Abstract
Successful infection by the opportunistic pathogen Legionella pneumophila requires the collective activity of hundreds of virulence proteins delivered into the host cell by the Dot/Icm type IV secretion system. These virulence proteins, also called effectors modulate distinct host cellular processes to create a membrane-bound niche called the Legionella containing vacuole (LCV) supportive of bacterial growth. We found that Ceg14(Lpg0437), a Dot/Icm substrate is toxic to yeast and such toxicity can be alleviated by overexpression of profilin, a protein involved in cytoskeletal structure in eukaryotes. We further showed that mutations in profilin affect actin binding but not other functions such as interactions with poly-L-proline or phosphatidylinositol, abolish its suppressor activity. Consistent with the fact the profilin suppresses its toxicity, expression of Ceg14 but not its non-toxic mutants in yeast affects actin distribution and budding of daughter cells. Although Ceg14 does not detectably interact with profilin, it co-sediments with filamentous actin and inhibits actin polymerization, causing the accumulation of short actin filaments. These results reveal that multiple L. pneumophila effectors target components of the host cytoskeleton.
Keywords: Profilin, Type IV secretion system, Intracellular growth, Vesicle trafficking
1. Introduction
Legionella pneumophila a Gram-negative facultative intracellular bacterium that is naturally found in aquatic environments, where it is able to infect and replicate within a wide range of protozoan hosts. In nature, these bacteria parasitize fresh water protozoa, which are considered the training site for the acquisition of its virulence factors necessary for surviving and replicating in mammalian macrophages. When water contaminated with L. pneumophila is aerosolized and inhaled by humans, the bacterium is also capable of infecting lung alveolar macrophages. In healthy human hosts, L. pneumophila is usually asymptomatically cleared by the immune system, but in immunocompromised individuals L. pneumophila infection can lead to a potentially fatal pneumonia called Legionnaire’s Disease, or the milder febrile illness Pontiac Fever [53].
After internalization by a phagocyte, the bacterium remains within a phagosome and embarks a unique intracellular trafficking route differing from inert particles or non-pathogenic bacteria. In the end, the bacterium resides in a membrane bound compartment with features resembling those of the rough endoplasmic reticulum (ER), where it replicates until nutrient depletion and is primed for next round of infection [8,44].
The biogenesis of the niche supportive of bacterial replication in phagocyte by L. pneumophila requires the activity of Dot/Icm, a specialized protein translocation system that delivers hundreds of virulence factors (also called effectors) into infected cells [2,33,46,62]. Some of these effectors interfere with host signaling cascades essential for its defense against infection. For example, at least six proteins are known to target the ubiquitination (Ub) pathway [55]. However, largely due to the lack of information about the bona fide cellular targets of these effectors, and the pleiotropic effects caused by interfering with the Ub signaling cascades, the exact roles of most of these proteins in L. pneumophila infection remain unknown. Effectors that directly modulate the metabolism of lipids, both phosphtidyinositol and phosphotidylacid that participate in diverse metabolism and signaling pathways in eukaryote have been found. For examples, SidF is a phosphatidylinositol polyphosphate 3-phosphatase that among other potential activities, functions to enrich PI(4)P on the bacterial vacuole to facilitate the anchoring of effectors [47]. LegS2, LecE and LpdA manipulate phospholipids biosynthesis, potentially to interfere with host signaling or metabolism mediated by lipids [11,57]. On the other hand, the mechanisms underlying the targeting of cellular pathways such as phagosome maturation and autophagy are much better understood. By sophisticated biochemical mechanisms, SidM, LepB, SidD, AnkX and Lem3 completely hijack the activation cycle of Rab1, the small GTPase important for multiple aspects of the secretory pathway [13,25,26,28,31,43]. The study of these effectors has lead to the discovery of novel enzymatic activities, such as de-AMPylation [13,25] and reversible phosphorylcholination [14,29], which potentially are utilized by eukaryotic cells under normal physiological conditions. Similarly, RavZ has been shown to inhibit autophagy by functioning as a protease that deconjugates ATG8 from the lipid anchor [59].
Because of the lack of defects in intracellular replication by mutants missing one or more effector genes, it is widely accepted that significant functional abundance exists among effectors delivered by the Dot/Icm transporter as well as the host processes involved [24,42]. For example, based on their effects on host secretory pathway in eukaryotic cells upon ectopic expression, it is believed that many more Dot/Icm substrates are involved in the modulation of this host pathway essential for membrane remodeling [48,58]. Clearly, assigning biochemical functions to these effectors will make it possible to group them according to activities, which is essential in determining their collective roles in the biogenesis of the bacterial phagosome.
Yeast genetics has been proven to be a useful tool in the identification and functional characterization of Dot/Icm substrates [19,48,61]. In particular yeast toxicity conferred by some effectors under normal or a specific condition has been exploited to study the activity of these proteins. For example, we have identified SidK as a regulator for the vacuolar ATPase by its toxicity to yeast in neutral medium [7], and the de-AMPylase SidD and the dephosphorylcholinase Lem3 by their ability to suppress yeast toxicity conferred by the corresponding bacterial enzyme that inhibits essential host functions [13,14]. Similarly, the biochemical functions of two Dot/Icm substrates involved in modulating host lipid metabolism were uncovered by clever use of yeast genetics [11].
One L. pneumophila hypothetical protein that our lab has previously identified as toxic when expressed in yeast is Ceg14/SidL(Lpg0437) (referred to as Ceg14 in the entire text) [1]. This protein was originally identified by the presence of a sequence putatively recognized by the response regulator PmrA, which regulates several known Dot/Icm effectors [1]. It was reidentified by both computational analyses and Cya-based translocation assay as a substrate of the Dot/Icm transporter [62]. Our initial studies with purified recombinant Ceg14 suggested that this protein inhibits protein synthesis in a reaction using lysates of rabbit reticulocyte and mRNA coding for luminescence [52], but the biochemical mechanisms underlying the toxicity as well as the protein synthesis inhibition remain mysterious. In this study, we exploited yeast genetics to analyze the mechanism of action of Ceg14 by screening for yeast genes capable of suppressing its toxicity. We found that overexpression of profilin, a multifunctional protein central to the cytoskeletal structure of eukaryote, completely alleviates the toxicity of Ceg14 to yeast. We further demonstrate that Ceg14 directly interact with actin and such interactions lead to inhibition of actin polymerization. Together with the recent discovery of the actin nucleation protein VipA [51], these results suggest that modulation of the host cytoskeleton is an important component of the interactions between L. pneumophila and its hosts.
2. Materials and Methods
2.1 Bacterial strains and plasmid construction
Bacterial and yeast strains used in the study were listed in Table S1. L. pneumophila strains used in this study were derived from Lp02, a derivative of strain Philadelphia 1 [64]. L. pneumophila was cultured in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) medium or on buffered charcoal yeast extract (BCYE) agar. When necessary, the media were supplemented with thymidine at 0.1 mg/ml for growing thymidine auxotrophic strains. E. coli strains were grown and maintained in liquid Luria broth (LB) or on LB agar. For E. coli, antibiotics were used at the following concentrations: ampicillin, 100 µg/ml, kanamycin, 30 µg/ml, chloramphenicol, 30 µg/ml. In-frame deletion of L. pneumophila genes was performed following an established procedure [56]. Briefly. DNA fragments amplified with the appropriate primers were inserted into appropriately digested pSR47s [22] to produce pSB14ΔSidL and pZH14ΔVipA and the mutants were generated as described [56]. Plasmids and primers used in this study were listed in Table S2 and Table S3, respectively. Genes were inserted into pQE30 (Qiagen) and pET28a-Sumo [47] to express His6-tagged or His6-sumo-tagged proteins and pGEX6p-1(Qiagen) was used to express GST-tagged proteins. Eukaryotic genes were amplified from cDNA clones purchased from Thermo Fisher Scientific (Waltham, MA) or from a yeast genomic DNA library [13]. Site-directed mutagenesis was performed with the Quikchange Kit (Agilent) using the Pfu UltraII (Agilent) hybrid high fidelity DNA polymerase. The sequences of all inserts and mutations were verified by DNA sequencing analysis.
2.2 Yeast manipulation
All yeast strains were derivatives of W303 [13] or PJ69-4A [41] (Table S1). Yeast was grown at 30°C in YPD medium or in synthetic media with appropriate amino acid dropout supplemented with 2% glucose, raffinose or galactose as the sole carbon source. Yeast transformation was performed using the standard lithium acetate protocol [50]. For inducible yeast lethality assay, we cloned ceg14 into pSB157 [13], which carries the galactose inducible Pgal promoter and the resulting plasmid was linearized and transformed into strain W303. To isolate non-toxic mutants, DNA of pSB11 (Table S2) derived from pGBKT7 (Clontech) treated with hydroxylamine [32] was transformed into yeast strain PJ694A. Colonies survived on Trp−/glucose medium were purified and cells grown in 5-ml medium were used to determine the ability to code for full-length Ceg14 with antibodies specific for Myc or Gal4 fused to the N-terminus of the protein (Clontech). Plasmids from clones capable of coding for full-length Ceg14 were rescued and the mutations were mapped by DNA sequencing. Plasmid p425GPD (Table S1) [27] was used for constitutive expression of suppressor proteins in yeast and when needed, a Flag tag was added to N-terminal end of the gene of interest.
2.3 Identification of yeast genomic clones that suppress the toxicity of Ceg14
Plasmid DNA of the Yep13-based yeast genomic library (ATCC no. 37323) was transformed into yeast strain W303 (pGal:ceg14) which consistently exhibit galactose-dependent toxicity. Plasmid rescued from colonies appeared on Ura−/Leu−/galactose medium were verified for the ability to suppress Ceg14 toxicity and the sequences of those that reproducibly gave the suppressor phenotype were determined by DNA sequencing with primers from both ends of the inserts as described [14]. The entire sequence of the inserts was retrieved from the yeast genome database (http://yeastgenome.org). The ORFs of the relevant genes were cloned into p425GPD [27], and the resulting plasmids were then transformed into W303(Pgal:ceg14) for further analysis.
2.4 Protein expression and purification
Plasmids directing the expression of proteins of interest were transformed into E. coli strain BL21(DE3) for protein production. A 1-Liter culture was established from a 20-ml saturated E. coli culture in LB and IPTG (isopropyl β-D-1-thiogalactopyranoside) was added to 0.3 mM when the culture grew to OD600 of 0.6~0.8. The induction was allowed to proceed for 16~18 hours at 18°C, and the cells were collected by centrifugation at 5000g for 10 min at 4°C. Cells were lysed with 30-ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole) containing 1 mM PMSF, 5 mM benzamidine and 1 mM EDTA, and the soluble fraction obtained by centrifugation at 12,000g for 20 min was incubated with lysis buffer-equilibrated Ni-NTA resins at 4°C for 2 hrs. Protein was eluted with 250 mM imidazole after washing the resin 3 times (50-ml each time) with the lysis buffer supplemented with 20 mM imidazole. GST-tagged proteins were purified with the Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) following the protocol suggested by the manufacturer. Soluble cell lysates were prepared with PBS buffer and the resin was washed with PBS containing 0.25% triton X-100. Bound protein was eluted with an elution buffer (50 mM Tris·HCl pH8.0, 150 mM NaCl, and 50 mM reduced glutathione). The purity of all recombinant proteins was more than 90% as estimated by SDS-PAGE and Coomassie brilliant blue staining (Fig. S1). Proteins were dialyzed in a storage buffer (50 mM Tris·HCl, 150 mM NaCl, 10% glycerol) at 4°C, the buffer was changed one time after about 12 hrs.
2.5 Cell culture
HEK239T cells were grown in Dulbecco’s modified minimum Eagle’s medium (DMEM) supplemented with 10% FBS. Primary bone marrow-derived macrophages (BMDM) from A/J mice were prepared and cultured following a standard protocol [15]. D. discoideum strains were cultured as described previously [18].
2.6 Antibodies and immunoblot
A Ceg14-specific antibody was raised with His6-Ceg14 following a standard protocol (Pocono Rabbit Farm and Laboratory, Canadensis, PA). Antibody purified with antigen covalently coupled to an Affigel matrix [56] was used at 1:2000. The antibodies specific for ICDH (isocitrate dehydrogenase)(1:5000), GFP (1:10,000) and GST (1:5000) were described before [7]. Anti-Flag (1:2000) was from Sigma. Anti-actin (1:2000) and anti-PFN1 (ab50667) (1:2000) were from Abcam. Anti-Gal4 (1:1000) (sc-577) and anti-Myc (Sc-40) were from Santa Cruz Biotechnology. Samples resolved by SDS-PAGE were transferred onto nitrocellulose membranes. The membranes were blocked for 2 hrs with 5% milk at RT, and were incubated with the relevant primary antibody in the blocking buffer. After 3x washes with PBS containing 0.1% Tween-20, the membranes were incubated with appropriate IRDye infrared-conjugated secondary antibody (Li-Cor’s Biosciences Lincoln, Nebraska, USA) and the signals were detected by an ODYSSEY® (LI-COR) imaging system.
The expression of Ceg14 in L. pneumophila was evaluated with total lysates prepared from relevant bacterial strains grown to the indicated density as described [36]. Lysates of 50-ml bacterial culture grown the indicated phases were subjected to immunoprecipitation with the Ceg14 specific antibody and the presence of the protein in the precipitates was detected by immunoblot. For yeast samples in which the protein of interest needs induction, cells from saturated culture grown in 5-ml Ura−/raffinose medium were washed once with sterile water and were resuspended in 5-ml Ura−/galactose medium and induction was allowed to proceed for 6 hrs. Cells were then collected, washed and resuspended in 100-µl yeast cracking buffer [40 mM Tris·Cl (pH 6.8), 5% SDS, 0.1 mM EDTA, 8 M urea, bromothymol blue 0.4 mg/ml] with glass beads (Sigma), and were lysed by incubating at 70°C for 10 min followed by vortexing for 2 min.
2.7 Affinity pull-down and immunoprecipitation
Affinity pull-down was performed by mixing 50µg of actin with 50µl beads coated with GST or GST-tagged protein in 700-µl of binding buffer (50 mM Tris·Cl, pH 8.0, 150 mM NaCl). The reactions were incubated at 4°C for 5 hrs before removing the unbound proteins by 4x washes with lysis buffer containing 0.25% NP-40. Proteins associated with beads were solubilized with 60-µl Laemmli buffer before SDS-PAGE separation and subsequent detection. Immunoprecipitation of yeast cell lysates was performed as described [7] with beads coated with the Flag antibody (M2, Sigma).
2.8 Intracellular bacterial growth assays
4×105 bone marrow-derived macrophages and 5×105 D. discoideum were seeded in 24-well plates, respectively and cells were challenged at an MOI of 0.05 with relevant L. pneumophila strains grown to the post-exponential phase. At the indicated time points, cells were lysed with 0.02% saponin and the bacterial growth was determined by enumerating the colony-forming-unit (CFU) of appropriately diluted cell lysates plated on bacteriological medium.
2.9 Actin co-sedimentation and polymerization assay
G-actin and pyrene-labeled actin (Cytoskeleton, 99% purity) were prepared following the instructions by the manufacturer. Prior to use, the actin was cleared by ultracentrifugation at 100,000g for 30 min. The co sedimentation assay was performed using an established protocol [3]. Briefly, 20µg G-actin was mixed with GST-PFN, GST-SipA, or His6-Sumo-Ceg14 in the G-actin buffer (5 mM Tris·HCl, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT) and the mixture was incubated for 30 mins at RT. Actin polymerization was initiated by adding 5-µl of 10x actin polymerization buffer (APB) (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM ATP,) and was allowed to proceed for 60 min. The samples were subjected to ultracentrifugation at 100,000g for 40 min. An aliquot of the supernatant was saved and the pellet was dissolved in 30µl ddH2O. The presence of actin in these two phases of samples was analyzed by SDS-PAGE. Actin polymerization kinetics was performed as described [51]. Briefly, 3.5µM G-actin (with 10% pyrene-labeled actin) was mixed with different testing proteins in G-actin buffer. After 5 min incubation at RT, actin polymerization was initiated with 14-µl of 10xAPB and was monitored by measuring the fluorescence intensity at 60 or 20 sec interval, using a fluorescence spectrophotometer (SpectraMAX Gemini). The excitation and emission wavelength was set at 365 nm and 410 nm, respectively. Data were collected and processed in Excel (Microsoft).
2.11 Immunostaining and visualization of actin filaments
To stain yeast actin, cells were induced to express Ceg14 with galactose for 6 hrs, fixed with formaldehyde and the actin was stained with Texas Red-X phalloidin (T7471, Life Technologies) using an established procedure [45,65]. Stained cells were applied on coverslips coated with 0.01% poly-L-lysine (Sigma) and the images were acquired using an Olympus IX-81 microscope. In vitro actin staining was performed with Alexa Fluor-488 Phalloidin (Life Technologies) according to a published protocol [45]. Briefly, samples containing 4µM actin received the testing proteins were incubated at RT for 30 min, and polymerization was initiated by adding 2µl 10xAPB, phalloidin was simultaneously added to a final concentration of 3 µM. After 1 hr incubation, 2 µl of the sample was diluted with 400 µl fluorescence dilution buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 100 mM DTT, 100 µg/ml glucose oxidase, 15 mg/ml glucose, 20 µg/ml catalase, 0.5% methylcellulose). Two microliter diluted samples were applied onto coverslips coated with 0.01% poly-l-lysine (Sigma). Actin filaments were visualized using an Olympus IX-81 microscope and the images were acquired and processed with the IPlab software package (Scanalytic, Inc. Fairfax, VA).
2.12 Statistical analyses
Statistical significance for relevant data was calculated using the unpaired Student t test.
3. Results
3.1 Ceg14 is toxic to yeast and residue G234 or T623 is important for such toxicity
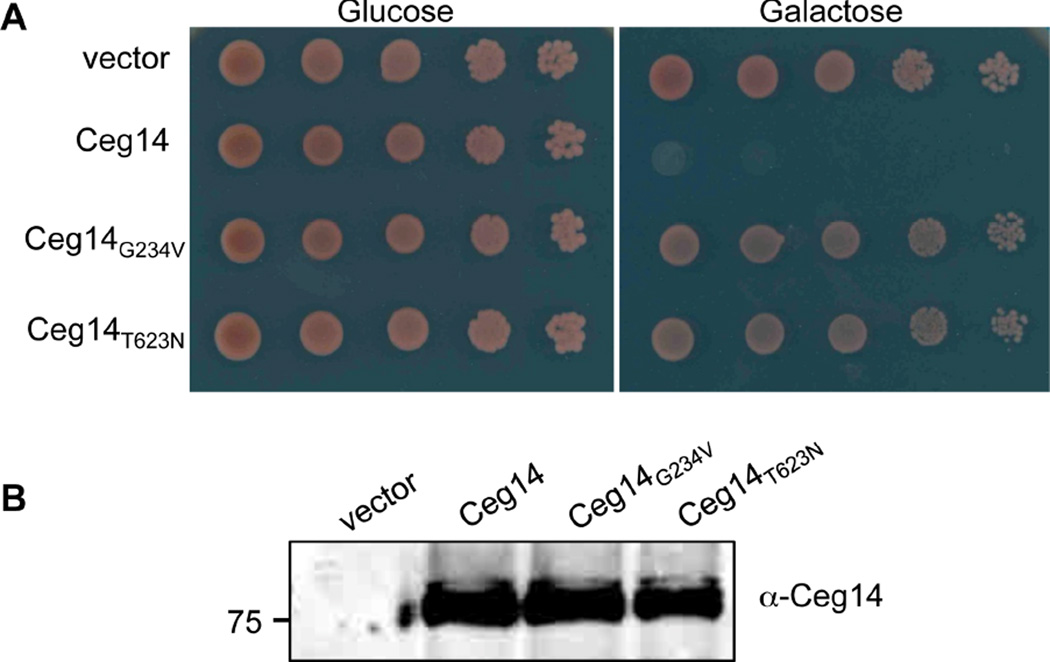
In our continuing efforts to study the function of Dot/Icm substrates by exploiting their toxicity to yeast, we found that transformation of yeast with a plasmid that allows the expression of Ceg14 from the alcohol dehyrogenase promoter (PADH) yielded no transformants, suggesting potential toxicity of this protein to yeast [21]. We further analyzed the toxicity by expressing Ceg14 from the galactose inducible promoter (PGAL) from the integration plasmid pSB157 [13]. On glucose medium, a yeast strain harboring chromosomally encoded PGAL::ceg14 grew at rates comparable to one harboring only the vector (Fig. 1). On inducing medium with galactose as the sole carbon source, whereas the control strain grew robustly, the strain harboring PGAL::ceg14 completely failed to form visible colony even after extended incubation (Fig. 1). These results indicate that Ceg14 is highly toxic to yeast, probably by targeting one or more essential host processes.
Fig. 1. The cytotoxicity of Ceg14 to yeast depends on residue Glycine 234 or Threonine 623.
A. Yeast strains derived from W303 harboring chromosomally integrated ceg14 or the mutants under the control of the galactose inducible promoter Pgal were grown in glucose medium to saturation. Cells diluted in sterile water were spotted onto synthetic medium with glucose or galactose as the sole carbon source. Images were acquired after 3-day incubation at 30°C. A strain harboring the vector was established as a control (top lane). B. Expression of the Ceg14 and its mutants in yeast. Yeast cells grown in raffinose medium were induced to express the proteins by galactose for 6 hrs and the total proteins of the cells were resolved by SDS-PAGE before probing for Ceg14 with a specific antibody. Reference of protein size markers (in kDa) is labeled in the left lane of the blot.
Single substitution mutants losing specific activity such as toxicity to eukaryotic cells are useful not only in mapping the region of a protein important for its potential biochemical function, but also in subsequent biochemical assays as important controls. We thus isolated non-toxic Ceg14 mutants by random mutagenesis. From more than 10 candidates capable of coding for full-length proteins in surviving yeast clones harboring PADH::ceg14, we obtained two substitution mutants Ceg14G234V and Ceg14T623N that no longer were toxic to yeast (Fig. 1). The loss of toxicity was not caused by lower protein stability in yeast as these mutants were expressed from the PGAL promoter at levels similar to the wild type (Fig. 1B).
We also examined the effects of Ceg14 in mammalian cells by expressing its GFP fusion in HEK293T cells, but no expression of the wild type protein can be detected (Fig. S2). Although the signals were weak, GFP fusions of both Ceg14G234V and Ceg14T623N were detectably expressed 24 hrs after transfection. In both cases, both mutants appeared to localize to the cytoplasm of the cells similar to GFP itself (Fig. S2). Consistent with the weak GFP signals, the protein levels of the GFP fusions of the mutants were low when detected by immunoblot (Fig. S2). Interestingly, an early termination non-toxic mutant missing the last 43 residues can be expressed at high levels in this cell line and showed a stronger cytoplasmic GFP signal (Fig. S2). Because factors attributing to the phenotypes associated with a truncation mutant are more likely to be multiple, we did not pursue the Ceg14.C43 mutant further. Taken together, these results suggest that the toxicity of Ceg14 to yeast is caused by a specific activity that requires residue G234 or T623. Further, the fact that we can express wild type Ceg14 in yeast allows us to examine its effects to eukaryotic cells by using this model organism.
3.2 Profilin suppresses the toxic effect of Ceg14 in yeast
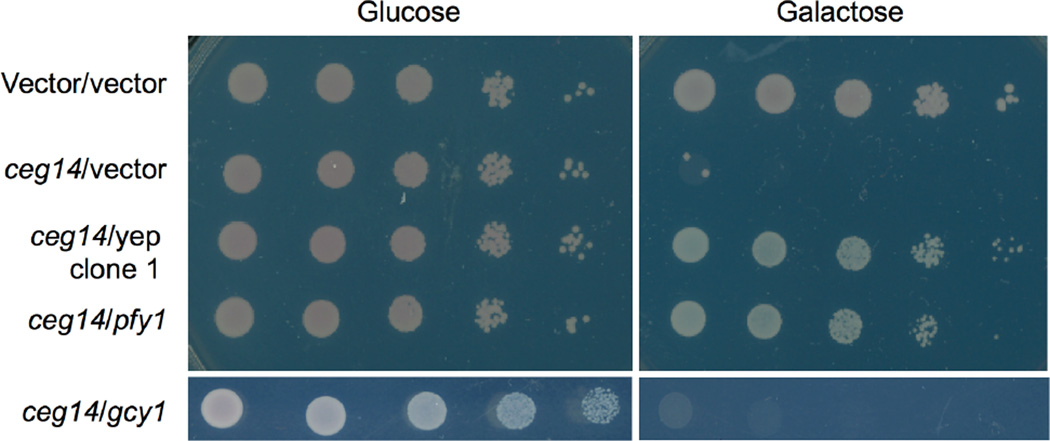
To analyze the mechanism of action of Ceg14, we first set to identify the host process targeted by this toxic effector. The highly inducible toxicity of Ceg14 to yeast prompted us to search for the proteins it potentially affects by screening for yeast genomic clones capable of suppressing the toxicity. We then transformed a yeast genomic DNA library [14] into strain W303(PGAL::ceg14) and selected for colonies capable of growing on galactose medium. A total of 9 independent Yep13 clones reproducibly exhibiting the suppressor activity were obtained and each was sequenced to determine the insert. We found that all of these genomic clones contained a DNA fragment from roughly the same locus on Chr. XV of S. cerevisiae (Fig. S3). Among these, clone #15 contained the smallest insert with only two genes: pfy1 that codes for the yeast profilin, and gcy1 that encodes a glycerol dehydrogenase (Fig. S3). By constructing plasmids that direct the expression of a single gene, we found that the pfy1 but not the gcy1 was able to suppress the toxicity (Fig. 2). The suppression by profilin was specific to Ceg14 toxicity, as it failed to rescue the toxicity of AnkX, a L. pneumophila effector that targets the small GTPase Rab1 (Fig. S4) [14]. These results indicate that Ceg14 targets host processes relevant to the functions of profilin.
Fig. 2. The yeast profilin suppresses the cytotoxicity of Ceg14.
Yeast strain W303(Pgal::ceg14) was transformed with an empty vector, a yeast genomic clone on pYep13 originally isolated in this study or a plasmid that carries the yeast profilin or the gcy1 gene and the resulting strains were grown in glucose medium before spotted onto repression (glucose) and induction (galactose) medium, respectively. Images were acquired after 3-day incubation at 30°C. A strain harboring only the Pgal vector transformed with an empty vector was used as a control (top lane). Note the robust growth of this control strain and strains harboring the profilin gene.
3.3 The ability to interact with actin is critical for the suppressor activity of profiling
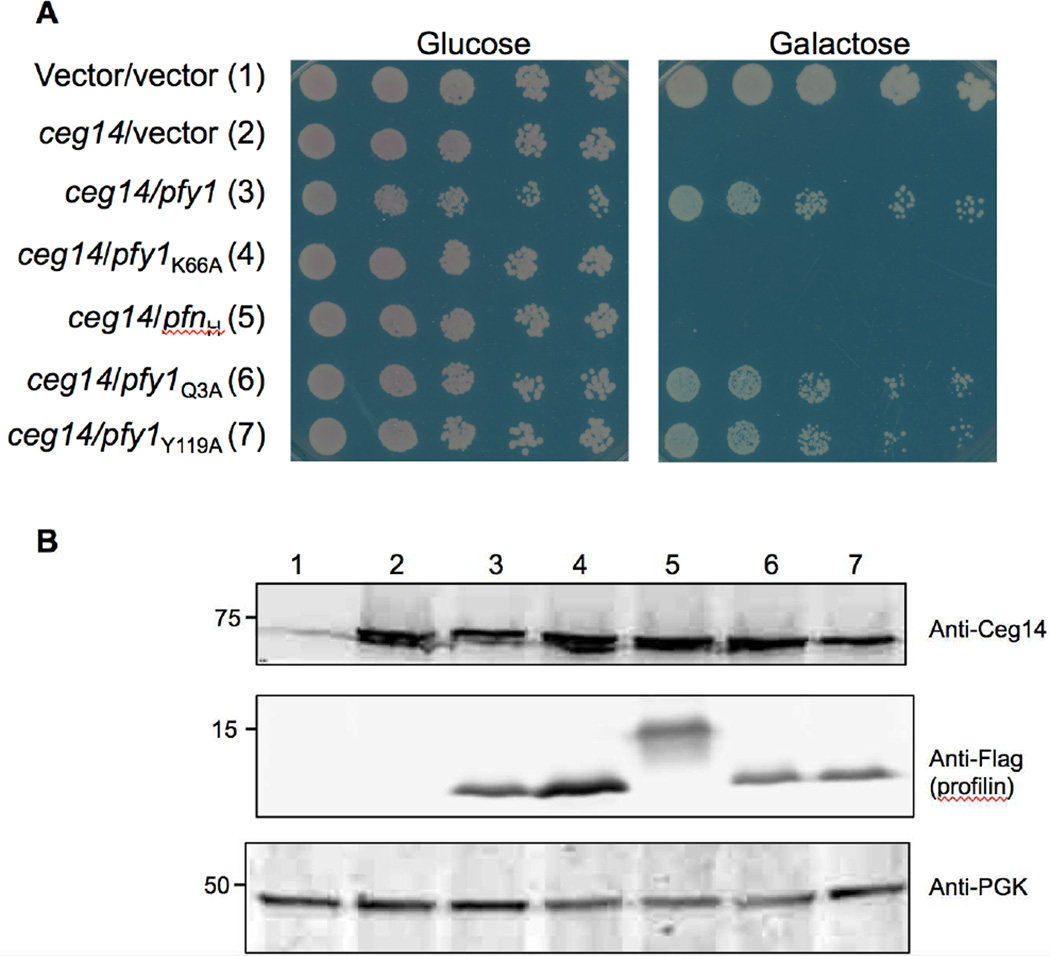
In yeast, profilin (PFY) is a central player in cytoskeletal structure that in general is a major factor that promotes the growth of actin filaments in vivo [40]. It interacts with monomeric actin at 1:1 ratio and catalyzes ADP to ATP nucleotide exchange on actin monomers [30]. Besides, it also binds phosphatidylinositol 4,5-biphosphate [PI(4,5)P2] [54] and numerous protein ligands with the poly-l-proline binding regions [9]. To determine the specific function of profilin important for its suppression of Ceg14 toxicity, we expressed Flag-tagged profilin or its mutants defective in binding to different ligands in yeast strain W303(PGAL::ceg14). Like the native protein, Flag-PFY strongly suppressed the toxicity of Ceg14 (Fig. 3). Similarly, mutants PFYQ3A and PFYY119A, which are defective in binding PI(4,5)P2 and poly-l-proline, respectively [34,37,60], exhibited similar suppressor activity (Fig. 3). On the other hand, the PFYK66A mutant defective in binding actin was unable to suppress the toxicity (Fig. 3). All of these mutants expressed at similar levels in these yeast strains, indicating that the lack of suppressor activity by PFYK66A is not a result of lower protein stability (Fig. 3B). Interestingly, human profilin I was unable to alleviate the toxicity imposed by Ceg14 (Fig. 3), suggesting that functions of profilin from these two organisms are not completely identical. Together, these results suggest that Ceg14 attacks one or more components of host cytoskeletal structure.
Fig. 3. A profilin mutant defective in binding actin has lost its ability to suppress the toxicity of Ceg14.
A. The yeast strain inducibly expressing Ceg14 was transformed with plasmids that direct the production of Flag-tagged human profilin, yeast profilin or its mutants defective in binding actin, phosphoinositol or poly-l-proline. The strains were then grown in glucose medium before spotting dilutions onto glucose and galactose plates. Images were acquired after 3-day incubation at 30°C. B. The expression of Ceg14, profilin and its mutants were probed in yeast cells induced by galactose for 6 hrs. Ceg14 was detected with a specific antibody and the two profilin proteins and mutants were detected by using the Flag-specific antibody. The 3-phosphoglycerate kinase (PGK) was also probed as a loading control (lower panel). Protein size makers (in kDa) are on the left lane of the blot.
3.4 Ceg14 is dispensable for proficient intracellular bacteria growth and is constitutively expressed by L. pneumophila
To determine the potential role of Ceg14 in L. pneumophila infection, we constructed an in-frame deletion mutant of this gene and examined intracellular replication of the mutant in bone marrow-derived macrophages and the protozoan host Dictyostelium discoideum. Deletion of Ceg14 did not affect bacterial uptake by either macrophages or D. discoideum, and like most of examined Dot/Icm substrates, ceg14 is not important for intracellular replication (Fig. S5). Similarly, a mutant lacking both ceg14 and its homolog lnaB [35] was not defective in intracellular growth in these hosts (Fig. S5). The fact that profilin suppressed the toxicity of Ceg14 suggests that this protein targets the host cytoskeleton, which is also targeted by VipA, a Dot/Icm substrate that directly interacts with actin [51]. We thus set to examine the potential functional redundancy of these two proteins by constructing a L. pneumophila mutant lacking both vipA and ceg14. This double mutant grew as proficiently as the wild type strain in two hosts examined (Fig. S6A–B). Furthermore, almost equal amounts of Ceg14 can be detected in bacterial cells grown at the exponential phase (OD600=2.0) or the postexponential phase (OD600=3.6) (Fig. S6C), indicating that the bacterium expresses Ceg14 independent of growth phase, which suggests that modulation of host cytoskeleton by this protein likely occurs throughout the entire intracellular growth cycle.
3.5 Ceg14 disrupts the actin distribution in yeast and inhibits its cell budding
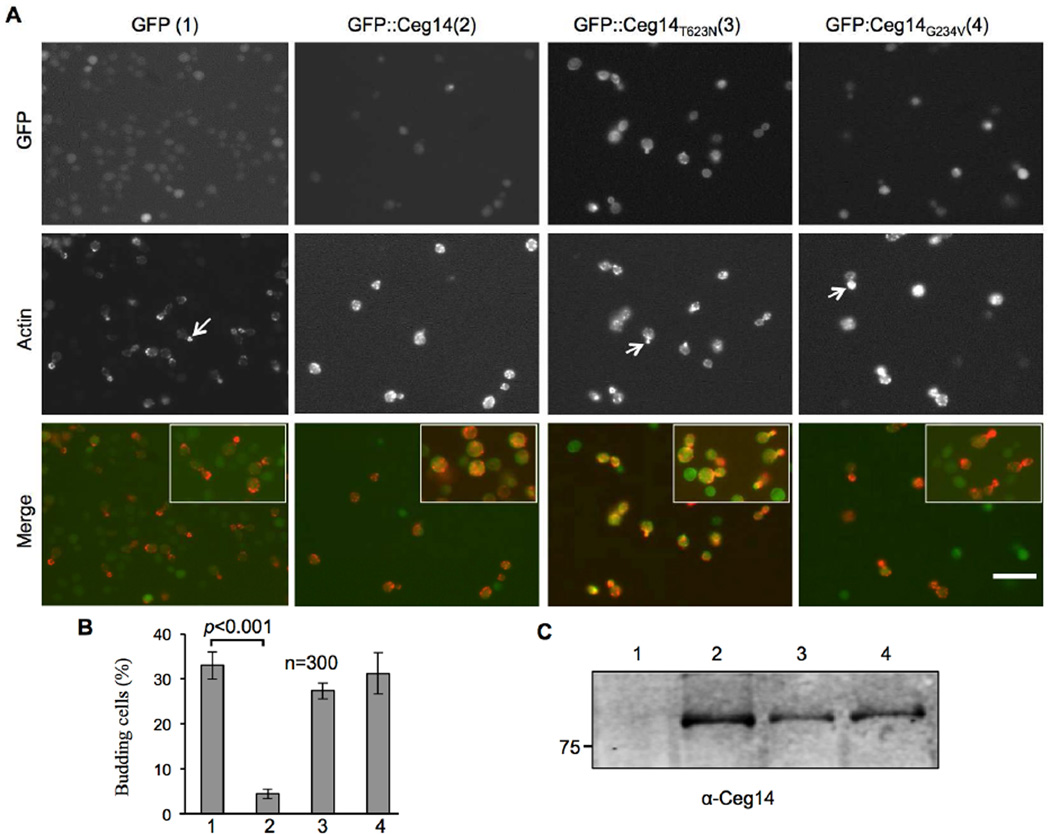
Since Ceg14 cannot be ectopically expressed in mammalian cells, we used yeast as a model to determine its cellular effects. To accomplish this, we constructed yeast strains inducibly expressing GFP, GFP-Ceg14, GFP-Ceg14G234V or GFP-Ceg14T623N. Six hrs after induction by galactose, we fixed the cells with formaldehyde and stained them with Texas Red-phalloidin (Life Technologies) for observation of the actin cytoskeleton. The GFP signals of wild type or mutant fusions evenly distributed in the cytoplasm, which is similar to the pattern observed in mammalian cells with the two non-toxic mutants (Figs. 4 & S2). We found that expression of Ceg14 led to significant reduction of budding cells (Fig. 4), which is consistent with the toxicity phenotype. Further, labeling of actin with Texas Red-X phalloidin revealed that in samples expressing GFP, actin in cells not actively dividing exhibited an uneven or polar distribution, and in dividing cells actin was mostly concentrated in the apical end of the budding daughter cells (Fig. 4A, 1st column). On the other hand, in cells expressing GFP-Ceg14, punctuate of actin appeared to concentrate in the cytoplasm or at the plasma membrane (Fig. 4A, 2nd column). Expression of neither of the two non-toxic mutants caused defects in cell budding, and the distribution of actin in these samples was similar to those expressing GFP (Fig. 4A 3rd and 4th columns). Thus, Ceg14 affects the cytoskeletal structure of eukaryotic cells, which very likely contributes to the lethal phenotype associated with this protein upon ectopic expression.
Fig. 4. Ceg14 interferes with daughter cell budding in yeast.
A. Yeast strains harboring GFP, GFP-Ceg14, GFP-Ceg14T623N, or GFP-Ceg14G234V under the control of the Pgal promoter were induced by galactose for 6 hrs. Fixed, permeabilized cells were stained with Texas Red-X phalloidin prior to image acquisition and scoring of budding cells. Note that in cells expressing GFP or GFP fusions of the mutants, the actin staining signals mostly localized to the growing tips of the daughter cells (arrows in the middle panel of the first, third and fourth column of images), whereas in cells expressing GFP-Ceg14, actin was lining along the plasma membrane (arrows in the middle of the second column of images). Bar, 10 µm. B. The number of budding cells was quantitated under a microscope in stained samples done in triplicate, 300 cells were scored in each sample. Similar results were obtained in multiple independent experiments and data shown were from a representative experiment. C. Expression of the fusion proteins in the samples. Total protein from a portion of the cells used for immunostaining resolved by SDS-PAGE was probed with a Ceg14 specific antibody.
3.6 Ceg14 inhibits actin polymerization and co-precipitates with filamentous actin
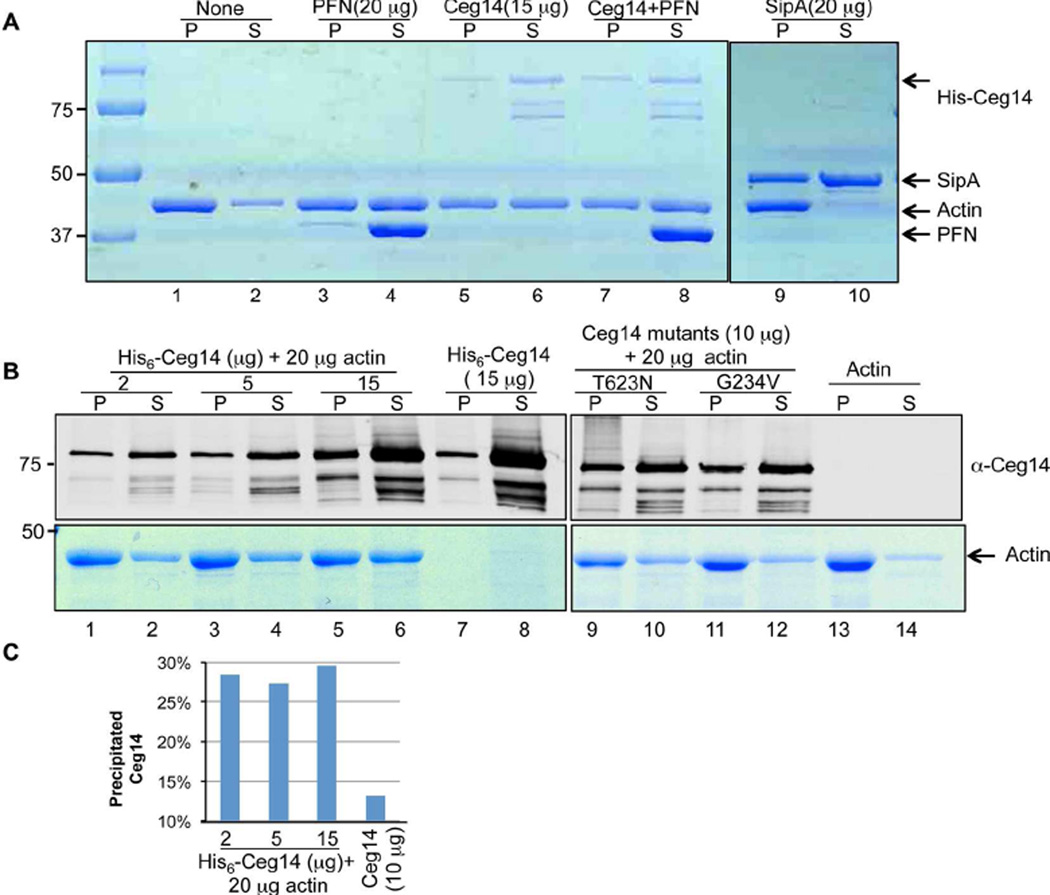
Our genetic evidence strongly suggested that profilin is the direct target of Ceg14. However, despite extensive efforts, we did not detect any biochemical effects such as binding or posttranslational modification of Ceg14 on profilin. Similarly, the interactions between profilin and actin in yeast cells expressing Ceg14 were indistinguishable from those expressing GFP (Fig. S7). We thus considered the possibility that the suppressor activity by profilin was a result of indirect effects on the cytoskeleton and other components of the eukaryotic cells are the direct target of Ceg14. To this end, we examined whether Ceg14 interferes with the property of actin, a common target for virulence factors that attack host cytoskeleton [49]. Actin spontaneously polymerizes in a polymerization buffer, leading to sediments after high-speed centrifugation (Fig. 5A, lanes 1 and 2). In reactions received His6-Ceg14, the amount of precipitated actin was significantly reduced, indicating an inhibitory effect of Ceg14 on actin polymerization (Fig. 5A, compare lane 1 to lane 5). Under this reaction condition, PFN appeared to inhibit actin polymerization and consistent with its preference to bind monomeric actin, most of the PFN fractioned into the soluble fraction, which is consistent with previous observations [4,5] (Fig. 5A, lanes 3 and 4). Further, inclusion of PFN and Ceg14 simultaneously in the reaction did not cause further inhibition of actin polymerization (Fig. 5A, compare lane 5 and lane 7). This is in agreement with the notion that Ceg14 did not directly interfere with the activity of profilin. As a control, the actin binding and stapling protein SipA from Salmonella typhimurium [3] strongly induced actin precipitation and preferably associated with filamentous actin (Fig. 5A, lanes 9 and 10).
Fig. 5. The effects of Ceg14 on actin polymerization.
A. Ceg14 inhibits actin polymerization. Polymerization in reactions containing 20µg G-actin and indicated recombinant proteins was initiated by adding the APB buffer and was allowed to proceed for 60 min. After centrifugation, filamentous actin (pellet) and monomeric actin (supernatant) were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining. Note the higher amount of filamentous actin in the sample with actin alone than those with His6-Ceg14 (compare lane 1 to lane 5 or 7). The actin bundling protein SipA from S. typhimurium was used as a control (lane 9). Also note the association of PFN with monomeric actin (lane 4) and the fact that PFN did not interfere with the inhibitory effects of Ceg14 (compare lane 5 to lane 7). B. Dose-dependent inhibition of actin polymerization by Ceg14. Indicated amounts of recombinant proteins were mixed with actin and assembled actin was evaluated after centrifugation. The amount of actin was assessed by Commassie brilliant blue staining and 10% of His6-Ceg14 resolved by SDS-PAGE was detected by immunoblot (upper panel). Note that 2 µg of His6-Ceg14 is able to exert significant inhibition of actin polymerization (compare lane 2 to lane 14) and a higher amount (15 µg) caused more significant inhibition (compare lane 2 to lane 6). Also note is the attenuated inhibitory effect of the two non-toxic mutants (compare lane 6 to lane 10 or 12). C. Quantitation of His6-Ceg14 associated with filamentous actin. The intensity of protein bands was determined by using the Li-cor Odyssey imaging system. Note the quick saturation of precipitated His6-Ceg14 and the significantly lower amount of this protein in the pellet in the absence of actin (the last column). Similar results were obtained in multiple experiments under the same experimental conditions. Reference of protein size was indicated on the left of the gels.
The relatively weak inhibitory effect of Ceg14 on actin assembly prompted us to further examine this activity. We found that in a reaction containing 20 µg actin, inclusion of 2 or 5 µg of His6-Ceg14 led to detectable but very marginal inhibitory effects of actin polymerization and such effects did not seem to further increase when 15 µg His6-Ceg14 was added, suggesting that under this experimental condition, 2µg of His6-Ceg14 almost saturated its inhibitory effects (Fig. 5B, compare lanes 2, 4 and 6). Importantly, although the mutations that abolished the toxicity of Ceg14 also abolished its ability to inhibit actin polymerization, these mutations did not appear to interfere with the weak interactions between Ceg14 and actin (Fig. 5B lanes 9–14). Consistently, we observed little increase in the amount of His6-Ceg14 associated with filamentous actin when more recombinant protein was included in the reactions (Fig. 5C). These results indicate that Ceg14 interferes with the dynamics of actin by inhibiting its spontaneous polymerization. Our results also suggest direct binding between actin and Ceg14, albeit apparently at a low affinity.
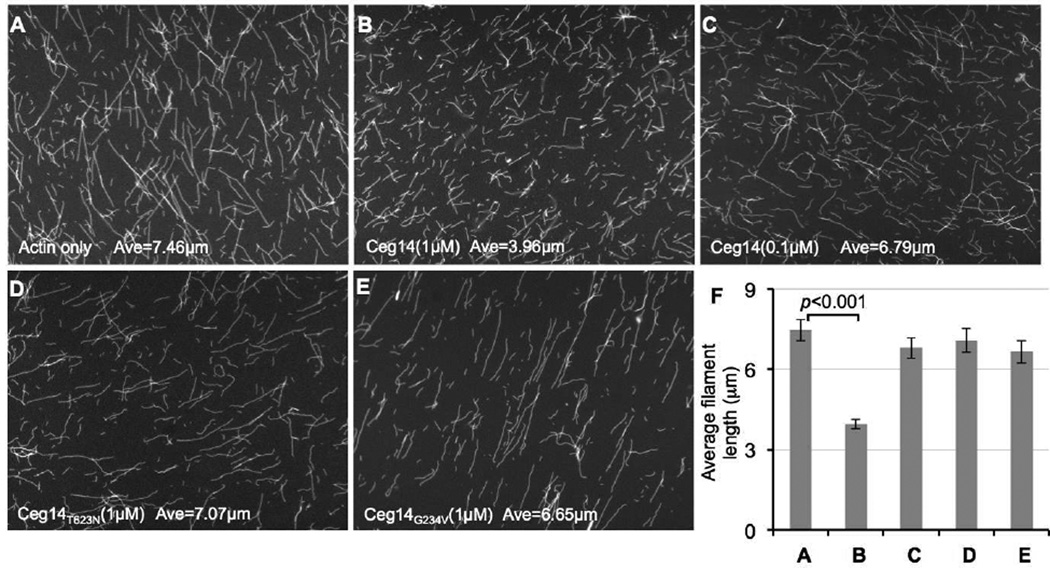
At least two mechanisms can account for the reduced actin polymerization caused by Ceg14 manifested in the precipitation assay: Inhibition of the nucleation or the growth of the actin filaments. We thus distinguished these two possibilities by directly visualizing actin filaments in the presence of Ceg14. His6-Ceg14 did not detectably affect the amount of filamentous actin, but the lengths of the actin filaments in reactions received 1 µM His6-Ceg14 were significantly shorter than the controls (Fig. 6, A, C and F). Consistent with the weak inhibitory effects observed in the actin polymerization assay, inclusion of 0.1 µM His6-Ceg14 in the reactions did not lead to the formation of significantly shorter actin filaments (Fig. 6, A, B and F). Again, in agreement with the genetic data, none of the two non-toxic mutants retained the inhibitory effects on actin polymerization (Fig. 6, D–F). Taken together, these results indicate that the observed inhibitory effects of Ceg14 on actin polymerization are caused by its ability to inhibit the growth of actin filaments.
Fig. 6. Ceg14 caused the formation of shorter actin filaments.
A. Reactions containing 4µM actin and indicated testing proteins were induced to polymerize for 1 hr. Samples were stained with Alexa Fluor-488 Phalloidin, diluted, and applied to poly-L-lysine coated coverslips. Samples were analyzed using an Olympus IX-81 microscope for image acquisition. Bar, 10µm. B. The average length of actin filaments was measured under microscope. At least 100 randomly chosen filaments were scored in experiments done in triplicate and data shown are from one representative experiment.
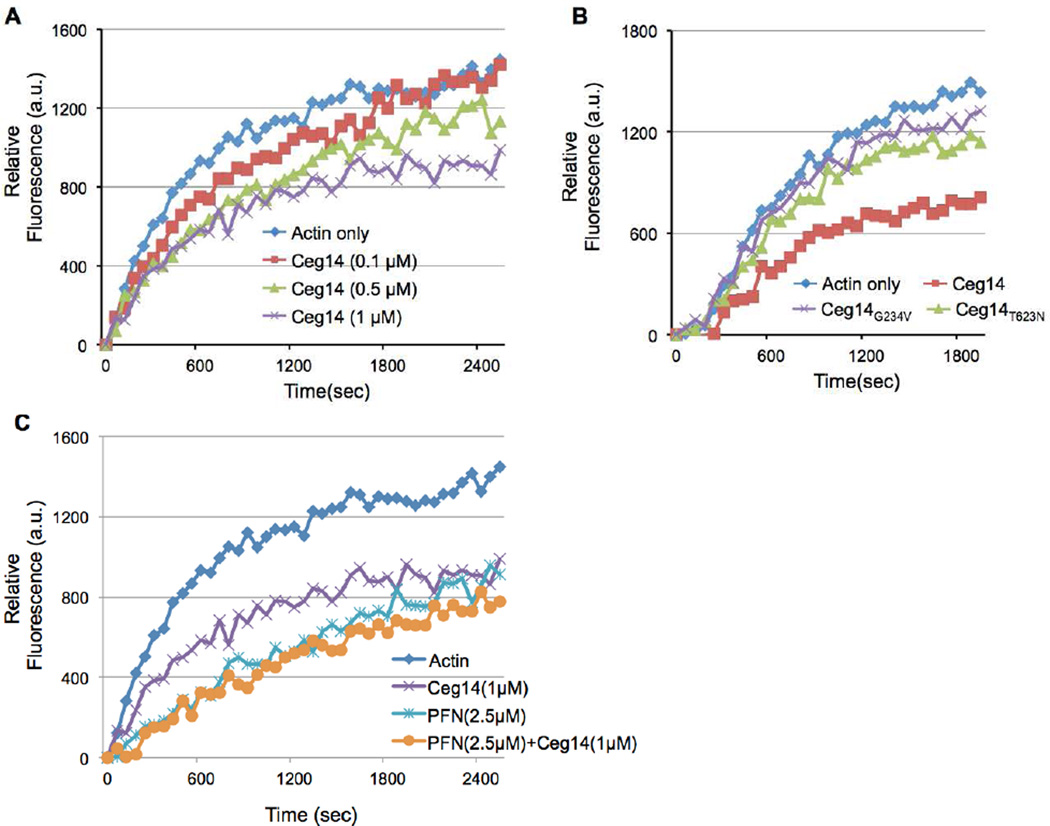
3.7 Ceg14 interferes with the actin dynamics in polymerization
To further determine the mechanism of action of Ceg14, we examined its effects on actin dynamics by using 10% pyrene-labeled G-actin. The inhibitory effects of His6-Ceg14 were not apparent until when 1µM protein was included in the reactions (Fig. 7A). Similar to results obtained in other assays, both Ceg14G234V and Ceg14T623N had lost the ability to inhibit actin polymerization (Fig. 7B). We also found that 2.5 µM of PFN exhibited a stronger inhibitory effect than 1µM of His6-Ceg14, but simultaneous inclusion of these two proteins in the reactions did not lead to further inhibition of actin polymerization or reverse of the effect of Ceg14 (Fig. 7C). These results further validate the conclusion that Ceg14 directly interferes with actin dynamics.
Fig. 7. Kinetics of the inhibition of actin polymerization by Ceg14.
Monomeric actin containing 10% pyrene-labeled actin was mixed with indicated testing proteins the general actin buffer. The intensity of fluorescence signals was monitored by a fluorescence spectrophotometer after initiating actin polymerization. A. Dose-dependent inhibition of actin polymerization by Ceg14. B. Non-toxic mutants of Ceg14 have lost the ability to inhibit actin polymerization. C. PFN did not interfere with the inhibitory effect of Ceg14. Note that addition of PFN did not alleviate the activity of Ceg14.
4. Discussion
Actin cytoskeleton governs many fundamental functions of a cell, such as its shape, movement, phagocytosis, and the distribution of organelles and it is a common target exploited by numerous successful pathogens to facilitate their entry, replication or dissemination [49]. Here we present results suggesting that the Dot/Icm substrate Ceg14 modulates the cytoskeleton of host cell.
Because the primary in vivo function of profilin is to promote actin filament growth [9,17,23], its strong suppressor activity points to a negative role of Ceg14 in host cytoskeletal structure. Several activities can fulfill such a negative role. For example, Ceg14 could target profilin by direct biochemical modification to abolish its activity in promoting actin polymerization. However, despite extensive efforts, we did not obtain any evidence that points to direct effects imposed by Ceg4 on this multifunctional host protein. Similarly, Ceg14 could be a capping protein, which binds to the barded end of growing actin filaments with high affinity and inhibits its growth [10]. Both actin and profilin are associated with the LCV membrane [12], whether Ceg14 locally targets one or both of these proteins remains to be determined. Differing from the property of a capping protein, Ceg14 does not interact with actin with high affinity and it is still toxic to a yeast mutant lacking the capping protein, suggesting of a non-capping activity (Fig. S8A). Instead, our collective results showed that Ceg14 directly inhibits actin polymerization and its inhibitory effects were correlated with its toxicity toward eukaryotic cells because non-toxic mutants have lost the inhibitory activity. These results are consistent with the fact that the primary in vivo function of profilin is to promote actin filament growth [9,17,23]. Thus, it is likely that the alleviation of Ceg14 toxicity by profilin results from an indirect and compensatory mechanism.
Unlike virulence factors from pathogens such as Listeria, Salmonella and Shigella that drastically alter the biochemical properties of the cytoskeletal components of host cells [49], the effects of Ceg14 on the activity of actin are moderate. Several reasons can account for the mild effects of Ceg14 on actin dynamics. First, the natural hosts of L. pneumophila are phagocytes, which actively engulf the bacterium, making its entry into the host cell a passive process. The role of Ceg14 in L. pneumophila infection more likely is in the biogenesis of the bacterial vacuole, which is similar to that of VipA, an actin nucleation factor that also appears to have moderate activity on actin [51]. Instead of targeting such critical regulatory molecules as the small GTPases Cdc42 and Rac1, these Legionella effectors directly target actin, which may allow more subtle and fine-tuned regulation of the function of the cytoskeleton. Second, the fact that VipA and Ceg14 have opposite effects on actin polymerization suggests balanced modulation of host cytoskeleton by the bacterium. However, VipA cannot suppress the toxicity of Ceg14 (Fig. S8B), suggesting that such balanced regulation is achieved by an indirect and compensatory mechanism, which differs from the regulation of Rab1 activity by effectors with opposite enzymatic activities [8]. Finally, it is possible that we have not established an optimal experimental condition to analyze the activity of Ceg14. For example, the recombinant protein used in the in vitro experiments may not be fully active. Nevertheless, the genetic results and the biochemical effects we consistently observed in different assays established a role Ceg14 plays in modulating host cytoskeletal structure.
BLAST searches identified lpg2527 (LnaB) as a paralog of Ceg14 (E value=3×10−16). LnaB is a Dot/Icm substrate that potently activates NFκB in mammalian cells [35]. In an assay using rabbit reticulocyte lysate and pre-synthesize mRNA, recombinant His6-Ceg14 was able to inhibit protein translation, which contributes to the induction of a subset of cytokines in mouse macrophages by inhibiting the re-synthesis of the labile IκB and allowing prolonged NFκB activation [52] (Fig. S9). Targeting of host cytoskeleton has recently been recognized as signals that induce immune response. For examples, stimulation of Rho-family GTPases by SopE and SopB of S. typhimurium leads to activation of both the MAPK and NFκB signaling pathways [63]; similarly, the actin-bundling protein SipA activates NFκB-dependent immune responses via the Nod1/Nod2 pathway [39]. It is possible that LnaB also targets host cytoskeletal structure, an activity that may account for its NFκB activation activity, which contributes to the immune responses induced by L. pneumophila infection.
Accumulating evidence has revealed a clear linkage between protein translation and the cytoskeletal structure [38]. For example, eEF1A, the major component of the ribosome involved in the addition of charged amino acids to the growing polypeptide, binds to actin and can sever microtubules [6,20]. Reciprocally, an intact F-actin system is required for efficient protein synthesis [16]. Thus, the seemingly remotely related activities associated with Ceg14 could be a result of its biochemical function towards one or more proteins in host cytoskeletal structure such as actin.
Although our data suggest that Ceg14 modulates host cytoskeleton, the detailed mechanism awaits further analysis. The fact that single substitution mutations can abolish the toxicity of Ceg14 to yeast suggests a biochemical activity associated with this protein. Such activity may be revealed by further careful mass spectrometry analysis of its potential targets such as actin and profilin or by structural study that may discover motifs for known biochemical function. The elucidation of such activity and its effects on the host target is the ultimate goal of future functional analysis of Ceg14. Furthermore, it is necessary to establish methods to examine the effects of Ceg14 on eukaryotic cells more relevant to L. pneumophila infection such as amoebae (i.e. D. discoideum) and macrophages. However, because of its toxicity, it may be necessary to use some unconventional methods such as microinjection of active recombinant protein to address these challenges.
Supplementary Material
Acknowledgements
We thank Drs. Chris Staiger and Daoguo Zhou (Purdue University, West Lafayette, Indiana, USA) for reagents and for helpful discussion and Dr. Peter Hollenbeck for critical reading of the manuscript. This work was supported by NIH-NIAID grants R01AI069344, K02AI085403, R21AI092043 and R56AI103168 (Z.-Q.L) from the National Institutes of Health and by an Oversea Collaboration Grant (No. 31228023) from Chinese Nature Science Foundation (Z.-Q.L and S.-J. Z). Z. Guo was supported by a fellowship from the China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou D, Mooseker MS, Galan JE. Role of the S typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 4.Yarmola EG, Bubb MR. How depolymerization can promote polymerization: the case of actin and profilin. Bioessays. 2009;31:1150–1160. doi: 10.1002/bies.200900049. [DOI] [PubMed] [Google Scholar]
- 5.Yarmola EG, Bubb MR. Profilin: emerging concepts and lingering misconceptions. Trends Biochem Sci. 2006;31:197–205. doi: 10.1016/j.tibs.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature. 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Luo ZQ. Cell biology of infection by Legionella pneumophila. Microbes Infect. 2013;15:157–167. doi: 10.1016/j.micinf.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Viner R, Chetrit D, Ehrlich M, Segal G. Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 2012;8:e1002988. doi: 10.1371/journal.ppat.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic. 2009;10:76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- 13.Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y, Arnold RJ, Luo ZQ. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A. 2011;108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stapulionis R, Kolli S, Deutscher MP. Efficient mammalian protein synthesis requires an intact F-actin system. J Biol Chem. 1997;272:24980–24986. doi: 10.1074/jbc.272.40.24980. [DOI] [PubMed] [Google Scholar]
- 17.Staiger CJ, Blanchoin L. Actin dynamics: old friends with new stories. Curr Opin Plant Biol. 2006;9:554–562. doi: 10.1016/j.pbi.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Solomon JM, Isberg RR. Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol. 2000;8:478–480. doi: 10.1016/s0966-842x(00)01852-7. [DOI] [PubMed] [Google Scholar]
- 19.Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1 alpha. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, Banga S, Liu Y, Xu L, Gao P, Shamovsky I, Nudler E, Luo ZQ. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rankin S, Isberg RR. Identification of Legionella pneumophila promoters regulated by the macrophage intracellular environment. Infect Agents Dis. 1993;2:269–271. [PubMed] [Google Scholar]
- 23.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor TJ, Boyd D, Dorer MS, Isberg RR. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 27.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 28.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mockrin SC, Korn ED. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5'-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- 31.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Luo ZQ, Smyth AJ, Gao P, Qin Y, Farrand SK. Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J Biol Chem. 2003;278:13173–13182. doi: 10.1074/jbc.M210035200. [DOI] [PubMed] [Google Scholar]
- 33.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C. Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem. 2002;3:12. doi: 10.1186/1471-2091-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 39.Keestra AM, Winter MG, Klein-Douwel D, Xavier MN, Winter SE, Kim A, Tsolis RM, Baumler AJ. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. MBio. 2011;2 doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 41.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 44.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 45.Huang S, Blanchoin L, Chaudhry F, Franklin-Tong VE, Staiger CJ. A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J Biol Chem. 2004;279:23364–23375. doi: 10.1074/jbc.M312973200. [DOI] [PubMed] [Google Scholar]
- 46.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. The E Block motif is associated with, Legionella pneumophila translocated substrates. Cell Microbiol. 2010;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu F, Zhu W, Brennan L, Tao L, Luo ZQ, Mao Y. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci U S A. 2012;109:13567–13572. doi: 10.1073/pnas.1207903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 2009;11:230–248. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 51.Franco IS, Shohdy N, Shuman HA. The Legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog. 2012;8:e1002546. doi: 10.1371/journal.ppat.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo ZQ, Vance RE. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fedorov AA, Magnus KA, Graupe MH, Lattman EE, Pollard TD, Almo SC. X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates. Proc Natl Acad Sci U S A. 1994;91:8636–8640. doi: 10.1073/pnas.91.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dumenil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- 57.Degtyar E, Zusman T, Ehrlich M, Segal G. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 2009;11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 58.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaudhary A, Chen J, Gu QM, Witke W, Kwiatkowski DJ, Prestwich GD. Probing the phosphoinositide 4,5-bisphosphate binding site of human profiling I. Chem Biol. 1998;5:273–281. doi: 10.1016/s1074-5521(98)90620-2. [DOI] [PubMed] [Google Scholar]
- 61.Campodonico EM, Chesnel L, Roy CR. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2005;56:918–933. doi: 10.1111/j.1365-2958.2005.04595.x. [DOI] [PubMed] [Google Scholar]
- 62.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 65.Amatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.