Significance
Metabolism of l-carnitine, a compound abundant in human diet, to trimethylamine by human microbiota has been shown to promote atherosclerosis and subsequent development of heart disease. However, the underpinning molecular and biochemical mechanisms remain unknown. In this study, we reveal that a previously unidentified Rieske-type protein is responsible for carnitine transformation to trimethylamine from human microbiota. Knowledge gained in our study provides the opportunity not only to explore Rieske protein inhibitors in preventing trimethylamine formation in animal studies and clinical trials, but also for its use as a functional genetic marker to better understand human microbiota and their dynamics in our health and disease in future epidemiological studies and dietary interventions.
Keywords: methylated amine metabolism, comparative genomics, gut microbiota
Abstract
Dietary intake of l-carnitine can promote cardiovascular diseases in humans through microbial production of trimethylamine (TMA) and its subsequent oxidation to trimethylamine N-oxide by hepatic flavin-containing monooxygenases. Although our microbiota are responsible for TMA formation from carnitine, the underpinning molecular and biochemical mechanisms remain unclear. In this study, using bioinformatics approaches, we first identified a two-component Rieske-type oxygenase/reductase (CntAB) and associated gene cluster proposed to be involved in carnitine metabolism in representative genomes of the human microbiota. CntA belongs to a group of previously uncharacterized Rieske-type proteins and has an unusual “bridging” glutamate but not the aspartate residue, which is believed to facilitate intersubunit electron transfer between the Rieske center and the catalytic mononuclear iron center. Using Acinetobacter baumannii as the model, we then demonstrate that cntAB is essential in carnitine degradation to TMA. Heterologous overexpression of cntAB enables Escherichia coli to produce TMA, confirming that these genes are sufficient in TMA formation. Site-directed mutagenesis experiments have confirmed that this unusual “bridging glutamate” residue in CntA is essential in catalysis and neither mutant (E205D, E205A) is able to produce TMA. Taken together, the data in our study reveal the molecular and biochemical mechanisms underpinning carnitine metabolism to TMA in human microbiota and assign the role of this novel group of Rieske-type proteins in microbial carnitine metabolism.
It is increasingly clear that the human microbiota plays an essential role in our health and disease (1–5). Understanding the metabolic potential of the human microbiota and its interaction with and regulation by the host and the environment holds the key to unravel the dynamic relationship between ourselves and our associated microbes (4–6). Over the last decade, our knowledge of the human microbiota has been significantly improved thanks to technological advances, including high-throughput sequencing, development of powerful bioinformatics, and the use of germ-free animal models (7–13). We can now not only characterize the taxonomic composition, species richness and dynamics, but combine the genetic potential encoded in human microbiota and establish the core metabolic pathways enabled by direct sequencing of the human microbiome (14, 15).
Advances in our understanding of the microbiome functions, however, do not match the pace of taxonomic characterization of species diversity and dynamics (16–18). Fully resolving the functional capacity encoded in the human microbiome and the dynamic effects on health and disease still remains a great challenge (11, 16–18). Direct sequencing of the human microbiome generates datasets dominated by genes encoding pathways for central metabolism (e.g., transcriptional and translational machinery, ATP synthesis, and so forth) (14, 15), therefore contributing little to our understanding of the variable functional capacity encoded in human microbiota between individuals (7, 8). Furthermore, a large fraction of the genes encoded in the human microbiome remain to be functionally characterized (14, 15). Assigning functions encoded in the human microbiome using existing databases can be problematic. For example, the Pfam protein database currently contains over 25% of protein families with no assigned functions (release 26.0) (19).
Lack of functional characterization of key microbial functions in our microbiota is exemplified by very recent studies on cardiovascular diseases (20–23). These studies have shown that the human microbiota is responsible for the production of trimethylamine N-oxide (TMAO), which is believed to promote atherogenesis through its interaction with macrophages and lipid metabolism (20–23). TMAO is derived from the microbial metabolism of dietary quaternary amines [e.g., choline, l-carnitine, glycine betaine (GBT), and phosphatidylcholine] to trimethylamine (TMA), which is subsequently oxidized to TMAO by the host hepatic flavin monooxygenases (21, 22). l-Carnitine (hereafter referred to as carnitine, unless otherwise specified) is considered an important nutrient for human health, playing a key role in mitochondrial fatty acid β-oxidation (24, 25). Although carnitine can be acquired through endogenous biosynthesis, our daily demand is largely met through dietary intake from carnitine-containing food (26). It is known that a significant proportion of dietary carnitine can be further metabolized by microbiota before absorption (20, 27). This microbial-mediated metabolic pathway not only diverts carnitine away from the host, causing conditional carnitine deficiency in certain human populations, but promotes TMAO formation and subsequent increased risk of atherosclerosis (20, 21). However, the underlying genetic and biochemical mechanisms of carnitine-dependent TMA production in human microbiota have not been uncovered.
In this report, we describe the identification of the genetic and biochemical mechanisms for TMA production from carnitine in representative human microbiota (Gammaproteobacteria, Betaproteobacteria, Firmicutes) through a synthesis of bioinformatics, genetic, and biochemical approaches. These in vivo experiments with microbial isolates prove valuable for exploring the metabolic attributes of a particular group of microbes in carnitine metabolism guided by bioinformatics (10). This recently identified carnitine-to-TMA enzyme is composed of an oxygenase component (CntA) and a reductase component (CntB). CntA belongs to a previously uncharacterized group of Rieske-type proteins, which are best known for ring-hydroxylation of aromatic hydrocarbons (28). CntA has an unusual alteration of the so-called “bridging” aspartate (substituted by a glutamate residue, E205), which plays an essential role in electron transfer in catalysis in the Rieske-type protein family (29–32).
Results and Discussion
Discovery of a Carnitine Utilization Gene Cluster and Identification of a Candidate Carnitine Oxygenase (cntAB).
We used the Human Microbiome Project (HMP) reference genomes (15) for mining for carnitine-degrading enzymes guided by the following hypotheses. Microbial conversion of carnitine to TMA has been studied in bacteria isolated from humans, Acinetobacter calcoaceticus (33) and Serratia marcescens (34). It is known that the cleavage of the carbon (C)–nitrogen (N) bond of carnitine produces TMA and a four-carbon (C4) molecule, which likely enters the central tricarboxylic acid cycle in the form of malate or succinate. We therefore hypothesize that the enzyme responsible for splitting the C-N bond is clustered in the genome with enzymes responsible for the synthesis of malate and succinate because the C4 unit is further used as a carbon source in these bacteria (33, 34).
We further reasoned that a transporter is needed for microbial carnitine transport. Two types of bacterial carnitine transporters are known, a BCCT-type permease (35) and an ABC-type active transporter of the choline/betaine/carnitine family (36). We therefore used a BLASTP algorithm to search the HMP reference genomes (754 were available as of February 2013) for genes encoding either CaiT (a BCCT type carnitine-specific antiporter) or CaiX (the carnitine-specific substrate binding protein of the ABC transporter cassette). Our BLASTP search data revealed that many gammaproteobacterial HMP genomes contain CaiT but not CaiX homologs (Table S1). CaiT is also found in some Betaproteobacteria and Firmicutes.
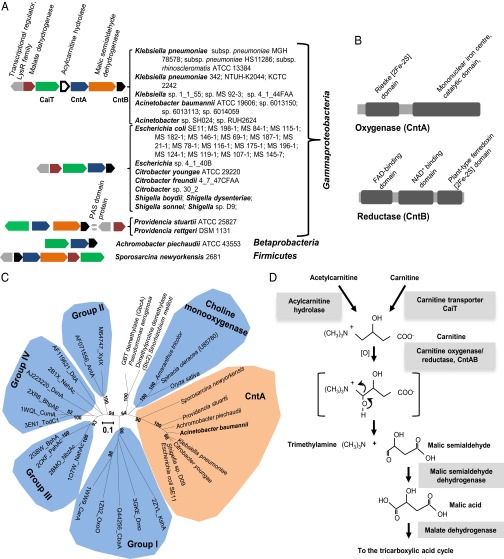
We then inspected the neighborhood of caiT for genes involved in malate or succinate metabolism and this resulted in the identification of five groups of gene clusters in 39 HMP reference genomes (Fig. 1A), with representative isolates from various sites of the body, including skin, airway, gastrointestinal tract, and feces (Table S2). We focused our analyses on Acinetobacter spp. because they are known to degrade carnitine to TMA (33). A close investigation of the caiT neighborhood revealed the presence of genes likely to be involved in C4 metabolism (Fig. 1A). These include genes coding for a malic semialdehyde dehydrogenase and a malate dehydrogenase, together channeling the C4 carbon into the central tricarboxylic acid cycle. A conserved lysR-type transcriptional regulator is present in these gene clusters. Immediately downstream of the carnitine transporter caiT is the gene encoding an acylcarnitine hydrolase, which is required for growth and hydrolysis of acylcarnitine to carnitine (37). The functions of two other genes—which are always present in the caiT gene cluster—are not explicit; we designate these as cntA and cntB, respectively.
Fig. 1.
Identification of the putative Rieske-type protein from human microbiota in carnitine-dependent TMA formation. (A) Putative carnitine-to-TMA gene cluster in representative genomes of human microbiota. CaiT, carnitine transporter; CntA, A Rieske-type oxygenase protein; CntB, a predicted reductase with a plant-type ferridoxin domain. (B) Analyses of conserved domains in CntA and CntB. FAD: flavin adenine dinucleotide; NAD+: nicotinamide adenine dinucleotide. (C) An unrooted phylogenetic tree (∼305 aa) of CntA, microbial Rieske-type terminal oxygenases (groups I–IV) and eukaryotic Rieske-type choline monooxygenases. Microbial Rieske-type terminal oxygenases (groups I–IV) are classified based on the nomenclature system of Nam et al. (57). These sequences are identified by a unique GenBank or PDB accession number followed by the gene name; other sequences are identified by the name of the species. Bootstrap values greater than 50 are shown (100 replicates). The scale bar represents one substitution per 10 amino acids. (D) Predicted pathway of carnitine catabolism via CntA/B.
Bioinformatic analyses predicted that cntB encodes a NAD(P)-dependent reductase, containing a flavin-binding domain as well as a plant-type ferredoxin [2Fe-2S] domain (Fig. 1B and Fig. S1). CntA is predicted to be a member of the Rieske-type protein family, which is characterized by two histidine and two cysteine residues coordinating the [2Fe-2S] cluster (28). Sequence analyses of CntA from HMP bacterial genomes showed conserved Rieske motif and a catalytic mononuclear iron domain (Fig. 1B). To date, most characterized microbial Rieske-type proteins are involved in the catabolism of ring-structured aromatic compounds (Table S3) (38). However, recent bioinfomatic and evolutional analyses suggest that Rieske-type proteins are much more diverse than previously thought (38–40). Indeed, eukaryotic Rieske-type terminal oxygenases have also been identified [e.g., choline monooxygenase (41, 42)], and recent studies have confirmed that Rieske-type proteins can carry out nonring hydroxylation reactions, such as oxidative demethylation (43) and oxidative carbocyclization (44). Furthermore, many novel Rieske-type proteins have been identified with no assigned function, many of which originated from newly sequenced microbial genomes (38). Our phylogenetic analyses reveal that CntA forms a distinct group in the Rieske-type protein family, which is more closely related to the eukaryotic choline monooxygenases (Fig. 1C). Our bioinformatic and phylogenetic analyses therefore suggest that cntAB may encode a novel enzyme that catalyses the initial step of carnitine degradation to TMA (and a C4 compound), although C-N bond cleavage by Rieske and Rieske-type enzymes has not previously been reported. The predicted pathway for carnitine metabolism to TMA is shown in Fig. 1D.
Deletion of Either cntA or cntB Abolishes TMA Formation from Carnitine in Acinetobacter baumannii.
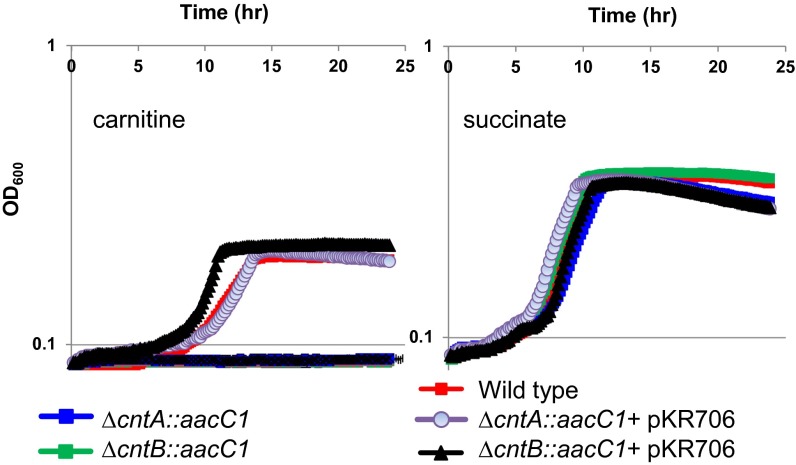
To test whether this gene cluster is indeed involved in carnitine transformation, we used Acinetobacter baumannii ATCC19606 as a model. The genetics for A. baumannii has been established and Acinetobacter spp. are known to degrade carnitine to TMA (33). This bacterium was cultivated in a defined medium with carnitine as the sole carbon source to establish whether or not it could produce TMA from carnitine. As predicted from the bioinformatics analyses, this strain could grow on carnitine as the sole carbon source (Fig. 2). TMA production was observed in the culture supernatant supplemented with carnitine but not succinate. We then carried out marker-exchange mutagenesis to investigate if cntA/cntB genes are essential in carnitine-dependent TMA production in this strain. The results shown in Fig. 2 demonstrate that the mutants lacking either cntA or cntB, which was replaced by a gentamicin-resistance gene cassette (aacC1), could no longer grow on carnitine as a sole carbon and energy source, whereas the growth on succinate was not affected. Furthermore, when the mutants were complemented with the native copy of the carnitine degradation gene cluster, their ability to grow on carnitine was restored (Fig. 2).
Fig. 2.
Growth of A. baumannii ATCC19606 wild-type, mutants (∆cntA::aacC1, ∆cntB::aacC1), and complemented mutants with plasmid pKR706 on carnitine (10 mM) or succinate (20 mM) as the sole carbon and energy source. The error bars represent SD from experiments run in eight replicates.
We performed further experiments to validate the mutants by quantifying TMA production and carnitine consumption from the wild-type, the mutants, and the complemented mutants of A. baumannii ATCC19606 (Fig. 3). Because the mutants did not grow on carnitine alone, we supplemented the medium with carnitine and succinate. As predicted, both mutants lost the ability to catalyze TMA formation from carnitine although they could grow on succinate as the sole carbon and energy source, whereas the complemented mutants restored the ability to convert carnitine to TMA. Taken together, the experiments confirmed that cntA and cntB are essential in TMA formation from carnitine.
Fig. 3.
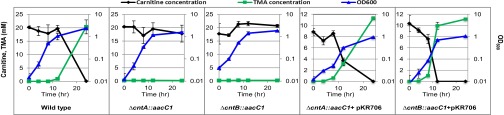
Quantification of TMA and carnitine of wild-type, mutants (∆cntA::aacC1, ∆cntB::aacC1), and complemented mutants with plasmid pKR706 in the culture medium supplemented with carnitine and succinate. The error bars represent SD from experiments run in triplicate.
Heterologous Expression of cntAB in Escherichia coli.
To complement our experiment in vivo, we performed further experiments in vitro by heterologous expression of cntAB in Escherichia coli. We cloned these two genes (cntA, cntB) from A. baumannii into an inducible T7 promoter-specific expression system to assess whether they are sufficient to perform carnitine-dependent TMA production heterologously. Using ion-exchange chromatography, we quantified TMA formation from carnitine using the supernatant of recombinant E. coli containing either overexpressed CntA or CntB, and no TMA production was seen (Fig. 4). However, when the two genes were coexpressed, TMA was detected from carnitine degradation using the cell-free culture extracts.
Fig. 4.
Quantification of TMA formation in vitro using the supernatant of recombinant E. coli containing overexpressed CntA, CntB, or CntAB, respectively. The error bars represent SD from experiments run in triplicate.
To further confirm that CntAB is sufficient for carnitine-dependent TMA production, we purified CntA and CntB by affinity chromatography and successfully reconstituted the activity of carnitine-to-TMA degradation using the purified recombinant proteins (Figs. S2 and S3). The identity of TMA produced from carnitine oxidation by CntAB was further confirmed by gas chromatography–mass spectrometry using authentic TMA standards (Fig. S3). Overall, our experiments demonstrate that CntA and CntB are necessary and sufficient for in vitro carnitine degradation to TMA.
The Unusual “Bridging Glutamate” Residue Is Essential in Carnitine Oxidation.
Phylogenetic analyses place the CntA protein within the Rieske-type protein family (Fig. 1C). This finding is confirmed by the presence of the highly conserved Rieske sequence motif [-CXHX15–17CXXH-] in CntA in HMP reference genomes (Fig. 5A). A close investigation of the catalytic mononuclear iron center revealed the conserved two-histidine-one-carboxylate facial triad motif in these CntA proteins (45). However, a key difference between CntA and other Rieske-type terminal oxygenases lies in the unusual substitution of the previously identified, highly conserved aspartate residue located immediately in front of the first histidine residue of the mononuclear iron center (E205) (Fig. 5A). This glutamate residue is, however, strictly conserved in all CntA proteins identified from the 39 genomes of HMP reference strains. Although the aspartate-to-glutamate substitution is conservative, it is rather unusual. X-ray crystal structures of Rieske oxygenases (e.g., naphthalene 1,2-dioxygenase, biphenyl 2,3-dioxygenase, nitrobenzene 1,2-dioxygenase) showed that the mononuclear iron and the Rieske center on the same subunit are too far (>40 Å) to allow direct electron transfer between them (46–49). The mononuclear iron centers, however, are located only ∼12 Å from the Rieske centers on the adjacent subunits of the homotrimers (46–49). It is believed that intersubunit electron transfer occurs between the ligating histidine residues which are coordinated by a so-called “bridging aspartate” residue (29–32) (Fig. S4), and substitutions of this aspartate residue to glutamate (D205E in naphthalene dioxygenase) severely diminished its catalytic activity (29).
Fig. 5.
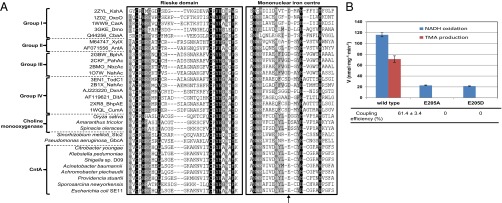
The unusual substitution of “bridging” aspartate to “bridging” glutamate (E205 in CntA) abolished catalytic activity for carnitine-dependent TMA formation. (A) Multiple sequence alignment of CntA from representative human microbiota and related Rieske-type proteins. The two boxes highlight the conserved Rieske domain and the mononuclear iron center domain. The arrow indicates the unusual but conserved glutamate residue in CntA. Each sequence has a unique identifiable label as shown in the legend of Fig. 1C. (B) Quantification of NADH oxidation and TMA production in vitro using purified CntA and site-directed mutants (E205A, E205D) in combination with purified CntB. Coupling efficiency is determined as the ratio of the total amount of TMA formed to the amount of NADH consumed. The error bars indicate SD from experiments run in triplicate.
To gain more insight into the role of this glutamate residue in CntA, we performed site-directed mutagenesis experiments, changing this glutamate residue to either aspartate (E205D) or alanine (E205A). The mutants were purified from recombinant E. coli and activity assays were performed by quantifying NADH oxidation and TMA production. The results shown in Fig. 5B demonstrate that this glutamate residue is essential in electron transfer between the Rieske center and the mononuclear iron center because electron transfer (as determined by coupling efficiency and TMA formation) was completely abolished in the mutants. Experiments using circular dichroism and native-PAGE demonstrated that site-directed mutagenesis of CntA neither altered its secondary structure nor the oligomeric state of the protein (Fig. S5). Our data, therefore, indicate that in CntA, a glutamate—but not aspartate—residue in this position is crucial in catalysis because neither of the mutants (E205A, E205D) could catalyze carnitine degradation to TMA (Fig. 5B). It is interesting to note that the glutamate residue is found in at least two other enzymes of the Rieske-type protein family, dimethylproline demethylase (Stc2) (43) and GBT demethylase (GbcA) (50), although no further data were available on the role of this residue in these proteins. Our study therefore suggests that caution needs to be taken when interpreting structure-function relationships of Rieske-type proteins using existing structures that are dominated by ring-hydroxylating oxygenases. Further X-ray structures are clearly warranted to reveal the function-structure relationships of an expanded Rieske-type protein family as revealed by bioinformatics approaches (38–40).
Concluding Remarks
The last decade has witnessed unparalleled progress in research on the human microbiota and its complex and dynamic relationship to our health and disease. It has become increasingly evident that understanding the functional capacity encoded in the human microbiota and its variation between individuals is necessary for future personalized healthcare and targeted medication. A call for a community response to work collaboratively on the functional annotation of uncharacterized proteins was made almost a decade ago (51), and yet proteins of unknown function in prokaryotic and eukaryotic genomes are still increasingly accumulating (18, 19). Indeed, a large proportion of the proteins encoded in the human microbiome have not yet been functionally characterized (14, 15), leaving a serious gap in our knowledge for complete understanding of the role of our microbiota. On the other hand, a number of specific activities of human microbiota have been discovered, with the corresponding encoding genes and biochemical mechanisms remaining unknown (reviewed in refs. 4 and 11).
The key role played by human microbiota in TMA production has been known for more than a century, but we have just begun to understand the underpinning molecular and biochemical mechanisms (52). The vital importance of the microbiota-mediated metabolic pathway for TMA formation has been convincingly highlighted by several recent studies, linking increased levels of plasma TMAO, the metabolite produced by hepatic oxidation of TMA, with elevated risk of atherosclerosis and associated acute cardiovascular diseases in humans (20–23). Demystifying the genetic and biochemical mechanisms in TMA formation can yield novel targets for future diagnosis and form baseline knowledge for personalized therapeutic strategies targeting an individual’s microbiota. In this study, we report the identification of a unique Rieske protein involved in TMA formation from carnitine in human microbiota. Coincidently, Rieske proteins have been extensively studied over the last few decades and many crystal structures of Rieske proteins are readily available. Our finding now offers the opportunity to explore previously studied Rieske protein inhibitors in preventing TMA formation in animal studies and clinical trials (53). Furthermore, knowledge gained in our study and others (52) now offers the use of functional genetic markers (e.g., cntA, cutC), in addition to the taxonomic ribosomal RNA markers, to better understand our microbiota and their dynamics in human health and disease in large-scale epidemiological and dietary intervention studies. What’s more, beyond the specifics of this study, one can also envisage that the synthesis of genetic, biochemical, and bioinformatics approaches can be a valuable tool for future functional genomic studies of human microbiota, not only addressing emerging issues in microbiota-host cross-talk, but also starting to fill in the gap in our knowledge of proteins of unknown functions in human microbiome.
Materials and Methods
Bioinformatic Identification of the Carnitine Oxygenase Gene Cluster.
We used sequenced microbial genomes from the HMP as the database for mining the genes involved in carnitine-to-TMA degradation. HMP references genomes were selected and analyzed through the IMG program on the Joint Genome Institute Web site (www.hmpdacc-resources.org/cgi-bin/imgm_hmp/main.cgi). Searching for homologs encoding either the carnitine-specific ABC transporter or the BCCT-type permease was carried out through the BLASTP algorithm (E value −50) using the following queries sequences CaiX (PA5388) (36) and CaiT (CAA52110) (54). No CaiX homologs were found in the HMP reference genomes, including Serratia spp. and Acinetobacter spp. which have been previously shown to catalyze carnitine degradation to TMA (33, 34). We found 122 close homologs (E ≤ −50) of CaiT in 91 unique genomes of Gammaproteobacteria, Betaproteobacteria, and Firmicutes, including Acinetobacter species. Because cleavage of the trimethylammonium molecule from carnitine results in the release of a C4 carbon, the neighborhood of caiT was manually inspected for genes encoding enzymes involved in the metabolism of C4 molecules, including malate, succinate, fumarate, and oxaloacetate. Sequence alignment and phylogenetic analyses were performed as described previously (55).
Marker Exchange Mutagenesis of cntA/cntB in A. baumannii.
Cultivation of A. baumannii was carried out in a defined medium supplemented with either succinate or carnitine (or both) as the sole carbon source. Targeted deletion of cntA/cntB was carried out by marker exchange mutagenesis, as described previously (56). The mutants (∆cntA; ∆cntB) were complemented with plasmid pKR706 by cloning the native carnitine oxygenase gene cluster into the vector pMQ300 (SI Materials and Methods). Bacterial strains, plasmids and primers used in this study are listed in Tables S4 and S5.
Heterologous Overexpression of cntA/cntB, Site-Directed Mutagenesis and Characterization of the CntA Mutants.
The cntA gene was amplified from A. baumannii and inserted into the vector pET28a (Novagen). Coexpression of cntA/cntB was achieved by insertion into pCOLADuet-1 under the BamHI/HindIII and the NdeI/KpnI sites, respectively. The CntA mutants (E205D, E205A) were chemically synthesized by GenScript and inserted into the expression vector pET28a under the NdeI/HindIII sites. The resulting plasmids were then transformed into the expression host E. coli BLR(DE3) pLysS (Merck Biosciences). Cultivation of recombinant E. coli, protein induction by isopropyl β-d-1-thiogalactopyranoside and further purification using His-tag affinity chromatography are detailed in SI Materials and Methods. Enzyme assays were performed at room temperature (∼22 °C) by quantifying NADH oxidation and carnitine-dependent TMA production. A 1-mL enzyme assay mixture contained 10 mM Hepes buffer (pH 7.6), 60 μg purified CntA and CntB, respectively, 0.25 mM carnitine, and 0.25 mM NADH. Coupling efficiency was determined as the ratio of the total amount of TMA formed to the amount of NADH consumed.
Analytical Methods.
Carnitine and TMA were quantified by a cation-exchange ion chromatography (Metrohm 881 Compact IC Pro) equipped with a Metrosep C4/250 mm separation column and a conductivity detector (Metrohm). NADH oxidation was quantified using a Shimadzu UV-VIS 1800 spectrophotometer by following decrease of absorbance at 340 nm (ε = 6.2 mM−1·cm−1).
Supplementary Material
Acknowledgments
This work is supported by the Royal Society (RG2011/R1) and partially by the Natural Environment Research Council (NE/I027061/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316569111/-/DCSupplemental.
References
- 1.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 5.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 9.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489(7415):250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 11.Marchesi JR. Human distal gut microbiome. Environ Microbiol. 2011;13(12):3088–3102. doi: 10.1111/j.1462-2920.2011.02574.x. [DOI] [PubMed] [Google Scholar]
- 12.Arumugam M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 15.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proctor LM. The human microbiome project in 2011 and beyond. Cell Host Microbe. 2011;10(4):287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013;29(1):51–58. doi: 10.1016/j.tig.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin MY, Koonin EV. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010;28(8):398–406. doi: 10.1016/j.tibtech.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett BJ, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17(1):49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman S, Fraenkel G. Reversible enzymatic acetylation of carnitine. Arch Biochem Biophys. 1955;59(2):491–501. doi: 10.1016/0003-9861(55)90515-4. [DOI] [PubMed] [Google Scholar]
- 25.Fritz IB, McEwen B. Effects of carnitine on fatty-acid oxidation by muscle. Science. 1959;129(3345):334–335. doi: 10.1126/science.129.3345.334. [DOI] [PubMed] [Google Scholar]
- 26.Rebouche CJ. Carnitine function and requirements during the life cycle. FASEB J. 1992;6(15):3379–3386. [PubMed] [Google Scholar]
- 27.Rebouche CJ, Chenard CA. Metabolic fate of dietary carnitine in human adults: Identification and quantification of urinary and fecal metabolites. J Nutr. 1991;121(4):539–546. doi: 10.1093/jn/121.4.539. [DOI] [PubMed] [Google Scholar]
- 28.Ferraro DJ, Gakhar L, Ramaswamy S. Rieske business: Structure-function of Rieske non-heme oxygenases. Biochem Biophys Res Commun. 2005;338(1):175–190. doi: 10.1016/j.bbrc.2005.08.222. [DOI] [PubMed] [Google Scholar]
- 29.Parales RE, Parales JV, Gibson DT. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J Bacteriol. 1999;181(6):1831–1837. doi: 10.1128/jb.181.6.1831-1837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto A, Tarasev M, Ballou DP. Substitutions of the “bridging” aspartate 178 result in profound changes in the reactivity of the Rieske center of phthalate dioxygenase. Biochemistry. 2006;45(30):9032–9041. doi: 10.1021/bi060216z. [DOI] [PubMed] [Google Scholar]
- 31.Jiang H, Parales RE, Lynch NA, Gibson DT. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178(11):3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beharry ZM, et al. Histidine ligand protonation and redox potential in the rieske dioxygenases: Role of a conserved aspartate in anthranilate 1,2-dioxygenase. Biochemistry. 2003;42(46):13625–13636. doi: 10.1021/bi035385n. [DOI] [PubMed] [Google Scholar]
- 33.Seim H, et al. Splitting of the C-N bond in carnitine by an enzyme (trimethylamine forming) from membranes of Acinetobacter calcoaceticus. FEMS Microbiol Lett. 1982;15(3):165–167. [Google Scholar]
- 34.Unemoto T, Hayashi M, Miyaki K, Hayashi M. Formation of trimethylamine from DL-carnitine by Serratia marcescens. Biochim Biophys Acta. 1966;121(1):220–222. doi: 10.1016/0304-4165(66)90382-5. [DOI] [PubMed] [Google Scholar]
- 35.Ziegler C, Bremer E, Krämer R. The BCCT family of carriers: From physiology to crystal structure. Mol Microbiol. 2010;78(1):13–34. doi: 10.1111/j.1365-2958.2010.07332.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol. 2010;75(1):29–45. doi: 10.1111/j.1365-2958.2009.06962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meadows JA, Wargo MJ. Characterization of Pseudomonas aeruginosa growth on O-acylcarnitines and identification of a short-chain acylcarnitine hydrolase. Appl Environ Microbiol. 2013;79(11):3355–3363. doi: 10.1128/AEM.03943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capyk JK, Eltis LD. Phylogenetic analysis reveals the surprising diversity of an oxygenase class. J Biol Inorg Chem. 2012;17(3):425–436. doi: 10.1007/s00775-011-0865-9. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt CL, Shaw L. A comprehensive phylogenetic analysis of Rieske and Rieske-type iron-sulfur proteins. J Bioenerg Biomembr. 2001;33(1):9–26. doi: 10.1023/a:1005616505962. [DOI] [PubMed] [Google Scholar]
- 40.Kweon O, et al. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem. 2008;9:11. doi: 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rathinasabapathi B, et al. Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: Prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci USA. 1997;94(7):3454–3458. doi: 10.1073/pnas.94.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsuya S, et al. Isolation and characterization of a novel peroxisomal choline monooxygenase in barley. Planta. 2011;234(6):1215–1226. doi: 10.1007/s00425-011-1478-9. [DOI] [PubMed] [Google Scholar]
- 43.Daughtry KD, et al. Quaternary ammonium oxidative demethylation: X-ray crystallographic, resonance Raman, and UV-visible spectroscopic analysis of a Rieske-type demethylase. J Am Chem Soc. 2012;134(5):2823–2834. doi: 10.1021/ja2111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sydor PK, et al. Regio- and stereodivergent antibiotic oxidative carbocyclizations catalysed by Rieske oxygenase-like enzymes. Nat Chem. 2011;3(5):388–392. doi: 10.1038/nchem.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hegg EL, Que L., Jr The 2-His-1-carboxylate facial triad—An emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur J Biochem. 1997;250(3):625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 46.Karlsson A, et al. Crystal structure of naphthalene dioxygenase: Side-on binding of dioxygen to iron. Science. 2003;299(5609):1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 47.Ferraro DJ, et al. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct Biol. 2007;7:10. doi: 10.1186/1472-6807-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friemann R, et al. Structural insight into the dioxygenation of nitroarene compounds: The crystal structure of nitrobenzene dioxygenase. J Mol Biol. 2005;348(5):1139–1151. doi: 10.1016/j.jmb.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 49.Martins BM, Svetlitchnaia T, Dobbek H. 2-Oxoquinoline 8-monooxygenase oxygenase component: Active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure. 2005;13(5):817–824. doi: 10.1016/j.str.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Wargo MJ, Szwergold BS, Hogan DA. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol. 2008;190(8):2690–2699. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts RJ. Identifying protein function—A call for community action. PLoS Biol. 2004;2(3):E42. doi: 10.1371/journal.pbio.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Link TA, Haase U, Brandt U, von Jagow G. What information do inhibitors provide about the structure of the hydroquinone oxidation site of ubihydroquinone: Cytochrome c oxidoreductase? J Bioenerg Biomembr. 1993;25(3):221–232. doi: 10.1007/BF00762584. [DOI] [PubMed] [Google Scholar]
- 54.Eichler K, Bourgis F, Buchet A, Kleber HP, Mandrand-Berthelot MA. Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli. Mol Microbiol. 1994;13(5):775–786. doi: 10.1111/j.1365-2958.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA. 2011;108(43):17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kochar M, et al. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother. 2012;56(4):1845–1853. doi: 10.1128/AAC.05334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nam JW, et al. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci Biotechnol Biochem. 2001;65(2):254–263. doi: 10.1271/bbb.65.254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.