Significance
Myosins function as groups of motors anchored to a cellular scaffold that interacts with neighboring actin filaments patterned in complex three-dimensional networks. Although much is known about the function of individual myosin molecules, the contribution of intermotor interactions to collective movement is poorly understood. We used a precisely engineered biomimetic system to investigate how the mechanics of a structural element in the individual myosin influences the collective motion of groups of motors. Stochastic simulations suggest that the interplay between intramolecular strain on the motor lever arm and intermotor tension dictates trajectory shapes in groups of identical and antagonistic motors. Our study provides a general approach to dissecting emergence in cellular processes with applications for the efficient synthetic design of molecular transport systems.
Keywords: motor proteins, single molecule biophysics, synthetic biology, DNA nanotechnology, collective decision-making
Abstract
The molecular motor myosin teams up to drive muscle contraction, membrane traffic, and cell division in biological cells. Myosin function in cells emerges from the interaction of multiple motors tethered to a scaffold, with surrounding actin filaments organized into 3D networks. Despite the importance of myosin function, the influence of intermotor interactions on collective motion remains poorly understood. In this study, we used precisely engineered myosin assemblies to examine emergence in collective myosin movement. We report that tethering multiple myosin VI motors, but not myosin V motors, modifies their movement trajectories on keratocyte actin networks. Single myosin V and VI dimers display similar skewed trajectories, albeit in opposite directions, when traversing the keratocyte actin network. In contrast, tethering myosin VI motors, but not myosin V motors, progressively straightens the trajectories with increasing myosin number. Trajectory shape of multimotor scaffolds positively correlates with the stiffness of the myosin lever arm. Swapping the flexible myosin VI lever arm for the relatively rigid myosin V lever increases trajectory skewness, and vice versa. A simplified model of coupled motor movement demonstrates that the differences in flexural rigidity of the two myosin lever arms is sufficient to account for the differences in observed behavior of groups of myosin V and VI motors. In accordance with this model trajectory, shapes for scaffolds containing both myosin V and VI are dominated by the myosin with a stiffer lever arm. Our findings suggest that structural features unique to each myosin type may confer selective advantages in cellular functions.
Collective motion of a group is often influenced by interactions between individual entities, leading to emergence not evident in the individual (1). Stellar streaming (2), segregation of pedestrian traffic (3), and the migration of biological cells (4) are examples that span the size spectrum. Stellar streaming emerges from the gravitational interactions between ancient orbiting stars and the entire galaxy (2). Pedestrians adjust their movements on the basis of visual cues, such as the distance to an obstruction, giving rise to spontaneous unidirectional lanes for faster migration through a crowd (3). In migrating cells, local remodeling of the cytoskeleton is sufficient to effect global changes in the shape and persistence of movement direction (4). Although the importance of emergence in nature is generally appreciated, defining the underlying parameters that influence collective motion remains a challenge. In biology, the cell is packed with proteins that undergo relatively weak interactions in spatially segregated groups that give rise to large-scale intracellular structure, cellular migration, and tissue-level phenomena such as muscle motion and memory. Despite its importance, studies of emergence in cell biology have been limited, until recently, by the lack of engineered systems at the nanoscale.
The cellular function of the myosin family of cytoskeletal motors emerges from the interaction of several myosins tethered to a scaffold with the surrounding actin filaments (5). Actin architecture in cells is inherently 2D or 3D, as witnessed in dense cortical meshworks (6), filament bundles in filopodia (7), and parallel hexagonal arrays in muscle sarcomeres (8, 9). In concert, collective myosin function in cells varies with actin organization. Whereas multiple membrane-tethered myosin VI motors interact with actin bundles to anchor stereocilia (10), myosin VI motors on uncoated endosomes are necessary for their timely transport through a dense actin cortex, and myosin VI localized on the Golgi functions as a tether to maintain organelle shape and size (11). In muscle, the collective interaction of myosins patterned on rod-like structures with actin filaments arranged in hexagonal lattices forms the molecular basis of muscle contraction (8, 9). In each of these instances, the contribution of intermotor interactions, patterned in different geometric configurations, on collective myosin remains unexplored.
Myosin function in a cellular context has been examined either as the interaction of single molecules with cellular actin networks (12, 13) or the movement of two motors tethered together on single actin filaments (14, 15). Single myosin studies have identified unique structural features in the protein that select for processive movement on certain actin topologies (13, 16, 17). Tethering two identical myosins enhances travel distance along single actin filaments, but at lower speeds (15). Tethering myosins that move toward the opposite ends of an actin filament results in unidirectional motion, with the two motors coordinating their stepping movements (14). The intermotor interactions in these systems have also been investigated with theoretical approaches that, combined with experimental observations, have yielded insights into the underlying parameters that govern collective movement (18–23). Although there have been studies on the collective movement of myosins on cellular actin networks (24, 25), they have relied on polystyrene beads as scaffolds, which do not provide control over the number, type, and organization of myosins (24, 25). Here, we use 2D DNA origami scaffolds (26–28) to precisely pattern a combination of myosin V and VI and systematically dissect the role of intermotor interactions on collective function.
In this study, we report the emergence of collective motion in myosins that is dependent on the myosin lever arm. DNA origami scaffolds were used to precisely engineer groups of myosin V and VI motors and study their interactions with a model 2D cellular actin network. A simple model suggests that tuning the flexural rigidity of the lever arm relative to the stiffness of intermotor linkages is sufficient to influence collective trajectories in groups of both identical and antagonistic motors. Our findings suggest that structural features unique to myosin V and VI confer selective advantages to their cellular functions.
Results
Single Myosin V and Myosin VI Move Processively on the Dense Keratocyte Actin Network with Similar Meandering Trajectories.
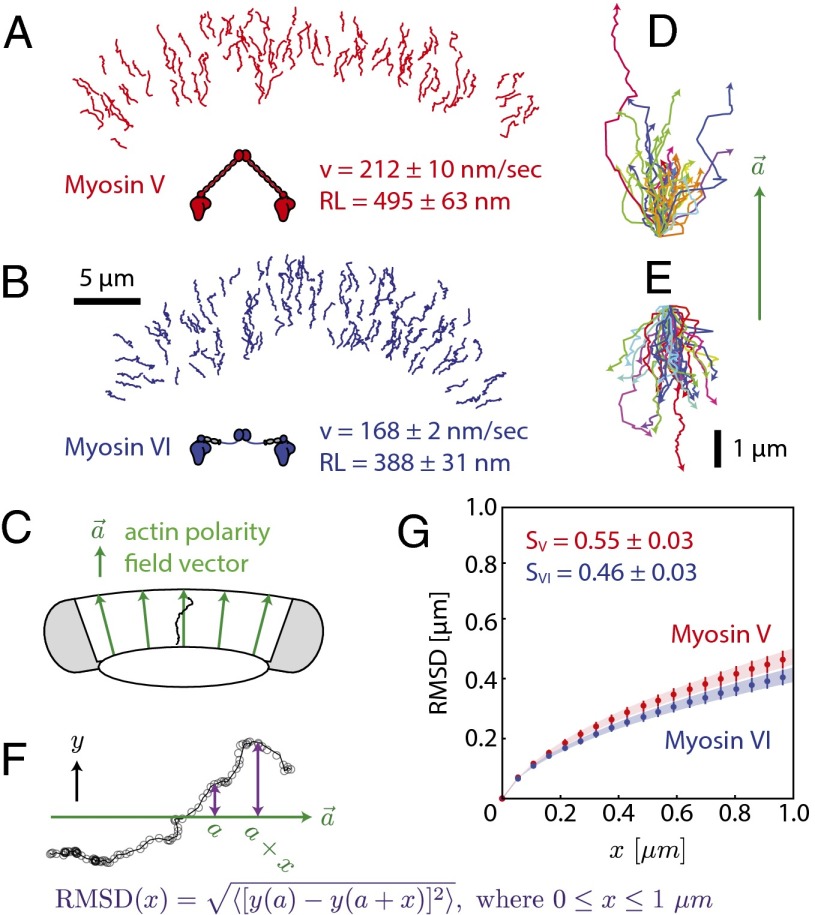
The movement of individual myosin V or VI on dense actin networks (SI Appendix, Fig. S1) was first assessed at the single-molecule level with total internal reflection fluorescence (TIRF) microscopy. Myosin V (Fig. 1A) moved toward the cell periphery, whereas myosin VI (Fig. 1B) traveled toward the cell center, consistent with previously reported actin network polarity (25). The mean speed 〈v〉 and mean run length 〈RL〉 of a single myosin V (212 ± 10 nm/s; 495 ± 63 nm) or myosin VI (168 ± 2 nm/s; 388 ± 31 nm) are in agreement with previous reports for movement of these myosins on single actin filaments (15, 29, 30). To visually compare the shapes of myosin trajectories, each trajectory was rotated to align their local actin polarity field vectors (Fig. 1C; SI Appendix, Fig. S2), followed by translation to match starting coordinates. The local actin polarity field vector is the shortest vector that passes through the center of mass of the trajectory, connects the inner and outer boundaries of the keratocyte actin network, and is directed from the cell center to the cell periphery (Fig. 1C; SI Appendix, Fig. S2). The aligned trajectories show a broad distribution of trajectory shapes for both myosin V and VI (Fig. 1 D and E). The mean trajectory shape was quantified using the root mean squared displacement of the population relative to the local actin polarity field vector (Fig. 1 F and G). The mean trajectory shape was also used to quantify the lateral skewness of the trajectories in terms of a shape factor (S; SI Appendix, Fig. S2), which quantifies the mean lateral deviation (μm) of a population of trajectories as the myosin moves a distance of 1 μm toward the cell center or periphery. Trajectories of single myosin V and single myosin VI had comparable shape factors (0.55 ± 0.3 μm/μm and 0.46 ± 0.3 μm/μm, respectively) that were statistically indistinguishable (P = 0.11), suggesting similar meandering movement for both myosin types.
Fig. 1.
Single myosin V and VI exhibit similar meandering trajectories. Trajectories of single myosin V (A) or myosin VI (B) on the keratocyte actin network, colored by direction of movement (red, toward cell periphery; blue, toward cell center). (C) The actin polarity field vector (green; SI Appendix, Fig. S2) is determined by the shortest distance between the cell center and periphery at any location. The vector points toward the cell periphery. The trajectories on the edges of the keratocyte (gray shaded regions) were excluded from further analysis. (D and E) Trajectories of single myosin V (D) or myosin VI (E) aligned with respect to their local actin polarity field vectors. (F) Root mean squared displacement calculation of a representative aligned trajectory (black; SI Appendix, Fig. S2). (G) Root mean squared displacement plots for the aligned trajectories of myosin V (red; n = 168) and VI (blue; n = 203). The shape factors of single myosin V and myosin VI are statistically indistinguishable (P = 0.11), with uncertainties (± SEM) estimated by bootstrapping (SI Materials and Methods).
Engineering 2D Actin–Myosin Interactions.
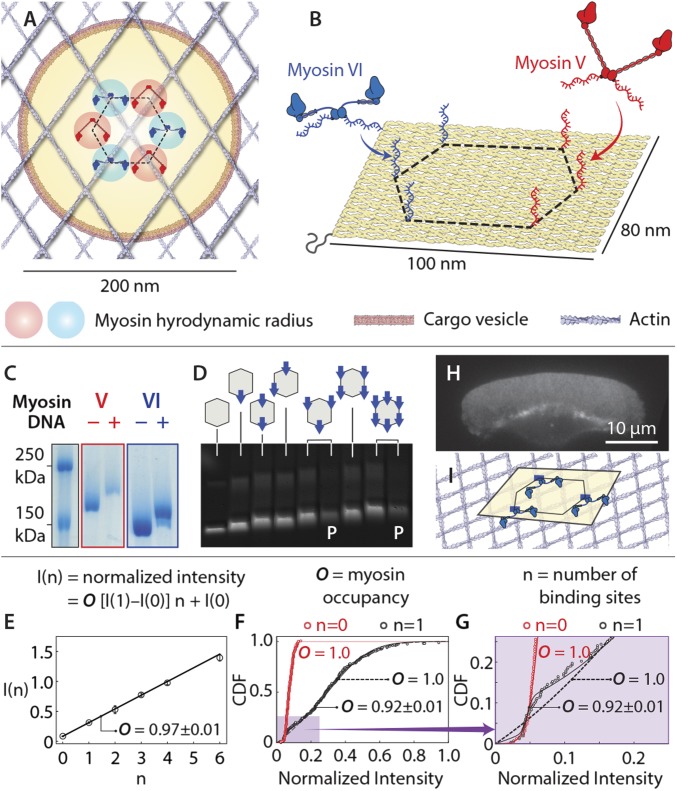
To systematically dissect the collective motion of multiple myosin motors, we designed a programmable biomimetic scaffold using a ∼100 nm × ∼80 nm flat rectangular DNA origami (27, 28). The myosin pattern on the origami surface models the interaction at the interface between a myosin-coated vesicle and the cortical actin meshwork that enmeshes it (Fig. 2A). Myosin was arranged in a hexagonal configuration, with the side length comparable to the myosin hydrodynamic diameter (Fig. 2 A and B; SI Appendix, Figs. S3 and S4; details in SI Notes S1). Myosin V and VI dimers were engineered with a SNAP-tag (alkyl-guanine-transferase) to facilitate the covalent attachment of an oligonucleotide. The myo-sin-linked oligo is complementary to a scaffold extension (Fig. 2B). The high efficiency of myosin labeling with oligo (>95%; Fig. 2C) was confirmed by a gel-shift assay. Origami scaffolds were designed with a biotinylated strand to facilitate removal of excess myosin (see Materials and Methods; SI Appendix, Fig. S5) while preserving the origami shape and myosin attachment (SI Appendix, Fig. S6). Precise control of myosin number on each scaffold was evident in defined gel-shifts of the origami scaffold in 1% agarose 0.1% SDS gels (94 ± 1% occupancy; Fig. 2D; SI Appendix, Fig. S6) and a photon counting assay (>92% occupancy; Fig. 2 E–G). Intact 2D actin networks with net polarity and large surface area were obtained from detergent extracted keratocytes, as previously reported (25) (Fig. 2H; SI Appendix, Fig. S1). The origami scaffolds move predominately on the surface of the actin network, whose ∼30-nm mean pore size (25) is significantly smaller than the origami scaffold but is comparable to the ∼36-nm step size of myosin V (29) and VI (30) (Fig. 2I).
Fig. 2.
Scaffolds precisely patterned with myosin V and/or VI. (A) Schematic depicting the interaction between a myosin-coated vesicle (yellow) and the cortical actin meshwork. In this model, myosins are closely packed in a hexagonal pattern (dashed hexagon) on the surface of a vesicle. (B) Illustration of a flat, rectangular DNA scaffold indicating positions of myosin attachment along the vertices of a hexagon (35-nm side). (C) Coomassie staining of SDS/PAGE gel of myosin V and VI before and after conjugation to benzylguanine-DNA. (D) SDS-agarose gel showing bands (Cy3 emission) corresponding to scaffold–myosin complexes. Myosin number and pattern are indicated. P, complexes purified by strand displacement (SI Appendix, Fig. S5). (E–G) More than 92% of myosin binding sites are occupied, as assessed with Cy3-labeled oligos. DNA scaffolds were labeled with Cy5, and myosin-binding sites were labeled with complementary Cy3 oligos (SI Materials and Methods). (E) Normalized intensities (Cy3/Cy5) increase linearly with number of myosin-binding sites, with 97 ± 1% occupancy at each site. (F and G) Cumulative distribution functions of normalized intensities for scaffolds with 0 (n = 0) and 1 (n = 1) binding sites. Intensity of scaffolds with no binding sites (n = 0) follow a single Gaussian distribution, whereas those with a single binding site (n = 1) are consistent with two populations with 92 ± 1% occupancy. (H) Representative detergent-extracted keratocyte actin network stabilized with Alexa488-phalloidin (SI Appendix, Fig. S1). (I) Schematic of scaffold-myosin movement on the surface of the keratocyte actin network. Mesh size of network (∼30 nm) (43) is smaller than the size of scaffold (∼100 nm). The keratocyte actin network is depicted by actin filaments oriented at ±35° (44). Error bars represent ± SEM.
Multimyosin Scaffolds Move Farther at Lower Speeds.
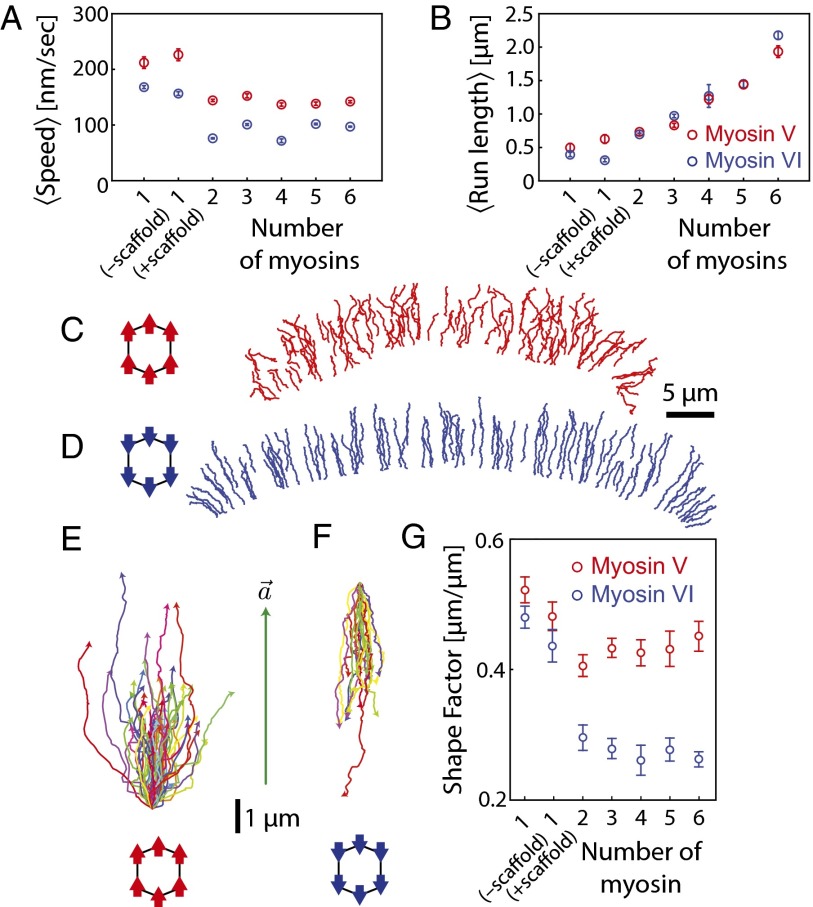
The mean speed of scaffolds with 2–6 myosin V or myosin VI motors is significantly lower than that of a single myosin V or VI, respectively (Fig. 3A; SI Appendix, Fig. S8). This observation is similar to previous reports for 2 myosin V motors on a single actin filament (15) but is in contrast to kinesin molecules that show essentially no change in speed with increasing motor number (26, 31). Multimyosin scaffold speed, however, does not depend on motor number (n = 2–6 myosins; Fig. 3A). Origami scaffolds with myosin V or VI show a characteristic increase in mean apparent run-length with increasing motor number (15, 32), consistent with the engagement of each additional motor with the actin networks (Fig. 3B; SI Appendix, Fig. S9). Given the limited width of the keratocyte actin network (∼5 μm), the myosin-dependent run length is substantially larger (SI Appendix, Fig. S10) than the observed run length (Fig. 3B). In both analyses, conjugating a DNA scaffold to a single myosin V or VI does not alter its speed (P ≥ 0.07; Fig. 3A) and mean apparent run length (P ≥ 0.09; Fig. 3B).
Fig. 3.
Collective movement of multimyosin scaffolds. Variation in (A) mean end-to-end speed and (B) mean run length as a function of myosin number (SI Appendix, Figs. S7–S9). (C and D) Movement trajectories of indicated scaffold–myosin complexes on the keratocyte actin network (Movie S1), with directionality specified by color (red, toward cell periphery; blue, toward cell center). (E and F) Movement trajectories of scaffolds with 6 myosin V (E) or 6 myosin VI (F) aligned relative to the local actin polarity field vector. (G) Shape factors as a function of myosin number (myosin VI, blue; myosin V, red; n = 58–636). Error bars are ± SEM.
Emergent Linear Trajectories in Myosin VI Groups.
Trajectories of multiple myosin V-driven scaffolds (Fig. 3 C and E; Movie S1, Upper) are qualitatively similar to those of a single myosin V (Fig. 1 A and D). In contrast, myosin VI-driven trajectories are highly linear (Fig. 3 D and F; Movie S1, Lower) and are strikingly different from the corresponding single-molecule movement (Fig. 1 B and E). Correspondingly, the shape factors for myosin VI, but not myosin V, trajectories decrease with increasing motor number (Fig. 3G). The similar trajectory shapes for single myosin V or VI molecules (Fig. 1 A and B) support a role for myosin structure rather than asymmetric organization of the actin network in the observed difference in the collective behavior of groups of motors.
Stochastic Simulation Suggests the Flexibility of the Lever Arm Influences Collective Myosin Movement.
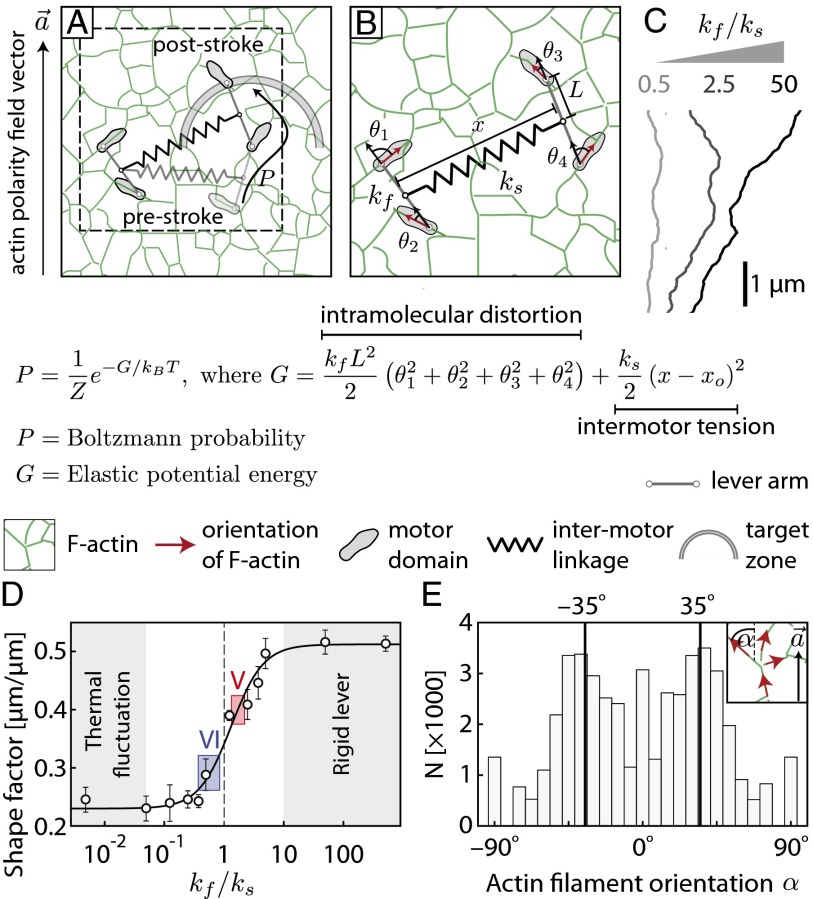
To understand the underlying mechanisms that give rise to the difference in movement patterns, a biophysical model was used to study the potential contribution of intra- and intermolecular forces to collective movement. In this model, the stepping process of an individual myosin within an ensemble (Fig. 4A; SI Appendix, Figs. S11 and S12) is guided by the interplay between the intramolecular strain on the lever arm and the intermolecular tension caused by the stretching of intermotor links (Fig. 4B). The flexural rigidity of the lever arm (kf) imposes an intramolecular penalty for misalignment of the lever relative to the actin filament where the corresponding myosin head is anchored. In contrast, the extension of structural elements in the motors and linkers between them increases the intermotor tension in proportion to the net spring stiffness (ks; Fig. 4B). Therefore, the Boltzmann probability of a given poststroke state derives from the relative magnitudes of the elastic potential energies associated with intramolecular strain and intermotor tension. Stochastic simulations based on this model showed that for an ensemble with defined ks, trajectory shapes can be tuned by varying kf/ks (Fig. 4C). Simulated trajectory shapes were quantified in terms of shape factors (Figs. 1G and 3G; SI Appendix, Fig. S2). For myosins with highly flexible lever arms (kfL2 < kBT; left shaded region in Fig. 4D and SI Appendix, Figs. S13 and S14), thermal fluctuations dictate both individual and collective motion. In this regime, the myosin motor freely steps off-axis to relieve intermotor tension, resulting in highly linear trajectories (low shape factor). For rigid levers (kf/ks ≫ 1, right shaded region in Fig. 4D and SI Appendix, Figs. S13 and S14), the motor steps minimize intramolecular strain. Thus, after release of the trailing head from an actin filament, it preferentially binds to an actin filament with the same spatial orientation as the one bound to the leading head. Subsequent steps by either motor continue to align the heads relative to each other, further minimizing intramolecular strain. The resulting trajectories meander about the local actin polarity field vector with a shape factor that increases with kf/ks. The orientation of trajectories for rigid levers (kf/ks ≫ 1) depends on the local orientations of actin filaments. Quantitative analysis of transmission electron microscopy (TEM) images of keratocyte actin networks (SI Appendix, Fig. S11) yields a bimodal distribution for orientation of actin filaments relative to the local actin polarity field vector, with peaks at ±35° (Fig. 4E; Materials and Methods). This nonrandom distribution of the underlying actin network contributes to the observed skewed trajectories for kf/ks ≫ 1. Between the extremes dominated by thermal fluctuations and rigid levers, the shape factor steadily increases as (kf/ks) increases (Fig. 4D, solid line). According to this model, the experimentally observed shape factors are consistent with a flexible lever for myosin VI (kf/ks < 1) and a relatively rigid lever for myosin V (kf/ks > 1). For single myosin molecules (ks = 0), the shape factor is solely dependent on the rigidity of the myosin lever relative to thermal fluctuations (kfL2/kBT, where L is the length of the lever arm; SI Appendix, Fig. S13). The experimentally observed shape factors for single myosins suggest that both motor types operate outside the regime dominated by thermal fluctuations (SI Appendix, Fig. S14).
Fig. 4.
Model and stochastic simulations of collective myosin movement. (A) Simplified model of collective myosin movement on a digitized keratocyte actin network (green; SI Appendix, Figs. S11 and S12). The motor domains (gray), lever arms, intermotor linkage (spring), and digitized actin network are drawn approximately to scale. The dashed box corresponds to a zoomed schematic (B). The Boltzmann probability of stepping to a target site within the target zone (shaded arc) is a function of the stored potential energy (G), which is a function of the net stiffness of the intermotor linkage (ks) and flexural rigidity of the lever arm (kf). θi is the angle between the lever and the actin filament bound to the motor domain, and xi is the intermotor distance. (C) Representative trajectories generated from the stochastic simulation (SI Appendix, Fig. S12) for kf/ks = 0.1, 0.5, and 50. (D) Shape factor as a function of kf/ks. Solid line is the least squares fit based on a sigmoidal curve (SI Appendix, Figs. S12–S14). Gray shaded regions indicate the two regimes in which the shape factor is not sensitive to kf/ks. The experimentally measured shape factors for scaffolds with 2 myosin V (red shaded box) or 2 myosin VI (blue shaded box) yield kf/ks values of 1.8 ± 0.35 and 0.50 ± 0.15, respectively. (E) Distribution of local actin orientation relative to the actin polarity field vector (α; Inset) in the digitized TEM image of keratocyte actin network (SI Appendix, Fig. S11). Solid lines corresponds to the characteristic Arp2/3 branch angle of the keratocyte actin networks (±35°), with respect to the actin polarity field vector (44). Error bars represent ± SEM.
Swapping Lever Arms Switches Trajectory Shape.
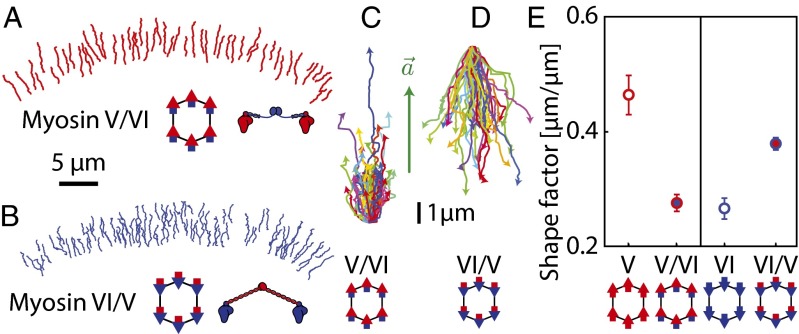
Our model demonstrates that the structural properties of the myosin lever arm form a parameter that can account for observed differences in the collective motion of the two myosins. This prediction was tested by experiments involving myosin V and VI chimeras with swapped lever arms. More specifically, chimeras involved the myosin V motor domain with the flexible myosin VI lever arm (myosin V/VI; Fig. 5A) and the myosin VI motor domain with the rigid myosin V lever arm (myosin VI/V; Fig. 5B). Although the direction of movement is dictated by the motor domain (Fig. 5 A and B), providing myosin V with a flexible lever arm decreases the shape factors, and vice versa (Fig. 5 C–E; n ≥ 391; P < 0.0001). This dramatic reversal supports the idea that trajectory shape emerges from the interplay between intermotor tension and lever arm-dependent intramolecular strain.
Fig. 5.
Exchanging lever arms switches trajectory shapes. (A and B) Trajectories of scaffolds with indicated myosin chimeras moving along the keratocyte actin network. (A) Scaffolds with 6 myosin V/VI (myosin V motor domain with myosin VI lever arm) move toward the cell periphery (red). (B) Scaffolds with 6 myosin VI/V (myosin VI motor domain with myosin V lever arm) move toward the cell center (blue). (C and D) Trajectories of scaffolds with 6 myosin V/VI chimeras (C) or 6 myosin VI/V chimeras (D) aligned relative to their local actin polarity field vectors. (E) Introduction of the myosin VI lever arm decreases the shape factor of the myosin V/VI chimera. In contrast, introduction of the myosin V lever arm increases the shape factor for myosin VI. Error bars represent ± SEM (n ≥ 391).
Trajectory Shape Is Dominated by Myosin V.
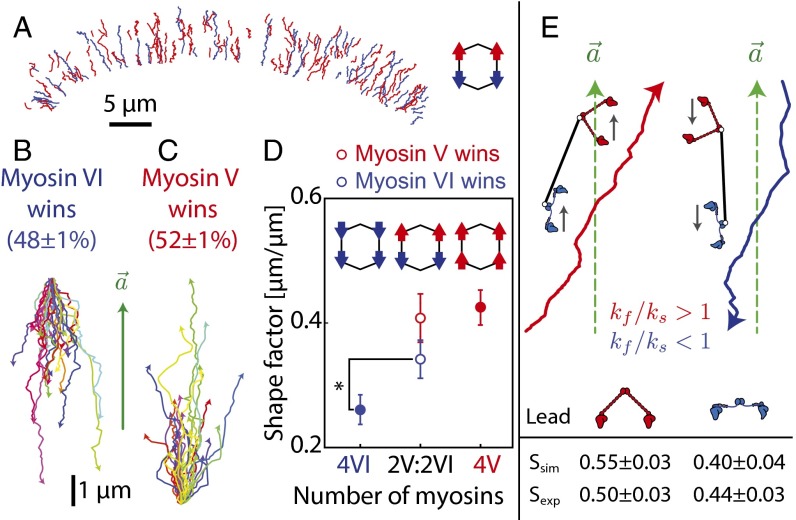
Myosin V and VI have been shown to colocalize to vesicles in neuronal growth cones (33). Hence, we investigated origami scaffolds with antagonistic motors to test the ability of myosin V and VI to influence each other’s movement. In accordance with previous reports, scaffolds with both myosin V and VI commit to one direction of movement (14, 26) and are sorted almost equally into movement toward cell periphery (52 ± 1%) and cell center (48 ± 1%) (Fig. 6A; Movie S2; n = 546). The shape factor for trajectories with myosin V leading was statistically unchanged compared with those with myosin V alone (P = 0.37; Fig. 6D). In contrast, the shape factor of trajectories with myosin VI leading was significantly higher than those with myosin VI alone (P = 0.02; Fig. 6D). This asymmetry suggests that for both inward and outward directed trajectories, the intramolecular strain in the rigid lever arm of myosin V dominates the collective movement, whereas the flexible myosin VI lever follows the path set by myosin V (Fig. 6E).
Fig. 6.
Trajectory shapes are dominated by myosin V. (A) Movement trajectories of scaffolds with 2 myosin V and 2 myosin VI (n = 546; Movie S2), with directionality indicated by color (red, toward cell periphery; blue, toward cell center). (B and C) Percentage of runs to cell periphery (red) and to cell center (blue) is indicated. Trajectories of scaffolds with 2 myosin V and 2 myosin VI moving toward the cell center (B) and cell periphery (C), aligned relative to the local actin polarity field vector. (D) Shape factors for trajectories moving toward the cell periphery (open red circle) or cell center (open blue circle) for scaffolds with 2 myosin V and 2 myosin VI compared with scaffolds with either 4 myosin V or VI. Error bars represent ± SEM. (E) Sample simulated trajectories of scaffolds with 1 myosin V (kf/ks = 1.8) and 1 myosin VI (kf/ks = 0.5), with either myosin V (left) or myosin VI (right) in the lead (ks = 0.2 pN/nm). The simulated shape factors are significantly higher when myosin V leads (P < 0.0001). This asymmetry is also observed in experimental trajectories moving toward the cell periphery or the cell center for matched myosin scaffolds (n = 425). Error bars represent ± SEM.
Discussion
Our study illustrates the importance of intermotor interactions in determining collective motion and suggests the need to similarly reconstitute cellular processes to understand the contribution of emergence to higher-order function. Myosin V and VI play important roles in diverse cellular processes, including membrane transport, tethering of organelles, cytokinesis, and actin organization (5). The skewed movement of multimyosin V scaffolds reported here is consistent with the myosin V-driven dispersive motion of pigment granules in the melanocyte cortex (34). In contrast, linear-directed trajectories, rather than meandering motion, are likely to reduce endocytic transport times, as observed in the presence of myosin VI (35). Myosin V and VI move toward the opposite ends of single actin filaments, albeit with similar stepping kinetics (29, 30). Our study shows that the myosin lever arm can influence trajectory shapes in groups of motors. The structural elements that constitute the myosin lever are unique to each member of the myosin family (36). Myosin V has a relatively rigid lever arm consisting of a series of IQ motifs wrapped by six calmodulin light chains (37). In contrast, myosin VI has only two calmodulin binding domains followed by a semiflexible single ER/K α-helix domain (36). The flexible myosin VI lever is necessary and sufficient to straighten trajectories in groups of myosin VI motors. A parallel observation is that structural elements in myosin X lever arm extension selectively enable processive movement on parallel actin bundles because of the poor processivity of this motor on single actin filaments (17). Accordingly, myosin X preferentially associates and moves along filopodia because of its increased residence time on this actin architecture. Further, single molecules of myosin V, VI, and X display preferential processive movement on distinct actin architecture (13). These observations are consistent with our findings that structural features within the myosin molecule influence cellular behavior by selectively influencing interactions with the actin network. This is in contrast to functional differences in myosins attributed to their distinct cargo binding domains, which are well-established determinants of subcellular localization through the selective binding of membranes or adaptor proteins (38).
Unraveling the rules of interaction between the individuals of a group is an essential step for understanding and controlling emergent behavior (1, 3). Hence, we paired experiments with stochastic simulations that incorporate detailed information on myosin stepping derived from single-molecule studies (37). Such simulations have been previously used to determine the relative population of different kinetic states during the processive movement of two-motor assemblies on single actin filaments (15) or microtubules (20). Our measurements complement a recent report on two-myosin assemblies (15), in that the multimyosin scaffolds move modestly longer distances, at reduced speeds, compared with single myosin molecules. Transition state models have suggested that two-myosin assemblies substantially populate states in which both motors are bound to an actin filament (20). For our multimotor scaffolds, population of states in which multiple motors are bound to an actin filament should decrease the probability that no motor is bound to an actin filament, resulting in longer runs. Correspondingly, we find that scaffold run length linearly increases with myosin number, supporting the concept that each additional motor can interact with actin filaments. The frequent population of states with multiple motors bound to actin filaments is also supported by the trend in speed with increasing motor number. Assuming the motors step asynchronously, we would expect that the stepping kinetics of each myosin are influenced by whether or not the scaffold is anchored to the actin filament by another motor, rather than the number bound. Accordingly, although multimyosin scaffolds move slower than ones with single myosin, their speeds do not depend on motor number.
Given that the trajectory skewness of multimotor assemblies is dependent on the type of lever arm (V or VI), our simulations incorporated the structural differences in the levers in terms of their flexural rigidity. This single parameter is sufficient to capture the observed differences in trajectory shapes for groups of myosin V and VI. The stiffer lever favors binding of myosin heads to actin filaments aligned with each other. Given the nonrandom distribution of local actin orientations, stiffer levers favor skewed orientation of trajectories relative to the actin polarity field vector. Our simplified model suggests that the balance between intermotor tension of the motor linkages and intramolecular strain in the lever arm dictates trajectory shapes. It must be noted that the model substantially simplifies both the actin architecture and computation of intra- and intermolecular interactions and does not account for the potential effects of intermotor forces on stepping kinetics. Hence, although the model can explain the observed behavior, it is entirely possible that additional parameters can similarly influence intermotor interactions to effect similar outcomes. Nonetheless, our findings suggest an elegant design principle for linear transport in a complex, 2D landscape. Regardless of motor type, tuning the balance between intra- and intermolecular interaction energies can control collective movement. This principle can be applied to designing efficient, long-range transport systems at the nanoscale.
Materials and Methods
Myosin Expression and Purification.
Recombinant myosin protein was expressed and purified from Sf9 insect cells. Myosin constructs contain an N-terminal FLAG tag, followed by a myosin, leucine zipper (GCN4) to ensure dimerization, alkyl-guanine-transferase (AGT), and His6 tag. Myosin VI consisted of residues 1-992 of Sus Scrofa myosin VI; myosin V is constructed from residues 1-1,0099 of Gallus gallus myosin V. Both myosin VI and myosin V/VI chimera were cloned into pBiex-1 (Novagen), whereas myosin V and myosin VI/V were cloned into pFastBac dual (coexpresses calmodulin). Protein was expressed by transient transfection (pBiex-1; Escort IV, Sigma) or baculovirus infection of Sf9 cells (Invitrogen). Expressed proteins were affinity purified at 72 h with Anti-FLAG resin (Sigma), using established procedures (39, 40).
Scaffold-Myosin Preparation and Purification.
Single-stranded M13mp18 DNA (scaffold strand; N4040S; NEB) were mixed with fourfold excess of short staple strands (unpurified; IDT), followed by 2-h-long annealing, as previously described (27, 28) (SI Materials and Methods). Intact scaffolds were separated from excess staple strands, and improperly folded scaffolds by gel purification (0.8% agarose with SyberGreen) and recovered in 30% sucrose, 1× TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA), 12.5 mM MgCl2 (41). Purified scaffolds were mixed with an excess of benzyl-guanine-conjugated myosin (SI Materials and Methods) and blocking oligos (mixture of 42-nt oligos with randomized sequence) and incubated for 20 min at 37 °C. Scaffold–myosin complexes were separated from excess myosin by affinity purification followed by strand displacement (SI Appendix, Fig. S5).
Photon Counting Assay.
Fractional occupancy of myosin binding sites on each DNA scaffold were quantified using a photon counting assay. Cy5-labeled DNA scaffolds were incubated with an excess of Cy3-labeled DNA strand for 30 min at 37 °C, followed by 30 min at room temperature. The scaffolds were diluted by a factor of 25,000 in 1× assay buffer (AB) (25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 25 mM Imidazole) + 1 mg/mL BSA, and subsequently immobilized in a neutravidin-coated flow chamber. Unbound Cy3 was removed by extensive washes with 1× AB⋅BSA. Scaffolds were imaged in 1× AB + [1 mg/mL BSA, 25 µg/mL glucose-oxidase, 45 µg/mL catalase, 1% (wt/vol) glucose] at room temperature, using a TIRF microscope (Olympus IX81; 60× NA 1.48 Apo TIRF objective), a 2× image magnifier (EMCCD iXON Ultra; Andor), a 532-nm laser (Crystalaser CL532-150mW-L), and a 640-nm laser (CUBE 640–100). For each field of view, successive images of Cy3 (excitation at 532 nm) and Cy5 (excitation at 640 nm) emissions were obtained with 2 s exposure time and ∼100-nm penetration depth (Olympus TIRF Illuminator). Cy3 and Cy5 intensities were quantified using custom Mathematica and MATLAB algorithms. Briefly, individual scaffolds were located using a particle tracking algorithm based on Cy5 intensity. Cy3 intensity for each scaffold was normalized by corresponding Cy5 intensity. Cy3 and Cy5 intensities were individually integrated over a 13 × 13-pixel region. Normalized intensity (I) was defined as the ratio between the intensity values in the Cy3 and Cy5 images (Fig. 2 E–G).
Motility Assays.
Keratocytes were derived from scales of Thorichthys meeki (Firemouth Cichlids), as previously described (25). Detergent-extracted keratocytes were washed into buffer AB + 1 mg/mL BSA, followed by incubation with myosin-scaffolds in imaging buffer AB + [1 mg/mL BSA, 2 mM ATP, 9.0 µM calmodulin, 1 mM phosphocreatine, 0.1 mg/mL creatine-phospho-kinase, 25 µg/mL glucose-oxidase, 45 µg/mL catalase, 1% (wt/vol) glucose, 1 µM blocking oligos] at room temperature. Time-lapse imaging was performed on an epi-fluorescence microscope (SI Materials and Methods), with the exception of single-molecule motility assays in Fig. 1. Single-molecule imaging was obtained using a TIRF microscope (SI Materials and Methods).
Data Analysis.
Trajectories of individual myosin labeled scaffolds were analyzed using custom MATLAB Particle Tracking software (42) and Imaris (Bitplane). A 2D-Gaussian fit was used to estimate scaffold position with subpixel resolution. Intensity of scaffold was used to exclude doublets and aggregates (<10%). The scaffold positions were used to compute run length, end-to-end speed, and local trajectory angle (SI Materials and Methods).
Supplementary Material
Acknowledgments
The authors thank M. Ritt and T. Tigney for technical assistance. We thank K. Verhey, B. Yurke, A. Dunn, C. Swanson, E. Winfree, A. Gopinath, P. Rothemund, S. Woo, D. Woods, R. Schulman, R. Malik, and R. Sommese for useful discussions and manuscript review. The authors thank E. Winfree and L. Qian for hosting the atomic force microscopy experiments. Research was funded by the American Heart Association Scientist Development Grant (13SDG14270009) and National Institutes of Health Director’s New Innovator Award (1DP2CA186752-01). The authors also thank the Single Molecule Analysis in Real-Time Center of the University of Michigan (seeded by National Science Foundation MRI-R2-ID award DBI-0959823).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315923111/-/DCSupplemental.
References
- 1.Vicsek T, Zafeiris A. Collective motion. Phys Rep. 2012;517(3-4):71–140. [Google Scholar]
- 2.Helmi A, White SD, de Zeeuw PT, Zhao H. Debris streams in the solar neighbourhood as relicts from the formation of the Milky Way. Nature. 1999;402(6757):53–55. [Google Scholar]
- 3.Moussaïd M, Helbing D, Theraulaz G. How simple rules determine pedestrian behavior and crowd disasters. Proc Natl Acad Sci USA. 2011;108(17):6884–6888. doi: 10.1073/pnas.1016507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacayo CI, et al. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5(9):e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–155. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verkhovsky AB, et al. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol Biol Cell. 2003;14(11):4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooseker MS, Tilney LG. Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol. 1975;67(3):725–743. doi: 10.1083/jcb.67.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Khayat HA, Morris EP, Kensler RW, Squire JM. Myosin filament 3D structure in mammalian cardiac muscle. J Struct Biol. 2008;163(2):117–126. doi: 10.1016/j.jsb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper SL, Hobbs KH, Thuma JB. Invertebrate muscles: Thin and thick filament structure; molecular basis of contraction and its regulation, catch and asynchronous muscle. Prog Neurobiol. 2008;86(2):72–127. doi: 10.1016/j.pneurobio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasson T, et al. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137(6):1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169(2):285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy S, et al. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105(28):9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc Natl Acad Sci USA. 2009;106(24):9685–9690. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali MY, et al. Myosin Va and myosin VI coordinate their steps while engaged in an in vitro tug of war during cargo transport. Proc Natl Acad Sci USA. 2011;108(34):E535–E541. doi: 10.1073/pnas.1104298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu H, et al. Collective dynamics of elastically coupled myosin V motors. J Biol Chem. 2012;287(33):27753–27761. doi: 10.1074/jbc.M112.371393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oguchi Y, et al. Robust processivity of myosin V under off-axis loads. Nat Chem Biol. 2010;6(4):300–305. doi: 10.1038/nchembio.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285(34):26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp S, Lipowsky R. Cooperative cargo transport by several molecular motors. Proc Natl Acad Sci USA. 2005;102(48):17284–17289. doi: 10.1073/pnas.0507363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller MJ, Klumpp S, Lipowsky R. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc Natl Acad Sci USA. 2008;105(12):4609–4614. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driver JW, et al. Productive cooperation among processive motors depends inversely on their mechanochemical efficiency. Biophys J. 2011;101(2):386–395. doi: 10.1016/j.bpj.2011.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamison DK, Driver JW, Rogers AR, Constantinou PE, Diehl MR. Two kinesins transport cargo primarily via the action of one motor: Implications for intracellular transport. Biophys J. 2010;99(9):2967–2977. doi: 10.1016/j.bpj.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driver JW, et al. Coupling between motor proteins determines dynamic behaviors of motor protein assemblies. Phys Chem Chem Phys. 2010;12(35):10398–10405. doi: 10.1039/c0cp00117a. [DOI] [PubMed] [Google Scholar]
- 23.Jamison DK, Driver JW, Diehl MR. Cooperative responses of multiple kinesins to variable and constant loads. J Biol Chem. 2012;287(5):3357–3365. doi: 10.1074/jbc.M111.296582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheetz MP, Spudich JA. Movement of myosin-coated fluorescent beads on actin cables in vitro. Nature. 1983;303(5912):31–35. doi: 10.1038/303031a0. [DOI] [PubMed] [Google Scholar]
- 25.Sivaramakrishnan S, Spudich JA. Coupled myosin VI motors facilitate unidirectional movement on an F-actin network. J Cell Biol. 2009;187(1):53–60. doi: 10.1083/jcb.200906133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338(6107):662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 28.Woo S, Rothemund PWK. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat Chem. 2011;3(8):620–627. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 29.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400(6744):590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 30.Rock RS, et al. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98(24):13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuta K, et al. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci USA. 2013;110(2):501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindler TD, Chen L, Lebel P, Nakamura M, Bryant Z. Engineering myosins for long-range transport on actin filaments. Nat Nanotechnol. 2014;9(1):33–38. doi: 10.1038/nnano.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suter DM, Espindola FS, Lin CH, Forscher P, Mooseker MS. Localization of unconventional myosins V and VI in neuronal growth cones. J Neurobiol. 2000;42(3):370–382. [PubMed] [Google Scholar]
- 34.Tuma MC, Gelfand VI. Molecular mechanisms of pigment transport in melanophores. Pigment Cell Res. 1999;12(5):283–294. doi: 10.1111/j.1600-0749.1999.tb00762.x. [DOI] [PubMed] [Google Scholar]
- 35.Aschenbrenner L, Naccache SN, Hasson T. Uncoated endocytic vesicles require the unconventional myosin, Myo6, for rapid transport through actin barriers. Mol Biol Cell. 2004;15(5):2253–2263. doi: 10.1091/mbc.E04-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spudich JA, Sivaramakrishnan S. Myosin VI: An innovative motor that challenged the swinging lever arm hypothesis. Nat Rev Mol Cell Biol. 2010;11(2):128–137. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trybus KM. Myosin V from head to tail. Cell Mol Life Sci. 2008;65(9):1378–1389. doi: 10.1007/s00018-008-7507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buss F, Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochem Biophys Res Commun. 2008;369(1-3):165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney HL, et al. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J Biol Chem. 1998;273(11):6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 40.Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci USA. 2007;104(3):772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bellot G, McClintock MA, Lin C, Shih WM. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat Methods. 2011;8(3):192–194. doi: 10.1038/nmeth0311-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci USA. 2005;102(5):1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J Struct Biol. 1995;115(3):290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- 44.Maly IV, Borisy GG. Self-organization of a propulsive actin network as an evolutionary process. Proc Natl Acad Sci USA. 2001;98(20):11324–11329. doi: 10.1073/pnas.181338798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.