Abstract
Since its inception, Bacterial Locomotion and Signal Transduction (BLAST) meetings have been the place to exchange and share the latest developments in the field of bacterial signal transduction and motility. At the 12th BLAST meeting, held last January in Tucson AZ, researchers from all over the world met to report and discuss progress in diverse aspects of the field. The majority of these advances, however, came at the level of atomic level structures and their associated mechanisms. This was especially true of the biological machines that sense and respond to environmental changes.
Introduction
All organisms must respond to environmental changes. The world is not static and, unless an organism can adapt to change, it risks death and ultimate extinction. Prominent examples include the ability to adapt to nutrient depletion or toxin influx. At the heart of any adaptation mechanism, irrespective of organism or habitat, is the ability to sense environmental changes and act upon them. In this regard, life has solved many problems that engineers face when designing autonomous systems. Imagine designing a simple robot capable of navigating between two points. Any design would involve sensors to guide the robot, a motor to move it, and a set of rules to determine its velocity and direction of motion. How the engineer specifically designs these components would be determined by the specific terrain to be navigated and the obstacles to be avoided. The engineer also would be concerned with the accuracy and range of the sensors, the efficiency of the motors, the fidelity of communication between and among the sensors and motors, and the robustness of the integrated system.
The questions posed by the biologist are often analogous to those of the engineer. Unlike the engineer, the biologist performs the design problem in reverse, determining the mechanisms used by living organisms to orient within and adapt to their environment. In other words, the biologist and engineer are effectively tackling the same problem from different ends. This is best exemplified in the study of bacterial chemotaxis, the process whereby cells migrate in response to changes in their environment – such as increased or decreased nutrients, toxins, pH or temperature. Similar to the simple robot example, this biological system provides a simple example of a sensor (a set of chemoreceptors) linked to an actuator (the flagellar motor) within an intervening logic layer (the chemotaxis pathway). Over fifty years of investigation have now provided us with a molecular-level view into the workings of chemotaxis, with exquisite detail especially in the case of the enteric bacteria Escherichia coli and Salmonella enterica. Not only has this work enabled us to peer into the inner workings of the sensor and the motor but also has allowed us to address many of the engineering questions enumerated above.
However, chemotaxis is not the only way organisms respond to change nor do all motile bacteria employ the same mechanisms. This is not surprising given that bacteria inhabit nearly every possible environment, ranging from deep-sea vents to the human gut. To live in such diverse environments, bacteria have evolved a wide range of adaptation mechanisms. Just as the engineer would employ a different design for an aerial robot than s/he would for a terrestrial robot, so do bacteria when foraging in liquids versus on surfaces. Likewise, a successful adaptive mechanism must sense the proper signals, which are as diverse as the environments occupied by bacteria and which range from small chemicals to specific wavelengths of light. In fact, what makes this field so exciting is the range and diversity of mechanisms that bacteria have evolved to deal with changes in their environment. Yet, with all this diversity, it is striking how much of the core machinery is conserved. Indeed, a major theme that has developed over the years is the modularity of these adaptive systems. This is seen not only at the level of the proteins, but also with the supramolecular assemblies that comprise the sensors and motors, and the pathways the connect them. Just as the engineer prefers off-the-shelf components to custom-designed ones, so does evolution.
The 12th Bacterial Locomotion and Signal Transduction Meeting gathered 149 scientists from 13 countries in Tucson, Arizona, from January 20th to 25th, 2013, to exchange the latest developments in the fields of chemotaxis, motility and sensory perception. BLAST XII was chaired by Urs Jenal (Biozentrum, Basel, Switzerland) and Karen Ottemann (University of California, Santa Cruz) and organized by Bob Bourret (University of North Carolina, Chapel Hill), Sandy Parkinson (University of Utah), Joe Falke (University of Colorado), and Michael Manson (Texas A&M University). A hallmark of these meetings is the priority given to talks by graduate students and postdoctoral fellows with 26 giving talks and 41 presenting posters.
Although the major focus of all BLAST conferences has been bacterial chemotaxis and motility, the specific theme of each conference has wandered, often in response to perceptive insights or technological advances. BLAST XII is no exception. Recent advances in diverse technologies have let researchers delve more deeply into the structure and function of the chemoreceptor signaling complex and the flagellar motor; as such, BLAST XII can be viewed as a story of these two magnificent machines. Yet, the field is much richer than chemotaxis and motility, and this was evident by the richness and diversity of this year’s presentations. Thus, despite a strong focus on the inner workings of those machines, much also was made of the intervening logic and the networks that regulate their expression.
Sensors
Chemotactic bacteria utilize a highly sensitive and adaptive sensory network that informs them of their environment so that they migrate properly, i.e., towards attractants and away from repellents. Attractants and repellents are perceived by chemoreceptors that are usually, but not always, integrated into the cytoplasmic membrane (Fig. 1). Chemoreceptors, also known as methyl accepting chemotaxis proteins (MCPs), form trimers-of-dimers in hexagonally ordered arrays that include the homodimeric histidine kinase (HK) CheA and the coupling protein CheW. Upon sensing a decrease in attractant or an increase in repellent, the now excited (active) receptors induce CheA to autophosphorylate, using ATP as the phosphoryl donor. Phospho-CheA serves as the phosphodonor to the cognate response regulator (RR) CheY, a soluble protein that diffuses through the cytoplasm to the switch complex on the cytoplasmic face of the flagellar basal body. Binding of phospho-CheY increases the probability of clockwise flagellar rotation, inducing reorientation maneuvers that vary from bacterium to bacterium. For example, the peritrichously flagellated enterics E. coli and S. enterica tumble, whist the polar-flagellated Sinorhizobium and Helicobacter can reverse direction of their translational movement (Hazelbauer et al., 2008).
Figure 1. Sensing of chemical stimuli and signal processing.
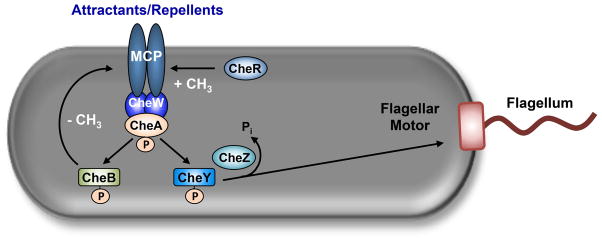
A chemical stimulus (either an attractant or a repellent) is detected by a transmembrane chemoreceptor, also known as a methyl-accepting chemotactic protein (MCP). Via the coupling protein CheW, the signal is transduced to the HK CheA. In response to an increase in the concentration of a repellent or a decrease in the concentration of an attractant, CheA autophosphorylates a conserved histidine residue. Phosphorylated CheA stimulates a rapid excitatory and a slow adaptive signal by donating its phosphoryl group to the RRs CheY and CheB, respectively. Excitation occurs when phosphorylated CheY diffuses through the cytoplasm and binds to the switch at the cytoplasmic face of the flagellar motor, increasing the probability of clockwise (CW) rotation. Excitation is modulated by CheZ, a specific phosphatase that dephosphorylates phosphorylated CheY. Adaptation occurs because phosphorylated CheB functions as a methylesterase that specifically removes methyl groups constitutively placed on the MCP by the methyltransferase CheR.
This excitatory pathway is balanced by two different mechanisms: dephosphorylation of phospho-CheY and adaptation (Fig. 1). Phosphorylated aspartyl residues, like that of phospho-CheY, are inherently unstable, having a short half-life of a few seconds. In the enterics, an additional protein CheZ accelerates dephosphorylation, so that half-life of phospho-CheY is about 0.1 seconds. In other organisms, non-CheZ phosphatases perform this function. Adaptation occurs simultaneously with excitation and also depends on CheA autophosphorylation; however, the pathways bifurcate at the level of the phospho-acceptor: CheY is the acceptor for excitation, whilst another RR (CheB) is the acceptor for adaptation. Phospho-CheB is a methylesterase, removing methyl groups from the methylation helix bundle of the chemoreceptor. Methylation of specific residues occurs constitutively via the CheR methyltransferase. Whereas excitation is rapid, adaptation is slow, permitting a rapid response followed by a slow return to the inactive or adapted state (Porter et al., 2011).
Electron microscopic maps generated by sub-volume averaging of wild-type chemoreceptor arrays in the adapted (inactive) state, together with crystal structures, have revealed the protein arrangement of the core chemotaxis proteins (Vu et al., 2012). Trimers of receptor dimers lie at each vertex of a hexagon, arranged such that one dimer points towards the center of the ring (Fig. 2A). The receptors (purple) surround rings of alternating regulatory subdomains (P5) of CheA (blue) and the coupling protein CheW (green). The kinase subdomains (P4) of CheA (dark grey) project toward the cytoplasm and away from the receptors, whereas the dimerization subdomains (P3) of CheA (light grey) link neighboring rings to form a stable, extended lattice. Although the principal structure of chemotaxis protein arrays is becoming clear, their dynamic nature and operation are not yet understood.
Figure 2. Chemoreceptor arrays.
A. Proposed chemoreceptor array model.
This model is based on the crystal structure 3UR1 and the EM map of wild-type E. coli arrays (Briegel et al., 2012). Receptor trimer-of-dimers, purple; CheW, green; CheA-P5, blue; CheA-P4, dark grey; and CheA-P3, light grey.
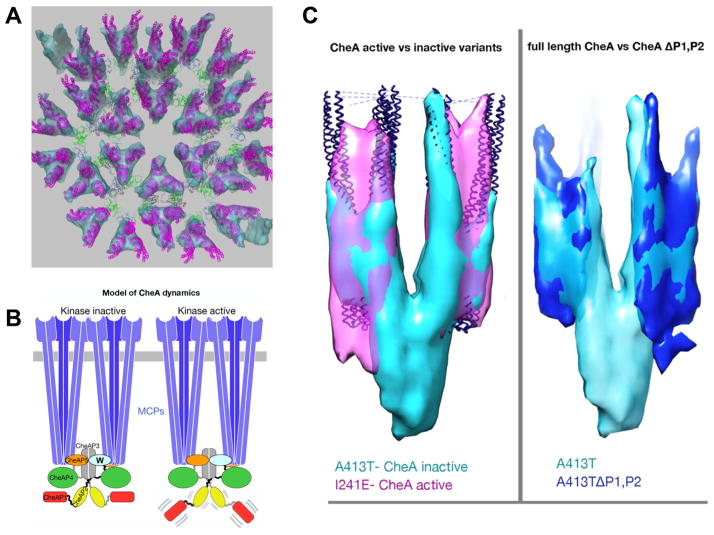
B. Proposed model for signaling through mobility control of the kinase domains P1 AND P2. In a kinase-off signaling state, the CheA domain P1 is trapped in a static, nonproductive interaction with P4, constraining the P1 and P2 domains to the keel volume. In the kinase-on signaling state, increased mobility of the P1 and P2 domains promotes cycles of P1 engagement, phosphorylation and release.
C. The mobility of CheA P1 and P2 domains differs between kinase activity states. Left:Overlay of chemoreceptor Tsr variants locked in the CheA inactivating state (A413T, turquoise) and CheA activating state (I241E, magenta). For reference and alignment, receptor crystal structures (blue) were fitted into the sub-tomogram averages by molecular-dynamics-based-flexible-fitting (MDFF). A prominent ‘keel density’ is visible in the kinase inactive state underneath the chemoreceptors, but absent in the kinase active state indicating increased mobility of the keel forming domains. Right: Overlay of Tsr A413T variants with full length CheA (turquoise) and CheA lacking the P1 and P2 domains (blue) reveals CheA domains P1 and P2 as the major components of the keel.
Ariane Briegel (Grant Jensen’s Lab, California Institute of Technology) reported the use of electron cryotomography to visualize structural differences between chemoreceptor variants with well-defined kinase output states (Ames et al., 2008; Briegel et al., 2013; Zhou et al., 2009). The new results support a model in which the signaling state does not alter the hexagonal arrangement of the chemoreceptors. Although chemoreceptors retain identical center-to-center spacing in all activation states, CheA exhibits significant structural differences (Fig. 2B). In receptor variants that lock CheA in the inactive state, the domains P1 and P2 of the CheA dimer pack stably in a ‘keel-like’ density (turquoise) beneath two adjacent receptor trimers (Fig. 2C, left). This density was confirmed to be P1 and P2, as a mutant that lacks these two domains was missing this density (Fig. 2C, right). When locked in the active state, this density is less prominent, suggesting signaling-dependent dynamics of these two CheA domains, which perform the phosphoryl transfer from CheA to CheY. This activity-dependent dynamic behavior of P1 and P2 domains also was observed biochemically as a difference in protease susceptibility.
Although the receptors and CheA clearly interact, the functional structures of the protein-protein interfaces that define the interaction remain unclear. Multiple structures have been solved; however, a molecular view of protein-protein contacts in the active, membrane-bound signaling array has proven elusive. Two recent crystal structures solved in the Crane Lab reveal two distinct possibilities for the interface between the receptor and the CheA kinase regulatory domain (Briegel et al., 2012; Li et al., 2013). To determine which structure best predicts the protein-protein interface, Kene Piasta (Joseph Falke’s Lab, University of Colorado) described the mapping of the receptor-kinase regulatory domain interface in functional, membrane-bound arrays using disulfide mapping in vitro and tryptophan and alanine mutagenesis to identify docking sites (TAM-IDS) in vivo. The results identify the receptor-kinase regulatory domain structure described in Li et. al. (Li et al., 2013) as the best representation of the interface in active, membrane-bound signaling arrays. Moreover, the findings yield a new model for signal transduction from receptor to kinase during on-off switching. In this model, the kinase regulatory domain (KRD) forms a tight complex with a specific receptor helix, and this KRD-helix complex undergoes a concerted rotational or translational motion relative to the rest of the receptor 4-helix bundle during attractant-triggered signaling that switches the kinase off (Piasta et al., 2013). Together, these results reveal the correct protein-protein interface between CheA and receptor in active, membrane-bound signaling arrays, as well as new insights into receptor-mediated kinase on-off switching.
To identify differences in the conformations and dynamics of the CheA-activating and CheA-inactivating signaling states of the receptor, Lynmarie Thompson (University of Massachusetts) described the use of hydrogen exchange mass spectrometry (Percy et al., 2012) to analyze ‘kinase-on’ and ‘kinase-off’ states of the receptor cytoplasmic domain assembled at high and low density on membrane vesicles in functional complexes with CheA and CheW. Relative to the kinase-off state, the kinase-on state exhibited slower hydrogen exchange in the methylation region of the receptor, perhaps due to closer receptor interactions. In contrast, very fast exchange occurred in the C-terminus, consistent with current views that it is a highly dynamic connector to the CheR binding site. Finally, the protein interaction domain exhibited protection from exchange that was greater in the kinase-on state, perhaps due to a larger interaction interface with CheA and CheW. Hydrogen exchange mass spectrometry is a promising tool for detecting structural and dynamic changes in functional membrane-bound, multi-protein complexes.
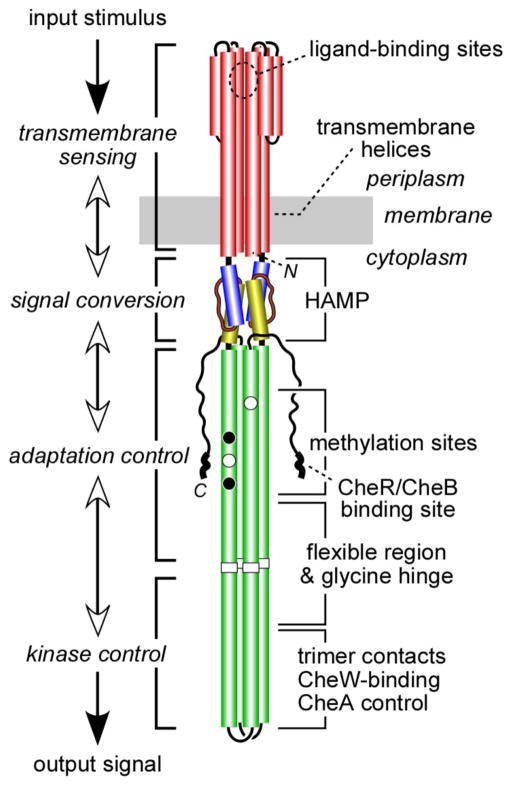
The dynamic bundle model (Zhou et al., 2009) proposes that chemoreceptor signaling involves opposed shifts in packing stability of the four-helix HAMP bundle and the adjacent methylation helix bundle, which contains the covalent modification sites for sensory adaptation (Fig. 3). To test aspects of this model, Sandy Parkinson and co-workers (University of Utah) used in vivo FRET to analyze the signaling behaviors of mutant variants of the Tsr receptor with structural alterations in HAMP and/or MH bundles. Each methylated position made equivalent free energy contributions to kinase-on and kinase-off transitions and HAMP lesions increased the response threshold. These data imply that structural interactions between different amino acid positions could restore response sensitivity by matching HAMP packing stability to the opposing structural effects of receptor methylation.
Figure 3.
Schematic of the domain structure of a chemoreceptor dimer with corresponding functions.
Ady Vaknin (Hebrew University) described the use of fluorescence-polarization measurements of tagged receptor-clusters to study receptor dynamics in living cells. When exposed to attractants, the packing of receptors within clusters slowly changed, perhaps by introducing disorder into the cluster. Consistent with reduced packing of the receptors, cluster-regulated kinase activity slowly evolved, altering the effective cooperativity of the response. Such an effect can be viewed as an effective ‘plasticity’ of the receptor cluster, which changes connectivity upon stimulation (Frank and Vaknin, 2013).
The dynamics of the bacterial chemotactic response have been characterized primarily in the model strain E. coli K12 (Shimizu et al., 2010; Sourjik and Berg, 2002). Yet, E. coli chemotaxis represents a streamlined example (Wuichet and Zhulin, 2010). To begin to characterize other more complex systems, Milena Lazova (Tom Shimizu’s lab, FOM Institute for Atomic and Molecular Physics, Amsterdam) studied chemotaxis properties in S. enterica, whose chemoreceptor species and structures differ from those of E. coli (Biemann and Koshland, 1994; Frye et al., 2006; Lazova et al., 2012; Wang et al., 2006). By using quantitative physiology approaches, such as real-time FRET (Sourjik et al., 2007), she showed that S. enterica response to the non-metabolizable attractant methylaspartate exhibits three-fold less cooperative receptor response, ten-fold broader dynamic range, and three-fold faster adaptation. Striking differences in the drift velocity were observed using microfluidics (Ahmed et al., 2010a, b). Differences in physiology and chemotactic performance could be explained by the existence of more receptor species and their different binding properties.
Although the previous study strengthened the hypothesis that sensing dynamics can depend on the number and characteristics of different receptor species, the following reports describe novel sensor functions in receptor-rich organisms. Whereas E. coli strains express five or fewer chemoreceptors, other bacteria express many more receptor species; for example, the Vibrio fischeri genome encodes 43 distinct receptors (Brennan et al., 2013). V. fischeri forms a monospecific symbiosis with the Hawaiian bobtail squid, colonizing a dedicated light organ (reviewed in (McFall-Ngai et al., 2012)). A major unanswered question is how the bacterium recognizes its niche. A related, methodological, question is how can researchers identify the stimuli of one receptor among many? Caitlin Brennan (Ned Ruby’s lab, University of Wisconsin-Madison) described efforts to answer both questions. The ligands for most V. fischeri receptors remain unknown. To identify those ligands, Brennan and co-workers screened receptor mutants in soft agar motility assays and found that one mutant (vfcA) did not respond to serine (Brennan et al., 2013). Further examination using capillary assays showed that the vfcA mutant exhibited altered chemotaxis towards multiple amino acids. To test if VfcA responds directly to amino acids, they collaborated with the Parkinson lab, using in vivo FRET kinase assays (Sourjik and Berg, 2002) to monitor the behavior of a chimeric chemoreceptor composed of the extracytoplasmic sensory domain of VfcA and the cytoplasmic signaling domain of E. coli Tar. Using CheZ (donor) and CheY (acceptor) as the FRET interaction partners, they showed that VfcA-Tar chimera responds both positively and negatively to distinct groups of amino acids. Using a similar strategy, they showed that a second V. fischeri chemoreceptor, VfcB, responds to hexoses, including some found in the host. In vivo, several receptor mutants showed reduced fitness, whilst others had increased fitness, suggesting that chemoreception contributes to light organ colonization efficiency.
Sinorhizobium melioti expresses an intermediate number of chemoreceptors (9) that permit it to recognize and respond to plant seed exudate components, allowing the bacterium to adapt to a soil and plant root environment (Currier and Strobel, 1976; Meier et al., 2007). To ask how chemotaxis affects the bacteria-plant symbiosis and to identify the participating receptors, Benjamin Webb and co-workers (Birgit Scharf’s lab, Virginia Tech) used mass spectrometry of legume seed exudate, soft agar assays of receptor mutants, and isothermal titration calorimetry to learn that the chemoreceptor McpU senses the amino acid proline. McpU and the V. cholerae receptor McpN possess significant homology in their N-terminal sensory domains, and McpN had been co-crystallized with its ligand alanine (PDB ID 3C8C). Using this information, they modeled the McpU structure and used that model to make and test predictions concerning proline binding.
Although all known chemoreceptors possess a common signaling domain, the input domains vary. For example, Aer-2 is a soluble PAS-HAMP type receptor found in several pathogens with somewhat different architectures (Watts et al., 2011). Kylie Watts (Loma Linda University) reported the analysis of the Aer-2 receptors of Pseudomonas aeruginosa (PaAer-2) and V. cholerae (VcAer-2). PaAer-2 contains a single PAS domain, binds heme, and responds to various oxy-gases (O2, CO, NO). In contrast, VcAer-2 contains two PAS domains that each bind heme and responds only to oxygen. To identify the VcPAS domain that senses O2 and to determine the roles of two conserved amino acid residues, substitutions were engineered into both truncated and full-length Aer-2 receptors and spectral and behavioral studies were performed. The data suggest that VcPAS-2 functions in signaling, whereas VcPAS-1 inhibits that function. The data also support the hypothesis that VcPAS-1 does not signal O2 binding and suggests that it stabilizes O2-binding by a mechanism that differs from the O2-sensing PAS domain.
One talk described a novel mechanism for sensing the neurotransmitter norepinephrine (NE) as a chemoattractant. Host stress causes NE to be secreted in the gastrointestinal (GI) tract. This NE could be an in vivo signal to guide motile bacteria through the ~200 mM thick mucus layer to the gastrointestinal (GI) epithelium. Matt Sears (Mike Manson’s lab, Texas A&M University) reported that the ability of the enteric bacterium E. coli to perform NE chemotaxis depends on its ability to metabolize NE to 3,4-dihydroxymandelic acid (DHMA). Using microfluidic assays and GFP-labeled E. coli, Sears and co-workers tested the response to NE by wild-type cells and an isogenic tsr mutant. They found that priming with NE enhanced the NE response, suggesting that the system is induced. They found that the primed cells did not respond to NE if they lacked the QseC sensor HK or the enzymes TynA and FeaB, which, based on their known activities as a monoamine oxidase and aromatic aldehyde dehydrogenase, respectively, may be able to metabolize NE to DHMA. Quantitative RT-PCR showed that NE induces transcription of tynA and feaB in a QseC-dependent manner. Finally, they identified Tsr as the chemoreceptor responsible for sensing DHMA. Peak chemotactic responses were observed to 10–1000 nM DHMA, concentrations that could be physiologically relevant in the GI tract. A model for how Tsr might sense DHMA also was discussed. In the proposed mechanism, one DHMA molecule would inhibit Tsr activation of the CheA kinase by binding to one of the serine-binding sites of the Tsr homodimer. Binding of a second DHMA molecule to the other serine-binding site, albeit with lower affinity because of negative cooperativity, would cancel out the attractant signal. Such a mechanism could explain why the response to DHMA peaks at low concentrations rather than saturates at high concentrations.
Although the enterics express only membrane-bound chemoreceptors that localize in arrays to the cell pole, other organisms also express receptors that form cytoplasmic clusters. The phototrophic bacterium Rhodobacter sphaeroides possesses a complex chemosensory network containing multiple homologues of each component of the paradigm chemosensory pathway of the enterics, such that it possesses two complete pathways: a classic system with membrane-spanning chemoreceptors located at the poles, and another system with cytoplasmic chemoreceptors located around the midcell (Wadhams et al., 2003). For vertical maintenance of the latter system, a mechanism must exist to ensure that each daughter cell receives a portion of the chemoreceptor cluster. Christopher Jones (Judith Armitage’s Lab, University of Oxford) showed that segregation of this cytoplasmic cluster depends on the ParA ATPase homologue PpfA. Par systems segregate bacterial chromosomes and plasmids during cell division. They typically consist of ParA proteins that polymerize into filaments and ParB proteins that associate with centromere-like DNA sequences and activate ParA depolymerization. PpfA is encoded within the operon that encodes the cytoplasmic chemosensory pathway. It interacts with the N-terminal part of the cytoplasmic chemoreceptor TlpT, which acts as the ParB component. Together, these proteins use the chromosome to segregate the chemosensory clusters upon division. PpfA localizes over the chromosome, but forms foci around the cytoplasmic chemosensory cluster and acts like a scaffold. In a PpfA deletion strain, it was shown that the chemosensory cluster was highly mobile within the cell compared to the wild type strain.
Actuators
Flagella are complex, cell envelope-anchored, proteinaceous nanomachines that both secrete proteins and propel the bacterium through its milieu. Each flagellum is built from over 30 proteins and has three architectural domains: basal body, hook and filament (Fig. 4A). Not surprisingly, multiple mechanisms control and direct flagellar biogenesis. Hierarchically ordered transcriptional and post-transcriptional networks work together, ensuring that the flagellar structural proteins are only synthesized when they are required and when the timing is right. Furthermore, the process by which the protein building blocks assemble onto each other is a highly ordered and interaction-dependent process (Fig. 4B). In the best-characterized enteric model, FliF subunits are first inserted as the MS ring into the cytoplasmic membrane. Proteins that comprise the rest of the flagellar type III secretion system (T3SS) then are assembled onto the periplasmic and cytoplasmic faces of the MS ring. This completed structure (called the basal body) provides a narrow channel through which the more external components of the machine are secreted such that they can assemble onto the distal end of the elongating structure. These external components include the hook (a flexible linker assembled from about 100 identical subunits) and the filament (a semi-rigid propeller built from thousands of flagellin subunits). Once assembled, the flagellum rotates, a process powered by the proton (or sodium) motive force via motor proteins inserted into the cytoplasmic membrane. The direction of rotation is dictated by a switching device that assembles onto the cytoplasmic face of the basal body in close proximity to the motor. As described in the previous section, this switch receives information from the chemotactic signaling network in the form of phospho-CheY, whose binding to the switch increases the probability that the flagellum will rotate clockwise. In the enterics, counterclockwise (CCW) rotation corresponds to forward translational motion, a behavior called a run. Runs occur when multiple flagella rotate CCW and, in the process, coalesce into a flagellar bundle that drives the cell forward. Reorienting tumbles occur when some of those flagella rotate CW, which causes a change in filament waveform from the ‘normal’ left-handed helix to a right-handed one that disrupts the bundle (Erhardt et al., 2010; Minamino et al., 2008; Terashima et al., 2008; Wolfe and Visick, 2008).
Figure 4. Flagellar structure and assembly.
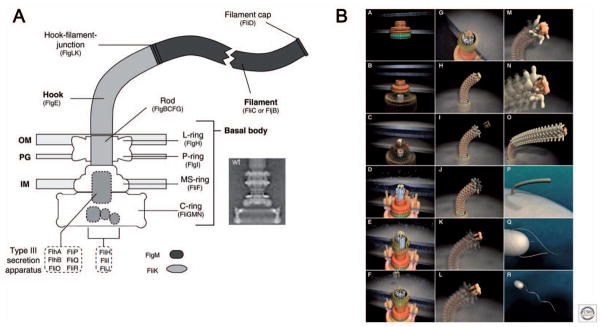
A. Schematic overview of the Salmonella flagellum. The flagellum consists of three parts: 1) a basal body with a flagellar-specific type III secretion system (T3SS) within the inner membrane ring, 2) a flexible hook that acts as a universal joint to 3) the rigid filament. Dashed boxes illustrate proteins with functions in flagellar type III secretion. OM = outer membrane; PG = peptidoglycan; IM = inner membrane. The inlay EM (electron micrograph) picture shows an isolated hook-basal-body complex of Salmonella (Thomas et al., 2001).
B. Steps in assembly of the bacterial flagellum. The self-assembly process of the bacterial flagellum starts from the top left (A) and proceeds to the bottom right (R). Assembly starts with the formation of the MS-ring in the cytoplasmic membrane (A). Afterward, the cytoplasmic C-ring is attached to the MS-ring and the flagellar-specific T3SS assembles within a central pore of the MS-ring (B). Flagellar secretion substrates are now secreted specifically via the T3SS. When flagellar proteins reach the distal end of the elongating structure, they self-assemble onto the existing structure with the help of distal cap proteins (shown as pentamers at the tips of the axial structures) (C–O). The rod, which acts as a driveshaft, is assembled beneath the rod-cap, which also acts as a muramidase to allow penetration through the peptidoglycan layer (C). Motor force generators that couple proton flow to torque generation assemble in the cytoplasmic membrane and interact with C-ring components (D). Rod assembly ends with the formation of the PL-ring bushing, outer membrane penetration, and the replacement of the rod scaffold with the hook scaffold (E–G). Hook subunits are then secreted until the hook grows to a final length of 55 nm (Hirano et al., 1994). This triggers a secretion specificity switch from rod-hook-type substrates to late-secretion substrates (H). Upon completion of the hook-basal-body complex, hook-filament junction proteins (I–J), the filament cap (K–L), and filament subunits are secreted, and the filament grows to a maximal length of about 10–15 μM (M–R). Panel A reprinted with permission from CSH Perspectives in Biology (Erhardt et al., 2010). Panel B reprinted with permission from Royal Society of Chemistry (Minamino et al., 2008).
A similar situation seems to occur when swimming cells are put under high pressure. Masayoshi Nishiyama (Kyoto University) and Yoshiyuki Sowa (Hosei University) described the performance of E. coli flagellar motors at high hydrostatic pressure. First, they described the relationship between pressure and motility, which correlates inversely such that translational motion stopped at about 80 MPa. The process is reversible, as reducing the pressure restored motility. To understand the underlying mechanism, Nishiyama and Sowa monitored the rotation of cells tethered by a single flagellum and found that motors continue to rotate at 80 MPa, but only at 60% the rate observed at ambient pressure (0.1 MPa). This result implies that reduced rotational speed suffices to prevent translational motion (Nishiyama and Kojima, 2012; Nishiyama and Sowa, 2012; Nishiyama et al., 2013). Since other physicochemical environmental changes are reported to affect flagellar filament waveform (Calladine et al., 2013), the filament waveform was monitored as a function of pressure. At 0.1 MPa, all filaments formed the normal left-handed structure. As pressure increased, the percentage of filaments in the normal waveform decreased, whilst the percentage of left-handed coiled and right-handed curly waveforms increased. Above 50 MPa, the coiled waveform decreased. Above 80 MPa, the curly waveforms increased exponentially. These results support the hypothesis that high pressure favors waveforms that inhibit bundle formation and thus translational movement (Nishiyama and Sowa, 2012).
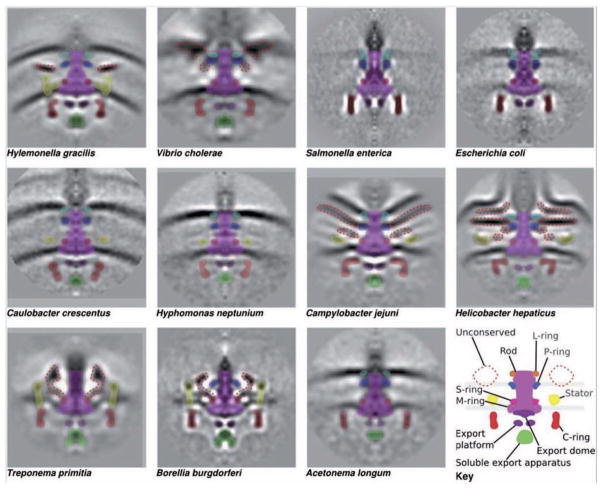
Although the enteric flagellum has been characterized comprehensively, the structure, assembly and function of similar machines in diverse model species await exploration and comparison to the enteric paradigm. To fill this knowledge gap, Morgan Beeby (Grant Jensen’s lab, California Institute of Technology) described the use of cryo-electron tomography to compare the structures of the flagellar motor and T3SS across a wide range of evolutionarily distinct bacteria in situ (Abrusci et al., 2013; Chen et al., 2011). A key finding was that the size of the C-ring varies in bacteria of different phylogenetic lineages (Fig. 5). Based on these results, Beeby and co-workers proposed that bacteria that swim in more viscous environments have evolved larger C-rings. The implication is that larger C-rings help generate the greater torque required to translate through more viscous environments. They further proposed the existence of densities of unknown structure that likely represent novel protein subunits or domains with novel functions in the flagellar basal body.
Figure 5. Flagellar basal body core and motor structures from different flagellated bacterial species obtained by electron cryotomography (ECT) and subtomogram averaging.
For each species, hundreds of cryotomograms of whole cells were collected. Subtomograms containing motors were computationally extracted from the data, mutually aligned, averaged and cylindrically symmetrized to obtain resolutions of a few nanometres (Chen et al., EMBO J 2011). Axial slices through average reconstructions of each flagellar basal body from ECT and the corresponding assignments of densities in motors from the different species are shown. Manual segmentation of conserved (solid colours) and unconserved (dotted lines) motor components based on visual inspection. The conserved components from bottom to top are soluble export apparatus (FliH, FliI and FliJ); export platform and dome (FlhA, FlhB, FliO, FliP, FliQ and FliR); C-ring (FliG, FliM and FliN); MS-ring (FliF); stators (MotA and MotB in the H+-dependent stators or PomA and PomB in the Na+-dependent stators); rod (FliE, FlgB, FlgC, FlgF and FlgG); P-ring (FlgI); and L-ring (FlgH). Scale bar, 10 nm. Reprinted with permission from (Chen et al., 2011).
Whereas Morgan Beeby’s approach was more focused on the completed machinery, Xiaowei Zhao (Jun Liu’s lab, University of Texas Houston Medical School) described the collaborative efforts of several labs, including those of Steven Norris (UT Houston), Nyles Charon (West Virginia), Md Motaleb (East Carolina Unversity) and Chunhao Li (SUNY Buffalo), to reconstruct the flagellar assembly process of the spirochete Borrelia burgdorferi (Charon et al., 2012), which causes Lyme disease. Specifically, they used high-throughput cryo-electron tomography to generate three-dimensional structures of flagella from an isogenic set of key assembly mutants. The result is one of the first direct visualizations at the molecular level of flagellar assembly intermediates. One novel and surprising finding was that the export channel, which traverses the axial center of the MS ring, appears to be closed until the start of rod assembly.
To function, the assembled flagellum must rotate. This requires a rotor and a stator, whose interaction permits ion flux through the stator, which generates torque. The rotor includes the basal body, the hook, and the filament. The stator is a ring of proteins inserted into the cytoplasmic membrane; it is thought to bind peptidoglycan (PG), enabling stable anchoring of the flagellum as it rotates around its central axis (Fig. 3B). Two basic types of stator are known: a proton-driven motor encoded by MotA and MotB and a sodium-driven motor encoded by the homologous proteins PomA and PomB (Minamino et al., 2008; Terashima et al., 2008). Whilst MotB is predicted to bind PG (Chun and Parkinson, 1988; De Mot and Vanderleyden, 1994; Muramoto and Macnab, 1998), the crystal structure of its folded form predicts that it should only span 50Å or about ⅓ of the projected distance between MotA and the PG (Kojima et al., 2009; O’Neill et al., 2011; Roujeinikova, 2008). To explain how MotB could span the gap, Anna Roujeinikova and her group (Monash University) engineered a soluble chimeric MotB protein, in which a leucine zipper replaced the two transmembrane helices of the MotB dimer (Andrews et al., 2013). Biochemical and structural analyses of this chimera provided evidence that it is properly folded and that an extended MotB could span the gap. It also supports a model, in which MotB remains folded, and thus unable to bind PG, until it assembles into the stator/motor, an event that would trigger the unfolding of the flexible linker that connects the MotA-binding domain to the PG-binding domain.
The stator subassembly of the sodium-driven Vibro alginolyticus motor is formed by PomA and PomB. On the basis of homology to the proton-driven motor of S. enterica, it had been proposed that assembly of a subunit into the stator causes PomB to undergo a large conformational change that activates the subunit for sodium flux (Kojima et al., 2009). Shiwei Zhu, from the labs of Michio Homma and Seiji Kojima (Nagoya University) and in collaboration with Katsumi Imada (Osaka University), described an elegant test of that hypothesis. With the crystal structure of the PomB periplasmic region (PomBc) as a guide, Zhu and co-workers sought residues in the a1 helix and the core domain that could be replaced by cysteines. Two such pairs of cysteine substitution mutations were identified, constructed and tested. Disulfide cross-linkage within each pair reduced stator activity without jeopardizing stability. Reduction of the cross-linkage restored activity. These results suggest that conformational restraint in the PomBc inhibits stator function.
A change in direction of flagellar rotation leads to reorientation of the cell body in three-dimensional space, such as by tumbling. This change in rotational direction depends on the docking of phospho-CheY to FliM, a major component of the C-ring that resides on the cytoplasmic face of the basal body (Barak and Eisenbach, 1996). Although the binding of phospho-CheY to FliM is thought to favor CW rotation, this event had never been observed directly. Hajime Fukuoka (Akihiko Ishijima’s lab, Tokoku University) described the use of CheY-GFP and TIRF microscopy (Terasawa et al., 2011) to observe phospho-CheY binding or dissociation at flagellar motors. Simultaneous recording of the rotational direction of each bound or unbound motor established a clear correlation with a lag of less than 20 msecs between the two events. The average number of bound phospho-CheY molecules differed for motors in the two different rotational states: three during CCW rotation, but 13 during CW rotation. Hence, CW rotation occurs when phospho-CheY binds a fraction of the proposed 34 FliM subunits per motor. Thus, as proposed previously, reversals of rotational direction must depend upon strong cooperativity between FliM subunits.
Although 34 FliM subunits are proposed per motor, it appears that the number of FliM subunits in rotating motors is not static but rather correlates with rotational direction. This finding derives from an attempt to understand an old observation - that cheRB mutants, which lack the methylation-dependent adaptation machinery, still adapt albeit slowly and partially (Segall et al., 1986). The Berg lab recently reported that this partial adaptation results from structural changes in the flagellar motor. Specifically, they found that the number of FliM subunits correlates with phospho-CheY concentration (Yuan et al., 2012). This remarkable result shows that the flagellum itself can adapt by changing basal body subunit number. Although not directly involved in gradient sensing per se, this mechanism explains how the motor can match its set point to basal phospho-CheY concentrations. Pushkar Lele (Howard Berg’s lab, Harvard University) reported that the number of FliM molecules in the motor and the fraction of FliM molecules that exchange depend on the direction of flagellar rotation, and not directly on phospho-CheY binding. They specifically found that motors rotating in the CCW direction contain on average 44 FliM subunits, whereas motors rotating CW have an average of 34 subunits, consistent with a model in which structural differences associated with the direction of rotation modulate the strength of FliM binding (Lele et al., 2012).
A long-standing question is whether individual flagellar motors work independently or not. Years ago, Berg and coworkers demonstrated that switching of different motors does not correlate when the distance between them is greater than 3 μm, the average length of an E. coli cell (Ishihara et al., 1983). More recently, Ishijima and coworkers showed that switching is strongly correlated between more closely adjacent motors, such as those in the same cell (Park et al., 2011; Terasawa et al., 2011). This strong correlation between motors could be extrinsic, whereby the motors respond to fluctuations of the same signal. Alternatively, the motors could be intrinsically, physically coupled such that synchronized switching is energetically favored. Bo Hu (Yuhai Tu’s lab, IBM) described a model of coordinate switching of flagellar motors in single cells developed to distinguish between these two hypotheses. Using this model, Hu and Tu explored two mechanisms: correlated fluctuations in phospho-CheY concentrations, and direct coupling between the motors due to proposed hydrodynamic interactions (Hu and Tu, 2013). Their analysis supports the latter, intrinsic, mechanism, whose underlying molecular causes remain as yet unknown.
Phosphatases increase the dynamic response of the chemotaxis pathway of bacteria, enabling them to sense and respond rapidly to small changes in attractant and repellent concentrations. The main phosphatase of the enteric system, CheZ, is a stand-alone protein. In contrast, FliY, the main phosphatase in the Bacillus subtilis chemotaxis system, is an integral component of the flagellar rotor. FliY is a multidomain protein with an N-terminal CheY binding domain, a central CheC-like phosphatase domain, and an N-terminal domain homologous to FliN. Ria Sircar (Brian Crane’s lab, Cornell University) solved the structure of the FliY middle domain from Thermotoga maritima. Multi-angle light scattering analysis suggested that the C-terminal domain of FliY promotes dimerization. Mutational analysis verified that two active sites can independently dephosphorylate CheY, although one is more reactive. Pull-down experiments with other rotor proteins did not reveal an interaction between FliY and FliG. This suggests that FliY does not mimic FliM, but instead is localized to the bottom of the C-ring by its FliN-like domains. With the structure of FliY (Sircar et al., 2013), we now have a better understanding of the rotor architecture of Gram-positive bacteria.
Recent developments in computational modeling are instructing experimental work on inter- and intra-molecular protein interactions of the nanomachine and improving our comprehensive view of the connectivity of receptors and actuators. FliG is a component of the C-ring, along with FliM and FliN (Fig. 4A). Shahid Khan (Molecular Biology Consortium at Lawrence Berkeley National Laboratory) presented work that applies correlated mutation analysis (CMA) (Lee and Kim, 2012; Taylor and Sadowski, 2011; Taylor et al., 2013) to investigate the coupling of FliG interaction surfaces to FliM and MotA. The CMA algorithm, analogous to direct coupling analysis (DCA; presented by Szurmant, see below), maps co-evolved amino acid residues onto protein crystal structures to characterize functional couplings. Using CMA, Khan proposed a model for an allosteric signaling network at the FliG C-terminal domain.
Bacteria can swim through liquid or swarm across solid surfaces (Partridge and Harshey, 2013a). The actuator of the latter behavior varies widely. For flagellated bacteria, swarming motility correlates with an increase in flagellar number. Some bacteria use type IV pili to execute twitching motility, whilst others use various types of gliding devices. Salmonella is both capable of swimming in liquids and swarming over agar surfaces. However, in contrast to some bacteria (see below), Salmonella does not increase overall flagella numbers to achieve the latter behavior (Partridge and Harshey, 2013b; Wang et al., 2006). FliL is a ubiquitous, but functionally enigmatic, flagellar basal body-associated protein. In Salmonella, the absence of FliL leads to small defects in swimming, but to the complete elimination of swarming due to rod fracture (Attmannspacher et al., 2008). However, the exact function of FliL remains unknown. Jonathan Partridge (Rasika Harshey’s lab, University of Texas at Austin) reported that FliL interacts with itself, the stator components MotA and MotB, and FliF, a component of the C-ring. They also showed that FliL and MotAB overexpression increases swarming on harder agar surfaces, likely because FliL helps MotAB incorporate into the structure, stabilizing it and generating faster motor speeds.
Like Salmonella, Proteus mirabilis swims in liquids and swarms over surfaces. The swimmer cells are short (1.5–2 μm) with few flagella, whereas the swarmer cells are elongated (10–80 μm) with many flagella. When swimmer cells encounter a highly viscous environment or surface, they differentiate into swarmer cells. P. mirabilis senses these changes by monitoring flagellar rotation: when flagellar rotation is inhibited, swimmer cells differentiate into swarmer cells. The viscosity- and surface-detecting mechanism involves FliL; fliL null mutants are elongated (Elo+); they can swim (Swm+), but cannot swarm (Swr-) (Cusick et al., 2012). Yi-Ying Lee (Robert Belas’ Lab, University of Maryland Baltimore County) reported progress towards understanding the manner in which FliL regulates this sensory system. Specifically, she reported that loss of FliL reduces the transcription of flaA, which encodes flagellin, despite increased transcription of both flhD and fliA, two positive regulators of flagellar gene expression. Although the underlying mechanism remains unknown, intriguingly, complementation of the Swr- phenotype requires the fliL promoter, the fliL coding region, and a portion of fliM DNA (Lee et al., 2013).
Pseudomonas aeruginosa swarms over surfaces in groups, where they often form branched tendril patterns (Caiazza et al., 2005; Kamatkar et al., 2011). Josh Shrout (University of Notre Dame) showed that P. aeruginosa swarmer cells propagate in high-density waves that spread symmetrically as rings within the swarm towards the extending tendrils. Shrout provided evidence that wave propagation and tendril formation depend on a competition between the changing viscosity of the bacterial liquid suspension and liquid film boundary expansion caused by Marangoni forces. Simulations predicted that P. aeruginosa efficiently colonizes surfaces by controlling the physical forces responsible for expansion of thin liquid films at the swarm edge and by swarming towards the tendril tips (Du et al., 2012). Type IV pili interfered with swarming. This behavior was predicted because these pili limit cell-cell alignment, which is required for optimal swarming.
Bacillus subtilis exhibits flagella-mediated swarming across surfaces on semisolid media (Kearns and Losick, 2003). B. subtilis swarmer cells appear hyper-flagellated, though how these cells become hyper-flagellated remains unknown. Sampriti Mukherjee (Daniel Kearns’ Lab, Indiana University Bloomington) used 3D-Structured Illumination Microscopy to quantify swarmer cell flagella. Hyper-flagellation was confirmed and found to be required for swarming in a strain whose flagellar number was artificially controlled. Hyper-flagellation also was correlated with the intracellular level of the flagellar master regulator SwrA: SwrA protein levels increased in swarmer cells, whilst SwrA overexpression in swimmer cells resulted in hyper-flagellation. SwrA protein levels are not regulated at the level of expression. Instead, B. subtilis regulates hyper-flagellation through SwrA stability mediated by the LonA protease, as a lonA mutant exhibits increased SwrA protein levels and hyper-flagellated swimmer cells. The Kearns lab hypothesizes that LonA degrades SwrA in swimmer cells, but not in swarmer cells. Swarming motility is regulated by Lon protease in diverse species such as Proteus mirabilis and Vibrio parahaemolyticus (Claret and Hughes, 2000; Stewart et al., 1997). Thus although the master regulators of flagella biogenesis are distinct in different bacteria, the mechanism to regulate them appears to be similar.
Flavobacterium johnsoniae and other members of the phylum Bacteroidetes glide over surfaces, a mechanism that involves neither flagella nor pili. Instead, these bacteria express a novel Gld/Spr gliding machine that doubles as a secretory apparatus. A key protein in this gliding machine is SprB, a large cell-surface adhesin. Daisuke Nakane (Koji Nakayama’s Lab, Nagasaki University) used electron microscopy to visualize SprB, which appeared as thin long filaments extending from the cell surface. Fluorescence microscopy revealed SprB proteins moving at ~2 μm/s around the cell in what seemed to be a closed left-handed helical loop. The mechanism seems to require the proton motive force, as CCCP rapidly and reversibly blocked SprB movement. Some SprB protein appeared to attach to the substratum, about which the cell body moved forward and rotated. Upon reaching the rear of the cell, the attached SprB often released from the substratum, apparently recycling to the front of the cell along a helical path. The results suggest a model for Flavobacterium gliding, supported by mathematical analysis, in which adhesins, such as SprB, are propelled along a closed helical loop track, generating rotation and translation of the cell body (Nakane et al., 2013).
Myxococcus xanthus possesses two independent gliding machines: the adventurous (A) system and the social (S) system. A-motility allows single cell movement, whereas S-motility uses cell-cell contact to move groups of cells. By reversing the polarity of these motility systems, M. xanthus reverses translational direction. Reversals are induced by the Frz chemosensory system, which functions upstream of MglA and MglB, a Ras-like GTPase and its cognate GTPase activating protein. During reversals, MglA-GTP and MglB switch poles (Leonardy et al., 2010; Zhang et al., 2012). Daniela Keilberg (Lotte Søgaard-Andersen’s Lab, Max Planck Institute for Terrestrial Microbiology Marburg) reported the identification of the RR RomR, which directly interacts with MglA and MglB to regulate swarming (Keilberg et al., 2012). She and co-workers combined fluorescence microscopy and biochemical methods with epistasis analysis, a classic genetic approach that permits determination of dependence relationships of genes and their products. This approach produced the following model: RomR directs MglA and MglB to opposite cell poles and stimulates motility by receiving information from FrzZ, the output RR of the chemosensory system. The exact mechanism of information transfer remains unknown. Thus, RomR connects a classic bacterial signaling system (Frz) to a classic eukaryotic polarity module (MglA/MglB).
Beiyan Nan from the Zusman lab investigated the mechanism of gliding motility in M. xanthus by tracking the movements of single molecules of AglR, a homologue of the E. coli flagellar stator protein MotA, by photoactivatable localization microscopy (PALM). They found that AglR, unlike MotA/MotB, uses the proton motive force to move in helical trajectories within the membrane. AglR molecules slow down at surface contact regions and accumulate in clusters, consistent with the hypothesis that these clusters exert force on the slime surface, pushing cells forward (Nan et al., 2013; Nan et al., 2011).
Whilst Mycoplasma mobile also glides over surfaces, its gliding machinery, composed of three large proteins designated Gli521, Gli349, and Gli123, is completely novel. Yuhei Tahara (Makoto Miyata’s lab, Osaka city University) reported the use of electron microscopy and high-speed atomic force microscopy to investigate this machine, in the process learning that these three proteins function as a retractable foot, in which Gli123 functions as the mount, Gli521 the crank, and Gli349 the leg and foot.
The Intervening Logic
The HK CheA and the RRs CheY and CheB form a bifurcated two-component signal transduction system (TCS) linked at the sensory end to chemoreceptors and at the effector end to actuators. Not all HKs receive sensory cues through separate chemoreceptors, but instead receive input directly through their own sensory domain. Upon receipt of a signal, these sensory HK autophosphorylate a conserved histidinyl residue, using ATP as the phosphoryl donor. The phosphorylated HK then donates its phosphoryl group to the RR, which autophosphorylates a conserved aspartyl residue. The HK is usually, but not always, an integral cytoplasmic membrane protein. The RR is often a transcription factor (Bourret and Silversmith, 2010). Although most RRs receive their phosphoryl group from their cognate HK, some RR can receive the phosphoryl group from a low molecular weight phosphodonor, e.g. acetyl phosphate or phosphoramidate (Wolfe, 2010).
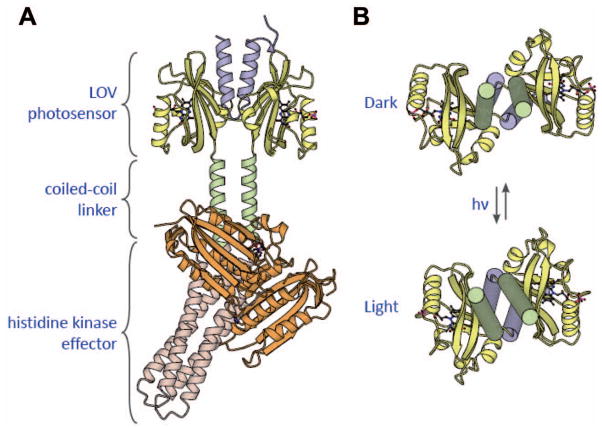
Currently, no atomic structure of an intact sensor HK containing both sensor and effector (catalytic) modules has been elucidated. With the aim to uncover unifying mechanisms for sensory activation of sensor HKs, Andreas Möglich (Humboldt-Universität zu Berlin) reported the full-length crystal structure at 2.3 Å resolution of the active form of the dimeric, blue-light-regulated sensor HK YF1 (Fig. 6) (Diensthuber et al., 2013). This functional chimeric sensor HK is comprised of two N-terminal light-oxygen-voltage (LOV) photosensors linked by a coiled coil to the C-terminal effector modules of FixL, including the DHp and CA domains. Möglich and co-workers propose that light signals detected at the FMN cofactor rings within the LOV domains are propagated through the beta sheet of the sensor to the central coiled-coil interface. Mutations in the N-terminal LOV domains profoundly affected light sensing; certain mutations even inverted the light response of a YF1-based two-component system (Ohlendorf et al., 2012). A structural model for the inactive sensor was generated and aligned to the crystal structure of the active conformation. This model highlights signal-induced quaternary structural changes within the N-terminal domains that propagate downstream as torque motions within the coiled-coil linker.
Figure 6. Structure of sensory domain of sensor histidine kinase (HK) chimera YF1 in light and dark conditions.
A. Complete structure of the functional YF1 (dark-adapted, 2.3 Å resolution), which is comprised of two N-terminal light-oxygen-voltage (LOV) photosensors linked by a coiled coil to the C-terminal effector modules of FixL, including the DHp and CA domains (Diensthuber et al., 2013).
B. Light and dark conformations of YF1 LOV sensory domain. The YF1 LOV photosensor domains in their dark-adapted state as determined by x-ray crystallography (left) are compared to a homology model of their light-adapted state (right) that is based on the structure of the Pseudomonas putida SB1 LOV protein (Circolone et al., 2012). Light absorption could induce quaternary structural rearrangements that culminate in a supertwist of the C-terminal coiled coil.
Why do non-photosynthetic bacteria possess photosensory proteins? For a few non-photosynthetic bacteria, photosensing bacteriophytochromes and LOV domain-containing HKs (LOV-HK) function in several different cellular processes, including intercellular attachment (Purcell et al., 2007) and biofilm formation (Bonomi et al., 2012). Their associated signal transduction pathways, however, remain unknown. In Pseudomonas syringae, light represses swarming (Wu et al., 2013). This plant pathogen encodes two bacteriophytochromes (BphP1 and BphP2) and one LOV-HK. BphP1 senses red/far-red light using biliverdin as a chromophore (Bhoo et al., 2001), whereas the LOV-HK senses blue light by means of a flavin mononucleotide-binding LOV domain (Swartz et al., 2007). BphP2 remains uncharacterized because its resists purification. Liang Wu (Gwyn Beattie’s lab, Iowa State University) reported genetic evidence showing that Bph1 and LOV-HK regulate swarming. The presented data support a model in which BphP1 represses swarming in response to red/far-red light and blue light, whilst LOV-HK promotes swarming by suppressing the BphP1-mediated blue light repression (Wu et al., 2013).
The TCS TodS/TodT of Pseudomonas putida controls the expression of the enzymes of the toluene dioxygenase (TOD) pathway that mineralizes exclusively toluene, benzene and ethylbenzene. Interestingly, the sensor HK TodS recognizes a wide range of different mono- and biaromatic compounds including the three TOD pathway substrates (Busch et al., 2009; Lacal et al., 2006). Some of these compounds function as agonists, whilst others act as antagonists (Busch et al., 2007). Tino Krell (CSIC, Granada, Spain) reported that the binding of antagonists competitively inhibits agonist-induced TodS activation (Busch et al., 2007). This concerted action of agonists and antagonists to regulate a HK appears to be a general feature of the family of TodS-like HKs (Silva-Jiménez et al., 2012) and may explain the failure of in situ bioremediation experiments of complex hydrocarbon mixtures, e.g. crude oil, that are mixtures of agonists and antagonists. The autokinase activity of TodS also is inhibited by the oxidative stress agent menadione, which acts on TodS via the conserved Cys320 (Silva-Jimenez, submitted). Reduction of TodS autokinase activity led to reduced tod gene expression in vivo. Because the presence of aromatic hydrocarbons generated an oxidative stress, the observed menadione-mediated TodS inhibition can be understood as a feedback mechanism.
Previous studies showed that phosphorylation of the chemotaxis RR CheY by small-molecule phosphodonors exhibits non-saturable kinetics (Da Re et al., 1999; Schuster et al., 2001; Thomas et al., 2013). This behavior suggests weak binding by the phosphodonor. CheY, however, does not dimerize like most RRs (Galperin, 2010) and therefore might not be the best model for the typical RR. Thus, Rachel Creager-Allenand co-workers (Robert Bourret’s Lab, University of North Carolina) monitored the ability of PhoB, a RR that dimerizes (Bachhawat et al., 2005; Mack et al., 2009; Sola et al., 2006; Solá et al., 1999), to use phosphoramidate as its phosphodonor (Creager-Allen et al., 2013). This reaction appeared both cooperative and saturable, characteristics that depended on maintenance of the dimer interface. Using simulations, they determined that these data could be explained by formation of a heterodimer that consists of one phosphorylated monomer and one non-phosphorylated monomer. These results challenge the notion that RR dimers form primarily between two phosphorylated monomers, and raise the possibility that RR heterodimers containing a single phosphoryl group may participate in gene regulation.
Like most RRs, CheY is activated by phosphorylation of its conserved aspartyl residue (Barak and Eisenbach, 1992), but CheY also is subject to Ne-lysine acetylation (Barak et al., 1992; Barak et al., 1998; Yan et al., 2008). CheY acetylated on multiple lysines has reduced affinity for its interaction partners CheA, CheZ and FliM (Liarzi et al., 2010); however, it remains unknown how each individual acetylation impacts CheY function. Particularly enigmatic is the seemingly conflicting observations that acetylation of CheY enhances CW rotation, whilst simultaneously disfavoring the binding of CheY to FliM. Milana Fraiberg (Michael Eisenbach’s lab, Weizmann Institute) reported that CheY is acetylated in vivo, and that according to preliminary results there is a quantitative hierarchy of acetylation (Ac-Lys91 > Ac-Lys92 > other lysines). Thus, they focused on the roles of Lys91 and Lys92, using mass spectrometry, genetics and biochemistry to understand how these lysines and their modifications affect flagellar rotation. It appears that unmodified Lys91 reduces CheY binding to FliM and thus inhibits CW rotation, whilst acetylation of Lys92 seems to increase CheY-FliM binding and thus supports CW rotation. Thus, Fraiberg proposed that a steady state exists between the strong repressor (unmodified Lys-91) and the weak activator (Lys-92), such that clockwise rotation requires deacetylation of acetylLys-91. It is now clear that Ne-lysine acetylation is common in all domains of life, including bacteria (Hu et al., 2010; Soppa, 2010; Thao and Escalante-Semerena, 2011). It is also clear that many posttranslational modifications exist, often on the same protein (Soufi et al., 2012). What remains to be learned is how these modifications interact with each other. CheY is an excellent model for such studies.
Clostridium difficile is the major causative organism of antibiotic-associated diarrhea, which is a huge problem in hospitals and other health care facilities. C. difficile is resistant to cationic antimicrobial peptides (CAMPs) produced by the host and by the indigenous microbiota. Shonna M. McBride (Emory University School of Medicine Atlanta) had previously shown that C. difficile responds to CAMP exposure by inducing expression of genes that lead to CAMP resistance. A hunt for resistance mutants had identified a locus that encodes a lantibiotic transport system (CprABC) and a typical HK (CprK), but not a RR (McBride and Sonenshein, 2011). Her group has now identified an orphan RR CD3320 (renamed CprR) that interacts with CprK to induce cpr transcription, specifically in response to eight different lantibiotics. In vivo experiments indicated the functional importance of the phosphorylation sites in CprK/CprR (Suarez et al., 2013).
Bce-type detoxification modules, composed of a TCS and an ATP-binding-cassette (ABC) transporter, mediate resistance to antimicrobial peptides (AMP). The TCS regulates expression of the transporter, but the HK of the TCS cannot detect the AMP without an active transporter (Gebhard, 2012). Since the HK does not contain a sensory domain and phylogenetic analysis indicates co-evolution of transporters (Dintner et al., 2011) and Bce-type HKs, Susanne Gebhard (Ludwig-Maximilians-Universität Munich) hypothesized that the transporter forms a sensory complex with the HK and serves as its co-sensor. In vivo and in vitro analyses of the bacitracin-resistance module BceRS-BceAB of B. subtilis support this hypothesis, revealing the importance of a functional active transporter for signaling activity (Kallenberg et al., 2013; Rietkotter et al., 2008). Thus, Bce-type signaling functions much like chemotaxis, wherein a receptor (the transporter or the chemoreceptor) receives a signal and in response induces its cognate HK to generate a cytoplasmic signal that elicits an output. Gebhard and co-workers used this knowledge to identify the cognate TCS for two orphan Enterococcus faecalis transporters. In the process, they learned that one transporter signals whereas the other confers resistance.
Like many HKs, Staphylococcus aureus WalK can function as a kinase or a phosphatase (Delauné et al., 2012). A major unanswered question is how the switch between these two functional states controls cellular physiology (Kenney, 2010). Tarek Msadek (Institut Pasteur Paris) reported the use of WalR-D55E, a constitutively active variant of the cognate RR, to show that WalK’s ability to switch activities influences gene expression and thus physiology. By manipulating the concentrations of WalK and WalR-D55E, he and co-workers obtained evidence that WalK acts as a net kinase during exponential growth, phosphorylating WalR, which activates cell wall degradation and turnover. During transition to stationary phase, once cell division stops, WalK acts as a net phosphatase, inactivating WalR and halting cell wall turnover. Intriguingly, WalR-D55E also significantly increased biofilm formation and a-hemolytic activity. A detailed transcriptome analysis indicated increased expression of 108 genes, including many encoding major virulence proteins involved in host matrix interactions, cytotoxicity, and innate immune defense evasion. Also up-regulated was the saePQRS operon, which encodes a TCS that controls expression of many of these virulence genes, suggesting that phosphorylated WalR controls virulence via the Sae TCS. Intriguingly, the WalR-D55E mutant was attenuated in mice. This appears related to high levels of released peptidoglycan fragments, likely due to the inability of the constitutively active RR to be inactivated by WalK. Peptidoglycan fragments are known to bind to Toll-like receptors, whose activation enhances the host inflammatory response. These findings reveal the importance of fine-tuning WalKR activity during the transition between S. aureus commensal and pathogenic lifestyles.
A less common bacterial signal transduction system consists of eukaryotic-like serine/threonine kinases (eSTKs) and phosphatases (eSTPs) (Pereira et al., 2011). Lindsie Goss (Jonathan Dworkin’s Lab, Columbia University) described two such systems: one that controls cell wall metabolism in B. subtilis and one that controls Enterococcus vancomycin resistance, also linked to cell wall biosynthesis. Intriguingly, each system interacts with and influences the activity of a TCS. In B. subtilis, the eSTK PrkC and eSTP PrpC reversibly phosphorylate Thr101 of WalR, regulating its activity, likely by influencing dimerization but not aspartyl phosphorylation. Since PrkC binds peptidoglycan, this activity explains, at least in part, how WalR can ‘sense’ the status of the cell wall. In the Enterococci, the PrkC homologue IreK phosphorylates a threonine residue of VanS, an HK that induces vancomycin resistance by modifying the cell wall. How VanS detects vancomycin has remained unknown. Vancomycin causes the release of peptidoglycan fragments that are recognized by IreK, and since full activation of the VanRS regulon requires IreK-dependent VanS phosphorylation, it appears that VanS ‘senses’ vancomycin via IreK. These results help to understand the complexity of TCS regulation, and identify new antibiotic targets, e.g., IreK, which is inhibited by the known eukaryotic Ser/Thr kinase inhibitor staurosporine.
Regulation and networks
Jürgen Lassak (Kirsten Jung’s Lab, Ludwig-Maximilians-Universität Munich) presented a novel translational regulation mechanism that links numerous cellular functions. This mechanism is mediated by elongation factor P (EF-P), which allows tight adjustment of protein copy numbers of receptors, e.g., for the pH-responsive sensor CadC of E. coli. EF-P is conserved in all bacteria and is orthologous to the archaeal and eukaryotic initiation factor 5A (a/eIF-5A). Fascinatingly, E. coli EF-P is β-lysinylated, a novel post-translational modification necessary for EF-P function. Recently, Lassak and co-workers reported a physiological role for β-lysinylated EF-P: it relieves ribosome stalling at polyproline stretches (Doerfel et al., 2013; Ude et al., 2013). Within the E. coli proteome, about 100 proteins possess a polyproline stretch, including the HKs EnvZ and PhoR, as well as the flagella master regulator FlhC. Thus the findings rationalize the motility defects and attenuated virulence exhibited by efp mutants of Salmonella (Navarre et al., 2010).
Since the completion of the first bacterial genome-sequencing project in 1995 more than 2305 bacterial genomes have been completely sequenced. Encoded by these genomes are about 130,000 TCS proteins, whose sequences have been sampled in evolutionary space to accomplish perfect functionality. Hidden within this sequence variation is a wealth of information that can reveal molecular mechanisms underlying the signal transduction cascade: How does the HK modulate activity in response to signals? How does the HK identify its cognate RR from amongst all the other RRs of similar structure? How does the HK physically interact with its cognate RR? To address these questions, Hendrik Szurmant (The Scripps Research Institute, La Jolla) described a computational method, called Direct Coupling Analysis (DCA), based on the concept that amino acid residues on opposing protein surfaces co-evolve (Dago et al., 2012; Schug et al., 2009). The method, like CMA (see above), calculates correlations between all amino acid residues across many homologues. A major advance in this approach permits calculation of the Direction Interaction (DI), which disentangles direct from indirect correlations. Using DCA, Szurmant and co-workers have predicted interaction surfaces between two proteins or between two domains of the same protein, including identifying the contact sites within an individual HK domain that change upon activation. Using crystal structures and genetic approaches, they have tested these predictions and found them to be highly accurate (Szurmant and Hoch, 2013). This is an exciting new tool to identify and characterize protein interaction surfaces and complex protein structures for which no structural information exists.
Transcriptional crosstalk occurs between the S. typhimurium flagellar regulon and the SP1 pathogenicity island. The regulation of the master flagellar regulator FlhD4C2 is itself quite complex. The flhDC regulatory region includes six transcription initiation sites, and transcription is controlled positively or negatively by multiple regulatory factors, including RcsB, LrhA, SlyA, DskA, EcnR, HNS, cAMP/CRP, CsrA, PefI-SrgD, HilD, RtsB and QseB (Wozniak et al., 2010). Chakib Mouslim (Kelly Hughes’ Lab, University of Utah) examined the role of each initiation site, their relation to each other, and the ability of some of these regulators to control their activity. The resultant data support a model in which certain promoters predominate. These promoters drive flhDC transcription either alone or in coordination with each other. Together with the function of certain transcription factors, these interactions generate regulatory circuits that control transcription of the flagellar regulon and the virulence genes encoded by SPI1.
The Lyme disease spirochete Borrelia burgdorferi alternates between arthropods and mammals. In each host, the bacterium must adapt to the available carbohydrates. For example, in the tick, chitobiose is available, and B. burgdorferi converts it to N-acetylglucosamine (GlcNAc). Ching Wooen Sze (Chunhao Li’s Lab, The State University of New York at Buffalo) showed that the RR Rrp1, which synthesizes cyclic diguanylate (c-di-GMP), regulates the chbC gene, which encodes a chitobiose transporter. An rrp1 mutant could not be transmitted via tick bite, a phenotype that could be partially rescued by exogenous supplementation of GlcNAc. This work (Sze et al., 2013) implicates chitobiose metabolism during spirochaete transmission.
The above work was closely related both to environmental regulation of phenotypic traits and to colonization and infection in different host by spirochetes. The following talk expanded on the role of motility in virulence and infection by B. burgdorferi in mouse and tick models. Tao Lin (UT Medical School at Houston) described the use of signature-tagged mutagenesis to dissect the mechanism of chemotaxis and motility in B. burgdorferi and explore the relationship between motility/chemotaxis and virulence (Lin et al., 2012). They found that nearly all chemotaxis and flagellar genes were required for mouse infection. Many mutants (including fliH, fliI and flhA) had elongated morphology and reduced motility. The fliH and fliI also exhibited slow growth and exhibited protein profiles that were different relative to their wild type parent and to each other. These divergent proteins are currently being analyzed. This represents the first identification of components of the flagellar export apparatus in B. burgdorferi.
Leptospira are spirochetes with a unique morphology (i.e., hook-shaped ends). Their motility depends upon coiled flagella contained entirely within the periplasm (Charon et al., 2012). The role of motility in leptospiral virulence has not been evaluated. Elsio Wunder (Yale School of Public Health) in collaboration with Nyles Charon, Mathieu Picardeau, and Albert Ko investigated a new flagellar sheath protein from the human pathogenic spirochete L. interrogans. They identified this sheath protein, called Fcp-1, by studying a small colony, non-motile mutant isolated from a patient. The individual mutant cells had straight terminal ends, their flagella were thinner than those of wild type, and the purified flagella were straight instead of coiled. A previous report showed that the leptospiral flagellar sheath does not consist of FlaA (Lambert et al., 2012). LC-MS analysis of the purified flagella associated Fcp1 with flagella of coiled morphology and thus with motility. Fcp1 was detected on the surface of flagellar filaments and thus can now be regarded as the major flagellar sheath component. Molecular Koch’s postulate analysis indicated that the presence of Fcp1, and thus motility, are essential for virulence, both for dissemination and persistence, but mostly for the initial bacterial entry process of tissue penetration.
Do flagella constitute an advantage or disadvantage during development of an E. coli biofilm? To address this question, Birgit Prüß (North Dakota State University) followed the genetic status of and transcription from the flhDC regulatory region within developing biofilms. Her data suggest that optimal development of E. coli biofilms may need both parental bacteria and flhD mutants. This may be due to the temporal and spatial regulation of flhD (Samanta et al., 2013).
Conclusions
The ability to sense and respond to environmental change is common to all organisms. The talks at this year’s BLAST meeting illustrated not only the breadth and complexity of these adaptation systems in bacteria, but also how closely we can now observe the intricate details of the exquisite machines that govern their behavior. While the BLAST XII meeting was predominantly a tale of two machines, we are fast uncovering the molecular blueprints and regulatory circuitry for many additional adaptation systems, including the remarkable engines involved in alternate, non-flagellated motility systems. Going forward, we foresee that the field will increasingly focus on the engineering-like principles that govern their organization. The language is not all that dissimilar, e.g. rotors, stators and clutches. The next step will be to understand how these complex systems evolved and to identify how these engineering-like principles emerged from the evolutionary process.
Acknowledgments
The authors thank Peggy O’Neil and Tara Bollinger for kind assistance with data collection, Ariane Briegel, Sandy Parkinson, and Andreas Möglich for the kind gift of unpublished illustrations in Figures 2, 3 and 6, respectively. We also thank our BLAST colleagues for ongoing stimulating discussions. The authors were supported by the German Research Foundation (JO344/3-2, SFB900) and the German Ministry for Education and Research (HeldivPAT) to awarded C.J., the German Research Foundation (Exc114/2, JU270/9-1) awarded to K.J., National Institutes of Health grant GM054365 awarded to C.V.R., and National Institutes of Health grant GM066130 awarded to A.J.W.
References
- Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Shimizu TS, Stocker R. Microfluidics for bacterial chemotaxis. Integrative Biology. 2010a;2:604–629. doi: 10.1039/c0ib00049c. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Shimizu TS, Stocker R. Bacterial Chemotaxis in Linear and Nonlinear Steady Microfluidic Gradients. Nano Letters. 2010b;10:3379–3385. doi: 10.1021/nl101204e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Zhou Q, Parkinson JS. Mutational Analysis of the Connector Segment in the HAMP Domain of Tsr, the Escherichia coli Serine Chemoreceptor. Journal of Bacteriology. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews DA, Xie M, Hughes V, Wilce MC, Roujeinikova A. Design, purification and characterization of a soluble variant of the integral membrane protein MotB for structural studies. Journal of The Royal Society Interface. 2013:10. doi: 10.1098/rsif.2012.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attmannspacher U, Scharf BE, Harshey RM. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Molecular Microbiology. 2008;68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- Bachhawat P, Swapna GVT, Montelione GT, Stock AM. Mechanism of Activation for Transcription Factor PhoB Suggested by Different Modes of Dimerization in the Inactive and Active States. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Regulation of interaction between signaling protein CheY and flagellar motor during bacterial chemotaxis. Curr Top Cell Regul. 1996;34:137–158. doi: 10.1016/s0070-2137(96)80005-7. [DOI] [PubMed] [Google Scholar]
- Barak R, Abouhamad WN, Eisenbach M. Both Acetate Kinase and Acetyl Coenzyme A Synthetase Are Involved in Acetate-Stimulated Change in the Direction of Flagellar Rotation in Escherichia coli. Journal of Bacteriology. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- Biemann HP, Koshland DEJ. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- Bonomi HR, Posadas DM, Paris G, Carrica MdC, Frederickson M, Pietrasanta LI, Bogomolni RA, Zorreguieta A, Goldbaum FA. Light regulates attachment, exopolysaccharide production, and nodulation in Rhizobium leguminosarum through a LOV-histidine kinase photoreceptor. Proceedings of the National Academy of Sciences. 2012;109:12135–12140. doi: 10.1073/pnas.1121292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Silversmith RE. Two-component signal transduction. Current Opinion in Microbiology. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, DeLoney-Marino CR, Mandel MJ. Chemoreceptor VfcA Mediates Amino Acid Chemotaxis in Vibrio fischeri. Applied and Environmental Microbiology. 2013;79:1889–1896. doi: 10.1128/AEM.03794-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proceedings of the National Academy of Sciences. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Molecular Microbiology. 2013 Jun 26; doi: 10.1111/mmi.12309. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Lacal J, Martos A, Ramos JL, Krell T. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proceedings of the National Academy of Sciences. 2007;104:13774–13779. doi: 10.1073/pnas.0701547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Guazzaroni ME, Lacal J, Ramos JL, Krell T. The Sensor Kinase TodS Operates by a Multiple Step Phosphorelay Mechanism Involving Two Autokinase Domains. Journal of Biological Chemistry. 2009;284:10353–10360. doi: 10.1074/jbc.M900521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Shanks RMQ, O’Toole GA. Rhamnolipids Modulate Swarming Motility Patterns of Pseudomonas aeruginosa. Journal of Bacteriology. 2005;187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine CR, Luisi BF, Pratap JV. A “Mechanistic” Explanation of the Multiple Helical Forms Adopted by Bacterial Flagellar Filaments. Journal of Molecular Biology. 2013;425:914–928. doi: 10.1016/j.jmb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. The Unique Paradigm of Spirochete Motility and Chemotaxis. Annual Review of Microbiology. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. Structural diversity of bacterial flagellar motors. EMBO. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun SY, Parkinson JS. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Circolone F, Granzin J, Jentzsch K, Drepper T, Jaeger KE, Willbold D, Krauss U, Batra-Safferling R. Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida. Journal of Molecular Biology. 2012;417:362–374. doi: 10.1016/j.jmb.2012.01.056. [DOI] [PubMed] [Google Scholar]
- Claret L, Hughes C. Rapid Turnover of FlhD and FlhC, the Flagellar Regulon Transcriptional Activator Proteins, during Proteus Swarming. Journal of Bacteriology. 2000;182:833–836. doi: 10.1128/jb.182.3.833-836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager-Allen RL, Silversmith RE, Bourret RB. A Link between Dimerization and Autophosphorylation of the Response Regulator PhoB. Journal of Biological Chemistry. 2013;288:21755–21769. doi: 10.1074/jbc.M113.471763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier WW, Strobel GA. Chemotaxis of Rhizobium spp. to Plant Root Exudates. Plant Physiology. 1976;57:820–823. doi: 10.1104/pp.57.5.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick K, Lee YY, Youchak B, Belas R. Perturbation of FliL Interferes with Proteus mirabilis Swarmer Cell Gene Expression and Differentiation. Journal of Bacteriology. 2012;194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Re SS, Deville-Bonne D, Tolstykh T, Véron M, Stock JB. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Letters. 1999;457:323–326. doi: 10.1016/s0014-5793(99)01057-1. [DOI] [PubMed] [Google Scholar]
- Dago AE, Schug A, Procaccini A, Hoch JA, Weigt M, Szurmant H. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proceedings of the National Academy of Sciences. 2012;109:E1733–E1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Molecular Microbiology. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- Delauné A, Dubrac S, Blanchet C, Poupel O, Mäder U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon JM, Adib-Conquy M, Msadek T. The WalKR System Controls Major Staphylococcal Virulence Genes and Is Involved in Triggering the Host Inflammatory Response. Infection and Immunity. 2012;80:3438–3453. doi: 10.1128/IAI.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber RP, Bommer M, Gleichmann T, Moglich A. Full-Length Structure of a Sensor Histidine Kinase Pinpoints Coaxial Coiled Coils as Signal Transducers and Modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S. Coevolution of ABC Transporters and Two-Component Regulatory Systems as Resistance Modules against Antimicrobial Peptides in Firmicutes Bacteria. Journal of Bacteriology. 2011;193:3851–3862. doi: 10.1128/JB.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P Is Essential for Rapid Synthesis of Proteins Containing Consecutive Proline Residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Du H, Xu Z, Anyan M, Kim O, Leevy WM, Shrout JD, Alber M. High Density Waves of the Bacterium Pseudomonas aeruginosa in Propagating Swarms Result in Efficient Colonization of Surfaces. Biophysical Journal. 2012;103:601–609. doi: 10.1016/j.bpj.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Namba K, Hughes KT. Bacterial Nanomachines: The Flagellum and Type III Injectisome. Cold Spring Harbor Perspectives in Biology. 2010:2. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank V, Vaknin A. Prolonged stimuli alter the bacterial chemosensory clusters. Molecular Microbiology. 2013;88:634–644. doi: 10.1111/mmi.12215. [DOI] [PubMed] [Google Scholar]
- Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. Identification of New Flagellar Genes of Salmonella enterica Serovar Typhimurium. Journal of Bacteriology. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Diversity of structure and function of response regulator output domains. Current Opinion in Microbiology. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard S. ABC transporters of antimicrobial peptides in Firmicutes bacteria -phylogeny, function and regulation. Molecular Microbiology. 2012;86:1295–1317. doi: 10.1111/mmi.12078. [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends in Biochemical Sciences. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. Journal of Bacteriology. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Tu Y. Coordinated Switching of Bacterial Flagellar Motors: Evidence for Direct Motor-Motor Coupling? Physical Review Letters. 2013;110:158703. doi: 10.1103/PhysRevLett.110.158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Molecular Microbiology. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara A, Segall JE, Block SM, Berg HC. Coordination of flagella on filamentous cells of Escherichia coli. Journal of Bacteriology. 1983;155:228–237. doi: 10.1128/jb.155.1.228-237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenberg F, Dintner S, Schmitz R, Gebhard S. Identification of Regions Important for Resistance and Signalling within the Antimicrobial Peptide Transporter BceAB of Bacillus subtilis. Journal of Bacteriology. 2013;195:3287–3297. doi: 10.1128/JB.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatkar NG, Sarna MJ, Shrout JD. Population dynamics during swarming of Pseudomonas aeruginosa. Commun Integr Biol. 2011;4:689–691. doi: 10.4161/cib.17109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Molecular Microbiology. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Keilberg D, Wuichet K, Drescher F, Sogaard-Andersen L. A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in Myxococcus xanthus. PLoS Genet. 2012;8:e1002951. doi: 10.1371/journal.pgen.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LJ. How important is the phosphatase activity of sensor kinases? Current Opinion in Microbiology. 2010;13:168–176. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, Homma M, Namba K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Molecular Microbiology. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- Lacal J, Busch A, Guazzaroni ME, Krell T, Ramos JL. The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins. Proceedings of the National Academy of Sciences. 2006;103:8191–8196. doi: 10.1073/pnas.0602902103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A, Picardeau M, Haake DA, Sermswan RW, Srikram A, Adler B, Murray GA. FlaA Proteins in Leptospira interrogans Are Essential for Motility and Virulence but Are Not Required for Formation of the Flagellum Sheath. Infection and Immunity. 2012;80:2019–2025. doi: 10.1128/IAI.00131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazova MD, Butler MT, Shimizu TS, Harshey RM. Salmonella chemoreceptors McpB and McpC mediate a repellent response to L-cystine: a potential mechanism to avoid oxidative conditions. Molecular Microbiology. 2012;84:697–711. doi: 10.1111/j.1365-2958.2012.08051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim D. Large-scale reverse docking profiles and their applications. BMC Bioinformatics. 2012;13:S6. doi: 10.1186/1471-2105-13-S17-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Patellis J, Belas R. Activity of Proteus mirabilis FliL Is Viscosity Dependent and Requires Extragenic DNA. Journal of Bacteriology. 2013;195:823–832. doi: 10.1128/JB.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele PP, Branch RW, Nathan VSJ, Berg HC. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proceedings of the National Academy of Sciences. 2012;109:20018–20022. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 2010;29:2276–2289. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. The 3.2 A Resolution Structure of a Receptor:CheA:CheW Signaling Complex Defines Overlapping Binding Sites and Key Residue Interactions within Bacterial Chemosensory Arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarzi O, Barak R, Bronner V, Dines M, Sagi Y, Shainskaya A, Eisenbach M. Acetylation represses the binding of CheY to its target proteins. Molecular Microbiology. 2010;76:932–943. doi: 10.1111/j.1365-2958.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack TR, Gao R, Stock AM. Probing the Roles of the Two Different Dimers Mediated by the Receiver Domain of the Response Regulator PhoB. Journal of Molecular Biology. 2009;389:349–364. doi: 10.1016/j.jmb.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Sonenshein AL. Identification of a Genetic Locus Responsible for Antimicrobial Peptide Resistance in Clostridium difficile. Infection and Immunity. 2011;79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Heath-Heckman EAC, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Seminars in Immunology. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier VM, Muschler P, Scharf BE. Functional Analysis of Nine Putative Chemoreceptor Proteins in Sinorhizobium meliloti. Journal of Bacteriology. 2007;189:1816–1826. doi: 10.1128/JB.00883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Current Opinion in Structural Biology. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Muramoto K, Macnab RM. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Molecular Microbiology. 1998;29:1191–1202. doi: 10.1046/j.1365-2958.1998.00998.x. [DOI] [PubMed] [Google Scholar]
- Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proceedings of the National Academy of Sciences. 2013;110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Bandaria JN, Moghtaderi A, Sun I-H, Yildiz A, Zusman DR. Flagella stator homologues function as motors for myxobacterial gliding motility by moving in helical trajectories. PNAS. 2013:E1508–1513. doi: 10.1073/pnas.1219982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. PNAS. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, Fang FC. PoxA, YjeK, and Elongation Factor P Coordinately Modulate Virulence and Drug Resistance in Salmonella enterica. Molecular Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Kojima S. Bacterial motility measured by a miniature chamber for high-pressure microscopy. Int J Mol Sci. 2012;13:9225–9239. doi: 10.3390/ijms13079225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Sowa Y. Microscopic Analysis of Bacterial Motility at High Pressure. Biophysical Journal. 2012;102:1872–1880. doi: 10.1016/j.bpj.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Sowa Y, Kimura Y, Homma M, Ishijima A, Terazima M. High Hydrostatic Pressure Induces Counterclockwise to Clockwise Reversals of the Escherichia coli Flagellar Motor. Journal of Bacteriology. 2013;195:1809–1814. doi: 10.1128/JB.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. Journal of Molecular Biology. 2012;416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- O’Neill J, Xie M, Hijnen M, Roujeinikova A. Role of the MotB linker in the assembly and activation of the bacterial flagellar motor. Acta Crystallographica. 2011;67:1009–1016. doi: 10.1107/S0907444911041102. [DOI] [PubMed] [Google Scholar]
- Park H, Oikonomou P, Guet CC, Cluzel P. Noise Underlies Switching Behavior of the Bacterial Flagellum. Biophysical Journal. 2011;101:2336–2340. doi: 10.1016/j.bpj.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Harshey RM. Swarming: Flexible Roaming Plans. Journal of Bacteriology. 2013a;195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Harshey RM. More than Motility: Salmonella Flagella Contribute to Overriding Friction and Facilitating Colony Hydration during Swarming. Journal of Bacteriology. 2013b;195:919–929. doi: 10.1128/JB.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-A review. Analytica Chimica Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- Pereira SFF, Goss L, Dworkin J. Eukaryote-Like Serine/Threonine Kinases and Phosphatases in Bacteria. Microbiology and Molecular Biology Reviews. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. Defining a Key Receptor-CheA Kinase Contact and Elucidating Its Function in the Membrane-Bound Bacterial Chemosensory Array: A Disulfide Mapping and TAM-IDS Study. Biochemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proceedings of the National Academy of Sciences. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietkotter E, Hoyer D, Mascher T. Bacitracin sensing in Bacillus subtilis. Molecular Microbiology. 2008;68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: Implications for peptidoglycan recognition. Proceedings of the National Academy of Sciences. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta P, Clark E, Knutson K, Horne S, Prüß B. OmpR and RcsB abolish temporal and spatial changes in expression of flhD in Escherichia coli Biofilm. BMC Microbiology. 2013;13:182. doi: 10.1186/1471-2180-13-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug A, Weigt M, Onuchic JN, Hwa T, Szurmant H. High-resolution protein complexes from integrating genomic information with molecular simulation. Proceedings of the National Academy of Sciences. 2009;106:22124–22129. doi: 10.1073/pnas.0912100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proceedings of the National Academy of Sciences. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proceedings of the National Academy of Sciences. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. 2010:6. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Jiménez H, García-Fontana C, Cadirci BH, Ramos-González MI, Ramos JL, Krell T. Study of the TmoS/TmoT two-component system: towards the functional characterization of the family of TodS/TodT like systems. Microb Biotechnol. 2012;5:489–500. doi: 10.1111/j.1751-7915.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar R, Greenswag AR, Bilwes AM, Gonzalez-Bonet G, Crane BR. Structure and Activity of the Flagellar Rotor Protein FliY: A Member of the CheC Phosphatase Family. Journal of Biological Chemistry. 2013;288:13493–13502. doi: 10.1074/jbc.M112.445171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola M, Drew DL, Blanco AG, Gomis-Ruth FX, Coll M. The cofactor-induced pre-active conformation in PhoB. Acta Crystallographica. 2006;62:1046–1057. doi: 10.1107/S0907444906024541. [DOI] [PubMed] [Google Scholar]
- Solá M, Gomis-Rüth FX, Serrano L, González A, Coll Ml. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. Journal of Molecular Biology. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010;2010:820681. doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Soares NC, Ravikumar V, Macek B. Proteomics reveals evidence of cross-talk between protein modifications in bacteria: focus on acetylation and phosphorylation. Current Opinion in Microbiology. 2012;15:357–363. doi: 10.1016/j.mib.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proceedings of the National Academy of Sciences. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Vaknin A, Shimizu TS, Berg HC. In Vivo Measurement by FRET of Pathway Activity in Bacterial Chemotaxis Methods in Enzymology. In: Simon MI, Crane BR, Crane A, editors. Two-Signaling Systems, Part B. Academic Press; 2007. pp. 363–391. [DOI] [PubMed] [Google Scholar]
- Stewart BJ, Enos-Berlage JL, McCarter LL. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. Journal of Bacteriology. 1997;179:107–114. doi: 10.1128/jb.179.1.107-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez JM, Edwards AN, McBride SM. The Clostridium difficile cpr Locus Is Regulated by a Noncontiguous Two-Component System in Response to Type A and B Lantibiotics. Journal of Bacteriology. 2013;195:2621–2631. doi: 10.1128/JB.00166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-Light-Activated Histidine Kinases: Two-Component Sensors in Bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- Sze CW, Smith A, Choi YH, Yang X, Pal U, Yu A, Li C. Study of the Response Regulator Rrp1 Reveals Its Regulatory Role in Chitobiose Utilization and Virulence of Borrelia burgdorferi. Infection and Immunity. 2013;81:1775–1787. doi: 10.1128/IAI.00050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Hoch JA. Statistical analyses of protein sequence alignments identify structures and mechanisms in signal activation of sensor histidine kinases. Molecular Microbiology. 2013;87:707–712. doi: 10.1111/mmi.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Sadowski MI. Structural constraints on the covariance matrix derived from multiple aligned protein sequences. PLoS One. 2011;6:e28265. doi: 10.1371/journal.pone.0028265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Hamilton RS, Sadowski MI. Prediction of contacts from correlated sequence substitutions. Current Opinion in Structural Biology. 2013;23:473–479. doi: 10.1016/j.sbi.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Terasawa S, Fukuoka H, Inoue Y, Sagawa T, Takahashi H, Ishijima A. Coordinated Reversal of Flagellar Motors on a Single Escherichia coli Cell. Biophysical Journal. 2011;100:2193–2200. doi: 10.1016/j.bpj.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kojima S, Homma M. In: Chapter 2 Flagellar Motility in Bacteria: Structure and Function of Flagellar Motor International Review of Cell and Molecular Biology. Jeon KW, editor. Academic Press; 2008. pp. 39–85. [DOI] [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Control of protein function by reversible N(epsilon)-lysine acetylation in bacteria. Current Opinion in Microbiology. 2011;14:200–204. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Morgan DG, DeRosier DJ. Structures of Bacterial Flagellar Motors from Two FliF-FliG Gene Fusion Mutants. Journal of Bacteriology. 2001;183:6404–6412. doi: 10.1128/JB.183.21.6404-6412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Immormino RM, Bourret RB, Silversmith RE. Nonconserved Active Site Residues Modulate CheY Autophosphorylation Kinetics and Phosphodonor Preference. Biochemistry. 2013;52:2262–2273. doi: 10.1021/bi301654m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation Elongation Factor EF-P Alleviates Ribosome Stalling at Polyproline Stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Vu A, Wang X, Zhou H, Dahlquist FW. The Receptor-CheW Binding Interface in Bacterial Chemotaxis. Journal of Molecular Biology. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Warren AV, Martin AC, Armitage JP. Targeting of two signal transduction pathways to different regions of the bacterial cell. Molecular Microbiology. 2003;50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mariconda S, Suzuki A, McClelland M, Harshey RM. Uncovering a Large Set of Genes That Affect Surface Motility in Salmonella enterica Serovar Typhimurium. Journal of Bacteriology. 2006;188:7981–7984. doi: 10.1128/JB.00852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts KJ, Taylor BL, Johnson MS. PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Molecular Microbiology. 2011;79:686–699. doi: 10.1111/j.1365-2958.2010.07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the Message Out: Cyclic-Di-GMP Regulates Multiple Levels of Flagellum-Based Motility. Journal of Bacteriology. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Current Opinion in Microbiology. 2010;13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak CE, Chevance FFV, Hughes KT. Multiple Promoters Contribute to Swarming and the Coordination of Transcription with Flagellar Assembly in Salmonella. Journal of Bacteriology. 2010;192:4752–4762. doi: 10.1128/JB.00093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, McGrane RS, Beattie GA. Light Regulation of Swarming Motility in Pseudomonas syringae Integrates Signaling Pathways Mediated by a Bacteriophytochrome and a LOV Protein. mBio. 2013:4. doi: 10.1128/mBio.00334-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Zhulin IB. Origins and Diversification of a Complex Signal Transduction System in Prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Barak R, Liarzi O, Shainskaya A, Eisenbach M. In Vivo Acetylation of CheY, a Response Regulator in Chemotaxis of Escherichia coli. Journal of Molecular Biology. 2008;376:1260–1271. doi: 10.1016/j.jmb.2007.12.070. [DOI] [PubMed] [Google Scholar]
- Yuan J, Branch RW, Hosu BG, Berg HC. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T. A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus. PLoS Genet. 2012;8:e1002872. doi: 10.1371/journal.pgen.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Molecular Microbiology. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]