Abstract
The HADDOCK score, a scoring function for both protein–protein and protein-nucleic acid modeling, has been successful in selecting near-native docking poses in a variety of cases, including those of the CAPRI blind prediction experiment. However, it has yet to be optimized for small molecules, and in particular inhibitors of protein–protein interactions, that constitute an “unmined gold reserve” for drug design ventures. We describe here HADDOCK2P2I, a biophysical model capable of predicting the binding affinity of protein–protein complex inhibitors close to experimental error (∼2-fold larger). The algorithm was trained and 4-fold cross-validated against experimental data for 27 inhibitors targeting 7 protein–protein complexes of various functions and tested on an independent set of 24 different inhibitors for which Kd/IC50 data are available. In addition, two popular ligand topology generation and parametrization methods (ACPYPE and PRODRG) were assessed. The resulting HADDOCK2P2I model, derived from the original HADDOCK score, provides insights into inhibition determinants: while the role of electrostatics and desolvation energies is case-dependent, the interface area plays a more critical role compared to protein–protein interactions.
Introduction
Protein–protein interactions (PPIs) define most of the cellular processes in the cell,1 such as differentiation, proliferation, signal transduction, and apoptosis. Being able to design PPI inhibitors will drastically catalyze the development of novel therapeutics for diseases, such as cancer.2 Such inhibitors are currently considered “an unmined gold reserve”3 for drug design in general. Consequently, novel software tools are being developed and made publicly available that target the design of inhibitor of PPIs, such as PocketQuery.4−6 The chemical space of PPI inhibitors is, however, rather unique.7−9 Protein–protein interfaces have been manually curated and collected in several dedicated databases, such as iPPi-DB,10 a database that includes associated pharmacological data, and the 2P2I database.11,12 Recent studies on a structure-based benchmark11 of PPI inhibitors (collected from the 2P2I database12) showed that the size of the ligands targeting the interface of protein–protein complexes is substantially larger than that of normal inhibitors that target the active site of single molecules like enzymes9 and suggest that PPI inhibitors are mainly large, lipophilic, and aromatic compounds. Because of the different nature of the target interface and ligands, dedicated biophysical models are needed to predict the binding affinity of PPI inhibitors. Although current biophysical models have proven to reasonably approximate the affinity of protein–ligand complexes,9,13 these have yet to be optimized to predict the affinity of PPI inhibitors. A drop by more than 10% in docking success rates is reported when inhibitors of protein–protein interactions are put to test in comparison to normal inhibitors.14 To stimulate the development of new models, data sets of binding affinities are required. Such a data set has recently been compiled for protein–protein complexes,15,16 but no such data set is available for PPI inhibitors. Although docking and binding affinity prediction have been performed with success for specific systems,17−19 a generic binding affinity prediction model for PPIs would be a welcome addition.
In this work, we report the optimization and performance of the HADDOCK score20,21 on the prediction of the binding affinity of PPI inhibitors. This was performed on a binding affinity benchmark consisting of Kd and Ki values for 27 complexes from the 2P2I database.11,12 In addition, we also compiled an independent set of PPI inhibitors for further validation, consisting of 19 protein-inhibitor complexes with available IC50 data. Finally, we predicted the affinity of five different inhibitors that target the interaction between bromodomains and acetylated histones. We additionally investigate experimental ambiguity in affinity measurement and propose an acceptable prediction error on this basis. The original HADDOCK score, which is a linear combination of van der Waals and electrostatics energy terms calculated using the OPLS force field,22 together with an additional empirical desolvation term,23 was originally optimized for discriminating near-native poses in protein–protein docking.21 We present here the PPI-optimized score, HADDOCK2P2I, which is shown to predict, close to experimental error (∼2-fold larger), the binding affinities of PPI inhibitors. Its components provide valuable new insights into the determinants of the inhibition of PPIs.
Materials and Methods
Benchmark Compilation
Information about available structural data for protein–protein interaction inhibitors was retrieved from the 2P2I database11 (see Table 1) and the corresponding coordinates downloaded from the Protein Data Bank (www.pdb.org).24 Binding affinity data (dissociation constants, Kd values, and inhibition constants, Ki values) were manually procured from literature (see Table 1 and associated references in Supporting Information Table S1). In total, data for 7 different protein–protein complexes were identified, for which 27 structures and binding affinity data of complexes with various PPI inhibitors were available.
Table 1. Ki and Kd Binding Affinity Data Set of Protein–Protein Interaction Inhibitorsa.
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| Bcl-xL/Bak | programmed cell death | 1bxl | 3.4 × 10–7 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 1 | N3B | 4369509 | Bcl-xL | 1ysi | 1.2 × 10–7 |
| 2 | ABT-737 | 11228183 | Bcl-xL | 2yxj | 5.0 × 10–10 |
| 3 | 4FC | 2782689 | Bcl-xL | 1ysg | 3.0 × 10–5 |
| 4 | TN1 | 68258 | Bcl-xL | 1ysg | 4.3 × 10–3 |
| 5 | LIU | 15991562 | Bcl-2 | 2o22 | 6.7 × 10–8 |
| 6 | W1191542 | 44182311 | Bcl-xL | 3inq | 1.1 × 10–8 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| MDM2/p53 | transcription regulation | 1ycr | 6.0 × 10–7 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 7 | HDM2 | 656933 | MDM2 | 1t4e | 8.0 × 10–8 |
| 8 | WK23 | 44825260 | MDM2 | 3lbk | 9.2 × 10–7 |
| 9 | MI-63 | 72200152 | MDM2 | 3lbl | 3.6 × 10–8 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| XIAP-BIR3/CASPASE-9 | programmed cell death | 1nw9 | 2.0 × 10–8 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 10 | 998 | 4369343 | XIAP-BIR3 | 1tfq | 1.2 × 10–8 |
| 11 | 997 | 5388929 | XIAP-BIR3 | 1tft | 5.0 × 10–9 |
| 12 | 9JZ | 72199974 | XIAP-BIR3 | 3hl5 | 3.4 × 10–5 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| XIAP-BIR3/SMAC | programmed cell death | 1g73 | 4.2 × 10–7 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 13 | BI6 | 72199334 | XIAP-BIR3 | 2jk7 | 6.7 × 10–8 |
| 14 | AoxSPW | 24916924 | XIAP-BIR3 | 2opy | 3.0 × 10–5 |
| 15 | Smac005 | 25011737 | XIAP-BIR3 | 3clx | 1.2 × 10–7 |
| 16 | Smac005 | 25011737 | XIAP-BIR3 | 3cm7 | 1.2 × 10–7 |
| 17 | Smac010 | 25011738 | XIAP-BIR3 | 3cm2 | 4.2 × 10–7 |
| 18 | Smac037 | 25058143 | XIAP-BIR3 | 3eyl | 2.2 × 10–7 |
| 19 | CZ3 | 72199333 | XIAP-BIR3 | 3g76 | 2.3 × 10–7 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| ZipA/FtsZ | cell cycle regulation/cellular structure | 1f47 | 2.0 × 10–5 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 20 | WAI | 656967 | ZipA | 1y2f | 1.2 × 10–5 |
| 21 | CL3 | 5287936 | ZipA | 1y2g | 8.3 × 10–5 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| HPV-E2/E1 | viral infection | 1tue | n/d | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 22 | BILH 434 | 5287508 | HPV-E2 | 1r6n | 4.0 × 10–8 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| IL-2/IL-2R | immune system regulation | 1z92 | 1.0 × 10–8 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | Ki∥Kd (modulator) (M) | |
|---|---|---|---|---|---|
| 23 | FRG | 5288250 | IL-2 | 1m48 | 2.2 × 10–5 |
| 24 | FRB | 23586028 | IL-2 | 1pw6 | 7.0 × 10–6 |
| 25 | SP-1985 | 5287951 | IL-2 | 1m49 | 7.5 × 10–6 |
| 26 | FRH | 5288251 | IL-2 | 1py2 | 1.0 × 10–7 |
| 27 | SP-4160 | 656989 | IL-2 | 1qvn | 1.4 × 10–6 |
Original references for the affinity data are provided in the Supporting Information (Table S1).
Structure Refinement
The crystal structures of the inhibitors of PPIs in complex with their respective proteins were refined in HADDOCK20,21,25 and subsequently scored. This ensured that all potentially missing side-chains were properly built and the interface of the complex was optimized using the OPLS force field.22 For this, the standard water refinement setup in HADDOCK was used, which starts by solvating the complexes in an 8 Å shell of water molecules (TIP3P) and consists of the following steps:
(1) Energy minimization (EM) of the water, 40 steps with the protein fixed (Powell minimizer), followed by 2 × 40 EM steps with harmonic position restraints on the protein heavy atoms (krest = 5 kcal mol–1 Å–2).
(2) Gentle simulated annealing protocol using molecular dynamics in Cartesian space consisting of
(a) a heating period with 500 MD steps at 100, 200, and 300 K with position restraints (krest = 5 kcal mol–1 Å–2) on the protein heavy atoms except for the side-chains at the interface,
(b) a sampling stage with 1250 MD steps with weak (krest = 1 kcal mol–1 Å–2) position restraints on the protein heavy atoms except for the backbone and side chains at the interface, and
(c) a cooling stage with 500 MD steps at 300, 200, and 100 K with weak (krest = 1 kcal mol–1 Å–2) position restraints on the protein backbone heavy atoms not at the interface. A time step of 2 fs was used for the integration of the equation of motions and the temperature was maintained constant by weak coupling to a reference temperature bath using the Berendsen thermostat.26
(3) Final 200 EM steps without any position restraints.
Nonbonded interactions were calculated with the OPLS force field22 using a cutoff of 8.5 Å. The electrostatic energy (Eelec) was calculated using a shift function while a switching function (between 6.5 and 8.5 Å) was used for the van der Waals energy (Evdw). This procedure generated 20 models for each complex, starting from different random velocities. As is default in the HADDOCK protocol, the average score of the top 4 models was considered.
All calculations were performed with HADDOCK, version 2.1/CNS,27 version 1.2, through the refinement interface of the HADDOCK web server25 (http://haddock.science.uu.nl/services/HADDOCK/haddockserver-refinement.html).
The same protocol can be run manually on a local installation of HADDOCK by turning off randomization of starting orientations, rigid body minimization and random removal of restraints and setting all steps for the semiflexible refinement stage (it1) to 0. This was used for the refinement runs using the ACPYPE parametrization of the ligands (see Results).
HADDOCK2P2I Model Development and Evaluation
Multiple linear regression analysis was used to create the optimized HADDOCK2P2I score based on the following equation:
| 1 |
where Evdw and EElec denote the intermolecular van der Waals and electrostatics energies, Edesolv an empirical desolvation energy,23 and BSA the buried surface area. log Kpred denotes the logarithm of the predicted binding affinity of the inhibitors.
The βi and c coefficients were optimized using 4-fold cross-validation by minimizing the χ2 function (eq 2) for the 27 complexes shown in Table 1.
| 2 |
where Kexp corresponds to the experimentally measured Kd and Ki constants. During optimization, the various HADDOCK score components were switched on and off to assess their usefulness in the final development of the model. Similar parametrization methods have already been employed successfully for various protein–ligand systems to date.28−30 The final coefficients were taken as the average of the 4-fold cross-validation optimization runs.
Independent Sets for Assessing the Prediction Performance of the HADDOCK Score and HADDOCK2P2I
For compiling the training set and performing the 4-fold cross-validation, we carefully selected only competitive inhibitors, avoiding any noncompetitive modulators of the interactions. Data from interaction inhibition are often reported not as Ki (or, equivalently, Kd), but instead as IC50, the latter denoting the concentration of ligand that reduces interaction activity by 50%. IC50 measures inhibitor binding in competition with another binding partner; consequently, it depends on the concentration of the competitive molecule and its affinity for the target protein. IC50s are usually larger than Ki values, but when the concentration of the substrate is very low, they should become essentially equal to Ki.31 Since data about protein concentration in the assays were rather scarce we chose to omit complexes with only IC50 values from the training/cross-validation set. These were, however, included as an independent set for prediction, because of their different physicochemical but related nature. A list of 19 inhibitors of protein–protein interactions with known molecular structures and IC50 values was manually procured from the literature using 2P2I database and iPPI-DB as starting points (Table 2).
Table 2. IC50 Binding Affinity Data Set of Protein–Protein Interaction Inhibitorsa.
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| Bcl-xL/Bak | programmed cell death | 1bxl | 3.4 × 10–7 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 28 | HI0 | 24798804 | Bcl-xL | 3qkd | 3.0 × 10–09 |
| 29 | 0Q5 | 56973540 | Bcl-xL | 4ehr | 1.3 × 10–08 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| LEDGF/75-integrase | programmed cell death | 2b4j | 1.1 × 10–8 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 30 | 723 | 921795 | 75-integrase | 3lpt | 1.2 × 10–5 |
| 31 | 976 | 45281242 | 75-integrase | 3lpu | 1.4 × 10–6 |
| 32 | TQ2 | 44199170 | 75-integrase | 4e1m | 2.2 × 10–7 |
| 33 | TQX | 44198672 | 75-integrase | 4e1n | 1.9 × 10–8 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| MDM2/p53 | transcription regulation | 1ycr | 6 × 10–7 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 34 | IMY | 49867154 | MDM2 | 1ttv | 1.6 × 10–7 |
| 35 | YIN | 5594130 | MDM2 | 3jzk | 1.2 × 10–6 |
| 36 | 0R2 | 56591324 | MDM2 | 4ere | 4.2 × 10–9 |
| 37 | 0R3 | 56965957 | MDM2 | 4erf | 1.1 × 10–9 |
| 38 | BLF | 56951871 | MDM2 | 4dij | 3.0 × 10–8 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| TNF/TNF receptor | inflammation | n/d | n/d | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 39 | 703 | 4470566 | TNF receptor | 1ft4 | 2.7 × 10–7 |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| bromodomain/histone | inflammation | n/d | n/d | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 40 | P9M | 53054259 | BRD2 | 4a9m | 5.0 × 10–7 |
| 41 | A9N | 72200779 | BRD2 | 4a9n | 1.5 × 10–6 |
| 42 | JQ1 | 46907787 | BRD2 | 3oni | 1.3 × 10–7b |
| 43 | JQ1 | 46907787 | BRDT | 4flp | 1.9 × 10–7b |
| 44 | JQ1 | 46907787 | BRD4 | 3mxf | 4.9 × 10–8b |
| 45 | EAM | 46943432 | BRD4 | 3p5o | 5.1 × 10–8b |
| 46 | I-BET151 | 52912189 | BRD4 | 3zyu | 1.0 × 10–7b |
| interaction | biological role | PDB (complex) | Kd (complex) (M) | ||
|---|---|---|---|---|---|
| integrin α-L/ICAM1 (CD-54) | host–virus interaction | 1nw9 | 2.0 × 10–8 | ||
| modulator (common name) | PubChem compound identifier (CID) | binding partner | PDB (complex) | IC50 (modulator) (M) | |
|---|---|---|---|---|---|
| 47 | LA1 | 5326914 | integrin α-L | 1xuo | 6.9 × 10–8 |
| 48 | 2O7 | 16040268 | integrin α-L | 2o7n | 1.5 × 10–8 |
| 49 | BQM | 11712628 | integrin α-L | 3bqm | 1.7 × 10–9 |
| 50 | E2M | 24875322 | integrin α-L | 3e2m | 0.4 × 10–10 |
| 51 | BJZ | 11699447 | integrin α-L | 3m6f | 2.5 × 10–9 |
Original references for the IC50 data were retrieved from the relevant PDB entries in the Protein Data Bank (www.pdb.org).
Kd data, retrieved from the Protein Data Bank (www.pdb.org).
Additionally, affinity predictions of inhibitors that target a protein–protein complex, not included in the training/cross-validation set were performed with HADDOCK2P2I. These inhibitors were designed to disrupt the interaction between bromodomains that specifically recognize ε-N-acetylation of lysine residues, a post-translational modification common on histone tails. All interactions include measured Kd data and have available molecular structures in the Protein Data Bank (www.pdb.org).
Estimation of Experimental Uncertainty and Qualitative Comparison to Prediction Error
To assess the physicochemical relevance of the prediction error in the cross-validated set and the test sets used, we collected from the iPPI-DB10 all interactions that fulfill the following criteria:
(1) The same small molecule must inhibit the same interaction.
(2) Two or more binding affinity measurements using different experimental methods should be available, but affinity constants must be of the same nature (e.g., Kd, Ki, or IC50).
We, therefore, compiled a data set of 72 interactions that match the above-mentioned criteria (Supporting Information Table S3). Estimation of experimental ambiguity of the derived data is performed using descriptive statistics and distribution analysis, and is empirically compared to the prediction accuracy.
Assessment of the Structural Variability between Ligands and Proteins in the Data Set
To verify that the protein–inhibitor complexes selected for both training and prediction were not structurally redundant, which meant the predictive ability of our biophysical model would be limited to a few classes of complexes, we carried out an analysis of the similarity between the ligands and the protein binders of each data sets. The protein similarity was assessed using sequence similarity following a pairwise global alignment of the sequences (as taken from the RCSB PDB Web site) calculated with the Needleman–Wunsch algorithm available at the EBI Web site (www.ebi.ac.uk/Tools/psa/emboss_needle/).32,33 Features such as histidine tags from the purification protocol were not included in these calculations. Ligands were compared using a substructure key-based 2D Tanimoto similarity,34 which was calculated via the PubChem score matrix web service (http://pubchem.ncbi.nlm.nih.gov//score_matrix/score_matrix.cgi).35
Results
We investigated the protein–protein complexes from the 2P2I database and collected from the literature affinity data for 27 inhibitors targeting 7 different protein–protein complexes. Ki and Kd data were combined into a single data set, excluding any IC50 measurements, which was manually curated and assembled from two different databases. These IC50 values and their respective complexes were used as a test set, to assess the performance of the developed functions in unknown cases, with fuzzier data.
Compilation of a Structure-Based Data Set of PPI Inhibitors with Known Ki and Kd Data
The resulting binding constants include Kd and Ki values describing the modulation/inhibition of the 7 protein–protein complexes (see Table 1). They span the range from very low (mM) to very high affinity (nM), covering thus a broad spectrum of potencies. For two of the complexes (Bcl-xL/Bak and SMAC-DIABLO/XIAP-BIR3), inhibitors with a large range of affinities have been cocrystallized. For example, the survival protein Bcl-xL in complex with the death-promoting region of the Bcl-2-related protein Bak36 is a medium affinity complex (Kd = 340 nM), whereas designed PPI inhibitors against it exhibit binding affinities from 4.3 mM to 0.5 nM. Another complex reported in this study is Smac/DIABLO bound to the third BIR domain (BIR3) of XIAP, a complex critical for cellular apoptosis.37 Its reported affinity is 420 nM, whereas inhibitors designed to disrupt this interaction cover a broad range of Ki, from 300 mM to 67 nM. For the other complexes reported in Table 1, fewer inhibitors with known Ki or Kd data have been reported; they exhibit various binding potencies.
Evaluation of Force Field Topologies and Parameters for PPI Inhibitors
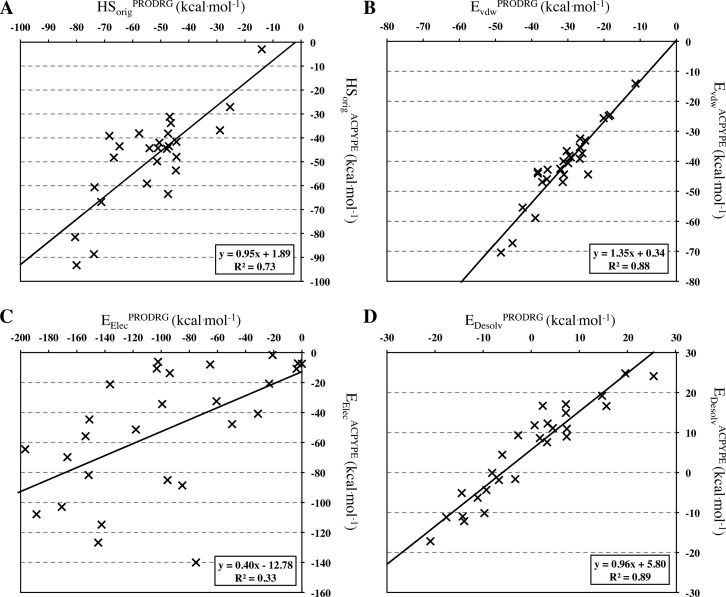
The crystal structures of the PPI inhibitors in complex with their respective proteins were subjected to short refinement and scored using both the HADDOCK refinement web interface25 and a local installation (see Methods). Force field topologies and parameters of all PPI inhibitors were generated using both PRODRG38 and ACPYPE.39 The main difference between the two tools is that, while PRODRG is based on a database search method to parametrize the molecules using OPLS-like parameters,22 ACPYPE uses ANTECHAMBER40 with a semiempirical quantum calculation method for the partial charges. A comparison of the original HADDOCK scores (HSorig = Evdw + 0.2Eelec + Edesol) of the complexes using these two parameter sets reveals differences (Figure 1A) (r2 = 0.73, N = 27, p-value < 0.0001), showing that ligand parameters play a substantial role in defining the interaction energy of each complex. While the desolvation (Edesolv) and van der Waals energies (EvdW) are essentially identical (r2 = 0.89, N = 27, p-value <0.0001 and r2 = 0.88, N = 27, p-value <0.0001, respectively) (Figure 1B and D), the electrostatic component (Eelec) changes dramatically (r2 = 0.33, N = 27, p-value = 0.0017) (Figure 1C) because of differences in partial charges. The buried surface area (BSA), which depends on the van der Waals radii, is however almost identical (r2 = 0.98, N = 27, p-value <0.0001, data not shown).
Figure 1.
Correlation plots between energetic components calculated using ACPYPE and PRODRG-derived parameters. (A) Original HADDOCK score (Evdw + 0.2Eelec + Edesol), (B) van der Waals energy, (C) electrostatic energy, and (D) desolvation energy.
Performance of HADDOCK Score in PPI Inhibitor Binding Affinity Prediction. Training and Cross-Validation of HADDOCK2P2I
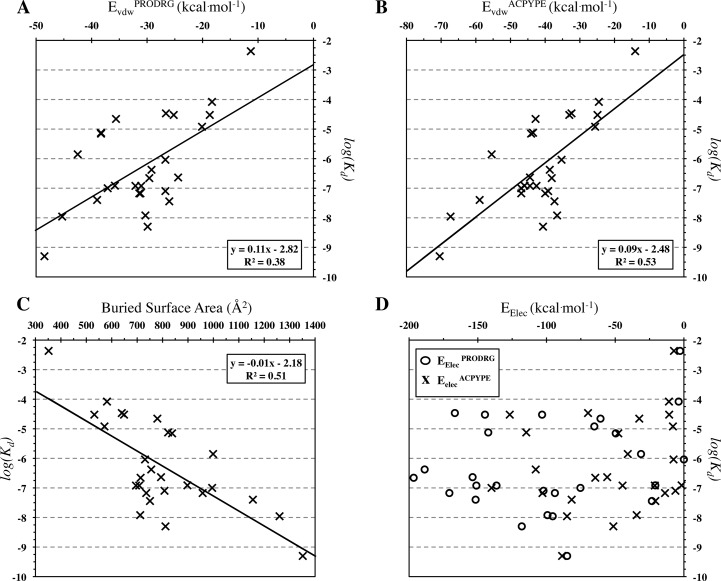
The individual components of HSorig show different contributions to the binding affinity with the van der Waals energy being the most significant contributor (r2 = 0.53, N = 27, p-value <0.0001 for EvdWACPYPE and r2 = 0.38, N = 27, p-value = 0.0006 for EvdWPRODRG) (Figure 2 A and B). The BSA also shows a strong positive correlation with binding affinity independently of the parametrization tool used (r2 = 0.51, N = 27, p-value <0.0001) (Figure 2C). The electrostatic energy, in contrast, does not correlate to binding affinity (r2 = 0.07, N = 27, p-value = 0.1823 for Eelec and r2 = 0.04, N = 27, p-value =0 .3172 for EelecPRODRG) (Figure 2D).
Figure 2.
Correlation plots of experimentally determined binding affinities of PPI modulators with (A, B) the van der Waals energy calculated with the two ligand parametrization schemes, (C) the buried surface area (calculated using PRODRG parameters for the ligands), and (D) the electrostatic energy.
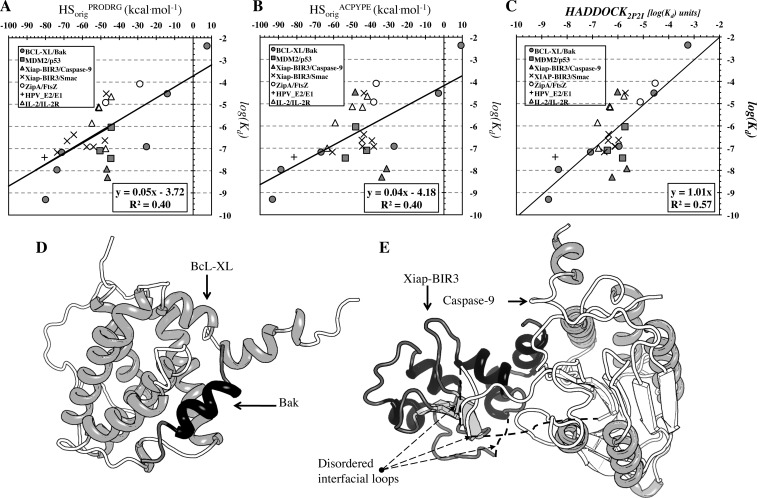
In terms of binding affinity prediction using HSorig, both parametrization schemes exhibit comparable results (r2 = 0.40, N = 27, p-value = 0.0004) (Figure 3A and B).
Figure 3.
Correlation plots of experimentally determined binding affinities with the original HADDOCK score (Evdw + 0.2Eelec + Edesol) calculated with (A and B) the two parametrization schemes and (C) the optimized HADDOCK2P2I score. (D and E) Cartoon representation of (D) the near-rigid Bcl-xL/Bak and (E) the flexible Xiap-BIR3/caspase-9 protein–protein complex.
The components of the original HADDOCK score, together with the BSA, which is used in HADDOCK in the scoring of the initial models at the rigid body stage, were optimized (see Methods) leading to the optimized HADDOCK score, termed HADDOCK2P2I. It is described by the following equation:
| 3 |
Note that the parameters are averages from the 4-fold cross-validation. As can been seen from the standard deviations given as subscripts, there can be quite some variation in some parameters, in particular the weight of the desolvation term. The values of these coefficients may very well vary depending on the data set used for cross-validation; only a much larger data set (unfortunately not available) would allow a better convergence. Still, the model selected (eq 3) outperforms all other models tested (see Supporting Information Table S2), reaching an r2 = 0.57, N = 27, p-value < 0.0001 (r2 = 0.53, N = 27, p-value < 0.0001 after cross-validation) for complexes parametrized with PRODRG (Figure 3C) with a mean absolute error (MAE) of 0.8 ± 0.6 kcal mol–1. It includes two of the original components of the HADDOCK score with an additional BSA term which substitutes the original van der Waals energy term. For complexes parametrized with ACPYPE, the model still retains its predictive capacity (r2 = 0.45, N = 27, p-value = 0.0001 after cross-validation), albeit to a lesser extent (see Supporting Information Table S2). Note that the HADDOCK2P2I scores from both parametrization schemes are very similar (r2 = 0.88, N = 27, p-value < 0.0001). The most notable difference compared to the original HADDOCK score, next to the difference in coefficients, is that the van der Waals energy term has been replaced by the Buried Surface Area term.
A protein–protein complex of central medical relevance is the one formed between the survival protein Bcl-xL and the death-promoting region of the Bcl-2-related protein Bak (Figure 3D). Proteins in the Bcl family are critical regulators of apoptosis and contain members that inhibit programmed cell death, such as Bcl-xL and Bcl-2. These proteins are overexpressed in many cancers and contribute to tumor initiation, progression, and resistance to therapeutics. Successful inhibition of this protein–protein interaction will have major consequences in cancer therapeutics.41 Both the original (r2 = 0.86, N = 6, p-value < 0.001) and optimized (r2 = 0.94, N = 6, p-value < 0.001) HADDOCK scores are able to relate the scores of all six designed inhibitors for this particular complex to their potency, with the optimized HADDOCK2P2I performing slightly better (Figure 3 solid circles, compare A–C).
Although the model can, in general, efficiently predict affinity data for different inhibitors of protein–protein complexes, the prediction performance is directly influenced by conformational changes taking place upon binding: indeed, while for the rigid interface of the BcL-XL/Bak complex discussed above (Figure 3D) predicted affinities are lying close to the regression line (solid circles in Figure 3C), in contrast, for the more flexible interface region of the Xiap-BIR3, that is also flexible in the interaction with Caspase 9 (Figure 3E), predicted affinities show a poor, or even lack of, correlation to experimental values (solid triangles in Figure 3C).
Last but not least, when we calculate the simple correlation between molecular weight of the inhibitor and its corresponding affinity, significant correlations emerge (r2 = 0.43, N = 27, p-value = 0.0002), albeit weaker compared to the derived HADDOCK2P2I score (r2 = 0.51, N = 27, p-value < 0.0001), indicating that 3D information is needed to more efficiently predict ligand affinities.
Compilation of a Structure-Based Data Set for PPI Inhibitors with IC50 Values. Prediction of IC50 Data with the HADDOCK Score and HADDOCK2P2I
To assess the performance of both the original HADDOCK score and HADDOCK2P2I, we compiled a data set of related affinity data that include a total of 19 protein-inhibitor complexes for 6 different protein–protein interactions (Table 2). Four complexes are new and have not been incorporated in the training/cross-validation set, including inhibitors of the interaction between human transcriptional coactivator p75 (LEDGF) and viral integrase. The derived inhibitors have corresponding IC50s ranging from 12 μM to 19 nM. Tighter binding is exhibited by inhibitors of another host–virus interaction, that between a viral integrin and CD54, also known as Intercellular Adhesion Molecule 1 (ICAM 1), where IC50s span from 69 nM to 400 pM. Overall, the derived test set comprises different complexes with different binding constants, making this data set significantly distinct compared to the test/cross-validation set.
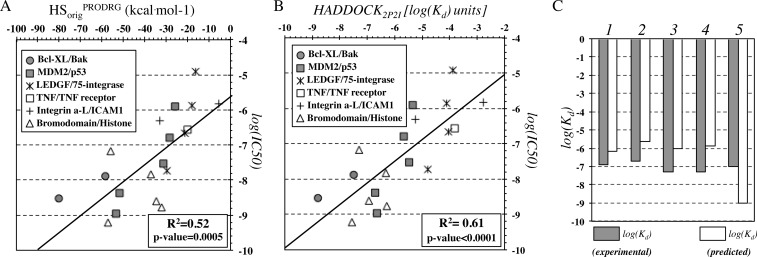
The best performance in predicting IC50 data is obtained for HADDOCK2P2I (r2 = 0.61, N = 19, p-value < 0.0001) but predictions with the original HADDOCK score are also reliable (r2 = 0.52, N = 19, p-value = 0.0005) (Figure 4A and B). Note, however, that these results should be treated with caution because the method was developed to reproduce Kd measurements in a quantitative manner and not IC50 values. Unfortunately, we could not compile an independent test set of Kd or Ki data due to the absence of combined structural and affinity data for protein–protein interaction inhibitors. There is however a clear trend for HADDOCK2P2I to relate to IC50 values for different complexes. The molecular weight of these ligands also correlates with affinity (r2 = 0.37, N = 19, p-value = 0.006), but the correlations with HADDOCK2P2I are much better (r2 = 0.61, N = 19, p-value < 0.0001), and those by the original HADDOCK score are also higher (r2 = 0.52, N = 19, p-value = 0.0005). This confirms the need of structural information to gain additional insights into ligand affinity.
Figure 4.
Correlation plots of experimentally determined binding affinities with (A) the original HADDOCK score (Evdw + 0.2Eelec + Edesol) and (B) the optimized HADDOCK2P2I score. (C) Prediction of affinities for inhibitors of bromodomains that recognize acetylated histone tails for complexes 1–5 corresponding to PDB IDs 3ONI, 4FLP, 3MXF, 3P5O, and 3ZYU, respectively.
HADDOCK2P2I was also used to predict Kd values for 5 known complexes of the same family and initially found in iPPI-DB. These data concern different bromodomains (BRD4, BRD2, BRDT), for which small-molecule inhibitors have been developed to disrupt their interaction with the ε-N-acetylated histone tails. Overall, the prediction of Kd for this set is reasonable, and the absolute mean error of the prediction is ∼1.6 kcal mol–1 (Figure 4C). Note that the highest prediction error (∼1.9 kcal mol–1) corresponds to the complex between BRD4 and I-BET151 (GSK1210151A) inhibitor. The complex buries an unusually large surface area (1351 Å2) that directly leads to higher predicted affinity. Note that large buried surface area in macromolecular complexation could be the outcome of extended conformational changes upon binding and, therefore, hamper accurate affinity estimation.16
Data Obtained from Different Experimental Methods Profoundly Limits the Prediction Accuracy
The iPPI-DB10 has the largest collection of affinity data for modulators/inhibitors of protein–protein interactions to date. It also includes affinity data for compounds targeting the same protein–protein interaction that were measured using different experimental methods. Multiple Kd data are rather scarce. Inhibition of the bromodomain 3 activity (BRD3) by compound entry 1603 in iPPI-DB was measured both by isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR). A 4-fold difference between affinities was observed (KdITC = 50 nM, Kd = 200 nM). For the same compound targeting the bromodomain 4 (BRD4) activity, a 6-fold difference in affinities was observed (KdITC = 55 nM, Kd = 324 nM). Even measurements with the same biophysical methods can vary – as an example, compound entry 713 in iPPI-DB inhibits the XIAP-Smac interaction in the μM range, but measured data with fluorescence polarization (FP) reach a 1.3-fold difference in Kd (KdFP1 = 0.275 μM, Kd = 0.209 μM).
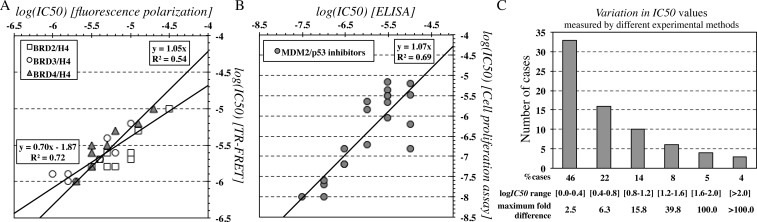
IC50 values obtained from various cellular assays are more abundant in the database. We have compiled a list that contains multiple IC50 measurements for 72 interactions (Supporting Information Table S3). In particular, FP and fluorescence resonance energy transfer (TR-FRET) were used to measure inhibition of different bromodomains for a variety of ligands (Figure 5A). Correlation between methods is strong for individual bromodomains (BRD1 r2 = 0.76, N = 9, p-value = 0.002, BRD2 r2 = 0.80, N = 9, p-value = 0.001; BRD3 r2 = 0.88, N = 8, p-value = 0.001), but when considered together, the relation is less profound, explaining 72% of the data (r2 = 0.72, N = 26, p-value < 0.0001). What is also clear is that IC50s obtained with TR-FRET are always of better affinity, reaching a 5-fold decrease in IC50, corresponding to 0.7 in −log scale. Therefore, a correction of the relationship with an additional β parameter is needed (y = 0.7x + 1.9, β = 1.9) (Figure 5A); When modeling affinity, however, the biophysical data are not corrected by this additional parameter because we assume a 1:1 relation between affinities measured from different experimental methods. In that case, the derived relationships lead to much lower correlations, albeit still significant (r2 = 0.54, N = 26, p-value < 0.0001) (Figure 5A). Similar conclusions have been made by experimentalists themselves, showing that 36 sulfonamides aiming to inhibit BRD4 activity exhibit similar IC50 values, but not identical between cell-based and biochemical assays (r2 = 0.57, N = 36, p-value < 0.0001).42
Figure 5.
Assessment of experimental uncertainty in the determination of IC50 for PPI inhibitors using different assays. (A) Inhibitors of various bromodomains, measured by both fluorescence polarization and fluorescence resonance energy transfer (TR-FRET). Two regression lines are shown (y = αx + β and y = αx), highlighting that absence of the β coefficient substantially lowers r2. (B) Inhibitors of the MDM2/p53 interaction, measured by both ELISA and cell proliferation assays. (C) Distribution of fold differences in IC50 for 72 inhibitors targeting various protein–protein interactions that have been measured by two (or more) different experimental methods.
Another example is the inhibition of the MDM2/p53 interaction using different ligands and measured by both ELISA and cell proliferation assays (CPA) (Figure 5B); we observe that IC50 values do indeed correlate without adding a correction coefficient β to their linear relation (r2 = 0.68, N = 21, p-value < 0.0001), but measurements may differ by more than 2 orders of magnitude (e.g., for entry 1143 of iPPI-DB, IC50ELISA = 0.1 μM, and IC50CPA = 158 μM).
The distribution of the overall ambiguity in affinity measurements is shown in Figure 5C for all 72 entries with multiple experimental measurements collected from iPPI-DB (Supporting Information Table S3). Nearly half of the interactions measured have an ambiguity of <0.4 in −log IC50 units, which corresponds to less than 2-fold changes in IC50. The other half of the data exhibit deviations in IC50s by more than 2-fold, reaching a maximum of 1650-fold change, the latter representing changes in −log IC50 by 3.2 units. Overall, the average experimental uncertainty for these data is rather low, 0.7 ± 0.7 −log IC50 units, with a median of 0.5 −log IC50 units and a maximum of 3.2 −log IC50 units.
Considering such deviations in −log IC50 units for a single system using two independent measurements, the prediction error of HADDOCK2P2I on the validation set (0.8 ± 0.6 kcal mol–1), on the “fuzzy” IC50 test set (1.6 ± 0.9 kcal mol–1) and the Kd set concerning inhibition of different bromodomains (1.3 ± 0.5 kcal mol–1), is in the same range of accuracy as experimental uncertainty.
Discussion
HADDOCK2P2I is a biophysical model optimized to predict the binding affinities of inhibitors of PPIs. By optimizing the weights of the original HADDOCK score and including a BSA term, the resulting model is able to predict binding affinities of PPI inhibitors close to experimental error (∼2-fold larger). To test this, we have compiled from iPPI-DB and analyzed a set of binding affinities obtained with various experiments for the same system. We have estimated an experimental uncertainty for IC50 values of 0.7 ± 0.7 in −log IC50 units (based on 72 data points), with a maximum of 3.2 −log IC50 units, the latter seemingly rare. Experimental conditions can also influence binding affinity determination, especially changes in pH. However, modeling the effect of pH was outside the scope of this study; this might well, if properly accounted for, lower the prediction error. Although the effect of pH on the binding affinity of PPI inhibitors is unclear at this time, it is known that for protein–protein complexes, changing the pH by three units, changes the Kd by a factor of 10–50, and ΔG by 1.4–2.3 kcal mol–1.16
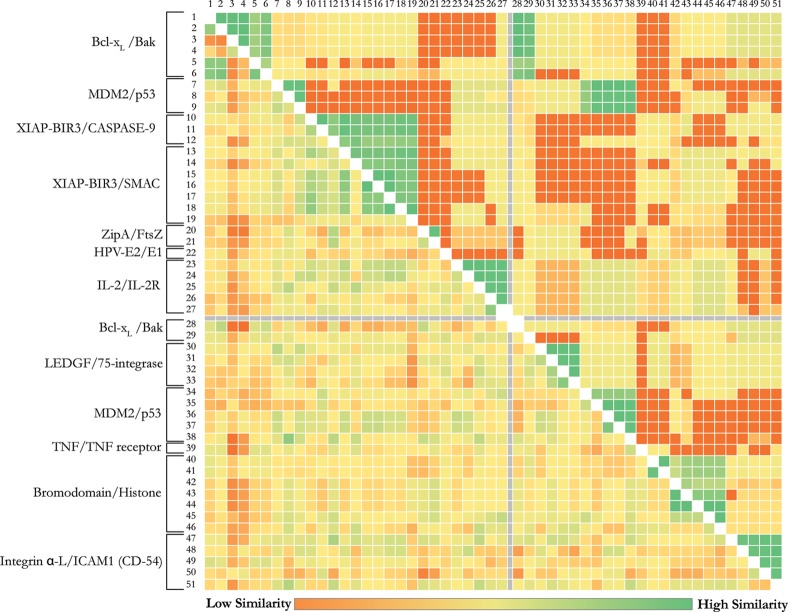
Despite those limitations, our algorithm reasonably reproduces a variety of experimental affinities of different nature (IC50, Ki, Kd) for distinct protein–protein interaction inhibitors. Since new PPI inhibitors are regularly published and crystallized with associated biophysical measurements for their interaction, this leaves room for further optimization of our HADDOCK2P2I binding affinity predictor. One could possibly argue that, because of the limited size of the data set, the prediction capacity of HADDOCK2P2I is not generalizable. Previous studies on scoring functions for “classical” protein–ligand complexes have shown that such limited amount of training data leads to a bias, which could only be surpassed when more than 100 cases are available in a data set.43 The diversity of the data as well as number of predictor variables used may also influence the results. To exclude a potential lack of diversity, we performed a similarity analysis of the proteins and ligands included in both training/cross-validation and test sets; this highlights the diversity of the studied systems and reflects their nonredundancy (Figure 6). Even for systems that have highly homologous protein structures, single mutations in the sequence, being directly at the interface or not, are often observed that could have implications in the binding energies of the ligands.
Figure 6.
All-versus-all similarity analysis for the proteins33 and their bound inhibitors34 of the used data set, highlighting the diversity in the systems under study (shown at the left of the matrix). The upper-right and the lower-left halves of the matrix represent the all-versus-all similarity for the proteins and their bound inhibitors, respectively. Rows and columns 1–27 and 28–51 correspond to the training/cross-validation set and the independent test set, respectively (following numbering introduced in Tables 1 and 2).
In this work, two different ligand parametrization tools were compared: a semiquantum mechanical approach (ACPYPE), and the faster, database-driven PRODRG. Both parametrization schemes yield similar performance in terms of binding affinity prediction using the optimized HADDOCK score, with PRODRG slightly outperforming ACPYPE. The main difference between the two sets of parameters resides in the electrostatics partial charges. While this might not affect much affinity prediction based on refined crystal structures, it might well have a much more profound impact on docking results, something that should be evaluated in the future.
The prediction performance for PPI inhibitors seems somewhat better than that of the most recent protein–ligand/drug design program. The latter, when tested against new blind data sets, showed a predictive capacity ranging from r2 = 0.30 to 0.40.44,45 For a fair comparison, HADDOCK2P2I and other small ligand binding affinity models should be tested against similar data sets. One test set used in this study contains IC50 data; these cannot be related to actual Kd or Ki measurements from biophysical methods, since substrate Michaelis constant (Km) and related concentration must be reported (S), assuming the inhibitor is competitive. If S ≪ Km, then Ki ≈ IC50, but again, verification from classical biophysical methods is advisable.
PPI inhibitors differ in nature from small molecule inhibitors that target enzymes and it remains to be seen how well the optimized function presented here will predict small molecule binding affinities (which was outside the scope of this work).
Next to yielding an optimized function for binding affinity prediction, this study also provides new insights into the determinants of the binding affinity of PPI inhibitors. In particular, affinity prediction deteriorates as a function of conformational freedom of the system under study, directly pointing to missing entropic contributions (Figure 3C–E). These results are similar in nature with the ones we derived for protein–protein complexes and with the impact of conformational change on binding affinity prediction.15,16 Nevertheless, the buried surface area (BSA) and van der Waals interactions show moderate-to-high correlations with binding affinity data for all complexes tested. The two are of course directly correlated. The desolvation and electrostatic energies show more complicated profiles than just plain correlation. This does not mean that they do not contribute per se: both the original and optimized models for the HADDOCK score clearly show a direct contribution of these two components, albeit to a smaller extent than the BSA: The BSA contributes on average, 3.6 ± 1.3 kcal mol–1 to the overall HADDOCK2P2I score, whereas the contributions of electrostatics and desolvation only reach 0.5 ± 0.4 and 0.1 ± 0.2 kcal mol–1, respectively, when calculated over the entire validation data set.
The BSA and van der Waals interactions are features that were already known to be critical factors for the affinity of protein–ligand46 and protein–protein complexes.47,48 The correlations calculated in this study for the BSA of inhibitors of protein–protein interactions are, however, substantially higher than the ones calculated for protein–protein complexes,16 but smaller than for standard protein–ligand complexes.46 This indicates that, in designing highly affine inhibitors of PPIs, one should aim at optimizing the complementarity of the inhibitors with the protein interface and always consider conformational changes as these are a limiting factor for accurate prediction.49 In conclusion, the newly designed score, HADDOCK2P2I, should facilitate and guide the design of PPI inhibitors, especially for the less flexible interfaces.
Acknowledgments
This work was supported by the Dutch Foundation for Scientific Research (NWO) through a VICI grant (No. 700.56.442) and a Focus and Massa grant from Utrecht University. The authors thank the many biochemists who measured the binding constants used in this article.
Supporting Information Available
Details of the data set of binding affinity data for protein–protein interaction inhibitors (Table S1), details of the optimization of the HADDOCK score (Table S2), and associated affinities for systems that were measured by two of more methods (Table S3). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† Panagiotis L. Kastritis: EMBL Heidelberg, Meyerhofstraße 1, 69117 Heidelberg, Germany, Tel. +49 6221 387–8107.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Costanzo M.; Baryshnikova A.; Bellay J.; Kim Y.; Spear E. D.; Sevier C. S.; Ding H.; Koh J. L.; Toufighi K.; Mostafavi S.; Prinz J.; St Onge R. P.; VanderSluis B.; Makhnevych T.; Vizeacoumar F. J.; Alizadeh S.; Bahr S.; Brost R. L.; Chen Y.; Cokol M.; Deshpande R.; Li Z.; Lin Z. Y.; Liang W.; Marback M.; Paw J.; San Luis B. J.; Shuteriqi E.; Tong A. H.; van Dyk N.; Wallace I. M.; Whitney J. A.; Weirauch M. T.; Zhong G.; Zhu H.; Houry W. A.; Brudno M.; Ragibizadeh S.; Papp B.; Pal C.; Roth F. P.; Giaever G.; Nislow C.; Troyanskaya O. G.; Bussey H.; Bader G. D.; Gingras A. C.; Morris Q. D.; Kim P. M.; Kaiser C. A.; Myers C. L.; Andrews B. J.; Boone C. The genetic landscape of a cell. Science 2010, 327, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M.; Cusick M. E; Barabasi A. L. Interactome networks and human disease. Cell 2011, 144, 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Protein–protein interaction inhibitors get into the groove. Nat. Rev. Drug Discovery 2012, 11, 173–175. [DOI] [PubMed] [Google Scholar]
- Koes D. R.; Camacho C. J. PocketQuery: Protein–protein interaction inhibitor starting points from protein–protein interaction structure. Nucleic Acids Res. 2012, 40, W387–W392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes D.; Khoury K.; Huang Y.; Wang W.; Bista M.; Popowicz G. M.; Wolf S.; Holak T. A.; Domling A.; Camacho C. J. Enabling large-scale design, synthesis and validation of small molecule protein–protein antagonists. PLoS One 2012, 7, e32839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes D. R.; Camacho C. J. Small-molecule inhibitor starting points learned from protein-protein interaction inhibitor structure. Bioinformatics 2012, 28, 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes C.; Host H.; Camproux A. C.; Laconde G.; Leroux F.; Mazars A.; Deprez B.; Fahraeus R.; Villoutreix B. O.; Sperandio O. Designing focused chemical libraries enriched in protein–protein interaction inhibitors using machine-learning methods. PLoS Comput. Biol. 2010, 6, e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villoutreix B. O.; Labbe C. M.; Lagorce D.; Laconde G.; Sperandio O. A leap into the chemical space of protein-protein interaction inhibitors. Curr. Pharm. Des. 2012, 18, 4648–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli X.; Bourgeas R.; Roche P. Chemical and structural lessons from recent successes in protein–protein interaction inhibition (2P2I). Curr. Opin. Chem. Biol. 2011, 15, 475–481. [DOI] [PubMed] [Google Scholar]
- Labbe C. M.; Laconde G.; Kuenemann M. A.; Villoutreix B. O.; Sperandio O. iPPI-DB: A manually curated and interactive database of small non-peptide inhibitors of protein–protein interactions. Drug Discovery Today 2013, 18, 958–968. [DOI] [PubMed] [Google Scholar]
- Bourgeas R.; Basse M. J.; Morelli X.; Roche P. Atomic analysis of protein–protein interfaces with known inhibitors: the 2P2I database. PLoS One 2010, 5, e9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse M. J.; Betzi S.; Bourgeas R.; Bouzidi S.; Chetrit B.; Hamon V.; Morelli X.; Roche P. 2P2Idb: A structural database dedicated to orthosteric modulation of protein–protein interactions. Nucleic Acids Res. 2013, 41, D824–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffery P.; Derreumaux P. Flexibility and binding affinity in protein–ligand, protein–protein and multi-component protein interactions: limitations of current computational approaches. J. R. Soc., Interface 2012, 9, 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger D. M.; Jessen G.; Gohlke H. How good are state-of-the-art docking tools in predicting ligand binding modes in protein-protein interfaces?. J. Chem. Inf. Model. 2012, 52, 2807–2811. [DOI] [PubMed] [Google Scholar]
- Kastritis P. L.; Bonvin A. M. Are scoring functions in protein–protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J. Proteome Res. 2010, 9, 2216–2225. [DOI] [PubMed] [Google Scholar]
- Kastritis P. L.; Moal I. H.; Hwang H.; Weng Z.; Bates P. A.; Bonvin A. M.; Janin J. A structure-based benchmark for protein–protein binding affinity. Protein Sci. 2011, 20, 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.; Macias A. T.; MacKerell A. D. Jr. Computational identification of inhibitors of protein–protein interactions. Curr. Top. Med. Chem. 2007, 7, 63–82. [DOI] [PubMed] [Google Scholar]
- Metz A.; Pfleger C.; Kopitz H.; Pfeiffer-Marek S.; Baringhaus K. H.; Gohlke H. Hot spots and transient pockets: predicting the determinants of small-molecule binding to a protein-protein interface. J. Chem. Inf. Model. 2012, 52, 120–133. [DOI] [PubMed] [Google Scholar]
- Dezi C.; Carotti A.; Magnani M.; Baroni M.; Padova A.; Cruciani G.; Macchiarulo A.; Pellicciari R. Molecular interaction fields and 3D-QSAR studies of p53-MDM2 inhibitors suggest additional features of ligand–target interaction. J. Chem. Inf. Model. 2010, 50, 1451–1465. [DOI] [PubMed] [Google Scholar]
- de Vries S. J.; van Dijk A. D.; Krzeminski M.; van Dijk M.; Thureau A.; Hsu V.; Wassenaar T.; Bonvin A. M. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 2007, 69, 726–733. [DOI] [PubMed] [Google Scholar]
- Dominguez C.; Boelens R.; Bonvin A. M. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Tirado-Rives J. T. The OPLS force field for proteins. Energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Fernandez-Recio J.; Totrov M.; Abagyan R. Identification of protein–protein interaction sites from docking energy landscapes. J. Mol. Biol. 2004, 335, 843–865. [DOI] [PubMed] [Google Scholar]
- Dutta S.; Burkhardt K.; Swaminathan G. J.; Kosada T.; Henrick K.; Nakamura H.; Berman H. M. Data deposition and annotation at the worldwide protein data bank. Methods Mol. Biol. 2008, 426, 81–101. [DOI] [PubMed] [Google Scholar]
- de Vries S. J.; van Dijk M.; Bonvin A. M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 8, 3684–3690. [Google Scholar]
- Brunger A. T.; Adams P. D.; Clore G. M.; DeLano W. L.; Gros P.; Grosse-Kunstleve R. W.; Jiang J. S.; Kuszewski J.; Nilges M.; Pannu N. S.; Read R. J.; Rice L. M.; Simonson T.; Warren G. L. Crystallography NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Gilson M. K.; Zhou H. X. Calculation of protein–ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 21–42. [DOI] [PubMed] [Google Scholar]
- Singh N.; Warshel A. Absolute binding free energy calculations: On the accuracy of computational scoring of protein–ligand interactions. Proteins 2010, 78, 1705–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R.; Friesner R. A.; Ghosh A.; Rizzo R. C.; Jorgensen W. L.; Levy R. M. New linear interaction method for binding affinity calculations using a continuum solvent model. J. Phys. Chem. B 2001, 105, 10388–10397. [Google Scholar]
- Cheng Y.; Prusoff W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- Rice P.; Longden I.; Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Needleman S. B.; Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [DOI] [PubMed] [Google Scholar]
- Rogers D. J.; Tanimoto T. T. A computer program for classifying plants. Science 1960, 132, 1115–1118. [DOI] [PubMed] [Google Scholar]
- Bolton E.; Wang Y.; Thiessen P. A.; Bryant S. H. PubChem: Integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 2008, 4, 217–241. [Google Scholar]
- Sattler M.; Liang H.; Nettesheim D.; Meadows R. P.; Harlan J. E.; Eberstadt M.; Yoon H. S.; Shuker S. B.; Chang B. S.; Minn A. J.; Thompson C. B.; Fesik S. W. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science 1997, 275, 983–986. [DOI] [PubMed] [Google Scholar]
- Shiozaki E. N.; Chai J.; Rigotti D. J.; Riedl S. J.; Li P.; Srinivasula S. M.; Alnemri E. S.; Fairman R.; Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 2003, 11, 519–527. [DOI] [PubMed] [Google Scholar]
- Schuttelkopf A. W.; van Aalten D. M. PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [DOI] [PubMed] [Google Scholar]
- Sousa da Silva A. W.; Vranken W. F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A.; Cheatham T. E. 3rd; Darden T.; Gohlke H.; Luo R.; Merz K. M. Jr.; Onufriev A.; Simmerling C.; Wang B.; Woods R. J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessene G.; Czabotar P. E.; Colman P. M. BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug. Discovery 2008, 7, 989–1000. [DOI] [PubMed] [Google Scholar]
- Bamborough P.; Diallo H.; Goodacre J. D.; Gordon L.; Lewis A.; Seal J. T.; Wilson D. M.; Woodrow M. D.; Chung C. W. Fragment-based discovery of bromodomain inhibitors part 2: optimization of phenylisoxazole sulfonamides. J. Med. Chem. 2012, 55, 587–596. [DOI] [PubMed] [Google Scholar]
- Wang R.; Liu L.; Lai L.; Tang Y. SCORE: A new empirical method for estimating the binding affinity of a protein-ligand complex. J. Mol. Model. 1998, 4, 379–394. [Google Scholar]
- Cheng T.; Li X.; Li Y.; Liu Z.; Wang R. Comparative assessment of scoring functions on a diverse test set. J. Chem. Inf. Model. 2009, 49, 1079–1093. [DOI] [PubMed] [Google Scholar]
- Warren G. L.; Andrews C. W.; Capelli A. M.; Clarke B.; LaLonde J.; Lambert M. H.; Lindvall M.; Nevins N.; Semus S. F.; Senger S.; Tedesco G.; Wall I. D.; Woolven J. M.; Peishoff C. E.; Head M. S. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [DOI] [PubMed] [Google Scholar]
- Olsson T. S.; Williams M. A.; Pitt W. R.; Ladbury J. E. The thermodynamics of protein–ligand interaction and solvation: Insights for ligand design. J. Mol. Biol. 2008, 384, 1002–1017. [DOI] [PubMed] [Google Scholar]
- Murphy K. P.; Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 1992, 43, 313–361. [DOI] [PubMed] [Google Scholar]
- Kastritis P. L.; Bonvin A. M. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J. R. Soc., Interface 2013, 10, 20120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastritis P. L.; Bonvin A. M. Molecular origins of binding affinity: seeking the Archimedean point. Curr Opin Struct Biol 2013, 23, 868–877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.