Significance
Many bacterial species engage in a form of cell–cell communication known as quorum sensing (QS). Despite great progress in unravelling the molecular mechanisms of QS, controversy remains over its functional role. There is disagreement over whether QS surveys bacterial cell density or rather environmental properties like diffusion or flow, and moreover there is no consensus on why many bacteria use multiple signal molecules. We develop and test a new conceptual framework for bacterial cell–cell communication, demonstrating that bacteria can simultaneously infer both their social (density) and physical (mass-transfer) environment, given combinatorial (nonadditive) responses to multiple signals with distinct half-lives. Our results also show that combinatorial communication is not restricted solely to primates and is computationally achievable in single-celled organisms.
Keywords: diffusion sensing, bacterial signaling, efficiency sensing, collective behavior, bacterial cooperation
Abstract
Quorum sensing (QS) is a cell–cell communication system that controls gene expression in many bacterial species, mediated by diffusible signal molecules. Although the intracellular regulatory mechanisms of QS are often well-understood, the functional roles of QS remain controversial. In particular, the use of multiple signals by many bacterial species poses a serious challenge to current functional theories. Here, we address this challenge by showing that bacteria can use multiple QS signals to infer both their social (density) and physical (mass-transfer) environment. Analytical and evolutionary simulation models show that the detection of, and response to, complex social/physical contrasts requires multiple signals with distinct half-lives and combinatorial (nonadditive) responses to signal concentrations. We test these predictions using the opportunistic pathogen Pseudomonas aeruginosa and demonstrate significant differences in signal decay between its two primary signal molecules, as well as diverse combinatorial responses to dual-signal inputs. QS is associated with the control of secreted factors, and we show that secretome genes are preferentially controlled by synergistic “AND-gate” responses to multiple signal inputs, ensuring the effective expression of secreted factors in high-density and low mass-transfer environments. Our results support a new functional hypothesis for the use of multiple signals and, more generally, show that bacteria are capable of combinatorial communication.
Bacteria must often make regulatory decisions on the basis of limited information about their external world (1). In many bacteria, these decisions are aided by the secretion and detection of small diffusible molecules, in a process called quorum sensing (QS) (2, 3). QS controls a variety of traits, including pathogen virulence (3) leading to “QS interference” (2–6) emerging as a control strategy for several bacterial pathogens.
The mechanisms underlying production, uptake, and response to these signal molecules are well-understood, but relatively little is known about how quorum sensing contributes to bacterial fitness. Put another way, why do bacteria use QS? The classic adaptive interpretation of QS is that cells produce signal molecules to serve as a proxy for cellular density: more signal implying more bacteria (7–10). Others have argued for a “diffusion-sensing” interpretation, with more signal implying lower rates of mass transfer (diffusion or flow) (11). However, low mass transfer and high cellular density both lead to high signal concentrations, and so these two unknowns—information on the social parameter of cellular density and the asocial mass-transfer rate—are inextricably linked when only one signal is used (12, 13). It is possible that a signal molecule can still provide a reliable indicator of the achievable density of a more costly secreted factor (“efficiency sensing”) (13). However, where investigated, the majority of QS-regulated genes encode nonsecreted gene products (14), and QS is known to control an array of traits in various bacteria that are not directly impacted by mass transfer, such as luminescence, conjugation, or type-3 secretion (10). Even among secretion-related phenotypes, accumulation of a single autoinducer cannot reliably predict the dynamics of secreted products differing in rates of mass transfer or chemical decay or in how they interact with the environment to form beneficial compounds. Here, we argue that, by combinatorially responding to multiple signal molecules with the appropriate molecular properties (differing in chemical decay rates), bacteria can potentially infer the properties of their social and physical environment simultaneously (Fig. S1).
Results
We first analyze a simple mathematical model capturing the extracellular dynamics of auto-inducing signal molecules in a well-mixed environment (Fig. 1 and Fig. S2). The environment contains stationary-phase bacteria at density N contributing to signal production and a mass-transfer rate m causing signal removal (by flow). The extracellular dynamics of signal Sk are given by 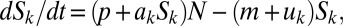 where
where 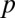 is baseline production,
is baseline production, 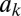 is the increased production due to autoinduction, and
is the increased production due to autoinduction, and 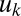 is decay rate. This system leads to a simple threshold behavior defined by the diagonal contours in Fig. 1
is decay rate. This system leads to a simple threshold behavior defined by the diagonal contours in Fig. 1
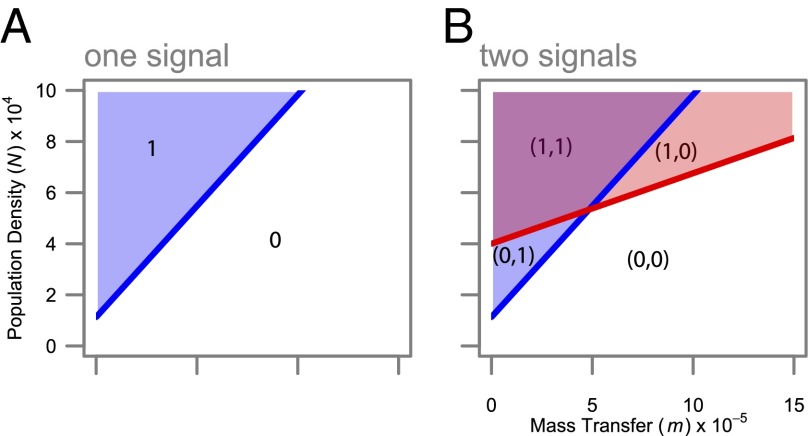
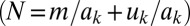 , above which production of the molecule increases and below which it does not. Fig. 1A illustrates the simple scenario where only one autoinducer molecule is used. In the shaded region (labeled “1”) the regulon is on, otherwise (labeled “0”) it is off. With one molecule, there is ambiguity between cellular density and mass-transfer properties, represented by the diagonal threshold contour. In the case of a set of genes best expressed at high bacterial density, variation in mass transfer can cause maladaptive regulation, due to conflation between the effects of density and mass transfer. Now, consider a system of two signal molecules. If the properties of decay
, above which production of the molecule increases and below which it does not. Fig. 1A illustrates the simple scenario where only one autoinducer molecule is used. In the shaded region (labeled “1”) the regulon is on, otherwise (labeled “0”) it is off. With one molecule, there is ambiguity between cellular density and mass-transfer properties, represented by the diagonal threshold contour. In the case of a set of genes best expressed at high bacterial density, variation in mass transfer can cause maladaptive regulation, due to conflation between the effects of density and mass transfer. Now, consider a system of two signal molecules. If the properties of decay 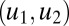 and autoinduction
and autoinduction 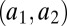 have positive covariance (
have positive covariance (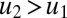 and
and 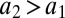 ), then the space of potential environmental states (m, N) can be split into four rather than two distinct sectors (Fig. 1B), improving resolution of both the physical and social environment.
), then the space of potential environmental states (m, N) can be split into four rather than two distinct sectors (Fig. 1B), improving resolution of both the physical and social environment.
Fig. 1.
Multiple signals allow greater environmental resolution. (A) With one auto-inducing signal molecule, populations can discriminate two environmental states in density/mass-transfer space (0,1). (B) With two signals, populations can, in principle, discriminate four states [(0,0),(0,1),(1,0), and (1,1)], if one signal (red) is more fragile than the other (blue). Signal-molecule concentration dynamics are given by by dSk/dt = (p + akSk)N − (m + uk)Sk, where N is cell density, 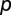 is baseline production,
is baseline production, 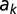 is the increased production due to autoinduction,
is the increased production due to autoinduction, 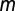 is mass-transfer rate, and
is mass-transfer rate, and 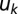 is decay rate. We define a signal to be “ON” (autoinduced) when
is decay rate. We define a signal to be “ON” (autoinduced) when 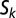 is above an unstable equilibrium
is above an unstable equilibrium 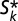 = Np/(m − akN + uk), which occurs when
= Np/(m − akN + uk), which occurs when 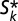 is negative: that is, when Nak
> m + uk. See SI Text for details.
is negative: that is, when Nak
> m + uk. See SI Text for details.
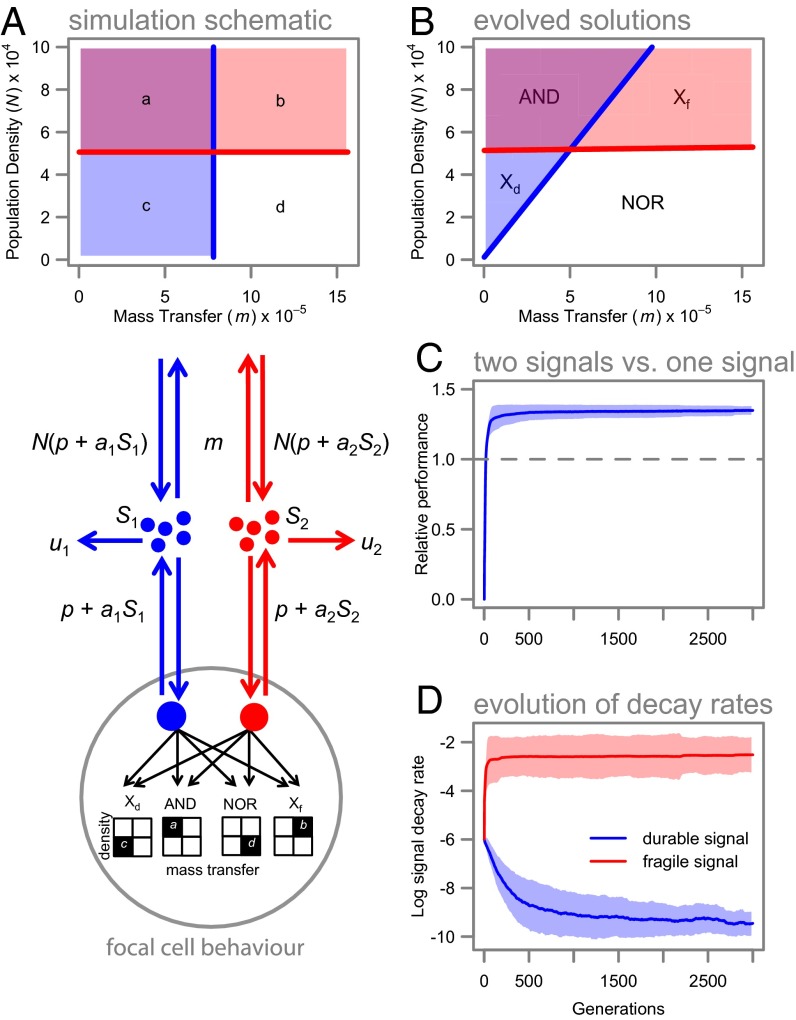
We next ask whether appropriate autoinduction and decay-parameter combinations (as in Fig. 1B) can emerge from an evolutionary process. Using individual-based simulations (in silico populations of evolving bacteria) (Fig. 2A), we test whether bacteria can evolutionarily tune a two-signal system to resolve variation in density and mass transfer into four sectors and respond with appropriate gene expression (Fig. 2A; and see SI Text for details). As predicted, two-signal populations rapidly outperformed the best one-signal strategy (Fig. 2C) by evolving distinct and positively covarying signal half-lives and autoinduction constants (Fig. 2D and Fig. S3), allowing an effective matching of gene expression (Fig. 2B) to environmental targets (Fig. 2A). Fig. 2B illustrates that, in the absence of any constraints on the rate of decay, a sufficiently fragile signal molecule can provide a robust discrimination of density (independent of fluctuations in mass transfer). On the other hand, estimates of mass transfer with a single signal are inherently confounded by variation in density (contrast blue and red thresholds in Fig. 2B). Therefore, in challenges requiring the discrimination of density and mass transfer, combinatorial responses to two physically distinct signals are likely to provide greater resolution. The association between variable environmental challenges and multisignal complexity is supported by the particularly complex multisignal QS found in highly generalist bacteria such as Pseudomonas aeruginosa and Burkholderia thailandensis (3, 15).
Fig. 2.
Multiple signals enhance fitness in evolving in silico simulations. (A) Schematic model of agent-based simulation structure. In silico populations evolve in environments characterized by four distinct regimes (a–d) of density and mass transfer, each requiring a distinct pattern of gene expression. (B) The behavior (gene-expression pattern) of the best performing clone in our simulations, with colors corresponding to the target environments in A. The use of multiple signals allows individuals to approximately map their gene expression to the different environments. Logic gates for each regulon are also indicated (Xf indicates “exclusive fragile” and Xd indicates “exclusive durable” gates). (C) The performance of a two-signal system, relative to the theoretical maximal performance of a single-molecule system (dashed line) when environments are fluctuating. Values shown are means ± 1 SD. (D) Signal-decay properties quickly diverge in the two-signal case. Values are log10 of the signal-decay rate ± 1 SD. See SI Text, Simulation Model for full details of the simulation procedure.
We test our principal predictions (differential decay of, and combinatorial responses to, multiple signals) using the opportunistic pathogen P. aeruginosa (16). In this organism, QS regulates a large fraction of the genome, including many secreted factors responsible for virulence (3, 17). To test for differences in signal durabilities, we measured signal decay in a variety of distinct growth environments. The two primary signal molecules of P. aeruginosa are the homoserine lactones N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) and N-butyryl-homoserine lactone (C4-HSL). Although the absolute rates of decay of these molecules differ nearly 10-fold across environments, in each environment, C4-HSL had a half-life approximately twice that of 3-oxo-C12-HSL (Table 1).
Table 1.
The primary P. aeruginosa signal molecules show a robust near twofold difference in decay rate across a diverse set of growth environments
| Environment | Decay Constants, h−1 (95% CI) |
Ratio | P value | |
| 3-Oxo-C12-HSL | C4-HSL | |||
| BHI | 0.34–0.39 | 0.17–0.22 | 1.87 | 7.3E−15 |
| KB | 0.04–0.05 | 0.02–0.03 | 1.70 | 6.0E−13 |
| LB | 0.07–0.08 | 0.04–0.05 | 1.69 | 4.4E−11 |
| M9 | 0.15–0.17 | 0.09–0.10 | 1.66 | 3.4E−18 |
The absolute decay rates of 3-oxo-C12-HSL and C4-HSL vary nearly 10-fold across environments, but the rates are tightly correlated between signals, so that C4-HSL is consistently around 1.7-fold more fragile. Decay constants were measured across rich laboratory growth media, plus a defined minimal medium: BHI, KB, LB, and M9, respectively (See SI Text, Signal Half-Life Experiments). Decay rates are represented as the 95% confidence intervals (CI) of proportional decay per hour. The P values indicate a significant difference between the decay rates of 3-oxo-C12-HSL and C4-HSL in that environment (ANOVA Interaction term in a log-linear model). Decay rates were measured using engineered bioreporters, the luminescence of which indicated the concentration of an AHL solution compared with a calibration of known concentrations (Figs. S4 and S5).
To examine responses to distinct environmental signal distributions, we assessed gene-expression level via microarray in a double-signal synthase mutant strain (PAO-JG1) (SI Text) under four signal-addition environments: both signals, one signal (3-oxo-C12 HSL or C4 HSL), or neither. As discussed above, our simulation model had the challenge of discriminating four distinct environments (Fig. 2A), resulting in four distinct regulons (sets of coregulated genes), each controlled by a distinct logic gate (Fig. 2B). We now ask: How many distinct regulons does PAO1 display in combinatorial response to two signal inputs? Using a model-selection approach based on information criteria (SI Text), we identified 14 quantitatively distinct regulons [95% confidence interval (CI) 12–15] featuring seven qualitatively distinct logic gates (Fig. 3A), highlighting the abundance and diversity of combinatorial responses to multiple signal inputs.
Fig. 3.
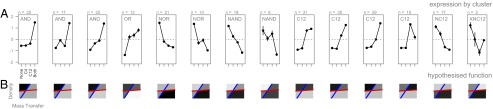
P. aeruginosa responds combinatorially to multiple signal inputs. (A) The set of QS-regulated genes in P. aeruginosa is partitioned into 14 distinct regulons, differentiated by distinct expression patterns across the four signal-addition treatments. The line plots represent the mean-centered and scaled expression profiles for each cluster of genes. Coarse-graining the expression data to discrete on/off states allows assignment of discrete logic-gate families, highlighting the prevalence and diversity of combinatorial processing rules. (B) Combining the distinct combinatorial responses to dual signal inputs with knowledge of relative signal stability (Table 1), we use our model to infer under which population-density and mass-transfer regimes each gene cluster would be expressed. The region plots (B) represent the inferred density/mass transfer target. Gene expression was measured using microarray. Cultures of PAO-JG1 were initiated with either (i) no signals, (ii) 15 µM C4-HSL, (iii) 15 µM 3-oxo-C12-HSL, or (iv) both 15 µM C4-HSL and 15 µM 3-oxo-C12-HSL in shaken cultures of LB broth for 8 h before RNA extraction. Regulon partitioning was achieved via k-means clustering, with selection of cluster number by BIC minimization (Fig. S6). Regulon gene content is detailed in Fig. S7.
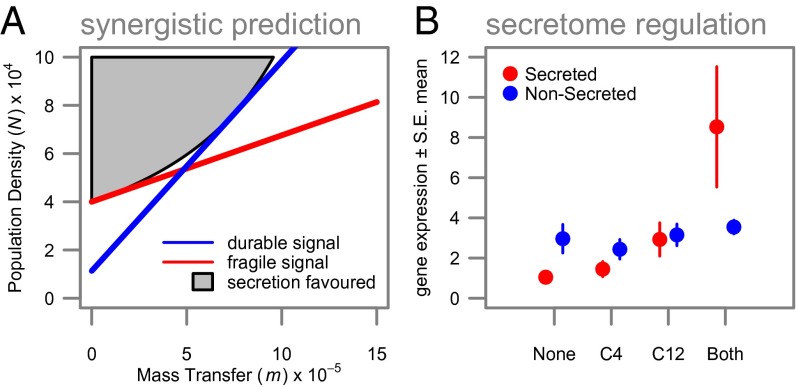
By combining the relative decay properties of the two signal molecules (Table 1) with the combinatorial responses to both signals (Fig. 3A), we can use our model assumptions to infer the environmental targeting for each regulon (Fig. 3B). QS is commonly associated with the control of secreted factors (13), and recent work has demonstrated that secreted factors are likely to be most beneficial under conditions of high density (17). We consider a mathematical model for the dynamics of a secreted digestive enzyme catalyzing the production of a beneficial digestive product and demonstrate (details in SI Text) that, if the digestive product is also subject to removal by mass transfer, then the production of the secreted factor will be most beneficial under high densities, but increasing mass-transfer rates will take an accelerating toll on the concentration of the digestive product (Fig. 4A). We therefore predict that secretome genes will preferentially be under synergistic “AND” gate control, turning on preferentially under the joint influence of both signal molecules to restrict expression to high-density/low-removal environments. In accordance with our functional prediction, we found that, whereas nonsecretome genes show evidence of interference when both signals are present (β= –0.297, SE = 0.061, P < 0.0001), secretome genes show synergistic expression in the presence of both signals (β = 0.522, SE = 0.241, P = 0.0309) (SI Text, Fig. 4B, and Table S1).
Fig. 4.
Secretome genes are under adaptive synergistic (AND-gate) control. (A) Secreted factors are predicted to be beneficial in high-density, low mass-transfer environments. The (gray) region where there is enough product to be advantageous is upward-curved and can be better approximated with two signals (versus one), with a synergistic response. The concentration of secreted molecules is described by dX/dt = PN − (m + f)X, where P and f are the production and decay rates; the formation of the beneficial product is described by dY/dt = qX − (cN + m + e)Y, where q is the rate of conversion, c is the rate of per-cell consumption, and e is its decay rate (SI Text and Fig. S8). (B) Gene expression among QS-regulated genes for secreted proteins (red) and nonsecreted proteins (blue). Expression of nonsecretome genes shows interference between signals; expression is lower when both signals are present than the sum of the individual effects of the signals. However, as predicted, secretome genes show synergistic expression in response to the combination of signals: i.e., expression is higher when both signals are present than the sum of the individual signal effects. Data shown are means ± SEM.
Discussion
Despite detailed mechanistic understanding of the intracellular genetic architecture of QS, the functional roles of QS remain controversial (12, 19). We propose a new functional model of “combinatorial QS” and demonstrate theoretically that bacteria can infer both their social (density) and physical (mass-transfer) environment, given combinatorial (nonadditive) responses to multiple signals with distinct half-lives. In support of our theoretical models, we show that P. aeruginosa displays diverse combinatorial gene-expression responses to two signals with differential rates of decay and uses a specific AND-gate response rule to limit the expression of costly secreted factors to the most beneficial high-density, low mass-transfer environments.
Our results support the hypothesis that bacteria use combinatorial processing of multiple QS signals to simultaneously match gene expression to both social and physical properties of their environment. However, other hypotheses exist for the functional role of multiple QS signal molecules. The first, inspired by the widespread production of the QS molecule AI-2 across distantly related bacteria, suggests that the ratio between differing QS molecules provides bacteria with information on community composition, as some molecules are produced only by conspecific individuals whereas others are common among species (20). This argument offers a rationale for individuals to monitor generic QS molecules. However, unlike our model, it does not offer a rationale for investment in these molecules, which are arguably better interpreted as by-product cues that may advertise a cell’s presence to competitors instead of being a signal in an evolutionary sense (21). More recently, it has been proposed that, if autoinducers naturally accumulate in a sequential order during single-species population growth, bacteria can distinguish phases in population development from total signal concentration (22, 23). A last possibility is that multiple signals simply allow for multiple thresholds for gene expression if bacteria require information on multiple-density thresholds (24). The latter two alternative adaptive explanations are unlikely whenever there is strong combinatorial integration as we demonstrate for P. aeruginosa (Fig. 3), but, to our knowledge, no such analysis of combinatorial responses has been conducted on a whole-genome scale in other bacteria.
We have focused in this manuscript on a general functional argument for combinatorial quorum sensing and have addressed the question of why many bacterial species produce multiple signal molecules. In our general analysis, we sacrificed several system-specific mechanistic details. Most significantly, we have assumed that the two autoinduction pathways are independent of each other. In fact, a significant degree of information processing in P. aeruginosa could occur by interactions between signaling pathways during autoinduction (25); whether a bistable threshold is reached or not may depend on the quantities of both molecules. Also, many bacteria use more than two signals (Table S2). It is possible that these bacteria are able to resolve more axes of environmental ambiguity (for instance, advection versus diffusion) by combinatorially processing signals that differ in distinct physical dimensions (for instance, diffusion constants). We have also simplified the ecological context, assuming shared interest (i.e., clonality) among interacting bacteria. Bacterial groups are often nonclonal, making the evolution of QS a social dilemma (24, 26). When multiple signals are combinatorially processed, we expect the range of possible QS strategies to be significantly greater.
Bacteria sense a significant amount of environmental information directly (27), but some environmental dimensions require indirect, signal-mediated sensing mechanisms. When multiple molecules are secreted and differentially affected by the environment, the information a bacterium can acquire about its social and physical environment can be greatly increased. The combinatorial use of multiple signals is a hallmark of human language and has recently been recognized in one other primate species (28, 29). Our results show that combinatorial communication has a much broader taxonomic distribution and is computationally achievable in single-celled organisms.
Materials and Methods
Simulation Model.
We consider a population of 1,000 strains interacting in clonal, well-mixed groups of variable cellular densities and mass-transfer rates. These in silico bacteria were set the task of matching the expression of four different regulons to appropriate levels of cellular density and mass transfer. Both the decay and autoinduction parameters of each strain’s two signals were allowed to evolve with mutation using a genetic algorithm, with strain fitness decided by the match between gene expression and the environments they experienced. We ran 100 replicate simulations for 3,000 generations of the genetic algorithm, recording strain performance and the evolutionary trajectories of signal-decay rates and autoinduction parameters (see SI Text for further details and parameter values).
Signal Decay.
The activity of synthetic N-Acyl homoserine lactones (AHLs) was tracked through time in distinct media, using appropriate biosensor strains (pSB536 and pSB1075 for C4-HSL and 3-oxo-C12 HSL, respectively). Biosensor luminescence per cell was calculated as relative light units per optical density at 600nm and compared with a calibration curve of known 2× serially diluted concentrations to determine AHL concentrations. Decay rates were determined by fitting log-linear models to AHL concentrations over time.
Microarray Experiments.
The effects of the different signal molecules were assessed using a double QS synthase mutant of P. aeruginosa PAO1 lasI/rhlI grown at 37 °C in 25 mL of LB broth and 250-mL flasks with shaking at 200 rpm (approximately 2.2 × g) in four treatments: (i) no addition; (ii) 3-oxo-C12-HSL; (iii) C4-HSL; and (iv) both 3-oxo-C12-HSL and C4-HSL. RNA was extracted from each culture after 8 h incubation (late exponential/early stationary phase of growth). Following Quantile normalization, differential expression was identified using Bayesian adjusted t statistics with false discovery rate correction for multiple testing.
Bioinformatic Analysis.
The genes in which differential expression was observed were then clustered across the four treatments using k-means clustering of mean standardized expression values. The most likely number of clusters was determined using the Bayesian information criterion (BIC). For each cluster, the best fitting on/off (i.e., Boolean) logic gate was identified.
Further details of experimental, simulation, and mathematical methods are given in SI Text.
Supplementary Material
Acknowledgments
We thank Rosalind Allen, Bartek Waclaw, Freya Harrison, and Iain Couzin for comments, and Victoria Wright for performing the microarray experiment. We thank the Wellcome Trust (WT082273 and WT095831), the Engineering and Physical Sciences Research Council (EP/H032436/1), the Natural Environment Research Council (NE/J007064/1), the Royal Society, the University of Edinburgh School of Biological Sciences, and the Centre for Immunity, Infection and Evolution for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE55110).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319175111/-/DCSupplemental.
References
- 1.Perkins TJ, Swain PS. Strategies for cellular decision-making. Mol Syst Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11):a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 4.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 5.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: A new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 6.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nature Reviews Microbiology. 2014 doi: 10.1038/nrmicro3232. 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swift S, Throup JP, Williams P, Salmond GP, Stewart GS. Quorum sensing: A population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21(6):214–219. [PubMed] [Google Scholar]
- 9.Salmond GP, Bycroft BW, Stewart GS, Williams P. The bacterial ‘enigma’: Cracking the code of cell-cell communication. Mol Microbiol. 1995;16(4):615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 10.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 11.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10(8):365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 12.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20(12):586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5(3):230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73(6):1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams P, Winzer K, Chan WC, Cámara M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schurek KN, Breidenstein EBM, Hancock REW. In: Antibiotic Discovery and Development. Dougherty TJ, Pucci MJ, editors. New York: Springer; 2012. pp. 679–715. [Google Scholar]
- 17.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185(7):2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109(21):8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt TG, Fuqua C. What’s in a name? The semantics of quorum sensing. Trends Microbiol. 2010;18(9):383–387. doi: 10.1016/j.tim.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 21.Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long T, et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7(3):e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anetzberger C, et al. Autoinducers act as biological timers in Vibrio harveyi. PLoS ONE. 2012;7(10):e48310. doi: 10.1371/journal.pone.0048310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SP, Johnstone RA. Cooperation in the dark: Signalling and collective action in quorum-sensing bacteria. Proc Biol Sci. 2001;268(1470):961–965. doi: 10.1098/rspb.2001.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12(2):182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 27.Cornforth DM, Foster KR. Competition sensing: The social side of bacterial stress responses. Nat Rev Microbiol. 2013;11(4):285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 28.Fitch WT. The Evolution of Language. Cambridge, UK: Cambridge Univ Press; 2010. [Google Scholar]
- 29.Hurford JR. The Origins of Grammar. Oxford: Oxford Univ Press; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.