Abstract
Co-evolution of fungal pathogens with their host species during the domestication of modern crop varieties has likely affected the current genetic divergence of pathogen populations. The objective of this study was to determine if the evolutionary history of the obligate rust pathogen on wheat, Puccinia triticina, is correlated with adaptation to hosts with different ploidy levels. Sequence data from 15 loci with different levels of polymorphism were generated. Phylogenetic analyses (parsimony, Bayesian, maximum likelihood) showed the clear initial divergence of P. triticina isolates collected from Aegilops speltoides (the likely B genome donor of modern wheat) in Israel from the other isolates that were collected from tetraploid (AB genomes) durum wheat and hexaploid (ABD genomes) common wheat. Coalescence-based genealogy samplers also indicated that P. triticina on A. speltoides, diverged initially, followed by P. triticina isolates from durum wheat in Ethiopia and then by isolates from common wheat. Isolates of P. triticina found worldwide on cultivated durum wheat were the most recently coalesced and formed a clade nested within the isolates from common wheat. By a relative time scale, the divergence of P. triticinia as delimited by host specificity appears very recent. Significant reciprocal gene flow between isolates from common wheat and isolates from durum wheat that are found worldwide was detected, in addition to gene flow from isolates on common wheat to isolates on durum wheat in Ethiopia.
Keywords: Uredinales, phylogenetics, fungi, coalescence, host adaptation, obligate parasite
Introduction
The beginning of agriculture 12 000 years ago in the Fertile Crescent made possible the rise of human culture and civilization, and changed the genetic makeup of cereal populations through extensive domestication (Salamini et al., 2002). Hybridization of different genomes leading to ploidy changes is a landmark of wheat evolution. Archeological, genetic and molecular evidence revealed that modern cultivated hexaploid common wheat, Triticum aestivum L. with A, B, and D genomes is derived from the hybridization of tetraploid emmer wheat, T. turgidum ssp. dicoccum L. that has A and B genomes, and the wild diploid D genome species Triticum tauschii (Feldman, 2001) ca. 8000 years ago. In emmer wheat, the A genome donor is derived from T. urartu and the B genome donor is likely from Aegilops speltoides. Coupled with the intriguing history of wheat evolution is the corresponding adaptation of fungal pathogens from wild grass hosts to the founding genotypes of present day modern wheat cultivars. We are interested in determining if the historical divergence of an obligate parasite with different forms on diploid, tetraploid and hexaploid wheat hosts is congruent with the history of wheat evolution.
Documentation of the destructive nature of the rust diseases on wheat can be traced back to classical Greek and Roman eras and also to references in the Old Testament (Chester, 1946). Distinct species of the causal pathogens were recognized only in the 19th century when the individual species were identified (Bolton et al., 2008). Among the three rusts of wheat, leaf rust caused by Puccinia triticina Erikss. is the most prevalent (Chester, 1946), causing regular economic losses due to reduced number and size of kernels and decreased quality of flour. With a macrocyclic-heteroecious life cycle, P. triticina produces pycniospores and aeciospores on Thalictrum speciosissimum L., known as common meadow rue, and urediniospores, teliospores and basidiospores on wheat and wheat-related hosts (Mains and Jackson, 1921). The uredinial/telial hosts include hexaploid common wheat worldwide, tetraploid durum wheat (T. turgidum ssp. durum L., A and B genomes) in the Middle East, Ethiopia, Europe, Mexico and South America (Ordoñez and Kolmer, 2007); tetraploid emmer wheat in the Middle East (Manisterski et al., 2000) and Ethiopia (Andenow et al., 1997) and diploid A. speltoides in Israel (Yehuda et al., 2004). Puccinia triticina also occurs on A. cylindrica in the USA, but no occurrence has been reported in natural stands of other wild grasses such as A. sharonensis, A. tauschii and T. timopheevi, or the ancient diploid wheat T. monococcum (Bolton et al., 2008) although infections on these hosts can be obtained in artificial greenhouse inoculations (Anikster et al., 2005b). Isolates of P. triticina collected from modern hexaploid wheat are also virulent to wild emmer wheat, T. turgidum ssp. dicoccoides (Dinoor et al., 1991; Manisterski et al., 2000; Anikster et al., 2005a) in Israel but are generally avirulent to cultivated durum wheat that was derived from emmer wheat. Isolates virulent to A. speltoides are avirulent to hexaploid and tetraploid wheats. Sexual reproduction of P. triticina on T. speciosissimum and other Thalictrum spp. was sporadically reported in Portugal (D'Oliveira and Samborski, 1966), Italy (Casulii and Siniscalco, 1987), Spain (Young and D'Oliveira, 1982) and the United States (Levine and Hildreth, 1957). However, molecular marker data in population genetic studies indicate that in worldwide populations the clonal reproduction of urediniospores is highly predominant, with little if any evidence of sexual recombination (Goyeau et al., 2007; Kolmer and Ordoñez, 2007; Ordoñez and Kolmer, 2009; Ordoñez et al., 2010; Kolmer et al., 2011).
Molecular markers have been used to assess genetic variation within and between populations of P. triticina. Genotypes of P. triticina generated with random amplified polymorphism of DNA markers demonstrated a general correlation with the virulence phenotypes (Kolmer and Liu, 2000). Genotypes of P. triticina based on amplified fragment length polymorphism identified the possible introduction of new races in North America (Kolmer, 2001). Simple sequence repeat (SSR) markers (Duan et al., 2003; Szabo and Kolmer, 2007) have been used to characterize P. triticina populations from different continents (Kolmer and Ordoñez, 2007; Ordoñez and Kolmer, 2007; Ordoñez and Kolmer 2009; Ordoñez et al., 2010; Kolmer et al., 2011). SSR genotypes of P. triticina collected from A. speltoides were highly divergent from P. triticina genotypes from wheat. Comparison of P. triticina isolates indicated that isolates from tetraploid durum wheat are less variable than isolates from hexaploid common wheat (Ordoñez and Kolmer, 2007; Goyeau et al., 2011) for both virulence and SSR genotypes. The goal of the present study was to reconstruct the evolutionary history of P. triticina divergence as related to its adaptation to hosts with different ploidy levels. Our initial hypotheses are that P. triticina on A. speltoides represents the earliest form of P. triticina, and that P. triticina isolates with virulence to tetraploid durum wheat are derived from genotypes with virulence to hexaploid common wheat.
Materials and methods
Puccinia triticina isolates
Seventy P. triticina isolates were selected based on a neighbor-joining tree constructed from SSR genotypes from a worldwide collection (663 total isolates) in order to include isolates representing all major genotype groups and geographic regions. Among those, 48 isolates were from common wheat that originated from Central Asia, Europe, the Middle East, North America, South America, South Africa and New Zealand and 20 isolates from durum wheat that originated from the Middle East, Ethiopia, Europe, North America and South America (Table 1). Two samples collected from A. speltoides were from Israel. Genomic DNA of the isolates used in previous studies was used for obtaining single nucleotide polymorphism (SNP) data. In addition, we used DNA from two isolates of P. persistens, (Markova and Urban, 1998) a sister species to P. triticina, that were collected in the Czech Republic and Sweden from Elymus repens, as the outgroup in phylogenetic and coalescence analyses. Isolate numbers, countries of origin and SSR genotype group designations are listed in Table 1.
Table 1. Origins and hosts of Puccinia triticina isolates used for phylogenetic and coalescence study.
| Species—isolatea | Haplotypes | Country | Population—SSR group | Reference |
|---|---|---|---|---|
| Puccinia triticina | ||||
| On Aegilops speltoidesa | ||||
| ISR850 | H8, H9 | Israel | — | Yehuda et al., 2004 |
| ISR1400 | H7, H10 | Israel | — | Yehuda et al., 2004 |
| On Triticum turgidum | ||||
| ARG9_3 | H12, H12 | Argentina | — | Ordoñez and Kolmer, 2007 |
| CA1_2 | H12, H14 | USA | — | Ordoñez and Kolmer, 2007 |
| CH14_21 | H5, H12 | Chile | — | Ordoñez and Kolmer, 2007 |
| M11_1 | H12, H16 | Mexico | — | Ordoñez and Kolmer, 2007 |
| M14_3 | H12, H15 | Mexico | — | Ordoñez and Kolmer, 2007 |
| ES1_1 | H11, H18 | Spain | EU_3 | Kolmer et al., 2012 |
| ES27 | H11, H14 | Spain | — | Ordoñez and Kolmer, 2007 |
| F2_2 | H12, H13 | France | — | Ordoñez and Kolmer, 2007 |
| F5_3 | France | Ordoñez and Kolmer, 2007 | ||
| ITA1_1 | H11, H18 | Italy | EU_3 | Kolmer et al., 2012 |
| ETH26_2 | H12, H14 | Ethiopia | ME_4 | Kolmer et al., 2011 |
| ETH4114_7 | Ethiopia | ME_3 | Kolmer et al., 2011 | |
| ETH4_1 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E1_1 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E6_1 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E10_4 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E11_1 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E16_1 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E16_4 | Ethiopia | — | Ordoñez and Kolmer, 2007 | |
| E4090_3 | Ethiopia | ME_3 | Kolmer et al., 2011 | |
| On Triticum aestivum | ||||
| AZB 1_2 | H17, H19 | Azerbaijian | CA_1 | Kolmer and Ordoñez, 2007 |
| AZB6_1 | Azerbaijian | CA_1 | Kolmer and Ordoñez, 2007 | |
| NKAZ51_2 | Kazakhstan | CA_2 | Kolmer and Ordoñez, 2007 | |
| S KAZ 44_3 | Kazakhstan | CA_2 | Kolmer and Ordoñez, 2007 | |
| AZB5_1 | Azerbaijian | CA_3 | Kolmer and Ordoñez, 2007 | |
| S KAZ 16B | H17, H30 | Kazakhstan | CA_3 | Kolmer and Ordoñez, 2007 |
| TJK7_1 | H6, H25 | Tajikistan | CA_4 | Kolmer and Ordoñez, 2007 |
| UZB5_2 | Uzbekistan | CA_4 | Kolmer and Ordoñez, 2007 | |
| CZ20_09 | Czech_Slovakia | EU_1 | Kolmer et al., 2012 | |
| GBR1_1_09 | H17, H23 | UK | EU_1 | Kolmer et al., 2012 |
| GBR15_3_09 | UK | EU_1 | Kolmer et al., 2012 | |
| CZ4_09 | H29, H29 | Czech_Slovakia | EU_2 | Kolmer et al., 2012 |
| TUR29_1_09 | H21, H22 | Turkey | EU_2 | Kolmer et al., 2012 |
| CZ_SK95_9_2 | Czech_Slovakia | EU_4 | Kolmer et al., 2012 | |
| CZ18_09 | Czech_Slovakia | EU_4 | Kolmer et al., 2012 | |
| CZ10_9 | H3, H4 | Czech_Slovakia | EU_5 | Kolmer et al., 2012 |
| HG95_10_1 | Hungary | EU_6 | Kolmer et al., 2012 | |
| HG95_3_2 | H17, H30 | Hungary | EU_6 | Kolmer et al., 2012 |
| ES9_1 | Spain | EU_7 | Kolmer et al., 2012 | |
| ITA14_1 | H18, H34 | Italy | EU_7 | Kolmer et al., 2012 |
| CZ11_09 | Czech_Slovakia | EU_8 | Kolmer et al., 2012 | |
| TUR1_1_09 | H17, H31 | Turkey | EU_8 | Kolmer et al., 2012 |
| EGY_154_A | H26, H32 | Egypt | ME_1 | Kolmer et al., 2011 |
| EGY_195_A | Egypt | ME_2 | Kolmer et al., 2011 | |
| EGY_195_B | Egypt | ME_2 | Kolmer et al., 2011 | |
| ISR_L_2 | Israel | ME_2 | Kolmer et al., 2011 | |
| 99 NC | H24, H27 | United States | NA_1 | Ordoñez and Kolmer, 2009 |
| Race_1 | H17, H17 | Canada | NA_1 | Ordoñez and Kolmer, 2009 |
| 01_AL_21 | H1, H17 | United States | NA_2 | Ordoñez and Kolmer, 2009 |
| MI 91_2 | United States | NA_2 | Ordoñez and Kolmer, 2009 | |
| 04 TX 67 | H17, H31 | United States | NA_3 | Ordoñez and Kolmer, 2009 |
| 04_TX_32 | H17, H35 | United States | NA_3 | Ordoñez and Kolmer, 2009 |
| CAN 71_2_96 | Canada | NA_4 | Ordoñez and Kolmer, 2009 | |
| Race_9 | Canada | NA_4 | Ordoñez and Kolmer, 2009 | |
| 04 MN 485 | United States | NA_5 | Ordoñez and Kolmer, 2009 | |
| 99_NC | H17, H26 | United States | NA_5 | Ordoñez and Kolmer, 2009 |
| NZ97_13_3 | New Zealand | — | Kolmer, unpublished | |
| NZ97_8_1 | H17, H20 | New Zealand | Kolmer, unpublished | |
| PER95_82 | Peru | SA_1 | Ordoñez et al., 2010 | |
| URG6_2 | H33, H33 | Uruguay | SA_1 | Ordoñez et al., 2010 |
| BRA19_3 | H2, H28 | Brazil | SA_2 | Ordoñez et al., 2010 |
| URG4_1 | H17, H25 | Uruguay | SA_2 | Ordoñez et al., 2010 |
| ARG512_1 | Argentina | SA_3 | Ordoñez et al., 2010 | |
| URG17_3 | H17, H31 | Uruguay | SA_3 | Ordoñez et al., 2010 |
| CHL08_3_3 | H29, H29 | Chile | SA_4 | Ordoñez et al., 2010 |
| PER87_4107_2 | Peru | SA_4 | Ordoñez et al., 2010 | |
| SF96_4_1 | South Africa | SAF | Kolmer, unpublished | |
| SF98_1_2 | South Africa | SAF | Kolmer, unpublished | |
| P. persistens | ||||
| HSZ0689 | Sweden | — | Liu et al., 2013 | |
| HSZ0695 | Czech republic | — | Liu et al., 2013 | |
Isolates in bold font are included for compatible data set for GENETREE analysis, and converted into haplotypes listed in next column.
Primer design
To screen for hypervariable DNA loci, the intron regions of house-keeping genes and the flanking regions of SSR markers were examined. In addition, an in silico approach was pursued to select SNPs in hypervariable anonymous regions based on the comparison of Illumina reads of four P. triticina genomes, that is, isolates Race 1, Race 161, Race 9 and PRTUS 35 through the Integrative Genomics Viewer (IGV) (Robinson et al., 2011).
DNA sequences of targeted genes including RNA polymerase II largest subunit (RPB1), the 2nd largest unit (RPB2) and elongation factor 1 alpha (EF1-α) were extracted from the complete genome of Puccinia graminis f. sp. tritici published by the Broad Institute (http://www.broadinstitute.org/annotation/genome/puccinia_group/Regions.html). The extracted sequences were submitted into BLAST searches against the non-annotated genome sequence of Puccinia triticina 1-1 Race 1 in order to obtain the homolog sequences from P. triticina. The DNA sequence segments including multiple intron regions of individual homologous genes from P. triticina were input separately into Primerquest on the Integrated DNA Technologies (IDT) website (http://www.idtdna.com/Scitools/Applications/Primerquest/) as template to design appropriate PCR primers.
To design primers for SSR flanking regions, 16 pairs of SSR primers (Duan et al., 2003; Szabo and Kolmer, 2007) were used to BLAST search the P. triticina genome to locate loci containing targeted SSRs. DNA sequence 1 kb up and down stream from targeted SSR loci were downloaded and input as templates into Primerquest to design appropriate primers. All the output primers were compared with the P. triticina whole genome by BLAST to screen for the candidates that did not match with unspecified regions in the genome. The alignment of Illumina reads of the four P. triticina genomes were examined through the Integrative Genomics Viewer (Robinson et al., 2011). Thirty-six sites of 1.5 kb with 1–92 putative SNPs were selected randomly. Primer design was attempted for these sites based on uniqueness of the site in the genome, specificity of the primers and sequencing cost efficiency. BLAST search was conducted to determine the uniqueness of the site in the genome and specificity of the primers. To be cost efficient, the length of amplified DNA fragment was adjusted to 500–900 bp, including at least 1 SNP.
Polymerase chain reaction and sequencing
PCR was performed in 10 μl reactions containing 0.1 mM dNTPs, 2.0 pmoles of each primer, 1 μl 10 × Titanium Taq buffer, 0.1 μl 50 × Titanium Taq DNA Polymerase (BD Biosciences, Mississauga, ON, USA). Touch-down thermal cycling conditions were used initially (except for locus Ctg84-1, for which the annealing temperature was set as 56 °C): 95 °C for 3 min, followed by 10 cycles of 95 °C for 30 s, 63 °C (decreasing 0.5 °C per cycle) for 20 s, and 72 °C for 2 min, then 26–30 cycles of 95 °C for 30 s, 58 °C for 20 s, 72 °C for 2 min, followed by a final extension of 72 °C for 8 min. In the case when unspecified fragments were amplified, the annealing temperature was kept at 63 or 65 °C to minimize the unspecified amplification (Supplementary Table 1). E-Gel CloneWell SYBR Safe system (Invitrogen, Carlsbad, CA, USA) was used to purify the specified band when unspecified bands could not be eliminated by increasing annealing temperatures. Otherwise, PCR products were cleaned by ExoSAP-IT (USB Products Affymetrix, Inc., Cleveland, OH, USA) before being sent to the BioMedical Genomics Center (University of Minnesota, St Paul, MN, USA) for sequencing. PCR primers were used for sequencing most of the loci, except for SSR flanking loci where internal primers were designed to enhance the sequence quality.
DNA sequence examination and heterozygous site recognition
DNA fragments sequenced by forward and reverse primers were assembled using Sequencher 4.10 (Gene Codes Corporation, MI, USA). Heterozygous sites are common in the dikaryotic P. triticina genome. To recognize the heterozygous sites, the chromatograms of the two strands (sequenced by forward and reverse primers) were examined by eye and only the sites that met at least one of the following three criteria were accepted: (1) sites showed two equal height peaks in at least one high-quality strand (confidence of surrounding bases >40, max. 60, Supplementary Figures 1A and 1B); (2) sites showed alternate bases of high quality (Supplementary Figure 1C) from two strands; (3) sites showed the secondary peaks higher than 50% of the primary peaks (Supplementary Figure 1D) in both strands.
Haplotype inference, gene diversity, neutrality test and phylogentic network
DNA sequences from 15 selected loci were analyzed for polymorphisms and neutrality using DnaSP 5.10 (Rozas et al., 2003) locus–by-locus for 70 isolates. Due to the significant genetic difference between the two isolates from A. speltoides with the other isolates from common and durum wheat, we conducted separate polymorphism analyses for P. triticina isolates on common and durum wheat only, and for all P. triticina isolates, including the isolates from A. speltoides. Diploid sequence data (dikaryotic) were converted to haplotype data by PHASE v2.1 (Stephens et al., 2001) through a coalescence-based Bayesian method. Neutrality was evaluated using parameters Fu and Li's D*, Fu and Li's F* (Fu and Li, 1993) and Tajima's D (Tajima, 1989).
A phylogenetic network was generated by SplitsTree 4.13 (Huson and Bryant) for 70 P. triticina isolates with a total of 140 alleles. The NeighborNet was performed with variance as ordinary least squares (Bryant and Moulton, 2004). To accommodate incompatible and ambiguous signals in the data, the split network implies evolutionary history by parallel edges and branches. When the network is not significantly non-tree like, tree-based phylogenies can be performed. The presence of recombination within and among 15 loci was evaluated with the PHIw test (Bruen et al. 2006) incorporated in SplitsTree 4.13 (Huson and Bryant 2006). This test is very general in that a single population is not assumed, and it determines only if a recombination signal is present in the sequence data. This approach is not sensitive to mutation rate correlation due to mutation hot spots that would falsely imply recombination.
Phylogenetic analyses
Phylogenetic analyses were conducted through the three different algorithms of parsimony, Bayesian and maximum likelihood. Parsimony analyses for each of the 15 selected loci and the concatenated DNA sequence matrix were conducted using PAUP*4.0b10 (Swofford, 1998). Heuristic searches with random stepwise addition of 100 replicates and tree bisection-reconnnection branch swapping were performed. A limit of 1 000 000 rearrangements was set for each replicate. Bootstrapping analyses were set with 200 replicates with full heuristic search of random step-wise addition of 20 replicates and limit of 10 000 rearrangements per replicate. A Bayesian phylogenetic analysis was conducted using MrBayes 3.1, with a GTR+G+I model of evolution. (Huelsenbeck and Ronquist, 2001). DNA substitution rates were set as a flat Dirichlet (1.0, 1.0, 1.0, 1.0), which allows the program to estimate the parameters while mixing among Markov chain Monte Carlo (MCMC) chains. Four chains of MCMC were performed and finished at 7 500 000 generations when the average deviation was lower than 0.01; sampling frequency was every 100 generations. The equilibrium of posterior probability was checked by plotting posterior probability value with generations in Excel. The first 1250 trees (1.67%) before equilibrium were discarded (burn-in). Maximum likelihood analyses were performed using GARLI 0.96 (Zwickl, 2006) with unspecified models. Two independent runs each with 10 000 000 generations were performed. Bootstrap replicates were 1000 with default settings. The two Puccinia persistens isolates were used as an outgroup.
Coalescence analysis
The coalescence-based program GENETREE 9.0 (Griffiths, http://www.maths. monash.edu.au/∼mbahlo/mpg/gtree.html) estimates most recent common ancestors for subdivided populations using a Markov chain simulation under an infinite-many-sites model. With the assumption that there is no recombination within and among loci, the DNA segregating sites must be screened for compatibility. Therefore, the isolates with gaps in sequence were eliminated and the Clade and Matrix (http://snap.cifr.ncsu) incorporated in SNAP workbench (Price and Carbone, 2005) was used to eliminate incompatible sites. An analysis of haplotypes of 35 selected isolates from common wheat, durum wheat and the two isolates from A. speltoides with a total of 88 compatible segregating sites was conducted. The matrix was submitted to GENETREE module in SNAP workbench. Two independent simulations were performed each with 100 000 steps; θ was obtained by maximum likelihood estimates. Initial backward migration matrix was inferred by the MIGRATE v. 3.0 module in SNAP workbench. Parameters were set as: short-chain 20, short-increase 200, short-sample 500 000; long-chain 10, long-increase 200, long-sample 500 000 and burn-in 10 000 000 (10%). Seven independent runs were performed in order to reach stabilized estimation.
The coalescence-based genealogy sampler BEAST reconstructs rooted and time-measured phylogenies based on strict or relaxed molecular clock models through Bayesian MCMC analysis for molecular sequences of multiple loci (Drummond et al., 2012). For the comparison with the results from GENETREE and general parsimony analyses, we analyzed two data sets: the 35 isolates (haplotypes) with 88 compatible segregating sites used for GENETREE, and 72 isolates (genotypes) for 10 395 nucleotides from 15 loci. BEAUti v 1.7.1 was used to generate XML input file for BEAST v 1.7.1. Since no time points are available for references, the strict clock model was adopted. HKY and GTR substitution models were attempted in alternate analyses for comparison; 10 000 000 generations and sample frequency 1000 were set for MCMC. The output was analyzed with TreeAnnotater v1.7.1 with burn-in as 1000 (10%). The final tree with highest probability was visualized with FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Isolation and migration
Low levels of genetic divergence among populations can be a result of recent divergence or due to recurrent gene flow among populations. An isolation-with-migration model (Nielsen and Wakeley, 2001) implemented in the IMa2 program developed by Hey and Nielsen (2007) can discriminate between these two different models among multiple populations through an MCMC approach. The migration with isolation model was tested for four P. triticina populations based on host and geographic origins, which were recognized based on the phylogenetic analyses mentioned above. Three independent runs were performed for simulation with IMa2; parameters were set as: metropolis coupling implemented using 80 chains, heating scheme by geometric increment model (–hfg –hn80 –ha0.99 –hb0.5) and burn-in 1 000 000 steps; migration prior follows exponential distribution with mean set as 10 (-j 7, -m 10), population size scalar 5 (-q 5), splitting time (-t 10), all the priors were set based on preliminary analyses; running time as determined by checking the output files for stationarity stage.
Results
Screening for DNA polymorphic loci
Initially, sixteen P. triticina isolates were used to test primers designed for three house-keeping genes (EF1-α, RPB1 and RPB2), 16 SSR flanking loci and 17 anonymous hyper variable regions. Among a total of 83 pairs of primers, 15 pairs were selected for the present study as a result of the uniqueness of the site in the genome, specificity of the primers, success rate of PCR and sequencing cost efficiency. The selected loci included: one house-keeping gene (RPB2), four SSR flanking regions (two up and down streams amplified by two pair of primers) and ten anonymous hypervariable regions located in supercontigs 1, 5, 9, 10, 12, 34, 47, 67 and 84 (Supplementary Table 1). Although amplification was attempted with 70 isolates with all 15 loci, for certain isolates the fragments could not be amplified or sequenced with high quality. For each locus, from 50 to 69 isolates were amplified (Table 2). The non-amplified fragments were treated as missing data in the phylogenetic analyses. DNA sequences for 15 loci were submitted into GeneBank; accession numbers are JN592765–592776 and JX543322–543364 for RPB2; JN592805–592816 and JX543275–543321 for FPtssr91U; JN592817–592829 and JX543227–543274 for FPtssr91D; JN592830–592844 and JX542944–542985 for FPtssr161U; JN592845–592859 and JX543187–543226 for FPtssr161D; JN592876–592890 and JX542836–542885 for Ctg1-3; JN592891–592906 and JX542617–542671 for Ctg5-1; JN592907–592920 and JX543146–543186 for Ctg9-2; JN592921–592936 and JX542727–542783 for Ctg10-2; JN592937–592950 and JX542886–542943 for Ctg10-3; JN592951–592963 and JX542986–543040 for Ctg12-2; JN592964–592978 and JX543041–543095 for Ctg34-1; JN592979–592992 and JX543096–543145 for Ctg47-1; JN592993–593007 and JX542672–542726 for Ctg67-1; JX542784–542835 for Ctg84-1.
Table 2. Polymorphic sites, DNA variation and neutrality tests of 15 loci for Puccinia triticina isolates.
| Polymorphic sites | Neutrality testsa | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Number of samples |
Number of sequences analyzed |
Total sites |
Sites excluding gaps and missing data |
Variable sitesb |
Singleton variable sites |
Parsimony-informative sites |
Variable sites in regions with gap |
Number of polymorphic sites per kb |
Fu and Li's D* |
Fu and Li's F* |
Tajima's D |
PHIw
recombination test (P value)c |
| RPB2 | 51 | 102 | 677 | 486 | 6 | 2 | 2 | 2 | 9 | −1.47720 | −1.01350 | 0.43399 | NS (0.5) |
| FPtssr91U | 57 | 114 | 842 | 617 | 5 | 1 | 1 | 3 | 6 | −1.1–168 | −0.69385 | 0.53891 | NA |
| FPtssr91D | 60 | 120 | 848 | 705 | 5 | 0 | 2 | 3 | 6 | −0.62657 | −0.25902 | 0.064935 | NS (1.0) |
| FPtssr161U | 54 | 108 | 577 | 410 | 8 | 1 | 3 | 4 | 14 | −0.28165 | −0.62160 | −1.04443 | NS (0.5) |
| FPtssr161D | 53 | 106 | 612 | 456 | 18 | 8 | 5 | 5 | 29 | 3.30563** | 3.25343** | −1.71255 | NS (0.3871) |
| Ctg1-3 | 64 | 128 | 758 | 735 | 26 | 4 | 19 | 3 | 34 | 0.11648 | 0.58632 | 1.07481 | NS (0.0512) |
| Ctg5-1 | 67 | 134 | 588 | 521 | 10 | 2 | 6 | 2 | 17 | −0.44298 | −0.11003 | 0.56497 | NS (1.0) |
| Ctg9-2 | 50 | 100 | 788 | 648 | 4 | 1 | 3 | 0 | 5 | −0.26472 | 0.00422 | 0.56915 | NS (1.0) |
| Ctg10-2 | 67 | 134 | 799 | 721 | 4 | 0 | 4 | 0 | 5 | 0.91610 | 0.90854 | 0.46869 | NS (1.0) |
| Ctg10-3 | 64 | 128 | 751 | 672 | 6 | 1 | 5 | 0 | 8 | 0.11315 | 0.36717 | 0.69720 | NS (1.0) |
| Ctg12-2 | 63 | 126 | 425 | 288 | 19 | 1 | 11 | 7 | 45 | 0.80631 | 1.44164 | 1.92003 | NS (1.0) |
| Ctg34-1 | 64 | 128 | 516 | 414 | 12 | 1 | 11 | 0 | 23 | 0.80394 | 1.22323 | 1.41667 | NS (1.0) |
| Ctg47-1 | 57 | 114 | 606 | 403 | 28 | 2 | 16 | 10 | 42 | 0.69961 | 0.71309 | 0.42508 | NS (0.7687) |
| Ctg67-1 | 64 | 128 | 650 | 379 | 32 | 3 | 8 | 21 | 49 | −0.66012 | −0.59934 | −0.22203 | NS (0.7148) |
| Ctg84-1 | 64 | 128 | 664 | 545 | 8 | 1 | 7 | 0 | 12 | 0.40629 | 0.26341 | −0.12577 | NS (1.0) |
| P. triticina on wheat and Aegilops speltoides | |||||||||||||
| RPB2 | 53 | 106 | 677 | 486 | 13 | 3 | 7 | 3 | 19 | −0.76132 | −0.92160 | −0.80240 | NS (0.2095) |
| FPtssr161U | 55 | 110 | 577 | 406 | 11 | 1 | 5 | 5 | 19 | 0.14364 | −0.43817 | −1.39400 | NS (0.5) |
| Ctg5-1 | 68 | 136 | 588 | 521 | 21 | 11 | 8 | 2 | 36 | −3.69668** | −3.27151** | −1.17507 | NS (1.0) |
| Ctg9-2 | 52 | 104 | 788 | 648 | 7 | 1 | 6 | 0 | 9 | 0.31181 | 0.16301 | −0.20288 | NS (1.0) |
| Ctg10-2 | 69 | 138 | 799 | 712 | 8 | 3 | 5 | 0 | 10 | −1.29576 | −1.32406 | −0.77360 | NS (1.0) |
| Ctg10-3 | 66 | 132 | 751 | 672 | 15 | 1 | 14 | 1 | 20 | 1.01080 | 0.42118 | −0.75956 | NS (1.0) |
| Ctg12-2 | 65 | 130 | 425 | 288 | 28 | 3 | 16 | 9 | 66 | 0.25324 | 0.39635 | 0.45730 | NS (1.0) |
| Ctg34-1 | 66 | 132 | 516 | 414 | 12 | 1 | 11 | 0 | 23 | 0.79930 | 1.22017 | 1.41568 | NS (1.0) |
| Ctg67-1 | 66 | 132 | 650 | 379 | 32 | 3 | 8 | 21 | 49 | −0.67164 | −0.54633 | −0.07807 | NS (0.7024) |
For the tests labeled with **, P<0.02, indicating the null hypothesis of neutrality can be rejected at 2% level; *, 0.05<P<0.1, indicating the null hypothesis of neutrality can be rejected at 10% level, but not at 5%. For all other tests, P>0.1, indicating the null hypothesis of neutrality cannot be rejected at 10% level.
Analyzed sites exclude gaps and missing data.
PHIw recombination test was performed in SplitsTree 4.13.1, NA stands for not available due to too few informative characters.
DNA polymorphism of 15 loci in P. triticina isolates
SNPs from 15 loci examined in P. triticina isolates on common and durum wheat varied from 5–49 SNPs per kilobase. Two anonymous regions on contig 9 (Ctg9-2) and contig 10 (Ctg10-2) had the overall lowest polymorphism rates, whereas anonymous contig 67 (Ctg67-1) had the highest rate (Table 2). Neutrality tests resulted in P values larger than 0.1 for most of the loci, indicating neutral evolution, except for microsatellite PtSSR161 down stream region (FPtssr161D), where Fu and Li's D* and Fu and Li's F* P was lower than 0.02, suggesting the mutations in this locus deviated from selective neutrality. However, Tajima's D tests suggested that all loci did not deviate significantly from neutral evolution.
Nine loci were examined for the DNA variation among isolates on wheat and A. speltoides; Ctg12-2 had the highest polymorphism rate of 66 per kilobase. In neutrality tests, Fu and Li's D* and Fu and Li's F* P was lower than 0.02 for locus ctg5-1, indicating non-neutral selection pressure. The haplotype inference by PHASE was confirmed for one locus (Ctg12-2) through sequencing different alleles by using allele specific primers (data not shown).
Test for recombination between and within loci were negative. The PHI test for the concatenated sequence data of the 15 loci in SpiltsTree 4.13 for recombination between loci was non-significant (P=0.0949) and also did not detect any statistically significant evidence for recombination within each locus (Table 2). The split network generated by SplitsTree 4 was generally tree-like (Supplementary Figure 2). The grouping was congruent with that from the other phylogenetic approaches (see below), in which isolates from Ethiopian durum wheat split next to the isolates from Aegilops, followed by isolates from common wheat, and then isolates from cultivated durum wheat imbedded in the isolates from common wheat. This indicated that the sampling error was not significant and that tree-based phylogenetic approaches could be applied.
Phylogenetic analyses
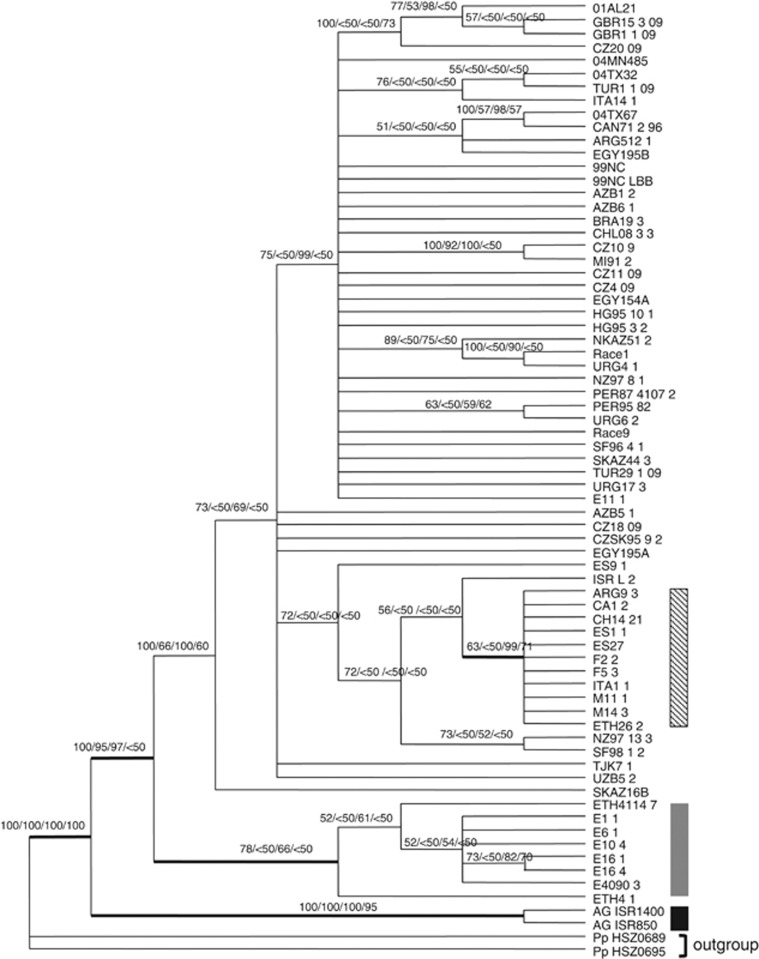
Phylogenetic analyses based on individual loci determined that P. triticina isolates from A. speltoides were genetically distinct from P. triticina on wheat, but the relationships between the subpopulations on wheat were variable from locus to locus. The low inference is possibly because of homoplasy and limited variation for some loci (Supplementary Figure 3, Supplementary Table 2). With relatively little phylogenetic information available, stochastic or sampling error would result in the different tree topologies. As a result, a holistic approach was used to maximize the phylogenetic information (Smith, 2000). The phylogenetic trees that resulted from parsimony majority rule consensus, Bayesian and maximum likelihood analyses based on concatenated DNA sequences of 15 loci separated the P. triticina isolates with various degrees of support into four groups that correlated with host specificities and geographic locations (Figure 1): P. triticina on A. speltoides (indicated as Aegilops form in the following paragraphs), P. triticina on worldwide cultivated durum wheat (cultivated durum form), P. triticina on durum wheat from Ethiopia (Ethiopian durum form) and P. triticina on worldwide common wheat (common wheat form). Rooted with P. persistens, the phylogenies indicated that the Aegilops form diverged initially and appeared as a strongly supported clade with high support values 100/100/100/95 (majority consensus frequency/parsimony bootstrap/Bayesian posterior probability/maximum likelihood bootstrap), followed by the separation between the common wheat form (100/66/100/60) and the Ethiopian durum form. The Ethiopian durum form appeared as a weakly supported clade in the parsimony majority rule consensus tree and bootstrap support values of both parsimony and maximum likelihood analyses were lower than 50% (78/<50/66/<50). The cultivated durum form grouped as a clade nested within the common wheat form with varied support (63/<50/99/71). The matrix of concatenated DNA sequences from 15 loci and phylogenetic tree were submitted into TreeBASE. Study accession no. S13628. Reviewer access URL: http://purl.org/phylo/treebase/phylows/study/TB2: S13628?x-access-code=f8d52f18514ed95babae6dedf2d6f924 &format=html.
Figure 1.
Majority consensus of 20 854 most parsimonious trees based on DNA sequences of 15 loci. L=392, CI=0.786, RI=0.924, RC=0.726, HI=0.214, G-fit=−202.450. Values on branches are majority consensus frequency/Parsimony bootstrap/Bayesian posterior probability/maximum likelihood bootstrap. Hedged bar indicates P. triticina on durum wheat, gray bar indicates P. triticina from Ethiopian durum wheat, black bar indicates P. triticina on Aegilops speltoides, non-labeled isolates are P. triticina from common wheat.
Coalescence analysis
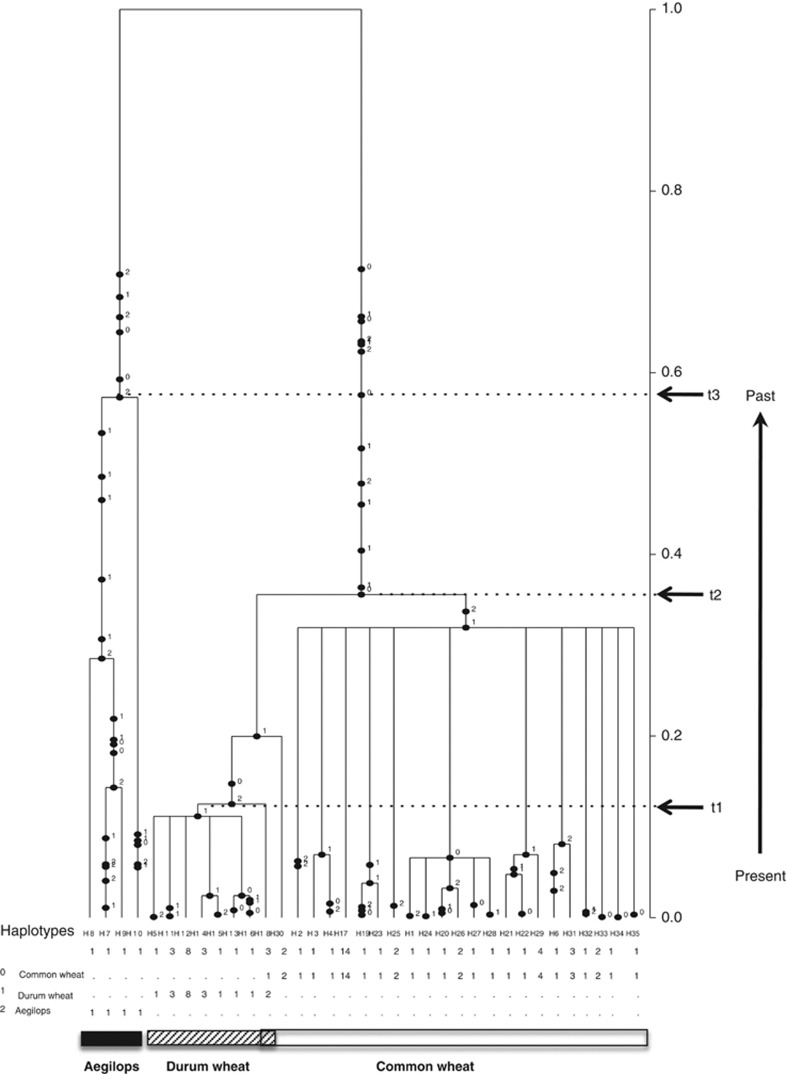
The coalescence tree by GENETREE with highest root probability for 35 isolates (35 haplotypes, Table 1) in three populations (the Ethiopia durum form was not included due to poor resolution that caused the program to crash) determined that the cultivated durum form coalesced the most recently and represented the most recently derived population, whereas the Aegilops form coalesced the earliest and represented the most ancestral population (Figure 2). The common wheat population was intermediate in coalescence between the Aegilops and cultivated durum forms.
Figure 2.
Coalescence tree with highest root probability. The vertical axis on the right stands for relative time scale, from the bottom to the top reflects from the present to the past. At the bottom are the haplotypes. Dots on the branches are the number of mutations. Haplotypes of cultivated durum form coalesced at time t1, common wheat form at t2 and Aegilops form at t3.
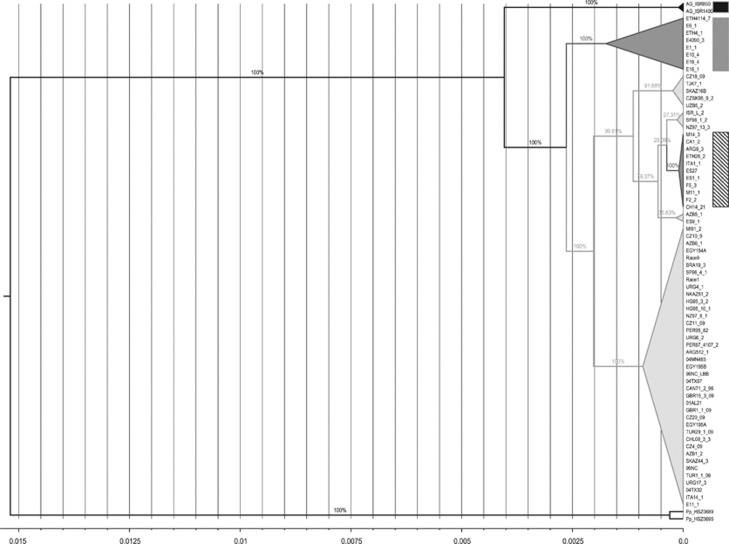
Time-scaled coalescence trees that resulted from BEAST analyses for 35 isolates (haplotypes) with 88 compatible segregating sites and 72 isolates (genotypes) for 10 395 nucleotides based on HKY and GTR substitution models revealed the same order of divergence for the P. triticina host-specified populations (only the tree of 72 isolates with HKY model is shown in Figure 3). The form on A. speltoides diverged first, followed by the Ethiopian durum form and then the forms on common wheat and cultivated durum wheat worldwide. The cultivated durum wheat form was recently derived than the common wheat form. Compared with the outgroup P. persistens, the most closely related species to P. triticina, the divergence of P. triticina populations are very recent (Figure 3). A distinction between P. triticina and P. persistens was not evident based on partial ITS and EF1-α sequence data, indicating that they are recently diverged species (M. Liu et al. 2013). By the relative scale, the divergence among P. triticina populations started 0.004 timescale ago, which is much later than the divergence between P. triticina and P. persistens, 0.015 timescale past.
Figure 3.
Rooted time-scaled phylogeny of 72 isolates by BEAST 1.7.1 based on 15 loci and HKY model. Time scale at the bottom from right (0.0) to left (0.015) indicates present to the past. The hedged bar shows P. triticina on cultivated durum wheat, the gray bar shows P. triticina on Ethiopian durum wheat, the black bar shows P. triticina on Aegilops speltoides, non-labeled isolates are P. triticina on common wheat.
Isolation and migration
The levels of gene flow among four populations detected through three runs of Ima2 analyses suggested current significant (P<0.001) reciprocal gene flow between the common wheat form and the cultivated durum wheat form (Table 3). There was also detectable gene flow from the common wheat form to the Ethiopian durum form. One of the three runs detected gene flow from the Ethiopian durum form to the cultivated durum form, but other two longer runs indicated the gene flow was not significant. There was no detectable gene flow to and from the Aegilops form.
Table 3. Gene flow analyses using IMa2, runtime information and migration rates for four populations of P. triticina.
| Populations | Runs | Steps (after burn-in) | Migration rate 2NM (LLRtest)a | ||
|---|---|---|---|---|---|
|
0 |
1 |
|
|
0>1b |
1>0 |
| CW | DW | Run 1 | 1 000 000 | 0.349 (26.389***) | 0.156 (307.336***) |
| Run 2 | 3 034 032 | 0.502 (29.254***) | 0.168 (105.039***) | ||
| Run 3 | 4 324 321 | 0.476 (30.177***) | 0.179 (105.804***) | ||
| CW | ED | Run 1 | 1 000 000 | 0.073 (1.399 NS) | 0.189 (8.139***) |
| Run 2 | 3 034 032 | 0.050 (0.649 NS) | 0.117 (8.068**) | ||
| Run 3 | 4 324 321 | 0.096 (0.619 NS) | 0.154 (8.805**) | ||
| CW | AG | Run 1 | 1 000 000 | 0.000 (−0.000 NS) | 0.081 (0.972 NS) |
| Run 2 | 3 034 032 | 0.000 (−0.000 NS) | 0.030 (0.101 NS) | ||
| Run 3 | 4 324 321 | 0.000 (−0.000 NS) | 0.052 (0.305 NS) | ||
| DW | ED | Run 1 | 1 000 000 | 0.119 (7.423**) | 0.026 (−0.202 NS) |
| Run 2 | 3 034 032 | 0.008 (0.299 NS) | 0.099 (1.397 NS) | ||
| Run 3 | 4 324 321 | 0.007 (1.103 NS) | 0.025 (−0.014 NS) | ||
| DW | AG | Run 1 | 1 000 000 | 0.000 (−0.000 NS) | 0.113 (0.079 NS) |
| Run 2 | 3 034 032 | 0.000 (−0.000 NS) | 0.119 (0.690 NS) | ||
| Run 3 | 4 324 321 | 0.000 (−0.000 NS) | 0.041 (0.338 NS) | ||
| ED | AG | Run 1 | 1 000 000 | 0.000 (−0.000 NS) | 0.009 (0.040 NS) |
| Run 2 | 3 034 032 | 0.005 (0.121 NS) | 0.000 (−0.000 NS) | ||
| Run 3 | 4 324 321 | 0.040 (0.039 NS) | 0.002 (−0.005 NS) | ||
*P<0.05, **P<0.01, ***P<0.001.
0>1 stands for migration from population 0 to population 1 in the coalescence sense (backwards in time); as time moves forward, the migration is from population 1 to population 0.
Discussion
Divergence of P. triticina populations
In asexual populations, accumulated mutation, genetic drift and selection are recognized as the main forces that affect divergence of lineages (Kohn, 2005). Host specificity and adaptation is an additional key factor driving the evolution and divergence of clonal obligate plant pathogens. Among the diploid ancestors of wheat, P. triticina is found only on A. speltoides in natural stands. The isolates were found in two locations at Ahihud and Qazir in northern and central Israel. Morphological examination and DNA content study revealed the two collections to be P. triticina (Yehuda et al., 2004). Inoculation experiments showed that the collections from A. speltoides were avirulent to common and durum wheat. However, the isolates from A. speltoides and from cultivated durum and common wheat were sexually compatible, indicating that these could be considered as a biological single species. Yehuda et al. (2004) recognized the isolates from A. speltoides as a new forma specialis of P. triticina. Our study based on phylogenetic analysis, BEAST and GENETREE coalescence indicated that the P. triticina on A. speltoides was the earliest diverged lineage, which is in accordance with A. speltoides as a wheat progenitor.
Eight of the P. triticina isolates from durum wheat in Ethiopia were grouped separately from the durum wheat isolates from Mexico, Europe and South America and also from the isolates from common wheat. The wheat population in Ethiopia is unique which may account for the early divergence of this set of isolates. Emmer wheat was introduced to Ethiopia via the Nile from Egypt 5000 years ago (Zaharieva et al., 2010), and Ethiopia is one of the few countries where it is still grown. Emmer wheat is grown on marginal land throughout Ethiopia, and is grown by farmers in mixtures with durum landrace cultivars and common wheat in the Bale, Tigray and Welo regions. Ethiopia is considered as a center of diversity for tetraploid wheats (Payne et al., 2001). The P. triticina population has likely been selected on emmer and tetraploid durum landraces for thousands of years. Since emmer and durum wheat predate common hexaploid wheat, the P. triticina population on tetraploid wheat in Ethiopia may be a remnant of the P. triticina population before the widespread cultivation of common hexaploid wheat. The Bayesian phylogeny and the BEAST coalescence analysis strongly indicated that the Ethiopian durum isolates are ancestral to the isolates from common wheat and to the other isolates from durum wheat. The consensus phylogeny tree weakly supported the separation of these isolates in a single clade. Isolates from durum wheat in Ethiopia were also previously noted to have SSR genotypes distinct from isolates from cultivated durum wheat and common wheat in other parts of the world (Ordoñez and Kolmer, 2007; Kolmer et al., 2011).
P. triticina on common hexaploid wheat is found worldwide, nearly wherever wheat is grown. On a worldwide scale P. triticina is highly diverse for virulence to leaf rust resistance genes in wheat (Kolmer and Liu, 2000) and also for distinct groups of SSR genotypes (Ordoñez and Kolmer, 2009; Kolmer et al., 2012). Although P. triticina isolates from common wheat are generally virulent to wild and cultivated emmer wheat (Anikster et al., 2005a), these isolates are usually avirulent to the modern durum wheat cultivars that are grown in North America, Europe, the Middle East and North Africa. The BEAST and GENETREE analyses all indicated that 12 of the isolates with virulence to durum wheat from Europe, Mexico, South America, California and Ethiopia were derived from P. triticina with virulence to common wheat. Isolates adapted to durum wheat were closest in SNP variation to isolates from common wheat from Central Europe, Central Asia, Israel, New Zealand, South Africa and Spain, based on phylogenetic and BEAST analyses. The cultivated durum form is recently diverged from the common wheat form and adaptation of P. triticina to modern durum wheat cultivars is relatively recent. Isolates of P. triticina from cultivated durum wheat in France (Goyeau et al., 2011) and worldwide (Ordoñez and Kolmer, 2007) had very limited SSR genotype variation and virulence variation to leaf rust resistance genes compared to isolates from common wheat, supporting the hypothesis of recent adaptation to cultivated durum wheat. An isolate of P. triticina with virulence to durum wheat that was collected in Arizona, USA in 2009 had an SSR genotype and virulence very similar to isolates collected from common wheat in western Europe (J. Kolmer, unpublished data). One of the isolates from Ethiopia (E11_1) collected from durum wheat was grouped with the isolates from common wheat in the BEAST and phylogenetic analyses. Shortly after the expansion of domesticated emmer wheat ca. 10 000 years ago, durum wheat appeared in the Near East and replaced emmer as the leading tetraploid wheat by 2000 BC (Zohary and Hopf, 2000). Durum wheat may have originated from a small population of emmer wheat in southeastern Europe (Oliveira et al., 2012). Since emmer wheat is generally susceptible to P. triticina, leaf rust resistance in durum wheat may have only been recently acquired, perhaps during selection for grain quality and agronomic traits.
Phylogenetic and coalescence approaches and interpretation
The strength of the groupings of the P. triticina isolates differed among the analyses employed. In general, the three phylogenetic approaches strongly separated the isolates from Aegilops, but the parsimony and maximum likelihood methods provided weak support for separation of the Ethiopian durum isolates, the common wheat isolates and the isolates from cultivated durum wheat worldwide. This is likely due to low levels of sequence variation overall, resulting in low levels of signal for each separate region. Such stochastic or sampling error would explain the different tree topologies that were observed for the individual regions. As there were only 319 variable sites in total, any phylogenetic inference or groupings will be less robust. Also, since the groups are relatively recently derived, any phylogenetic separation would be expected to be weak in comparison to separation of P. persistens and the isolates from Aegilops, which were strongly supported. In contrast the Bayesian phylogeny and the coalescence approaches used in GENETREE and BEAST strongly supported the separation of the different isolate types. The GENETREE analysis was limited to only compatible sites while BEAST utilized complete sequence data, with compatible and incompatible sites. Both methods strongly placed the isolates from cultivated durum wheat that were found worldwide in a single group that was very recently derived from the isolates from common wheat, while BEAST also strongly grouped the isolates from durum wheat in Ethiopia in an earlier derived group. The differing results from the phylogenetic and coalescence approaches may be due to underlying differences in evolutionary models. BEAST in particular can be used for rigorous statistical inference and hypothesis testing of evolutionary models, in addition to generating phylogenies. BEAST is especially useful for dealing with heterogeneous data and incongruence among data sets (Drummond et al. 2012). The Bayesian inference employed with GENETREE and BEAST also performs better when dealing with missing data (Yang and Rannala, 2012) as occurred in each of the 15 loci examined.
Gene flow among populations
Reciprocal gene flow between the common wheat form and the cultivated durum wheat form and uni-directional gene flow from the common wheat form to the Ethiopian durum form were consistently detected through multiple analyses. These results seem logical when compared with the phylogenetic trees and coalescence-based genealogies: between the two most closely related lineages, the common wheat form and the cultivated durum wheat form, there were high levels of gene flow. There was no detectable gene flow between the Aegilops form and the other forms since it diverged the earliest. These results are consistent with the results from infection experiments that almost all A. speltoides accessions are highly resistant to the P. triticina isolates from common and durum wheat (Anikster et al., 2005b). However, the forms on Aegilops and wheat are sexually compatible, so potential gene flow between the Aegilops form and isolates from wheat cannot be excluded.
In conclusion, our study demonstrated the ability of the obligate plant pathogen P. triticina to expand its host range from the wheat progenitor host species Aegilops speltoides to the modern-day tetraploid and hexaploid wheat varieties. The P. triticina isolates with virulence to durum cultivars that have been found in Europe, South America, Mexico and the Middle East very recently arose from isolates from hexaploid common wheat, indicating a rapid adaptation to a genetically distinct host population. Finally, coalescence analyses indicated that isolates from tetraploid wheats in Ethiopia may be the oldest derived form of P. triticina found on wheat, predating the adaptation of P. triticina to the hexaploid wheats that are grown worldwide.
Data archiving
Sequence data of 15 P. triticina loci were deposited in GenBank with individual accession numbers listed in results section.
Acknowledgments
We thank Drs Les Szabo, John Fellers and Christina Cuomo for facilitating ML to access IGV and Pt whole genome database; Kun Xiao, Jerry Johnson and Kim Phuong Nguyen for technical help.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andenow Y, Hulluka M, Belay G, Tesemma T. Resistance and tolerance to leaf rust in Ethiopian tetraploid wheat landraces. Plant Breed. 1997;116:533–536. [Google Scholar]
- Anikster Y, Manisterski J, Long DL, Leonard KJ. Leaf and stem rust resistance in Triticum dicoccoides populations in Israel. Plant Dis. 2005;89:55–62. doi: 10.1094/PD-89-0055. [DOI] [PubMed] [Google Scholar]
- Anikster Y, Manisterski J, Long DL, Leonard KJ. Resistance to leaf rust, stripe rust, and stem rust in Aegilops spp. in Israel. Plant Dis. 2005;89:303–308. doi: 10.1094/PD-89-0303. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Garvin DF. Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol. 2008;9:563–575. doi: 10.1111/j.1364-3703.2008.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V. NeighborNet: an agglomerative algorithm for the construction of planar phylogenetic networks. Mol Biol Evo. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Casulii F, Siniscalco A. Proceedings of the 7th Congress of the Mediterranean Phytopathology Union. Granada: Spain; 1987. Thalictrum flavum L. as an alternate host of Puccinia recondita f. sp. tritici in Southern Italy; pp. 91–93.pp. 1984 [Google Scholar]
- Chester KS. The Nature and Prevention of the Cereal Rusts as examplified in the Leaf Rust of Wheat. Chronica Botanica: Waltham, MA, USA; 1946. [Google Scholar]
- D'Oliveira BD, Samborski DJ.1966Aecial Stage of Puccinia recondita on Ranunculaceae and Boraginaceae in PortugalIn: Macer RC, Wolfe MS (eds). Proceedings of the First European Brown Rust Conference, Cereal Rust Conferences 133–150.Cambridge: UK; 1964 [Google Scholar]
- Dinoor A, Eshed N, Ecker R, Grerechter-Amitai Z, Solel Z, Manisterski J, et al. Fungal diseases of wild tetraploid wheat in a natural stand in northern Israel. Isr J Bot. 1991;40:481–500. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Bio Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Enjalbert J, Vautrin D, Solignac C, Giraud T. Isolation of 12 microsatellite loci, using an enrichment protocol, in the phytopathogenic fungus Puccinia triticina. Mol Ecol Notes. 2003;3:65–67. [Google Scholar]
- Feldman M.2001Origin of cultivated wheatIn: Bonjean AP, Angus WJ (eds). The World Wheat Book, A History of Wheat Breeding Lavoisier Publishing: Paris; 1–56. [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyeau H, Berder J, Czerepak C, Gautier A, Lanen C, Lannou C. Low diversity and fast evolution in the population of Puccinia triticina causing durum wheat leaf rust in France from 1999 to 2009, as revealed by an adapted differential set. Plant Pathol. 2011;61:761–772. [Google Scholar]
- Goyeau H, Halkett F, Zapater MF, Carlier J, Lannou C. Clonality and host selection in the wheat pathogenic fungus Puccinia triticina. Fungal Genet Biol. 2007;44:474–483. doi: 10.1016/j.fgb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Nat Acad Sci. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kohn LM. Mechanisms of fungal speciation. Annu Rev Phytopathol. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Kolmer JA. Molecular polymorphism and virulence phenotypes of the wheat leaf rust fungus Puccinia triticina in Canada. Can J Bot. 2001;79:917–926. [Google Scholar]
- Kolmer JA, Hanzalova A, Goyeau H, Bayles R, Morgounov A. Genetic differentiation of the wheat leaf rust fungus Puccinia triticina in Europe. Plant Pathol. 2012;62:21–31. [Google Scholar]
- Kolmer JA, Liu JQ. Virulence and molecular polymorphism in international collections of the wheat leaf rust fungus Puccinia triticina. Phytopathology. 2000;90:427–436. doi: 10.1094/PHYTO.2000.90.4.427. [DOI] [PubMed] [Google Scholar]
- Kolmer JA, Ordoñez ME. Genetic differentiation of Puccinia triticina populations in Central Asia and the Caucasus. Phytopathology. 2007;97:1141–1149. doi: 10.1094/PHYTO-97-9-1141. [DOI] [PubMed] [Google Scholar]
- Kolmer JA, Ordoñez ME, Manisterski J., Anikster Y. Genetic differentiation of Puccinia triticina populations in the Middle East and genetic similarity with populations in Central Asia. Phytopathology. 2011;101:870–877. doi: 10.1094/PHYTO-10-10-0268. [DOI] [PubMed] [Google Scholar]
- Levine MN, Hildreth EC. A natural occurrence of the aecial stage of Puccinia rubigo-vera var. tritici in the United States. Phytopathology. 1957;47:110–111. [Google Scholar]
- Liu M, Szabo LJ, Hambelton S, Anikster Y, Kolmer JA. Molecular phylogentic relationships of the brown leaf rust fungi and on wheat, rye, and other grasses. Plant Dis. 2013;97:1408–1417. doi: 10.1094/PDIS-02-13-0152-RE. [DOI] [PubMed] [Google Scholar]
- Mains EB, Jackson HS. Aecial stage of the orange leaf rust of wheat, Puccinia triticina Eriks. J Agric Res. 1921;22:151–172. [Google Scholar]
- Manisterski J, Eyal Z, Ben-Yehuda P, Kosman E. Comparative analysis of indices in the study of virulence diversity between and within populations of Puccinia recondita f. sp. tritici in Israel. Phytopathology. 2000;90:601–607. doi: 10.1094/PHYTO.2000.90.6.601. [DOI] [PubMed] [Google Scholar]
- Markova JV, Urban Z. The rust fungi of grasses in Europe. 6. Puccina persistens Plow., P. perplexans Plow., and P. elymi Westend. Acta Univ Carolinae. 1998;41:329–402. [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira HR, Campana MG, Jones H, Hunt HV, Leigh F., Redhouse DI, et al. Tetraploid wheat landraces in the Mediterranean basin: taxonomy, evolution and genetic diversity. PLos One. 2012;7:e37063. doi: 10.1371/journal.pone.0037063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordoñez ME, German SE, Kolmer JA. Genetic differentiation within the Puccinia triticina population in South America and comparison with the North American population suggests common ancestry and intercontinental migration. Phytopathology. 2010;100:376–383. doi: 10.1094/PHYTO-100-4-0376. [DOI] [PubMed] [Google Scholar]
- Ordoñez ME, Kolmer JA. Simple sequence repeat diversity of a world-wide collection of Puccinia triticina from durum wheat. Phytopathology. 2007;97:574–583. doi: 10.1094/PHYTO-97-5-0574. [DOI] [PubMed] [Google Scholar]
- Ordoñez ME, Kolmer JA. Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in North America. Phytopathology. 2009;99:750–758. doi: 10.1094/PHYTO-99-6-0750. [DOI] [PubMed] [Google Scholar]
- Payne TS, Wanjema JK, Girma B.2001Eastern Africa Wheat PoolIn: Bonjean A, Angus W (eds). The World Wheat Book A History of Wheat Breeding Lavoisier: Paris; 901–938. [Google Scholar]
- Price EW, Carbone I. SNAP: workbench management tool for evolutionary population genetic analysis. Bioinformatics. 2005;21:402–404. doi: 10.1093/bioinformatics/bti003. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- Smith JF. Phylogenetic signal common to three data sets: Combining data which initially appear heterogeneous. Plant Syst Evol. 2000;221:179–198. [Google Scholar]
- Stephens M., Smith NJ, Donnelly P. A new statistical method for haplotye reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL.1998PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods)Version 4 (Computer Program)Sinauer Associates: Sunderland, MA [Google Scholar]
- Szabo LJ, Kolmer JA. Development of simple sequence repeat markers for the plant pathogenic rust fungus Puccinia triticina. Mol Ecol Notes. 2007;7:708–710. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Molecular phylogenetics: principles and practice. Nat Rev Genet. 2012;13:303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- Yehuda PB, Eilam T, Manisterski J, Shimoni A, Anikster Y. Leaf rust on Aegilops speltoides caused by a new forma specialis of Puccinia triticina. Phytopthology. 2004;94:94–101. doi: 10.1094/PHYTO.2004.94.1.94. [DOI] [PubMed] [Google Scholar]
- Young HC, D'Oliveira B. A further study of race populations of Puccinia recondita f. sp. tritici. Garcia de orta, estudos agron micos. 1982;9:37–52. [Google Scholar]
- Zaharieva M, Ayanna NG, Hakimi AA, Misra S, Monneveux P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: a review. Genet Resour Crop Evol. 2010;57:937–962. [Google Scholar]
- Zohary D, Hopf M. Domestication of Plants in the Old World. Oxford University Press: New York; 2000. [Google Scholar]
- Zwickl DJ. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. University of Texas: Austin; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.