Abstract
Activation of steroid receptors results in global changes of gene expression patterns. Recent studies showed that steroid receptors control only a portion of their target genes directly, by promoter binding. The majority of the changes are indirect, through chromatin rearrangements. The mediators that relay the hormonal signals to large-scale chromatin changes are, however, unknown. We report here that APRIN, a novel hormone-induced nuclear phosphoprotein has the characteristics of a chromatin regulator and may link endocrine pathways to chromatin. We showed earlier that APRIN is involved in the hormonal regulation of proliferative arrest in cancer cells. To investigate its function we cloned and characterized APRIN orthologs and performed homology and expression studies. APRIN is a paralog of the cohesin-associated Pds5 gene lineage and arose by gene-duplication in early vertebrates. The conservation and domain differences we found suggest, however, that APRIN acquired novel chromatin-related functions (e.g. the HMG-like domains in APRIN, the hallmarks of chromatin regulators, are absent in Pds5). We show that in interphase nuclei APRIN localizes in the euchromatin/heterochromatin interface and we also identified its DNA-binding and nuclear import signal domains. Our results indicate that APRIN, in addition to its Pds5 similarity, has the features and localization of a hormone-induced chromatin regulator.
Keywords: APRIN, hormonal regulation, proliferative arrest, chromatin, cancer, HMG domain, AT-hook, Pds 5
1. Introduction
The ability of cells to undergo proliferative quiescence is critical in development and it emerged in multicellular organisms [1]. A major component of its regulation is hormonal [2, 3] and it is part of the differentiation program in reproductive tissues [4, 5]. Steroid hormones play a critical role in differentiation and induce massive changes in gene expression patterns. Microarray analysis showed that approximately 3.2- 4.3% of the genes tested were hormone regulated, both in vitro [6] and in vivo [7]; these changes were also observed at the protein level [8]. Consequently, androgen regulation may affect a total of ∼ 1,200-4,000 genes in the human genome, on the basis of ∼28,000-30,000 coding- and ∼100,000 non-coding genes by recent estimates [9]. In contrast, analyses of receptor-promoter complexes and confocal microscopy detected only 250-300 androgen receptor sites per nucleus [10] and only a handful has been characterized [7].
These data together with recent chromatin immune-crosslinking analyses strongly suggest that steroid receptors control only a portion of the target genes directly [11]. The global changes in gene expression patterns are mostly the results of indirect mechanisms [12] including non-genomic membrane receptor effects and chromatin changes. The long-term, differentiation-related hormonal changes, however, argue against the participation of plasma membrane-based mechanisms because molecules involved in these pathways reach a peak in a few minutes and their effects last only for a couple of hours [13]. Chromatin involvement, on the other hand, was confirmed by receptor localization studies [14, 15], receptor interaction assays [16] and by detection of global rearrangements of the chromatin architecture [17]. Hormone-induced chromatin changes, in turn, can regulate the expression of large sets of genes [18]. The mediators and the mechanisms that translate the hormonal signals into large-scale chromatin changes are mostly unknown.
To explore these mechanisms, we used the hormone-sensitive LNCaP cell line [19, 20, 21] in which the transition to hormone-induced proliferative arrest [22, 23] has been shown to coincide with chromatin changes [17]. We isolated proliferation arrest-specific genes [23] and showed that a newly identified gene, APRIN (formerly AS3) has a critical role in proliferative arrest in the G0/G1 phase of cell cycle [24, 25]. We found that loss of heterozygosity (LOH) in the D13S171 marker within the APRIN genomic sequence [26, 27] linked this protein to a variety of cancers, including prostate cancer [28-31]. The molecular mechanisms of the APRIN-mediated proliferative arrest and its association with cancer, however, remain unknown.
A commonly used approach to assess the molecular mechanisms of proteins is to establish their functional domains [32, 33]. Phylogenetic sequence analysis is a conservation-based method to identify functional domains and related proteins [34, 35]. Conservation correlates with functional significance and identifies biologically relevant structural units [36]. We cloned and sequenced human and rodent ortholog APRIN cDNAs, computed the local conservation differences within small subdomains and established putative functional units. APRIN shares similarities with the ancient Pds5 (precocious dissociation of sister chromatids) gene lineage. Proteins of this family are involved in chromatid cohesion, so a cohesion-related function was proposed for APRIN [37]. The physical association of APRIN with chromatin has been demonstrated, but its cohesin association showed low affinity and a mechanistic role in cohesion remains to be confirmed [38].
The domain and structural differences we report here, however, turned the APRIN functional model to a new direction. We show that APRIN is only a paralog, and it diverged from the Pds5 lineage by gene duplication. The accumulated conservation differences, domain acquisitions in APRIN and its chromatin localization indicate a new functional entity that shares features with chromatin architectural regulators.
2. Materials and Methods
2.1. Tissue culture and cell lines
The MCF7-AR1 cell line, a human androgen receptor-transfected MCF7 cell line was established in this laboratory [39]. This cell line and the LNCaP human prostate cancer cells were routinely maintained in DMEM (Dulbecco's modified Eagle's medium) medium supplemented with 5% fetal bovine serum (FBS). We kept the cells in 5% hormone-stripped FBS media for 3 days and treated them with 10 nM methyltrienolone (R1881) (NEN Life Science Products, Boston, MA), a synthetic androgen. For transfection experiments we used Lipofectamine 2000 following the manufacturer's instructions (Invitrogen, San Diego, CA).
2.2. Computer methodology
For similarity searches we used GenBank softwares (PairwiseBlast, Nucleotide Blast and Protein Blast) and the GCG program package (Genetics Computer Group, Madison, WI) including BestFit, GrowTree, Frames and Translate programs; for motif search, the Motifs, ProfileScan, CoilScan, and the HTHScan programs were used. In the alignment BLASTN program (version BLASTN 2.2.6), the default values were used in the score calculations: a) 1 and -2 matrix values for match and mismatch, respectively; b) for scoring gap penalties 5 and 2 were used for existence and extension; c) the x_dropoff value was 15 and d) for expect number and wordsize values we used 10 and 11, respectively. For domain search we used the HmmerPfam program and other web-based resources (www.sanger.ac.uk/Software/Pfam/).
2.3. Isolation and cloning of the mouse and rat APRIN cDNAs
We used RT-PCR methodology to isolate the full length mouse and rat APRIN cDNA open reading frames. Two-step nested primers were designed using mouse-and rat-specific sequence information generated by our database searches. For amplification of the mouse APRIN, the following primers were used: mF1: 5′gaggggtacagacatttccatcatg, mF2: 5′atggctcattcaaagacaaggaccaac, mR1: 5′aatagaagttaatgacgtgttcatcg, mR2: 5′gttcatcgtctctctcgtttggag. For rat APRIN amplification the following primers were used: rF1 (same as mF1), rF2: 5′atggctcattcaaagacaagaaccaacg, rR1: 5′cataaagaaagttaatggcatgttcatc, rR2: 5′gcatgttcatcgtctctctcgtttgg. We used Elongase (Invitrogen) to amplify Marathon-Ready cDNA preparations from mouse and rat brains, respectively (Clontech, Palo Alto, CA). The PCR products (4340 bp for the mouse and 4334 bp for the rat) were cloned into the pCRII vector (Invitrogen) and sequenced in the Automatic Sequencing Core Facility at Tufts University.
2.4. Antibody reagents and expression studies
We generated two affinity purified polyclonal anti-APRIN antibody reagents. The first was raised against an oligopeptide between positions 1370-1387 (anti-APRIN-1370) [24]. Subsequent functional analyses revealed, however, that the epitope overlapped with the second HMG-like (High Mobility Group) DNA binding domain (AT-hook) and potentially interfered with APRIN expression studies and DNA binding assays. The recently developed second antibody targeted an epitope between positions 1434-1447 and was also affinity purified (anti-APRIN-1434). Both antibodies recognized their epitopes in both rat [25] and mouse APRIN proteins. Protein extractions and Western blots were performed according to standard procedures [25] using ventral and dorsal prostate lobes from 18 week old CD-1 mice. For immunohistochemistry analyses, cell cultures were seeded on coverslips, fixed in cold 50% methanol-50% acetone for 10 min and permeabilized by 0.1% Triton X100 for 10 min. Formaldehyde-fixed, paraffin-embedded prostate sections were treated for antigen retrieval using sodium citrate buffer [25] and sequentially reacted with anti-APRIN-1370 antibody (1:100), biotin-conjugated goat anti-rabbit IgG (1:500) (Zymed Laboratories, San Francisco, CA) and streptavidin-peroxidase conjugate with diaminobenzidine (DAB) as chromogen (Sigma, St Louis, MO). Harris' hematoxylin was used as counterstain.
2.5. FLAG-fusion expression studies
The APRIN open reading frame was cloned in frame with the N-terminal FLAG epitope in the EcoRI-BamHI sites of the pFLAG-CMV vector (Eastman-Kodak, Rochester, NY). The MCF7-AR1 cells were seeded on coverslips and transfected as described above. A C-terminal-truncated APRIN protein (a stop codon upstream of the nuclear localization signal in domain 8, not shown) was also transfected and expressed. Twenty four hours later the cells were fixed (methanol:acetone, 1:1) and stained using 1:300 dilution of anti-FLAG monoclonal antibody (Eastman-Kodak, Rochester, NY). Biotin-conjugated anti-mouse secondary antibody (1:400 dilution) was visualized using FITC-streptavidin for confocal analyses (propidium iodide counterstain), or Alexa 594-streptavidin for fluorescent microscopy (Hoechst 33258 counterstain). Fluorescence microscopy (NIKON Eclipse E400) and confocal analyses (Leica, TCS SP2) were performed at the Tufts University Core Facility.
2.6. APRIN immunohistochemistry for chromatin localization studies
The MCF7-AR1 cells were grown on coverslips as described above. After 3 days in 5% hormone-stripped FBS media, the cell were treated with 10 nM R1881 for two days and harvested. After fixation and permeabilization as above, we used anti-APRIN-1434 in 1:2000 dilution following standard procedures. The staining was developed by using Alexa 488-conjugated anti-rabbit secondary antibody (1:500 dilution) (Invitrogen, Molecular Probes) and visualized using fluorescent microscopy.
2.7. APRIN expression in human prostate cancer
To study APRIN downregulation and the changes in the nuclear chromatin architecture, archived paraffin embedded human prostate cancer samples were stained with the anti-AS3 antibodies using standard methodology as described above. The prostate tissue samples were obtained by the approval of the local Institutional Review Board following Informed Consent and HIPAA regulations.
3. Results
3.1. Cloning, sequencing and genomic mapping of the mouse APRIN
Conservation analysis of the human APRIN protein required ortholog APRIN sequences from other species. We identified the mouse APRIN genomic sequence in a PAC clone (exons 1-11 on pPAC417G6, see Figure 1 panel A) and from a supercontig in the mouse database (Mm5_WIFeb01_97, Acc # NW_000249). We used the genomic sequence to design mouse-specific primers and isolated the physical cDNA of APRIN (Figure 1 panel B). A major open reading frame (between position 147 and the UGA stop codon at position 4491) is indicated. The complete mouse APRIN cDNA sequence is available under GenBank accession number AY102267.
Figure 1.
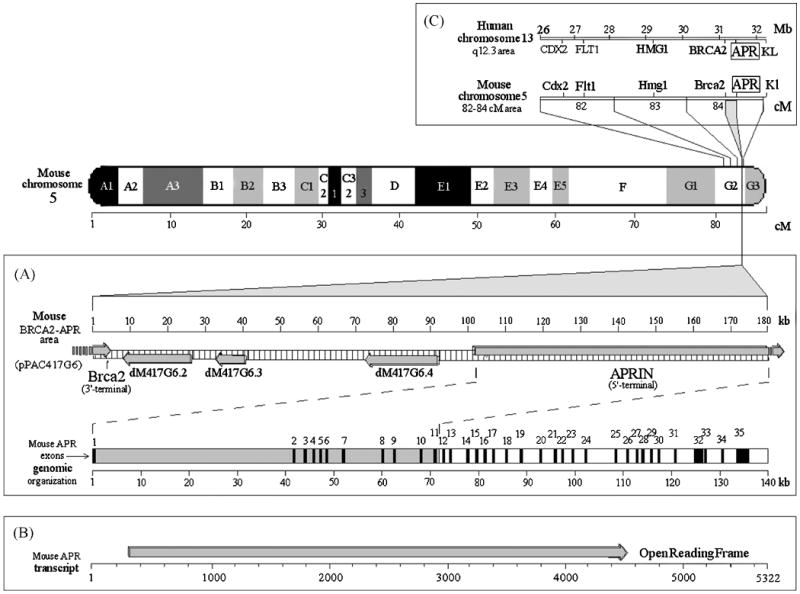
The chromosomal, contig and exon maps of the mouse APRIN gene. Mouse chromosome 5 is depicted with G-banding, the scale is in centimorgans (cM). Panel A, The full pPAC417G6 clone map (“mouse BRCA2-APR area” map line), the identified genes are depicted on a kilobase scale. The arrows indicate the direction of transcription. Exons were mapped on the mouse genomic supercontig, Mm5_WIFeb01_97. Panel B, APRIN mRNA and the open reading frame are shown. Panel C, the human and mouse APRIN chromosomal areas are compared (human chromosome 13 q12.3 and the corresponding mouse chromosome 5 region, 82-84 cM). The corresponding gene names and positions are indicated. Positions are given in million base pair units in the human chromosome segment and in centimorgan units in the mouse chromosome 5 segment
The PAC417G6 clone also showed a genomic link between the genes for APRIN and Brca2 in the mouse (Figure 1 panel A). Brca2 had been mapped on mouse chromosome 5 [40]. In our human studies APRIN was mapped between BRCA2 and the KL (Klotho gene) on (human) chromosome 13 [27, 41], (Figure 1). By using other genes in the region (CDX2, FLT1), the mouse gene was mapped at the 84 cM or G2 area on the long arm (q) on mouse chr. 5 and established that this area is syntenic with human chr. 13 (Figure 1 panel C) [42]. By using the cDNA we also identified the correct mouse APRIN exon pattern (Figure 1 panel A and Table 1). In contrast to the highly conserved exons, intron sequences are significantly reduced in size in the mouse (∼140 kbp v.∼200 kbp in human).
3.2. Expression and nuclear localization of the mouse APRIN gene product
Western blot analyses with both anti-APRIN antibodies we generated detected a 158-162 kDa polypeptide band from the mouse prostate (Figure 2, panel a), identical in size to the human [24] and the rat APRIN proteins [25]. Immunohistochemistry on mouse prostate sections revealed the predominantly nuclear localization of the APRIN protein in epithelial cells (Figure 2, panels b and c) and in most of the basal and fibromuscular stromal cells. APRIN expression levels in the normal prostate epithelial cells were uniformly high and nearly homogeneous.
Figure 2.
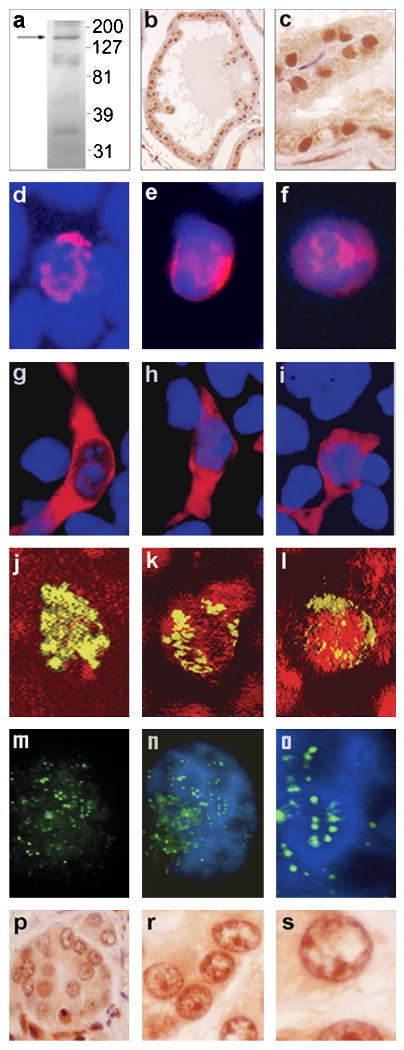
APRIN protein expression, nuclear transport and chromatin localization. Panel “a”, Western blot analysis of mouse prostate proteins using the anti APRIN-1370 antibody. The protein markers on the right are in kDa, the arrow indicates the mouse APRIN protein band. Panels “b” and “c”, anti-APRIN-1370 immuno histochemistry of mouse prostate sections, peroxidase-DAB staining, 100× (“b”) and 400× (“c”) magnifications. Panels “d”-“i”, expression and nuclear localization of transiently transfected FLAG-tagged APRIN protein. Panels “d”-“f”, nuclear immunofluorescence of MCF7-AR1 cells, transfected with an N-terminal FLAG-tag APRIN fusion construct. They were stained with mouse anti-FLAG and Alexa 594-labeled secondary antibody (red). Hoechst33258 was used as counterstain (blue) (2000×). Panels “g”-“i”, transfections with C-terminal truncated APRIN constructs (cytoplasmic). The host cell line and staining were the same as in the previous panels (1000×). Panels “j”-“l”, confocal microscopy of MCF7-AR1 cell nuclei transfected with the N-terminal FLAG-tag APRIN fusion construct. The anti-FLAG/FITC-labelled secondary antibody staining is green and the DNA counterstaining with propidium iodide is red. The overlap of FLAG expression and DNA staining indicates co-localization of APRIN with the nuclear DNA (yellow). The three panels show various compartmentalization patterns of the APRIN-FLAG fusion protein in the nucleus (2000×). Panels “m”-“o”, high resolution fluorescence immunohistochemistry of the native APRIN protein using the anti APRIN-1434 antibody. A single MCF7-AR1 nucleus is shown (6000×). Panel “m”, Anti APRIN-1434 only (green). Panel “n”, colocalization of APRIN with the chromatin. The DAPI counter-stain (blue) labels the heterochromatin. Panel “o”, the middle section of panel “n” is electronically magnified to show APRIN colocalization with the euchromatin/ heterochromatin interface. Panels “p”-“s”, APRIN expression in vivo, in prostate cancer. The panels depict formaldehyde-fixed, paraffin-embedded human prostate tissue sections in 400×, 1000× and 2000× magnifications, respectively. The sections were stained with anti-APRIN-1434 and with anti-rabbit peroxydase-DAB, using hematoxylin counter staining. The left panel shows a disorganized acinus in neoplasia, the middle and right panels show the same nuclei in increasing magnifications.
3.3. Unique high conservation in the polypeptide sequence among ortholog APRIN genes
Sequence comparisons between the human and murine APRIN polypeptide sequences showed 96 % amino acid identity. The two species diverged about 80-100 million years ago [43, 44] so the enormous conservation during ∼150 million generations (∼12 weeks per mouse generation) was intriguing. To assess the general conservation levels between mouse and human proteins, we calculated the conservation of 38 other proteins representing various functional groups. The results are depicted in Figure 3 in a tagged graph format. The results indicate that some of the chromatin-, structural- and differentiation-related proteins are extremely conserved, in the 95-100% range. These proteins are usually short, in average 200-300 residues. APRIN, however, stands out in this group by its complexity (1447 amino acids) and shows higher conservation than some of the most ancient histones, tubulins and cyclophilins. Elements of androgen signaling, prostate specific proteins, cell-cycle, chromatin remodeling, etc. are typically conserved at the 80-86 % level or less. The extreme conservation of APRIN, particularly considering its size, may indicate that APRIN has some fundamental functions beyond its role in androgen signal transduction.
Figure 3.
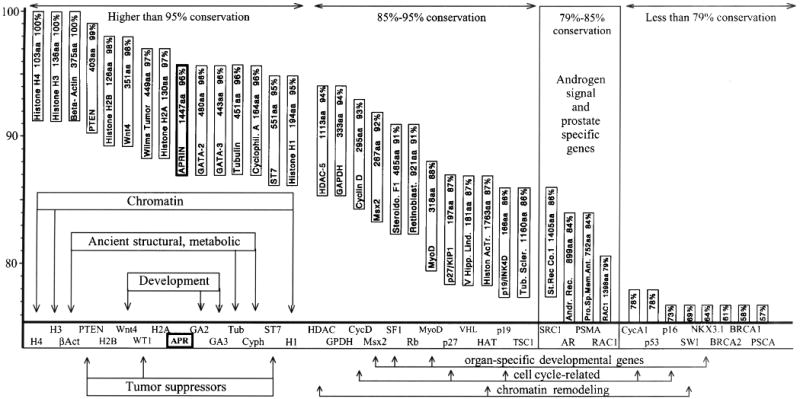
Protein conservation analysis between mouse and human proteins; APRIN is highly conserved. The positions of the boxes reflect the percentage of conservation between mouse and human in a decreasing order. The gene-boxes are aligned with the conservation scale on the y-axis. The x-axis shows the abbreviations of the names of genes. The boxes also indicate the sizes of the polypeptides in amino acids and the percent identities between the murine and human sequences. Groups of genes of particular functions are indicated, APRIN is highlighted.
3.4. Conservation differences identified the putative functional domain map of the APRIN protein
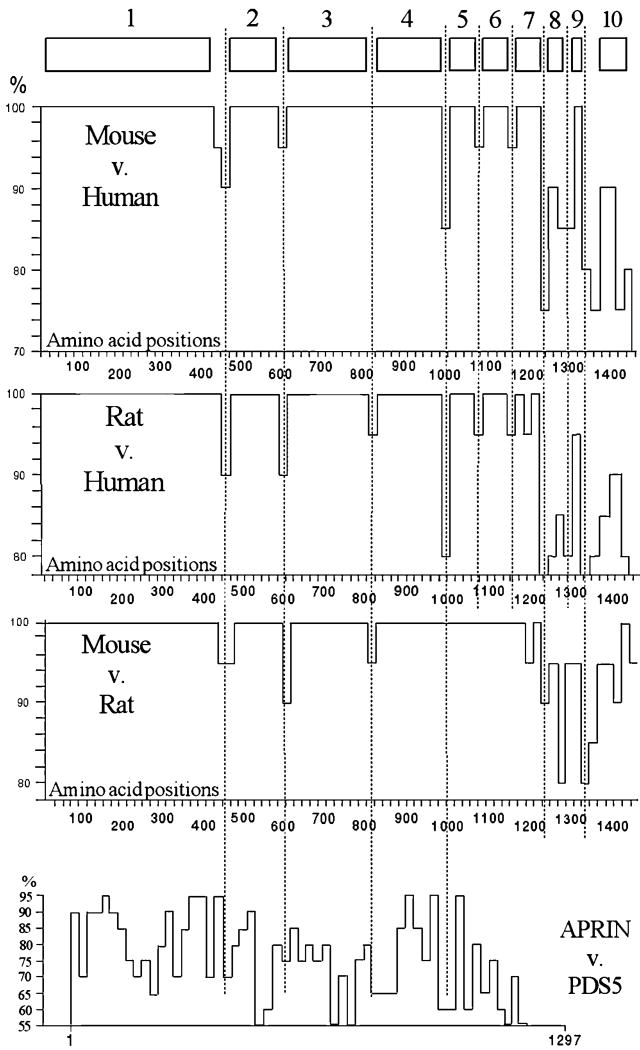
Based on evolutionary considerations [36], a pattern of highly conserved domains reflects the functional domain pattern of a protein. We aligned the mouse and human APRIN polypeptide sequences by using the BestFit (GCG) comparison program. To plot the differences topologically, we divided the sequences into 20 amino acid units (Figure 4). This helped to achieve high mapping resolution (74 units), while maintained a functionally meaningful size (that of transmembrane domains, nuclear localization signals, hinge domains, epitope sizes, alpha helices, etc.). The particular conservation index of each unit was then individually calculated and expressed as percent similarity in Figure 4.
Figure 4.
Conservation-based domain map of the APRIN orthologs. The three large panels represent the graphic displays of conservational calculations between the species indicated. The numbered boxes at the top represent the identified conservation domains. The vertical scale shows protein identity in percents, the horizontal scale represents the amino acid positions along the APRIN polypeptide, in 20 amino acid units. The dotted lines identify the major domain or subdomain boundaries. The graph at the bottom (APRIN v. Pds5) represents the conservation map of the human paralog Pds5. The comparison is based on combined sequences from the KIAA0648 [72], (Acc#: AB014548) and the SCC-112 [73] (Acc#: NP_065015) sequences, which overlap and represent different isolates of the human Pds5 cDNA
Although the overall conservation between the mouse and human APRIN was extremely high, it was not uniform, and areas of variability outlined seven highly conserved major domains. Three highly conserved smaller units were also identified in the less conserved C-terminal region. We also analyzed the partial rat APRIN sequence generated in our laboratory (98% complete, sequencing in progress). It revealed a pattern nearly identical to the mouse-human arrangement (Figure 4). A similar pattern of variability was also observed within the rodent group. Superposition of the established variable regions clearly outlined an evolutionarily conserved domain architecture, as indicated on the top of Figure 4.
The conservation map between APRIN and another APRIN-related human protein, Pds5 is shown at the bottom of Figure 4 [37]. The human Pds5 and APRIN proteins displayed much less similarity to each other than the human and rodent APRIN sequences. Our results show that after the divergence of the Pds5 and APRIN lineages, the two genes evolved independently. The lack of conservation indicates the lack of functional constraints between these genes during their evolution. In addition, the unique acquisition of HMG domains by APRIN (see later) suggests further functional differences between these two proteins.
3.5. The APRIN conserved domain map is consistent with the ancestral Pds5 domain pattern in the Pfam database (Protein Families Domain Database)
The Pfam Database contains multiple sequence alignments of common protein domains [45] (www.sanger.ac.uk/Software/Pfam/). The Pfam-A database includes functional annotations, while the functions of Pfam-B domains are unknown. Our search of the APRIN protein (pfam identifier: Q9Y451) resulted in a number of hits in the Pfam-B database and the alignments are shown in Figure 5. The Pfam domains were consistent with the conservation-based domain architecture we identified. The core unit in the evolution of the APRIN/Pds5 ancestry is a combination of three domains: Pfam # 3566, #4398 and #8120. They are observed in extant unicellular eukaryotes, suggesting that they evolved first in ancient eukaryotes. The domain organization and map positions precisely co-align with the domains we identified and confirm the validity of our conservation analysis.
Figure 5.
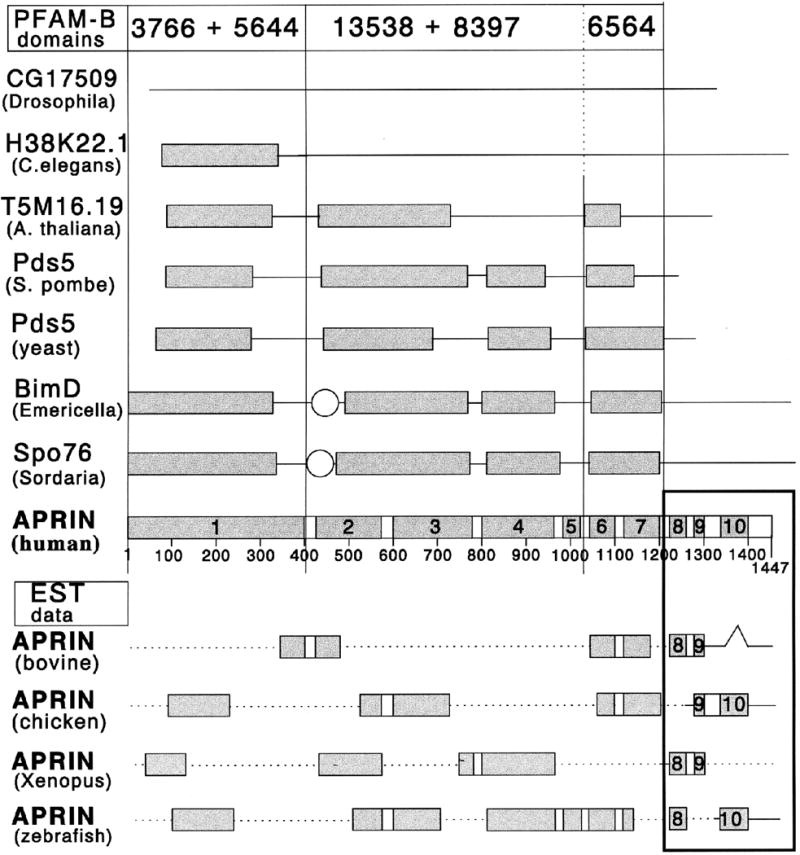
Comparison of the APRIN conserved domain pattern with APRIN-related sequences in the Pfam domain database. Genes names (in bold) and organisms (in parentheses) are indicated on the left. The numbers at the top label the three major Pfam domain families recognized in the APRIN sequence. The scale and the conserved domains in the AS3 line are the same as in Figure 4. The boxes below the APRIN domains are EST data representing partial vertebrate sequences with homology to the human APRIN sequence. (EST accession numbers from N-terminal to C-terminal direction: bovine, BE756042, AW483768 and BF043498; chicken: BG712953, BG625340, BG625239 and AJ392316; Xenopus, BI448976, BG360016+BG346620, BG016343 and BI095461; zebrafish, BG515319, AI558326, AW175091, AW128709 and BI472753). The interruptions within the EST sequences signal regions of low conservation. The dotted lines between the boxes represent regions where sequence information was not available.
3.6. Vertebrate APRIN proteins have three unique C-terminal domains not found in proteins of the Pds5 lineage
We found that the human and rodent C-terminal ∼250 amino acid region (conserved domains #8, #9 and #10) was present in all vertebrate APRIN orthologs, but it was missing from the Pds5 lineage and their ancient forms in invertebrates. In the EST databases all vertebrate sequences we searched (mammalian, avian, amphibian and fish) contained the unique C-terminal domains and the arrangements were identical to that of the human (Figure 5). In several vertebrate species we found two AS3-related sequences, consistent with the pattern of the two human genes (APRIN and Pds5) (not shown).
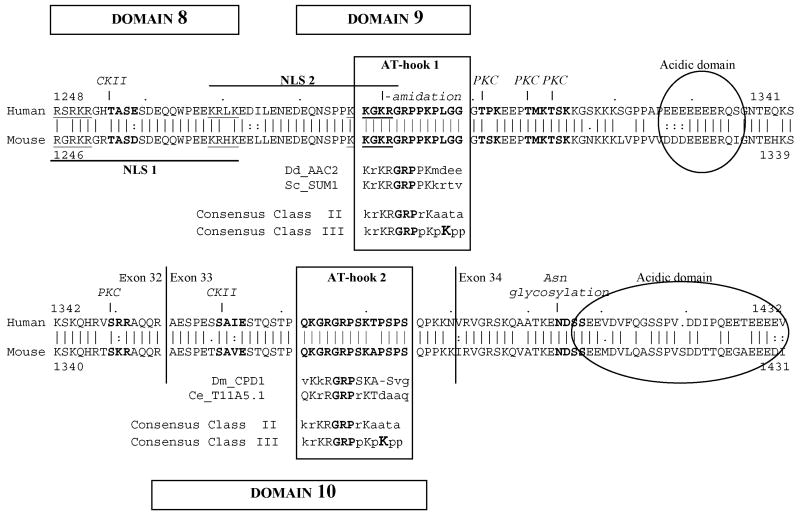
3.7. The unique C-terminal sequence of APRIN has two HMG box AT-hook domains
A search on the Pfam-A database using the unique C-terminal sequence recognized two AT-hook type HMG box motifs (High Mobility Group protein domains) [46] (Figure 6). Our conserved domains #9 and #10 accurately represent AT-hook 1 (positions 1287-1300) and 2 (positions 1372-1385), respectively. The domains show great similarities to known functional AT-hook sequences (AAC2, SUM1, CPD1 and T11A5, for details see Figure 6 legend). The core of all AT-hook sequences is a glycine-arginine-proline (“GRP”) sequence flanked by various basic and proline residues. Of the three general classes of AT-hooks [46], both APRIN AT-hooks belong to Class II and share the features of other known AT-hook domains. The two AT-hooks are usually separated by >20 amino acids [47], they are typically on separate exons [48] with a characteristic C-terminal acidic domain [47, 49] and usually regulated by phosphorylation through protein kinase C (PKC) and casein kinase II (CKII) target sequences [50] (Figure 6). The kinase targets on APRIN correspond to sites on the human High Mobility Group A1a protein [51] (data not shown). The amidation and asparagine glycosylation sites indicate complex post-translational modifications.
Figure 6.
AT-hook domains and nuclear localization signals in the unique C-terminal sequence of APRIN. The C-terminal APRIN polypeptide sequences of the human (top sequence) and the mouse (bottom sequence) are shown in alignment, the numbers indicate amino acid positions. Domains #8, #9 and #10 are marked. The identified AT-hook motifs are boxed and labeled. The boxes also contain sequences of high similarity with other DNA binding proteins (Dd_AAC2, “AAC-rich mRNA” gene from Dictyostelium discoideum, acc.# P14196; Sc_SUM1, “suppressor of mar1 mutation” gene from Saccharomyces cerevisiae, acc # P46676; Dm_CPD1, “chromosomal protein D1” gene from Drosophila melanogaster, acc # P22058; and Ce_T11A5.1, “protein 1 from region T11A5” from Caenorhabditis elegans). The two classes of AT-hook domains (Class II and Class III) with close similarity to the APRIN elements are also shown [46]. The lysine (K) which is critical in Class III (highlighted) is missing in the APRIN sequence, classifying it to a Class II motif. The acidic domains adjacent to the AT-hooks are indicated by the ellipses. The sequences indicated by the bold long lines below and above the alignment represent the two bipartite nuclear localization signals (NLS1 and NLS2). The two positively charged elements within the localization signals are underlined. The single arrows above the human sequence indicate casein kinase II (CKII) and protein kinase C (PKC) targets. The amino acids in the recognition sequences are in bold. The double arrows indicate putative amidation and asparagine glycosylation sites.
Further searches on the PROSITE database [52] (www.expasy.org/tools/scanprosite/) identified two high-score nuclear localization sequences (NLS). One coincides with conserved domain 8, while the second is in conserved domain 9, overlapping AT-hook 1 (Figure 6). The bipartite NLS sequences share similarities with the androgen receptor and DNA polymerase NLS motifs [53, 54]. Notably, both putative NLS motifs are potential targets for postranslational modifications by CKII or amidation.
3.8. APRIN expression studies established that the putative NLS motifs are functional and localized APRIN in discrete nuclear compartments
To investigate the putative nuclear localization sequences, we transfected MCF7-AR1 cells with full length and C terminal (NLS) truncated APRIN variants. Since C-terminal truncations also eliminate the epitopes for the anti-APRIN antibodies, we expressed FLAG-tagged APRIN constructs and used the anti-FLAG antibody for localization. Immunofluorescence cytochemistry detected nuclear localization of the full construct in about 25 % of transiently-transfected cells (Figure 2, panels “d”, “e” and “f”). The expression level and localization of the FLAG-APRIN signal showed variability, but it was found mostly in discrete nuclear compartments. Truncation of the C-terminal portion upstream of the NLS sequence in domain 8 eliminated nuclear transport and the FLAG-APRIN fusion protein accumulated only in the cytoplasm (Figure 2, panels “g”, “h” and “i”).
3.9. The nuclear pattern of transient APRIN expression showed partial heterochromatin-association
Confocal microscopy to detect the FLAG-tagged APRIN construct further confirmed that APRIN was nuclear (Figure 2 panels “j”, “k” and “l”). We found the APRIN construct in discrete internal compartments and in nuclear membrane-proximal regions. Since the latter is characteristically occupied by heterochromatin, APRIN appeared to associate with heterochromatin, but only partially, suggesting that APRIN is not a heterochromatic structural element. The characteristic punctate, non-random distribution indicated that APRIN may be specific to particular states of the chromatin and potentially involved in regulation.
3.10. High resolution fluorescence imaging localized APRIN in the heterochromatin-euchromatin interface
The specificity of the anti-APRIN-1434 antibody allowed high-resolution fluorescence localization of APRIN in the interphase nuclei of androgen-treated MCF7-AR1 cells. Figure 2, Panel “m” shows APRIN in a fine, non-randomly distributed punctate pattern in the interphase nuclei. DAPI is known to bind AT-rich heterochromatin and in combination with our APRIN antibody (Figure 2, Panel “n”) the localization pattern revealed that APRIN followed the heterochromatin, but did not appear to be an integrate part of it. This is more clearly shown in Figure 2, Panel “o”, where a portion of Panel “n” is highlighted. APRIN localization can be best described as non-random punctate foci at the interface between the heterochromatin and the euchromatin, with occasional minor populations of larger or smaller foci within the euchromatin.
3.11. The expression pattern of APRIN in vivo, in the human prostate confirms heterochromatin localization and its downregulation correlates with lost structural integrity in the chromatin
To detect the native APRIN protein in a human tissue, human prostate cancer sections were stained with the anti-APRIN antibody. We found that in neoplastic epithelial cells, APRIN expression also co-localized with the heterochromatin in the nucleoplasm and at the periphery of the nucleus. (Figure 2, Panels “p”, “r” and “s”). APRIN expression, however, was significantly downregulated in about 60% of the cancer samples. We also noticed that APRIN downregulation coincided with disorganized nuclear structures in enlarged, swollen nuclei.
4. Discussion
Cell proliferation is stringently controlled in multicellular organisms. Differentiation and proliferative arrest are largely hormone-regulated in reproductive tissues, but the mechanisms are poorly understood. Hormone regulation induces global changes in gene expression patterns, but only a minority of the genes is controlled directly by the hormone receptors (see INTRODUCTION). The global changes in gene expression patterns, therefore, are believed to be the results of indirect mechanisms, including chromatin effects. Very little is known, however, about the mechanisms that convey the hormonal signals to the chromatin.
The LNCaP prostate epithelial cell line undergoes proliferative arrest at physiological androgen concentrations with major changes in chromatin structure. We searched for the mediators, identified the APRIN protein and showed that it was involved in the regulation of proliferative arrest. We report here that the basic features of APRIN (lineage, domain architecture, unique HMG domains and localization) are consistent with a chromatin regulator.
To study the functional domains of the APRIN protein, we applied a novel conservation-based approach. We used only basic computational resources that do not require the complex algorithms of other published protocols [56-58]. The method is based on the generally accepted principle that in evolution, substitution rate differences within a protein reflect the differences in selective constraints. Domains of critical functions, therefore, are more conserved than non-essential regions [36]. To determine the conservation parameters, we cloned and mapped the mouse APRIN cDNA. Sequence comparisons indicated an unusually high degree of protein conservation (96 % identity). This conservation is not uncommon in small proteins with long evolutionary history (histones, tubulins, etc), but highly unusual for large polypeptides. We also analyzed conservation at the level of the nucleotide sequence. The silent (synonymous) nucleotide substitutions (Ks) were compared to the ones that altered the code (Ka) [36]. In areas under functional constraints the code-altering substitutions are eliminated or suppressed. Therefore, in molecular evolution studies the Ka/Ks ratio is a key parameter. If Ka/Ks=1, there has been no selective pressure to eliminate the changed residues, so the domain has no functional constraints. If Ka/Ks < 1, the altered residues have been purged by “purifying selection”. This is indicative of selective pressure to maintain a function and the corresponding domain [59]. We found Ka/Ks=0.13 in the entire coding region (57 code altering changes/420 silent changes). In the highly conserved first 1200 amino acid area the Ka/Ks value was 0.03 (10 code altering changes/345 silent changes) and in the first extremely conserved 420 amino acid area the Ka/Ks value was 0.009 (1 code altering change/112 silent changes). These findings suggest that APRIN has been under enormous purifying selection in the last 100 million years and implicates that the protein has a very critical function.
We found that the highly conserved regions were interrupted by short less-conserved sequences and outlined the conservation-based domain pattern of APRIN. The resulting domain architecture is in agreement with our earlier computer maps of putative functional motifs, namely 1-400: coiled-coil; 400-600: ATP-binding, catalytic; 600-1000: low score DNA-binding motifs [26].
The domain architecture of APRIN is also in agreement with data in the Pfam B domain database. The core domains of APRIN are identical to an ancient domain pattern. This pattern is found in unicellular eukaryotic and invertebrate proteins that belong to the Pds5 lineage, namely BimD in Emericella [60], Spo76 in Sordaria [61] and Pds5p in Saccharomyces [62, 63]. These proteins [63], as well as their human ortholog, the Pds5 protein [37] were shown to form complexes with cohesins. Cohesins prevent sister chromatid separation after DNA replication [64]. Experimental efforts to demonstrate a role of APRIN in cohesion, however, have yet to show its function in this process [38].
We found critical differences between the APRIN and the Pds5 proteins. The low conservation between the APRIN and Pds5 lineages, the presence of the two related sequence variants (APRIN v. Psd5) in all vertebrate species we tested and the novel HMG domains in all genes in the APRIN lineage in vertebrates indicate the following evolutionary scenario. The data are consistent with the occurrence of a gene duplication event at the time of early vertebrate divergence, when APRIN branched out from the ancient Pds5p-Spo76-BimD lineage. In addition, the early ancestor of the APRIN lineage integrated three novel C-terminal exons. Our results clearly classify APRIN as a paralog of Pds5, with common ancestry in the Pds5 lineage.
Further analyses of the acquired C-terminal element in APRIN identified two perfect matches to AT-hook consensus sequences. We show that they are highly conserved in other vertebrates, suggesting that they are critical for APRIN function. The AT-hook motifs of the small non-histone nuclear proteins (High Mobility Group proteins, HMGI/Y) bind AT-rich sequences in the minor groove of DNA [47]. The acidic domains (Figure 6) are believed to bind and stabilize the core sequences [49, 50]. Minor groove binding enables these proteins to bind DNA in a nucleosomal complex and participate in opening and remodeling chromatin [47, 65].
The domain arrangement with C-terminal HMG units is not unprecedented. Some of the AT-hook domains became integrated by exon acquisitions [46] by several large (>1000 residue) DNA-binding nuclear polypeptides, like the yeast SNF2 protein [66], the C. elegans lin-15b polypeptide [67], the D. melanogaster TAFII-230 factor [68], the human BR140 protein [69], etc. Notably, these proteins regulate gene expression by controlling heterochromatin-euchromatin partitioning, chromatin remodeling and they have critical functions in differentiation, development and proliferation. The inactive heterochromatin is in densely packed nuclear matrix-associated compartments in the nucleus [70]. The active euchromatin is arranged in large 50-100 kb loops with clusters of genes which are attached to the matrix [71]. The sequences that anchor the loops to the matrix consist of AT-rich elements (S/MAR regions) [71], which are recognized by regulators with AT-hook domains. These regulators can control global gene expression patterns by affecting the chromatin architecture. We presented evidence that APRIN shares functional domains and chromatin localization with these factors.
In summary, the present report turns the APRIN functional model toward a new direction. Although APRIN emerged from the Pds5 family of cohesin-associated proteins, its independent evolution, its novel domain architecture and its localization in the interphase chromatin suggest a new functional entity. In the light of our findings we propose that APRIN, in addition to its putative Pds5-associated role in the chromosomes, may function as a chromatin architectural regulator and a candidate mediator of hormone induced chromatin changes in differentiation and cancer.
Supplementary Material
Acknowledgments
We thank Dr Sandro Rusconi, Dr Paul Planet, Cheryl Schaeberle and Janine Calabro for their critical evaluation of this manuscript and for their helpful suggestions. This work was supported by the National Institute of Health Grant NCI 55574 (A.S., P.G.); DOD-DAMD17-03-1-0266 (P.G.) and The Susan G. Komen Breast Cancer Foundation BCTR0403214 (P.G.). The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sonnenschein C, Soto AM. The Society of Cells: Cancer and Control of Cell Proliferation. Springer Verlag; New York: 1999. [Google Scholar]
- 2.Takahashi K, Ohmichi M, Yoshida M, Hisamoto K, Mabuchi S, Arimoto-Ishida E, Mori A, Tsutsumi S, Tasaka K, Murata Y, Kurachi H. Both estrogen and raloxifene cause G1 arrest of vascular smooth muscle cells. J Endocrinol. 2003;178:319–329. doi: 10.1677/joe.0.1780319. [DOI] [PubMed] [Google Scholar]
- 3.Kofler R, Schmidt S, Kofler A, Ausserlechner MJ. Resistance to glucocorticoid-induced apoptosis in lymphoblastic leukemia. J Endocrinol. 2003;178:19–27. doi: 10.1677/joe.0.1780019. [DOI] [PubMed] [Google Scholar]
- 4.Ling MT, Chan KW, Choo CK. Androgen induces differentiation of a human papillomavirus 16 E6/E7 immortalized prostate epithelial cell line. J Endocrinol. 2001;170:287–296. doi: 10.1677/joe.0.1700287. [DOI] [PubMed] [Google Scholar]
- 5.Udayakumar TS, Jeyaraj DA, Rajalakshmi M, Sharma RS. Culture of prostate epithelial cells of the rhesus monkey on extracellular matrix substrate: influence of steroids and insulin-like growth factors. J Endocrinol. 1999;162:443–450. doi: 10.1677/joe.0.1620443. [DOI] [PubMed] [Google Scholar]
- 6.Oosterhoff JK, Grootegoed JA, Blok LJ. Expression profiling of androgen-dependent and -independent LNCaP cells: EGF versus androgen signalling. Endocr Relat Cancer. 2005;12:135–148. doi: 10.1677/erc.1.00897. [DOI] [PubMed] [Google Scholar]
- 7.Nantermet PV, Xu J, Yu Y, Hodor P, Holder D, Adamski S, Gentile MA, Kimmel DB, Harada S, Gerhold D, Freedman LP, Ray WJ. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. Journal of Biological Chemistry. 2004;279:1310–1322. doi: 10.1074/jbc.M310206200. [DOI] [PubMed] [Google Scholar]
- 8.Waghray A, Feroze F, Schober MS, Yao F, Wood C, Puravs E, Krause M, Hanash S, Chen YQ. Identification of androgen-regulated genes in the prostate cancer cell line LNCaP by serial analysis of gene expression and proteomic analysis. Proteomics. 2001;1:1327–1338. doi: 10.1002/1615-9861(200110)1:10<1327::AID-PROT1327>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 10.Tomura A, Goto K, Morinaga H, Nomura M, Okabe T, Yanase T, Takayanagi R, Nawata H. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J Biol Chem. 2001;276:28395–28401. doi: 10.1074/jbc.M101755200. [DOI] [PubMed] [Google Scholar]
- 11.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nature Genetics. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 12.O'Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Molecular Endocrinology. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 13.Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem. 2002;277:50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- 14.Sasano H, Fukushima K, Sasaki I, Matsuno S, Nagura H, Krozowski ZS. Immunolocalization of mineralocorticoid receptor in human kidney, pancreas, salivary, mammary and sweat glands: a light and electron microscopic immunohistochemical study. J Endocrinol. 1992;132:305–310. doi: 10.1677/joe.0.1320305. [DOI] [PubMed] [Google Scholar]
- 15.Kyprianou N, Gingell JC, Davies P. Intranuclear distribution of androgen receptors in human prostate carcinoma. J Endocrinol. 1987;112:161–169. doi: 10.1677/joe.0.1120161. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai A, Ichikawa K, Hashizume K, Miyamoto T, Yamauchi K, Ohtsuka K, Nishii Y, Yamada T. Possible role of histones in the organization of rat liver thyroid hormone receptors in chromatin. J Endocrinol. 1989;121:337–341. doi: 10.1677/joe.0.1210337. [DOI] [PubMed] [Google Scholar]
- 17.Schatten H, Ripple M, Balczon R, Weindruch R, Chakrabarti A, Taylor M, Hueser CN. Androgen and taxol cause cell type-specific alterations of centrosome and DNA organization in androgen-responsive LNCaP and androgen-independent DU145 prostate cancer cells. J Cell Biochem. 2000;76:463–477. [PubMed] [Google Scholar]
- 18.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 19.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 20.English HF, Santen RJ, Isaacs JT. Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate. 1987;11:229–242. doi: 10.1002/pros.2990110304. [DOI] [PubMed] [Google Scholar]
- 21.English HF, Drago JR, Santen RJ. Cellular response to androgen depletion and repletion in the rat ventral prostate: autoradiography and morphometric analysis. Prostate. 1985;7:41–51. doi: 10.1002/pros.2990070106. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenschein C, Olea N, Pasanen ME, Soto AM. Negative controls of cell proliferation: human prostate cancer cells and androgens. Cancer Res. 1989;49:3474–3481. [PubMed] [Google Scholar]
- 23.Geck P, Szelei J, Jimenez J, Lin TM, Sonnenschein C, Soto AM. Expression of novel genes linked to the androgen-induced, proliferative shutoff in prostate cancer cells. J Steroid Biochem Molec Biol. 1997;63:211–218. doi: 10.1016/s0960-0760(97)00122-2. [DOI] [PubMed] [Google Scholar]
- 24.Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM. Androgen-induced proliferative quiescence in prostate cancer: the role of AS3 as its mediator. Proc Nat Acad Sci USA. 2000;97:10185–10190. doi: 10.1073/pnas.97.18.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maffini MV, Geck P, Powell CE, Sonnenschein C, Soto AM. Mechanism of androgen action on cell proliferation AS3 protein as a mediator of proliferative arrest in the rat prostate. Endocrinology. 2002;143:2708–2714. doi: 10.1210/endo.143.7.8899. [DOI] [PubMed] [Google Scholar]
- 26.Geck P, Szelei J, Jimenez J, Sonnenschein C, Soto AM. Early gene expression during androgen-induced inhibition of proliferation of prostate cancer cells: a new suppressor candidate on chromosome 13, in the BRCA2-Rb1 locus. J Steroid Biochem Molec Biol. 1999;68:41–50. doi: 10.1016/s0960-0760(98)00165-4. [DOI] [PubMed] [Google Scholar]
- 27.Geck P, Sonnenschein C, Soto AM. The D13S171 marker, misannotated to BRCA2, links the AS3 gene to various cancers. Am J Hum Genet. 2001;69:461–463. doi: 10.1086/321968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards SM, Dunsmuir WD, Gillett CE, Lakhani SR, Corbishley C, Young M, Kirby RS, Dearnaley DP, Dowe A, Ardern-Jones A, Kelly J, Spurr N, Barnes DM, Eeles RA. Immunohistochemical expression of BRCA2 protein and allelic loss at the BRCA2 locus in prostate cancer. CRC/BPG UK Familial Prostate Cancer Study Collaborators. Int J Cancer. 1998;78:1–7. doi: 10.1002/(sici)1097-0215(19980925)78:1<1::aid-ijc1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 29.Beckmann MW, Picard F, An HX, van Roeyen CR, Dominik SI, Mosny DS, Schnurch HG, Bender HG, Niederacher D. Clinical impact of detection of loss of heterozygosity of BRCA1 and BRCA2 markers in sporadic breast cancer. Br J Cancer. 1996;73:1220–1226. doi: 10.1038/bjc.1996.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada H, Uchida N, Shimada Y, Kumimoto H, Shinoda M, Imamura M, Ishizaki K. Polymorphism and allelic loss at the AS3 locus on 13q12-13 in esophageal squamous cell carcinoma. Int J Oncol. 2001;18:1003–1007. doi: 10.3892/ijo.18.5.1003. [DOI] [PubMed] [Google Scholar]
- 31.Huang XP, Wei F, Liu XY, Xu X, Hu H, Chen BS, Xia SH, Han YS, Han YL, Cai Y, Wu M, Wang MR. Allelic loss on 13q in esophageal squamous cell carcinomas from northern China. Cancer Lett. 2002;185:87–94. doi: 10.1016/s0304-3835(02)00234-3. [DOI] [PubMed] [Google Scholar]
- 32.Siffroi-Fernandez S, Delom F, Nlend MC, Lanet J, Franc JL, Giraud A. Identification of thyroglobulin domain(s) involved in cell-surface binding and endocytosis. J Endocrinol. 2001;170:217–226. doi: 10.1677/joe.0.1700217. [DOI] [PubMed] [Google Scholar]
- 33.Byun D, Mohan S, Baylink DJ, Qin X. Localization of the IGF binding domain and evaluation of the role of cysteine residues in IGF binding in IGF binding protein-4. J Endocrinol. 2001;169:135–143. doi: 10.1677/joe.0.1690135. [DOI] [PubMed] [Google Scholar]
- 34.Seasholtz AF, Valverde RA, Denver RJ. Corticotropin-releasing hormone-binding protein: biochemistry and function from fishes to mammals. J Endocrinol. 2002;175:89–97. doi: 10.1677/joe.0.1750089. [DOI] [PubMed] [Google Scholar]
- 35.Dai G, Wang D, Liu B, Kasik JW, Muller H, White RA, Hummel GS, Soares MJ. Three novel paralogs of the rodent prolactin gene family. Journal of Endocrinology. 2000;166:63–75. doi: 10.1677/joe.0.1660063. [DOI] [PubMed] [Google Scholar]
- 36.Li WH. Molecular Evolution. Sinauer Association, Inc.; Sunderland: 1997. [Google Scholar]
- 37.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. Journal of Cell Science. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 39.Szelei J, Jimenez J, Soto AM, Luizzi MF, Sonnenschein C. Androgen-induced inhibition of proliferation in human breast cancer MCF7 cells transfected with androgen receptor. Endocrinology. 1997;138:1406–1412. doi: 10.1210/endo.138.4.5047. [DOI] [PubMed] [Google Scholar]
- 40.Sharan SK, Bradley A. Murine BRCA2: sequence, map position, and expression pattern. Genomics. 1997;40:234–241. doi: 10.1006/geno.1996.4573. [DOI] [PubMed] [Google Scholar]
- 41.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 42.Mouse Genome Sequencing Consortium, Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 43.Thornton WJ, Kelley DB. Evolution of the androgen receptor: structure-function implications. BioEssays. 1998;20:860–869. doi: 10.1002/(SICI)1521-1878(199810)20:10<860::AID-BIES12>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 44.Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proceedings of the National Academy of Science of the United States of America. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 48.Friedmann M, Holth LT, Zoghbi HY, Reeves R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21:4259–4267. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padmanabhan S, Elias-Arnanz M, Carpio E, Aparicio P, Murillo FJ. Domain architecture of a high mobility group A-type bacterial transcriptional factor. J Biol Chem. 2001;276:41566–41575. doi: 10.1074/jbc.M106352200. [DOI] [PubMed] [Google Scholar]
- 50.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 51.Diana F, Sgarra R, Manfioletti G, Rustighi A, Poletto D, Sciortino MT, Mastino A, Giancotti V. A link between apoptosis and degree of phophorylation of high mobility group A1a protein in leukemic cells. J Biol Chem. 2001;276:11354–11361. doi: 10.1074/jbc.M009521200. [DOI] [PubMed] [Google Scholar]
- 52.Bairoch A, Bucher P, Hofmann K. The prosite database and its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]
- 54.Chung DW, Zhang JA, Tan CK, Davie EW, So AG, Downey KM. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc Nat Acad Sci USA. 1991;88:11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 57.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 58.Simon AL, Stone EA, Sidow A. Inference of functional regions in proteins by quantification of evolutionary constraints. Proc Nat Acad Sci USA. 2002;99:2912–2917. doi: 10.1073/pnas.042692299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner A. Selection and gene duplication: a view from the genome. Genome Biology. 2002;3:1012. doi: 10.1186/gb-2002-3-5-reviews1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Heemst D, Kafer E, John T, Heyting C, van Aalderen M, Zickler D. BimD/SPO76 is at the interface of cell cycle progression, chromosome morphogenesis, and recombination. Proc Nat Acad Sci USA. 2001;98:6267–6272. doi: 10.1073/pnas.081088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Heemst D, James F, Poggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 62.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Current Biology. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 63.Wang SW, Read RL, Norbury CJ. Fission yease Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J Cell Sci. 2002;115:587–598. doi: 10.1242/jcs.115.3.587. [DOI] [PubMed] [Google Scholar]
- 64.Losada A, Hirano T. Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Current Biology. 2001;11:268–272. doi: 10.1016/s0960-9822(01)00066-5. [DOI] [PubMed] [Google Scholar]
- 65.Maher JF, Nathans D. Multivalent DNA-binding properties of the HMG-1 proteins. Proc Nat Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Nat Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark SG, Lu X, Horvitz HR. The Caenorhabiditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kokubo T, Gong DW, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y. Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 1993;7:1033–1046. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 69.Thompson KA, Wang B, Argraves WS, Giancotti FG, Schranck DP. BR140, a novel zinc-finger protein with homology to the TAF250 subunit of TFIID. Biochem Biophys Res Commun. 1994;198:1143–1152. doi: 10.1006/bbrc.1994.1162. [DOI] [PubMed] [Google Scholar]
- 70.Li F, Chen J, Izumi M, Butler MC, Keezer SM, Gilbert DM. The replication timing program of the Chinese hamster beta-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J Cell Biol. 2001;154:283–292. doi: 10.1083/jcb.200104043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishikawa K, Nagase T, Suyama M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Research. 1998;5:169–176. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- 73.Kumar D, Sakabe I, Patel S, Zhang Y, Ahmad I, Gehan EA, Whiteside TL, Kasid U. SCC-112, a novel cell cycle-regulated molecule, exhibits reduced expression in human renal carcinomas. Gene. 2004;328:187–196. doi: 10.1016/j.gene.2003.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.