUN5), these analyses of PIF-target genes provide an initial definition of the complex regulatory network downstream of the PIFs. Interestingly, several transcription factors that are induced by the PIFs (PRE1, PRE2, ATHB-2, ATHB4, HAT2, HAT3, BEE1, BIM2, and SOM) promote hypocotyl growth and/or repress photomorphogenesis under different light conditions, whereas others that are PIF repressed (GLK2, GNL/CGA1, and GNC) act to promote different aspects of photomorphogenesis (Supplemental Table 1), suggesting that the initial PIF signal is diversified through downstream transcriptional circuits. Also, as detailed below, hormone-related genes are well represented in this table, especially genes related to auxin biosynthesis, transport, and signaling (YUCCA8, YUCCA9, TAA1, CYP79B2, PIN3, WAG2, IAA29, IAA19, and SAUR19) known to be involved in growth responses to shade, diurnal, or high temperature conditions and in hook formation or phototropic responses. Some of these genes participate in the regulation of organ-specific responses (MIDA10/BBX23 and MIDA9), suggesting branching of PIF signaling (Sentandreu et al., 2011, 2012). Intriguingly, several PIF-induced targets are negative regulators of PIF-regulated processes (Supplemental Table 1), including some well-known repressors of growth (HFR1, GAI, and PAR1), whereas several PIF-repressed genes are positive regulators of PIF-regulated processes (e.g., MIDA9, STO, and JAZ9). These data suggest a complex scenario whereby PIFs induce negative feedback loops that provide a molecular brake for the processes initiated by the PIFs possibly to avoid an overresponse and thus providing reinforcement for robustness of the system.
Together, these studies provide biological relevance to the transcriptional network targeted by the PIFs and provide evidence that PIF-regulated traits are implemented by a vast network of PIF targets that contribute collectively, possibly to allow multiple points of integration with other signals. Complementary to the gene-by-gene approach presented in Supplemental Table 1, future gain- and loss-of-function mutant screens aimed at identifying suppressors of pif mutants should provide additional valuable information on how PIFs implement growth and development.
Modes of Transcriptional Regulation by the PIFs
The view that PIFs act predominantly as transcriptional activators is reinforced by the observation that PIF1, PIF3, PIF4, PIF5, and PIF7 show intrinsic transcriptional activation activity in transfection or heterologous systems (Huq et al., 2004; Al-Sady et al., 2008; de Lucas et al., 2008; Leivar et al., 2008a; Hornitschek et al., 2009; Oh et al., 2012). Interestingly, evidence for the converse scenario of PIFs acting as transcriptional repressors of light-induced genes has also been presented. First, although they appear to represent a minority, a significant number of PIFq-bound genes are upregulated in pif mutants especially during the deetiolation response (Figure 2C; Oh et al., 2009, 2012; Zhang et al., 2013), suggesting that PIFs act as transcriptional repressors of these genes. Second, evidence for PIFs acting as intrinsic transcriptional repressors of specific genes has been provided in a number of cases, such as PIF1 repression of PSY expression in the dark (Toledo-Ortiz et al., 2010), PIF7 repression of CBF2 expression under diurnal conditions (Kidokoro et al., 2009; Lee and Thomashow, 2012), and PIF1 and/or PIF3 repression of genes associated with chlorophyll biosynthesis, photosynthesis, and reactive oxygen species (ROS) responses during deetiolation (Chen et al., 2013; Liu et al., 2013b). Collectively, the data are consistent with a model whereby in environments with low or absent phy activation, PIFs accumulate and either activate the expression of PIF-induced genes or inhibit the expression of PIF-repressed genes, and this activity is rapidly reversed by light through phy-induced degradation of the PIFs (Figures 3A and 3B).
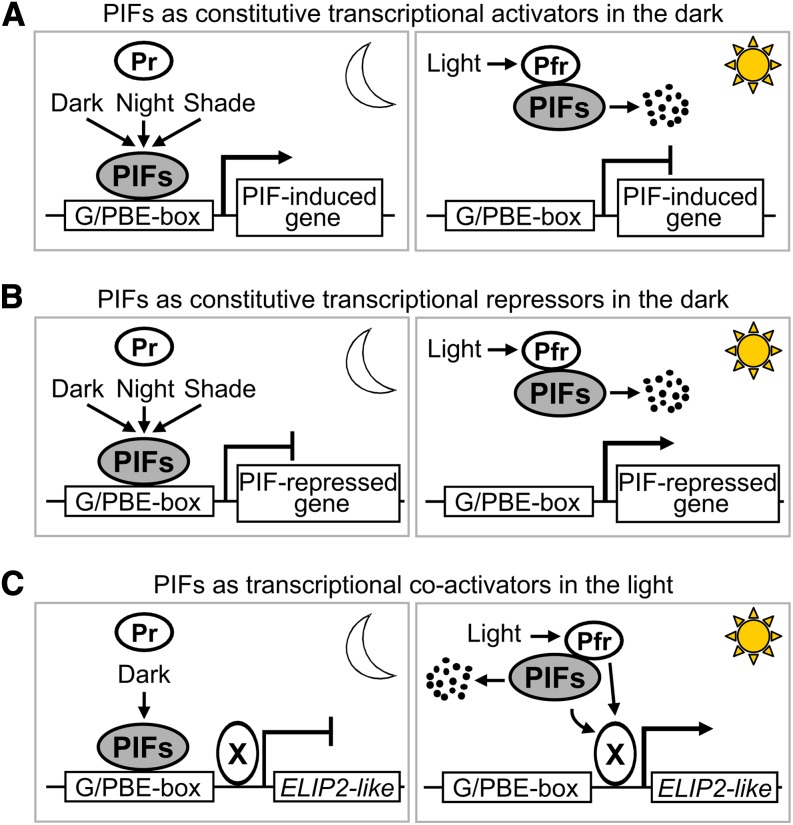
Figure 3.
Modes of Transcriptional Regulation by the PIFs.
(A) PIFs predominantly act as constitutive transcriptional activators of genes like PIL1 in the dark (etiolated seedlings), in response to shade ,or at night under diurnal conditions, and light reverses this activity through phy-induced removal or inactivation of the transcription factors.
(B) PIFs also act as constitutive transcriptional repressors of a relatively smaller subset of light-induced genes, such as PSY, especially during deetiolation, suggesting that PIFs may have a dual activity depending on the promoter and developmental context.
(C) PIFs act as constitutive coactivators of transiently light-induced genes like ELIP2 during deetiolation. This model implies the participation of an additional unknown transcriptional coactivator (represented as X) that is activated by light.
Although the vast majority of PIF-regulated genes show reciprocal regulation by light and PIFs during deetiolation (i.e., PIF-induced genes in the dark are light repressed, and PIF-repressed genes in the dark are light-induced) (Figure 2C; Leivar et al., 2009), closer examination of the gene expression profiles show alternative modes of transcriptional regulation by the PIFs. For example, ELIP1 and ELIP2 are early light–induced genes that are not repressed by the PIFs in the dark, but instead PIFs are required for their rapid light induction (Al-Sady et al., 2008; Leivar et al., 2009). The fact that ELIP2 is a PIF1-, PIF4-, and PIF5-bound gene (Oh et al., 2009, 2012; Hornitschek et al., 2012) provides support for the model postulated by Al-Sady et al. (2008), whereby PIFs act as coactivators of a transcription factor that is activated by light to induce a rapid and transient expression of ELIP2 (Figure 3C). In the dark, PIFs alone cannot activate the expression of ELIP2. During early light exposure, an unknown factor (represented as X in Figure 3C) is activated and induces the expression of ELIP2 together with the PIFs. This effect is transient due to the subsequent light-induced reduction in PIF levels. Interestingly, several early light–induced genes that are not PIF regulated in the dark, such as SIGE (AT5G24120) or ABS2 (AT2G36080), show expression patterns similar to ELIP (Leivar et al., 2009), suggesting that this mechanism might operate for a broader spectrum of genes. Consistent with this possibility, the subset of early light–induced genes that are not PIF regulated in the dark are also enriched in PIFq-bound genes (44%).
Together, a complex scenario is emerging whereby PIFs have the potential to implement a diversity of light-regulated expression profiles. Current evidence suggests that PIFs may do so either by acting alone or in combination with other transcriptional regulators, as discussed below.
CROSSTALK OF PIF SIGNALING WITH OTHER PATHWAYS
It is increasingly evident that regulation of PIF transcriptional activity within transcription modules represents an integration hub of light development with other signals. Recent work has also started to unravel how transcriptional regulation of PIF expression provides an additional layer of integration of PIF function with other pathways.
Regulation of PIF Transcriptional Activity
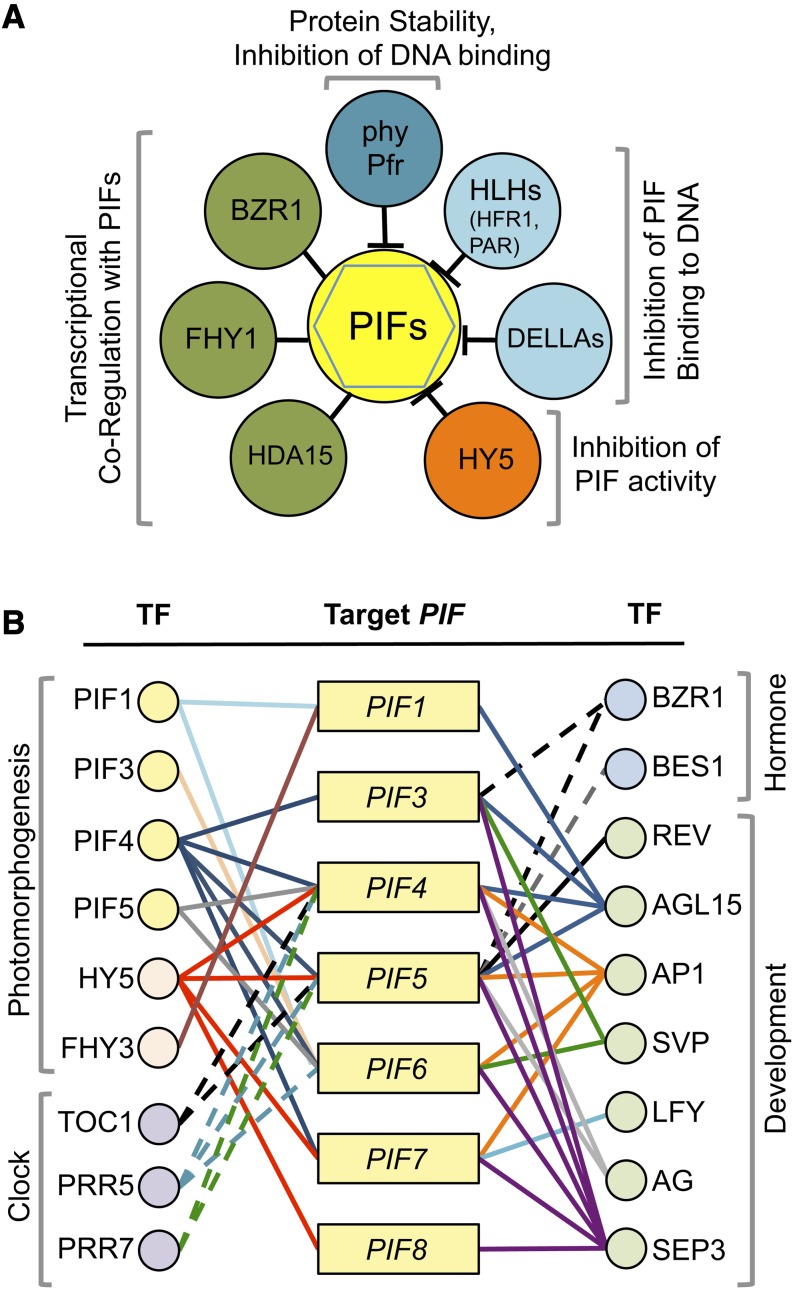
Several PIF binding partners identified by yeast two-hybrid screens or direct protein–protein analyses coupled with functional assays are shedding light on how the transcriptional activity of the PIFs is regulated within transcriptional modules that allow integration of light and other cellular pathways (Figure 4A).
Figure 4.
Transcriptional and Posttranslational Regulation of the PIFs.
(A) Posttranslational regulation of PIF transcriptional activity. PIF binding proteins exert regulation of PIF transcriptional activity by acting as coregulators (green circles), blocking their ability to bind to DNA (blue circles), or inhibiting their intrinsic capacity to activate transcription (orange circle). In addition, interaction with phy Pfr triggers PIF degradation or inhibits PIF activity.
(B) Direct transcriptional regulation of the PIF genes. PIFs potentially are targeted by photomorphogenic-, clock-, hormone-, and development-related transcription factors (TF). Based on ChIP-seq and ChIP-chip data, the regulatory regions of PIF genes (rectangles) are bound by PIFs and other factors (circles). Lines connecting TF with the PIFs depict the binding that has been verified experimentally (see text for details).
Inhibition of PIF Binding to DNA
In addition to the capacity of phyB to regulate PIF binding to DNA (Park et al., 2012), other PIF interacting partners, including DELLAs and HLH proteins, also regulate PIF target binding. PIFs integrate light and gibberellin (GA) signaling through their capacity to interact with the growth repressors called DELLAs, which comprise a family of five members in Arabidopsis (GAI, RGA, RGL1, RGL2, and RGL3). In the dark, GAs accumulate and induce the degradation of DELLAs by the 26S proteasome pathway. In the light, GA synthesis is repressed and GA levels decrease allowing DELLAs to accumulate. DELLAs have been shown to interact with at least PIF3 and PIF4 (de Lucas et al., 2008; Feng et al., 2008) and possibly with other PIFs (Gallego-Bartolomé et al., 2010) through the bHLH DNA binding motif of the PIFs. This interaction blocks PIF binding to DNA and contributes to DELLA repression of hypocotyl growth (de Lucas et al., 2008; Feng et al., 2008). HLH proteins lack the basic DNA binding domain present in bHLH proteins and can interact and form non-DNA binding heterodimers with bHLH factors. After exposure to shade, PIFs accumulate rapidly to promote growth by directly binding to and inducing the expression of growth-promoting genes as part of the SAS. Later in the shade response, the HLH protein HFR1 accumulates and negatively regulates elongation. The mechanism proposed for HFR1 function involves interaction with PIF4 and PIF5 to form non-DNA binding heterodimers that limit the action of the PIFs (Fairchild et al., 2000; Hornitschek et al., 2009). HFR1 inhibition of PIF4 and PIF5 has also been suggested to regulate deetiolation under FR light (Lorrain et al., 2009), and HFR1 also forms non-DNA binding heterodimers with PIF1 to antagonize PIF1 function during germination (Shi et al., 2013). A similar mechanism presumably operates with the HLH factors PAR1 and PAR2. PAR1 is a negative regulator of growth in light and shade (Roig-Villanova et al., 2007; Hao et al., 2012) and is proposed to function by interacting with PIF4 to form non-DNA binding heterodimers (Hao et al., 2012). These PIF–HLH interactions have been proposed to create a negative feedback loop that acts to limit the response of plants to light and shade and prevent an elongation overresponse. Further complexity in this bHLH/HLH network might be achieved by additional layers of competitive inhibition. For example, the small HLH proteins KIDARI and PRE1 have been proposed to attenuate HFR1 and PAR1 activity, respectively, by competitive interaction to form nonfunctional HLH-HLH heterodimers, causing liberation of PIF4 from the transcriptionally inactive HLH-PIF4 complex (Hao et al., 2012; Hong et al., 2013), which might help fine-tune PIF4 activity under fluctuating light conditions.
PIF Transcriptional Coregulators
PIFs have been shown to bind to several factors (BZR1, FHY1, and HDA15) that are proposed to function as transcriptional coregulators. PIFs integrate light and brassinosteroid (BR) signaling through their capacity to interact with the BR-activated transcription factor BZR1. Whereas light does not significantly affect BR or BZR1 levels, BZR1 has been shown in genome-wide analysis to bind to many light-regulated genes (Sun et al., 2010). Identification and comparison with PIF4 direct targets showed that BZR1 and PIF4 bind to overlapping genomic targets that include ∼50% of PIF4 targets. The mechanism proposed for BZR1 function involves BZR1 interaction with PIF4 to coregulate common target genes in an interdependent fashion, a possibility that was supported by sequential ChIP-rechIP analysis showing co-occupancy of promoters in vivo (Oh et al., 2012). This mechanism is consistent with observations that the gain-of-function bzr1-1D mutant shows a constitutive etiolated phenotype in the dark (even in the absence of BR biosynthesis or signaling) that requires PIFs and that plants overexpressing PIF4 are dwarfed in a bri1 genetic background in the light, indicating that at least in the light PIFs need BRs to promote cell elongation. Based on the combination of genome-wide identification of binding sites and RNA-seq analysis, the model postulates that PIF4 and BZR1 promote growth by interdependently coactivating genes involved in auxin and GA responses and cell elongation, while repressing the transcription pathways for chloroplast development.
PIF3 is a positive component in PHYA-mediated induction of anthocyanin biosynthesis in FRc (Shin et al., 2007). FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and its less abundant homolog FHY1-LIKE mediate FR responses by facilitating light-induced phyA nuclear translocation and by interacting with transcription factors (Yang et al., 2009; Rausenberger et al., 2011). Direct interaction of PIF3 with the FHY1-phyA complex has been proposed to underlie the regulation of anthocyanin biosynthesis (Chen et al., 2012). Through this interaction, PIF3 recruits FHY1 and phyA to the CHALCONE SYNTHASE (CHS) promoter under FR light. Based on molecular data, FHY1 has been proposed to coactivate CHS transcription together with PIF3.
Histone acetylation of chromatin was suggested a decade ago as having an important role in light-regulated development processes (Chua et al., 2003). Loss of function of the histone deacetylase HDA15 leads to upregulation of a number of chlorophyll biosynthetic and photosynthetic genes accompanied with an increase of histone acetylation in their promoters. The proposed model of how HDA15 regulates light-responsive gene expression involves direct HDA15 binding to PIF3, which was recently shown to recruit HDA15 to the G-box elements of these genes in the dark (Liu et al., 2013b). PIF3 and HDA15 corepress gene expression by decreasing the acetylation levels and RNA polymerase II binding. Light-induced degradation of PIF3 lifts this corepressive action by promoting dissociation of HDA15 from the targets, allowing activation of light-responsive target genes.
Inhibition of PIF Transcriptional Activity
Finally, a recent example also illustrates how PIF activity can be inhibited by other transcription factors, such as HY5. PIFs and HY5/HYH display antagonistic functions in the regulation of protochlorophyllide (Pchlide) production and ROS-responsive genes during the dark-to-light transition in Arabidopsis (Chen et al., 2013). Thus, the PIFs and HY5/HYH together regulate chlorophyll biosynthesis and prevent overproduction of ROS during deetiolation, effectively integrating light and ROS signaling. The proposed model involves direct binding of these factors to the promoters of ROS-responsive genes through their G-box motifs, with PIF1/PIF3 acting as negative regulators and HY5/HYH as activators. Molecular evidence indicates that these transcription factors coexist and form bHLH/bZIP heterodimers. The model for the integration of light signal transduction and the production of ROS proposes that in the dark, HY5/HYH are unstable, whereas PIF1/PIF3 are abundant and repress ROS responsive gene expression. In the light, HY5/HYH are more abundant and form heterodimers with PIF1/PIF3, which might function as inactive forms, maintaining ROS-related transcripts at basal levels. Under high-light conditions, PIF1/PIF3 are almost completely degraded and HY5/HYH become more prevalent and increase ROS-responsive gene expression to activate their network and optimize deetiolation.
Transcriptional Regulation of the PIFs
The circadian clock has been shown to regulate the transcription of PIF4 and PIF5. In diurnal conditions, PIF4 and PIF5 transcript levels start to rise at midday through the night, with a peak at dawn (Nozue et al., 2007). The evening complex (EC) formed by ELF3, ELF4, and LUX is necessary for this expression pattern (Nusinow et al., 2011; Herrero et al., 2012; Lu et al., 2012). LUX has been found to directly bind to PIF4/5 promoters through the LUX binding site (GATA/TCG) (Helfer et al., 2011) and recruit ELF3 and ELF4. Whereas ELF3 does not appear to directly bind DNA, it is necessary and sufficient to form the EC. The expression of the EC is diurnally regulated and peaks at dusk. PIF4/5 expression in ELF3-deficient mutants is antiphasic, consistent with EC acting as repressor of PIF4/5 during the evening (Nusinow et al., 2011; Lu et al., 2012). CCA1 also participates in the control of PIF4/5 expression under diurnal conditions based on the finding that plants overexpressing CCA1 have constitutively high levels of PIF4 and PIF5 transcript (Nozue et al., 2007), although this might be indirect given that CCA1 has been shown to directly bind and repress ELF3 expression (Lu et al., 2012). TOC1, PRR5, and PRR7 also repress PIF4 and PIF5 expression (Yamashino et al., 2003; Niwa et al., 2009). Moreover, PIF7 oscillates in SD, long-day (LD), and free-running conditions, suggesting that expression of PIF7 is probably under clock regulation as well (Kidokoro et al., 2009; Lee and Thomashow, 2012).
In addition to the circadian clock, several hormonal signals have been reported to regulate PIF transcript levels. Ethylene suppresses hypocotyl length in the dark, while it promotes elongation in the light. Light reversion of ethylene function requires the direct activation of PIF3 expression by ETHYLENE-INSENSITIVE3 (EIN3) binding to EIN3 binding sites [(T)ACTTT and CTCTGCAT] (Solano et al., 1998; Kosugi and Ohashi, 2000) in the promoter region of PIF3 (Zhong et al., 2012). PIF3 induction by EIN3 has been proposed to be relevant only in the light, when PIF3 levels are limiting due to phy-induced degradation. Consistent with this idea, PIF3 is indispensable for ethylene-induced hypocotyl elongation in the light but not in the dark. PIF3 is also a BR-regulated gene that is directly repressed by BZR1 (Sun et al., 2010), although the relevance of this regulation is unknown. Recent reports suggest that nitric oxide (NO) can also regulate PIF levels, although the mechanisms involved are not understood. NO-deficient mutants display long hypocotyls under Rc and have a moderate increase in PIF1, PIF3, and PIF4 transcript levels (×1.5), suggesting that NO might regulate hypocotyl elongation in R light by decreasing the levels of PIFs. Consistent with this possibility, pifq showed reduced sensitivity to the NO donor sodium nitroprusside (Lozano-Juste and León, 2011).
Interestingly, database analyses have shown that PIFs are differentially expressed in response to environmental and endogenous stimuli (reviewed in Castillon et al., 2007; Jeong and Choi, 2013), suggesting that they might be differentially targeted by transcriptional regulators specific to particular signals. Recent publication of genome-wide binding sites for several Arabidopsis transcriptional regulators allows initial mapping of proteins that have been reported to associate with PIFs potentially to regulate their expression. A compilation of available data for photomorphogenesis-, clock-, hormone-, and development-related regulators is presented schematically in Figure 4B. This analysis indicates that each PIF member is targeted by a unique combination of transcriptional regulators, suggesting that differential regulation of PIFs expression might underlie, at least in part, their specificity in response to various stimuli. Interestingly, the PIFs themselves are PIF bound, which may be linked to the light-induced expression of some PIFs, such as PIF7 (Leivar et al., 2008a), PIF4, and PIF5 (Leivar et al., 2009), during deetiolation and suggests the existence of a complex autoregulatory feedback mechanism. PIF4/5 also emerge as direct targets of circadian clock proteins TOC1, PRR5, and PRR7 (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013a), which might contribute to the regulation of the oscillatory expression of PIF4/5 as a repression mechanism complementary to the EC. In addition to the described binding by BZR1 (Sun et al., 2010), BRs might also target PIF5 through direct binding by BES1 (Yu et al., 2011). Intriguingly, several transcription factors involved in the regulation of flowering (AP1, SVP, LFY, AG, and SEP3) have also been identified to bind PIFs (Kaufmann et al., 2009, 2010; Moyroud et al., 2011; Tao et al., 2012; Gregis et al., 2013; ÓMaoiléidigh et al., 2013), suggesting a direct link to the reported flowering phenotype of pif mutants (Brock et al., 2010; Nozue et al., 2011; Kumar et al., 2012). Finally, binding by REV and AGL15 (Zheng et al., 2009; Brandt et al., 2012) suggests that some of the PIFs might be involved in leaf patterning or embryogenesis, a possibility that needs to be explored.
PIFs AS SYSTEMS INTEGRATORS
Interface with Hormonal Pathways
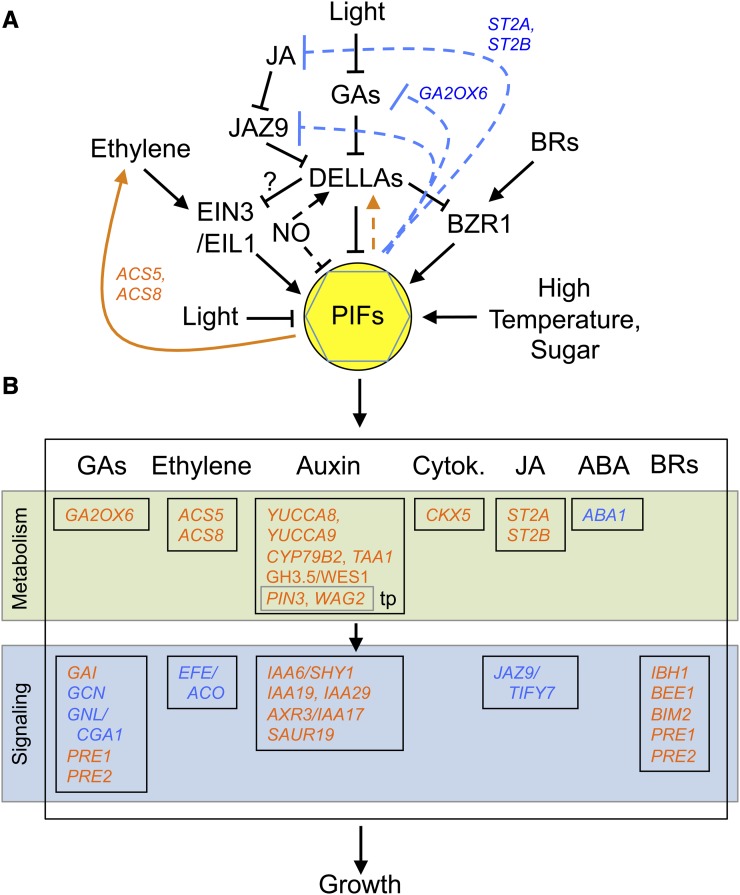
PIFs are emerging as integrators of signals from different hormone pathways during growth and development. Recent studies have shed light on how seedling deetiolation is controlled by the intricate integration of light and hormone transduction pathways, establishing hypocotyl growth as a paradigm of the integration of PIF action with multiple hormones involved in the regulation of cell elongation, including GAs, BRs, jasmonate (JA), ethylene, auxins, and NO (Figure 5).
Figure 5.
Interface of PIF and Hormonal Pathways to Regulate Seedling Photomorphogenesis.
Simplified model depicting hormone regulation of PIF levels and/or activity (A) and direct PIF transcriptional regulation of hormonal pathway components upstream (A) or downstream (B) of the PIF proteins to regulate growth. Solid lines originating from PIFs represent direct transcriptional events that have been proposed elsewhere, whereas dotted lines represent new direct potential connections based on ChIP-seq and transcriptomic data (Supplemental Table 1). The question mark indicates a possible connection based on data for hook development (An et al., 2012). Arrows indicate induction, whereas T-lines indicate repressive action. Colors indicate genes that are directly induced (orange) or repressed (blue) by the PIFs. Integration with other signals is represented by the effect of temperature and sugars. tp, transport.
GAs induce hypocotyl elongation in the dark, and seedlings deficient in GA synthesis or signaling or treated with the GA biosynthesis inhibitor paclobutrazol display a partially deetiolated phenotype when grown in darkness, indicating that GAs play a prominent role in the regulation of deetiolation (Alabadí et al., 2008). Accordingly, exposure of etiolated seedlings to light results in rapid reduction in bioactive GA content (Symons et al., 2008). GAs are perceived by the GID1 family of nuclear receptors (Sun, 2010). The GA-GID1 complex regulates development through targeting the DELLAs for ubiquitylation and degradation. In the absence of GAs, DELLAs accumulate and repress GA-regulated gene expression and growth. The repressive action of the DELLAs in hypocotyl elongation is demonstrated by reduced inhibition of growth in light-grown quadruple DELLA mutant seedlings (Achard et al., 2007), as well as by the suppression of the short hypocotyl phenotype of dark-grown GA-deficient ga1 seedlings by genetic removal of DELLAs (Alabadí et al., 2004; Achard et al., 2007). As explained above, recent findings have suggested that control of light-regulated hypocotyl elongation by PIFs and DELLAs is integrated, as DELLAs are able to interact with PIFs and inhibit their capacity to bind to DNA in the light (de Lucas et al., 2008; Feng et al., 2008). Based on the genetic and biochemical data presented in these two pivotal studies, a model was presented whereby in the dark, GAs accumulate and induce degradation of DELLAs allowing PIF-promoted growth, whereas in the light, GA synthesis is repressed and DELLAs accumulate and interact with the PIFs to inhibit their growth-promoting function by interfering with their binding to DNA. According to this model, the long hypocotyl of the della mutant is explained by higher PIF activity in the absence of the DELLA repressors and therefore should be suppressed by the pif mutations, a genetic validation of the model that is still lacking. This PIF/DELLA mechanism might also be operating in shade, since DELLAs have also been shown to constrain growth under these conditions (Djakovic-Petrovic et al., 2007).
BRs are also skotomorphogenesis-promoting hormones that induce hypocotyl elongation in the dark and negatively regulate photomorphogenesis. Seedlings deficient in BR biosynthesis or signaling or treated with the BR inhibitor brassinazole show a deetiolated phenotype in the dark (Li et al., 1996). At the molecular level, BRs regulate light-dependent development through the control of the phosphorylation status of the brassinazole-resistant transcription factor BZR1. When BR levels are high, a phosphorylation/dephosphorylation cascade is initiated by activation of the BRI1 receptor, leading to the promotion of active BZR1 accumulation in the dephosphorylated state and induction of BR-regulated gene expression and growth. When BR levels drop, BZR1 is inactivated by phosphorylation. However, BR levels are not significantly reduced when etiolated seedlings are exposed to light (Symons et al., 2008), which suggests that deetiolation might involve regulation of BR activity rather than levels. The gain-of-function mutant bzr1-1D, which constitutively accumulates unphosphorylated BZR1, displays an etiolated phenotype in the dark in the absence of BR synthesis or signaling (Tang et al., 2011), further supporting the role of BRs in the control of light-regulated development. An initial genomic mapping study of BR action revealed a regulatory network of BRZ1 targets that extensively overlapped with light signaling (Sun et al., 2010). ChIP-seq analysis indicated that PIF4 shares 50% of its targets with BZR1 at overlapping sites on chromatin, and PIF4 and BRZ1 were found to co-occupy the promoter of these common target genes in vivo (Oh et al., 2012). As explained above, PIF4 and BZR1 directly interact; thus, it was proposed that they might bind DNA as heterodimers. Based on the genomic, genetic, and biochemical data presented in this work, the current model proposes that PIF4 and active BZR1 accumulate in the dark to promote hypocotyl growth by interdependently activating the expression of common target genes involved in cell elongation and responses to auxin and GA. Consistent with this model, the pifq mutant displayed reduced sensitivity to exogenous BR compared with the wild type in the light, and overexpressing PIF4 yielded a dwarf phenotype in a bri1-116 genetic background in which BZR1 is phosphorylated and inactive, suggesting that PIFs need BRs to promote cell elongation. Likewise, bzr1-1D constitutive etiolated phenotype in the dark (even in the absence of BR biosynthesis or signaling) was lost in a PIFq-deficient background, suggesting that BZR1 function to promote elongation requires PIF activity. Interestingly, BZR1 has been found to bind directly to DELLAs (Bai et al., 2012; Gallego-Bartolomé et al., 2012). As previously shown for the PIFs, DELLAs also inhibited BZR1 binding to DNA and interaction with its target genes (Bai et al., 2012; Gallego-Bartolomé et al., 2012). Activated BZR1 was necessary for the elongation of seedlings upon GA treatment in the dark, indicating that BZR1 mediates GA growth promotion in the dark. This raises the possibility that PIF4 integrates both BR and GA signals at the promoter level of light-, GA-, and BR-regulated genes to regulate hypocotyl elongation. Consistent with this possibility, growth promotion of the bzr1-1D mutation was suppressed in the PIFq-deficient background pifq, and the pifq bzr1-1D mutant seedlings were insensitive to GA (Bai et al., 2012), indicating that PIFs are required for BZR1-mediated GA promotion of hypocotyl elongation in the dark.
Plant defense against stress often triggers significant growth inhibition. Activation of JA-mediated defense signaling restricts growth in what is a prominent example of growth-defense tradeoff in plants. Recently, a model was proposed to explain the coordination of growth and defense, which involves the action of PIFs (Yang et al., 2012). In the absence of JA, the transcriptional repressor JAZ9 accumulates to repress JA-responsive genes through interaction with transcription factors like MYC2. JAZ9 also interacts with DELLA proteins, and this interaction interferes with DELLA binding to PIFs, which remain active and promote growth (Yang et al., 2012). JA accumulation leads to proteasome-dependent degradation of JAZ9, which causes the induction of JA-responsive genes and the concomitant release of DELLA proteins that can now interact with PIFs to block their action and inhibit growth (Yang et al., 2012). Consistent with this model, overexpression of PIF3 partially overcomes JA-induced growth inhibition, whereas the pifq mutant does not respond to JA-mediated growth inhibition.
Ethylene participates in light-dependent seedling growth by suppressing elongation in the dark and inducing it in the light. Compared with the wild type, the ethylene-insensitive mutant ein2 is short, whereas the constitutive ethylene response mutant ctr1 affected in the ethylene receptor is longer in light (Smalle et al., 1997; Alonso et al., 1999). Ethylene promotion of hypocotyl growth in the light depends on the ethylene-induced transcription factors EIN3 and EIN3-LIKE1 (EIL1), and ein3 eil1 mutants are insensitive to treatment with 1-aminocyclopropane-1-carboxylic acid, a biosynthetic precursor of ethylene that promotes elongation of light-grown wild type (Zhong et al., 2012). Zhong et al. (2012) showed that ethylene-induced growth in light is PIF3 dependent. In accordance, pif3 was insensitive to 1-aminocyclopropane-1-carboxylic acid treatment in dark and ethylene-induced growth becomes increasingly more dependent on PIF3 under increasing photoperiods or intensities of light. As mentioned above, PIF3 transcription is directly and constitutively activated in this process by EIN3/EIL1. Interestingly, based on the recent finding that DELLAs are able to interact with the DNA binding domains of EIN3/EIL1 during hook formation leading to repression of HOOKLESS1 (HLS1), which is directly activated by EIN3/EIL1 (An et al., 2012), DELLAs might also interfere with the capacity of EIN3/EIL1 to induce PIF3 expression. If so, PIF3 might work to integrate both ethylene and GA signals at the promoter level of light- and GA and ethylene-regulated genes to regulate hypocotyl elongation, a possibility that needs to be tested.
Whereas etiolated seedlings almost completely lack auxin (Bhalerao et al., 2002), auxin synthesis is triggered by light in developing young leaves from where it is distributed throughout the seedling to regulate photomorphogenesis. Mutant seedlings defective in auxin biosynthesis, such as sav3/taa1, tend to be short, whereas mutants accumulating higher levels of auxin, like yucca, display long hypocotyls (reviewed in Halliday et al., 2009). Application of auxin promotes growth under a variety of photoperiods through the activation of a transcriptional response mediated by the TIR1/AFB family of F-box protein auxin receptors and the AUX/IAA and ARF families of transcriptional regulators. Based on recent reports, the relationship of PIFs with auxin signaling appears to be nonlinear and is likely to involve feedback regulation mechanisms. In response to shade and high temperatures, and possibly under diurnal conditions or in response to Suc (Lilley et al., 2012), PIFs induce auxin synthesis and responses (see below), placing PIFs upstream of auxin signaling. However, transcriptome analysis to identify auxin-regulated genes expressed in elongating hypocotyls (Chapman et al., 2012) suggested a model whereby auxin might promote growth partly through PIF-dependent pathways, potentially placing auxin upstream of PIFs, although this possibility needs further testing. Importantly, pif4 pif5 double mutants displayed altered responsiveness to exogenous auxin, suggesting that PIFs modulate auxin signaling (Nozue et al., 2011; Hornitschek et al., 2012).
Finally, NO has also been shown to have a role in light-regulated seedling growth. The NO-deficient double mutant nia1,2 noa1-2 was found to display longer hypocotyls under R light, whereas exogenous application of NO or endogenous NO overproduction in the nox1 mutant resulted in reduced hypocotyl length (Lozano-Juste and León, 2011). NO production increases during seedling deetiolation, and NO was shown to regulate the levels of PIF transcripts, as well as those of DELLA proteins. The proposed model of how NO regulates hypocotyl growth during deetiolation involves NO repression of PIF expression and concomitant activation of DELLA protein accumulation through an unknown mechanism (Lozano-Juste and León, 2011). Consistently, pifq was almost insensitive to NO-triggered inhibition of hypocotyl elongation, indicating that PIFs might integrate NO and GA signaling at the promoter level of light, GA, and NO-responsive genes to fine-tune deetiolation.
With the exception of auxin, for which most of the current evidence suggests functions downstream of the PIFs, hormones involved in the coordination of seedling hypocotyl elongation modulate the ability of PIFs to promote growth by regulating PIF transcription levels and/or PIF DNA binding capacity or transcriptional activity. Some of these mechanisms and transcriptional modules that operate during hypocotyl elongation (Figure 5A) have also been proposed to be active during other light-regulated developmental processes, such as photodamage protection and hook development. During seedling deetiolation, PIFs promote cotyledon greening and prevent photodamage by regulating the production of chlorophyll precursors and carotenoids in the dark. This action involves direct PIF repression of genes like PORC (which encodes the Pchlide reductase C enzyme that catalyzes the conversion of Pchlide to chlorophyll), CHLH, and PSY (Huq et al., 2004; Moon et al., 2008; Shin et al., 2009; Toledo-Ortiz et al., 2010; Cheminant et al., 2011). Evidence also indicates that, in etiolated seedlings, DELLAs accumulate in the cotyledons and work in antagonistic fashion with the PIFs to derepress chlorophyll and carotenoid biosynthesis, at least in part by inhibiting PIF binding to their targets (Cheminant et al., 2011). Consistently, ga1-3 seedlings with increased DELLA content display higher levels of CHLH, PORC, and PSY in the dark similar to the wild type in light (Cheminant et al., 2011). Photodamage protection is also regulated by ethylene. Another report established that ethylene activation of EIN3/EIL1 induces greening and prevents Pchlide accumulation by directly activating PORA and PORB (Zhong et al., 2009). EIN3/EIL1 activity seems to be independent of PIF1 but partly dependent on PIF3 based on the similar Pchlide accumulation found in pif3, ein3 eil1, and pif3 ein3 eil1 mutants and the failure of ethylene application or loss of CTR1 function to rescue the greening defect of pif3 (Zhong et al., 2010). This finding might be linked with the capacity of EIN3/EIL1 to directly activate PIF3 expression, as shown for hypocotyl elongation (Zhong et al., 2012). PIFs also regulate hook development during seedling deetiolation. PIFq deficiency results in absence of a hook (Leivar et al., 2008b), whereas overexpression of PIF5 induced an exaggerated closed hook (Khanna et al., 2007). GAs induce hook curvature, and the level of GA activity determines the speed of hook formation and the extent of curvature, probably by modulating auxin transport and response (Gallego-Bartolomé et al., 2011). To prevent hook opening, GAs are necessary in cooperation with the ethylene pathway to regulate HLS1 expression. HLS1 is directly activated by EIN3/EIL1, and EIN3/EIL1 physically interact with DELLAs leading to repression of HLS1 expression, possibly by blocking EIN3/EIL1 DNA binding (An et al., 2012). GAs increase hook curvature at least in part by destabilizing DELLAs, and the proposed model suggests that accumulation of DELLAs upon light exposure contributes to hook opening by repressing the function of EIN3/EIL1 and possibly the PIFs (An et al., 2012). Accordingly, DELLA mutants have an exaggerated hook (Gallego-Bartolomé et al., 2011; An et al., 2012). After hook formation, promotion of hook maintenance in the dark appears to involve PIF5 transcriptional activation of the AGCVIII kinase gene WAG2 (Willige et al., 2012). WAG2 has been proposed to phosphorylate PIN proteins to regulate auxin efflux and the auxin maximum at the concave side of the apical hook to prevent premature hook opening before light exposure (Willige et al., 2012). PIFs therefore might integrate GA, ethylene, and auxin signals to regulate hook formation and development, possibly sharing similar transcriptional modules as those participating in the control of hypocotyl elongation.
In addition to these conserved transcriptional modules, there is also evidence for alternative regulatory interactions between PIF-regulated light signaling and hormonal pathways. For example, during deetiolation, growth of hypocotyl and cotyledons is in opposite directions, but both are regulated by light signals in a PIF-dependent manner. Whereas cotyledon growth is repressed in the dark, both DELLAs and PIFs act in the same direction to promote cotyledon expansion in the light (Josse et al., 2011), suggesting that they regulate this process via a mechanism that is distinct from that proposed to regulate hypocotyl growth, greening, and hook development (see above). This might indicate organ or temporal specificity of some of these PIF-hormonal transcriptional modules. This possibility is also consistent with the described role of both PIFs and DELLAs as repressors of germination (Oh et al., 2007; reviewed in Lau and Deng, 2010). PIF1 inhibition of germination is DELLA dependent, as defects in germination in PIF1 overexpressing plants are suppressed in an rga background (Oh et al., 2007), indicating that they regulate germination via a mechanism that is distinct from that proposed to regulate hypocotyl elongation. The proposed model involves direct PIF1 induction of expression of the DELLA genes GAI and RGA in the dark and a concomitant indirect PIF1 reduction of GA levels, which results in stabilization of the DELLA proteins and suppression of GA responses in the seed like testa rupture and subsequent germination (Oh et al., 2007). In addition, PIF1 promotes seed dormancy indirectly by inducing abscisic acid accumulation in the dark. Light-induced phy-mediated degradation of PIF1 reverses these processes and the seeds eventually germinate.
Adding an additional layer of complexity to PIF integration of light and hormonal pathways, recent evidence establishes that PIFs are themselves direct regulators of hormone metabolism and/or responses (Figure 5B; Supplemental Table 1). Several PIFs induce de novo synthesis of auxin by directly inducing the expression of auxin biosynthetic genes, including YUCCA8 and YUCCA9 in response to shade (Hornitschek et al., 2012; Li et al., 2012), YUCCA8, TAA1, and CYP79B2 in response to high temperature (Franklin et al., 2011; Sun et al., 2012), and possibly YUCCA8 during growth in diurnal conditions (Nozue et al., 2011). In turn, PIFs are also proposed to regulate auxin signaling by inducing the expression of several AUX/IAA and SAUR genes during deetiolation, shade, diurnal growth, and high-temperature responses (Franklin et al., 2011; Nozue et al., 2011; Hornitschek et al., 2012; Leivar et al., 2012b; Li et al., 2012). Interestingly, IAA19 and IAA29 (Sun et al., 2013), as well as SAUR19 (Franklin et al., 2011), are shown to participate in phototropic and high-temperature responses, respectively, downstream of the PIFs. During hook formation, PIFs regulate auxin transport by directly inducing WAG2 (Willige et al., 2012) and ethylene levels by directly inducing ACS5 and ACS8 expression (Khanna et al., 2007; Gallego-Bartolomé et al., 2011). PIFs also regulate GA signaling by repressing GNL and GNC (Richter et al., 2010) and GA and BR signaling by inducing PRE1 and PRE2 (Hao et al., 2012; Oh et al., 2012). PIFs might also directly repress the JA-regulated transcriptional repressor JAZ9 (Supplemental Table 1), which suggests a regulatory loop between JAs and PIFs (Figure 5A). The relevance of the regulation of these genes by the PIFs is illustrated by SAUR19 and PRE1, which rescue the short hypocotyl phenotype of pif4 mutants in high temperature and of pifq mutants in Rc, respectively, when overexpressed (Franklin et al., 2011; Oh et al., 2012). In addition, genome-wide transcriptional analyses and binding site identification are uncovering potential PIF targets that are hormone-related genes with a described role in various aspects of photomorphogenesis, but as yet uncharacterized with respect to their function as components in PIF signaling (Supplemental Table 1; Figure 5B). These genes might represent additional sites of PIF integration of light and hormonal pathways to regulate hypocotyl growth (Figure 5).
Interface with the Clock
Whereas hypocotyl elongation is maximal in seedlings grown in continuous darkness, the extent of hypocotyl elongation in Arabidopsis seedlings growing under diurnal conditions depends on the duration of the dark period in a nonlinear fashion, and acceleration requires the longer nights of SD (Niwa et al., 2009). A seminal work established that seedlings display rhythmic growth in SD with maximal elongation rates at the end of the night (Nozue et al., 2007). Under these conditions, PIFs are prominent regulators of seedling development, and hypocotyl elongation is largely due to the combined actions of PIF1, PIF3, PIF4, and PIF5, which promote growth specifically at dawn (Nozue et al., 2007; Niwa et al., 2009; Soy et al., 2012, 2014). Growth in SD, where night and day alternate, is controlled through the combined action of not only light and hormone signaling but also the circadian clock, and PIFs are emerging as integrators of these different pathways to regulate and precisely time hypocotyl elongation (Figure 6).
Figure 6.
Interface of PIF, Clock, and Hormones to Regulate Diurnal Growth.
Simplified model depicting PIF-mediated hypocotyl growth under SD conditions integrating hormone and clock signals. PIF4 and PIF5 mRNA expression is repressed by the circadian clock during the beginning of the dark period and rises in the middle of the night to peak at dawn in SD-grown seedlings. By contrast, PIF3 and PIF1 mRNA remain constant throughout the day (top panel). phyA and phyB activities induce degradation of PIFs during the day, and phyB is active during the early night to keep PIF3 and probably PIF1 levels low. As the night proceeds, phyB activity decreases and PIF1 and PIF3 progressively accumulate and peak at the end of the night. For PIF4 and PIF5, coincidence of clock and light regulation ensures that protein accumulation peaks before dawn. DELLA proteins accumulate during the night-to-day transition and remain high during the day to block the DNA binding activity of PIFs during dawn and possibly throughout the day to inhibit residual PIF protein (second panel). The clock maintains light repression of hormonal pathway genes during early night and gates their expression at dawn. PIFs mediate the induction of GA-, auxin-, and BR-related genes at the end of the night and directly induce the expression of growth-related genes at the end of the night (exemplified by PIL1, HFR1, and XTR7) to induce hypocotyl growth before dawn (bottom panels). (Adapted from Soy et al. [2012], Figure 5.)
For PIF4 and PIF5, a coincidence mechanism has been described that combines regulation of PIF4 and PIF5 transcript levels by the circadian clock, at least in part through the transcriptional regulation of the EC, superimposed on the control of their protein accumulation by light (as detailed above) (Nozue et al., 2007; Nusinow et al., 2011; Yamashino et al., 2013). Accordingly, PIF4/5 mRNA expression is repressed during the beginning of the dark period and rises in the middle of the night to peak at dawn, whereas the protein is stabilized in the dark and degraded in the light. It is therefore only at the end of the night, when transcription coincides with protein stabilization, that PIF4/5 accumulate at maximum levels and are able to promote growth (Nozue et al., 2007; Yamashino et al., 2013). For PIF3, and possibly PIF1, transcript levels are relatively constant, and oscillations of protein abundance are imposed by the action of the phytochromes, which allow progressive accumulation throughout the night period to peak before dawn, at which time they accelerate growth together with PIF4 and PIF5 (Shen et al., 2005; Soy et al., 2012, 2014). Transition to the morning light destabilizes the PIFs and growth rate is rapidly reduced again (Nozue et al., 2007; Soy et al., 2012).
The transcriptional activity of the PIFs might also be targeted for regulation under SD. The circadian clock gates GA signaling through transcriptional regulation of the GA receptor GID1 (Arana et al., 2011). Combined with the light-induced reduction in GA levels (Symons et al., 2008), this results in a diurnal maximum in DELLA protein abundance in the hypocotyls during daytime and a minimum during the night (Arana et al., 2011). DELLAs might therefore contribute to the inhibition of PIF-promoted growth during early morning and throughout the day by blocking PIF binding to their targets. In agreement, quintuple della knockout (5xdella) mutants growing in SD exhibit arrhythmic hypocotyl elongation. However, genome-wide analysis of 5xdella mutants found only a limited number of genes that overlap with PIF-regulated genes (Arana et al., 2011), and Nozue et al. (2011) found no significant overlap between GA-responsive genes and PIF4- or PIF5-regulated genes, which suggests that GA contribution to diurnal growth might be largely independent of the PIFs. This apparent contradiction is currently not well understood.
As detailed above, PIFs regulate transcription of numerous hormone biosynthetic and signaling genes, which are in most cases additionally regulated by the circadian clock as output pathways, including GAs, BRs, and auxins (Covington and Harmer, 2007; Covington et al., 2008; Michael et al., 2008). Additionally, genes regulated by auxin, GAs, and BRs tend to peak in expression at dawn, coinciding with maximum growth (Michael et al., 2008). The current model proposes that the clock maintains light repression of hormonal pathway genes during the beginning of the dark period and gates their expression at dawn, allowing them to promote growth in this time window (Michael et al., 2008). Auxin responsiveness is greater at dawn and the auxin response promoter element AuxRE is implicated (Covington and Harmer, 2007), whereas the expression peak of hormone-associated genes correlates with overrepresentation of the Hormone Up at Dawn (HUD) element (Michael et al., 2008). Interestingly, the sequence of the HUD element (CACATG) is the same as the PBE identified as overrepresented in PIF direct targets (Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013). This coincidence suggests that light and clock signaling might converge at the promoter of diurnally regulated hormone-associated genes.
While we are still lacking a genome-wide analysis of PIF target genes under diurnal conditions to delineate how PIFs implement daily growth rhythms in SD, microarray analysis using pif4 pif5 double mutants and comparison with clock-regulated transcriptomes has provided a first glimpse of how PIFs integrate light, hormone, and clock signals (Nozue et al., 2011). This work found that auxin biosynthesis and response genes were enriched among PIF4/5-regulated genes, while seedling responses to auxin were affected in pif4 pif5 mutants and PIF5 overexpressor mutants, suggesting that PIF5 and possibly PIF4 modulate auxin sensitivity as a means to integrate information from the light pathway and the circadian clock. Analysis of a targeted selection of genes has established that PIFs also regulate other hormone- and growth-related genes in SD, including the PIF3 direct targets PIL1, HFR1, and XTR7 (Supplemental Table 1; Soy et al., 2012) and the potential PIF targets in SD BIM1, ACS8, CKX5, or GAI (Supplemental Table 1; Nomoto et al., 2012; Oh et al., 2012).
In addition to regulation of hypocotyl length under diurnal conditions, integration with the clock is important in other PIF-regulated responses, like SAS (reviewed in Casal, 2013), deetiolation, or cold. For SAS, pioneering work interrelating clock and growth by Salter et al. (2003) showed that the circadian clock gates long-term promotion of hypocotyl elongation in response to 2 h of low R/FR treatment under free-running conditions in a response that requires TOC1 and PIL1 and triggers maximum growth at subjective dusk. Similarly, under a modified SD of 10 h day + 14 h dark of natural radiation, shade induces elongation in the evening but not in the morning (Sellaro et al., 2012). The mechanism proposed under diurnal conditions involves LHY-CCA1 and is possibly mediated by the PIFs, which probably accumulate to higher levels during the night after the evening shade, similarly to what has been described for end-of-day FR conditions (Monte et al., 2004; Shen et al., 2005; Leivar et al., 2012a, 2012b; Soy et al., 2014). Seedling deetiolation also requires integration with the clock, as mutants in several clock components show phy-mediated deetiolation phenotypes, but the mechanism is not well understood (Ito et al., 2007). Interestingly, during deetiolation, daily heat shocks in etiolated seedlings gate hypocotyl growth upon exposure to R light, enhancing hypocotyl inhibition (Karayekov et al., 2013). The proposed mechanism in part involves heat induction of LHY and CCA1 rhythms, which in turn cause oscillations of PIF4 and PIF5 to modulate the seedling ability to respond to light (Karayekov et al., 2013). In the case of cold, the circadian clock regulates the expression and gates the cold induction of the C-repeat binding factors CBF1, 2, and 3, a pathway with a major role in plant cold acclimation and freezing tolerance. The mechanism involves direct CBF induction in the morning by CCA1 and LHY and evening repression by PRR5, 7, and 9 (Dong et al., 2011; Nakamichi et al., 2012). PIF7 and PIF4 also regulate the circadian and diurnal expression of CBFs at warm temperatures, thus potentially participating in the photoperiodic control of the CBF cold acclimation pathway (Kidokoro et al., 2009; Lee and Thomashow, 2012). The proposed model involves direct transcriptional repression of CBFs by PIF7 and presumably PIF4 during daytime in LDs, which is relieved in SDs through an unknown mechanism that might involve lower accumulation of PIF4 and PIF7 proteins during the day hours compared with LDs (Lee and Thomashow, 2012). Although this mechanism might underlie the enhanced freezing tolerance of SD-grown compared with LD-grown plants, the model needs to be verified by phenotypic studies of pif4 pif7 mutants. Nevertheless, CBF expression represents a good example of how PIF and clock signals converge at the promoter level to provide a coordinated response.
FUTURE PERSPECTIVES
Owing to the extraordinary progress made in our understanding of the role of the PIFs in plant development since they were first identified in 1998 (Ni et al., 1998), PIFs are emerging as required factors for growth thanks to their capacity to integrate multiple internal and external signals to optimize plant development. Although the framework for the PIF regulatory network has been established, many essential questions still need to be answered.
The early events leading to phosphorylation, ubiquitylation, and degradation of the PIFs, which take place immediately after light exposure and interaction with phy Pfr, are still not completely understood. Recent advances have identified multiple Ser/Thr residues that are phosphorylated in response to light and promote PIF3 degradation (Ni et al., 2013), as well as a CK2 kinase that phosphorylates PIF1 independently of light (Bu et al., 2011b), and the HEMERA protein, similar to the yeast multiubiquitin binding protein RAD23, shown to be required for nuclear speckle formation and PIF1 and PIF3 degradation (Chen et al., 2010; Galvão et al., 2012). However, the central components of the phy-induced PIF degradation pathway remain unknown, including the light-induced protein kinase(s) and the E3-ubiquitin ligase(s) presumably involved.
Another important missing aspect is the cell type, tissue, and temporal-specific distribution of each PIF and the associated signaling networks. High-resolution imaging at the cellular and subcellular levels, together with high-resolution temporal analysis of responses, and genomic analyses of cell type–specific responses are needed to acquire a deeper understanding of PIF-regulated development. Definition of the PIF regulatory networks in response to internal and external stimuli is also necessary to decipher how light and other cues are integrated. Still lacking is the identification of protein complexes where PIFs might be integrated, as well as their changing dynamics in response to variations in environmental conditions. Advances in imaging and proteomics technologies will provide the required platform to explore this dimension.
Finally, although our current knowledge of PIFs is mostly derived from work in Arabidopsis, PIF ortholog factors have been identified in other plants like rice (Oryza sativa) and Lotus japonicas (Nakamura et al., 2007; Carretero-Paulet et al., 2010; Ono et al., 2010). Given the central role of the PIFs in Arabidopsis, it is of interest to explore how PIFs contribute to growth and development in other plant species, particularly crops. Evidence showing the rice PIF-like protein Os-PIL1/PIL13 as a key regulator of reduced internode elongation in rice under drought conditions (Todaka et al., 2012) supports the view that PIFs likely play essential roles in crop species.
In summary, much work still lies ahead to fully comprehend the complexity of PIFs acting as integrators of diverse signaling pathways in Arabidopsis and other plant species, which will help decipher how light and other cues are integrated to optimize plant fitness and potentially impact important facets of crop growth and productivity.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Summary of a Selection of PIF Targets That Have Established or Potential Regulatory Morphogenic Functions Downstream of the PIFs.
Supplemental Data Set 1. List of PIF-Regulated Genes Shown in Figures 2B and 2C.
Supplementary Material
Acknowledgments
We thank lab members and Peter H. Quail for helpful discussions. We apologize for any work that has not been cited due to space constraints. This work was supported by the “Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa” fellowship from the Generalitat de Catalunya (Beatriu de Pinós program) and Marie Curie IRG PIRG06-GA-2009-256420 Grant to P.L., as well as by grants from the Spanish “Ministerio de Economía” BIO2012-31672, from the “Ministerio de Educación, Cultura y Deporte” PRX12/00631, and from the Generalitat de Catalunya 2009-SGR-206 to E.M.
AUTHOR CONTRIBUTIONS
P.L. and E.M. wrote the article.
Footnotes
Online version contains Web-only data.
References
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., Harberd N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Gil J., Blázquez M.A., García-Martínez J.L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Kikis E.A., Monte E., Quail P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.V., Marín-de la Rosa N., Maloof J.N., Blázquez M.A., Alabadí D. (2011). Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Brandt R., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Brock M.T., Maloof J.N., Weinig C. (2010). Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol. Ecol. 19: 1187–1199 [DOI] [PubMed] [Google Scholar]
- Bu Q., Castillon A., Chen F., Zhu L., Huq E. (2011a). Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 77: 501–511 [DOI] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Dennis M.D., Yu L., Lu S.X., Person M.D., Tobin E.M., Browning K.S., Huq E. (2011b). Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J. Biol. Chem. 286: 12066–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Casson S.A., Franklin K.A., Gray J.E., Grierson C.S., Whitelam G.C., Hetherington A.M. (2009). phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 19: 229–234 [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2009). Blue light induces degradation of the negative regulator phytochrome interacting factor 1 to promote photomorphogenic development of Arabidopsis seedlings. Genetics 182: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Greenham K., Castillejo C., Sartor R., Bialy A., Sun T.P., Estelle M. (2012). Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., Lin R. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Shi X., Chen L., Dai M., Zhou Z., Shen Y., Li J., Li G., Wei N., Deng X.W. (2012). Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell 24: 1907–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Galvão R.M., Li M., Burger B., Bugea J., Bolado J., Chory J. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Watson L.A., Gray J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T., Shokry A., Moffet M., Liu P., Faul M., Sharrock R.A. (2009). Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Harmer S.L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- de Wit M., Lorrain S., Fankhauser C. (September 6, 2013). Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol. Plant. http//dx..org/10.111/pp1.12099. [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T., de Wit M., Voesenek L.A., Pierik R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Johansson H., Hornitschek P., Josse E.M., Fankhauser C., Halliday K.J. (2011). Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65: 441–452 [DOI] [PubMed] [Google Scholar]
- Franklin K.A. (2008). Shade avoidance. New Phytol. 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Arana M.V., Vandenbussche F., Zádníková P., Minguet E.G., Guardiola V., Van Der Straeten D., Benkova E., Alabadí D., Blázquez M.A. (2011). Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 67: 622–634 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Galvão R.M., Li M., Kothadia S.M., Haskel J.D., Decker P.V., Van Buskirk E.K., Chen M. (2012). Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., et al. (2013). Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday K.J., Martínez-García J.F., Josse E.M. (2009). Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 1: a001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.Y., Seo P.J., Ryu J.Y., Cho S.H., Woo J.C., Park C.M. (2013). A competitive peptide inhibitor KIDARI negatively regulates HFR1 by forming nonfunctional heterodimers in Arabidopsis photomorphogenesis. Mol. Cells 35: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Nakamichi N., Nakamura Y., Niwa Y., Kato T., Murakami M., Kita M., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. (2007). Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 48: 971–983 [DOI] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Gan Y., Bou-Torrent J., Stewart K.L., Gilday A.D., Jeffree C.E., Vaistij F.E., Martínez-García J.F., Nagy F., Graham I.A., Halliday K.J. (2011). A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Ni M. (2006). Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayekov E., Sellaro R., Legris M., Yanovsky M.J., Casal J.J. (2013). Heat shock-induced fluctuations in clock and light signaling enhance phytochrome B-mediated Arabidopsis deetiolation. Plant Cell 25: 2892–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Keller M.M., Jaillais Y., Pedmale U.V., Moreno J.E., Chory J., Ballaré C.L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C.M., Tsuchisaka A., Theologis A., Schäfer E., Quail P.H. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S., Maruyama K., Nakashima K., Imura Y., Narusaka Y., Shinwari Z.K., Osakabe Y., Fujita Y., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Shin J., Lee S.H., Kweon H.S., Maloof J.N., Choi G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 108: 1729–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinmonth-Schultz H.A., Golembeski G.S., Imaizumi T. (2013). Circadian clock-regulated physiological outputs: dynamic responses in nature. Semin. Cell Dev. Biol. 24: 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2000). Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A., Yamashino T., Mizuno T. (2010). PHYTOCHROME-INTERACTING FACTORS PIF4 and PIF5 are implicated in the regulation of hypocotyl elongation in response to blue light in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 74: 2538–2541 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee C.M., Thomashow M.F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008a). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Cohn M.M., Quail P.H. (2012a). Phytochrome signaling in green Arabidopsis seedlings: Impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008b). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012b). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley J.L., Gee C.W., Sairanen I., Ljung K., Nemhauser J.L. (2012). An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 160: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Carlsson J., Takeuchi T., Newton L., Farré E.M. (2013a). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76: 101–114 [DOI] [PubMed] [Google Scholar]
- Liu X., Chen C.Y., Wang K.C., Luo M., Tai R., Yuan L., Zhao M., Yang S., Tian G., Cui Y., Hsieh H.L., Wu K. (2013b). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang Y., Liu R., Hao H., Wang Z., Bi Y. (2011). Phytochrome interacting factors (PIFs) are essential regulators for sucrose-induced hypocotyl elongation in Arabidopsis. J. Plant Physiol. 168: 1771–1779 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J., León J. (2011). Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis. Plant Physiol. 156: 1410–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H., Wang Z., Tobin E.M. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyroud E., Minguet E.G., Ott F., Yant L., Posé D., Monniaux M., Blanchet S., Bastien O., Thévenon E., Weigel D., Schmid M., Parcy F. (2011). Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23: 1293–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Kato T., Yamashino T., Murakami M., Mizuno T. (2007). Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 71: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Chalkley R.J., Pham T.N., Guan S., Maltby D.A., Burlingame A.L., Wang Z.Y., Quail P.H. (2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nomoto Y., Kubozono S., Yamashino T., Nakamichi N., Mizuno T. (2012). Circadian clock- and PIF4-controlled plant growth: A coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 53: 1950–1964 Erratum. Plant Cell Physiol. 54: 643. [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K., Harmer S.L., Maloof J.N. (2011). Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÓMaoiléidigh D.S., Wuest S.E., Rae L., Raganelli A., Ryan P.T., Kwasniewska K., Das P., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Ishida K., Yamashino T., Nakanishi H., Sato S., Tabata S., Mizuno T. (2010). Genomewide characterization of the light-responsive and clock-controlled output pathways in Lotus japonicus with special emphasis of its uniqueness. Plant Cell Physiol. 51: 1800–1814 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proveniers M.C., van Zanten M. (2013). High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18: 59–64 [DOI] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Reymond M.C., Brunoud G., Chauvet A., Martínez-Garcia J.F., Martin-Magniette M.L., Monéger F., Scutt C.P. (2012). A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 24: 2812–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Müller I.K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I., Novák O., Pěnčík A., Ikeda Y., Jones B., Sandberg G., Ljung K. (2012). Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sellaro R., Pacín M., Casal J.J. (2012). Diurnal dependence of growth responses to shade in Arabidopsis: Role of hormone, clock, and light signaling. Mol. Plant 5: 619–628 [DOI] [PubMed] [Google Scholar]
- Sentandreu M., Leivar P., Martín G., Monte E. (2012). Branching of the PIF3 regulatory network in Arabidopsis: Roles of PIF3-regulated MIDAs in seedling development in the dark and in response to light. Plant Signal. Behav. 7: 510–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentandreu M., Martín G., González-Schain N., Leivar P., Soy J., Tepperman J.M., Quail P.H., Monte E. (2011). Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Anwer M.U., Davis S.J. (2013). Phytochrome-interacting factors (PIFs) as bridges between environmental signals and the circadian clock: Diurnal regulation of growth and development. Mol. Plant 6: 592–595 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Smalle J., Haegman M., Kurepa J., Straeten D.V., Van Montagu, M (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., González-Schain N., Sentandreu M., Prat S., Quail P.H., Monte E. (2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., Monte E. (January 13, 2014). PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J. Exp. Bot. http//dx..org/10.1093/jxb/ert465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J.A., Gallego-Bartolomé J., Gómez M.D., Yoshida S., Asami T., Olsen J.E., García-Martínez J.L., Alabadí D., Blázquez M.A. (2009). Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2010). Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 154: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Smith J.J., Nomura T., Davies N.W., Yokota T., Reid J.B. (2008). The hormonal regulation of de-etiolation. Planta 227: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Todaka D., et al. (2012). Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. USA 109: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wigge P.A. (2013). Ambient temperature signalling in plants. Curr. Opin. Plant Biol. 16: 661–666 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ogiso-Tanaka E., Zourelidou M., Schwechheimer C. (2012). WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139: 4020–4028 [DOI] [PubMed] [Google Scholar]
- Yamashino T. (2013). From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana. Biosci. Biotechnol. Biochem. 77: 10–16 [DOI] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. (2003). A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629 [DOI] [PubMed] [Google Scholar]
- Yamashino T., Nomoto Y., Lorrain S., Miyachi M., Ito S., Nakamichi N., Fankhauser C., Mizuno T. (2013). Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal. Behav. 8: e23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Jang I.C., Henriques R., Chua N.H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xi Y., Guo H. (2010). Ethylene is crucial for cotyledon greening and seedling survival during de-etiolation. Plant Signal. Behav. 5: 739–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.