Abstract
Light excitation of humic matter generates reactive oxygen species (ROS) in surface waters of aquatic ecosystems. Abundant ROS generated in humic matter rich lakes include singlet oxygen (1O2) and hydrogen peroxide (H2O2). Because these ROS differ in half-life time and toxicity, we compared their effects on microbial activity (14C-Leucine incorporation) and bacterial community composition (BCC) in surface waters of humic Lake Grosse Fuchskuhle (North-eastern Germany). For this purpose, experiments with water samples collected from the lake were conducted in July 2006, September 2008 and August 2009. Artificially increased 1O2 and H2O2 concentrations inhibited microbial activity in water samples to a similar extent, but the effect of the respective ROS on BCC varied strongly. BCC analysis by 16S rRNA gene clone libraries and RT-PCR DGGE revealed ROS specific changes in relative abundance and activity of major bacterial groups and composition of dominating phylotypes. These changes were consistent in the three experiments performed in different years. The relative abundance of Polynucleobacter necessarius, Limnohabitans-related phylotypes (Betaproteobacteria), and Novosphingobium acidiphilum (Alphaproteobacteria) increased or was not affected by photo-sensitized 1O2 exposure, but decreased after H2O2 exposure. The opposite pattern was found for Actinobacteria of the freshwater AcI-B cluster which were highly sensitive to 1O2 but not to H2O2 exposure. Furthermore, group-specific RT-PCR DGGE analysis revealed that particle-attached P. necessarius and Limnohabitans-related phylotypes exhibit higher resistance to 1O2 exposure compared to free-living populations. These results imply that 1O2 acts as a factor in niche separation of closely affiliated Polynucleobacter and Limnohabitans-related phylotypes. Consequently, oxidative stress caused by photochemical ROS generation should be regarded as an environmental variable determining abundance, activity, and phylotype composition of environmentally relevant bacterial groups, in particular in illuminated and humic matter rich waters.
Introduction
Dissolved organic matter (DOM) is the major carbon and energy source for heterotrophic bacteria in aquatic ecosystems [1]. Humic lakes are characterized by a high content of allochthonous DOM with recalcitrant high-molecular-weight poly-aromatic compounds. Photochemical transformations of these compounds generate low-molecular-weight substances and thereby stimulate microbial activity and growth [2], [3]. On the other hand, photochemical processes lead to inhibitory effects including (i) photo-oxidation and (ii) transformation of labile compounds [4], [5] as well as (iii) generation of reactive intermediates such as reactive oxygen species (ROS) [6], [7], [8]. ROS generation in aquatic ecosystems occurs by light excitation of DOM, in particular humic matter, and subsequent formation of triplet excited states in poly-aromatic compounds [8]. Light-excited DOM transfers energy or electrons to molecular oxygen. Thereby, the transfer of energy generates singlet oxygen (1O2) and the incomplete reduction of oxygen leads to the formation of hydrogen peroxide (H2O2) and other ROS. Recent experiments strongly suggest that distinct structures in humic matter are linked to the formation of 1O2 or H2O2 [9] and that the reaction of 1O2 with DOM generates small amounts of H2O2 [9], [10].
Effects of photochemically altered DOM on microorganisms were mainly investigated via inoculation of pre-irradiated DOM with natural microbial assemblages [11], [12] including studies, which examined the effect of substrate availability on bacterial community composition (BCC) [13], [14], [15]. In a recent study, we showed that 1O2 has the potential to inhibit typical freshwater bacterial species and consequently affect BCC [16]. Singlet oxygen is highly reactive, exhibits a half-life time in water of ∼3.5 μs [17], and causes cell damage by oxidation of lipids, nucleic acids, and proteins [18], [19]. In contrast, H2O2 has a half-life time of up to 8 hours in freshwater [20]. Moreover, H2O2 diffuses through biological membranes and mainly reacts with iron-sulphur clusters leading to subsequent intracellular hydroxyl radical formation and damage of biomolecules [21]. Hence, potentials for cell damage caused by 1O2 and H2O2 differ substantially.
In a previous study, short and long term effects of 1O2 on BCC were investigated [16]. The present study compares effects of increased 1O2 and H2O2 concentrations on BCC and includes experiments on the activity of heterotrophic bacteria in the surface water of the humic matter rich south-west (SW) basin of Lake Grosse Fuchskuhle (North-eastern Germany). The experiments were designed to elucidate differences in sensitivity of dominating bacterial phylotypes towards naturally occurring ROS of different toxicity. We tested the following hypotheses: i) 1O2 and H2O2 exposure elicit specific changes in microbial activity and BCC and ii) ROS-induced changes differ between particle-attached and free-living bacterial communities. Investigation of the latter hypothesis is of particular importance since a higher generation of 1O2 has been observed in particles compared to the ambient water in humic matter rich ecosystems [22], [23].
Results
1O2 and H2O2 C°ncentrations in Surface Water Samples
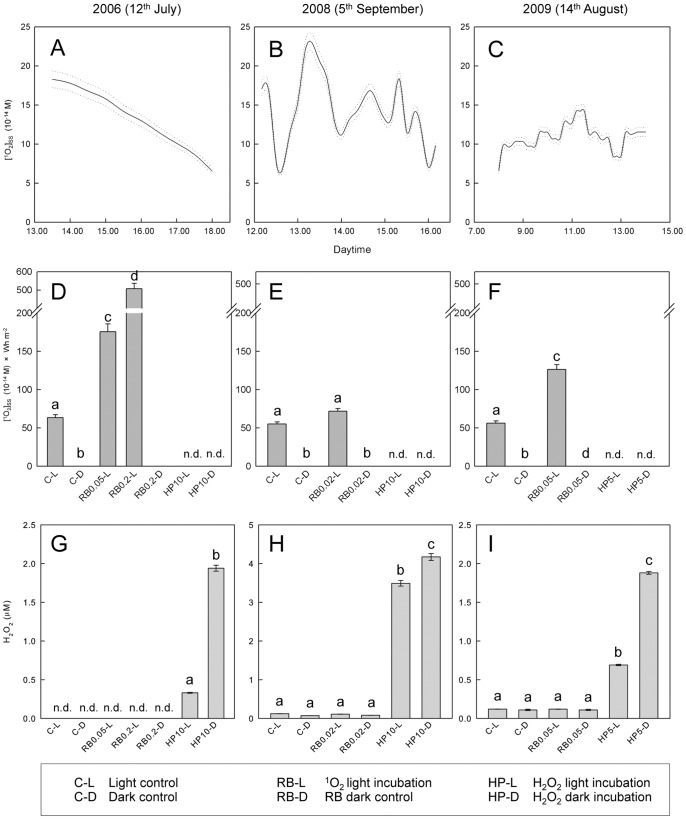
Three sets of in situ experiments were performed in July 2006, September 2008 and August 2009. For each experiment day, 1O2 steady state [1O2]SS concentrations and H2O2 formation were determined, because variations in sunlight intensity and in concentration of dissolved organic carbon (DOC) were observed (Table 1). By applying the furfuryl alcohol (FFA) method [24] we observed similar in situ [1O2]SS concentrations on all three experiment days that were in the range of 11.2 to 14.1×10−14 M in the surface water layer of the lake (Table 1). The kinetics of 1O2 formation differed between experiment days (Fig. 1A–C), but the dose of 1O2 exposure was very similar and ranged from 56.2 to 63.5×10−14 M Wh m−2 (C-Ls; Fig. 1D–F). Hydrogen peroxide concentrations were low in all water samples. During diurnal cycle studies ∼50 nM were detected on 11th July 2006 (data not shown), but in 2008 and 2009, H2O2 concentrations were in the range of 70 to 120 nM (Fig. 1H and I).
Table 1. Selected environmental parameters on experiment days in 2006, 2008 and 2009.
| Parameter | Sample | ||
| 2006 (12nd July) | 2008 (5th September) | 2009 (14th August) | |
| DOC (mg C L−1) | 23.3±1.8 | 34.0±0.1 | 28.4±1.1 |
| Average light intensity (W m−2) | 570 | 445 | 557 |
| In situ [1O2]SS (10−14 M) | 14.1±0.8 | 11.8±0.01 | 11.2 |
| In situ H2O2 (nM) | n.d. | 120±2.5 | 120±1.42 |
DOC concentration, average light intensity and subsequent [1O2]SS and H2O2 concentrations slightly differed between experiment days of the three studied years.
n.d.: not determined.
Figure 1. Formation of 1O2 and H2O2 during experiments in 2006, 2008 and 2009.
Kinetics of [1O2]SS in the surface water layer (A–C) were calculated from the rate of furfuryl alcohol decay and the light intensity according to Haag and Hoigne (1986). The formation of 1O2 largely depends on the light intensity (Table S1) and hence [1O2]SS kinetics depend on the weather conditions. A. 12nd July 2006: a clear sky during the afternoon led to a steady decrease in [1O2]SS concentrations from noon to late afternoon. B. 5th September 2008: a cloudy sky during the afternoon caused fluctuation in [1O2]SS concentrations. C. 14th August 2009: a slightly overcasted sky during the whole day led to reduced fluctuations in [1O2]SS concentrations compared to 2008. Values for solar radiation and rainfall within 30 days prior to the experiments were similar (Fig. S9) and hence all three experiments were conducted under comparable environmental situations. The addition of Rose Bengal (RB) increased the formation of 1O2 (D–F). D. 2.8 -fold for RB0.05-L and 8-fold for RB0.2-L in 2006, E. 1.3-fold in 2008, and F. 1.9-fold in 2009. Hydrogen peroxide concentrations were analysed in all samples at the end of the experiments (G–H). G. and H. 10 μM H2O2 were added in 2006 and 2008, respectively. I. 5 μM H2O2 were added in 2009. Numbers at RB and HP on the x-axis labels correspond to μM concentrations of RB or H2O2. Please note the different scale in panel H compared to panels G and I. n.d.: not determined. An overview of the abbreviations used for the experimental setups is given in the box at the bottom of the Figure. C–L/D: Light and dark control incubations, RB-L: Light incubation with increased [1O2]SS, RB-D: Dark control for RB, HP-L/D: Light and dark incubations with H2O2. Dotted lines in A–C and error bars in D–F represent the standard deviation of the FFA method where three distinct water samples were used to determine sample specific [1O2]SS concentrations. Error bars in G–H indicate the standard deviation of three analysed samples. Different letters at the top of the bars depict statistically significant differences (with p≤0.001) between values as determined by one-way ANOVA followed by pair-wise multiple comparison analysis with the Tukey’s test performed in Sigma Stat v. 2.0 (Systat Software). The same letters indicate that depicted values are not significantly different to each other.
Environmental conditions with respect to ROS concentrations may have varied throughout the years. In order to ensure that the reactivity of natural organic matter (NOM) was similar on each experiment day, 0.22 μm filtered water samples were further analysed (Materials S1). Normalization of ROS formation to mg DOC L−1 revealed that the specific 1O2 formation was similar between the experiment days. In contrast, the specific H2O2 formation was higher in 2009 compared to 2006 and 2008 (Table S1). Large variations of in situ 1O2 formation were not observed. In contrast, for H2O2 an up to 4–5 fold variation in formation rate was detected (Table S1), but concentrations measured in lake water samples remained similar (Fig. 1H and I).
Modification of 1O2 and H2O2 Concentrations
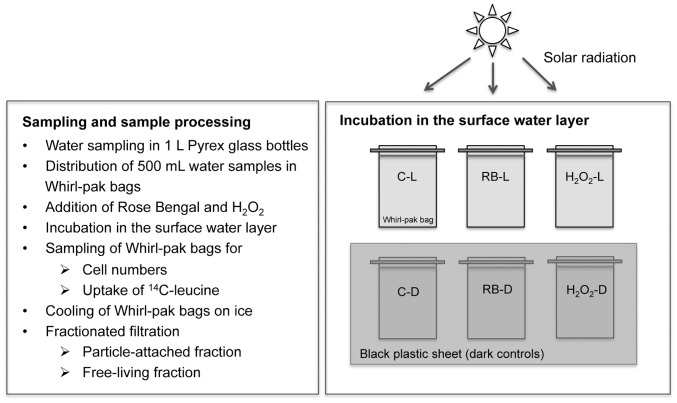
All in situ experiments performed in the summers of 2006, 2008 and 2009 were designed to test whether effects of increased 1O2 and H2O2 concentrations consistently differ in surface waters (hypothesis i). Respective field experiments (Fig. 2) were performed by obtaining water samples from the lake. Increased environmental ROS levels, particularly of H2O2, were obtained by adding the photosensitizer Rose Bengal (RB), a poly-aromatic compound that specifically generates 1O2 in the presence of light and oxygen or by H2O2 addition.
Figure 2. Design of field experiments.
Field experiments performed in 2006, 2008 and 2009 followed the same experimental outline as displayed in the flow chart. Whirl-pak bags were incubated in the surface water layer on large metal racks after addition of Rose Bengal and H2O2. Dark controls were covered with a black plastic sheet to avoid exposure to solar radiation. Abbreviations are given in Fig. 1.
Concentrations of 1O2 increased by 1.3 to 8-fold in light incubations after RB addition (Fig. 1D–F). Addition of 5 μM H2O2 in 2009 or 10 μM in 2006 and 2008 represented an increase in H2O2 concentrations by ∼45 to 200-fold, respectively. In experiments with H2O2 addition, the concentrations decreased during the time of incubation and ranged between 0.25 and 4.2 μM at the end of the experiments. Concentrations of H2O2 were lower in light incubations compared to dark controls (Fig. 1G–I) and H2O2 end concentrations were ∼3 to 33-fold higher compared to the non-amended controls. This notion is in line with the high capacity for H2O2 degradation found for humic matter rich water samples of the SW compartment (Materials S1).
Microbial Activity is Hampered by ROS Exposure
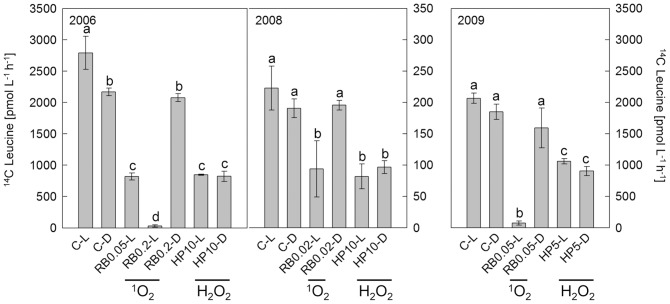
Activity of heterotrophic microorganisms, assessed by 14C-Leucine incorporation, was highest in the light controls (C–L) reaching 2800, 223, and 2100 pmol leucine L−1 h−1 in 2006, 2008, and 2009, respectively (Fig. 3). In 2006, microbial activity was significantly higher in the light than in the dark control. A similar trend occurred in 2008 and 2009, but it was not statistically significant. In all experiments, increased ROS levels caused inhibition of microbial activity. Precisely, generation of 1O2 (RB0.05-L) and addition of H2O2 (HP10-L/D) decreased microbial activity to 30% of that in the respective C–L in 2006. Similar treatments caused a decrease to 43% in 2008. In 2009, the addition of 5 μM H2O2 in light and dark treatments (HP5-L/D) resulted in a decrease of microbial activity to 51 and 44% of that in the respective C–L. Singlet oxygen generation in RB0.2-L in 2006 and RB0.05-L in 2009 decreased microbial activity to below 5% of the respective C–L. In 2009, particle-attached and free-living bacteria were assessed separately to investigate differences in their potential to incorporate leucine. In controls, particle-attached bacteria incorporated 2.3 to 2.6-fold more leucine than free-living bacteria. Exposure to ROS decreased the activity of both fractions to the same extent (Fig. S1), indicating an overall similar sensitivity of the microbial community to ROS exposure.
Figure 3. Activity of heterotrophic microorganisms after 1O2 and H2O2 exposure.
Microbial activity was measured as leucine incorporation during 1(with p≤0.001) between values as determined by one-way ANOVA followed by pair-wise multiple comparison analysis with the Tukey’s test performed in Sigma Stat v. 2.0 (Systat Software). The same letters indicate that depicted values are not significantly different to each other. Tests were done separately for each year. Abbreviations are given in Fig. 1.
Significant changes in cell numbers were not correlated with ROS exposure, except for the 1O2 exposure in 2008 (Fig. S2). As observed in earlier experiments [16] increased numbers of micrococcoid cells were responsible for elevated total cell numbers (Table S2).
Different concentrations of RB and H2O2 were used on the three experiment days (Fig. 3). Overall, we aimed for a similar inhibition of microbial activity by 1O2 and H2O2 in order to enable a direct comparison of changes in BCC within each experiment. Therefore, several RB concentrations were tested in 2006 and 2008 (data not shown) and for further analysis only those treatments were chosen which showed a similar inhibition of 14C-Leucine incorporation.
Relative Abundance of Bacterial Groups After 1O2 and H2O2 Exposure
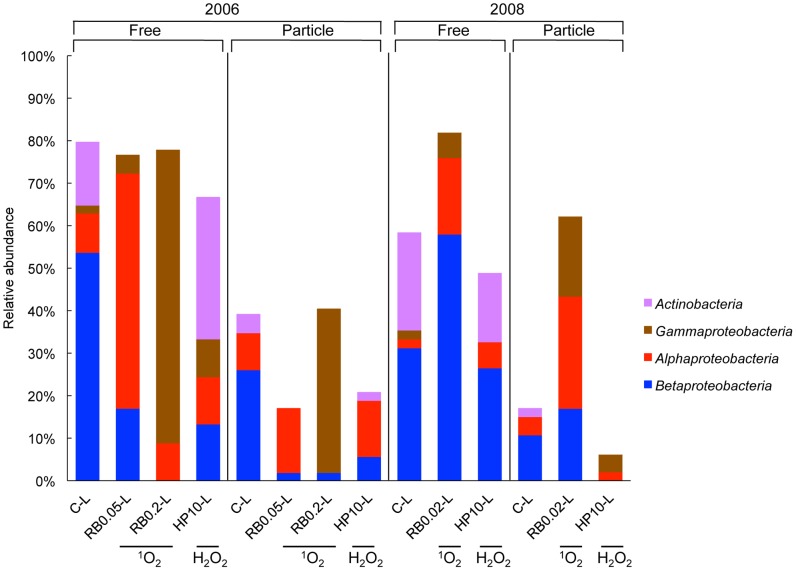
Clone libraries of free-living bacterial fractions in light controls (C-Ls) in 2006 and 2008 (Fig. 4) were dominated by Betaproteobacteria (54 and 31%), followed by Actinobacteria (15 and 23%) and Alphaproteobacteria (9 and 2%). In the respective particle-attached fractions, Betaproteobacteria (26 and 10%) and Bacteroidetes (11 and 13%, Table 2) dominated, followed by Alphaproteobacteria (9 and 4%), and Actinobacteria (4 and 2%). In both years, less abundant groups including Firmicutes, Chlorobii, Verucomicrobia, and Acidobacteria represented only 4 to 6% of free-living as well as 6 to 13% of particle-attached bacterial fractions (Table 2). In 2006 and 2008, chloroplast sequences accounted for 2 and 15% of free-living or 24 and 31% of the particle-attached fractions, respectively.
Figure 4. Relative abundance of major bacterial groups.
16S rRNA gene clone libraries generated with universal bacterial primers obtained from free-living (0.22–8 μm in 2006 and 0.22–5 μm in 2008) and particle-attached (>8 or >5 μm, respectively) bacterial fractions after 1O2 and H2O2 exposure. Clone libraries were generated for control (C-L), 1O2 (RB-L) and H2O2 (HP-L) light treatments of in situ experiments 2006 and 2008. The relative abundance represents fractions (%) of all investigated clones of each clone library. For abbreviations see Fig. 1. Colours indicate the phylogenetic affiliation: Actinobacteria (purple), Gammaproteobacteria (brown), Alphaproteobacteria (red), and Betaproteobacteria (blue).
Table 2. Relative abundance and phylogenetic affiliation of sequenced phylotypes.
| 2006 | 2008 | ||||||||||||||||||||
| 0.22–8 μm | >8 μm | 0.22–5 μm | >5 μm | ||||||||||||||||||
| OTUs | C-L | RB0.05-L | RB0.2-L | HP10-L | C-L | RB0.05-L | RB0.2-L | HP10-L | C-L | RB0.02-L | HP10-L | C-L | RB0.02-L | HP10-L | Freshwatercluster | RDP Naive Bayesian rRNAClassifier | BLAST results | ||||
| β-Proteobacteria | 1 | 31 | 17 | 4 | 9 | 2 | 2 | 4 | 13 | 30 | 8 | 4 | 2 | bet II, PnecC | Burkholderiaceae | 100% | 99% | P. necessariusQLW-P1-DMWA-1T | CP000655 | ||
| 2 | 4 | 2 | 6 | bet II, PnecA | Burkholderiaceae | 100% | 97% | P. acidophobusMWH-PoolGreenA3 | FM208180 | ||||||||||||
| 3 | 2 | 2 | 13 | 2 | 13 | 20 | 4 | 2 | 2 | bet I, Lhab-A4 | Comamonadaceae | 98% | 99% | Lake GrosseFuchskuhle clone FNE11-10 | DQ501302 | ||||||
| 4 | 19 | 2 | 2 | 2 | 2 | 2 | bet IV, RDP18A09 | Methylophilaceae | 100% | 98% | Parker river clonePRD18A09 | AY947994 | |||||||||
| 5 | 4 | bet III, betIII-A1 | Alcaligenaceae | 100% | 99% | Grosse Lacke isolateQLW-p2DMWB-4 | AJ938031 | ||||||||||||||
| 6 | 2 | 4 | 2 | 4 | bet I, Lhab-A4 | Comamonadaceae | 100% | 97% | Lake IJssel clone Stal-17 | AJ416187 | |||||||||||
| 7 | 4 | 6 | 4 | 6 | bet VII | Oxalobacteraceae | 100% | 97% | Lake Grosse Fuchskuhleclone NE45 | AJ575695 | |||||||||||
| α-Proteobacteria | 8 | 9 | 36 | 2 | 4 | 7 | 4 | 2 | 16 | 6 | 13 | alf IV-A, Novo-A1 | Sphingomonadaceae | 100% | 99% | N. acidiphilumFSW06-204dT | EU336977 | ||||
| 9 | 19 | 11 | 2 | Bradyrhizobiaceae | 15% | 96% | Lake Pohlseeclone Hv_38 | EF667926 | |||||||||||||
| 10 | 7 | Hyphomicrobiaceae | 36% | 90% | Mesorhizobium sp.CCBAU 33182 | GU433452 | |||||||||||||||
| 11 | 2 | 4 | 9 | 2 | alf II | Caulobacteraceae | 100% | 97% | Adriondack lake cloneADK-BTe02-51 | EF520395 | |||||||||||
| 12 | 4 | Hyphomicrobiaceae | 46% | 98% | Rhodomicrobium vannieliiE.Y. 33T | M34127 | |||||||||||||||
| 13 | 7 | 2 | alf I, alf I-B1 | Beijerinckiaceae | 69% | 98% | Lake Grosse Fuchskuhleisolate FSW06-301 | FJ798303 | |||||||||||||
| 14 | 2 | 9 | alf VIII | Acetobacteraceae | 100% | 95% | Asaia lannaensis BCC 15733T | AB286050 | |||||||||||||
| γ-Proteobacteria | 15 | 2 | 58 | 2 | 13 | close to gam III | Methylococcaceae | 100% | 95% | Hypertrophic freshwater lakeclone ML-9-70.2 | DQ520192 | ||||||||||
| 16 | 7 | 7 | 15 | Legionellaceae | 100% | 96% | Legionella longbeachaeATCC 33484 | AY444741 | |||||||||||||
| 17 | 4 | 4 | 6 | Legionellaceae | 99% | 92% | Legionella impletisoliOA1-1T | AB233209 | |||||||||||||
| 18 | 4 | Ectothiorhodospiraceae | 70% | 96% | Activated sludge cloneAS1o9 | AJ514448 | |||||||||||||||
| 19 | 2 | 6 | 19 | 4 | gam I | Methylococcaceae | 100% | 96% | Methylomonas rubraNCIMB 11913 | AF304194 | |||||||||||
| Actinobacteria | 20 | 15 | 33 | 4 | 2 | 23 | 16 | 2 | acI-B, scB-3 | Microbacteriaceae | 45% | 99% | Lake Grosse Fuchskuhleclone FSW11-16 | DQ316348 | |||||||
| Firmicutes | 21 | 2 | 4 | 2 | 17 | 27 | 8 | Paenibacillaceae | 95% | 93% | Paenibacillus polymyxa SC2 | CP002213 | |||||||||
| Chlorobii | 22 | 4 | 2 | 2 | 9 | 17 | 17 | Chlorobiaceae | 100% | 98% | Pelodictyon phaeoclathratiformeBU-1T | CP001110 | |||||||||
| Bacteroidetes | 23 | 2 | 11 | 4 | 4 | 13 | 4 | Chitinophagaceae | 100% | 98% | Lake Grosse Fuchskuhleclone FukuS59 | AJ290042 | |||||||||
| 24 | 4 | Sphingobacteriaceae | 100% | 96% | Tatachia forest soil cloneTSC56 | EU359966 | |||||||||||||||
| Verucomicrobia | 25 | 4 | 2 | Subdivision5 | 78% | 97% | Lake Kinneret sedimentclone d0-26 | AM409824 | |||||||||||||
| Acidobacteria | 26 | 4 | 2 | 8 | 6 | 4 | 2 | Holophagaceae | 100% | 96% | Geotrix fermentansATCC 700665 | U41563 | |||||||||
| Chloroplasts | 27 | 13 | 2 | 7 | 55 | 2 | 13 | Bacillariophyta | 87% | 97% | Parker river clonePRD18F11 | AY948053 | |||||||||
| 28 | 4 | 2 | 8 | Chlorarachniophyceae | 40% | 93% | Parker river clonePRD18D01 | AY948021 | |||||||||||||
| 29 | 2 | 20 | 4 | 15 | 12 | 2 | 6 | Chlorarachniophyceae | 68% | 93% | Adriondack lake cloneADK-HDe02-54 | EF520517 | |||||||||
| 30 | 9 | 8 | 17 | 29 | Cryptomonadaceae | 100% | 94% | Adirondack lake cloneADK-SGh02-76 | EF520521 | ||||||||||||
| 31 | 4 | 8 | 13 | 2 | 17 | Cryptomonadaceae | 100% | 98% | Parker river clonePRD18E12 | AY948043 | |||||||||||
| 32 | 8 | Chlorophyta | 100% | 91% | Polytoma oviformecloroplast | AF374188 | |||||||||||||||
| Rare OTUs (%)* | 6 | 2 | 2 | 7 | 7 | 2 | 8 | 4 | 4 | 4 | 2 | ||||||||||
| Single OTUs (%)# | 11 | 6 | 13 | 7 | 13 | 6 | 23 | 23 | 17 | 4 | 27 | 23 | 13 | 10 | |||||||
| Total No. ofclones | 54 | 47 | 45 | 45 | 46 | 53 | 52 | 53 | 48 | 50 | 49 | 47 | 53 | 48 | |||||||
| Coverage (%) | 83 | 91 | 84 | 87 | 80 | 92 | 69 | 77 | 79 | 92 | 69 | 74 | 87 | 90 | |||||||
*Rare OTUs: OTUs that occur only once in one clone library;
Single OTUs: OTUs that occur only once in at least two clone libraries.
Exposure to 1O2 and H2O2 induced specific shifts in BCC. Increased 1O2 exposure led to the disappearance of Actinobacteria and Bacteroidetes in both free-living and particle-attached fractions, whereas the effects on Beta-, Alpha-, and Gammaproteobacteria as well as Firmicutes depended on 1O2 dose and bacterial fraction (Fig. 4, Table 2). In 2006, a 2.8-fold increased 1O2 exposure decreased Betaproteobacteria by 37 and 24% in the free-living and particle-attached fraction, respectively. In contrast, Alphaproteobacteria increased by 46% in the free-living and by 6% in the particle-attached fraction, whereas Firmicutes increased by 15% only in the particle-attached fraction (Table 2). After an 8-fold increased 1O2 exposure, Gammaproteobacteria dominated and accounted for 69 and 38% of the free-living and particle-attached fraction, respectively. In contrast, Alphaproteobacteria disappeared in the particle-attached fraction, but did not change in the free-living one. Firmicutes strongly increased by 25% exclusively in the particle-attached fraction (Table 2). The much lower 1.3-fold elevated 1O2 exposure in 2008 increased Betaproteobacteria by 27 and 7% in the free-living and particle-attached fraction, respectively. In both fractions, Alphaproteobacteria increased by 16 and 22%, and Gammaproteobacteria by 4 and 19%.
After H2O2 exposure, BCC changed in a very different manner. The abundance of free-living Betaproteobacteria decreased by 41 and 4% in 2006 and 2008, but in both years they remained highly abundant (Fig. 4). Particle-attached Betaproteobacteria decreased after H2O2 exposure by 20% in 2006, and were not detected in 2008. The change in relative abundance of free-living Actinobacteria varied between an 18% increase (2006) and a 7% decrease (2008), but negative effects were less pronounced than after exposure to 1O2. Actinobacteria remained highly abundant and the relative abundance of further bacterial groups only slightly changed after H2O2 exposure (Table 2).
Changes in the Overall Bacterial Diversity by Clone Library Analysis
The coverage of the individual clone libraries ranged between 69 and 92%, with a mean coverage value of 82.4% (Table 1). Rarefaction analysis showed that rarefaction curves generated for each clone library did not reach complete saturation by a number of approx. 50 clones for each investigated clone library (Fig. S3). The lack of saturation was mainly due to single and rare OTUs, which ranged between 8 to 31%. The focus of our study, however, was on investigating ROS-induced changes in relative abundance of the most prominent freshwater bacterial groups or species. Therefore, such single and rare OTUs were not investigated by sequence analysis and our clone library analyses did not aim to cover the overall diversity within each treatment. The number of investigated OTUs was sufficient to depict distinct differences in phylotype abundance after increased 1O2 and H2O2 exposure. Especially for free-living bacteria, rarefaction curves were closer to saturation after exposure with 0.05 μM RB in the light (1O2 treatments) in 2006 and 2008, respectively (Fig. S3). This finding indicates that bacterial diversity after 1O2 treatments are lower than in C–L and H2O2 light treatments for experiments in 2006 and 2008.
Effects of 1O2 and H2O2 Exposure on Predominant Bacterial Phylotypes
Sequencing of clones representing the most abundant operational taxonomic units (OTUs) revealed those bacterial phylotypes causing major changes in BCC upon ROS exposure (Fig. 5–9, Table 2). In 2008, Polynucleobacter necessarius (PnecC sub-cluster) represented the most abundant Betaproteobacteria phylotype (OTU-1). Increased abundance of Betaproteobacteria after 1O2 exposure was mainly due to P. necessarius and a Limnohabitans-related phylotype (OTU-3). Both phylotypes decreased after exposure to H2O2. A second Polynucleobacter phylotype (OTU-2) representing the PnecA sub-cluster only occurred in the free-living fractions after H2O2 addition.
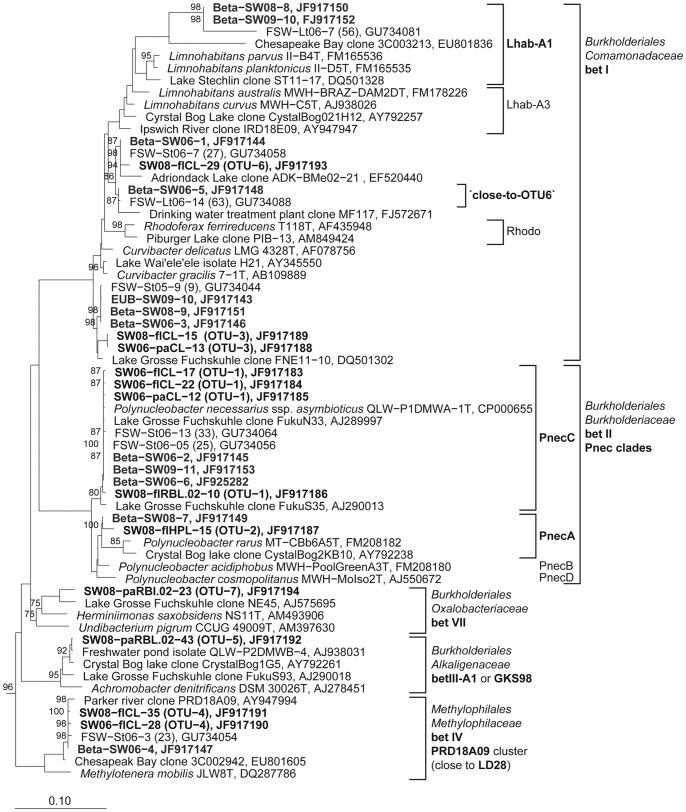
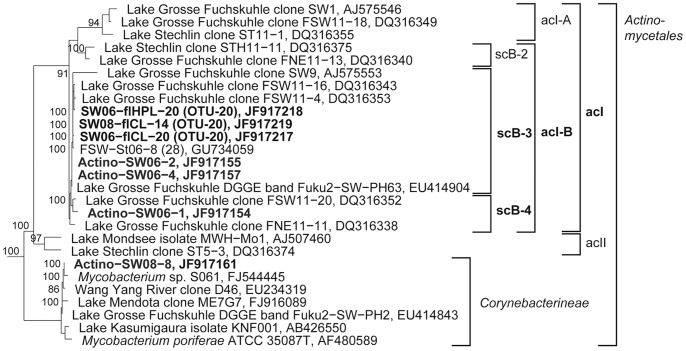
Figure 5. Phylogenetic affiliation of 16S rRNA gene sequences representing OTUs and DGGE bands to the Betaproteobacteria.
Maximum likelihood trees showing the phylogenetic affiliation of OTU and DGGE band sequences to the Betaproteobacteria. Sequences obtained from DGGE bands are depicted in bold letters. Numbers at roots represent bootstrap values (≥70%) of 100 re-samplings. Scale bars: 0.1 nucleotide substitutions per site. Sequences representing OTUs are assigned as follows: SW: South West basin, 06, 08: year of in situ experiment in 2006 or 2008, fl: free-living bacteria, pa: particle-attached bacteria. Sequences signed with EUB, Beta, or Actino are from Bacteria, Betaproteobacteria, or Actinobacteria-specific RT-PCR DGGE bands, respectively.
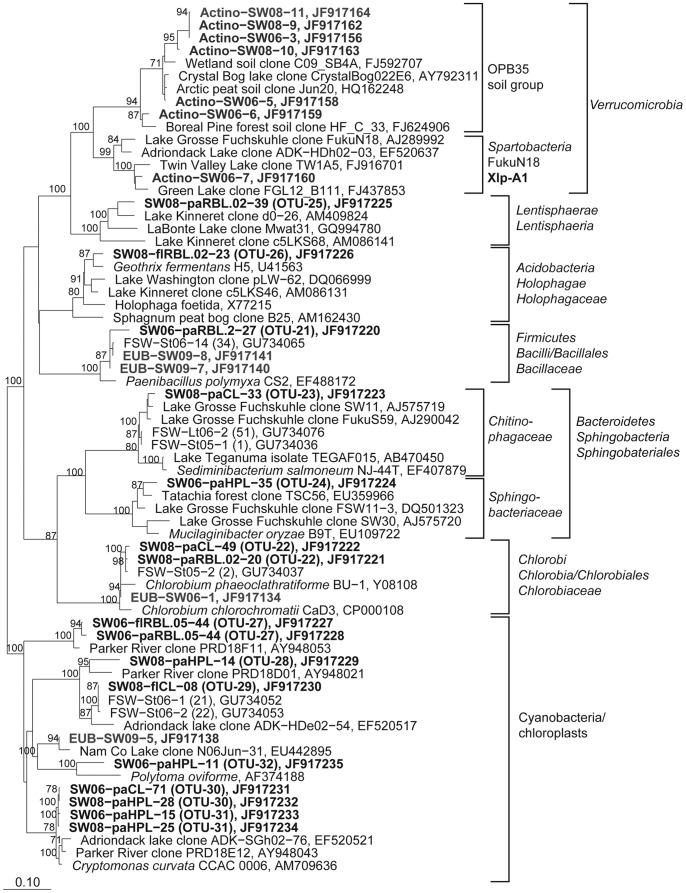
Figure 9. Phylogenetic affiliation of 16S rRNA gene sequences representing OTUs and DGGE bands to the less abundant bacterial groups and chloroplast sequences.
Maximum likelihood trees showing the phylogenetic affiliation of OTU and DGGE band sequences to less abundant bacterial groups and chloroplast sequences. Details and abbreviations are indicated in the legend to Figure 5.
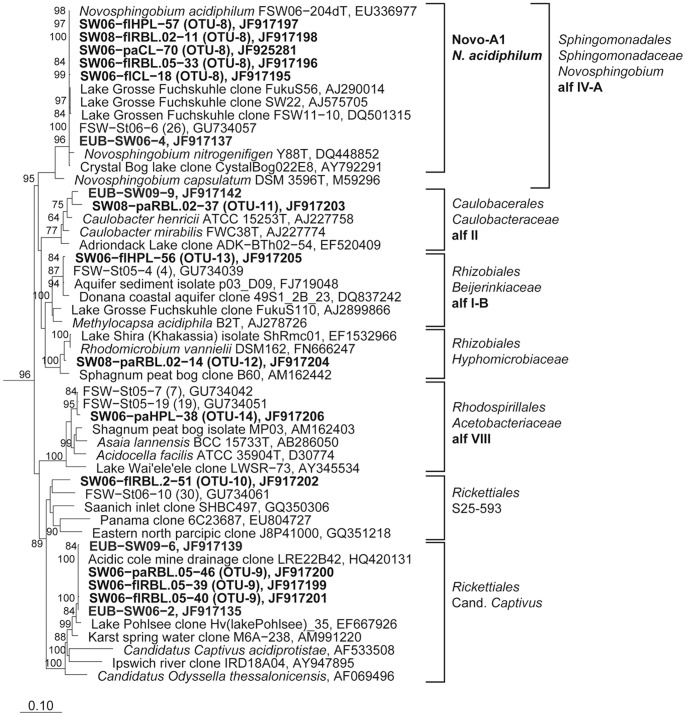
Figure 6. Phylogenetic affiliation of 16S rRNA gene sequences representing OTUs and DGGE bands to the Alphaproteobacteria.
Maximum likelihood trees showing the phylogenetic affiliation of OTU and DGGE band sequences to the Alphaproteobacteria. Details and abbreviations are indicated in the legend to Figure 5.
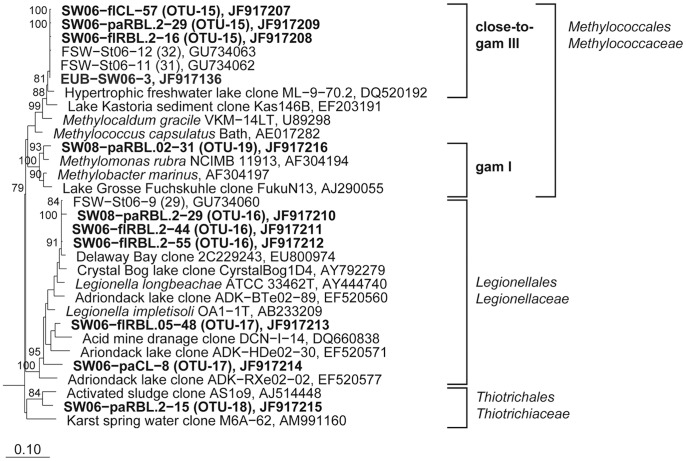
Figure 7. Phylogenetic affiliation of 16S rRNA gene sequences representing OTUs and DGGE bands to the Gammaproteobacteria.
Maximum likelihood trees showing the phylogenetic affiliation of OTU and DGGE band sequences to the Gammaproteobacteria. Details and abbreviations are indicated in the legend to Figure 5.
Increased abundance of Alphaproteobacteria after 1O2 exposure was mainly due to OTU-8 representing Novosphingobium acidiphilum (Table 2). In addition, increase of an uncultured phylotype (OTU-9) resulted in a highly increased Alphaproteobacteria abundance after 1O2 exposure in 2006. After H2O2 exposure, in the attached fraction, a Caulobacteraceae-related phylotype (OTU-11) increased in relative abundance in 2008 and two other Alphaproteobacteria phylotypes (OTU-13/14) in 2006 (Table 2).
Five different phylotypes were responsible for the increased abundance of Gammaproteobacteria after high 1O2 exposure in 2006 (OTU-15 to 19, Table 2). In contrast, only one freshwater-cluster AcI-B phylotype (OTU-20) was responsible for the high abundance of Actinobacteria in controls and after H2O2 exposure.
Changes in the Composition of Metabolically Active Bacteria
Analysis of metabolically active bacteria by unweighted pair-group method using arithmetic average (UPGMA) cluster analysis of Bacteria RT-PCR Denaturing Gradient Gel Electrophoresis (DGGE) patterns confirmed BCC changes after 1O2 and H2O2 exposure as observed by clone library analysis (Fig. 10 and S4). All in situ experiments performed in 2006, 2008 and 2009 were repeated within a few days (Fig. S5 A–C).
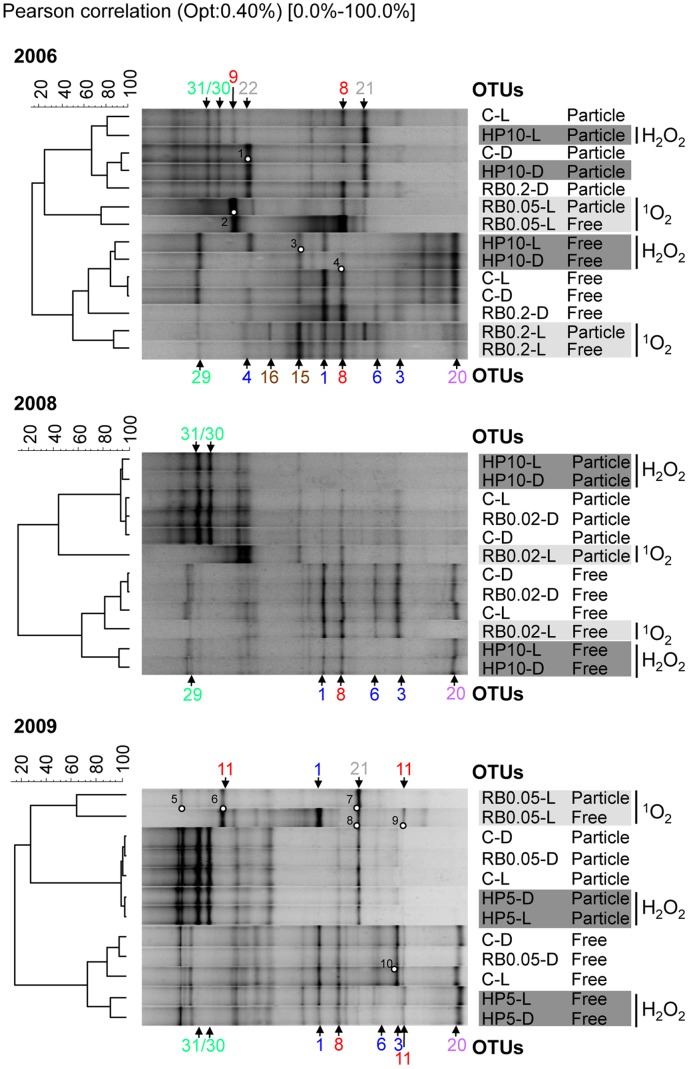
Figure 10. Cluster analysis of Bacteria RT-PCR DGGE patterns.
Cluster analysis and RT-PCR DGGE patterns of metabolically active free-living (0.22–8 μm in 2006 and 0.22–5 μm in 2008 and 2009) and particle-attached (>8 or >5 μm, respectively) Bacteria of in situ experiments 2006, 2008 and 2009. Universal Bacteria 16S rRNA gene targeting primers were used for analysis. Cluster analyses were performed in GelCompare II version 4.5 (Applied Maths) using unweighted pair-group method using arithmetic average (UPGMA) clustering based on the Pearson correlation which considers the intensity of DGGE bands. Distance matrices are shown in Fig. S4. DGGE bands marked with circles were sequenced. OTU numbers depicted next to the DGGE patterns point at DNA bands identical in DNA sequence (see Table 2). Colours of OTU numbers indicate the phylogenetic affiliation: Actinobacteria (purple), Gammaproteobacteria (brown), Alphaproteobacteria (red), and Betaproteobacteria (blue), cyanobacteria/chloroplasts (green), and other Bacteria (grey). Phylogenetic affiliations to sequenced DGGE bands are given in Fig. 5–9 and Table S6. Abbreviations are given in Fig. 1.
In UPGMA combining all experiments stable clusters were formed by patterns affiliated with experiments performed in the respective year (data not shown). Therefore, cluster analysis was performed separately for all three years, in which DGGE patterns of particle-attached and free-living bacteria formed separate clusters (Fig. 10) Within these clusters, control experiments (C-L/D, RB-Ds) and H2O2 treatments (HP-L/D) clustered with each other. In contrast, 1O2 exposure caused more pronounced changes in DGGE banding patterns. Particle-attached and free-living fractions in 2006 and 2009 were found in the same cluster after 2.8 and 1.9-fold (RB0.05-L, 2006 and 2009) and after 8-fold (RB0.2-L, 2006) 1O2 increase. After moderate 1O2 exposure (RB0.05-Ls), changes in DGGE bands representing the uncultured Alphaproteobacterium OTU-9 and the Firmicutes OTU-21 in both particle-attached and free-living fractions greatly affected cluster formation. At higher 1O2 exposure (RB0.2-L), however, DGGE banding patterns of the free-living fraction were similar to the respective controls (Fig. 10) represented by P. necessarius OTU-1, N. acidiphilum OTU-8, and Methylococcaceae OTU-15. In 2008, slightly increased 1O2 exposure (RB0.02-L) had a minor effect on BCC and the respective DGGE clusters were similar to the controls. In all three experiments, disappearance of the DGGE band representing AcI-B Actinobacteria OTU-20 comprised the most obvious change in community composition of free-living bacteria after 1O2 exposure (Table 2).
BCC changes after H2O2 exposure were generally caused by i) decreased intensity of DGGE bands representing P. necessarius OTU-1 and N. acidiphilum OTU-8 and ii) the absence of DGGE bands representing Limnohabitans-related OTU-3/6. These changes occurred in different extent in free-living fractions of all three experiments and also partially in the respective particle-attached fractions.
Phylotype-specific Changes within Major Bacterial Groups
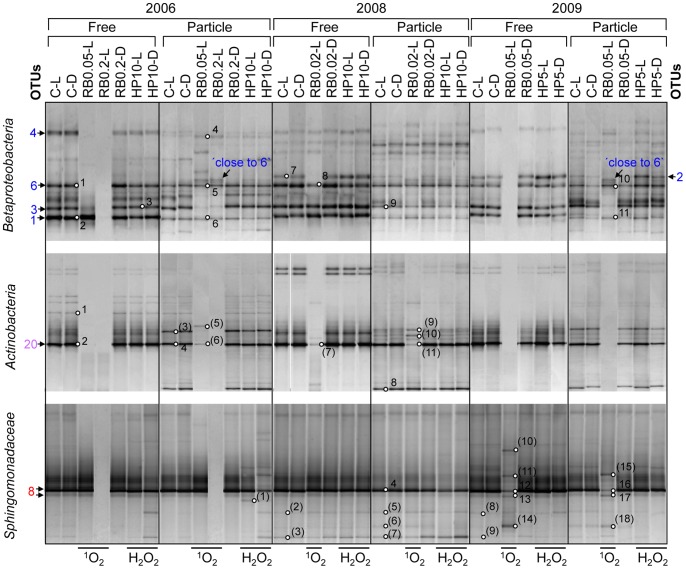
Betaproteobacteria, Actinobacteria, and Sphingomonadaceae-specific RT-PCR DGGE analysis increased the phylogenetic resolution of our study and revealed separate clusters for free-living and particle-attached bacteria by UPGMA analysis (Fig. S6). After 1O2 exposure (RB-Ls), DGGE banding patterns obtained for all three bacterial groups were separated from controls, whereas after H2O2 exposure, the DGGE bands always clustered together with controls.
Major DGGE bands of both Betaproteobacteria fractions represented P. necessarius OTU-1 and Limnohabitans-related OTU-3/6 (Fig. 11). In 2008 and 2009, the DGGE band representing PnecA OTU-2 was observed with higher intensities in the free-living fractions. Singlet oxygen exposure resulted in different effects on phylotype composition of free-living vs. particle-attached Betaproteobacteria. The 2.8-fold increased 1O2 exposure decreased diversity of free-living Betaproteobacteria to solely 2 DGGE bands in 2006 represented by P. necessarius OTU-1 and Limnohabitans-related OTU-3. The 8-fold increased 1O2 exposure diminished all free-living Betaproteobacteria, whereas DGGE bands of particle-attached Betaproteobacteria representing P. necessarius OTU-1 and Limnohabitans-related OTU-6 were not affected by 1O2 exposure. In the same treatment, an additional DGGE band representing a phylotype closely related to OTU-6 occurred (DGGE band 5, Fig. 11). In 2008, the much lower 1O2 exposure led to the disappearance of a DGGE band in the free-living fraction representing PnecA OTU-2. The same DGGE band became more intense after H2O2 exposure in both, particle-attached and free-living fractions of 2008 and 2009. In general, the effects of 1O2 exposure on Betaproteobacteria in 2006 were confirmed in 2009 whereby the 1.9-fold increased 1O2 exposure in 2009 had similar effects compared to the 8-fold increased 1O2 exposure in 2006.
Figure 11. Group specific RT-PCR DGGE analysis.
RT-PCR DGGE analysis of metabolically active free-living (0.22–8 μm in 2006 and 0.22–5 μm in 2008 and 2009) and particle-attached (>8 or >5 μm, respectively) Betaproteobacteria, Actinobacteria, and Sphingomonadaceae after 1O2 and H2O2 exposure. Group-specific 16S rRNA gene targeting primer-systems were used for analysis. All treatments of in situ experiments 2006, 2008 and 2009 were investigated. DGGE bands marked with circles were sequenced. DGGE band numbers in brackets were not affiliated to the investigated groups. Numbers with arrows show the assignment to respective OTUs (see Table 2). Abbreviations are given in Fig. 1.
The AcI-B OTU-20 represented the most abundant Actinobacteria DGGE band of free-living and particle-attached fractions. However, the relative abundance of Actinobacteria was low on particles as revealed by clone-library (Fig. 4) and Bacteria RT-PCR DGGE analysis (Fig. 10). After 1O2 exposure, Actinobacteria DGGE bands were lacking, except in 2008 when a DGGE band representing a Mycobacteria-related phylotype occurred (band 8, Fig. 8 and 11). Other DGGE bands present after 1O2 exposure belonged to the Verrucomicrobia (Fig. 9 and 11).
Figure 8. Phylogenetic affiliation of 16S rRNA gene sequences representing OTUs and DGGE bands to the Actinobacteria.
Maximum likelihood trees showing the phylogenetic affiliation of OTU and DGGE band sequences to the Actinobacteria. Details and abbreviations are indicated in the legend to Figure 5.
Sphingomonadaceae-specific RT-PCR DGGE analysis indicated that N. acidiphilum (OTU-8) was the pre-dominant Sphingomonadaceae in the SW basin. Only high 1O2 exposure affected the intensity of its respective DGGE band (Fig. 11).
Discussion
Comparison of 1O2 and H2O2 Toxicity
Moderately increased 1O2 and highly increased H2O2 concentrations caused similar inhibition of 14C-leucine incorporation suggesting different toxic potentials of 1O2 and H2O2. This finding also indicates that small changes of 1O2 generation (frequent during diurnal changes in sunlight intensity) may hamper microbial activity in surface waters of humic lakes. In contrast, only large changes in H2O2 concentrations may affect the activity of dominant bacterial species. However, the H2O2 concentrations applied in our experiments were not exaggerated and the natural potential of H2O2 formation in 0.22 μm filtered lake water of the SW basin was high (Fig. S7). In H2O2 depleted water samples, H2O2 concentrations in the μM range can be reached rapidly after irradiation with sunlight or UV-A/B which has been frequently observed for boreal lakes [25], [26]. Microorganisms strongly contribute to the decay of H2O2 [27]. This is indicated by 2.4-fold higher H2O2 decay rates in our unfiltered water samples compared to those filtered through 0.22 μm (Materials S1). Obviously, the bacterial community or at least some phylotypes can detoxify H2O2 and therefore balances H2O2 levels in their environment. This notion is in line with earlier findings that bacteria are involved in H2O2 degradation in marine surface waters [27] and that H2O2 degradation by some bacterial populations is important for growth of other bacteria in aquatic environments [28]. Hence, bacteria thriving in surface waters of humic lakes are well adapted to H2O2 exposure and may prevent accumulation of toxic H2O2 concentrations.
Contrasting Effects of 1O2 and H2O2 on Actinobacteria and Betaproteobacteria
AcI-B Actinobacteria and betII lineage Betaproteobacteria mainly of the PnecC sub-cluster are the most abundant bacterial groups in the SW basin [29], [30], [31]. Actinobacteria of the AcI-B cluster are low in abundance on particles [32]. Their high sensitivity to 1O2 and the finding that humic matter rich particles generate high amounts of 1O2 [23] could explain the obvious absence of AcI-B Actinobacteria from particles. Contrary, AcI-B Actinobacteria of the SW basin were more resistant to H2O2 exposure. Thus, it is likely that AcI-B Actinobacteria produce peroxidases to degrade recalcitrant organic matter and contribute to the high overall extracellular peroxidase activity in Lake Grosse Fuchskuhle [33]. This life-style requires increased resistance to peroxides and thus may explain the high relative abundance of AcI-B Actinobacteria at increased H2O2 concentrations. A recently analysed single cell genome of the AcI-B lineage supports this notion, because several genes encoding glutathione depended peroxiredoxins were identified that potentially account for the proposed resistance against peroxides [34].
In Lake Grosse Fuchskuhle and in other freshwater ecosystems the abundances of Actinobacteria and Betaproteobacteria are negatively correlated [14], [30], and Actinobacteria numbers are usually lower in summer months. The addition of photo-chemically modified DOM to water samples increased Actinobacteria abundance [14]. By irradiating DOM high amounts of H2O2 accumulate [11], and the subsequent incubation in the dark excludes formation of 1O2. Therefore, only effects of H2O2 on bacterial dynamics can be monitored by such assays. Actinobacteria had a high resistance against H2O2 in our study. In contrast, several Betaproteobacteria phylotypes detected in our study were H2O2 sensitive, but resistant to 1O2 exposure. Consequently, the negative correlation between Actinobacteria and Betaproteobacteria in the SW basin is at least partly the result of their contrasting resistance and sensitivity to 1O2 and H2O2.
High solar radiation causes high 1O2 exposure during the summer months and may result in reduced AcI-B Actinobacteria abundance. In contrast, P. necessarius was favoured by increasing 1O2 concentrations and generally shows highest abundance and activities in summer [35] and it also grows well on photodegradation products of humic matter, such as acetate [36], [37], [38]. AcI-B Actinobacteria are more abundant in autumn and early spring [30], [32] when input of unbleached NOM from the adjacent fen into the SW basin is high. This unbleached NOM generates much more H2O2 than 1O2 (Materials S1) and may be a key regulator of the observed opposing dynamics of AcI-B Actinobacteria vs. Betaproteobacteria.
Alpha- and Gammaproteobacteria Resist High 1O2 Doses
Alpha- and Gammaproteobacteria are two major lineages of freshwater bacteria, which have gained relatively little attention in the past [39]. Our data and previously published clone libraries [29], [30], [31] indicate the persistence of N. acidiphilum in the humic matter rich SW basin. Its relative abundance strongly increased during 1O2 exposure suggesting a high 1O2 resistance which can be explained by a high cellular carotenoid content [40]. In addition, Sphingomonadaceae are known to degrade aromatic compounds and N. acidiphilum was favoured by the addition of phenol that represents an important fraction of leached DOM [41]. Thus, cellular quenching of 1O2 by carotenoids and the use of aromatic compounds are features of N. acidiphilum, which may well explain its persistence in humic matter rich systems.
The increase in relative abundance of several Alpha- and Gammaproteobacteria after 1O2 exposure may be related to specific defence-systems protecting, for example, anoxygenic phototrophic Alphaproteobacteria against 1O2 damages [42], [43]. This is supported by the recent finding that anoxygenic phototrophic bacteria of the SW-basin mainly consist of Alphaproteobacteria [44]. The key regulators controlling such cellular responses include specific RNA polymerase sigma factors and have been found in the genomes of several Alpha- and Gammaproteobacteria lineages [45] including non-phototrophic Caulobacter crescentus [46]. Thus, induction of 1O2-specific defence-systems may explain the increased relative abundance of the Caulobacteraceae-related phylotype (OTU-11) in the present study.
Particle-attached Phylotypes are More Resistant to 1O2 Exposure
Particles represent hotspots of bacterial activity in aquatic environments [47]. Humic matter rich particles have been shown to generate higher 1O2 concentrations compared to the surrounding water by the application of hydrophobic 1O2 traps [23]. Recent studies could not verify a high 1O2 generation in humic particles [48] or suggest that 1O2 is quenched by certain reactive groups [49]. Our study revealed the existence of particle-associated phylotypes that were obviously more resistant to 1O2 exposure than their free-living counterparts. Particle-attached bacteria represented by P. necessarius OTU-1 and the Limnohabitans-related OTU-6 were indeed more resistant to 1O2 exposure than their free-living counterparts. Particle-associated bacteria exhibit different lifestyles and thus often represent different ecotypes [50], which requires also adaptation to different levels of oxidative stress. Alternatively, phylotypes in particle-attached and free-living fractions may represent the same ecotypes, whereby inducible response mechanisms against increased oxidative stress should allow for colonization of particles in the upper, well-illuminated water layers. Furthermore, it cannot be fully excluded that P. necessarius 16S rRNA gene sequences in the particle-attached fraction (>5 μm) originate from ciliate endosymbionts, namely Stentor amesthystinus (Dziallas and Grossart, unpubl. data). In contrast, highly 1O2 sensitive AcI-B Actinobacteria were absent from humic particles representing nutrient, but 1O2 rich microhabitats (see above).
Defence Mechanisms Against Environmental ROS Exposure
Details on the presence of molecular response mechanisms against environmental ROS exposure in typical freshwater bacteria are elusive. Recently, molecular defence systems against 1O2 exposure were found in bacteria [42], [43] and defence strategies against H2O2 generated in aerobic metabolism are known in detail for several bacterial model systems [21].
Carotenoids are inevitable in photosynthetic bacteria and in the chloroplasts of plants to prevent photosystem based generation of 1O2 [42], [43]. Non-photosynthetic bacteria also exhibit carotenoids, which likely serve as quenchers of 1O2 generated by cellular photosensitizers such as flavins [42] or by various extracellular sources. Cellular scavengers, which include amino acids such as L-histidine and trypotphan, reduced thiols (glutathione, thioredoxin), mycosoprine lysine and polyamines also minimize cellular damages by 1O2. Such scavengers need to be regenerated after their reaction with 1O2, and therefore enzymes involved in adjusting the cellular redox homeostasis need to be activated (reviewed in [43]).
In photosynthetic Alphaproteobacteria, response mechanisms to 1O2 exposure are controlled by the alternative sigmafactor RpoE, which is bound to the anti-sigmafactor ChrR under non-stress conditions. The release of RpoE from ChrR after 1O2 exposure triggers the induction of genes encoding stress response mechanisms and further regulatory factors, including RpoHII and several small regulatory RNAs [42]. Homologs of these sigmafactors are conserved in photosynthetic Alphaproteobacteria and have been found in several Beta- and Gammaproteobacteria lineages [45]. Genomes of species representing abundant freshwater bacterial clades did not harbour homologous genes. Hence, defence systems and their control in abundant freshwater bacteria may substantially differ from established bacterial model systems.
Very likely, individual bacterial lineages use different strategies to overcome natural 1O2 exposure, which could explain very well the species specific sensitivity to 1O2 exposure in our study.
Hydrogen peroxide is detoxified by cellular enzymes such as catalases and peroxidases (glutathione peroxidases and peroxiredoxin) [21]. Increased H2O2 concentrations lead to rapid cell death by the oxidation and disassembly of iron-sulphur clusters, which are common in electron transport chain components. Hydrogen peroxide together with free iron(II) leads to the formation of highly toxic hydroxyl radials by the Fenton reaction, which rapidly react with most cellular components and facilitate cell mortality. Therefore, cellular levels of H2O2 are tightly balanced and the cellular response is well regulated by, for example, OxyR or PerR which coordinate genes for H2O2 degradation, glutathione turnover, production of redox buffers as glutaredoxin and thioredoxin as well as genes involved in controlling iron metabolism. All bacteria with an aerobic metabolism, therefore, require defence systems against H2O2 exposure. This may explain, why H2O2 had a much smaller effect on BCC compared to 1O2 in the environment.
Niche Separation of Closely Related Species Caused by Exposure to Different ROS
Our experiments in 2008 indicate niche separation of closely related Polynucleobacter phylotypes by moderately increased 1O2 exposure. The Polynucleobacter phylotype of the PnecC sub-cluster (OTU-1) was highly resistant against exposure to 1O2, but negatively affected by H2O2. In contrast, the Polynucleobacter phylotype of the PnecA sub-cluster (OTU-2) was only detected after H2O2 exposure in clone libraries of free-living bacteria. Additionally, a corresponding DGGE band was observed in all free-living fractions by Betaproteobacteria-specific RT-PCR DGGE analysis, except after intense 1O2 exposure. Hence, ecological niches of those related phylotypes might be separated by variations in their sensitivity to 1O2 and H2O2. In line with our finding, occurrence of the Polynucleobacter sub-cluster PnecA and PnecC depends on lake colour [51], most likely because H2O2 formation largely depends on concentration and quality of NOM [25]. Moreover, the presence of various Polynucleobacter sub-clusters may also reflect the availability of different substrates since Polynucleobacter species assimilate low-molecular-weight substances [38] that can be also generated by photochemical NOM degradation.
We further observed ROS dependent niche separation for Limnohabitans-related phylotypes. Betaproteobacteria-specific RT-PCR DGGE patterns revealed the occurrence of a Limnohabitans-related phylotype closely related to OTU-6′ on particles after increasing 1O2 exposure in 2006 and 2009. This phylotype was also enriched after long-term exposure with moderately increased 1O2, whereas the OTU-6 phylotype only occurred in the respective controls [16] indicating a lower 1O2 resistance. Fortunately, we were able to isolate a respective strain from the SW basin and found an efficient adaptation to inhibitory 1O2 exposure by pre-incubation with non-inhibitory 1O2 concentrations (data not shown). This notion suggests that highly effective response mechanisms to increased 1O2 may be present in this specific Limnohabitans strain. Niche separation of coexisting closely related Limnohabitans strains has been shown recently [52], but in this case it was caused by differences in predation and virus infections. Niche separation of closely related phylotypes of Limnohabitans by 1O2 exposure underlines our hypothesis that different ROS affect BCC in a highly phylotype-specific manner, particularly in humic matter rich lakes.
Conclusions
From our data we conclude that differences in sensitivity to 1O2 and H2O2 may explain the negative correlation in abundance of Actinobacteria and Betaproteobacteria in the surface waters of Lake Grosse Fuchskuhle and elsewhere. The exclusion of specific bacterial lineages from humic matter rich particles and the presence of species-like taxa due to ROS specific separation of ecological niches should be regarded as an ecological factor shaping natural microbial communities. Hence, temporal and spatial differences in ROS generation, particularly in humic matter rich aquatic ecosystems, have the potential to affect major microbial processes and their rates. For example, niche separation by ROS has strong implications for bacterial adaptation and evolution in natural ecosystems. We propose that changes in 1O2 exposure have a larger impact on BCC than H2O2, because 1O2 is i) more toxic compared to H2O2 and ii) defence mechanisms against H2O2 are present in all aerobic organisms, whereas putative defences against singlet oxygen exposure may only occur in bacteria specifically adapted to cellular or environmental 1O2 formation. Further, insights into the molecular mechanisms of cellular defences against environmental ROS in general and singlet oxygen in particular are necessary to understand in detail the role of 1O2 and H2O2 for controlling activity and composition of aquatic microbial communities.
Materials and Methods
Study Site
All field studies were conducted in the humic acid rich south-west basin of the artificially divided dystrophic Lake Grosse Fuchskuhle [52]. Physico-chemical parameters of the lake were described previously [16], [29], [30] and are compiled for all experimental periods in Table S3.
The IGB is authorized by the Landkreis Oberhavel to obtain samples from Lake Grosse Fuchskuhle and to conduct mesocosm experiments as performed in our study. Our studies did not endanger protected wildlife in or around the lake.
Sampling and Experimental Conditions
Subsurface water samples were collected in autoclaved Pyrex-glass bottles on the same day prior to the start of in situ exposure experiments. All set ups were prepared in a dark shelter at the lake shore and water samples were subsequently incubated 10 cm below the water surface in the humic SW basin of Lake Grosse Fuchskuhle.
Generation of 1O2 was artificially increased by adding 0.02 to 0.2 μM of the photosensitizer Rose Bengal under sunlight exposure (RB-L). Concentrations of H2O2 were experimentally increased by adding 5–10 μM H2O2 to enhance peroxide stress in light and dark incubations (HP-L and HP-D). Controls included light and dark incubations of natural lake water (C-L and C–D) without addition of any chemicals and a RB dark control (RB-D).
For the first experiment on 12th July 2006 [16], 1 L water samples were incubated in polypropylene bags (Carl Roth, Karlsruhe, Germany) between 13∶30 and 18∶00. The light treatments were repeated in 2006 on 14th July (C-L), 15th July (RB0.2-L), 18th July (RB0.05-L) and 20th July (HP-L). In each experiment we compared the exposure to the untreated control obtained at the start of the experiment. The second experiment was performed on 5th September 2008 by incubating 400 mL water samples in polyethylene Whirl-Pak Bags (Nasco, Fort Atkinson, WI, USA) between 12∶15 and 16∶15. Prior to incubations, water samples were diluted with an equal volume of 0.22 μm pre-filtered surface water. A replicate of this experiment was performed on 4th September. In the third experiment on 14th August 2009, we incubated 500 mL water samples in Whirl-Pak Bags between 9∶00 and 13∶00. All Whirl-Pak bags were covered with UV-A/B absorbing polyester sheets 90 NR (Modulor, Berlin) to exclude effects of UV-A/B radiation. The experiment was repeated in triplicates on 17th August. Transmission spectra are given in Fig. S8 for plastic bags and sheets, respectively.
Solar radiation and rainfall affects the NOM reactivity in the lake. In order to monitor pre-experiment weather conditions, weather data for 30 day prior to the each experiments were obtained from the weather station in Menz (53°10′ N, 13°05′ E). Menz is closely located to Lake Grosse Fuchskuhle. The data were purchased from the Deutscher Wetterdienst (www.dwd.de) and depicted in the Figure S9.
Measurement of 1O2 and H2O2
ROS concentrations were determined in 0.22 μm filtered water samples. Singlet oxygen steady state concentrations ([1O2]SS) were measured using furfuryl alcohol [24] as described previously [16]. Concentrations of H2O2 were measured by using the Amplex Red method [53] with slight modifications (Materials S1). Analysis were performed in triplicates. Differences between treatments were analysed by one-way ANOVA followed by pair-wise multiple comparison analysis with the Tukey test (Sigma Stat version 2.0, Systat Software, Richmond, California, USA).
Bacterial Numbers and Microbial Activity
Bacteria cell numbers were determined by Sybr Green I staining and epifluorescence microscopy [16]. Microbial activity was measured by [14C]-leucine incorporation [54]. Sample-triplicates (5 mL) and formalin-fixed controls were incubated immediately after experiments with [14C]-leucine (1.15×1010 Bq mmol−1; Amersham) for 1 h at in situ temperature in the dark. Incubations were stopped by formalin addition.
Simultaneous DNA and RNA Extraction from Water Samples
All in situ experiments performed in 2006, 2008 and 2009 were repeated within a few days. Water samples were immediately put on ice prior to filtration. Particle-attached bacteria were collected on 8 μm cellulose-nitrate membranes (Satorius, Göttingen, Germany) in 2006 or on 5 μm sterile Minisart syringe filters (Sartorius) in 2008 and 2009. Free-living bacteria from the 5 μm filtrates were collected on 0.22 μm Sterivex™-GP filter units (Millipore, Schwalbach, Germany) and filters were immediately stored at −80°C. Triplicates from the second experiment performed in 2009 were pooled prior to the extraction of nucleic acids. DNA and RNA were extracted simultaneously as described by [55]. Reaction volumes were decreased for the use of 2-ml tubes. Precipitated nucleic acids were resuspended in 100 μl RNase/DNase-free water (Carl Roth). RNA extracts were treated with 1 U RQ1 DNase (Promega, Madison, WI, USA) and purified with phenol/chloroform (2006) or were incubated with 1 U DNase I (Fermentas, St. Leon-Rot, Germany), which was subsequently heat-inactivated (2008 and 2009).
16S rRNA Gene Clone Libraries and RT-PCR DGGE
Bacterial 16S rRNA gene clone libraries were generated with primers 8F and 1492R [56] and operational taxonomic units (OTUs) were defined by Amplified Ribosomal DNA Restriction Analysis (ARDRA) [57]. Community changes of metabolically active Bacteria, Actinobacteria, Betaproteobacteria, and Sphingomonadaceae were investigated by 16S rRNA targeting RT-PCR DGGE. Details are given in Materials S1 and Tables S4 and S5.
Phylogenetic Analysis of 16S rRNA Gene Sequences
Sequences were aligned with the SINA Web aligner (http://www.arb-silva.de/aligner/) and analysed in ARB [58] using the SILVA SSURef NR 104 database [59]. Maximum likelihood trees were constructed with using RAxML v7.04 [60] with GTR-GAMMA and rapid bootstrap analysis. Trees were generated with nearly full-length sequences (>1300 nt) spanning E. coli positions 56 to 1444 [61]. Tree topologies were confirmed by the generation of trees using Proteobacteria, Actinobacteria, and Bacteria 50% base frequency filters. Partial sequences were added with ARB parsimony without changing the overall tree topology. Sequences are deposited in GenBank with accession numbers JF917134–JF917235, JF925281, and JF925282.
Supporting Information
Activity of heterotrophic bacteria after 1O2 and H2O2 exposure.
(PDF)
Cell numbers in controls and in 1O2 and H2O2 treatments.
(PDF)
Rarefaction analysis of nearly full-length 16S rRNA gene clone libraries.
(PDF)
Distance matrices of Pearson correlation based UPGMA cluster analysis.
(PDF)
Robustness of changes in BCC observed by RT-PCR-DGGE analysis shown by repeats of the 2006 (A), 2008 (B) and 2009 (C) in situ experiments.
(PDF)
Cluster analysis of Betaproteobacteria, Actinobacteria, and Sphingomonadaceae- specific 16S rRNA gene based RT-PCR DGGE analysis.
(PDF)
Delayed formation of hydrogen peroxide (H2O2) in 0.22 μm filtered water samples exposed to natural sunlight.
(PDF)
Transmission scans of poly-propylene (PP) bags, poly-ethylene (PE) Whirl-Pak bags, and the UVA/B block sheet.
(PDF)
Weather data for 30 day prior to the experiments carried out in 2006, 2008, and 2009.
(PDF)
NOM concentrations and reactivity of surface water samples from the SW basin of Lake Grosse Fuchskuhle.
(PDF)
Cell numbers of in situ incubation experiments in 2006 and 2008.
(PDF)
Physico-chemical parameters of Lake Grosse Fuchskuhle SW compartment.
(PDF)
Sequences of 16S rRNA gene targeting oligonucleotide primer used for RT-PCR DGGE analysis.
(PDF)
PCR and RT-PCR programs for the amplification of 16S rRNA and 16S rRNA gene fragments of Bacteria and bacterial subgroups used for DGGE fingerprint analysis.
(PDF)
Phylogenetic affiliation of 16S rRNA gene sequences representing DGGE bands.
(PDF)
Characterization of water sample photo-reactivity, including NOM characteristics of south-west compartment samples, comparison of water sample photo-reactivity, in situ H2O2 formation and decay, and the potential photochemical effects of unbleached material from the acidic fen area. Investigation of bacterial community composition by 16S rRNA (gene) based methods by the generation and screening of 16S rRNA gene clone libraries, 16S rRNA targeting reverse transcriptase (RT)-PCR DGGE analysis and group-specific 16S rRNA targeting RT-PCR DGGE analysis.
(PDF)
Acknowledgments
We thank Elke Mach for leucine incorporation measurements. Ivette Salka helped in 2008 experiments, and Franziska Leunert and Kai Störkel in 2009 experiments. Ryan Newton kindly provided the ARB database generated for phylogenetic classification of freshwater bacteria [39]. Beate Lindenstruth is acknowledged for determining DOC concentrations. Meteorological information was kindly provided by the Neuglobsow measurement station of the German Environmental Agency (UBA).
Funding Statement
This work was supported by funds of the German Science Foundation (DFG) given to JG (JG-620/2-1) and to HPG (GR-1540/17-1 and PA- 1655/1-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pomeroy LR (1974) The ocean’s food web: A changing paradigm Bioscience. 24: 499–504. [Google Scholar]
- 2. Goldstone JV, Pullin MJ, Bertilsson S, Voelker BM (2002) Reactions of hydroxyl radical with humic substances: Bleaching, mineralization, and production of bioavailable carbon substrates. Environ Sci Technol 36: 364–372. [DOI] [PubMed] [Google Scholar]
- 3. Moran MA, Zepp RG (1997) Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol Oceanogr 42: 1307–1316. [Google Scholar]
- 4. Keil RG, Kirchman DL (1994) Abiotic transformation of labile protein to refractory protein in sea-water. Mar Chem 45: 187–196. [Google Scholar]
- 5. Bertilsson S, Tranvik LJ (2000) Photochemical transformation of dissolved organic matter in lakes. Limnol Oceanogr 45: 753–762. [Google Scholar]
- 6. Baxter RM, Carey JH (1983) Evidence for photochemical generation of superoxide ion in humic waters. Nature 306: 575–576. [Google Scholar]
- 7. Cooper WJ (1989) Sunlight induced photochemistry of humic substances in natural waters: major reactive species. Adv Chem Ser 219: 332–362. [Google Scholar]
- 8. Zepp RG, Wolfe NL, Baughman GL, Hollis RC (1977) Singlet oxygen in natural waters. Nature 267: 421–423. [Google Scholar]
- 9. Dalrymple RM, Carfagno AK, Sharpless CM (2010) Correlations between dissolved organic matter optical properties and quantum yields of singlet oxygen and hydrogen peroxide. Environ Sci Technol 44: 5824–5829. [DOI] [PubMed] [Google Scholar]
- 10. Cory RM, McNeill K, Cotner JP, Amado A, Purcell JM, et al. (2010) Singlet oxygen in the coupled photochemical and biochemical oxidation of dissolved organic matter. Environ Sci Technol 44: 3683–3689. [DOI] [PubMed] [Google Scholar]
- 11. Anesio AM, Granéli W, Aiken GR, Kieber DJ, Mopper K (2005) Effect of humic substance photodegradation on bacterial growth and respiration in lake water. Appl Environ Microbiol 71: 6267–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joux F, Jeffrey WH, Abboudi M, Neveux J, Pujo-Pay M, et al. (2009) Ultraviolet radiation in the rhone river lenses of low salinity and in marine waters of the northwestern mediterranean sea: attenuation and effects on bacterial activities and net community production. Photochem Photobiol 85: 783–793. [DOI] [PubMed] [Google Scholar]
- 13. Judd KE, Crump BC, Kling GW (2007) Bacterial responses in activity and community composition to photo-oxidation of dissolved organic matter from soil and surface waters. Aquat Sci 69: 96–107. [Google Scholar]
- 14. Pérez MT, Sommaruga R (2007) Interactive effects of solar radiation and dissolved organic matter on bacterial activity and community structure. Environ Microbiol 9: 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piccini C, Conde D, Pernthaler J, Sommaruga R (2009) Alteration of chromophoric dissolved organic matter by solar UV radiation causes rapid changes in bacterial community composition. Photochem Photobiol Sci 8: 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glaeser SP, Grossart H-P, Glaeser J (2010) Singlet oxygen, a neglected but important environmental factor: short-term and long-term effects on bacterioplankton composition in a humic lake. Environ Microbiol 12: 3123–3136. [DOI] [PubMed] [Google Scholar]
- 17. Ogilby PR (2010) Singlet oxygen: there is indeed something new under the sun. Chem Soc Rev 39: 3181–3209. [DOI] [PubMed] [Google Scholar]
- 18. Davies MJ (2004) Reactive species formed on proteins exposed to singlet oxygen. Photochem Photobiol Sci 3: 17–25. [DOI] [PubMed] [Google Scholar]
- 19. Ryter SW, Tyrrell RM (1998) Singlet molecular oxygen (O-1(2)): A possible effector of eukaryotic gene expression. Free Rad Biol Med 24: 1520–1534. [DOI] [PubMed] [Google Scholar]
- 20. Cooper WJ, Lean DRS (1989) Hydrogen-peroxide concentration in a northern lake - photochemical formation and diel variability. Environ Sci Technol 23: 1425–1428. [Google Scholar]
- 21. Imlay JA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77: 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grandbois M, Latch DE, McNeill K (2008) Microheterogeneous concentrations of singlet oxygen in natural organic matter isolate solutions. Environ Sci Technol 42: 9184–9190. [DOI] [PubMed] [Google Scholar]
- 23. Latch DE, McNeill K (2006) Microheterogeneity of singlet oxygen distributions in irradiated humic acid solutions. Science 311: 1743–1747. [DOI] [PubMed] [Google Scholar]
- 24. Haag WR, Hoigne J (1986) Singlet oxygen in surface waters. 3. Photochemical formation and steady-state concentrations in various types of waters. Environ Sci Technol 20: 341–348. [DOI] [PubMed] [Google Scholar]
- 25. Häkkinen PJ, Anesio AM, Granéli W (2004) Hydrogen peroxide distribution, production, and decay in boreal lakes. Can J Fish Aqua Sci 61: 1520–1527. [Google Scholar]
- 26. Scully NM, McQueen DJ, Lean DRS, Cooper WJ (1996) Hydrogen peroxide formation: The interaction of ultraviolet radiation and dissolved organic carbon in lake waters along a 43–75 degrees N gradient. Limnol Oceanogr 41: 540–548. [Google Scholar]
- 27. Petasne RG, Zika RG (1997) Hydrogen peroxide lifetimes in south Florida coastal and offshore waters. Mar Chem 56: 215–225. [Google Scholar]
- 28.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER (2011) Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLOS ONE 6. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed]
- 29. Allgaier M, Grossart H-P (2006) Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol 72: 3489–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burkert U, Warnecke F, Babenzien D, Zwirnmann E, Pernthaler J (2003) Members of a readily enriched betaproteobacterial clade are common in surface waters of a humic lake. Appl Environ Microbiol 69: 6550–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, et al. (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria . Appl Environ Microbiol 66: 5053–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Allgaier M, Grossart H-P (2006) Seasonal dynamics and phylogenetic diversity of free-living and particle-associated bacterial communities in four lakes in northeastern Germany. Aquat Microb Ecol 45: 115–128. [Google Scholar]
- 33. Buck U, Babenzien HD, Zwirnmann E (2008) Extracellular peroxidase activity in an experimentally divided lake (Grosse Fuchskuhle, northern Germany). Aquat Microb Ecol 51: 97–103. [Google Scholar]
- 34. Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, et al. (2013) Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J 7: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grossart HP, Jezbera J, Hornak K, Hutalle KML, Buck U, et al. (2008) Top-down and bottom-up induced shifts in bacterial abundance, production and community composition in an experimentally divided humic lake. Environ Microbiol 10: 635–652. [DOI] [PubMed] [Google Scholar]
- 36. Buck U, Grossart HP, Amann R, Pernthaler J (2009) Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a humic lake. Environ Microbiol 11: 1854–1865. [DOI] [PubMed] [Google Scholar]
- 37. Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T (2009) Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp nov and P. necessarius subsp asymbioticus subsp nov. Int J Syst Evol Micr 59: 2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe K, Komatsu N, Ishii Y, Negishi M (2009) Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS Microbiol Ecol 67: 57–68. [DOI] [PubMed] [Google Scholar]
- 39. Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S (2011) A Guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75: 14–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glaeser SP, Kampfer P, Busse H-J, Langer S, Glaeser J (2009) Novosphingobium acidiphilum sp. nov., an acidophilic salt-sensitive bacterium isolated from the humic acid-rich Lake Grosse Fuchskuhle. Int J Syst Evol Micr 59: 323–330. [DOI] [PubMed] [Google Scholar]
- 41. Hutalle-Schmelzer KML, Zwirnmann E, Kruger A, Grossart HP (2010) Enrichment and cultivation of pelagic bacteria from a humic lake using phenol and humic matter additions. FEMS Microbiol Ecol 72: 58–73. [DOI] [PubMed] [Google Scholar]
- 42. Glaeser J, Nuss AM, Berghoff BA, Klug G (2011) Singlet oxygen stress in microorganisms. In: Advances in Microbial Physiology, Vol Poole RK, editor. 58: 141–173. [DOI] [PubMed] [Google Scholar]
- 43. Ziegelhoffer EC, Donohue TJ (2009) Bacterial responses to photo-oxidative stress. Nat Rev Microbiol 7: 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salka I, Cuperova Z, Masin M, Koblizek M, Grossart HP (2011) Rhodoferax-related pufM gene cluster dominates the aerobic anoxygenic phototrophic communities in German freshwater lakes. Environ Microbiol 13: 2865–2875. [DOI] [PubMed] [Google Scholar]
- 45. Dufour YS, Landick R, Donohue TJ (2008) Organization and evolution of the biological response to singlet oxygen stress. J Mol Biol 383: 713–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lourenço RF, Gomes SL (2009) The transcriptional response to cadmium, organic hydroperoxide, singlet oxygen and UV-A mediated by the sigma(E)-ChrR system in Caulobacter crescentus . Mol Microbiol 72: 1159–1170. [DOI] [PubMed] [Google Scholar]
- 47. Grossart HP, Simon M, Logan BE (1997) Formation of macroscopic organic aggregates (lake snow) in a large lake: The significance of transparent exopolymer particles, phytoplankton, and zooplankton. Limnol Oceanogr 42: 1651–1659. [Google Scholar]
- 48. Minella M, Romeo F, Vione D, Maurino V, Minero C (2011) Low to negligible photoactivity of lake-water matter in the size range from 0.1 to 5 mu m. Chemosphere 83: 1480–1485. [DOI] [PubMed] [Google Scholar]
- 49. Carlos L, Pedersen BW, Ogilby PR, Martire DO (2011) The role of humic acid aggregation on the kinetics of photosensitized singlet oxygen production and decay. Photochem Photobiol Sci 10: 1080–1086. [DOI] [PubMed] [Google Scholar]
- 50. Grossart H-P (2010) Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ Microbiol Rep 2: 706–714. [DOI] [PubMed] [Google Scholar]
- 51. Jones SE, Newton RJ, McMahon KD (2009) Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ Microbiol 11: 2463–2472. [DOI] [PubMed] [Google Scholar]
- 52. Simek K, Kasalicky V, Hornak K, Hahn MW, Weinbauer MG (2010) Assessing niche separation among coexisting Limnohabitans strains through interactions with a competitor, viruses, and a bacterivore (vol 76, pg 1406, 2010). Appl Environ Microbiol 76: 3762–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tranvik L, Kokalj S (1998) Decreased biodegradability of algal DOC due to interactive effects of UV radiation and humic matter. Aquat Microb Ecol 14: 301–307. [Google Scholar]
- 54. Simon M, Azam F (1989) Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser 51: 201–213. [Google Scholar]
- 55.Eichler S, Weinbauer MG, Dominik D, Höfle MG (2004) Extraction of total RNA and DNA from bacterioplankton. In: Kowalchuk GA, Bruijn FJD, Head IM, Akkermans ADL, van Elsas JD, editors. Molecular microbial ecology manual, 2nd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers. 103–120.
- 56.Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester: John Wiley. 115–174.
- 57. Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiolol 63: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 61. Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide-sequence of a 16S ribosomal-RNA gene from Escherichia coli . Proc Nat Acad Sci USA 75: 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Activity of heterotrophic bacteria after 1O2 and H2O2 exposure.
(PDF)
Cell numbers in controls and in 1O2 and H2O2 treatments.
(PDF)
Rarefaction analysis of nearly full-length 16S rRNA gene clone libraries.
(PDF)
Distance matrices of Pearson correlation based UPGMA cluster analysis.
(PDF)
Robustness of changes in BCC observed by RT-PCR-DGGE analysis shown by repeats of the 2006 (A), 2008 (B) and 2009 (C) in situ experiments.
(PDF)
Cluster analysis of Betaproteobacteria, Actinobacteria, and Sphingomonadaceae- specific 16S rRNA gene based RT-PCR DGGE analysis.
(PDF)
Delayed formation of hydrogen peroxide (H2O2) in 0.22 μm filtered water samples exposed to natural sunlight.
(PDF)
Transmission scans of poly-propylene (PP) bags, poly-ethylene (PE) Whirl-Pak bags, and the UVA/B block sheet.
(PDF)
Weather data for 30 day prior to the experiments carried out in 2006, 2008, and 2009.
(PDF)
NOM concentrations and reactivity of surface water samples from the SW basin of Lake Grosse Fuchskuhle.
(PDF)
Cell numbers of in situ incubation experiments in 2006 and 2008.
(PDF)
Physico-chemical parameters of Lake Grosse Fuchskuhle SW compartment.
(PDF)
Sequences of 16S rRNA gene targeting oligonucleotide primer used for RT-PCR DGGE analysis.
(PDF)
PCR and RT-PCR programs for the amplification of 16S rRNA and 16S rRNA gene fragments of Bacteria and bacterial subgroups used for DGGE fingerprint analysis.
(PDF)
Phylogenetic affiliation of 16S rRNA gene sequences representing DGGE bands.
(PDF)
Characterization of water sample photo-reactivity, including NOM characteristics of south-west compartment samples, comparison of water sample photo-reactivity, in situ H2O2 formation and decay, and the potential photochemical effects of unbleached material from the acidic fen area. Investigation of bacterial community composition by 16S rRNA (gene) based methods by the generation and screening of 16S rRNA gene clone libraries, 16S rRNA targeting reverse transcriptase (RT)-PCR DGGE analysis and group-specific 16S rRNA targeting RT-PCR DGGE analysis.
(PDF)