Summary
Inflammation is associated with DNA damage, cellular senescence and aging. Cessation of the inflammatory cytokine response is mediated in part through cytokine mRNA degradation facilitated by RNA binding proteins, including AUF1. We report a major unrecognized function of AUF1 – it activates telomerase expression, suppresses cellular senescence and maintains normal aging. AUF1 deficient mice undergo striking telomere erosion, markedly increased DNA damage responses at telomere ends, pronounced cellular senescence and rapid premature aging that increases with successive generations, which can be rescued in AUF1 knockout mice and their cultured cells by resupplying AUF1 expression. AUF1 binds and strongly activates the transcription promoter for telomerase catalytic subunit Tert. In addition to directing inflammatory cytokine mRNA decay, AUF1 destabilizes cell cycle checkpoint mRNAs, preventing cellular senescence. Thus, a single gene, AUF1, links maintenance of telomere length and normal aging to attenuation of inflammatory cytokine expression and inhibition of cellular senescence.
Introduction
Telomeres cap the linear ends of chromosomes. Failure to maintain telomere integrity results in genomic instability, DNA damage responses particularly at the ends of chromosomes, increased cellular senescence and organ degeneration, which are key drivers of the natural aging process (De Lange 2005; Jaskelioff et al. 2010; Sahin and Depinho 2010). Telomere length shortens with successive cell divisions due to an inability of telomerase, a specialized reverse transcriptase that uses the non-coding RNA template TERC, to copy the most distal sequences (Collins 2006). Immune and other proliferating somatic cells, as well as germ cells, activate telomerase to elongate the telomeric DNA strand (Forsyth et al. 2002). Because telomeres of mice are much longer than human telomeres, defects in telomerase function have no immediate phenotypic effect in mice. However, later generation telomerase-deficient mice exhibit progressive telomere shortening, emergence of aging phenotypes, germ cell abnormalities, and reduced life-span, all of which increases in severity and manifests earlier with succeeding generations, a process known as genetic anticipation (Blasco et al. 1997; Rudolph et al. 1999). Mice heterozygous for mTERT or mTERC show haploinsufficiency in telomere length maintenance (Greider 2006), indicating that even partially reduced expression of mTERT or mTERC has profound effects. Diseases of aging and chronic inflammatory responses are both associated with telomere shortening and decline in organ function (Browner et al. 2004; Andrews et al. 2010; Sahin and Depinho 2010). We therefore sought to determine whether intrinsic molecular factors couple the regulation of the inflammatory cytokine response to the process of telomere loss-associated aging.
AUF1 (heterogeneous nuclear ribonucleoprotein D, hnRNP D) is an established attenuator of the inflammatory cytokine response, inducibly destabilizing inflammatory cytokine and other mRNAs containing specific AU-rich elements (AREs) in their 3′-untranslated regions (3′UTR) (Lu et al. 2006; Sadri and Schneider 2009). AUF1 consists of four highly related protein isoforms generated by alternate RNA splicing from a single genetic locus (Guhaniyogi and Brewer 2001). We previously demonstrated that knockout mice deficient in one or both copies of the AUF1 gene cannot properly degrade key inflammatory cytokine mRNAs induced by bacterial endotoxin and succumb to endotoxic (septic) shock, as well as development of a pruritic dermatitis similar to human psoriasis (Lu et al. 2006; Sadri and Schneider 2009). AUF1 levels are therefore important for attenuation of the inflammatory response.
As many inflammation-related diseases are associated with accelerated telomere erosion, DNA damage signaling and increased markers of aging (Browner et al. 2004; Andrews et al. 2010; Sahin and Depinho 2010), we sought to identify common intrinsic molecular drivers. Our current study reveals that AUF1, in addition to attenuating inflammatory cytokine responses, activates telomerase expression, thereby suppressing cellular senescence and maintaining normal telomere length, while also regulating levels of essential aging-linked cell cycle regulators. Thus, AUF1 is an intrinsic factor that regulates telomerase expression, telomere maintenance and markers of aging by two distinct mechanisms, coupling them to attenuation of the inflammatory response.
Results
Genetic anticipation, accelerated aging and cellular senescence in AUF1 deficient mice
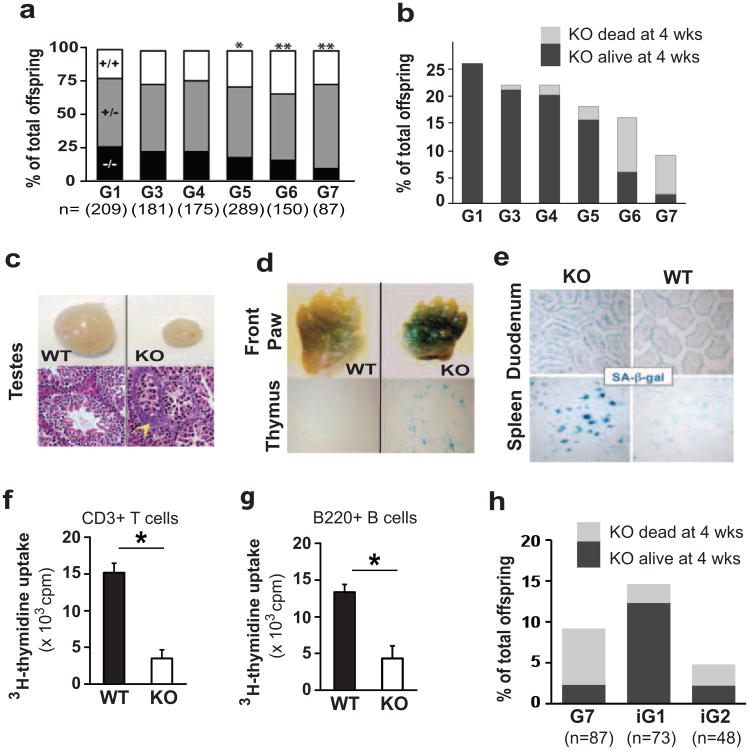
Auf1 chimeric founder males were generated as described (Lu et al. 2006) and bred with either C57Bl/6 (B6) or 129/SvEvTac (129S6) wild-type (WT) females to produce generation 0 (G0) Auf1+/- heterozygous (Auf1+/-) mice, and these mice then crossed to produce G1 offspring in a B6;129S6 or 129S6 background. Successive generations of mice in each background were generated by interbreeding of Auf1+/- mice using a cousin-mating strategy (Fig. S1a), as AUF1-/- crosses were not successful (Fig. S2a). Although 26% of G1 offspring in mixed B6;129S6 background were Auf1-/- mice, progeny yields progressively decreased to 9% by G7 (Fig. 1a), as did survival (Fig. 1b). Most G1 through G3 Auf1-/- mice survived beyond four weeks of age, compared to 87% of G5, 37% of G6 and 8% of G7 mice (Fig. 1b). Similar results were found in the 129S6 genetic background (Fig. S2b-c), indicating that AUF1 deficiency has a genetic anticipation phenotype. As with telomerase knockout mice (Herrera et al. 1999), there was a stronger phenotype and greater decrease in number and survival of Auf1-/- offspring in a congenic C57BL/6 background (Fig. S2d). G7 Auf1-/- mice demonstrated marked tissue atrophy, including severe reduction in size of the testes, and aberrant maturation and degeneration of germ cell epithelium (Fig. 1c). We therefore determined whether late-generation Auf1-/- mice display increased cellular senescence and organ degeneration by measuring expression of senescence-associated-β-galactosidase (SA-β-gal) in tissues (Itahana et al. 2007). Two-day old G7 Auf1-/- knockout (KO) mice exhibited widespread, strong staining of SA-β-gal in skin, thymus and other tissues compared to WT littermates (Fig. 1d). Twelve month-old Auf1-/- mice had strongly increased SA-β-gal expression in the spleen, gastrointestinal tract and other tissues compared to WT littermates (Fig. 1e). Cellular proliferation was 3-4 fold reduced in splenic CD3+ T cells from KO animals, determined by LPS stimulation of isolated T cells and uptake of 3H-thymidine into newly-synthesized DNA (Fig. 1f). Similar results were found for B220+ B cells in the splenocyte population (Fig. 1g).
Figure 1.
Decreased survival and increased markers of aging in late generation Auf1-/-mice. (a) Percentage of each genotype at live birth from Auf1+/- crossings for indicated generation. n= number of mice in each group. P-values based on expected Mendelian ratios, n=3. *P≤0.04; **P≤0.02, calculated by Chi-squared test. (b) Frequency and survival beyond 4 weeks for Auf1-/- KO mice per generation. (c) Top: Representative images of whole testes from 25-day old G7 WT (Auf1+/+) and KO littermates. Bottom: H&E sections showing aberrant maturation and degeneration of germ cell epithelium (yellow arrow head). (d) Representative images of SA-β-gal staining, a marker for cellular senescence, in indicated tissues from two day old WT and KO littermates. (e) Staining for SA-β-gal in the villi of the duodenum and white pulp of the spleen from 12-month-old mice. (f) and (g) Proliferation of isolated splenic CD3+ T cells and B220+ B cells by 3H-thymidine labeling of G6 animals. *P<0.01 by paired Student's t-test ± standard error of the mean (SEM), n=3). (h) Frequency of live births and survival of G7, G5.BC1.iG1 and G5.BC1.iG2 KO mice.
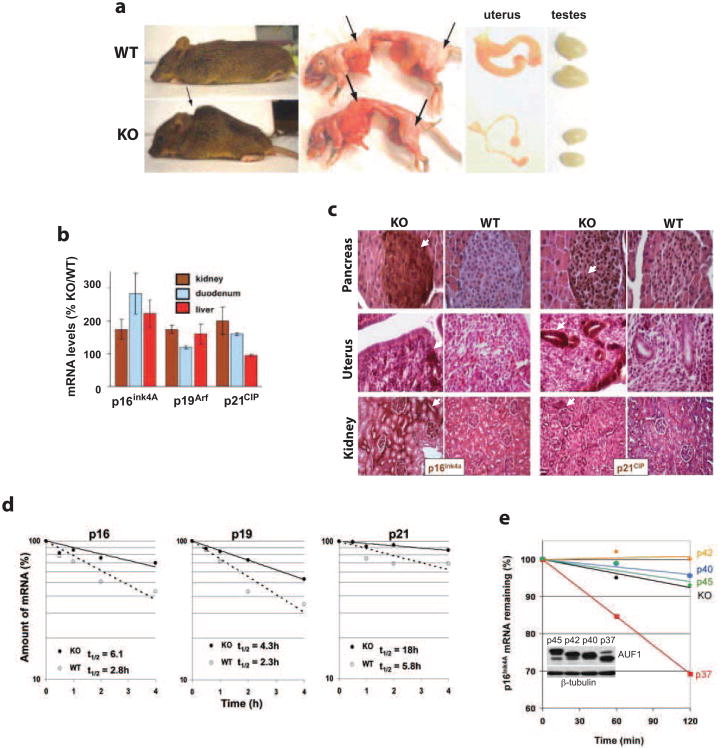
Auf1-/- mice in all genetic backgrounds displayed classic hallmarks of accelerated aging, including kyphosis (hunchback), reduced subcutaneous body fat and atrophy of the reproductive organs in most animals by 12 months of age (Fig. 2a). Increased expression of cell cycle regulators p16Ink4a, p19Arf and possibly p21CIP are markers of mammalian aging or cellular senescence (Krishnamurthy et al. 2004). Twelve month-old Auf1-/- mice displayed a 2.5-3 fold elevation of mRNA and protein levels in a variety of tissues surveyed (Fig. 2b, c). The p19Arf and p21CIP 3′UTRs have AU-rich elements, similar to that reported for AUF1-target p16Ink4a (Wang et al. 2005). Accordingly, decay rates for p16Ink4a, p19Arf and p21CIP mRNAs in Auf1-/- mouse embryo fibroblasts (MEFs) were 2-3-fold slower compared to WT (Fig. 2d). Exogenous expression of single AUF1 isoforms in Auf1-/-MEFs showed that only the p37 protein restored rapid destabilization of p16Ink4a mRNA (Fig. 2e), consistent with established p37 function (Sarkar et al. 2003b).
Figure 2.
Aging-related phenotypes and upregulation of p16Ink4A, p19Arf and p21CIP expression in Auf1-/- mice. (a) Representative images of kyphosis (hunchback) (arrow), presence (top panel) and absence (bottom panel) of subcutaneous body fat (arrows), and atrophied gonads typically observed in 12-month-old Auf1-/- (KO) mice but not in WT littermates. Images shown represent the majority of KO animals surveyed (>60%, kyphosis; >90%, reduced body fat; >90%, gonadal atrophy). (b) Increased relative expression of p16Ink4a, p19Arf and p21CIP mRNAs in tissues and organs from 12-month-old KO mice compared to WT littermates, determined by qRT-PCR. (c) Immunohistochemical staining for p16Ink4a and p21CIP (brown, arrows) in organ sections from 12-month-old KO mice compared to WT littermates. Sections counter-stained by H&E. (d) Decay plots of p16Ink4a, p19Arf and p21CIP mRNAs in MEFs from G2 embryos. Transcription was blocked with actinomycin D, cells were collected at indicated time points, RNA was extracted and mRNA levels were quantified by qRT-PCR. Plots are the mean of three independent experiments with calculated half-lives shown. (e) Decay plots showing recovery of p16Ink4a mRNA destabilization by individual isoforms of AUF1 in KO MEFs. Plot shown is an average of three independent experiments.
AUF1 is directly responsible for the genetic anticipation phenotype
While increased stabilization of cell cycle checkpoint mRNAs promotes cellular senescence, it cannot account for the genetic anticipation phenotype, previously associated with telomere dysfunction in mice (Herrera et al. 1999; Rudolph et al. 1999; d'Adda di Fagagna et al. 2003). To test whether AUF1 is directly responsible for the genetic anticipation phenotype, we restored AUF1 by crossing G5 heterozygous Auf1+/-mice with WT mice for one generation and then bred intergenerational (iG) offspring using a cousin-mating scheme (Fig. S1b). Compared to corresponding G7 Auf1-/- offspring, iG1 litters had an instant ∼2-fold increase in the proportion of offspring that were Auf1-/- and a 4-fold increase in their survival (Fig. 1h): Eighty-two percent of intergenerational offspring (G5.BC1.iG1) survived beyond four weeks compared to 20% for G7 Auf1-/- mice. Intergenerational Auf1-/- mice also displayed long-term survival comparable to G6 Auf1-/-mice (Fig. S2e). Increased survival was not maintained in subsequent intergenerational crosses lacking AUF1: G5.BC1.iG2 Auf1-/- mice showed a similar phenotype to G7 Auf1-/-mice (Figs. 1h, S1b). Backcrossing to WT mice for one generation therefore delayed the genetic anticipation phenotype by one generation, indicating that the genetic and biologic effects are a result of AUF1 deficiency.
Telomere erosion and chromosome fusion with AUF1 deficiency
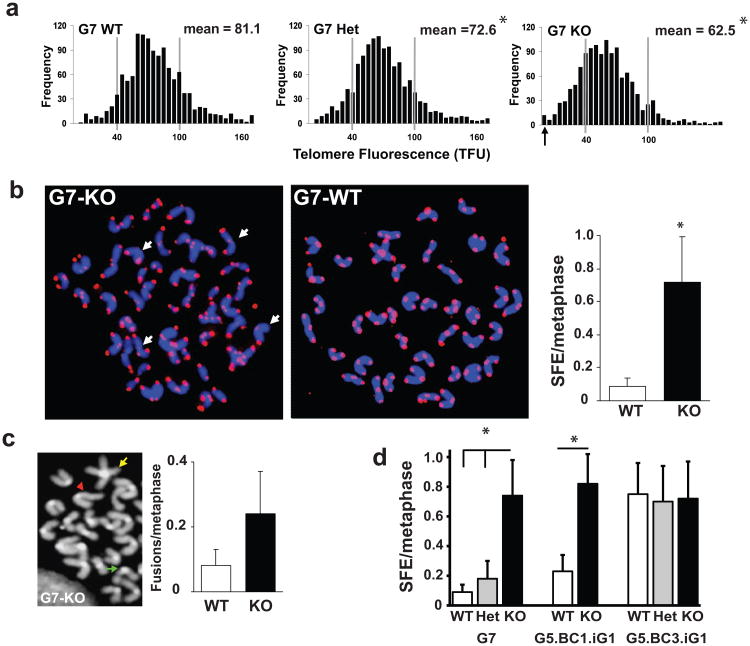
Since the genetic anticipation seen in late generation Auf1-/- mice is similar to that seen in telomerase-deficient mice, we directly characterized telomere lengths in splenocytes from G7 mice. We performed quantitative fluorescence in situ hybridization (Q-FISH; Fig. 3a) to quantify the relative number of telomere repeat sequences in individual chromosomes from metaphase cells (Poon et al. 1999). Average telomere length in splenocytes from G7 Auf1-/- mice was significantly decreased (62.5 TFUs) compared to WT littermates (81.1 TFUs). Heterozygous G7 Auf1+/- mice were intermediate in telomere length (72.6 TFUs), indicating that AUF1 displays haploinsufficiency in average telomere length maintenance (Fig. 3a), similar to telomerase heterozygous mice (Hao et al. 2005). Splenocytes from G7 Auf1-/- mice displayed increased heterogeneity in telomere signals, with a 7-fold increase in the number of chromosomes lacking a detectable telomere signal (signal-free ends, SFEs) (Fig. 3b), and a 4-fold increase in chromosomal fusions (Fig. 3c).
Figure 3.
Decreased telomere lengths in late generation Auf1-/- splenocytes. (a) Q-FISH telomere length analysis, frequency distribution of telomeric DNA signal intensities (telomere fluorescence units, TFU), ≥20 metaphase splenocyte spreads of 3 mice. Arrow, chromosome ends with no detectable telomere (Signal Free Ends, SFEs), *P<0.01. (b) Left, representative splenocyte metaphase spreads from G7 mice stained with Cy3-labeled PNA telomere probes (arrows, SFEs). Right, frequency of SFEs in G7 splenocyte metaphase spreads. *P<0.05, ± SEM, n=3. (c) Left, chromosomal abnormalities in KO spreads: representative fused chromosomes (yellow), chromosomal breaks (green), uneven translocated chromatids (red). Right, frequency of end-to-end fusions in G7 splenocyte metaphase spreads, ≥150 metaphase spreads from three mice each, ± SEM. (d) Frequency of SFEs in splenocyte metaphase spreads from G7 mice and corresponding mice from single WT backcrossed (G5.BC1.iG1) or triple WT backcrossed (G5.BC3.iG1) backgrounds. Values are mean ± SEM (n=4). *P<0.05. All statistics by paired Student's t test.
To examine whether AUF1 was directly responsible for the telomere erosion phenotype, AUF1 was restored by backcrossing heterozygous with WT mice and telomere lengths examined. G5 Auf1+/- mice were backcrossed sequentially three times to pure WT mice, then the heterozygous progeny of the final backcross mated to produce a mixed litter of all three Auf1 genotypes (denoted G5.BC3.iG1). Q-FISH analysis of splenocytes from these mice found that progeny G5.BC3.iG1 WT and KO animals had similar frequencies of SFEs, whereas corresponding G7 KO mice had a 7-fold increase compared to WT littermates (Fig. 3d). The higher overall SFE frequency in G5.BC3.iG1 vs. G7 animals is a result of the increased generation number in these animals. Progeny from mice resulting from only one backcross to pure WT mice, G5.BC1.iG1, were not rescued in SFEs (Fig. 3d). However, reintroduction of AUF1 via three subsequent rounds of backcrossing (Fig. 3d) was sufficient to elongate critically short telomeres, but not the average telomere length in Auf1-/- progeny (Fig. S3). This was expected and is similar to studies in telomerase deficient mice, in which the shortest telomeres have been shown to be the preferential substrates for extension (Mili et al. 2001). Thus, AUF1 acts in a dose-dependent manner to maintain telomere length.
AUF1 deficiency promotes DNA damage foci at telomeres
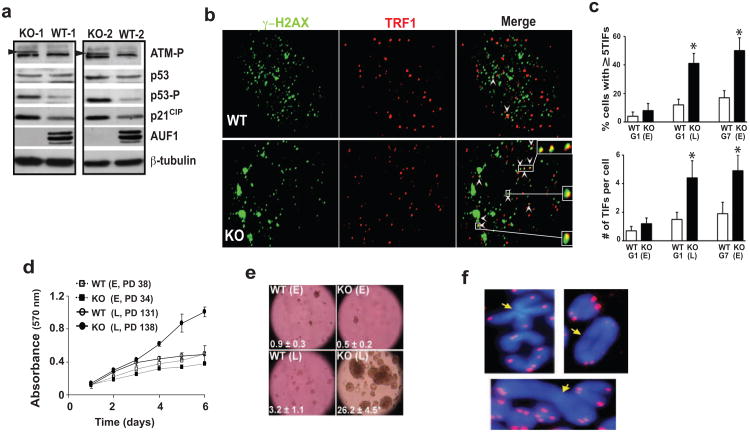
Dysfunctional short and uncapped telomeres activate p53-dependent DNA damage signaling responses which promote cellular senescence (d'Adda di Fagagna et al. 2003). Correspondingly, Auf1-/- MEFs show increased phosphorylation of ATM and p53, as well as increased p21CIP levels (Fig. 4a), at least partially through mRNA stabilization (Fig. 2d). We therefore investigated the DNA damage response at telomere ends as a measure of telomere dysfunction and contribution to senescence in Auf1-/- cells. Co-localization of DNA damage factor γ-H2AX with the telomere binding protein TRF1 at telomere ends in MEFs was quantified (Fig. 4b). A >3-fold increase in telomere dysfunction-induced foci (TIFs) was seen in late-passage G1 and early-passage G7 Auf1-/- MEFs (Fig. 4c). No significant difference in TIFs was found in early passage G1 WT or KO MEFs (Figs. 4c, S4c). Telomere end co-localization by TRF1 staining with DNA damage response factors 53BP1 or phosphorylated RAD17 was similarly increased in AUF1-/- MEFs (Fig. S4a-c). Thus, AUF1 maintains telomere length and integrity, with AUF1 deficiency resulting in increased DNA damage signaling at telomere ends associated with increased cell doublings or animal generational age.
Figure 4.
Increased DNA damage signaling at telomere ends and cell growth defects in Auf1-/- cells. (a) Levels of phospho-ATM (arrowhead), phospho-p53 (Ser15), and p21CIP determined by immunoblot analysis in two independent AUF1 WT and KO MEF cell lines. (b) Immunofluorescence staining for γ-H2AX and TRF1 co-localized at telomere ends. Arrowheads: telomere dysfunction induced foci (TIFs) (Takai et al. 2003). (c) Quantification of TIFs in MEFs from indicated generation, ≥30 nuclei from 3 MEF cell lines. Positive cells contain ≥ 5 TIFs. Values are mean ± SEM (n=3). P<0.01 by paired Student's t test. E, early passage. L, late passage. (d) MEF proliferation determined by MTT assay. E, early passage; L, late passage; PD, population doublings. (Values are P <0.01, ± SEM, n=4). (e) Anchorage-independent growth assayed by soft agar colony formation. Number of colonies shown. (f) Representative end-to-end chromosomal fusions lacking detectable telomere signal (arrow) in AUF1 KO splenocyte cell metaphase spreads stained with telomere-specific probes and DAPI counter-stained.
Telomerase deficiency promotes genomic instability (O'Hagan et al. 2002), resulting in increased cytogenetic abnormalities in older mice (Rudolph et al. 1999) and their cultured cells, a product of highly recombinogenic dysfunctional telomeres (Romanov et al. 2001). In contrast to WT and early passage Auf1-/- MEFs, proliferation of late passage Auf1-/- MEFs continued linearly with no evidence for contact inhibition (Fig. 4d) or anchorage dependence (Fig 4e). Extensive chromosomal abnormalities not seen in WT controls were evident in all late generation Auf1-/- splenocyte metaphase chromosome spreads, including fusions (Fig. 4f).
AUF1 binds the mTert promoter and activates transcription
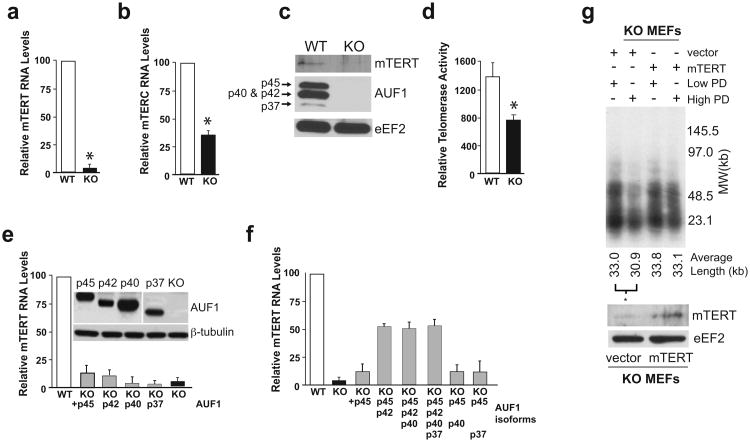
Loss of telomere length in Auf1-/- mice could indicate absence of an essential AUF1 function at telomeres or an inability to express functional telomerase in proliferating cells such as splenocytes or MEFs. We therefore investigated a role for AUF1 in expression of functional telomerase. In the absence of AUF1, mTERT RNA was reduced >20-fold and mTERC ∼3-fold compared to WT MEFs (Fig. 5a-b). AUF1 deficiency also greatly reduced levels of mTERT protein compared to wild type cells, as shown by immunoblot analysis (Fig. 5c). In addition, telomerase reverse transcriptase activity was impaired, as shown by a telomere repeat amplification protocol (TRAP) assay (Fig. 5d). The TRAP assay is an imprecise measure of telomerase activity, accounting for the difference in sensitivity between the two assays.
Figure 5.
Strongly decreased telomerase levels and activity in AUF1-deficient cells. (a) & (b) Relative mTert and mTerc RNA levels in WT and AUF1 KO MEFs, determined by qRT-PCR. *P<0.05 ± SEM, n=3. (c) Immunoblot showing mTERT protein levels in WT and KO MEFs. (d) Relative telomerase activity in extracts from WT and KO MEFs as determined by TRAP assay. Mean ± SEM of three independent experiments, *P<0.05. (e) Partial rescue of mTert mRNA expression in AUF1-/- (KO) MEFs by ectopic expression of individual AUF1 isoforms, determined by qRT-PCR. Representative immunoblot for AUF1 is shown. The p37 AUF1 panel was exposed 3× longer. (f) Rescue of mTERT mRNA expression by p45/p42 AUF1 isoforms in AUF1 KO MEF cells, carried out as described above. *P<0.05 ± SEM (n = 4). (g) Telomere restriction fragment analysis of serially-passaged KO MEF cells with or without ectopic mTERT expression. Autoradiograph indicates intensity of probe-hybridization to specific telomeric sequence. Typical results shown of 3 studies. Average telomere fragment length is indicated below. *P<0.05 by Student's t test, n=3.
Next, we determined which of the AUF1 protein isoforms is responsible for stimulating expression of mTERT mRNAs. All four AUF1 isoform cDNAs were cloned as independent expression vectors and tested singly and in combination in Auf1-/- MEFs for stimulation of mTERT and mTERC expression. Ectopic expression of p42 or p45 AUF1 isoforms stimulated expression of mTERT mRNA in Auf1-/- MEFs by ∼5-fold (15% of WT) (Fig. 5e). The p37 or p40 AUF1 isoforms did not stimulate mTERT mRNA expression at all. Co-expression of p45 plus p42 AUF1 increased mTERT mRNA expression levels by >17-fold (∼55% of WT) (Fig. 5f), whereas additional co-expression with p40 and/or p37 AUF1 isoforms had no effect. In contrast, no combination of AUF1 isoforms significantly stimulated expression of mTERC RNA levels (Fig. S5). Unlike p37 and p40 AUF1 isoforms, p45 and p42 AUF1 proteins have been shown to be more chromatin-associated (Mili et al. 2001), less involved in promoting rapid decay of AU-rich mRNAs, and to undergo less nuclear-cytoplasmic shuttling than the p37 and p40 AUF1 isoforms (Sarkar et al. 2003b; He and Schneider 2006), consistent with these results.
To determine whether decreased telomerase expression is the cause of telomere length erosion in Auf1-/- cells, mTert was stably expressed in Auf1-/-MEFs using retroviral transduction then cultures were serially passaged for >75 population doublings (PD). Genomic DNA was prepared from Auf1-/-cells expressing mTert or empty vector at both high (>75) and low (<5) serial passage cell population doublings. Telomere length was then measured by Telomere Restriction Fragment (TRF) analysis (Hsiao et al. 2006). Auf1-/- cells expressing only the empty vector demonstrated a significant decrease in average telomere fragment length in transition from low to high population doublings (33.0kb to 30.9kb, p<0.05), whereas with expression of ectopic mTert there was no statistical decrease in telomere length despite extensive cell population doublings (33.8kb to 33.1kb) (Fig. 5g). Importantly, the extensive telomere erosion in Auf1-/- cells can be prevented by complementation with ectopic mTert.
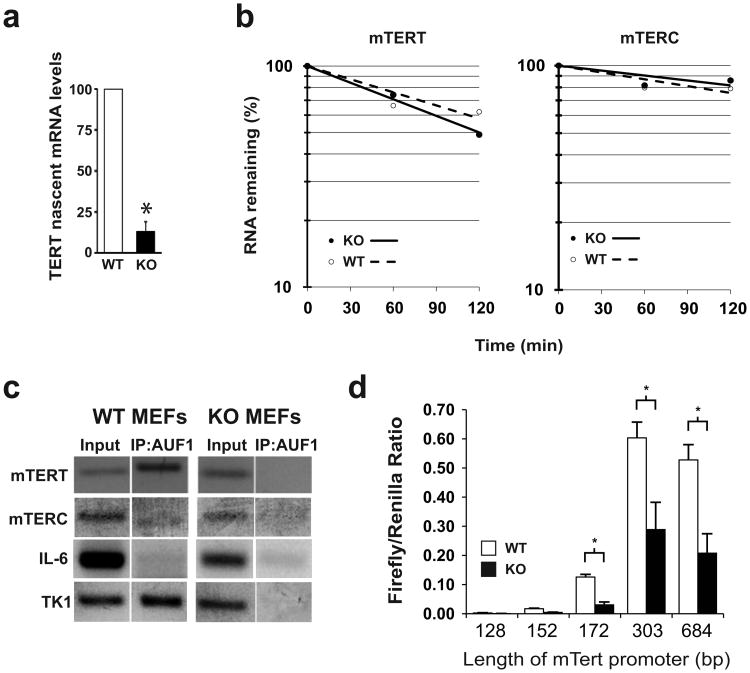
Mammalian telomerase (mTert) and its template RNA (mTerc) expression are well established to be controlled at the level of transcription (Greider 2006). We therefore measured mTert transcriptional activity by qPCR for nascent unprocessed introncontaining mTert transcripts. Auf1-/- MEFs had an 8-fold decrease in nascent mTert transcript levels as compared to WT (Auf1+/+) MEFs (Fig 6a). Analysis of mRNA decay rates showed no significant differences in stability of mTert or mTerc RNAs in comparison of WT to KO MEFs (Fig. 6b). We therefore next determined whether AUF1 is associated with promoter regions of mTert and mTerc genes by utilizing a chromatin immunoprecipitation (ChIP) assay (Fig. 6c). The mTert promoter region was strongly detected by PCR amplification of purified AUF1-bound chromatin, similar to thymidine kinase-1(Tk1), a reported AUF1-interacting promoter (Lau et al. 2000). In contrast, whereas the promoter of an established AUF1 non-responsive gene, cytokine IL-6 (Lu et al. 2006), was not detectably bound by AUF1 (Fig. 6c). The mTerc promoter was weakly associated with AUF1.
Figure 6.
AUF1 stimulates mTERT transcription. (a) Nascent mTert transcript levels determined by intron-exon qRT-PCR. *P<0.05. b) Decay rates of mTert and mTerc RNAs. Plots are the average of three independent experiments. (c) Promoter regions of mTert, mTerc, IL-6 and Tk1 genes were detected by semi-quantitative PCR from input and AUF1-bound immunoprecipitated chromatin samples from WT and KO MEFs. Representative negative images from ethidium-bromide stained agarose gels are shown. (d) Promoter-coupled luciferase constructs in WT and KO MEFs from mTert expression. Firefly luciferase activity was normalized to Renilla luciferase activity of a co-transfected null-promoter plasmid. Length of mTERT promoter in each construct is indicated. *P<0.05, ± SEM.
Several previous reports have identified the proximal region of ∼333 base-pairs of the mTert upstream region (relative to the initiating ATG codon) as the core transcriptional promoter (Nozawa et al. 2001; Si et al. 2011). Luciferase transcriptional reporter assays in Auf1+/+ and Auf1-/- MEFs using constructs containing different lengths of the mTert promoter confirmed that the proximal region of 303 base-pairs is responsible for basal mTert transcription (Fig. 6d). Adding additional upstream sequences did not increase transcriptional activity. Sequential deletions of portions of this region sharply decreased resultant mTert promoter transcription activity, suggesting that transcriptional activation by AUF1 occurs by binding within this core region (Fig. 6d). Finally, AUF1-/- MEFs show significantly reduced transcriptional activity compared to wild type MEFs for all constructs containing the core promoter region, indicating that AUF1 is responsible for the increased activity (Fig. 6d). Together with the ChIP data showing AUF1 binding to the mTert promoter region, we can conclude that AUF1 is a transcriptional activator of the mTert gene
Discussion
Aging at the organismal level is a multifaceted process involving many molecular pathways (Rubinsztein et al. 2011). In our current work, we show that AUF1, an RNA-binding protein/cytokine mRNA destabilizing protein, regulates several aging-associated pathways by two largely distinct mechanisms of action. Auf1-/- mice have increased levels of p16ink4a, p19Arf, and p21CIP cell-cycle regulator proteins and mRNAs, a result of failure to carry out rapid ARE-mRNA decay Fig. 2d). An increased level of p16ink4a in particular is a robust marker of mammalian aging (Krishnamurthy et al. 2004) and increased p16ink4a protein accumulation is associated with reduced cell and tissue viability (Krishnamurthy et al. 2004; Molofsky et al. 2006; Liu et al. 2011). Interestingly, it has been noted that human diploid fibroblasts have reduced levels of AUF1, particularly the p37 isoform which is best associated with ARE-mRNA decay, as well as decreased AUF1-p16ink4a mRNA interaction as fibroblasts approach replicative senescence (Wang et al. 2005). These data and those reported here suggest that an age-dependent decrease in p37 AUF1 levels may be an important mechanism by which p16ink4a levels increase in age-dependent senescence. Although recent work that examined AUF1 levels in human tissue sections by immunohistochemistry showed similar levels of AUF1 staining in both young and old tissues (Masuda et al. 2009), the different AUF1 isoforms could not be distinguished by this approach.
Aging is also associated with and thought to be partially caused by telomere shortening (Donate and Blasco 2011), as shown in hematopoietic niches (Allsopp et al. 2003). In our work, loss of AUF1 resulted in severely reduced average telomere length in splenocytes and other proliferative cells, as well as an inability to maintain the shortest telomeres (Figs. 3, S3). One allele of Auf1 was found to protect against development of critically short telomeres but was not sufficient to maintain average telomere length in successive animal generations or cell doublings, again similar to mice heterozygous for mTert or mTerc genes (Hemann et al. 2001; Erdmann et al. 2004; Greider 2006). Furthermore, we showed that AUF1 stimulates mTERT levels by activation of mTert transcription (Fig. 5, 6a,c,d), which is consistent with previous work identifying transcription as a primary regulatory point for control of mammalian telomerase expression (Greider, 2006). Interestingly, the AUF1 isoforms responsible for mTert transcriptional regulation, p42 and p45, differ from those involved in p16ink4a accelerated mRNA decay (p37, p40), supporting the hypothesis that there are distinct molecular roles for the four related AUF1 isoforms.
Previous studies demonstrated a link between inflammatory diseases, increased telomere erosion and DNA damage signaling (Browner et al. 2004; Andrews et al. 2010; Sahin and Depinho 2010). In this regard, AUF1 deficiency in mice causes an increase in pruritic skin inflammation (Sadri and Schneider 2009) and increased susceptibility to endotoxic shock upon LPS challenge (Lu et al. 2006). Our findings provide a physiological basis by which AUF1, well established as an intrinsic factor to attenuate inflammation, also prevents development of cellular senescence, loss of telomere maintenance and therefore regulates the mammalian aging process.
It is presently unclear how AUF1 stimulates mTert transcriptional expression. Structural studies imply that AUF1 might interact with G-quadruplex DNA structures (Enokizono et al. 2005), which are strongly enriched in certain promoter sequences (Brooks et al. 2010) including that of mTert (Palumbo et al. 2009). Unlike the mTert promoter, the mTerc promoter is not strongly enriched in putative G-quadruplex DNA (Brooks et al. 2010). Current work is investigating whether AUF1 interacts with the G-quadruplex and other promoters with similar motifs.
Experimental Procedures
MEFs
Mouse embryonic fibroblasts (MEFs) were generated from day 13.5 embryos from Auf1-/-and wild-type mice on a mixed B6;129S6 background, cultured in DMEM supplemented with 10% fetal bovine serum, and immortalized using the 3T3 protocol (Todaro and Green 1963). Immortalized MEFs were split at 1:10 dilution when confluent.
Plasmids, transfections and retrovirus transduction
Murine embryonic fibroblasts were transduced in the presence of polybrene (4μg/mL) with virus generated by co-transfection by CaPO4 method of one of the four AUF1 isoform constructs (pBabe-p37, pBabe-p40, pBabe-p42, pBabe-p45), pCMV-VSVG, and pCMV-Tat (gifts of D. Ron, NYU School of Medicine) into 293GP cells (gift of M. Pagano, NYU School of Medicine) which were harvested 48 and 72 h post transfection. The AUF1-containing pBabe constructs were generated by cloning each isoform PCR product into BamHI and SalI sites of pBabe-puro. 48 h post transduction the isoform expressing cells were selected with 2 μg/mL puromycin (InvivoGen). For ectopic mTERT expression, AUF1-/- KO MEFs were transduced and selected as above using the mTert-pBABE-puro plasmid (gift of J. Bidwell and F. Pavalko, Indiana University School of Medicine)
Murine embryonic fibroblasts were transfected with pFLAG-CMV-2 AUF1 constructs (Sarkar et al. 2003a; Sarkar et al. 2003b) using the TransIT-LT1 Transfection Reagent according to the manufacturer protocols (Mirus). Cells were harvested at 48 h post-transfection and RNA isolated as described below.
For Luciferase transcription quantification, plasmids were constructed by PCR-cloning various lengths of mTert promoter sequence from the full-length promoter-containing 5k- mTert-EGFP construct (gift of A, Gutiérrez-Adán, INIA, Spain) and inserting PCR products into the SacI and NheI sites of pGL3-Basic vector (Promega). WT and KO MEFs were transfected using the TransIT-LT1 Transfection Reagent according to manufacturer protocols (Mirus) with one of the five pGL3-mTert constructs (pGL3-mTert128, pGL3-mTert152, pGL3-mTert172, pGL3-mTert303, and pGL3-mTert684) and null Renilla luciferase plasmid pRL-null (Promega). Transfected cells were harvested at 48 h post-transfection with Passive Lysis Buffer (Promega) and assayed with Dual-Luciferase Reporter Assay System (Promega) according to manufacturer protocols.
Histology, Immunohistochemistry, and SA-β-gal staining
For histological analysis of tissues, organs and tumors, tissues were processed and stained with hematoxylin and eosin as described (Lu et al. 2006). For immunohistochemistry, frozen sections were stained according to standard protocols using antibodies to p16 (JC2, Lab Vision) and p21 proteins (ab9, Lab Vision) and the Vectastain ABC kit (Vector Labs). For cellular senescence analysis, frozen tissue sections and MEFs were processed and stained with the senescence marker β-galactosidase staining kit (Cell Signaling) according to manufacturer protocols.
Q-FISH analyses
Q-FISH was performed as previously described (Hsiao et al. 2006). Splenocyte single-cell suspensions were made by mincing samples through a 70-μm cell strainer (BD Biosciences). Splenocytes were cultured in DMEM + 10% fetal bovine serum (FBS) and were activated with lipopolysacharide (LPS) (5 μg/ml; Sigma), phorbol-12-myristate-13-acetate (2 μM; Calbiochem), and F(ab')2 anti-IgG antibody (1 μg/ml; Jackson ImmunoResearch) for 36 h before cells were arrested in metaphase by 6 h treatment with nocodazole (0.1 μg/ml; Sigma). Cells were collected, metaphase spreads were prepared and stained as described (Dynek and Smith 2004). Images were acquired on a Zeiss Axioplan 2 microscope with a Photometrix SenSyn camera. Photographs were processed using OpenLab software. TFL-TELO software (BC Cancer Agency) was used to quantify the telomere signal from at least 10 metaphase spreads for each sample.
Immunoblot analysis
Immunoblot analysis was performed according to standard protocols using ECL detection (PerkinElmer). Quantification was conducted by densitometry. Antibodies to the following proteins were used: p53 (no. 2524), phospho-p53 (no. 9284), p27KIP (no. 2552), and eEF2 (no. 2332) from Cell Signaling; phospho-ATM (ab2888) and p21CIP (ab7960) from Abcam; Rb (14-6743) from eBiosceince; β-tubulin (D66) from Sigma; mTERT (sc-7212) from Santa Cruz, and AUF1 (PcAb 995 antibody).
Immunofluorescence
Primary or immortalized MEFs grown on gelatin-coated coverslips were washed with cold phosphate buffered saline (PBS), fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 and then blocked in 5% normal mouse serum. Cells were stained with primary antibodies to TRF-1 (E-15; Santa Cruz), γ-H2AX (no. 2577; Cell Signaling), 53BP1 (nb 100-304, Novus Biologicals), and Phospho-Rad-17 (no.3421; Cell Signaling) in 5% normal mouse serum. Primary antibodies were detected with FITC-anti-rabbit antibody and TRITC-anti-goat antibody from Jackson ImmunoResearch. Cells were mounted using Vectashield medium containing DAPI (Vector Laboratories). Samples were examined and photographed using a Zeiss LSM510 Meta confocal microscope using an oil immersion 100× objective. For each cell examined, z-stacks of 0.5 μm were made throughout the entire nucleus to avoid bias and the total number of co-localized foci were counted.
MTT and soft agar assay
MTT assay was performed according to a previously described method (Mosmann 1983). In brief, MEFs were seeded in quadruplet at 103 cells per well in 96-well plates. At indicated times post-seeding, cultures were incubated with MTT (0.5 mg/ml; Sigma) for 4 h and then the crystals in each well were dissolved with 100 ml of dissolving buffer (10% SDS, 45% isopropanol, and 0.04 N HCl) for 4 h. The absorbance of each well was measured on a Sunrise (Tecan) microplate reader at a wavelength of 570 nm. For soft agar assays, 5 × 104 immortalized MEF cells were suspended in 1 ml of 0.3% agar in DMEM supplemented with 10% FBS and plated on 6-well plates containing a solidified bottom layer (1.5 ml of 0.5% agar in DMEM + 10% FBS). 1 ml of 0.3% agar in DMEM + 10% FBS was added every 3 days, and colony counts and pictures were taken on day 12.
ChIP analysis
Conventional chromatin immunoprecipitation was performed as previously described (Ren et al. 2002) on WT and KO MEFs. Immunoprecipitated DNA fragments were assayed via semi-quantitative PCR and products visualized by agarose gel-electrophoresis. Primer sequences are available upon request.
Splenic B and T cell proliferation assays
Splenic B cells were purified using the MACS CD45R microbead isolation kit and were verified to be ≥98% B220+ B cells by FACS analysis. Splenic T cells were purified using the MACS Pan T Cell Isolation kit and similarly verified by FACS to be ≥98% CD3+ T cells.
For proliferation analysis, 2 × 104 cells were plated in triplicate in 96 well plates and stimulated with 10 μg/ml lipopolysacharide (LPS) for 48 hours, pulsed for the last 8 hours with 1 μCi/well of 3H-thymidine. Radioactivity incorporated into DNA was quantified by scintillation counting.
Telomere Repeat Amplification Protocol (TRAP) Assays
TRAP assays were performed using equal numbers of WT and KO MEFs employing the TeloTAGGG Telomerase PCR ELISA Plus kit (Roche) according to manufacturer instructions.
Telomere Restriction Fragment (TRF) Analysis
For telomere restriction fragment analysis, equal numbers of indicated cell lines were embedded in agarose plugs and their DNA digested with HinfI and RsaI, DNA fragments were then separated by pulsed-field gel electrophoresis (CHEF DR-II apparatus, Bio-Rad), and telomere restriction fragments were detected by in-gel blotting with a 32P-labeled TTAGGG probe. Average telomere length was calculated from autoradiographs using ImageJ (NIH) and methods previously described (Haussmann et al. 2011).
Statistical analysis
All data are shown as means ± standard deviations (s.d). Two-sided Student's t-test was used to compare experimental groups. Survival was analyzed using the Kaplan-Meier method and the log-rank test was used for group comparisons. Other statistical analyses are described in the text.
Supplementary Material
Highlights for Pont et al.
AUF1 deficient mice undergo telomere erosion
AUF1 suppresses cellular senescence
AUF1 stimulates telomerase expression and normal aging
AUF1 links telomere maintenance to reduced inflammation
Acknowledgments
This work was supported by NIH grant GM R01 GM085693-03 to R.J.S. N.S and A.P. were supported by NIH grant T32 T32AI007180.
Footnotes
Author Contributions: N.S., A.R.P. and R.J.S. designed and interpreted the experiments. H.J.H. and S.S. helped design telomere studies and interpret the results. N.S. and A.R.P. conducted all studies. N.S., A.R.P. and R.J.S. wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allsopp RC, Morin GB, Horner JW, DePinho R, Harley CB, Weissman IL. Effect of TERT over-expression on the long-term transplantation capacity of hematopoietic stem cells. Nature Med. 2003;9:369–371. doi: 10.1038/nm0403-369. [DOI] [PubMed] [Google Scholar]
- Andrews NP, Fujii H, Goronzy JJ, Weyand CM. Telomeres and immunological diseases of aging. Gerontology. 2010;56:390–403. doi: 10.1159/000268620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browner WS, Kahn AJ, Ziv E, Reiner AP, Oshima J, Cawthon RM, Hsueh WC, Cummings SR. The genetics of human longevity. Am J Med. 2004;117:851–860. doi: 10.1016/j.amjmed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- Donate LE, Blasco MA. Telomeres in cancer and ageing. Phil Trans Royal Soc London Series B. 2011;366:76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- Erdmann N, Liu Y, Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc Natl Acad Sci USA. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb Symp Quant Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Hao LY, Armanios M, Strong MA, Karim B, Feldser DM, Huso D, Greider CW. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Haussmann MF, Salomons HM, Verhulst S. Telomere measurement tools: Telometric produces biased estimates of telomere length. Heredity (Edinb) 2011;107:371. doi: 10.1038/hdy.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadinanos J, Horner JW, Maratos-Flier E, Depinho RA. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2010;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JS, Baumeister P, Kim E, Roy B, Hsieh TY, Lai M, Lee AS. Heterogeneous nuclear ribonucleoproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J Cell Biochem. 2000;79:395–406. doi: 10.1002/1097-4644(20001201)79:3<395::aid-jcb50>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Liu Y, Johnson SM, Fedoriw Y, Rogers AB, Yuan H, Krishnamurthy J, Sharpless NE. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Marasa B, Martindale JL, Halushka MK, Gorospe M. Tissue-and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging (Albany NY) 2009;1:681–698. doi: 10.18632/aging.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Shu HJ, Zhao Y, Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nozawa K, Maehara K, Isobe K. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J Biol Chem. 2001;276:22016–22023. doi: 10.1074/jbc.M011181200. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon SS, Martens UM, Ward RK, Lansdorp PM. Telomere length measurements using digital fluorescence microscopy. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Sadri N, Schneider RJ. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J Invest Dermatol. 2009;129:657–670. doi: 10.1038/jid.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003a;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol. 2003b;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si SY, Song SJ, Zhang JZ, Liu JL, Liang S, Feng K, Zhao G, Tan XQ. Cloning of mouse telomerase reverse transcriptase gene promoter and identification of proximal core promoter sequences essential for the expression of transgenes in cancer cells. Oncol Rep. 2011;26:377–382. doi: 10.3892/or.2011.1303. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.