Ribosome ambiguity (ram) mutants cause miscoding during translation. This study analyzes two types of ram mutants, those that increase miscoding in a general way and those that alter the A site and increase miscoding in a codon-anticodon-dependent manner.
Keywords: ribosome, translation, tRNA, decoding, EF-Tu
Abstract
During decoding, the ribosome selects the correct (cognate) aminoacyl-tRNA (aa-tRNA) from a large pool of incorrect aa-tRNAs through a two-stage mechanism. In the initial selection stage, aa-tRNA is delivered to the ribosome as part of a ternary complex with elongation factor EF-Tu and GTP. Interactions between codon and anticodon lead to activation of the GTPase domain of EF-Tu and GTP hydrolysis. Then, in the proofreading stage, aa-tRNA is released from EF-Tu and either moves fully into the A/A site (a step termed “accommodation”) or dissociates from the ribosome. Cognate codon-anticodon pairing not only stabilizes aa-tRNA at both stages of decoding but also stimulates GTP hydrolysis and accommodation, allowing the process to be both accurate and fast. In previous work, we isolated a number of ribosomal ambiguity (ram) mutations in 16S rRNA, implicating particular regions of the ribosome in the mechanism of decoding. Here, we analyze a representative subset of these mutations with respect to initial selection, proofreading, RF2-dependent termination, and overall miscoding in various contexts. We find that mutations that disrupt inter-subunit bridge B8 increase miscoding in a general way, causing defects in both initial selection and proofreading. Mutations in or near the A site behave differently, increasing miscoding in a codon-anticodon-dependent manner. These latter mutations may create spurious favorable interactions in the A site for certain near-cognate aa-tRNAs, providing an explanation for their context-dependent phenotypes in the cell.
INTRODUCTION
Translation of the genetic code is highly accurate (error rate of 10−3–10−6), and the ribosome plays an active role in achieving this level of fidelity (Rodnina 2012). Decoding occurs through a two-stage selection process. During initial selection, aminoacyl-tRNA (aa-tRNA) binds the ribosome as part of a ternary complex (TC) with elongation factor Tu (EF-Tu) and GTP. Interactions between the codon and anticodon in the 30S A site lead to activation of EF-Tu and GTP hydrolysis. Then, during the proofreading stage, the aa-tRNA either moves into the A/A site (a step termed “accommodation”), where it can participate in peptide bond formation, or dissociates from the ribosome. Discrimination of cognate vs. near-cognate aa-tRNA relies on a number of key steps in the process (Pape et al. 1999; Gromadski and Rodnina 2004; Geggier et al. 2010). Cognate codon-anticodon pairing not only stabilizes aa-tRNA at both stages of decoding but also increases the forward rate constants for GTPase activation/GTP hydrolysis and accommodation. Acceleration of these forward steps in response to codon-anticodon pairing allows translation to be both accurate and fast.
High-resolution structures of ribosomal complexes show that cognate codon-anticodon pairing induces three universally conserved residues in the 16S rRNA (G530, A1492, and A1493) to rearrange and dock into the minor groove of the codon-anticodon helix (Ogle et al. 2001, 2002; Voorhees et al. 2010). The contacts formed by these 16S nucleotides require Watson-Crick base pair (bp) geometries at the first two positions of the codon-anticodon helix. The geometry of the third base pair is less stringently monitored, allowing, for example, the formation of wobble pairs. These structures help explain how the ribosome increases the binding specificity for cognate aa-tRNA (Ogle et al. 2002; Gromadski et al. 2006; Ledoux et al. 2009). How these interactions in the 30S A site stimulate GTP hydrolysis and subsequent accommodation remains less clear.
We previously isolated many ribosomal ambiguity (ram) mutations in the 16S rRNA (McClory et al. 2010). These mutations increase errors during translation elongation and cluster in several distinct regions of the ribosome. Based on the number of isolates obtained and the strength of phenotypes conferred, three regions stand out as most critical for decoding fidelity: (1) the A site; (2) helix h12, which lies on the solvent side of the subunit near S4 and S5; and (3) helices h8 and h14, which constitute the 30S portion of inter-subunit bridge B8. In line with this assessment, ram mutations isolated in r-proteins map to the S4/S5 interface and to L19, a component of the 50S portion of B8 (Olsson and Isaksson 1979; Bjorkman et al. 1999; Dahlgren and Ryden-Aulin 2000; Maisnier-Patin et al. 2002, 2007).
Analysis of a subset of these 16S ram mutations has shown that bridge B8 increases the stringency of decoding by negatively regulating GTPase activation (McClory et al. 2010; Fagan et al. 2013). Mutations predicted to disrupt bridge B8 (i.e., h8Δ3, a 3-bp truncation of h8; h14Δ2, a 2-bp truncation of h14; and G347U in h14)—as well as mutation G299A in h12—increase the rate of EF-Tu-dependent GTP hydrolysis, particularly in the near-cognate case. Crystal structures of mutant 70S ribosomes show that either G347U or G299A induces a distortion of h8/h14 that disrupts B8 (Fagan et al. 2013). These conformational changes in h8/h14 are virtually identical to those seen in cocrystal structures of wild-type 70S ribosomes bound with TC (stabilized with either kirromycin or GDPCP) (Schmeing et al. 2009; Voorhees et al. 2010; Fagan et al. 2013), suggesting that G347U and G299A act, at least in part, by promoting formation of the GTPase-activated state. Together, these observations indicate that disruption of bridge B8 is a critical aspect of GTPase activation and that h12 is allosterically linked to B8 (Fagan et al. 2013).
In this study, we characterize a larger subset of ram mutations in 16S rRNA, comparing their effects on initial selection, proofreading, RF2-dependent termination, and overall miscoding in multiple contexts. Our data show that bridge B8 contributes to both initial selection and proofreading and that mutations in or near the A site influence decoding in a codon-anticodon-dependent manner.
RESULTS
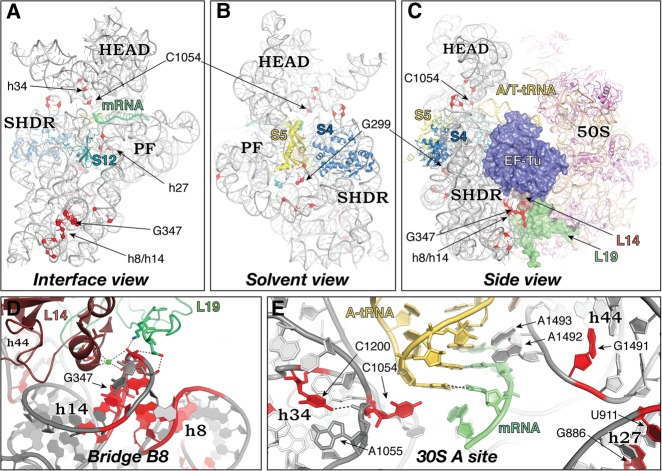
To better understand the roles of distinct regions of the ribosome in decoding, we selected a subset of mutations for further analysis. These mutations were chosen based on their phenotypes in the cell and their location in the ribosome (Fig. 1). Three mutations in the h8/h14 region (h8Δ3, h14Δ2, and G347U) directly destabilize bridge B8 (Fig. 1A,D). Mutation G299A is located in h12, on the solvent side of the subunit near the S4/S5 interface (Fig. 1B,C), and destabilizes bridge B8 allosterically (Fagan et al. 2013). All four of these mutations increase both missense and nonsense suppression in vivo (McClory et al. 2010). Mutation G886A is located in h27 (Fig. 1A), near the binding sites of several error-promoting antibiotics (Carter et al. 2000). While conferring more modest effects on missense and nonsense suppression (McClory et al. 2010), G886A was the only mutation to be additionally identified in a screen for translation initiation errors (Qin and Fredrick 2009). Four mutations (C1054U, C1054A, C1200U, and G1491A) are located in or near the A site (Fig. 1E) and have variable effects on missense and nonsense suppression. With our reporters, C1054U acts as a nonsense (UGA) suppressor but exhibits a restrictive (hyperaccurate) phenotype for Glu-tRNA misincorporation (McClory et al. 2010). C1200U and G1491A increase nonsense suppression to a greater degree than missense suppression, while C1054A strongly enhances both missense and nonsense suppression (McClory et al. 2010). In the ribosome, C1054 sticks out of h34 and packs against nucleotide (nt) 34 of the anticodon of A-tRNA (Fig. 1E). C1200 forms a base triple with A1055 and U1205 in h34 and forms a hydrogen bond with the phosphate of A1055. G1491 lies on the opposite side of the A site, next to A1492 and A1493 in h44 (Fig. 1E).
FIGURE 1.
Locations of ram mutations in 16S rRNA. Tertiary structure of the 16S rRNA, viewed from the subunit interface (A) and solvent (B) perspectives, and in the context of the 70S ribosome with bound ternary complex (C). Red spheres indicate positions of ram mutations. Small subunit proteins other than S4, S5, and S12 have been computationally removed for clarity. (SHDR) Shoulder domain, (PF) platform domain. (D) Zoomed-in view of intersubunit bridge B8. Hydrogen bonds that are lost upon bridge disruption are shown as dashed lines. (E) Zoomed-in view of the 30S A site showing the interactions of various 16S rRNA residues with A-tRNA and mRNA. For clarity, h18 and S12 are omitted from the foreground of this view. (Gray) 16S rRNA, (tan) 23S/5S rRNA, (magenta) unlabeled 50S proteins, other features as indicated. Figure based on PDB files 2WRN, 2WRO, 2WDG, and 2WDI.
Effects of 16S ram mutations on initial selection
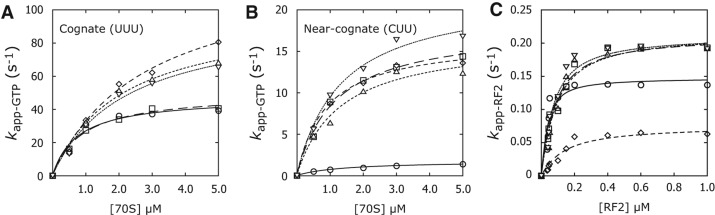
As mutations h8Δ3, h14Δ2, G347U, and G299A all cause defects in initial selection (McClory et al. 2010; Fagan et al. 2013), we tested whether the A-site ram mutations similarly affected EF-Tu-dependent GTP hydrolysis. Control and mutant 70S initiation complexes (70SICs), programmed with either a cognate UUU or near-cognate CUU codon in the A site, were rapidly mixed with EF-Tu•[γ32P]GTP•Phe-tRNAPhe under single-turnover conditions, and the reactions were quenched at various time points. Apparent rates obtained from individual time courses were plotted as a function of 70SIC concentration, and the data were fit to a hyperbolic function to determine the kcat and KM parameters (Fig. 2A,B; Table 1). Like previously characterized ram mutations (McClory et al. 2010; Fagan et al. 2013), those in or near the A site increased kcat for both cognate and near-cognate reactions, without strongly impacting KM (Table 1). The effect on kcat was considerably larger in the near-cognate case, reducing the selectivity of the reaction substantially. C1054U appeared somewhat unique in that the mutation failed to increase kcat in the cognate case. Nevertheless, each 16S ram mutation tested thus far causes similar defects in initial selection (Table 1).
FIGURE 2.
Effects of 16S A-site mutations on EF-Tu-dependent GTP hydrolysis and RF2-dependent termination. (A,B) 70S initiation complexes (70SIC) programmed with either cognate UUU or near-cognate CUU in the A site were rapidly mixed with EF-Tu•[γ32P]GTP•Phe-tRNAPhe, and rates of GTP hydrolysis were determined. Shown are apparent rates for cognate (A) and near-cognate (B) reactions plotted vs. 70SIC concentration. Data were fit to the equation kapp = kcat[70SIC]/(KM+[70SIC]), yielding the kinetic parameters shown in Table 1. (C) Ribosomes (0.2 µM) containing Ac[35S]Met-tRNAMet in the P site and codon UAA in the A site were rapidly mixed with RF2, and rates of Ac[35S]Met-tRNAMet hydrolysis were determined. Shown are apparent rates plotted as a function of RF2 concentration. Data were fit to the quadratic equation kapp = kcat{(A + B + KM) – [(A + B + KM)2 – 4AB]1/2}/(2A), where A and B represent the total concentrations of ribosomal complex and RF2, respectively, in the reaction. This yielded the kcat and KM parameters shown in Table 2. Control, ○ and solid lines; C1054U, □ and long-dashed lines; C1054A, ⋄ and medium-dashed lines; C1200U, △ and short-dashed lines; G1491A, ▽ and dotted lines.
TABLE 1.
Effects of 16S rRNA ram mutations on single-turnover EF-Tu-dependent GTP hydrolysis

Effects of 16S ram mutations on RF2 function
Certain A-site mutations (C1054U, C1200U, G1491A) were found previously to confer a notably stronger phenotype for nonsense (UGA) suppression than missense suppression (McClory et al. 2010), raising the possibility that these mutations act largely by impairing RF2 function. To investigate this, we measured the rate of single-turnover RF2-catalyzed hydrolysis of Ac[35S]Met-tRNAMet bound to the P site of 70S ribosomes programmed with a UAA codon in the A site (Fig. 2C; Table 2), as described previously (Devaraj and Fredrick 2010). (The stop codon UAA was chosen for technical reasons—an mRNA with AUG UGA would create GUG in the +2 frame, which could potentially compete with AUG for Ac[35S]Met-tRNAMet pairing in the P site.) Mutations C1054U, C1200U, and G1491A all increased kcat and KM modestly, resulting in similar two- to threefold decreases in kcat/KM. In contrast, mutation C1054A reduced kcat and increased KM, resulting in a ninefold drop in kcat/KM, consistent with earlier studies (Arkov et al. 1998, 2000). While this substantial termination defect of C1054A probably contributes to the high-level UGA read-through seen in vivo, C1054A also confers strong missense suppression, indicative of a large decoding defect (McClory et al. 2010). The small effects of C1054U, C1200U, and G1491A on RF2-dependent termination do not readily explain the differential effects of these mutations on nonsense and missense suppression in the cell.
TABLE 2.
Effects of 16S rRNA ram mutations on single-turnover RF2-dependent peptidyl-tRNA hydrolysis

Effects of 16S ram mutations on proofreading
To determine which 16S mutations influenced proofreading, we measured the extent of GTP hydrolysis vs. peptide bond formation in single-turnover decoding reactions (Table 3). EF-Tu• [γ32P]GTP•Phe-tRNAPhe was mixed with 70SICs containing P-site [35S]fMet-tRNAfMet and either codon UUU or CUU in the A site. After a 30-sec incubation at room temperature, the reaction was quenched, and the extent of both [γ32P]GTP hydrolysis and [35S]fMet-Phe formation was quantified. The ratio of peptide bonds formed to GTP molecules hydrolyzed (dip/Pi) was then calculated. This ratio is proportional to the probability of successful aa-tRNA accommodation after GTP hydrolysis. Finally, the fidelity of the proofreading phase of decoding (FP) was estimated as FP = (dip/Pi)cognate/(dip/Pi)near-cognate (Table 3), essentially as described (Gromadski and Rodnina 2004).
TABLE 3.
Effects of 16S rRNA ram mutations on the proofreading phase of decoding

Small variations in prepared TC made calculating absolute values of dip/Pi nontrivial; hence, we chose to normalize all dip/Pi values to that of the cognate control reaction, which was included in every experiment. For each mutant ribosome programmed with a cognate UUU codon, dip/Pi was near 1 (Table 3). Consistent with previous work (Gromadski and Rodnina 2004), control ribosomes programmed with a CUU codon in the A site gave a dip/Pi value of 0.09, indicating that rejection of Phe-tRNAPhe (Phe-tRNA) at the proofreading stage is 11 times more likely in the presence of this first position codon-anticodon mismatch (FP = 11). Ribosomes with G299A, h8Δ3, h14Δ2, or G347U all displayed proofreading defects, with FP values decreased to 3.7, 3.4, 2.6, and 2.2, respectively. All of these mutations disrupt bridge B8 (McClory et al. 2010; Fagan et al. 2013), indicating that B8 regulates not only initial selection but also proofreading. In contrast, substitutions of A-site residue C1054 showed no obvious defects in proofreading of Phe-tRNA. C1054A ribosomes gave an FP value similar to that of the control, while C1054U ribosomes gave an FP value of 21, indicating somewhat hyperaccurate proofreading. Mutation C1200U led to a proofreading defect, reducing FP to 3.7, similar to that of B8-disrupting mutations. Another A-site mutation, G1491A, conferred a smaller proofreading defect, indicated by an FP value of 5.5 (Table 3).
The unique behavior of C1054A and C1054U ribosomes prompted us to repeat the assay for another aa-tRNA, Tyr-tRNATyr (Tyr-tRNA), and ribosomes programmed with a cognate UAC or near-cognate UAG codon (Table 3). In this case, UAG-programmed control ribosomes rejected Tyr-tRNA 25 times more often than those programmed with the cognate UAC codon. Again, G299A and G347U ribosomes displayed proofreading defects, with FP values of 2.6 and 3.6, respectively. C1054A and C1054U ribosomes also displayed defects in proofreading of Tyr-tRNA, with respective FP values of 3.5 and 2.1, even though these ribosomes retained faithful proofreading in the Phe-tRNA case described above. Thus, substitutions of C1054 appear to differentially influence proofreading, depending on the particular tRNA and/or codon.
Effects of 16S ram mutations on miscoding in various contexts
The differences in proofreading by the C1054-substituted ribosomes raised the possibility that certain ram mutations promote only some types of errors. To investigate this, we measured apparent rates of miscoding with four different aa-tRNAs: Phe-tRNAPhe, Tyr-tRNATyr, Lys-tRNALys, and Glu-tRNAGlu. Various ribosome complexes programmed with a near-cognate codon were mixed with each TC under conditions that facilitate rapid reassembly of the TC (Johansson et al. 2012), giving aa-tRNA multiple chances for misincorporation.
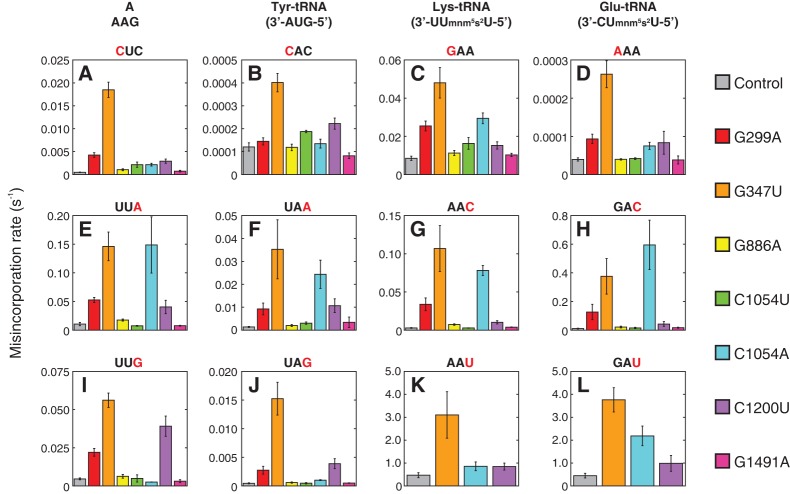
In control ribosomes, the apparent rate of miscoding varied dramatically depending on the nature of the tRNA and codon-anticodon mismatch (Fig. 3; note scales of y-axes). First-position mismatches generally produced slower rates of misincorporation than third-position mismatches. However, there was one excursion from this pattern—misincorporation of Lys-tRNALys (Lys-tRNA) occurred at higher frequency in response to GAA [(8.5 ± 1.1) × 10−3 sec−1] than to AAC [(2.9 ± 0.4) × 10−3 sec−1]. Notably, the rate of misincorporation for Glu-tRNAGlu (Glu-tRNA) at codon AAA (first-position mismatch) was more than 10,000-fold lower than at GAU (third-position mismatch) (Fig. 3D,L), exemplifying the wide range of rates observed in this set of experiments.
FIGURE 3.
Effects of 16S ram mutations on overall miscoding in various contexts. Control and mutant 70SICs (color-coded, as indicated) programmed with various near-cognate codons in the A site were mixed with the TC under conditions that facilitate reassembly of the TC, and apparent rates of miscoding were measured. Misincorporation rates for Phe (A, E, I), Tyr (B, F, J), Lys (C, G, K), and Glu (D, H, L) in the presence of various first-position (A–D) or third-position (E–L) codon-anticodon mismatches were compared. The codon programmed in the A site of each ribosome complex is indicated above each graph, with the mismatched base colored red. Anticodon sequences for each tRNA are indicated at the top of the figure. Data in panels A–J represent the mean ± SEM from three or more independent experiments. For panels K and L, rates were measured using a quench-flow machine, and reported values and their standard errors come from the curve fits.
Mutation G347U increased misincorporation to the greatest degree in almost all cases, from three- to 38-fold, and the magnitude of its effects did not follow any discernible pattern with regard to first-position or third-position mismatches (Fig. 3). Mutation G299A similarly increased misincorporation in every case but one (Tyr-tRNA with the codon CAC) (Fig. 3B) and showed no obvious bias between first and third position mismatches. These data suggest that G299A and G347U enhance miscoding in a general way.
Mutation C1054A, on the other hand, increased miscoding in a codon-anticodon-dependent manner (Fig. 3). C1054A modestly increased misincorporation of first-position mismatched Phe-tRNA (fourfold), Lys-tRNA (fourfold), and Glu-tRNA (twofold), and had no significant effect on first-position mismatched Tyr-tRNA (Fig. 3A–D). However, C1054A dramatically increased misincorporation for several third-position mismatched tRNAs (Fig. 3E–H). Interestingly, the magnitude of the C1054A effects correlate with the identity of the mismatched base pair at the third position. Phe-tRNA and Tyr-tRNA both form A-G mismatches with UUA- and UAA-programmed ribosomes, respectively, and C1054A increased misincorporation in these cases by 14- and 18-fold (Fig. 3E,F). For Lys-tRNA with an AAC codon and Glu-tRNA with a GAC codon, both forming C-mnm5s2U mismatches, C1054A increased misincorporation by 27- and 54-fold (Fig. 3G,H). These same two tRNAs create U-mnm5s2U wobble mismatches with AAU and GAU codons, and the observed increase was again similar but smaller, two- and fivefold (Fig. 3K,L). Finally, for G-G wobble mismatches, C1054A had the smallest effects, decreasing misincorporation for Phe-tRNA on UUG by twofold and increasing misincorporation for Tyr-tRNA on UAG by twofold (Fig. 3I,J).
Like C1054A, mutation C1200U influenced decoding in a context-dependent manner (Fig. 3). C1200U modestly increased misreading for first-position mismatched aa-tRNA, and strongly increased misincorporation for certain third-position mismatched reactions, depending on the identity of the third-position mismatch. The contexts where C1200U had the greatest effects were G-G pairings for Phe-tRNA and Tyr-tRNA, (Fig. 3I,J) followed by A-G pairings for the same two tRNAs (Fig. 3E,F), then C-mnm5s2U (Fig. 3G,H) followed by U-mnm5s2U pairings for Lys-tRNA and Glu-tRNA (Fig. 3K,L). This pattern of G-G > A-G > C-mnm5s2U > U-mnm5s2U is clearly distinct from the C-mnm5s2U > A-G > U-mnm5s2U > G-G pattern seen with C1054A.
Mutations G886A and C1054U increased misincorporation in several cases, although the effects were small (Fig. 3). Mutation G1491A had little or no effect on misincorporation, at least for the contexts tested in Figure 3.
DISCUSSION
In this study, we provide evidence that ram mutations in 16S rRNA fall into two functional classes—those that generally increase miscoding and those that influence decoding in a codon-anticodon-dependent manner. Mutations in the former group include G299A and G347U, which lie away from the A site and are known to disrupt bridge B8 (McClory et al. 2010; Fagan et al. 2013). These mutations consistently increase misreading, regardless of context. Mutations in the latter group include C1054A and C1200U, which are predicted to alter the structure of the 30S A site. These mutations cause high-level misreading in a manner that depends on the tRNA and/or nature of the mismatch.
C1054 contributes to the 30S A site, packing against nt 34 of the anticodon of A-tRNA. This interaction is seen whether tRNA occupies the A/T site or A/A site, suggesting that the contact is maintained throughout most of the decoding process (Ogle et al. 2001; Schmeing et al. 2009; Voorhees et al. 2009, 2010). The base of C1200 is oriented to form a hydrogen bond with the 5′-phosphate of A1055, stabilizing the position of C1054. Hence, mutations C1054A and C1200U are predicted to alter ribosomal contacts to nt 34 of A-tRNA, directly or indirectly. We propose that these mutations allow spurious favorable interactions to form, which stabilize near-cognate aa-tRNA in certain contexts. Because base-stacking interactions involving purines are generally more stable than those involving pyrimidines (Saenger 1984), an introduced purine at position 1054 may promote base stacking with nt 34 of A-tRNA, particularly for certain mismatched or non-Watson-Crick base pairs. Precedent for this idea comes from observed stacking between C1054 and base 34 of A-tRNA in crystal structures of ribosomes containing the ASL of tRNAArgICG (Murphy and Ramakrishnan 2004). When ASLArgICG is bound to a CGC codon, forming a Watson-Crick cytosine-inosine (C-I) base pair at the wobble position, C1054 packs normally against the ribose of I34. However, when ASLArgICG is bound to a CGA codon, the purine-purine A-I pair widens the codon-anticodon helix at the third position, and C1054 is reoriented to stack with the inosine base. In an analogous way, the geometry of an A-G mismatch at the third position may allow for favorable base stacking between G34 of Phe-tRNA or Tyr-tRNA and A1054, explaining the high misreading rate for C1054A ribosomes in such contexts. Mutations C1054A and C1054G confer much stronger miscoding phenotypes in vivo than C1054U (McClory et al. 2010), as one would expect if spurious stacking of 16S rRNA base 1054 and tRNA base 34 is responsible for the higher rates of miscoding. We had hoped to include C1054G in this study but were unable to generate the corresponding Escherichia coli Δ7 prrn strain, perhaps because the decoding defects conferred by the mutation are too large (McClory et al. 2010).
In both bacteria and yeast, substitutions of C1054 cause variable suppression phenotypes, depending on the particular reporter constructs employed (Hanfler et al. 1990; Prescott et al. 1991; Prescott and Kornau 1992; Chernoff et al. 1996; Pagel et al. 1997; Konstantinidis et al. 2006; McClory et al. 2010, 2011). The basis of this “phenotypic heterogeneity” has been a long-standing puzzle in the field. Here, we show that the effects on decoding of C1054A (and to a lesser degree, C1054U) vary depending on the codon and anticodon involved. This helps to explain the complex suppression patterns conferred by mutations of C1054 in the cell.
Nucleotide G1491 is immediately adjacent to the universally conserved A1492 and A1493, which dock into the minor groove of the codon-anticodon helix. While G1491 itself is not highly conserved (Cannone et al. 2002), mutation G1491A in E. coli leads to high-level nonsense suppression, with a considerably smaller effect on missense suppression (McClory et al. 2010). High UGA read-through does not seem to be due to a termination defect, as G1491A ribosomes are not particularly compromised in RF2 function (Table 2). This conclusion should be considered tentative, however, because our assay employed UAA rather than UGA, and mutations conferring UGA-specific defects in RF2-dependent termination have been reported (Ortiz-Meoz and Green 2011). G1491A does have significant effects on initial selection (Fig. 2), and to a lesser extent, on proofreading (Table 3), for Phe-tRNA. Puzzlingly, though, G1491A has little or no effect on the overall rate of misincorporation for any of the 10 contexts tested (Fig. 3A–J), even the case of Phe-tRNA misreading CUC (Fig. 3A). We propose that G1491A strongly enhances miscoding only in certain contexts (e.g., Trp-tRNA misreading of UGA, Phe-tRNA misreading of CUU), none of which were part of the Figure 3 data set. Consistent with this suggestion, a wider preliminary screen of contexts revealed that misreading of AAC by Tyr-tRNA is substantially increased (>20-fold) in G1491A ribosomes. Further work will be needed to elucidate the effects of G1491A on decoding.
There is considerable interest in methods to incorporate unnatural amino acids into proteins. Among the most successful of these are translation systems that employ orthologous aa-tRNA/aa-tRNA synthetase pairs in combination with nonsense and/or frameshift suppression (Liu and Schultz 2010). Our finding that the A-site ram mutations impact decoding in a codon-anticodon-dependent fashion raises the possibility that the corresponding mutant ribosomes might prove useful in these protein engineering systems. For example, a particular A-site ram mutation could substantially increase the efficiency of orthologous aa-tRNA incorporation events without causing a problematic reduction in overall translation fidelity. In line with this idea, selection experiments for 16S rRNA mutations that specifically enhance quadruplet decoding by Ser-tRNAUCUU identified mutations of nt 1195–1197 (Neumann et al. 2010), and A1196 normally contacts C1054 (Voorhees et al. 2009).
In control ribosomes, apparent rates of miscoding by Phe-tRNA, Tyr-tRNA, Lys-tRNA, and Glu-tRNA in response to a number of near-cognate codons varied over a wide range—four orders of magnitude (Fig. 3). With one exception (see below), the frequency of misincorporation was higher for third- than first-position mismatches. These data are in line with more comprehensive analyses of Lys-tRNA miscoding in vitro and in vivo (Kramer and Farabaugh 2007; Johansson et al. 2012) and argue against the idea that kinetic effects of single mismatches are similar regardless of their position (Gromadski et al. 2006). The exceptional case in our data set is that of Lys-tRNA, which seems to misread GAA at higher frequency than AAC, in contrast to previous reports (Kramer and Farabaugh 2007; Johansson et al. 2012). In retrospect, we noticed that the nucleotide following GAA in the mRNA for the experiment of Figure 3C is G. Hence, it is possible that the higher frequency of incorporation in this case stems from a +1 frameshift of fMet-tRNAfMet (to pair with UGG, one base different from the cognate start codon UUG) and presentation of the cognate Lys codon AAG in the A site. Consistent with this possibility, earlier toeprinting experiments show that ribosome complexes with P-site tRNAfMet paired to m291 (codons 1–2: AUG UUU) give a toeprint at position +16, whereas those with tRNAfMet paired to m292 (codons 1–2: AUG GUA) give toeprints at +16 and +17 (Shoji et al. 2006). Importantly, the experiment of Figure 3C is the only one in which this putative +1 frameshift event would present a cognate codon in the A site. Thus, we are confident that the measured incorporation rates for the other 11 contexts reflect misreading rates. Worth noting here is that, in our data set, misreading of GAU by Glu-tRNA and of AAU by Lys-tRNA occurred at the highest frequencies (Fig. 3K,L), and the corresponding missense mutations are among the few such mutations that are clearly suppressible in the cell (Kramer and Farabaugh 2007; McClory et al. 2010; Manickam et al. 2014).
In the proofreading stage of decoding, aa-tRNA is either accommodated into the A/A site or rejected from the ribosome. Cognate codon-anticodon pairing speeds accommodation and slows rejection (Pape et al. 1999; Geggier et al. 2010), greatly increasing the probability of productive aa-tRNA incorporation. While molecular dynamics simulations provide plausible paths for aa-tRNA transit from A/T to A/A sites (Whitford et al. 2010), how the ribosome impacts these tRNA movements that ultimately determine the selectivity of proofreading remains unclear. In this study, we find that mutations that disrupt bridge B8 (G299A, h8Δ3, h14Δ2, and G347U) reduce the stringency of not only initial selection but also proofreading. The proofreading defects of these mutant ribosomes, indicated by substantially higher levels of dipeptide formation in the near-cognate case, must stem from an increased rate of accommodation and/or a decreased rate of rejection. How might B8 influence proofreading? One possibility is that B8 affects proofreading indirectly by promoting EF-Tu•GDP dissociation. As discussed above, formation of the GTPase-activated state (with A/T aa-tRNA) involves disruption of B8. After GTP hydrolysis, EF-Tu undergoes a conformational change and dissociates from the ribosome. Reformation of B8 during these latter steps of decoding may facilitate release of EF-Tu•GDP. If so, mutations that disrupt B8 would be predicted to slow EF-Tu•GDP dissociation. As bound EF-Tu would present a steric obstacle to aa-tRNA release (Schmeing et al. 2009; Voorhees et al. 2010), a slower rate of EF-Tu•GDP release should effectively shunt aa-tRNA toward the productive pathway. Another possibility is that B8 allosterically destabilizes aa-tRNA and thereby increases the stringency of proofreading. Mutations that disrupt B8, then, would be predicted to stabilize aa-tRNA during the A/T-to-A/A transition and hence increase the probability of successful accommodation. Mutation G299A (in h12) allosterically disrupts B8 (Fagan et al. 2013), so there is precedent for the idea that B8 is conformationally linked to distal ribosomal sites. Clearly, further experiments are needed to test these hypotheses and elucidate the molecular mechanism through which B8 tunes proofreading.
Ram mutations away from the A site cluster largely to two regions—bridge B8 and the h12/S4/S5 region (Olsson and Isaksson 1979; Bjorkman et al. 1999; Dahlgren and Ryden-Aulin 2000; Maisnier-Patin et al. 2002, 2007; McClory et al. 2010). The recent finding that mutation G299A in h12 allosterically disrupts B8 raises the question of whether other ram mutations in the h12/S4/S5 region act in a similar way (Fagan et al. 2013). Zaher and Green (2010) studied the effects of mutation rpsD12 on decoding in some detail (Zaher and Green 2010). This is a frameshift mutation that causes a C-terminal truncation of S4, effectively removing numerous contacts at the S4-S5 interface. They found that ribosomes harboring this S4 truncation exhibit defects in initial selection but not in proofreading, suggesting a functional distinction between rpsD12 and G299A. One caveat here, though, is that Zaher and Green analyzed Leu-tRNA misreading of UUC as the near-cognate case, whereas we have used Phe-tRNA misreading of CUU. Clearly, it will be worthwhile to directly compare G299A, rpsD12, and other mutations in the h12/S4/S5 region to determine if and how their modes of action differ.
MATERIALS AND METHODS
Reagents
All experiments were done in polymix buffer (Ehrenberg et al. 1990; Shoji et al. 2006), unless otherwise noted. Purified E. coli MRE600 tRNAs (tRNAfMet, tRNAMet, tRNAPhe, tRNATyr, tRNALys, and tRNAGlu) were purchased from Chemical Block and aminoacylated as described (Walker and Fredrick 2008). For initial selection and proofreading experiments, mRNA was transcribed in vitro from pGENE32-based plasmids and gel-purified as described (Fredrick and Noller 2002). The mRNAs used for proofreading experiments have the sequence 5′-(N)42AAGGAAAUAAAAAUGNNNGUAUACAAAUCU(N)67-3′, where NNN corresponds to UUU, CUU, UAC, or UAG. Messages for the miscoding experiments of Figure 3 were synthesized by Sigma-Aldrich and have the sequence 5′-AAGGAAAUAAAAAUGNNNGUAUACAAAUCU-3′, where NNN is the indicated A site codon. Ribosomes, translation factors, and phenylalanyl-tRNA synthetase were purified as described (McClory et al. 2010). Tyrosyl-, lysyl-, and glutamyl-tRNA synthetases were purified with Talon Cobalt Affinity Resin (Clontech) from overexpression strains JW1629, JW2858, and JW2395, obtained from the ASKA collection (National BioResource Project-E. coli at the National Institute of Genetics, Japan). Pyruvate kinase and myokinase were purchased from Sigma-Aldrich.
Initial selection and termination assay
Initial selection and RF2-dependent peptide release experiments were carried out as described (Devaraj and Fredrick 2010; McClory et al. 2010).
Proofreading assay
70SICs were prepared by incubating 70S ribosomes (1 µM), IF1 (2 µM), IF2 (1 µM), IF3 (2 µM), mRNA (4 µM), [35S]-fMet-tRNAfMet (1 µM), and GTP (1 mM) in polymix buffer at 37°C for 1 h. Complexes were then purified by ultracentrifugation over 1.2 mL 1.1 M sucrose cushions made with polymix buffer and 15 mM MgCl2 in an SW50.2 rotor (Beckman) at 36,600 rpm for 2.5 h. Pelleted complexes were dissolved in polymix buffer, flash-frozen, and stored at −70°C. Ternary complex (TC) was made by incubating EF-Tu (10 µM), aa-tRNA (10 µM), phosphoenolpyruvate (PEP, 3 mM), pyruvate kinase (50 µg/mL), and [γ32P]-GTP (50 µM) in polymix buffer at 37°C for 15 min. Following incubation, TC was passed through P30 spin columns (Bio-Rad; pre-equilibrated in polymix) to remove unbound GTP and PEP, and then diluted to 0.5 µM. 70SICs were thawed and diluted in polymix to 2 µM. Equal volumes (10 µL) of 70SIC (2 µM) and TC (0.5 µM) were mixed and incubated at 20°C for 30 sec (Phe experiment) or 60 sec (Tyr experiment), at which point both GTP hydrolysis and peptide bond formation had reached plateaus. Reactions were quenched with 10 µL of 40% formic acid and centrifuged at 20,000g for 15 min to pellet tRNA. The extent of GTP hydrolysis was determined using TLC, by spotting the supernatant on PEI-cellulose TLC plates and developing with 500 mM potassium phosphate, pH 3.5. The extent of peptide bond formation was determined using electrophoretic TLC (Merryman and Green 2004), after washing the pellets with 15% formic acid, dissolving in 500 mM KOH, and spotting products on cellulose TLC plates. This method enables clear separation of unreacted formyl-methionine and the various dipeptides. Each experiment was done in parallel with control ribosomes, and the ratio of GTP hydrolyzed per peptide bond formed was normalized to the cognate control reaction. Experiments were performed at least three independent times.
Misincorporation experiments
For the experiments of Figure 3, 70SICs were prepared by incubating 70S ribosomes (0.2 µM), IF1 (0.4 µM), IF2 (0.4 µM), IF3 (0.4 µM), mRNA (1 µM), [35S]-fMet-tRNAfMet (0.2 µM), and 1× energy mix (1 mM ATP, 1 mM GTP, 10 mM PEP, 50 µg/mL pyruvate kinase, 5 µg/mL myokinase) in polymix buffer at 37°C for 15 min. TC was similarly prepared in polymix by incubating EF-Tu (3 µM), EF-Ts (3 µM), tRNA (2 µM), aa-tRNA synthetase (1.5 µM), amino acid (200 µM), and 1× energy mix. Because ATP, GTP, and PEP are Mg2+ chelators, buffer for the 70SIC and TC preparations was supplemented with 6.5 mM additional Mg(OAc)2 to adjust the free Mg2+concentration to ∼5 mM (Johansson et al. 2012). Equal volumes (25 µL) of 70SIC and TC were mixed and incubated at 20°C. At various time points, 4 µL of the reaction were removed and quenched with an equal volume of 500 mM KOH. Reactions were spotted on cellulose TLC plates subjected to electrophoretic TLC analysis. Dipeptide formation was plotted vs. time, and the data were fit to a single-exponential equation to obtain apparent rates.
ACKNOWLEDGMENTS
We thank Q. Liu for technical assistance with dipeptide formation and comments on the manuscript, J. Alfonzo and C. Dunham for helpful discussions, and NBRP-E. coli at NIG for protein expression strains. This work was supported by National Institutes of Health grant no. GM072528.
REFERENCES
- Arkov AL, Freistroffer DV, Ehrenberg M, Murgola EJ 1998. Mutations in RNAs of both ribosomal subunits cause defects in translation termination. EMBO J 17: 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Freistroffer DV, Pavlov MY, Ehrenberg M, Murgola EJ 2000. Mutations in conserved regions of ribosomal RNAs decrease the productive association of peptide-chain release factors with the ribosome during translation termination. Biochimie 82: 671–682 [DOI] [PubMed] [Google Scholar]
- Bjorkman J, Samuelsson P, Andersson DI, Hughes D 1999. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol Microbiol 31: 53–58 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348 [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Liebman SW 1996. The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: Genetic evidence in yeast. Proc Natl Acad Sci 93: 2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren A, Ryden-Aulin M 2000. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie 82: 683–691 [DOI] [PubMed] [Google Scholar]
- Devaraj A, Fredrick K 2010. Short spacing between the Shine-Dalgarno sequence and P codon destabilizes codon-anticodon pairing in the P site to promote +1 programmed frameshifting. Mol Microbiol 78: 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland CG 1990. Design and use of a fast and accurate in vitro translation system. In Ribosomes and protein synthesis—a practical approach (ed. Spedding G), pp. 101–129 IRL Press, Oxford [Google Scholar]
- Fagan CE, Dunkle JA, Maehigashi T, Dang MN, Devaraj A, Miles SJ, Qin D, Fredrick K, Dunham CM 2013. Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state. Proc Natl Acad Sci 110: 9716–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF 2002. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell 9: 1125–1131 [DOI] [PubMed] [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC 2010. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol 399: 576–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV 2006. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell 21: 369–377 [DOI] [PubMed] [Google Scholar]
- Hanfler A, Kleuvers B, Goringer HU 1990. The involvement of base 1054 in 16S rRNA for UGA stop codon dependent translational termination. Nucleic Acids Res 18: 5625–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Zhang J, Ehrenberg M 2012. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci 109: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis TC, Patsoukis N, Georgiou CD, Synetos D 2006. Translational fidelity mutations in 18S rRNA affect the catalytic activity of ribosomes and the oxidative balance of yeast cells. Biochemistry 45: 3525–3533 [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC 2009. A sequence element that tunes Escherichia coli tRNAAlaGGC to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Schultz PG 2010. Adding new chemistries to the genetic code. Annu Rev Biochem 79: 413–444 [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Berg OG, Liljas L, Andersson DI 2002. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol Microbiol 46: 355–366 [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Paulander W, Pennhag A, Andersson DI 2007. Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J Mol Biol 366: 207–215 [DOI] [PubMed] [Google Scholar]
- Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ 2014. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 20: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClory SP, Leisring JM, Qin D, Fredrick K 2010. Missense suppressor mutations in 16S rRNA reveal the importance of helices h8 and h14 in aminoacyl-tRNA selection. RNA 16: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClory SP, Devaraj A, Qin D, Leisring JM, Fredrick K 2011. Mutations in 16S rRNA that decrease the fidelity of translation. In Ribosomes (ed. Rodnina MV, et al. ), pp. 237–247 Springer-Verlag, Austria [Google Scholar]
- Merryman C, Green R 2004. Transformation of aminoacyl tRNAs for the in vitro selection of “drug-like” molecules. Chem Biol 11: 575–582 [DOI] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V 2004. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11: 1251–1252 [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW 2010. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464: 441–444 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Olsson MO, Isaksson LA 1979. Analysis of rpsD mutations in Escherichia coli. I. Comparison of mutants with various alterations in ribosomal protein S4. Mol Gen Genet 169: 251–257 [DOI] [PubMed] [Google Scholar]
- Ortiz-Meoz RF, Green R 2011. Helix 69 is key for uniformity during substrate selection on the ribosome. J Biol Chem 286: 25604–25610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel FT, Zhao SQ, Hijazi KA, Murgola EJ 1997. Phenotypic heterogeneity of mutational changes at a conserved nucleotide in 16 S ribosomal RNA. J Mol Biol 267: 1113–1123 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CD, Kornau HC 1992. Mutations in E. coli 16s rRNA that enhance and decrease the activity of a suppressor tRNA. Nucleic Acids Res 20: 1567–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott C, Krabben L, Nierhaus K 1991. Ribosomes containing the C1054-deletion mutation in E. coli 16S rRNA act as suppressors at all three nonsense codons. Nucleic Acids Res 19: 5281–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Fredrick K 2009. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol Microbiol 71: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV 2012. Quality control of mRNA decoding on the bacterial ribosome. Adv Protein Chem Struct Biol 86: 95–128 [DOI] [PubMed] [Google Scholar]
- Saenger W 1984. Principles of nucleic acid structure. Springer-Verlag, New York [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K 2006. Reverse translocation of tRNA in the ribosome. Mol Cell 24: 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Fredrick K 2008. Preparation and evaluation of acylated tRNAs. Methods 44: 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY 2010. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2010. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol Cell 39: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]