Abstract
Polyploidization is an evolutionarily rare but important mechanism in both plants and animals because it increases genetic diversity. Goldfish of the Carassius auratus species complex can be tetraploids, hexaploids and octaploids. Polyploidization events have occurred repeatedly in goldfish, yet the extent of this phenomenon and its phyletic history are poorly understood. We explore the origin, tempo and frequency of polyploidization in Chinese and Japanese goldfish using both mitochondrial (mtDNA) and nuclear DNA sequences from up to 1202 individuals including the outgroup taxon, Cyprinus carpio. Analyses of de novo nuclear gene data resolve two clusters of alleles and the pattern supports the prior hypothesis of an ancient allotetraploidization for Carassius. Alleles shared by tetraploid and hexaploid individuals indicate recent autoploidizations within the C. auratus complex. Sympatric tetraploids and hexaploids share mtDNA haplotypes and these frequently occur independently within six well-supported lineages and sublineages on a small spatial scale. Gene flow estimates (Fst values) indicate that hexaploids differ only slightly from sympatric tetraploids, if at all. In contrast, allopatric populations of tetraploids and hexaploids differ from one another to a far greater extent. Gene flow between sampled localities appears to be limited. Coalescence-based time estimations for hexaploids reveal that the oldest lineage within any sampled locality is around one million years old, which is very young. Sympatric, recurrent autoploidization occurs in all sampled populations of the C. auratus complex. Goldfish experience polyploidization events more frequently than any other vertebrate.
Keywords: goldfish, autopolyploidization, tetraploid, hexaploid
Introduction
The frequency and hence the evolutionary importance of polyploidization for diversification differs greatly between plants and animals. More than 70% of angiosperms experienced one or more episodes of polyploidization and about 95% of fern species are polyploids (Soltis and Soltis, 1999; Van de Peer et al., 2009). Notwithstanding, whole-genome duplications have contributed to the evolutionary success of vertebrates. Extensive comparative genomic work reveals three rounds of whole-genome duplication early in teleost diversification (Ohno, 1970; Vandepoele et al., 2004; Van de Peer et al., 2009). The genomes of all 25 000 or so species in this lineage of vertebrates reflect this whole-genome duplication event. In fact, whole-genome duplication appears to be causally related to their evolutionary success (Wittbrodt et al., 1998). Generally speaking, polyploidization occurs only rarely in animals, especially in vertebrates, in part because sex determination and development are disrupted by polyploidization (Mable, 2004; Mable et al., 2011). Among animals, polyploidization is common only in some lineages of invertebrates such as ostracods and insects. Some species of fishes, amphibians and reptiles are polyploids and in some evolutionary lineages of fishes polyploidization is not uncommon (Dawley, 1989; Otto and Whitton, 2000; Leggatt and Iwama, 2003). Cyprinid fishes are of special interest in this regard because they are unusually rich in polyploidy lineages, several of which are ancient (Otto and Whitton, 2000), including the subfamilies Barbinae (Tsigenopoulos et al., 2002), Cyprininae and Schizothoracinae (Yu et al., 1989; Buth et al., 1991; Le Comber and Smith, 2004). Full genomic duplications have occurred in at least 40 cyprinid species (Yu et al., 1989; Buth et al., 1991).
Recurrent polyploidization events occur either by interspecific hybridization and the fusion of two or more genomes (allopolyploidy) or by genome doubling within a species or a population (autopolyploidy). In plants, most cases of polyploidization occur via allopolyploidization, and this process is thought be an important mechanism for the rapid formation of species (Soltis and Soltis, 1999; Hegarty and Hiscock, 2008; Van de Peer et al., 2009; Hu et al., 2011). Polyploidization may be involved in about 2–4% of the speciation events in angiosperms and ferns, respectively (Otto and Whitton, 2000), but only a handful of these cases are autopolyploidization events (Segraves et al., 1999; Parisod and Besnard, 2007). In contrast, only in a single case of multiple autopolyploid origins among invertebrates is known: the lepidopteron Solenbia triquetrella (Psychidae) (Lokki and Saura, 1980). Little is known about the extent of multiple autopolyploid origins in vertebrates, and some possible cases are now believed to represent allopolyploidization (for example, Holloway et al., 2006).
The genus Carassius has a recent, allotetraploid origin (Buth, 1983; Risinger and Larhammar, 1993; Yang and Gui, 2004; Luo et al., 2006). Within Carassius, at least two species are recognized: Carassius carassius (the crucian carp) and C. auratus, a complex of several lineages (Table 1) that includes the silver crucian (gibel) carp and Japanese ginbuna (silver crucian carp), nagobuna, nigorobuna and gengorobuna; the latter is sometimes considered to be the species Carassius cuvieri (Takada et al., 2010 and therein). C. carassius occurs from the Ertis River in China westwards to Central and Eastern Europe and the British Isles (Luo and Yue, 2000; Wheeler, 2000); C. carassius may no longer occur in the Ertis River in China, based on our attempts to locate the species. C. auratus (Cypriniformes, Cyprinidae) is a complex of tetraploid and hexaploid fishes. Individuals within each lineage are morphologically similar yet their chromosome numbers and reproductive strategies vary. Except for exclusively tetraploid C. cuvieri, bisexual tetraploid and gynogenetic hexaploid and occasional octaploid forms occur sympatrically in China, Russia and Japan (Golovinskaya et al., 1965; Chen et al., 1996; Murakami et al., 2001; Brykov et al., 2002). Multiple levels of polyploidy occur in Fangzheng, Puan and Lake Dianchi, China (Zan, 1982; Zan et al., 1986 and therein; Chen et al., 1996 and therein).
Table 1. Breed, distribution, chromosome number and karyotype of the genus Carassius.
| Species, subspecies or local types | Distribution | Reproductive strategy | Male/female | Chromosome no. | Ploidy | Karyotype formula | Source |
|---|---|---|---|---|---|---|---|
| Carassius carassius (Crucian carp) | Central &Eastern Europe, British Isles | Sexual | Male, female | 100 | 4n | ? | Vinogradov, 1998 |
| C. auratus auratus (Goldfish) | Euro-Asia Continent | Sexual | Male, female | 100 | 4n | 22 M+30SM+48 T. ST 24 M+30SM+46ST-T | Zan et al., 1986; Wang et al., 1988 |
| C. auratus gibelio (Gibel carp) | Euro-Asia Continent | Gynogenesis | Female | 156± | 6n | 42 M+74SM+40ST-T | Shen et al., 1983 |
| C. auratus gibelio (Gibel carp) | Euro-Asia Continent | \ | Male, female | 162± | ? | 48 M+56SM+18ST+40 T | Shan et al., 1988 |
| C. auratus (back high type) | Yuannan | Gynogenesis | Female mainly | 162± | 6n | 33 M+53SM+76ST-T | Zan, 1982 |
| C. auratus (Suogu breed) | Hunan | Sexual | Female | 156 | 4n | 36 M+36SM+84ST | Yu et al., 1987 |
| C. auratus (Qihe breed) | Henan | Gynogenesis | ? | 156± | 6n | ? | Lou et al., 1989 |
| C. auratus (Puan breed, Form A) | Guizhou | Gynogenesis | Female | 156± | 4n | 38 M+28SM+18ST+72 T | Yu et al., 1992 |
| C. auratus (Pengze breed) | Jiangxi | Gynogenesis | Male, female | 166± | 4n | 32 M+40SM+18ST+76 T | Chen et al., 1996 |
| C. a. langsdorfi (Ginbuna) | Japanese Islands | Gynogenesis | Female | 152–158 | 6n | 34 M+62SM+60 T | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
| C.s a. langsdorfi (Ginbuna) | Japanese Islands | Sexual | Male, female | 100 | 4n | 12 M+36SM+52 T | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
| C. cuvieri (Gengorobuna) | Japanese Islands | Sexual | Male, female | 100 | 4n | 12 M+36SM+52 T | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
| C. auratus. buergeri (Nagabuna) | Japanese Islands | Sexual | Male, female | 100 | 4n | 12 M+36SM+52 T | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
| C. a.buergeri (Nagabuna) | Japanese Islands | Gynogenesis | Female | 150± | 6n | ? | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
| C. a. grandoculis (Nigorobuna) | Japanese Islands | Sexual | Male, female | 100 | 4n | 12 M+36SM+52 T | Kobayashi, 1978; Ueda and Ojima, 1978; Ojma and Yamano, 1980 |
Abbreviations: M, metacentric; SM, submetacentric; ST, subtelcentric; T, telcentric.
The occurrence of allopatric polyploids questions may owe to two phenomena: independent origins or a rather ancient, single origin followed by dispersal to different drainage systems within Eurasia (Table 1). For Carassius, preference for one alternative over the other involves biogeography as well as the tempo and mode of polyploidization. (Takada et al., 2010) and (Gao et al., 2012) detailed the biogeography of the complex. Risinger and Larhammar (1993) and (Buth et al., 1991 and therein) documented the mode of polyploidization; allozymic and paralog information identified allo- and autopolyploidization events. The tempo of polyploidization remains unknown.
Analyses involving both maternally inherited mitochondrial DNA (mtDNA) and nuclear genes can identify the maternal progenitor (Evans et al., 2004) of polyploids. If two paralogs cluster in different clades and with different species, then allopolyploidization provides the best explanation. When this pattern does not occur, we cannot reject the null hypothesis of autopolyploidization. The mtDNA gene-tree can identify the maternal progenitor within one of the nuclear gene clusters (Evans et al., 2004).
The origin hexaploids, their population genetic features and their relationship to tetraploids remains unclear. Although initial reports suggested that all hexaploid C. auratus are females, males have been recently discovered in several populations (Lu et al., 2006; our unpublished data). Hexaploids might have originated from tetraploid C. auratus auratus (Allendorf and Thorgaard, 1984). Normally, bisexual tetraploid fish do not exclude the second polar cell until fertilization (Gui et al., 1995), but hexaploids possibly originate when the second diploid polar cell is not excluded from a tetraploid egg as a consequence of either hydrostatic pressure shock (Gui et al., 1995) or heat/cold stock (Pandian and Koteeswaran, 1998). Most hexaploid individuals are females and they reproduce gynogenetically as sexual parasites; females require exogenic sperm to stimulate egg development (Gui, 2007). Because hexaploids are closely related to tetraploids, they can serve as a model system to explore the origin, history and mechanisms of autopolyploidization within species.
We use our published sequence data (Gao et al., 2012) and those for the Japanese goldfish from GenBank along with de novo ploidy information for Eurasian Carassius to investigate the history and possible multiple origins of polyploidy within the C. auratus complex. If hexaploidization is rare and originated only once or very few times in independent drainages, we would expect hexaploids to be restricted to one or a few matrilines (Supplementary Figure S1). Alternatively, if hexaploidization is not rare but rather independently reoccurs often, then hexaploids should be widespread on the tree (Supplementary Figure S1). We track the maternal history and population structure of tetraploid and hexaploid populations within the C. auratus complex using mtDNA sequence data, and then map polyploidization events onto the tree. Subsequently, we investigate the origin of the hexaploids by calculating population coalescence times for each lineage. We sequence three nuclear genes to investigate the characteristics of duplicated genomes and autopolyploidy within each species complex.
Materials and methods
Samples
Sampling included all taxonomically recognized forms of C. carassius and the C. auratus complex, the latter from its native distribution in mainland China. We downloaded all available mtDNA sequences in GenBank (Supplementary Tables S1–S3) and combined them with six de novo sequences to form two mtDNA data sets for constructing a matrilineal genealogy: control region (CR) and CR+cytb. The first data set consisted of 1202 mitochondrial CR sequences that comprised 237 unique haplotypes, including the outgroup taxa. This data set consisted of seven parts: (1) 340 sequences from Gao et al. (2012) plus six de novo sequences (total 346) that were accompanied by genome sizes and/or number of chromosomes (Supplementary Table S1, part 1 and 2); (2) 18 gibel carp studied by Li and Gui (2008) (Supplementary Table S1, part 1); (3) six Japanese sequences from the study by Gao et al. (2012) along with 725 sequences they used from GenBank that had ploidy information (Supplementary Table S2); (Murakami et al., 2001; Iguchi et al., 2003; Takada et al., 2010); (4) three samples of C. cuvieri (Supplementary Table S3); (5) three samples of C. carassius from the study by Gao et al. (2012); (Supplementary Table S3); (6) 94 haplotypes downloaded from GenBank without ploidy information; and (7) seven sequences from carp (C. carpio) for the outgroup (Supplementary Table S3). The second data set contained complete sequences of the gene encoding cytochrome b (cytb) plus CR for 180 individuals taken from the study by Gao et al. (2012) including all taxa of C. carassius and the C. auratus complex (Supplementary Table S3). Among these sequences, 1095 had ploidy information and these were used for population analyses. The remaining 107 sequences were used for constructing matrilineal trees only.
We investigated whether polyploidization in the goldfish occurred by either auto-or allopolyploidization at the nuclear genomic level (Woods and Buth, 1984). We sequenced three nuclear genes (see below) from 15 individuals as follows: four individuals of C. carassius, one silver crucian carp, one tetraploid and one hexaploid goldfish each from Lake Dianchi, Zhejiang and Heilongjiang (six total), three gengorobuna (C. cuvieri) and one common carp (C. carpio).
Ploidy determination
We used two methods to newly determine ploidy levels for 340 individuals from the study by Gao et al. (2012) plus six new individuals: karyotyping, following the study by Yu et al. (1989), and flow cytometry, as in the study by Luo et al. (2006). The chromosomal numbers of 167 individuals were visually determined from karyotypes either using photographs or under a microscope (Olympus, Tokyo, Japan). Flow cytometry used a fluorescence-activated cell sorter (FACS Vantage SE System, BD Biosciences, San Jose, CA, USA) to estimate ploidy levels of 179 individuals. We modified the method of Luo et al. (2006) by staining blood cells for 10 min and using 1:10 concentration of domesticated goldfish blood cells: target specimen cells to standardize genome size estimates.
DNA extraction, PCR amplification and nucleotide sequencing
MtDNA fragment amplification and sequencing for six individuals with known ploidy levels were performed as in the study by Gao et al. (2012). We targeted the CR because of its high level of variability.
We used the following primers for amplifying and sequencing three nuclear genes: a partial fragment of the gene encoding macrophage migration inhibitory factor (MIF), 4MIF-5′-RCGCCCAAAATAARCAATACT-3′, 4MIF-5′-FAAGATGTTGTCTGCTGTAAG-3′ a partial fragment of the gene encoding steroidogenic acuteregulatory protein (StAR), 2StAR-5′-RGTGATGCTGGAACAGAAGAC-3′, 2StAR-5′-FGCACAACGGACACTTACAAA-3′ and a partial fragment of the gene for growth hormone (GH) 5′-GH-U2TGCTGGTTAGTTTGTTGGTG-3′, GH-5′-L2GCTCYTCTGYGYTTCATCTTT-3′. Amplifications were performed using the Gene Amp PCR system 9700 (ABI, Foster City, CA, USA). The total PCR volume was 25 μl with final concentrations of 1x buffer containing 0.15 mM MgCl2 (TaKaRa, Dalian, China), 0.25 mM dNTPs (TaKaRa), 1U Taq DNA polymerase (TaKaRa) and 25–50 ng total DNA. Following an initial 5 min denaturing step at 94 °C, PCR comprised 35 cycles at 94 °C for 30 s, 58 °C for 30 s and 72 °C 30 s, followed by a final extension at 72 °C for 7 min. The corresponding PCR products were purified on agarose gels and extracted using a kit (Watson BioMedical Inc., Shanghai, China). Subsequently, the purified products were ligated and cloned with the pUC18 DNA (TaKaRa). Each clone was picked individually from a Luria broth (LB) plate and cultured in LB liquid medium. Plasmid DNA was manually isolated via the standard alkaline lysis miniprep method. The plasmid DNA was sequenced using an ABI 3730 with an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit according to the manufacturer's instructions. The following universal primers were used for sequencing the plasmids: BOO12 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and BOO13 5′-AACAGCTATGACCATG-3′.
Data analysis
We added 21 CR haplotypes from downloaded sequences to the 216 haplotypes of Gao et al. (2012) and constructed genealogies. Ploidy levels were then mapped onto the trees, and the proportions of ploidy were also shown.
MtDNA variation and divergence within and between populations of different ploidy levels were explored using several indices as implemented in ARLEQUIN 3.5 (Excoffier et al., 2005). Between population Fst (Reynolds et al., 1983; Slatkin, 1995) and within population haplotype diversity (H) and nucleotide diversity (π) (Nei, 1987) were assessed. Neutrality tests estimated population stability, expansion and bottlenecking (Tajima, 1989; Fu, 1997). Coalescence time (Tco) and the maternal effective population size (Nef) (Ruvolo, 1997) were calculated using a generation time (t) of 2 y because the sexual maturity of the wild goldfish was shown to be 1–2 y (Lorenzoni et al., 2007). The frequency of polyploidy events was estimated by evaluating the number of shared haplotypes and the coalescence time (Tco) between the most divergent haplotypes.
The three nuclear gene sequences were aligned with DNASTAR 5.0 (DNASTAR Inc. Madison, WI, USA) and refined by eye. DAMBE 4.1.19 (Xia and Xie, 2001) was used to identify shared haplotypes, and MEGA 4.1 (Tamura et al., 2007) was used to extract information on variable sites. These fragments were used to reconstruct polyploidization events in Carassius. Maximum parsimony (MP) analyses were implemented in PAUP* 4.0b10 (Swofford, 2002), maximum likelihood (ML) used RAxML (Stamatakis et al., 2008) and Bayesian inference (BI) was implemented in MrBayes 2.01 (Huelsenbeck and Ronquist, 2001). Best-fitting models for the BI and ML analyses used likelihood ratio tests (Goldman, 1993; Huelsenbeck and Crandall, 1997) as implemented in the jMODELTEST 0.0.1 (Posada, 2008). For BI, four independent MCMC chains were simultaneously run for 5 000 000 generations while sampling one tree per 500 replicates, Burnin=0, and Burninfrac=0.5 (0.1 or 0.2 and so on), with two runs conducted independently. Sampled trees were used to construct a 50% majority rule consensus tree. Frequency of nodal resolution was termed a Bayesian posterior probability (BPP) and these values were mapped onto the consensus tree. Nodal support in the MP and ML analyses was assessed using nonparametric bootstraps (BS) (Felsenstein, 1985) calculated for MP in PAUP* and for ML in RAxML with 1000 replicates each. We assessed the robustness of our genealogy by comparing trees obtained from MP, ML and BI analyses.
Results
Ploidy level and sequence variation
We successfully determined the ploidy levels of 346 fishes from mainland China and to this we added 18 samples from Li and Gui (2008) (total 364; Supplementary Table S1 part 1). C. auratus complex had 57 tetraploids and 146 hexaploids from Yunnan (Southwest China), 27 hexaploids from Guizhou (Southwest China), 23 tetraploids and 21 hexaploids from Zhejiang (East China), 5 tetraploids and 6 hexaploids from Hunan (East China), 14 hexaploids from Guangdong (East China) and 29 tetraploids and 36 hexaploids from Heilongjiang (Northeast China). In total, these included 114 tetraploids and 250 hexaploids (Supplementary Table S1). Further, the ploidy levels of 725 Japanese fish were previously determined (Supplementary Table S2; Murakami et al., 2001; Iguchi et al., 2003; Takada et al., 2010). For six Japanese individuals whose CR sequences were used by (Gao et al., 2012), we determined their ploidy levels.
For mitochondrial regions, the 320–426 nucleotide sites of partial CR sequences from 1202 fishes yielded 237 haplotypes (Supplementary Table S3). In the CR sequences, 130 nucleotide positions varied of which 106 were potentially parsimony-informative and 24 were invariant. Uncorrected pairwise p-distances between haplotypes ranged from 0.24 to 11.52%. Within Carassius, p-distances averaged 3.84%. The majority of substitutions were transitions and among ingroup representatives the transition/transversion ratio averaged 6.57. Inclusion of the outgroup dropped the transition/transversion ratio to 5.01. Pairwise p-distances between Carassius and Cyprinus ranged from 10.53 to 17.63%, averaging 13.72%. Both genera were distinct genetically.
Gao et al. (2012) sequenced cytb for 104 selected samples (accession no. GU135503–GU135605; NC_010768) to resolve deeper level relationships than those revealed from CR data. We mapped our ploidy information onto their matrilineal genealogy.
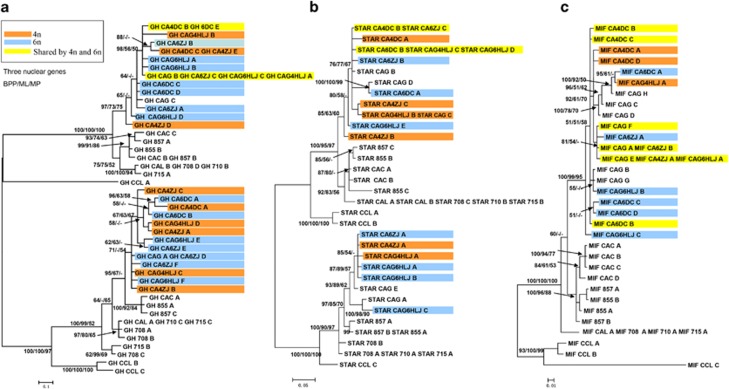
Sequencing of the three nuclear genes used a total of 15 individuals including one common carp as the outgroup. Among the 269 clones sequenced for GH, 43 alleles were identified (accession no. JX406582—406620, KC462749-KC462762), including 2–6 alleles belonging to two copies in each individual. Thirty-two alleles were identified from 272 clones of StAR (accession no. JX406621—406650, KC462770-KC462782), including 2–5 alleles belonging to two copies in each individual. Finally, for MIF, 32 alleles were identified from 269 sequenced clones (accession no. JX406651–JX406681, KC462763–KC462769), including 1–8 alleles belonging to at least two copies in each individual (Supplementary Table S4). There were 75/65, 62/62 and 92/23 potentially parsimony-informative/uninformative nucleotide positions in GH, MIF and StAR, respectively. Phylogenetic analyses recovered two major groups each for GH and StAR (Figure 1). In GH, the maximum and minimum distances within group I were 4.5% and 0.3%, respectively, and within group II 4.1% and 0.3%, respectively. In StAR, the maximum and minimum distances within group I were 5.3% and 0.3%, respectively, and within group II 4.6% and 0.3%, respectively. Only one group was recognized for MIF, and the maximum and minimum distances were 5.2% and 0.3%, respectively. Distances for GH between ingroup sequences and carp sequences varied from 13.7 to 11.3% in group I, and 13.2 to 6.1% in group II. For StAR, distances ranged from 12.2% to 5.0% in group I and from 13.4 to 3.5% in the group II. Finally, within from MIF distances ranged from 23.2 to 9.1%.
Figure 1.
BI 50% majority rule consensus trees for three nuclear genes, GH (a) StAR (b) and MIF (c) indicate autopolyploidization within the genus Carassius. Numbers represent nodal support inferred from BPPs, ML bootstrapping and MP bootstrapping, respectively. In all genes, most sequences from sympatric tetraploid and hexaploid fishes clustered together. GH and StAR had two major groups, which nested with carp alleles and indicated an earlier round of allotetraploidization in the ancestor of goldfish and carp. Within each cluster of alleles for GH and StAR, sympatric tetraploid and hexaploid individuals share the same alleles (in yellow), a result of autoploidization. A full color version of this figure is available at the Heredity journal online.
Autopolyploidization or allopolyploidization based on nuclear genes?
Analyses of GH and StAR obtained very similar sets of relationships. Both trees depicted two primary clusters and one or two outgroup alleles occurred in at least one of the ingroup clusters (Figure 1). Further, within one cluster (Figure 1), the topology was similar to that derived from the mtDNA data: C. carassius diverged first, and the Japanese gengorobuna, the gibel carp and the sympatric Chinese tetraploids and hexaploids followed this in sequence. Finally, for all sampling localities, alleles of hexaploid individuals did not cluster together, not even within the collecting sites including Yunnan (Lake Dianchi), Zhejiang and Heilongjiang (Figures 1a and b). Unlike for GH and StAR, the tree derived from sequences of MIF did not resolve two distinct groups (Figure 1c) even after either including or deleting gaps in the phylogenetic analyses. Within each locality, hexaploids independently nested with sympatric tetraploids.
Maternal genealogy of the C. auratus complex
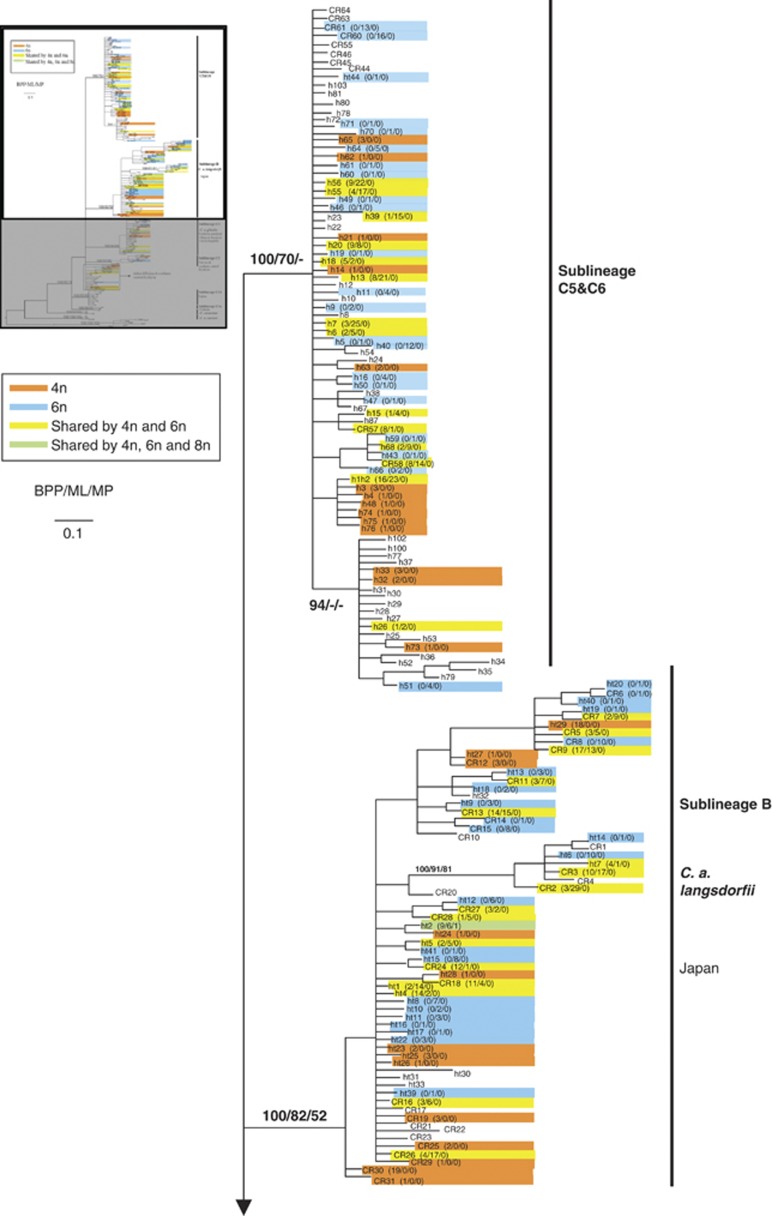
Matrilineal relationships of Carassius reconstructed using BI from 237 haplotypes of CR were used to visualize the distribution of the tetraploid and hexaploid lineages (Figure 2). In comparison, MP and ML analyses produced very similar topologies for the main lineages except for the relative positions of C. carassius and the Japanese gengorobuna. In the BI and ML trees, C. carassius formed the sister group of C. auratus, yet in the MP tree, the Japanese gengorobuna diverged first followed by C. carassius and then the other forms of C. auratus. The conflicting nodes were not well supported. The Japanese gengorobuna rooted at the base of the BI tree with strong support (Figure 2; BPP=100% BS=100%). Next, C. carassius from Central Europe and Russia branched off (Figure 2; BS=93–100%). The remaining samples of the C. auratus complex formed five highly supported groups.
Figure 2.
BI 50% majority rule consensus tree depicting the matrilineal relationships within Carassius based on 320–426 nucleotides positions of mtDNA CR sequences. Numbers represent nodal support inferred from BPPs, MLBS and MPBS, respectively. Branch support values are given for major lineages only. Colors indicate ploidy levels. Haplotype name is followed by the number of tetraploid, hexaploid and octaploid individuals. C. carpio formed the outgroup. A full color version of this figure is available at the Heredity journal online.
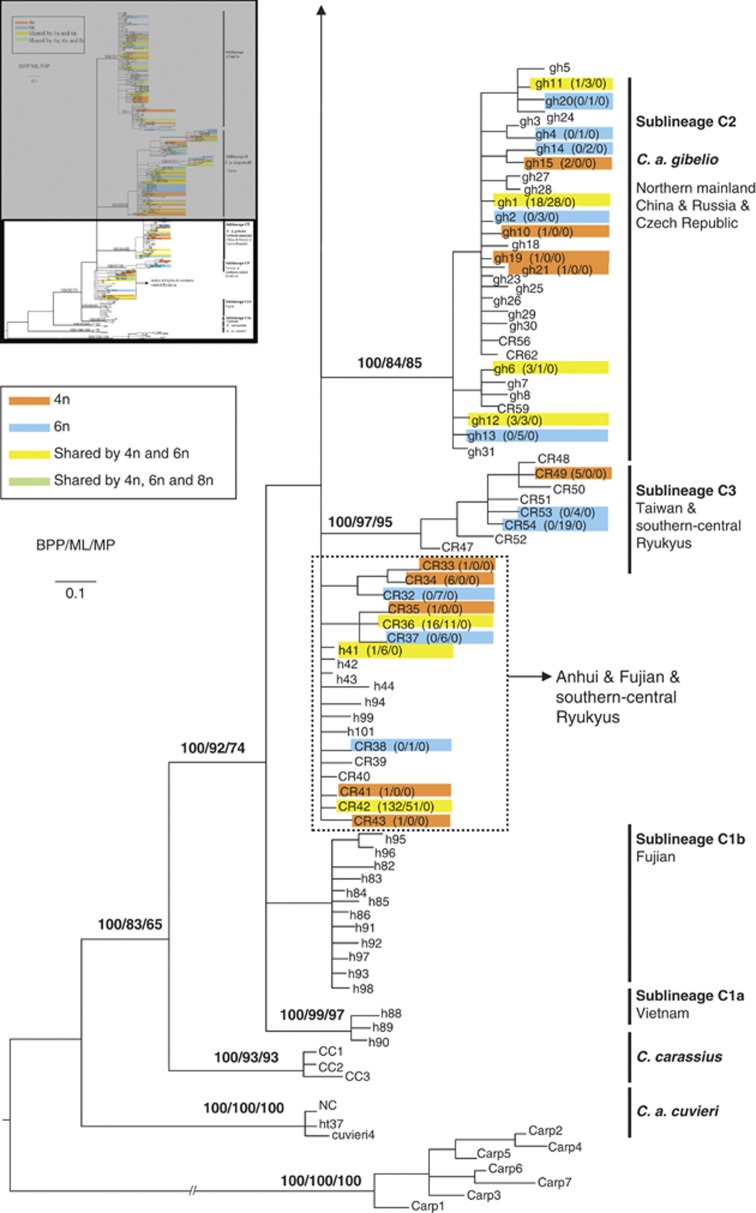
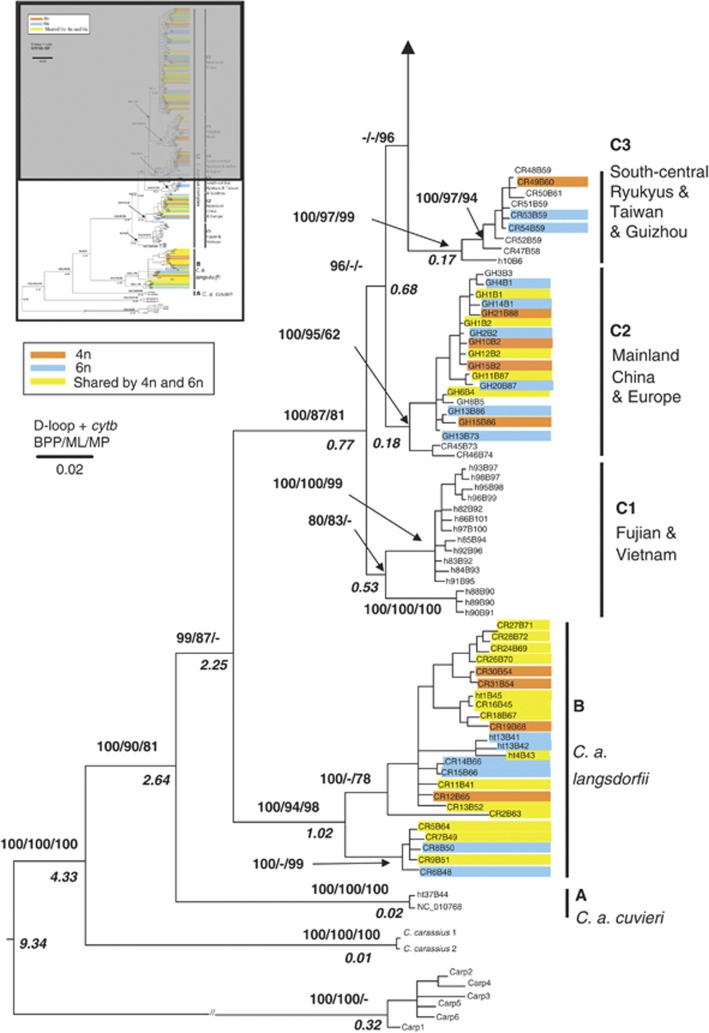
Analysis of the concatenated (CR+cytb) sequences resolved the main nodes of the CR tree and with stronger support (Figure 3). C. carassius split first and with high support (Figure 3; BPP=100% BS=100%). Sequentially, the next lineages included the Japanese gengorobuna and ginbuna (lineages A and B, Figure 3), both of which received high bootstrap support. Compared with the CR tree, six sublineages were revealed in lineage C (sublineages C1–C6, Figure 3), excluding the Japanese ginbuna.
Figure 3.
BI 50% majority rule consensus tree depicting the matrilineal relationships within Carassius based on 426 bp from the mitochondrial CRplus 1141 bp from the complete gene encoding cytochrome b. Numbers represent nodal support inferred from BPPs, MLBS and MPBS, respectively. Branch support values are given for major lineages only. Colors indicate ploidy levels. Haplotype name is followed by the number of tetraploid, hexaploid and octaploid individuals. C. carpio formed the outgroup. A full color version of this figure is available at the Heredity journal online.
Hexaploid lineages in the C. auratus complex
Hexaploids were scattered on the maternal genealogies based on both CR (Figure 2) and the concatenated data (CR+cytb) (Figure 3). In the Japanese goldfish, the hexaploids mapped onto two highly supported branches. Similarly, hexaploid C. auratus were scattered among the six sublineages. This distribution of hexaploids received high nodal support and various estimated dates of origin. Thus, hexaploids had multiple independent origins in both the C. auratus complex and the Japanese ginbuna; at least six polyploidization events were identified in well-supported matrilines.
The proportion of tetraploids/hexaploids was summarized on sublineages C2, C5, C6, Japanese ginbuna and goldfishes from Anhui, Fujian and south-central Ryukyus (Figure 2). The mapping depicted two clear features. First, tetraploids and hexaploids were intermixed among lineages. Hexaploidy occurred repeatedly. Second, tetraploids and hexaploids, and even octaploids, shared the same haplotypes. In sublineages C2, C5 and C6, 18 haplotypes (marked in yellow in Figure 2; Supplementary Tables S1 and S2) were shared among tetraploid and hexaploids and these different haplotypes were substantially diverged from one another. The most divergent haplotypes, h39 and h41, differed by 12 substitutions yet both contained tetraploid and hexaploid individuals. In total, 15.38% (18/117) of the haplotypes were shared by tetraploid and hexaploid samples in sublineages C2, C5 and C6 (Figure 2; Supplementary Tables S1 and S2). In Yunnan, China, 12 of 20 (60%) haplotypes of the hexaploids were shared with tetraploids, and 93.84% (137/146) of the hexaploid individuals shared one of the 12 haplotypes. In the Japanese samples, haplotype CR58 was shared between tetraploids and hexaploids in sublineages C5 and C6. Goldfish from Japan and the Kyusyu Islands exhibited a similar pattern; 22 of 72 haplotypes (30.56%) were shared by tetraploids and hexaploids, including one haplotype, ht2 occurred in tetraploid, hexaploid and octaploid individuals (Figure 2; Supplementary Table S2).
Genetic variation within/between populations of differing ploidy
Levels of variation in CR for tetraploid and hexaploid individuals were estimated after assigning populations to China, Kyusyu and Japan (Table 2). Among Chinese C. auratus, hexaploid haplotypes (40) were more common than tetraploid haplotypes (35) (Table 2). Similarly, hexaploid haplotypes (5) outnumbered tetraploid haplotypes (41) in Japan, including the Kyusyu Islands (Table 2; Supplementary Table S2). Similarly, hexaploid populations showed higher levels of polymorphism than did the tetraploids within China (H4n=0.9360±0.0109; H6n=0.9413±0.0048), Japan (H4n=0.9252±0.0097; H6n=0.9417±0.0074) and the Kyusyu Islands (H4n=0.6720±0.0325; H6n=0.8896±0.0134). The same patterns occurred when all data were combined.
Table 2. Genetic diversity of tetraploids and hexaploids in the C a. auratus complex based on mitochondrial control region, defined by lineage (1) and sympatric and non- sympatric tetraploid and hexaploid populations (2).
| N | Haplotype (HN) | Polymorphic sites (PN) | Gene diversity (H) | Nucleotide diversity (π)% | Tco (years) | Nef | |
|---|---|---|---|---|---|---|---|
| (1) | |||||||
| C5 and C6 4n | 91 | 27 | 18 | 0.9316±0.0122 | 0.8846±0.5233 | 3.77 × 105 | 9.43 × 104 |
| C5 and C6 6n | 322 | 35 | 25 | 0.9044±0.0102 | 0.8110±0.4834 | 4.24 × 105 | 1.06 × 105 |
| subB 4n | 162 | 30 | 25 | 0.9357±0.0065 | 1.9364±1.0265 | 6.60 × 105 | 1.65 × 105 |
| subB 6n | 215 | 38 | 25 | 0.9506±0.0054 | 2.0829±1.0944 | 6.60 × 105 | 1.65 × 105 |
| C2 4n | 30 | 8 | 7 | 0.6322±0.0959 | 0.2731±0.2024 | 2.36 × 105 | 5.89 × 104 |
| C2 6n | 47 | 9 | 8 | 0.6318±0.0766 | 0.2819±0.2068 | 2.83 × 105 | 7.08 × 104 |
| (2) | |||||||
| C. a. auratus 4n (CH) | 114 | 35 | 37 | 0.9360±0.0109 | 1.9912±1.0286 | 0.99 × 106 | 2.48 × 105 |
| C. a. auratus 6n (CH) | 250 | 40 | 39 | 0.9413±0.0048 | 1.7456±0.9069 | 0.94 × 106 | 2.36 × 105 |
| C. a. auratus 4n (KY) | 238 | 18 | 36 | 0.6720±0.0325 | 2.0887±1.0967 | 1.08 × 106 | 2.70 × 105 |
| C. a. auratus 6n (KY) | 193 | 18 | 34 | 0.8896±0.0134 | 3.2436±1.6543 | 1.13 × 106 | 2.83 × 105 |
| C. a. auratus 4n (JAP) | 114 | 23 | 24 | 0.9252±0.0097 | 2.1181±1.1159 | 1.18 × 106 | 2.95 × 105 |
| C. a. auratus 6n (JAP) | 185 | 38 | 39 | 0.9417±0.0074 | 2.2277±1.1642 | 1.18 × 106 | 2.95 × 105 |
| C. a. auratus 4n (All) | 466 | 76 | 53 | 0.9064±0.0110 | 3.1954±1.6195 | 1.13 × 106 | 2.83 × 105 |
| C. a. auratus 6n (All) | 628 | 93 | 57 | 0.9750±0.0017 | 3.7081±1.8612 | 1.13 × 106 | 2.83 × 105 |
Abbreviations: CH, China; JAP, Japan; KY, Kyusyu; Nef, estimated effective population size; Tco, estimated time to coalescence.
Estimated coalescence times for Chinese hexaploids were slightly younger than their sympatric tetraploids and the inverse pattern occurred for populations from the Kyusyu Islands (Table 2). Estimated coalescence times for tetraploids and hexaploids from Japan were the same as were those for all three groups combined. Thus, female effective population size (Table 2) was concordant with the estimated coalescence times.
We evaluated the possibility of population expansion by assuming that the 39 (17 in China plus 22 in Japan) incidences of haplotypes being shared by tetraploids and hexaploids represented independent polyploidization events. Subsequently, we estimated the frequency of polyploidization by coalescence times. The suggested coalescence time for the 17 Chinese populations with hexaploid individuals was Tco=0.94 × 106 years (Table 2), that is, a rate of at least 0.55 polyploidization events per 105 years. For the 14 populations on the main islands of Japan, the coalescence time was Tco=1.18 × 106 years (Table 2), or a minimum rate of 0.84 polyploidization events per 105 years. Finally, the eight occurrences of hexaploids on the Ryukyu Islands had Tco=1.13 × 106 year (Table 2), or 1.41 events per 105 years. Thus, population expansion was unlikely to explain the pattern. Further, a recent population expansion was not identified by using either the Tajima's D or Fu's neutrality test among the different ploidy levels of either the Chinese or the Japanese populations. Populations in the species complex appeared to be established and stable.
When individuals were assigned to one of three populations (China, Kyusyu and Japan), Fst values indicated that most sympatric tetraploid and hexaploid populations were more similar to each other than to individuals in non-sympatric populations (Table 3). The Fst values of sympatric tetraploid/hexaploid populations in China, Ryukyu Islands and Japan were less than 0.140. Except for Chinese tetraploids and hexaploids from the Kyusyu Islands, for which Fst=0.212, all other values of Fst of non-sympatric tetraploid/hexaploid exceeded 0.253, and most were greater than 0.318. Non-sympatric tetraploid/hexaploid populations showed higher pairwise divergence than those in sympatry.
Table 3. Pairwise FST between (1) different lineages and (2) sympatric and non-sympatric tetraploid and hexaploid populations.
| (1) | C5 and C6 4n | C5 and C6 6n | sub 4n | sub 6n | C2 4n | C2 6n |
|---|---|---|---|---|---|---|
| C5 and C6 4n | \ | |||||
| C5 and C6 6n | 0.05951** | \ | ||||
| subB 4n | 0.72781** | 0.77716** | \ | |||
| subB 6n | 0.70736** | 0.75823** | 0.03692** | \ | ||
| C2 4n | 0.83776** | 0.82471** | 0.66209** | 0.66848** | \ | |
| C2 6n |
0.84308** |
0.82348** |
0.67655** |
0.67913** |
−0.00281 |
\ |
| (2) |
China 4n |
China 6n |
Kyusyu 4n |
Kyusyu 6n |
Japan 4n |
Japan 6n |
| China 4n | \ | |||||
| China 6n | 0.01413 | \ | ||||
| Kyusyu 4n | 0.39061** | 0.41156** | \ | |||
| Kyusyu 6n | 0.21249** | 0.25333** | 0.14008** | \ | ||
| Japan 4n | 0.60135** | 0.62542** | 0.40157** | 0.31834** | \ | |
| Japan 6n | 0.59791** | 0.62181** | 0.40037** | 0.32612** | 0.03397** | \ |
Note: ** denotes P=0.000.
Discussion
Genealogical relationships and recurrent polyploidy
C. carassius (Linnaeus 1758) and C. auratus (Linnaeus 1758) are clearly distinguished only in the total evidence mtDNA tree (Figure 3); they are resolved as polyphyletic (polygenealogical sensu Murphy and Méndez de la Cruz, 2010) in the CR trees because the Japanese gengorobuna splits earlier than C. carassius (Figure 2). Analyses of nuclear GH and StAR resolve the Japanese gengorobuna as a unique lineage following after C. carassius (Figures 1a and b). In contrast, analyses of MIF suggest that the Japanese gengorobuna diverged before C. carassius (Figure 1c). These inconsistent results suggest that these two lineages might have diverged near simultaneously. Irrespective of the particular topologies of the trees, the Japanese gengorobuna has been treated as a species, C. cuvieri. C. carassius has 100 chromosomes (Vinogradov, 1998) as does the Japanese gengorobuna, along with its diagnostic C-banding pattern (Ueda and Ojima, 1978). Considering that both species are tetraploids and sequentially root at the base of the tree (Figures 2 and 3), their common ancestor appears to be a tetraploid (Vinogradov, 1998).
The taxonomic classification of the C. auratus complex is complicated (Figures 2 and 3). Other than the Japanese gengorobuna, two main lineages exist: lineage B (Japanese ginbuna) and a lineage C (mainly from China and the Kyusyu Islands). Lineage C contains six sublineages (Gao et al., 2012). Recurrent polyploidizations occur within both lineages and tetraploid and hexaploid individuals frequently share identical haplotypes. Every case implies the independent origin of a hexaploid. Several hexaploid lineages of C. auratus occur in China, for example, Lake Dianchi (Luo et al., 1999), and in these cases they are most closely related to local tetraploids (Zan et al., 1986 and therein; Zhang et al., 1998 and therein). Curiously, hexaploids from Guizhou cluster with both local tetraploid C. auratus and the gibel carp. This population contains mixed, divergent lineages. Regardless, at this locality the hexaploids are not monogenealogical (sensu Murphy and Méndez de la Cruz, 2010); the mtDNA data show that they do not share a single common maternal ancestor.
Hexaploid C. a. auratus show greater variation than tetraploids. Hexaploid individuals appear to be more common than tetraploids in nature, as suggested by previous collections (Xiao et al., 2011). Kleptogenesis—the temperature-dependent incorporation of sperm in otherwise gynogenetic polyploids (Bogart et al., 2007)—may be a contributing factor. Alternatively, unidirectional gene flow and recent expansion of the hexaploid populations of C. auratus might also promote higher levels of polymorphism in the hexaploid goldfish. These possibilities require further testing.
In Japan, both tetraploid and hexaploid forms exist in the ginbuna (Murakami et al., 2001; Takada et al., 2010 and therein). Because gene flow can only occur between tetraploids, the mixture of tetraploids and hexaploids indicates additional recurrences of polyploidization (Figures 2 and 3).
The tempo, mode and origin of tetraploid and hexaploid Carassius
The ancestor of Carassius is most likely a tetraploid carp (Buth et al., 1991 and therein; our analyses). All Carassius have at least 100 chromosomes, twice as many than the standard cyprinid karyotype, and their genome sizes are also about twice as large as diploid cyprinids (Ohno and Atkin, 1966; Ohno, 1970; Yu et al., 1989; Risinger and Larhammar, 1993). The initial tetraploidization might have been an alloploidization event that occurred about 14.2–14.5 Ma, as estimated from nuclear gene studies (Risinger and Larhammar, 1993; Yang and Gui, 2004; Luo et al., 2006). Our analyses from both mitochondrial and nuclear data place tetraploid C. carassius and Japanese C. cuvieri at the base of the tree for Carassius. Analyses of GH and StAR also support a model of allopolyploidization of the genus because at least one allele of each gene clusters within the outgroup carp, and two clusters are indicated (Figures 1a and b). Hence, our results confirm the assumption of allotetraploidization of the genus Carassius. Analyses of the nuclear gene data cannot reject the hypothesis of recurrent polyploidization via autoploidization because we did not detect either C. carassius or C. cuvieri within the C. auratus complex. For example, tetraploid and hexaploid individuals share localized alleles within clusters of both GH and StAR (Figures 1a and b) only in the C. auratus complex. Thus, ancient alloploidization and recent autopolyploidization are indicated for Carassius.
The tempo indicates that recurrent autopolyploidization occurred after the origin of the C. auratus complex. Gao et al. (2012) estimated divergence time of C. carassius and the C. auratus complex to be 3.6–4.0 Ma. Hexaploids in different geographic regions are younger than the two species themselves (Table 2) and no recurrent event occurred in their common ancestor. Analyses suggest a rate of about 0.55–1.41 mutation events per 105 years.
The most intriguing discovery lies in the ubiquitous, recurrent hexaploidization. The mapping of polyploids on the matrilineal genealogy precludes the possibility of one polyploidization event (Supplementary Figure S1). The nuDNA allele tree, matrilineal genealogy, coalescence times and scale of divergence in sympatric/non-sympatric tetraploid/hexaploid individuals indicate that the hexaploidization has several independent origins (Figures 2 and 3; Tables 2 and 3). Shared mtDNA haplotypes reject the hypothesis of a single origin for the hexaploids and in doing so they support the alternative hypothesis of multiple formations of polyploids. Convergent mutations can explain a single origin but two lines of evidence reject this possibility. First, the absence of saturated substitutions precludes convergence as an explanation. Second, because these unisexual fishes are gynogenetic, divergent 4n and 6n individuals should occur within both sister lineages if the common ancestor was a hexaploid; this does not occur in any of the incidences of sympatry. Shared haplotypes occur in divergent lineages that differ by up to 13 polymorphic sites. Independent polyploidization events best explain the numerous co-occurrences of tetraploids and hexaploids.
Laboratory experiments identify some possible mechanisms for hexaploidization. Hydrostatic pressure, cold and heat shock following fertilization results in the retention of the second polar cell. When this occurs, three nuclei combine yielding an additional set of chromosomes and a hexaploid individual (Gui et al., 1995; Pandian and Koteeswaran, 1998). Thus, possible environmental perturbations may promote the formation of hexaploids. Variation in hydrostatic pressure or temperature is a potential factor and environmental data and future analyses may confirm this possibility. Regardless, after formation, hexaploids reproduce gynogenetically.
The frequent dispersal of historically isolated populations might facilitate mixture of the tetraploids and hexaploids. This scenario does not appear to be true for Carassius. Geographic structure exists in both Japan and mainland China (Takada et al., 2010; Gao et al., 2012). Migration does not seem to have blurred the geographic structure and, again, the evidence suggests site-specific-independent polyploidization events. This mechanism may explain elevated levels of variation in hexaploids; hexaploids would have two paternal contributions.
Gynogenesis is common in hexaploid Carassius, and this might significantly influence population structure. It occurs sporadically among invertebrates (Smith, 1978), and in vertebrates it is especially widespread in polyploid teleosts (Gui, 1997; Le Comber and Smith, 2004). Smith (1978) argued that short-term selection can favor unisexuality. Gynogenetic reproduction forms a barrier to gene flow from hexaploid to tetraploid individuals. Hexaploid females only require sperm from tetraploid males to trigger egg development; egg and sperm nuclei do not fuse (Gui, 1997). Where tetraploids, hexaploids and, rarely, octaploids coexist in natural waters, hexaploid male C. auratus occur but they do not contribute genetically to tetraploid reproduction. A combination of hexaploid and tetraploid nuclei would form an irregular number of chromosomes (about 125) and no such individuals are known (Chen et al., 1996; Xiao et al., 2011). Thus, hexaploids depend on tetraploids even though the former do not fertilize eggs of the later and their syntopic occurrence is widespread.
Polyploid Carassius: a model system for investigations in vertebrates
With its large genome, Carassius possesses many duplicated genes. Ancestral carp appear to have about 50 chromosomes (Yu et al., 1989) and a genome size of about 2.0 pg per cell. In contrast, tetraploid Carassius have from 3.52 pg to 3.84 pg per cell, almost twice that of diploid barbs (http://www.genomesize.com/; Luo et al., 2006). Hexaploid Carassius have about 5.25 pg per cell (http://www.genomesize.com/). Our results indicate that tetraploids have 2–4 copies for each nuclear gene, and hexaploids normally have 3–6 copies (Supplementary Table S4). Thus, Carassius is likely to be a polyploid genus, and not a diploid/triploid system as sometimes claimed (for example, Takada et al., 2010).
The existence of both allo- and autopolyploidization events facilitates investigations into post-polyploidization events at both genetic and epigenetic levels. The origin of the allopolyploidization in Carassius is known (Yang and Gui, 2004; Luo et al., 2006). The importance of allopolyploidization and how its evolutionary effects differ from those of autopolyploidization (Parisod and Besnard, 2007) is a topic of considerable interest (Soltis and Soltis, 1999; Vandepoele et al., 2004; Van de Peer et al., 2009). This level of interest differs from that for autopolyploidization (Parisod and Besnard, 2007). Autopolyploidization might be advantageous under conditions of environmental instability because of genic redundancy and polysomic inheritance (Parisod and Besnard, 2007). Thus, this genus is an ideal model system for investigating genetic and epigenetic changes soon after polyploidizations, either allo- and autopolyploidy.
In summary, intraspecific polyploidization occurs multiple times in angiosperms, yet this phenomenon is less common and not yet fully understood in vertebrates. We document recurrent polyploidization events in the genus Carassius based on matrilineal history. The C. auratus complex has recurrent formations of hexaploids; tetraploids and hexaploids share identical haplotypes in three main lineages and within localities. The origin of the gibel carp likely owes to a single hexaploidization event in China within the C. auratus complex. The other two main lineages, the Chinese C. auratus complex and the ginbuna of Japan, have at least 22 independent hexaploidization events, plus one octaploidization. These events make the species of the genus Carassius an ideal model system for the study of recurrent polyploidization and the evolution of genomic complexity. How polyploidy contributes to adaptation and evolution, as well as speciation in vertebrates, is a very promising area of future research.
Data archiving
Sequences were deposited in GenBank: Accession numbers: JX406582-JX406681 and KC462749-KC462782.
Acknowledgments
This work was supported by grants from National Science Foundation of China (31360514 and 30870291 to J Luo; 30600065 to Y Gao; and 31061160189 to Y-p Zhang; 30930071 to SJ Liu), Bureau of Science and Technology of Yunnan Province and the State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, CAS. Manuscript preparation was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (3148) and by a Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences to RW Murphy. We thank CL Zhu, L Zhang, SG Fang, YF Ge, JH Liu and X Chen et al. for sample collection and technical help. We also thank the two anonymous reviewers for constructive suggestions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Thorgaard GH.1984Tetraploidy and the evolution of salmonid fishesIn: Turner BJ (ed) Evolutionary Genetics of Fishes Plenum Press: New York, NY, USA; 1–53. [Google Scholar]
- Bogart JP, Bi K, Fu JZ, Noble DWA, Niedzwieki J. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome. 2007;50:119–136. doi: 10.1139/g06-152. [DOI] [PubMed] [Google Scholar]
- Brykov VA, Poliakova NE, Skurikhina LA, Dolganov SM, Eliseikina MG, Mlu Kovalev.2002Mitochondrial DNA variation in goldfish Carassius auratus gibelio from Far Eastern water reservoirs Genetika 381387–1392.(in Russian). [PubMed] [Google Scholar]
- Buth DG. Duplicate isozyme loci in fishes: origins, distribution, phyletic consequences, and locus nomenclature. Isozymes: Curr Top Biol Med Res. 1983;10:381–400. [PubMed] [Google Scholar]
- Buth DG, Dowling TE, Gold JR.1991Molecular and cytological investigationsIn Winfield IJ, Nelson JS (eds)Cyprinid Fishes: Systematics, Biology, and Exploitation Chapman & Hall: London, UK; 83–126. [Google Scholar]
- Chen MR, Yang XQ, Yu XM, Chen HX. Karyotype studies on the bisexual natural gynogenetic crucian carp (Carassius auratus) of Pengze. Acta Hydrob Sinica. 1996;20:25–31. [Google Scholar]
- Dawley RM.1989An introduction to unisexual vertebratesIn: Dawley RM, Bogart JP (eds) Evolution and Ecology of Unisexual Vertebrates New York State Museum: Albany, NY, USA; 1–18. [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang SY, Luo J, Murphy RW, Du R, Wu SF, et al. Quaternary palaeoenvnvironmental oscillations drove the evolution of East Asian Carassius auratus complex (Carassius, Cypriniformes, Cyprinidae) J Biogeogr. 2012;39:2264–2278. [Google Scholar]
- Goldman N. Statistical tests of models of DNA substitution. J Mol Evol. 1993;36:182–198. doi: 10.1007/BF00166252. [DOI] [PubMed] [Google Scholar]
- Golovinskaya KA, Romashov DD, Cherfas NB.1965Unisexual and bisexual forms of silver crusian carp (Carassius auratus gibelio Bl.) Vopr Ikhtiol 5614–629.(in Russian). [Google Scholar]
- Gui JF.1997Retrospectus and prospects of studies on the mechanism of natural gynogenesis in silver crucian carp (Carassius auratus gibelio) Bull Nat Sci Found China 111–16.(in Chinese). [Google Scholar]
- Gui JF. Genetic Basis and Artificial Control of Sexuality and Reproduction in Fish. Science Press: Beijing, China; 2007. [Google Scholar]
- Gui JF, Xiao W, Liang S, Jiang Y. Preliminary study on the cytological mechanism of triploidy and tetraploidy induced by hydrostatic pressure shock in transparent colored crucian carp. Acta Hydrob Sinica. 1995;19:49–55. [Google Scholar]
- Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Curr Biol. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- Holloway AK, Cannatella DC, Gerhardt HC, Hillis DM. Polyploids with different origins and ancestors form a single sexual polyploid species. Am Nat. 2006;167:E88–101. doi: 10.1086/501079. [DOI] [PubMed] [Google Scholar]
- Hu T, Pattyn P, Bakker E, Cao J, Cheng J, Clark R, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Crandall KA. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu Rev Ecol Syst. 1997;28:437–466. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Iguchi K, Yamamoto G, Matsubara N, Nishida M. Morphological and genetic analysis of fish of a Carassius complex (Cyprinidae) in Lake Kasumigaura with reference to the taxonomic status of two all-female triploid morphs. Biol J Linn Soc. 2003;79:351–357. [Google Scholar]
- Kobayashi H.1978Classification of the genus Carassius and cellular genetics of gynogenesis of ginbuna (C. auratus langsdorfi) Genetics 3228–38.(in Japanese). [Google Scholar]
- Le Comber SC, Smith C. Polyploidy in fishes: patterns and processes. Biol J Linn Soc. 2004;82:431–442. [Google Scholar]
- Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fisheries. 2003;13:237–246. [Google Scholar]
- Li FB, Gui JF. Clonal diversity and genealogical relationships of gibel carp in four hatcheries. Anim Genet. 2008;39:28–33. doi: 10.1111/j.1365-2052.2007.01671.x. [DOI] [PubMed] [Google Scholar]
- Lokki J, Saura A.1980Polyploidy in insect evolutionIn: Lewis W (ed) Polyploidy: Biological Relevance Plenum Press: New York, NY, USA; 277–312. [Google Scholar]
- Lorenzoni M, Corboli M, Ghetti L, Pedicillo G, Carosi A.2007Growth and reproduction of the goldfish Carassius auratus: a case study from ItalyIn: Gheradi F (ed) Biological Invaders in Inland Waters: Profiles, Distribution, and Threats Springer-Verlag: New York, NY, USA; 259–273. [Google Scholar]
- Lou YD, Zhang YP, Weng ZH, Zhao L.1989Preliminary studies on cytogenetics and isoenzymes of crucian carp (Carassius auratus) in Qihe River of Henan province J Fish China 13254–258.(in Chinese). [Google Scholar]
- Lu CY, Yang YH, Tong GX, Hao J, Sun XW, Liang LQ, et al. Comparative studies of genetic diversity between C. auratus gibelio (Bloch) and C. auratus auratus (L.) in the same reservoir. Chinese J Fisheries. 2006;19:42–50. [Google Scholar]
- Luo J, Zhang YP, Zhu CL, Xiao WH, Huang SY. Genetic diversity in crucian carp (Carassius auratus) Biochem Genet. 1999;37:267–279. doi: 10.1023/a:1018751008848. [DOI] [PubMed] [Google Scholar]
- Luo J, Lang M, Salzburger W, Siegel N, Stolting KN, Meyer A. A BAC library for the goldfish Carassius auratus auratus (Cyprinidae, Cypriniformes) J Exp Zool Part B Mol Dev Evol. 2006;306:567–574. doi: 10.1002/jez.b.21112. [DOI] [PubMed] [Google Scholar]
- Luo YL, Yue PQ.2000CyprinidaeIn: Yue PQ (ed) Fauna Sinica. Osteichthyes Cypriniformes III Science Press: Beijing, China; 399–433.(in Chinese. [Google Scholar]
- Mable BK. Why polyploidy is rarer in animals than in plants: myths and mechanisms. Biol J Linn Soc. 2004;82:453–466. [Google Scholar]
- Mable BK, Alexandrou MA, Taylor MI. Genome duplication in amphibians and fish: an extended synthesis. J Zool. 2011;284:151–182. [Google Scholar]
- Murakami M, Matsuba C, Fujitani H. The maternal origins of the triploid ginbuna (Carassius auratus langsdorfi): phylogenetic relationships within the C. auratus taxa by partial mitochondrial D-loop sequencing. Gene Genet Syst. 2001;76:25–32. doi: 10.1266/ggs.76.25. [DOI] [PubMed] [Google Scholar]
- Murphy RW, Méndez de la Cruz FR.2010The herpetofauna of Baja California and its associated islands: a conservation assessment and prioritiesIn: Wilson LD, Townsend JH, Johnson JD (eds) Conservation of Mesoamerican Amphibians and Reptiles Eagle Mountain Publishing, LC: Eagle Mountain, UT, USA; 238–273. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA; 1987. [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer-Verlag: New York, NY, USA; 1970. [Google Scholar]
- Ohno S, Atkin NB. Comparative DNA values and chromosome complements of eight species of fishes. Chromosoma. 1966;18:455–466. doi: 10.1007/BF00332549. [DOI] [PubMed] [Google Scholar]
- Ojma Y, Yamano T. The assignment of the nuclear organizer in the chromosome of the fauna (Carassius, Cyprindae, Pisces) Proc Jpn Acad. 1980;56B:551–556. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pandian TJ, Koteeswaran R. Ploidy induction and sex control in fish. Hydrobiologia. 1998;384:167–243. [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Mol Ecol. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation for the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger C, Larhammar D. Multiple loci for synapse protein SNAP-25 in the tetraploid goldfish. Proc Natl Acad Sci USA. 1993;90:10598–10602. doi: 10.1073/pnas.90.22.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo M. Molecular phylogeny of the hominoids: inferences from multiple independent DNA sequence data sets. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- Segraves KA, Thompson JN, Soltis PS, Soltis DE. Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia. Mol Ecol. 1999;8:253–262. [Google Scholar]
- Shan SX, Jiang YG.1988Karyotype studies of crucian carp Carassius auratus gibelio Acta Hydrobiol Sin 12381–384.(in Chinese). [Google Scholar]
- Shen JB, Wang GR, Fan ZT.1983The ploidy of crucian carp and its geographical distribution in Heilongjiang main river J Fish China 779–94.(in Chinese). [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. The Evolution of Sex. Cambridge University Press: Cambridge, Great Britain; 1978. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and other Methods). Version 4.0b10* Sinauer Associates: Sunderland, MA, USA; 2002. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada M, Tachihara K, Kon T, Yamamoto G, Iguchi K, Miya M, et al. Biogeography and evolution of the Carassius auratus-complex in East Asia. BMC Evol Biol. 2010;10:7. doi: 10.1186/1471-2148-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsigenopoulos CS, Rab P, Naran D, Berrebi P. Multiple origins of polyploidy in the phylogeny of southern African barbs (Cyprinidae) as inferred from mtDNA markers. Heredity. 2002;88:466–473. doi: 10.1038/sj.hdy.6800080. [DOI] [PubMed] [Google Scholar]
- Ueda T, Ojima Y. Differential chromosomal characteristics in the funa subspecies (Carassius) Proc Jpn Acad. 1978;54B:283–288. [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry. 1998;31:100–109. doi: 10.1002/(sici)1097-0320(19980201)31:2<100::aid-cyto5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Wang RF, Shi LM, He WS.1988A comparative study of the Ag-NORs of Carassius auratus from different geographic districts Zool Res 9165–169.(in Chinese). [Google Scholar]
- Wheeler A. Status of the crucian carp, Carassius carassius (L.), in the UK. Fisheries Manage Ecol. 2000;7:315–322. [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish. Bioessays. 1998;20:511–515. [Google Scholar]
- Woods TD, Buth DG. High level of gene silencing in the tetraploid goldfish. Biochem Syst Ecol. 1984;12:415–421. [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xiao J, Zou TM, Chen YB, Chen L, Liu SJ, Tao M, et al. Coexistence of diploid, triploid and tetraploid crucian carp (Carassius auratus) in natural water. BMC Genet. 2011;12:20. doi: 10.1186/1471-2156-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Gui JF. Positive selection on multiple antique allelic lineages of transferrin in the polyploid Carassius auratus. Mol Biol Evol. 2004;21:1264–1277. doi: 10.1093/molbev/msh121. [DOI] [PubMed] [Google Scholar]
- Yu HX, Xu H, Guan HW.1992Preliminary study on the karyotypes of the natural gynogenetic crucian carp Carassius auratus (A form) of Puan, Guizhou province Acta Hydrobiol Sin 1687–89.(in Chinese with English abstract). [Google Scholar]
- Yu HX, Zhang HM, Lin LY.1987A preliminary study on the biology and culture experiment of the gynogenetic crucian carp Carassius auratus of Guangdong Acta Hydrobiol Sin 11278–288.(in Chinese with English abstract). [Google Scholar]
- Yu XJ, Zhou D, Li YC, Li K, Zhou M.1989Chromosomes of Chinese Fresh Water Fishes Science Press: Beijing, China; 135–143.(in Chinese with English Abstract). [Google Scholar]
- Zan RG.1982Studies on sex chromosomes and C-banding karyotypes of two forms of Carassius auratus in Kunming Lake Acta Genet Sinica 932–39.(in Chinese). [Google Scholar]
- Zan RG, Song Z, Liu WG.1986Studies on karyotypes and nuclear DNA contents of some cyprinoid fishes, with notes on fish polyploids in ChinaIn: Uyenno T, et al. (eds)Indo-Pacific Fish Biology: Proceedings of the Second International Conference on Indo-Pacific Fish Ichthyological Society of Japan: Tokyo, Japan; 877–885. [Google Scholar]
- Zhang H, Dong XH, Ye Y, Wu QJ. Comparative studies of the mtDNA from three strains of hexaploid and C. auratus auratus. Acta Genet Sinica. 1998;25:330–336. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.