This work described the use of a method for the specific isolation of ribosome-associated mRNAs of in vivo–grown pollen tubes to identify over 500 genes with transcripts highly enriched or specific to in vivo–grown pollen tubes. T-DNA insertion alleles of a number of these genes conferred defects in late stages of the pollination process by haploid gametes.
Abstract
Transcriptome profiling has been used to identify genes expressed in pollen tubes elongating in vitro; however, little is known of the transcriptome of in vivo–grown pollen tubes due to the difficulty of collecting pollen that is elongating within the solid maternal gynoecium. Using a pollen-specific promoter (ProLAT52) to generate epitope-tagged polysomal-RNA complexes that could be affinity purified, we obtained mRNAs undergoing translation (the translatome) of in vivo–grown pollen tubes from self-pollinated gynoecia of Arabidopsis thaliana. Translatomes of pollen grains as well as in vivo– and in vitro–cultured pollen tubes were assayed by microarray analyses, revealing over 500 transcripts specifically enriched in in vivo–elongating pollen tubes. Functional analyses of several in vivo mutants (iv) of these pollination-enhanced transcripts revealed partial pollination/fertilization and seed formation defects in siliques (iv2, iv4, and iv6). Cytological observation confirmed the involvement of these genes in specialized processes including micropylar guidance (IV6 and IV4), pollen tube burst (IV2), and repulsion of multiple pollen tubes in embryo sac (IV2). In summary, the selective immunopurification of transcripts engaged with polysomes in pollen tubes within self-fertilized florets has identified a cohort of pollination-enriched transcripts that facilitated the identification of genes important in in vivo pollen tube biology.
INTRODUCTION
In flowering plants, pollen grains are the haploid male gametophytes that play a critical role in fertilization and crop production. Seeds and grains are a major food supply around the world, and their production depends on successful double fertilization of female gametophytes by male gametophytes. This process starts with the adhesion of pollen on the stigma, followed by grain hydration, tube generation and penetration of the stigma, and tube elongation inside the style (Berger et al., 2008). After exiting the transmitting tract of the pistil, the pollen tube continues to elongate along the funiculus, reaches the micropyle, enters the female gametophyte, penetrates one of the synergid cells, and bursts within the female gametophyte to release the two male gametes (sperm cells), which target the egg and the central cell in a double fertilization event (Nowack et al., 2006). After fertilization, the egg and the central cell develop into the embryo and the endosperm, respectively, giving rise to a young seed (Kawashima and Berger, 2011). The interaction between pollen tube, the male gametophyte, and the maternal pistil during pollination constitutes an excellent system to study cell-to-cell signaling and crosstalk, which are key to diverse cellular events in eukaryotes (Cheung and Wu, 2008; Wang et al., 2010; Chae and Lord, 2011). Moreover, the double fertilization event and the highly functional specialized male gametophyte are two major innovations that contribute to the evolutionary success of angiosperms (Borg et al., 2009).
When compared with pollen tubes cultured in vitro, those grown in the pistil exhibit a faster growth rate and longer tube lengths (Taylor and Hepler, 1997). Therefore, it has been long hypothesized that specific compounds within the maternal pistil promote pollen tube elongation and guidance during pollination. Indeed, several Arabidopsis thaliana mutants defective in ovule development, such as bell1, short integuments1, 47H4, and 54D12 display defects in the guidance of pollen tubes to ovules (Fowler and Quatrano, 1997). Arabidopsis female gametophytic mutants magatama1 (maa1) and maa3 also show defective female gametophyte development and pollen tubes guidance toward the micropyle (Shimizu and Okada, 2000; Shimizu et al., 2008). Several studies have successfully identified different maternal factors involved in chemotropic guidance of the pollen tube, including the tobacco (Nicotiana tabacum) TRANSMITTING TISSUE GLYCOPROTEIN (Wu et al., 1995), the Arabidopsis γ-aminobutyric acid (Palanivelu et al., 2003), the lily (Lilium longiflorum) stylar chemocyanin (Kim et al., 2003), and defensin-like molecules secreted by the synergid cells of Torenia fournieri (Okuda et al., 2009, 2013). Another important factor is the Arabidopsis NON-TRANSMITTING TRACT PROTEIN, which is responsible for expression of the genes HALF FILLED, BRASSINOSTERIOD ENHANCED EXPRESSION1 (BEE1), and BEE3, which are required for the development of the transmitting tract and for the elongation of pollen tubes in an apical-to-basal direction (Crawford and Yanofsky, 2011).
After exiting the transmitting tract of the pistil, the pollen tube requires two types of guidance to continue its journey to the ovule. The first system is guidance to the funiculus and the second guidance is to the micropyle (Higashiyama and Hamamura, 2008; Takeuchi and Higashiyama, 2011). Funicular guidance initiates when the pollen tube emerges from the surface of the septum (placenta) and moves onto the funicular surface, while micropylar guidance refers to the process in which the pollen tube extends onto the funicular surface and penetrates into the micropyle of the ovule (Higashiyama et al., 2003). The Arabidopsis CATION/PROTON EXCHANGERS (CHX) double mutant chx21 chx23 shows defective pollen tube guidance to the funiculus for fertilization (Lu et al., 2011). Additionally, the transcriptional regulator of the R2R3 family MYB98 in synergid cells (Kasahara et al., 2005), the factor CENTRAL CELL GUIDANCE in the central cell (Chen et al., 2007), and the GAMETE EXPRESSED PROTEIN1 in the egg cell (Alandete-Saez et al., 2011) are all critical for pollen tube guidance toward the micropyle. However, very little is known about the mechanisms by which pollen tubes respond to maternal signals.
Diverse transcriptomic and proteomic approaches have been used to address this question in several model plants. For instance, proteomic studies in rice (Oryza sativa) and Arabidopsis have shown that mature pollen contains more than 100 unique proteins that show differences in expression during pollen development (Holmes-Davis et al., 2005; Noir et al., 2005). In Arabidopsis, large-scale transcriptomic studies have concluded that pollen-specific transcripts show different expression profiles during pollen development and during extrusion of the pollen tube (Honys and Twell, 2004). Nevertheless, most of these studies were conducted under in vitro conditions due to the difficulty in collecting enough in vivo–grown pollen tubes from the pistil for characterization. The mature pollen grain accumulates large amounts of proteins, rRNAs, mRNAs, bioactive small molecules, and metabolic reserves to support rapid elongation of the pollen tube within the maternal pistil (Mascarenhas, 1993). In addition, the pollen tube must dynamically adjust its growth pattern based on complicated cell-to-cell interactions with the female pistil and on its rapid and long journey toward the ovule. In lily pollen tubes, this task requires dynamic biosynthesis of mRNA transcripts and proteins during in vivo growth (Huang et al., 2011). To investigate the transcriptome changes of pollinated pollen tubes, a semi–in vivo approach was used, identifying 383 genes enriched in pollen tubes emerging from cut pistils compared with in vitro–cultured ones (Qin et al., 2009). Moreover, by taking advantage of the hollow styles of Easter lilies (L. longiflorum cv Avita and cv Snow Queen), we were able to collect in vivo–grown pollen tubes to identify novel pollen tube transcripts and proteins induced by pollination (Huang et al., 2011).
In this study, the pollen tube mRNA species engaged in translation during pollination of an intact gynoecium were identified with the goal of recognizing genes important in late stages in the pollination process. Transgenic Arabidopsis plants (LAT52:HF-RPL18) harboring an epitope-tagged ribosomal protein L18 driven by the pollen-specific ProLAT52 (Twell et al., 1989, 1990) were used for mRNA-ribosome complex isolation (Zanetti et al., 2005). The method provides the advantage of requiring no dissection for the isolation of a subset of the ribosome-associated transcripts of a multicellular organ (Bailey-Serres, 2013). After collection of polyribosomal (polysomal) complexes from self-pollinated, nonpollinated styles and in vitro–cultured pollen tubes, the actively translated mRNAs (the translatome) were purified and hybridized to microarrays. This enabled a systematic analysis of mRNAs in the different samples, relative to pollen grains within buds and in vitro–germinated pollen tubes, and identified 519 transcripts present in in vivo–grown pollen tubes, of which 41 were highly enriched in this sample. To validate the biological function of these transcripts in pollination, T-DNA mutants of candidate genes were surveyed for defective fertility due to impaired micropyle guidance during pollination. We conclude that isolation of the ribosome-associated mRNAs from pollen tubes of fertilized gynoecia in vivo allows for the identification of genes that regulate late stages of pollination in flowering plants and paves the way for better understanding of the pollen-based mechanisms that promote fertilization.
RESULTS
Polysome Purification and mRNA Isolation from Self-Pollinated or Unpollinated Styles and Cultured Pollen Tubes
Pollen tubes growing in the style engage in complicated cell-to-cell interactions. Therefore, it is of interest to investigate the systematic and genome-wide changes of the pollen tube transcriptome in response to female signals in a model plant. However, to date, there has been no noninvasive means to evaluate gene regulation within the pollen tube of a pollinated pistil. To achieve this goal, we generated LAT52:HF-RPL18 transgenic Arabidopsis that express the ribosomal protein L18 (RPL18) tagged with a His6-FLAG dual-epitope driven by the tomato (Solanum lycopersicum) pollen-specific ProLAT52 (Twell et al., 1989, 1990) (Figure 1A). These transgenic plants allowed the specific immunopurification (IP) of polysomes (polysome-mRNA complexes) from in vivo–grown pollen tubes (Zanetti et al., 2005), providing a facile strategy to overcome the major obstacle of in vivo pollen tube collection and allowing evaluation of pollen transcripts engaged with ribosomes, and therefore likely to be undergoing active translation within pollinated styles.
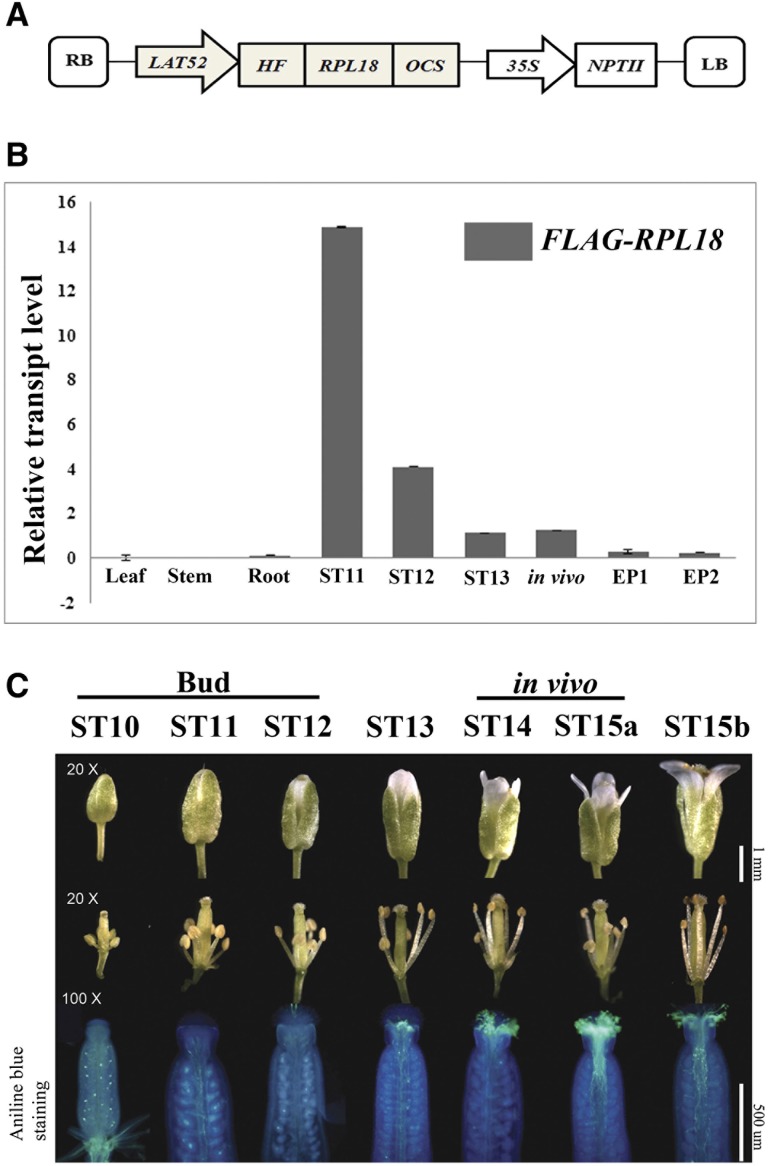
Figure 1.
Generation and Characterization of Arabidopsis LAT52:HF-RPL18 Transgenic Plants.
(A) Chimeric construct of the pollen-specific ProLAT52-driving His6-FLAG (HF) epitope-tagged 60S ribosomal subunit protein L18B, HF-RPL18, and a kanamycin resistance gene, NPTII, for immunoprecipitation of translation complexes and transgenic plant screening, respectively, used to generate transgenic Arabidopsis. Octopine Synthase (OCS) terminator; LB, left border of T-DNA; RB, right border of T-DNA.
(B) Validation of the floral stages in which FLAG-RPL18 mRNA accumulated in LAT52:HF-RPL18 transgenic plants. Quantitative RT-PCR analysis was performed with a primer set specific to FLAG-RPL18; ACT2 was used as the internal control. ST11-13: stage 11-13 flower buds; in vivo, stage 14 and stage 15a flower buds; EP1 and 2, two biological replicate samples of emasculated pistils.
(C) Photographs of all floral developmental stages used for collection of pollen grains and self-pollinated pollen tubes. Stages 10 to 12 (ST10-12) have developing microspores or mature pollen grains and are termed “bud” stage; stage 13 (ST13) flower buds are just about to open, the anthesis; stages 14 and 15a (ST14-15a) have pollen grains deposited on the stigma and are termed the “in vivo” pollination stage; stage 15b (ST15b) flowers have pollen tubes reaching the ovules indicating their fertilization. Aniline blue–stained gynoecia were used to visualize pollen tubes elongating in vivo.
We used the well-defined floral developmental stages 10-15b to select material at distinct stages of pollination (Smyth et al., 1990; Ferrándiz et al., 1999; Cai and Lashbrook, 2008; Cecchetti et al., 2008) (Figure 1C). Aniline blue staining showed there was no pollination of the developing microspore and mature grains at stages 10 to 12 (termed the “bud stage”). At the stages collectively termed the in vivo stages, anther dehiscence had occurred and many pollen tubes had reached the stigma (stage 14) and elongated one-third of the length of a style (stage 15a) (Figure 1C). Extension of the anthers above the stigma characterized floral stage 15a and the full expansion of the petals marked the later floral stage (stage 15b; Smyth et al., 1990).
Using this floral staging scheme, we harvested the buds/flowers and performed quantitative PCR analysis on LAT52:HF-RPL18 transgenic Arabidopsis to evaluate the spatiotemporal expression of FLAG-RPL18. FLAG-RPL18 transcript was detected at all of these floral stages with the highest accumulation at stages 11 and 12 (ST11 and ST12) (Figure 1B). The transcript was below the threshold of detection in vegetative organs and at very low level in pistils from emasculated pistils. These results confirm the strong temporal and specific expression of ProLAT52:FLAG-RPL18 at the targeted developmental stages. Furthermore, to define pollen maturity and germination at these floral stages, aniline blue staining was performed on pistils from self-pollinated ST10-ST15 flowers (Figure 1C). This confirmed that stigmata of bud stage florets contained the majority of mature pollen grains, whereas the ST14 and ST15a flowers possessed elongating pollen tubes, some of which had reached the gynoecia. At stage 15b (ST15b) the pollen tubes extended all the way to the ovules, as expected at or after fertilization. We termed the ST14-ST15a florets the “in vivo” pollination stage due to the enrichment in grains undergoing tube elongation and fertilization.
The ProLAT52:FLAG-RPL18 genotype was used in two different manners to purify polysome-mRNA complexes. In the first method, crude cell extracts (C) prepared with a polysome-stabilizing buffer were prepared from unpollinated bud stage flowers, in vivo stage flowers, and in vitro–cultured pollen grains/tubes (termed “in vitro” pollen) and used directly to affinity purify polysome-mRNA complexes by use of FLAG antibody–associated agarose beads (Zanetti et al., 2005). This yielded the crude extract-FLAG fraction (CF) samples. In the second method, the crude extracts were fractionated through Suc density gradients to obtain the polysome complexes from each tissue sample (Figure 2A; Supplemental Figure 1 and Supplemental Table 1). The pattern of polysome, monosome, and ribosome subunit peaks detectable in these absorbance profiles was typical for tissues with active protein biosynthesis. The polysome-enriched fractions were pooled and incubated with FLAG antibody–associated agarose beads to isolate the FLAG-tagged ribosome complexes of pollen. This yielded the crude extract-Suc gradient fraction-FLAG fraction (CSF). Both the CF and CSF fractions were used to obtain polysomal mRNA, and the crude cell extract was used to isolate total mRNA for each of the three tissue preparations.
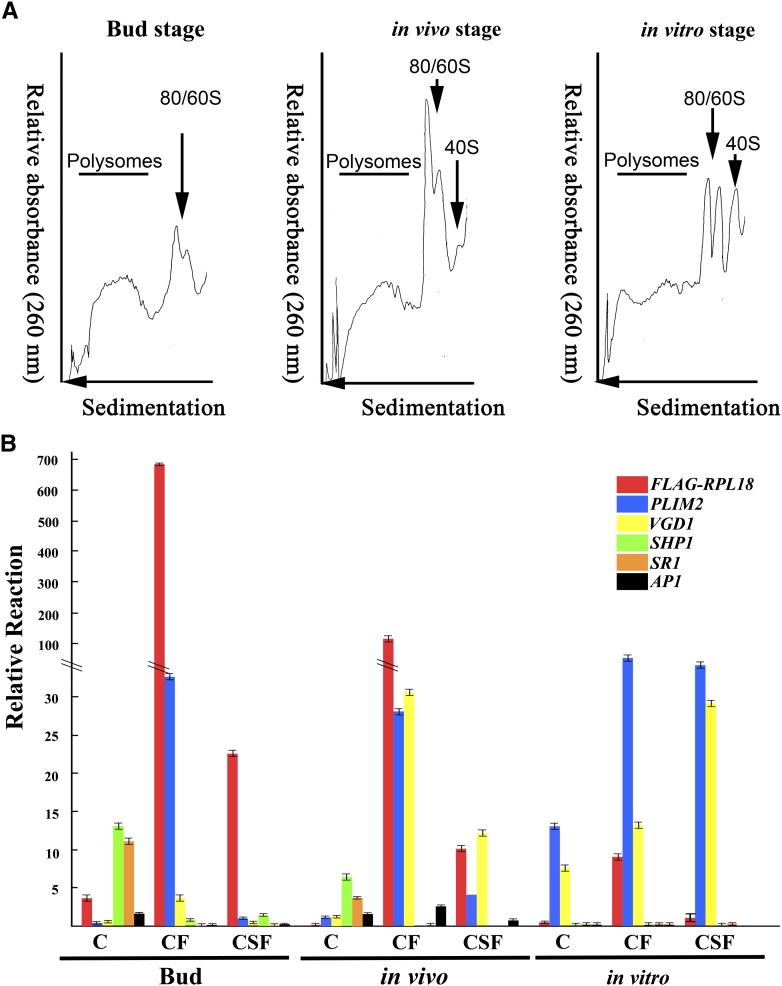
Figure 2.
Polysome UV Absorbance Profiles and Validation of the Specificity of IP of mRNAs Associated with Pollen from Pollinated Floral Buds, in Vivo–Pollinated Pollen Tubes, and in Vitro–Cultured Pollen Tubes of LAT52:HF-RPL18 Transgenic Plants.
(A) Typical Suc gradient absorbance (A260) profiles of ribosome complexes obtained from bud stage, in vivo stage, and in vitro–cultured pollen. Positions of peaks corresponding to polysomes (line), 40S ribosomal subunits, and 60S ribosomal subunit/80S monosomes (arrowheads) are indicated.
(B) The male gametophyte specificity of immunopurified polysomal mRNAs was examined by quantitative RT-PCR with primer sets targeting the FLAG-RPL18 transgene and organ-specific genes. Primers span both the His6-FLAG tag and the RPL18 sequence (FLAG-PRL18); pollen-specific PLIM2 and VGD1, female-specific SHP1 and SR, and petal/sepal-specific AP1. ACT2 was used as the internal control. Values mean ± se (n = three biological repeats).
The C, CF, and CSF fractions from bud, in vivo, and in vitro samples were examined by quantitative PCR analyses to evaluate enrichment of pollen- and pollen tube–specific mRNAs. The analysis included the FLAG-RPL18 transcript as well as Arabidopsis floral organ–specific mRNAs including pollen-specific LIM (PLIM2) (Arnaud et al., 2007; Wang et al., 2008) and VANGUARD1 (VGD1) (Pina et al., 2005), style-specific SHATTERPROOF1 (SHP1) (Alvarez et al., 2009), ovule-specific SERINE RACEMASE1 (SR1) (Fujitani et al., 2006; Boavida et al., 2011; Michard et al., 2011), and petal/sepal-specific APETALA1 (AP1) (Irish and Sussex, 1990). Compared with the total tissue mRNA, FLAG-RPL18 mRNA was significantly enriched in all of the CF and CSF samples (Figure 2B). The enrichment of the two pollen-specific transcripts, PLIM2 and VGD1, in these samples validated the utility of the ribosome IP approach to enrich for highly expressed pollen/pollen tube–specific mRNAs from intact floral tissue and isolated pollen. We also examined levels of floral organ–specific transcripts that are not expressed in pollen. This confirmed that SHP1, SR1, and AP1 mRNAs were abundant in the total mRNA (C) from bud and in vivo stage flowers but absent from that of in vitro–germinated pollen. These transcripts were significantly reduced in the immunopurified mRNAs from CF and CSF samples, except the in vivo CF sample (Figure 2B). Because of the detection of some AP1 mRNA in the CF from the in vivo sample, we decided to use the CSF in the IP of pollen polysomal mRNA in the subsequent experiments. Based on these results, LAT52:HF-RPL18 transgenic Arabidopsis can be used for IP of pollen-enriched polysomal transcripts from intact floral organs and germinating pollen.
Gene Cluster Evaluation of mRNAs Isolated from FLAG-Tagged Ribosomes of Florets and Pollen of LAT52:HF-RPL18
To evaluate the composition of polysomal mRNAs in the bud stage (IP-bud), in vivo stage (IP-in vivo), and in vitro stage (IP-in vitro) samples, their polysomal-mRNAs (translatome) were hybridized to Affymetrix GeneChips. As a first step, we performed a meta-analysis by pairwise comparisons of the translatomes of the four samples (bud [IP-bud], in vivo–stage floret [IP-in vivo], in vitro pollen [IP-in vitro], and seedlings [IP-seedling]; Branco-Price et al., 2008). This enabled identification of 5288 probe pair sets (genes) that showed a significant difference in transcript abundance in one or more of the five signal log2 ratio (SLR) comparisons (false discovery rate [FDR] < 0.05 and |SLR| ≥ 1) (Supplemental Data Set 1). The SLR values for the 5288 genes were used in k-means cluster analysis (k = 20) to identify coregulated transcripts. This analysis primarily recognized mRNAs enriched in the IP-in vitro sample relative to IP-bud and IP-seedling mRNAs with an average 2-fold or greater enrichment in clusters 10, 13, and 17 (Figure 3; Supplemental Data Set 1). Gene Ontology (GO) evaluation revealed that the genes in these clusters were enriched for biological functions involved in regulation of heat acclimation (adjusted P value 8.00E-04, cluster 10), autophagy (adjusted P value 6.05E-04, cluster 13), Golgi vesicle transportation (adj. P value 2.61E-03, cluster 13), protein import into peroxisome matrix (adjusted P value 3.09E-02, cluster 13), pollen tube development (adjusted P value 1.30E-11, cluster 17), plant-type cell wall modification (adjusted P value 1.24E-09, cluster17), actin filament–based process (adjusted P value 1.74E-05, cluster 17), and regulation of ARF GTPase activity (adjusted P value 3.60E-03, cluster 17). Many of these enriched genes are essential for pollen tube growth and fertilization (Supplemental Data Set 1) (Xiao and Mascarenhas, 1985; Goubet et al., 2003; Young et al., 2004; Song et al., 2006; Footitt et al., 2007; Suen and Huang, 2007; Harrison-Lowe and Olsen, 2008; Lycett, 2008; Szumlanski and Nielsen, 2009; Boavida et al., 2011; Peng et al., 2011; Zhang et al., 2011; Zhou et al., 2011). These results suggest that mRNAs from polysome-mRNA complexes of in vitro germinated pollen grains are enriched in transcripts previously validated as functioning in germination and tube growth of cultured pollen.
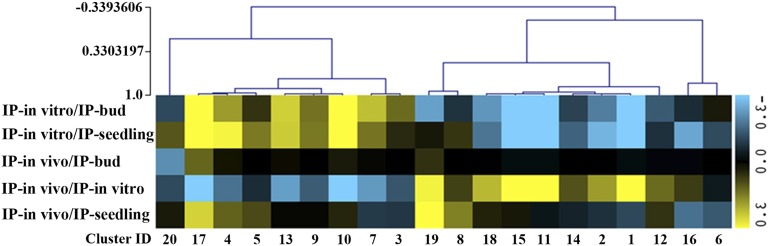
Figure 3.
Evidence of Distinctions between in Vivo– and in Vitro–Germinated Pollen Polysomal mRNA Populations in LAT52:HF-RPL18 Transgenic Plants.
Heat map for the 20 clusters displaying the average SLR for each of 20 k-means clusters identified by analysis of differentially expressed genes (probe pair sets) (|SLR| ≥ 1; FDR < 0.05; n = 5288) identified by pairwise comparisons of immunopurified polysomal (translatome) mRNA abundance determined by DNA microarray hybridization in this study or from publically available data for seedlings (Supplemental Data Set 1). IP-in vitro, immunopurified polysomal mRNA from in vitro–cultured pollen; IP-bud, immunopurified polysomal mRNA from pollen grains from bud stage florets; IP-seedling, immunopurified polysomal mRNA from 7-d-old seedlings. IP-in vivo, immunopurified polysomal mRNA from in vivo–germinated pollen tube samples.
For the in vivo–grown pollen tubes (IP-in vivo), comparisons of the mean SLR values with IP-bud, IP-in vitro, and IP-seedling identified mRNAs enriched in IP-in vivo, such as clusters 8 and 19 (Figure 3; Supplemental Data Set 1). This analysis also indicated mRNAs enriched in the IP-in vivo compared with IP-in vitro, with and an average 2-fold or greater enrichment in clusters 1, 11, 15, 18, and 19 (Figure 3; Supplemental Data Set 1). This demonstrates that the translatomes of self-pollinated pollen tubes (in vivo) were distinct from ones obtained from in vitro–germinated pollen grains/tubes (in vitro). Interestingly, the mRNAs with significant upregulation in the in vivo grown pollen tubes were enriched for ribosome biogenesis (adjusted P value 1.10E-14, cluster 1), suggesting that ribosome biogenesis is active during in vivo tube elongation. There were a number of other GO categories that were significantly enriched in the in vivo pollen translatomes: hydrogen peroxide catabolic process (adjusted P value 1.81E-03, cluster 1), covalent chromatin modification (adjusted P value 1.21E-05, cluster 8), heat acclimation (adjusted P value 2.47E-03, cluster 8), gluconeogenesis (adjusted P value 1.21E-03, cluster 11), defense response to bacterium (4.02E-03, cluster 11), pentose-phosphate shunt (adjusted P value 4.68E-03, cluster 11), response to cadmium ion (adjusted P value 7.48E-15, cluster 15), Golgi organization (adjusted P value 7.29E-06, cluster 15), catabolic process (adjusted P value 2.59E-12, cluster 18), response to wounding (adjusted P value 9.53E-13, cluster 19), and hyperosmotic salinity response (adjusted P value 2.97E-08, cluster 19) (Supplemental Data Set 1). At this level of analysis, the data suggest that there are distinctions between the in vitro– and in vivo–grown pollen tubes, but also demonstrate that the mRNAs obtained from LAT52:HF-RPL18 transgenic florets correspond to transcripts encoding proteins with roles in diverse processes to support rapid pollen tube elongation and pollination/fertilization.
Highly Expressed Transcripts in Pollen Tubes during Pollination
The polysomal mRNAs isolated from different samples were further analyzed to identify gene transcripts specifically enriched in the in vivo elongating pollen tubes. First, by systematic comparison of our data sets, we identified 3034 mRNAs enriched in immunopurified polysomal mRNA of bud stage florets as well as in vitro–germinated and in vivo–pollinated pollen (Figure 4A). There were 519 mRNAs exclusively present in the in vivo-IP sample (Figure 4A; Supplemental Data Set 2). Of these, 67 were also found in the 1386 gene transcripts that highly accumulated during pollen–pistil interactions (Boavida et al., 2011) (Supplemental Figure 2B). We hypothesize that these 519 in vivo–pollinated pollen-enriched genes may be important in pollen germination and tube elongation in response to a pistil signal for tube guidance. GO analysis indicated overrepresentation of proteins involved in heme (adjusted P value 1.24E-04) and iron (adjusted P value 3.0E-03) binding in the in vivo–pollinated pollen-enriched mRNAs (Supplemental Data Set 3). To validate the finding of in vivo pollen-enriched mRNAs, quantitative RT-PCR was performed for 16 genes randomly chosen from the set of 519. The abundance of these mRNAs in hand-pollinated styles and in vitro–germinated pollen tubes samples from wild-type Arabidopsis was compared. All 16 gene transcripts were more abundant in the in vivo-IP samples, and some of them (i.e., AT1G17190, AT1G66540, AT2G23590, AT2G27500, and AT4G18425) were exclusively detected in the in vivo-IP sample (Figure 4B). Among the 519 pollen-enriched genes, some have been reported previously to be involved in male gametophyte development, such as PIN-FORMED3 (PIN3, AT1G70940) and PIN4 (AT2G01420). These may regulate auxin polar distribution in elongating pollen tube, as Arabidopsis PIN8 regulates auxin homoeostasis and maintains the optimal auxin levels critical for pollen development and functionality (Ding et al., 2012).
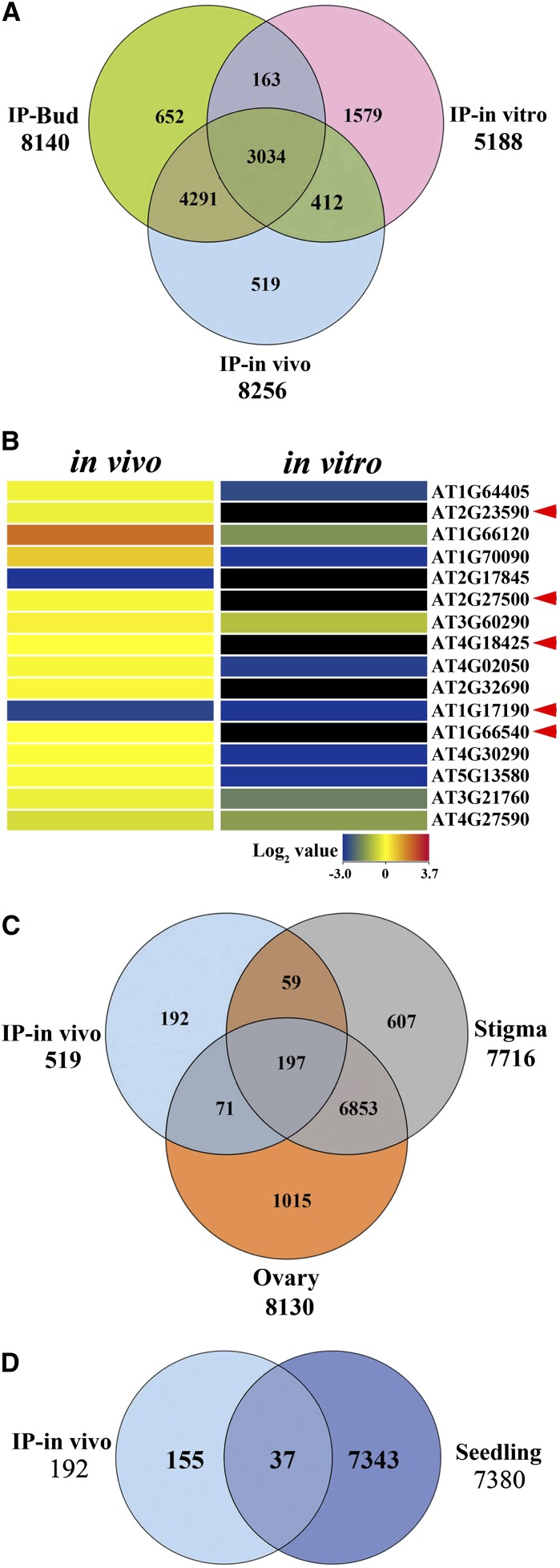
Figure 4.
Identification and Verification of Actively Translated Pollen Tube Transcripts Induced by Pollination in LAT52:HF-RPL18 Transgenic Plants.
(A) Comparison of genes (probe pair sets) with polysomal mRNA signal values >100 from IP-bud (bud, ST11-12, developing microspore, and mature pollen grains), IP-in vivo (ST14, elongating in vivo pollen tubes), and IP-in vitro (in vitro–cultured pollen tubes) by Venn diagram analysis identified 519 genes as candidates for specific involvement in pollen tube elongation.
(B) Heat map of in vivo (hand-pollination) and in vitro–cultured pollen tube total mRNA transcript levels for 16 randomly selected genes from the 519 IP-in vivo genes in (A), as determined by quantitative PCR of wild-type Arabidopsis. Each gene expression value is the mean of three biological replicate samples and duplicate technical replicates. Arrowheads indicate highly expressed transcripts in the in vivo pollen tubes.
(C) and (D) Venn diagram comparisons of IP-in vivo genes (519) with transcripts enriched in other cell types and tissues. Data obtained from enriched gene lists for stigma (Swanson et al., 2005), ovary (Swanson et al., 2005), and 7-d-old seedling (Swanson et al., 2005).
(C) Stigma-enriched genes (7716), ovary-enriched genes (8130), and IP-in vivo comparison.
(D) Seedling-enriched genes (7380) and IP-in vivo comparison.
To further recognize mRNAs specifically enriched in the polysomal mRNA of in vivo–grown pollen tubes, we compared the 519 in vivo–pollinated pollen-enriched mRNAs with transcriptomes of reproductive tissues (stigma and ovary) as well as seedlings (Swanson et al., 2005). This uncovered 192 transcripts highly enriched in elongating tubes in vivo compared with other reproductive tissues (Figure 4C) and 155 transcripts highly enriched in elongating tubes in vivo compared with vegetative tissue seedling (Figure 4D) (Swanson et al., 2005). Comparison of the 155 in vivo–pollinated pollen-specific transcripts to the 383 genes identified as pollen-enriched by semi–in vivo pollination (Qin et al., 2009) yielded only one common gene (Supplemental Figure 2A). This suggests a significant distinction between pollen tube expression profiles of semi–in vivo and true in vivo–germinated pollen when comparing transcriptome-enriched to translatome-enriched transcripts. This was confirmed by separate comparisons of the 519 in vivo–pollinated pollen-enriched mRNAs and the reported semi–in vivo pollination-enhanced mRNAs with reported pollination-induced mRNAs (Boavida et al., 2011). Whereas an overlap of 13% (67/519) was observed with the in vivo and pollination-induced mRNAs (Supplemental Figure 2C), there was only 4% (17/383) overlap between the semi–in vivo and the pollination-induced mRNAs (Supplemental Figure 2C).
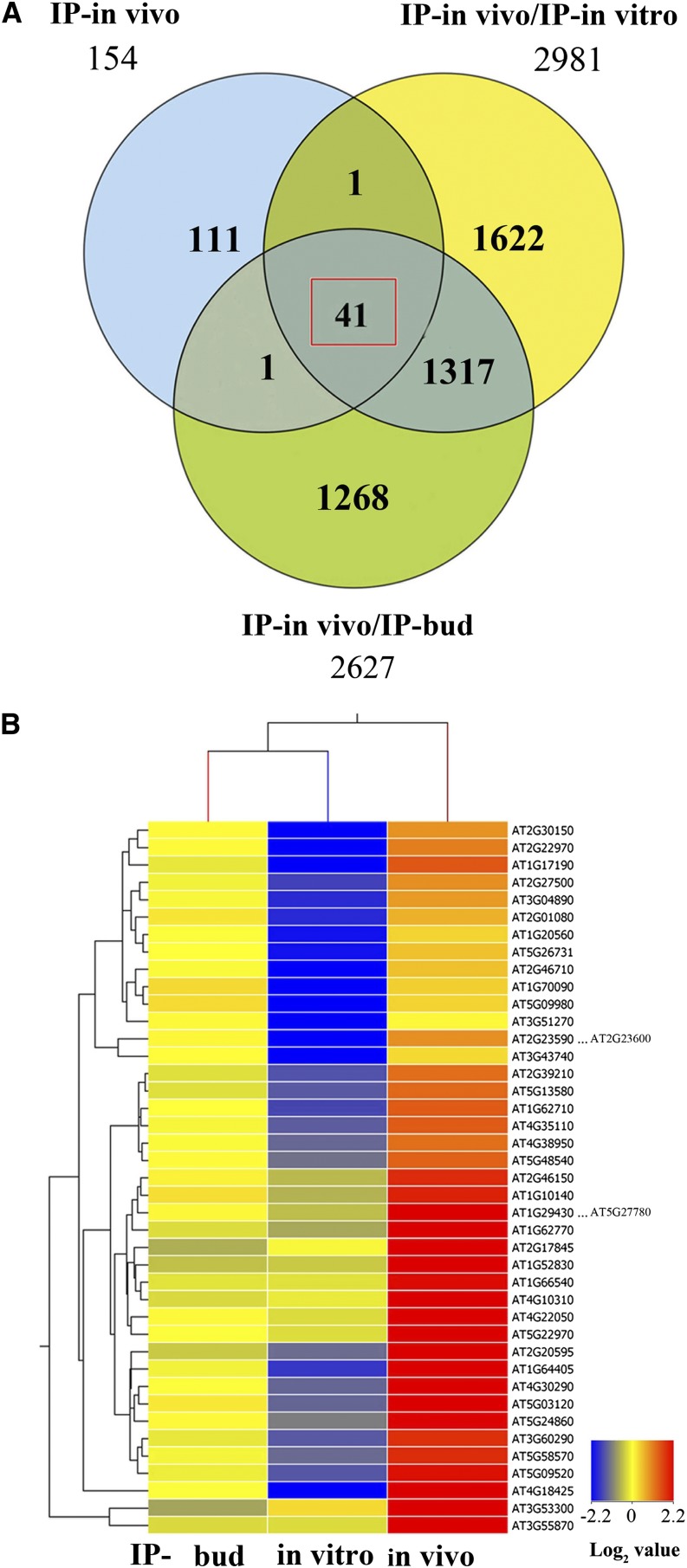
To further identify mRNAs highly enriched in the in vivo pollen translatome, a one-way ANOVA was performed. The use of a 5-fold enrichment as the threshold to parse the significantly upregulated in vivo pollen-specific genes from the IP-in vivo/IP-bud and IP-in vivo/IP-in vitro comparisons identified 41 mRNAs that were highly enriched in the in vivo–germinated pollen tubes (Figure 5A; Supplemental Data Set 2). A heat map display confirms the significantly enhanced accumulation of these transcripts in the in vivo sample compared with bud stage florets and in vitro–germinated pollen (Figure 5B). With few exceptions, these mRNAs were more abundant in bud stage flowers than in in vitro–germinated pollen. These results suggest that interaction of pollen grain/tube with the gynoecium, stigma and style may enhance transcriptional and/or translational gene regulation in male gametophytes. We also confirmed that these 41 mRNAs were recognized as in vivo pollen-enriched using the GeneSpring platform or robust multichip average (RMA) methods for differential gene expression analyses (R2 values 0.68 and 0.80 for the IP-in vivo/IP-bud and IP-in vivo/IP-in vitro comparisons, respectively) (Supplemental Figure 3). Five genes were selected for validation by quantitative RT-PCR and confirmed to have significantly higher abundance in the in vivo-IP pollen sample compared with bud stage flowers or in vitro–germinated pollen (Table 1). Altogether, these results confirm the identification of 41 transcripts that are highly enriched in the polysomal mRNA of pollen tubes of in vivo–fertilized florets, indicating they may be important during pollination.
Figure 5.
Identification of Actively Translated Transcripts Enriched in in Vivo–Grown Pollen Tubes.
(A) To identify transcripts highly expressed and enriched in the in vivo–germinated pollen tubes, the 154 transcripts identified after subtracting seedling and semi–in vivo expressed transcripts (Figure 4D; Supplemental Figure 2A) were compared with the signal value of the comparisons of in vivo-IP/in vitro-IP and in vivo-IP/bud-IP polysomal mRNA samples. This revealed 41 transcripts with signal changes >5-fold in either in vivo-IP/in vitro-IP or in vivo-IP/bud-IP comparisons.
(B) Heat map of 41 transcripts highly enriched in the in vivo–grown pollen tube translatomes. Values are the normalized averages generated with GeneSpring and presented as the log2 value in the heat map.
Table 1. Quantitative RT-PCR Validation of Five Specifically and Highly Enriched Transcripts in the Polysomal mRNA of Pollen Tubes of in Vivo–Fertilized Florets.
| Fold Enrichment in Pollen
Tubes | |||||||
|---|---|---|---|---|---|---|---|
| Relative Transcript Abundance
(in Vivo Pollen Tubes/Bud Comparison) |
Relative Transcript Abundance
(in Vivo Pollen Tubes/in Vitro Pollen Tubes) |
||||||
| Gene ID | Microarray | qRT-PCR | sd | Gene ID | Microarray | qRT-PCR | sd |
| AT3G53300 | 33.72 | 279.95 | 183.86 | AT4G18425 | 38.72 | >1000 | 0 |
| AT3G55870 | 13.63 | 19.23 | 13.49 | AT2G23590 | 33.34 | >1000 | 0 |
| AT2G17845 | 7.99 | 9.72 | 3.58 | AT1G64405 | 18.17 | >1000 | 0 |
| AT2G20595 | 7.88 | 3.77 | 0.3 | AT1G17190 | 16.39 | >1000 | 0 |
| AT5G22970 | 7.57 | 5.14 | 2.07 | AT3G53300 | 16.24 | >1000 | 0 |
qRT-PCR, quantitative RT-PCR.
Functional Studies of in Vivo Pollination-Enriched mRNAs Using T-DNA Insertion Mutants
To elucidate the possible roles of the genes encoded by the mRNAs that were highly enriched in pollen tubes of naturally pollinated florets, all available SAIL and SALK T-DNA insertion lines for 34 out of 41 genes were obtained and screened for abnormal seed set phenotypes. Our goal was to determine if the in vivo pollen translatome was enriched in genes involved in late stages of pollination that had been recalcitrant to identification in functional genomics studies. Among the 48 available T-DNA insertion lines for these 34 specifically and highly enriched in vivo pollen mRNAs, 12 (24%; iv1, 2, 3, 4, 6, 19, 20, 25, 34, 36, 37, and 39) showed obvious unfertilized seeds in mature siliques (Supplemental Figure 4 and Supplemental Table 2), but their flower and leaf development were similar to the wild type.
Prior to their use in functional studies, we confirmed that the lines from the SAIL collection had a single insertion in the indicated genes. Plants were self-crossed and the F1 progeny phenotype for BastaR was used to identify the lines with single T-DNA insertions. Three of the seven lines (iv2, 6, and 25) had a F1 BastaR frequency value below 75%, but only iv2 (χ2 = 10.54) and iv6 (χ2 = 7.17) had segregation ratios consistent with a single T-DNA insertion event (Table 2; Supplemental Figure 5A). Next, we examined the male fertility of iv2 and iv6 heterozygotes in reciprocal crosses. The BastaR frequency of the F1 progeny from wild-type pistils pollinated with pollen from the mutants was <50% for iv2 (46.34%, χ2 = 5.26) and iv6 (37.86%, χ2 = 17.73), suggesting reduced male gamete transmission for both mutants. By contrast, the BastaR frequency of the F1 progeny from heterozygous iv2 and iv6 mutant pistils pollinated with wild-type pollen was ∼50% for both iv2 (51.69%) and iv6 (50.35%) (Table 2), indicating no affect on female fertility. PCR-based genotyping of individual heterozygous mutants confirmed cosegregation of the T-DNA and reduced seed set phenotype (Table 2; Supplemental Figure 5B).
Table 2. Characterization of the Mutation Transmission Rates of Selected iv Mutants (SAIL Lines) by Self-Fertilization and Reciprocal Cross.
| Transmission of SAIL
Insertion Alleles | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant Lines | Self-Fertilized Heterozygous
Mutant |
Cross (Wild Type ♀
× Heterozygous Mutants ♂) |
Cross (Heterozygous Mutants
♀ × Wild Type ♂) |
||||||||
| % BastaR (Seedlings) | χ2 (75% BastaR) | P Value | % BastaR | Total | P Value | χ2 (50% BastaR) | % BastaR | Total | χ2 (50% BastaR) | P Value | |
| Wild type | 0 (459) | ||||||||||
| iv2 | 69.69 (739) | 10.97* | 9.26 × 10−4 | 46.34 | 986 | 5.26* | 2.18 × 10−2 | 51.69 | 445 | 0.51 | 0.475 |
| iv3 | 93.50 (139) | ||||||||||
| iv6 | 69.78 (493) | 7.17* | 7.41 × 10−3 | 37.86 | 309 | 17.73* | 2.55 × 10−5 | 50.35 | 143 | 0.43 | 0.511 |
| iv19 | 82.91 (392) | ||||||||||
| iv25 | 70.18 (218) | 2.7 | |||||||||
| iv36 | 87.18 (117) | ||||||||||
| iv37 | 100 (100) | ||||||||||
Asterisks indicate significant difference from expected (75% in self-cross or 50% in reciprocal crosses; χ2, P < 0.05).
We also investigated SALK mutants for the targeted genes (Supplemental Table 2) and selected those that produced <25% homozygous progeny when heterozygotes were self-fertilized (iv1 and iv4). The iv4 mutant met this criterion (Table 3), but the NPTII gene carried on the T-DNA was apparently silenced due to the lack of kanamycin resistance among the progeny. DNA gel blot analysis confirmed that the iv4 mutant had a single T-DNA insertion (Supplemental Figure 5C). We also used reciprocal crosses to examine the male gamete transmission. When wild-type pistils were fertilized with pollen grains from iv4 heterozygotes (Supplemental Figure 5B), only 45.35% (χ2 = 7.62) heterozygous F1 progeny were obtained; however, the reciprocal cross produced 57.14% iv4 heterozygotes (Table 3). Thus, iv4 reduced male gamete transmission and consequentially seed set, as seen for iv2 and iv6 (Figures 6F to 6H). We subsequently performed RT-PCR to determine the levels of transcripts produced in the iv2, 4, and 6 mutant homozygotes (Supplemental Figure 5D). This was performed with mRNA from stage 14 to stage 15a florets with results indicating that iv2, 4, and 6 are knockdown mutants. Altogether, these results indicate that the iv2, 4, and 6 affect pollen tube elongation and/or fertilization.
Table 3. Characterization of the Mutation Transmission Rates of Selected iv Mutants (SALK Lines) by Self-Fertilization and Reciprocal Cross.
| Transmission of SALK
Insertion Alleles | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Self-Fertilized Heterozygous
Mutant |
Cross (Wild Type ♀
× Heterozygous Mutants ♂) |
Cross (Heterozygous Mutants
♀ × Wild Type ♂) |
||||||||||||
| Mutant Lines | No. Wild Type (%) | No. Heterozygous (%) | No. Homozygous (%) | Total | No. Wild Type (%) | No. Heterozygous (%) | Total | χ2 (1:1) | P Value | No. Wild Type (%) | No. Heterozygous (%) | Total | χ2 (1:1) | P Value |
| iv1 | 21 (15.7%) | 62 (46.3%) | 51 (38%) | 134 | ||||||||||
| iv2 | 37 (25.9%) | 77 (53.8%) | 29 (20.3%) | 143 | 482 (54.65%) | 400 (45.35%) | 882 | 7.62* | 5.78 × 10−3 | 153 (42.86%) | 204 (57.14%) | 357 | 7.29* | 6.93 × 10−3 |
Asterisks indicate significant difference from expected (50% in reciprocal cross; χ2, P < 0.01). The progeny genotype was determined by PCR.
Figure 6.
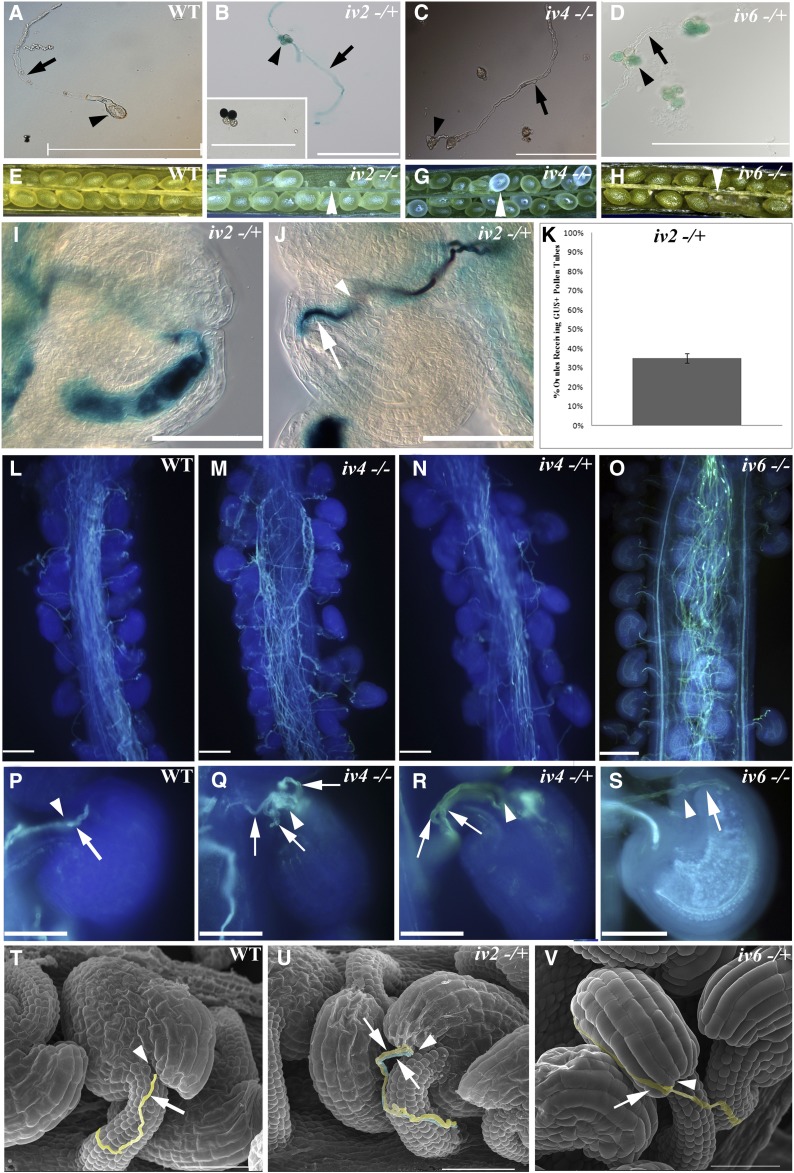
Defective Pollen Tube Guidance and Fertilization Found in the T-DNA Insertion Mutants of Several IV Genes.
(A) to (D) Compared with the wild type, several self-pollinated heterozygous and/or homozygous iv mutants show normal in vitro pollen tube elongation. iv2 and iv6 are SAIL mutants in the qrt1 genetic background containing a ProLAT52-driven GUS reporter gene that could be used to track tube elongation and fertilization of haploid mutant pollen (inset in [B]). The in vitro pollen germination and tube elongation of iv2−/+, iv4−/−, and iv6-/+ pollen grains show no significant difference to the wild type.
(B) and (D) Arrowheads and arrows indicate the tetrad pollen grains and the GUS-stained haploid mutant pollen tubes of iv2−/+ and iv6−/+ heterozygous pollen, respectively.
(C) Arrowheads and arrows indicate the pollen grain and pollen tube of iv4−/− homozygous pollen, respectively.
(E) to (H) Siliques of the wild type (E) are filled with seeds, whereas siliques of iv2−/− (F), iv4−/− (G), and iv6−/− (H) show severe seed abortion (arrowheads).
(I) and (J) Photographs of wild-type pistils pollinated with heterozygous iv2−/+ pollen. GUS staining revealed normal pollen tube micropylar penetration and bursting (I). In some ovules, iv2 mutant pollen tube (arrow) penetrates the micropyle (arrowhead) but fails to burst (J).
(K) Quantitative analysis of fertilization by GUS-stained iv2−/+ pollen tubes in 141 ovules reveals reduced fertilization by iv2 haploid mutant pollen.
(L) to (S) Aniline blue staining reveals pollen tube behavior and fertilization of SALK line iv4 and SAIL line iv6 pollen on wild-type styles. (L) and (P) are self-pollinated wild-type pistils containing a single pollen tube (arrow) that reaches the micropyle (arrowhead). Both homozygous (iv4−/− in [M] and [Q]) and heterozygous (iv4−/+ in [N] and [R]) iv4 pollen tubes elongate deeply in wild-type pistils but show multiple pollen tube reception by a single micropyle ([Q] and [R]). Some homozygous iv6−/−pollen tubes elongate deeply in wild-type styles (O) but fail to track toward the micropyle (S).
(T) to (V) Scanning electron micrographs of heterozygous iv2 (iv2−/+) or heterozygous iv6 (iv6−/+) pollen grains on wild-type gynoecia. In the wild type, only one pollen tube (arrow) penetrates the micropyle (arrowhead in [T]), whereas two heterozygous iv2 pollen tubes move toward the same micropyle (U) and heterozygous iv6 pollen tubes elongate along the surface of ovules, failing to targeting to the micropyle (V).
Since the SAIL mutants are in the quartet (qrt) background with a β-glucuronidase (GUS) reporter gene driven by ProLAT52, the pollen produced from a single meiosis can be monitored. Therefore, we tracked pollen germination and tube elongation by X-Gluc staining in the SAIL iv mutants. By comparison to wild-type pollen tubes (Figure 6A), iv2 (Figure 6B) and iv6 (Figure 6D) heterozygous pollen quartets include two GUS-positive pollen grains (Figure 6B, inset) and two GUS-negative germinated grains. These data also support the conclusion that iv2 and iv6 are single T-DNA insertion mutants. Compared with the wild type, seed abortions were frequently found in the siliques of iv2 and iv6 mutants (arrowheads in Figures 6F and 6H), strongly indicating their importance in pollination.
To further verify the roles of IV2 and IV6 in male gamete transmission, reciprocal pollinations between mutant pollen and wild-type pistils were performed, and fertilization was visually monitored. We observed that iv2 pollen tubes reached and penetrated the micropyle, but only some burst and released the sperm cells for fertilization (Figure 6I), whereas others did not (Figure 6J, arrow). To further quantitate the transmission defect of iv2 pollen, we analyzed 141 wild-type ovules pollinated with iv2 heterozygous pollen and confirmed only 35% GUS-positive ovules (Figure 6K). This solidly indicates a reduction in fertilization success of iv2 pollen. Using scanning electron microscopy, we confirmed that iv2 heterozygote pollen applied to wild-type florets could result in two pollen tubes that penetrate the same micropyle (Figure 6U). These results indicate that IV2 is important in the process of release of sperm cells into the embryo sac. In the case of iv6 heterozygous pollen grains, the GUS staining was weaker than normal, enabling us to use aniline blue staining to monitor the growth of iv6 pollen tubes in wild-type pistils. Aniline blue staining showed the pollen tubes of iv6 homozygous grew well in styles (Figure 6O) but some passed around the micropyle (Figure 6S). The failure in guidance to the micropyle and fertilization by iv6 pollen in wild-type pistils was confirmed by scanning electron microscopy (Figure 6V).
Our investigation of iv4 homozygous mutant pollen grains led to the finding that they germinated well in vitro (Figure 6C) but conferred transmission defects in vivo. Self-pollinated iv4 homozygotes produced siliques with etiolated seeds and reduced seed number compared with the wild-type control (Figure 6G). Both iv4 homozygous and iv4 heterozygous mutant pollen grains were used to fertilize wild-type pistils and examined after over 16 h by aniline blue staining. This confirmed normal appearing pollen tube elongation and guidance to the micropyle in both genotypes (Figures 6M and 6N), but multiple pollen tubes penetrated a single ovule (Figures 6Q and 6R). For most flowering plants, only one pollen tube is allowed to penetrate the micropyle for double fertilization by the two sperm of the pollen. Our finding suggests that IV4 is important for pollen tube perception/response to the fertilized micropylar signal associated with repulsion of additional pollen tubes. In conclusion, the identification of polysomal mRNAs that are highly enriched in pollen of naturally pollinated florets enabled identification of three mutants, iv2, iv4, and iv6, that exhibit altered pollen tube behavior and reduced male gamete transmission.
DISCUSSION
Pollen-Enriched Transcripts Can Be Isolated by Ribosome IP
In flowering plants, the pollen tube is essential for delivery of two sperm cells to the egg and central cells within the embryo sac for double fertilization. Functional studies of genes involved in pollen tube elongation and sperm entry provide new insights into the interaction with the female partner during fertilization, aspects that are crucial for plant breeding. The analysis of gene transcripts enriched in elongating pollen tubes provides a functional genomics approach to identify and explore the genes of the male gamete required for pollination. To date, most genes identified and shown to uniquely function in elongating pollen tubes have been ascertained by studying in vitro–germinating pollen grains. Nevertheless, cultured pollen tubes never reach the growth rate and length as those of in vivo–fertilized flowers. This perhaps results from the activation of specific genes in in vivo–grown pollen tubes in response to female signals during pollination. In support of this, the carbohydrate-rich extracellular matrix secreted by the pistil, the pistil exudates, changed the gene transcript profile of pollen tubes elongating in vitro (Guyon et al., 2000). Due to the challenge of collecting in vivo–grown pollen tubes from the style, the mRNAs specifically enriched in pollen during the fertilization process have remained poorly characterized. Recently, several approaches to overcome this barrier have been reported, including extraction and comparison of transcripts from pollinated Arabidopsis gynoecia during different pollination stages (Boavida et al., 2011) and collection of pollen tubes extending from the cut gynoecia in a semi–in vivo pollination system (Qin et al., 2009). Here, by taking advantage of a pollen-specific promoter (ProLAT52) and affinity purification of tagged polysomal-RNA complexes, we obtained actively translated mRNAs of in vivo pollen tubes from pollinated LAT52:HF-RPL18 transgenic Arabidopsis gynoecia (Figures 1 and 2; Supplemental Figure 1). After collecting the polysomal mRNAs from mature pollen grains (unpollinated gynoecia/prepollination stage buds) and in vivo–cultured (naturally pollinated gynoecia), and in vitro–cultured pollen tubes, microarray hybridization and subsequent bioinformatic analyses identified 41 transcripts that were highly enriched in in vivo pollen tubes (Figures 4 and 5; Supplemental Figure 2A). Functional studies of selected T-DNA insertion mutants corresponding to these in vivo pollination pollen-enriched mRNAs (called iv mutants) revealed their involvement in micropylar guidance (IV6 and IV4), sperm cell discharge into the embryo sac, and repulsion of other pollen tubes following fertilization (IV2) (Figure 6).
The LAT52 protein is essential for pollen hydration on the stigma and tube elongation in the style during pollination in tomato (Muschietti et al., 1994; Gerola et al., 2000), and it can interact with pollen-specific kinase for pollen tube guidance and fertilization in tomato (Tang et al., 2002). ProLAT52 is active during late pollen development (Twell et al., 1989, 1990) and its specificity to the vegetative pollen cell has made it a useful tool for driving pollen-specific gene expression in diverse plant species including Arabidopsis. For example, it has been used to drive GUS expression as a cell-autonomous tag to screen T-DNA inserted male gametophytes and revealed 32 hapless mutants with abnormal tube growth within the ovary (Johnson et al., 2004). In combination with the qrt mutation, an Arabidopsis T-DNA insertional resource generated by a ProLAT52-GUS construct has been used to explore pollen tube genes identified from different studies including the semi–in vivo system (Qin et al., 2009) and this study. In most plants, the mature pollen grain contains many mRNA species and proteins to support rapid pollen germination and tube growth, but diverse posttranscriptional processes significantly affect the stability of pollen mRNA (Mascarenhas, 1993; Wang et al., 2004). Therefore, gene transcript data generated from the steady state mRNA population is likely to be less reflective of proteins undergoing biosynthesis during pollen tube elongation. In yeast, it has been shown that the polysomal mRNA population is more representative of newly synthesized proteins (Gygi et al., 1999), and this is likely to be true in plants, especially when translation is compromised by limiting levels of nucleotide triphosphates. In Arabidopsis, mRNAs from epitope-tagged polysomal complexes have been used for global translatome analyses in response to environmental stress (Zanetti et al., 2005; Branco-Price et al., 2008; Juntawong and Bailey-Serres, 2012; Juntawong et al., 2013), responses to hormones (Ribeiro et al., 2012), or distinct cell types (Mustroph et al., 2009; Jiao and Meyerowitz, 2010). Here, by use of the ProLAT52 to drive expression of a FLAG-tagged ribosomal protein, we achieved IP of polysomal mRNAs, presumably actively undergoing translation from mature grains and elongating pollen.
Pollen-Enriched Transcripts of in Vivo–Germinated Tubes Provide New Gene Candidates for Functional Genomics Analyses
A systematic analysis of the purified polysome-mRNA complexes from bud-stage florets, as well as in vitro– and in vivo–germinated pollen tubes, revealed 519 mRNAs that were highly enriched or specifically present in the in vivo–germinated pollen tubes (Figure 4A). Comparison of these with the 1386 pollination-induced genes identified previously (Boavida et al., 2011) identified 67 enriched mRNAs in common (Supplemental Figure 2B). Additionally, quantitative RT-PCR analyses of transcripts of 16 selected genes from the 519 confirmed their enrichment in in vivo–germinated pollen tubes (Figure 4B). Five of these mRNAs (AT3G60290, AT4G30290, AT1G64405, AT1G70090, and AT1G66120) were also present in the group of 67. Further comparison of the 519 pollen-enriched mRNAs with mRNAs highly expressed in stigmata, ovules, and seedlings narrowed our study to a group of 155 genes that were highly enriched in the in vivo–grown pollen tubes (Figures 4C and 4D). Surprisingly, there was only one gene in this set that was highly enriched in the transcriptome identified in semi–in vivo–grown pollen tubes (Qin et al., 2009) (Supplemental Figure 2A). A similar comparison between the published semi–in vivo pollen tube–enriched (Qin et al., 2009) and the 1386 pollination-induced mRNAs (Boavida et al., 2011) also showed minimal overlap (Supplemental Figure 2C).
The striking differences between the pollen-enriched mRNAs of our study and that of Qin et al. (2009) could be due to a number of factors. First, the semi–in vivo–grown pollen tubes collected after emerged from the cut style from agar possibly experience a dramatic change in environment. Second, there may be differences between the transcriptome (the steady and actively translated mRNAs) versus translatome (actively translated mRNAs) populations of germinating pollen tubes, as shown in control-grown seedlings (Branco-Price et al., 2008). Third, other maternal tissues, including transmitting tract and ovule, are likely to be responsible for the upregulation of pollen tube transcripts during pollination. In fact, the phenotypes of the mutants characterized here illustrate defects in pollen tube guidance and reception (Figure 6) and seed set (Supplemental Figure 4) that may be directed by maternal signals to the pollen. Therefore, it will be interesting to explore the roles of other genes identified in this study in the perception of maternal signals during later pollination phases that facilitate pollen tube guidance and fertilization.
Looking more closely at the pollen-enriched transcripts (i.e., the 519 from in vivo–germinated pollen), we identified a number associated with functions that are relevant to pollination. For example, transcripts were enriched for molecular function associated with iron ion (adjusted P value 3.0E-03) and heme binding (adjusted P value 1.24E-04) (Supplemental Data Set 3). This may reflect the recent finding that heme may participate in pollen tube burst during fertilization. It is known that oxygen content is extremely low in ovules (Linskens and Schrauwen, 1966; Blasiak et al., 2001), which may reduce hemeoxygenase activity and result in heme accumulation in pollen tubes. In pear (Pyrus pyrifolia), pollen tube accumulation of heme restricts potassium channel transport and thereby increases the osmolarity in the pollen tube, resulting in pollen tube burst (Wu et al., 2011). This and other enrichments of GO categories may provide important hints about pollen biology.
Functional Characterization of Pollination-Enriched mRNAs Identifies Genes Involved in Late Stages of Fertilization
In most flowering plants, only one pollen tube enters the ovule micropyle to release the two sperm cells during fertilization (Berger et al., 2008). However, pollen tubes of several iv mutants identified in this study displayed defective micropylar guidance and/or repulsion of additional pollen tubes during pollination (Figure 6). Two of these encode proteins associated with cell wall composition. IV6 encodes a putative xyloglucan endotransglucosylase/hydrolase, an enzyme important for construction of the xyloglucan cross-links in the cell wall (Sasidharan et al., 2010) and required for successful automatic self-pollination in Arabidopsis (Kurasawa et al., 2009). Aniline blue staining and scanning electron microscopy observation of self-pollinated homozygous iv6 and reciprocally crossed iv6 heterozygous plants exposed defective micropyle targeting (Figures 6O, 6S, and 6V).
IV2 encodes a putative methylesterase (MES8). In tobacco, a pollen pectin methylesterase gene, PPME1, is responsible for maintenance of the equilibrium between strength and plasticity in the apical pollen tube cell wall, as antisense inhibition of Nt-PPME1 expression caused a mild but decreased in vivo pollen tube elongation (Bosch and Hepler, 2006). The germination and growth of iv2 pollen tubes, genotyped by GUS staining in the SAIL background, was normal under in vitro culture conditions (Figure 6B). However, iv2 pollen exhibited reduced micropylar targeting (Figures 6J and 6K) concomitant with reduced male transmission (Table 2). The inability of iv2 pollen to sense an ovule signal for micropylar targeting was also evident from the observation of two pollen tubes growing along the surface of iv2 heterozygous ovules (Figure 6U).
Control of the cellular oxidation state may also be important in fertilization as IV4 encodes a putative glutathione transferase (GSTU26), which functions to conjugate a toxic substrate with glutathione. These S-glutathionylated conjugates are then transported to the vacuole by specific transporters for degradation (Dixon et al., 2002). Reciprocal crosses of iv4 heterozygotes and the wild type demonstrated reduced male transmission of iv4 pollen (Table 3). Aniline blue staining of iv4 pollinated styles revealed multiple pollen tubes competing for the same ovule (Figures 6Q and 6R), implicating IV4 in the micropylar repulsion mechanism that blocks pollination by additional pollen tubes. Recently, several studies indicated that defective fusion of the sperm cell with egg cell results in the attraction of multiple pollen tubes targeting toward the same ovule (Beale et al., 2012; Kasahara et al., 2012). It is of interest to further explore whether iv4 and iv2 pollen tubes may lose the ability to discharge the sperm cells in the micropyle and further trigger synergid cell degradation. Compared with the wild type, all iv mutants characterized here (iv2, iv4, and iv6) showed partial male gametophyte transmission (Tables 2 and 3). This was consistent with the observation that these insertion events only reduced but did not abrogate the accumulation of the transcripts of the disrupted genes (Supplemental Figure 5). However, we cannot exclude the possibility that the partial phenotype is due to gene redundancy. Based on these results and the absence of transmission defects or our inability to obtain homozygous recessive mutants, it seems likely that critical male gametophytic genes may be either redundant or absolutely required for gamete transmission.
In summary, through use of a method for the specific isolation of the ribosome-associated mRNAs of pollen, we succeeded in identifying over 150 genes with transcripts highly enriched or specific to in vivo–grown pollen tubes. T-DNA insertion alleles of a number of these genes conferred defects in late stages of the pollination process by haploid gametes. Further study of the genes with transcripts highly enriched in the polysomes of pollen tubes during in vivo pollination by genetic and cellular approaches can provide new insights into pollen tube growth, guidance, and pollen–ovule interactions essential for plant reproduction.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia-0 was grown in a growth chamber under 18-h light (8500 LUX)/6-h dark at 22°C. The mRNA for the microarray experiments was extracted from different floral developmental stages and in vitro–germinated pollen tubes of LAT52:HF-RPL18 transgenic Arabidopsis. SALK and SAIL lines were obtained from the ABRC and analyzed as described in Supplemental Methods. The SAIL lines used for GUS staining were in the qrt1 mutant genetic background containing a T-DNA insertion of plasmid-pCSA110 with the GUS reporter gene driven by the ProLAT52 and resistance to Basta (Johnson et al., 2004).
Generation of Arabidopsis LAT52:HF-RPL18 Transgenic Plants
Vector pGATA-HF-RPL18, a binary T-DNA vector that has the Gateway recombination cassette att1-cmR-ccdb-att2 just 5′ of the tobacco (Nicotiana tabacum) mosaic virus omega 5′ leader (66 bp), which is fused to a six-His-FLAG epitope-seven Gly peptide [(M(H)6(G)3DYKDDDDK] in frame with the 187–amino acid coding sequence of RPL18B was recombined with a pENTR/D-TOPO vector (Invitrogen) containing ProLAT52 (Twell et al., 1990), as described for construction of other promoter-HF-RPL18 constructs (Mustroph et al., 2009). Following Arabidopsis Columbia-0 transformation and initial genetic and biochemical characterization, a homozygous line expressing LAT52:HF-RPL18 was established. The primer pair for cloning the ProLAT52 is listed in Supplemental Table 3.
Collection of Flowers and in Vitro Pollen Tubes for Polysome Extraction
Stage 11 (ST11) and stage 15 (ST15) developmental floral buds were collected according to aniline blue staining results. ST10 to ST12 floral buds were classified as the “bud stage,” and the ST14 to ST15a flowers were termed “in vivo stage.” To extract polysome from bud stage and in vivo–grown pollen tubes, 800 to 900 and 500 to 700 flowers from LAT52:HF-RPL18 Arabidopsis were collected, respectively. To collect in vitro pollen tubes, pollen grains from 7200 LAT52:HF-RPL18 Arabidopsis stage 15 (ST15) flowers were collected in 15% germination medium (1 mM KCl, 5 mM CaCl2, 0.8 mM MgSO4, 1.5 mM boric acid, 15% [w/v] Suc, 10 mM myoinositol, and 5 mM MES, pH 5.8) with vigorous shaking. The flowers and medium mixture were centrifuged at 11,200g for 5 min at room temperature to obtain pollen grains. After removing flower tissues from the supernatant, the mixture was recentrifuged at 11,200g for 5 min. The pollen pellet was resuspended in 15% (w/v) germination medium (100 μL) and transferred onto 19.8% solid germination medium (1 mM KCl, 5 mM CaCl2, 0.8 mM MgSO4, 1.5 mM boric acid, 1% [w/v] agarose, 19.8% [w/v] Suc, 0.05% (w/v) lactalbumin hydrolysate, 10 mM myoinositol, and 5 mM MES, pH 5.8) for 4.5 h. The elongating pollen tubes were observed by light microscopy (BX51; Olympus). Prior to polysome extraction, 30 μL of polysome extraction buffer (PEB; 200 mM Tris-HCl, pH 9.0, 200 mM KCl, 25 mM EGTA, pH 8.3, 36 mM MgCl2, 5 mM DTT, 50 mg mL−1 cycloheximide, 50 mg mL−1 chloramphenicol, 0.5 mg mL−1 heparin, 1% [v/v] Triton X-100, 1% [v/v] Tween 20, 1% [w/v] Brij-35, 2% [v/v] polyoxyethylene, and 1% [w/v] deoxycholic acid) was used to sluice the growing pollen tubes from the medium into a 2-mL tube and subjected to polysome extraction.
Polysome Extraction from Arabidopsis LAT52:HF-RPL18 Transgenic Plants
To isolate polysomal RNA from bud stage, in vivo–grown pollen tubes, and in vitro–cultured pollen tubes of LAT52:HF-RPL18 Arabidopsis, we followed procedures described previously (Zanetti et al., 2005; Mustroph et al., 2009) with minor modifications. For one method of extraction, crude extract from frozen bud stage or in vivo stage flower tissue or freshly harvested in vitro grown pollen tubes (Supplemental Table 1) was extracted in PEB (1250 μL) and centrifuged at 13,400g for 20 min at 4°C, and the supernatant passed through a shredder column (Qiagen) to remove remaining cell debris. A spectrophotometer (DU 648B; Beckman) was used to detect the OD260 units of the flow through. The purified crude extract (500 μL, 30 to 40 OD260 units) was loaded on a 4.2-mL 20 to 60% Suc density gradient and ultracentrifuged at 18,800g for 3.5 h at 4°C in a SW 41Ti rotor (Beckman). After centrifugation, the absorbance profile of the gradient was detected using a UV monitor (Monitor UVIS-920; GE Healthcare) at absorbance 260 nm as the gradient was fractionated. Fractions containing polysomes (disomes and larger complexes) were collected for immunoprecipitation of the FLAG-tagged ribosome complexes. These samples were referred to as the CSF. Alternatively, the samples referred to as the CF were the crude extract from bud stage or in vivo stage flower tissue or freshly harvested in vitro–grown pollen tubes described above used for direct immunoprecipitation of FLAG-tagged ribosome complexes without Suc gradient separation.
IP of mRNAs from Polysome-mRNA Complexes
IP of polysomes from LAT52-HFRPL18 Arabidopsis was described previously (Zanetti et al., 2005) with minor modifications. Polysome obtained (2000 μL) from the Suc gradient separation or crude extract (2000 µL) were incubated with the anti-FLAG agarose beads (Anti-Flag M2-Agarose from mouse; Sigma-Aldrich) (100 μL) in a 15-mL plastic Falcon tube with 5 mL PEB for 2 h at 4°C with gentle shaking. The beads were washed four times with wash buffer (200 mM Tris-HCl, pH 9.0, 200 mM KCl, 25 mM EGTA, pH 8.3, 36 mM MgCl2, 5 mM DTT, 1 mM PMSF, 50 mg mL−1 cycloheximide, 50 mg mL−1 chloramphenicol, and 20 units mL−1 RNasin Ribonuclease Inhibitor [Promega]), and the complexes were eluted by treatment of the agarose beads with 300 μL of competition buffer (40 ng μL−1 3× FLAG peptide [Sigma-Aldrich] and 50 units mL−1 RNasin) for 30 min at 4°C. The eluent was combined with 300 μL phenol/chloroform (1:1 [v/v]) and followed by centrifugation for 20 min at 13,400g at 4°C to collect the supernatant. After isopropanol precipitation, the RNA pellet was resuspended in 20 μL of RNase-free water (Invitrogen Life Technologies) and stored at −80°C.
RNA Preparation for Hybridization with Gene Chips
Polysomal RNA of the bud stage, in vivo–grown pollen tubes, and in vitro–cultured pollen tubes samples were further processed with an RNA purification kit (RNeasy Mini Kit; Qiagen) and a single linear amplification (MessageAmp Premier RNA amplification kit) to generate the antisense RNA (aRNA). The aRNA quality and quantity were determined by use of a NanoDrop 2000/2000c spectrophotometer (Thermo Scientific). The aRNAs from three independent biological replicate samples of bud, in vivo, and in vitro polysomal mRNA were labeled with biotin by in vitro transcription in amplification kit and their integrity was examined by use of an Agilent 2100 Bioanalyzer (Agilent Technologies). The labeled aRNA was fragmented to 35 to 200 nucleotides, and bud, in vivo, and in vitro samples were hybridized to GeneChip Arabidopsis ATH1 genome arrays (Affymetrix) for 16.5 h at 45°C. The array signal was scanned by use of the Affymetrix GeneChip Scanner 3000 (Agilent Technologies). Normalization for each chip was performed with Microarray Suite version 5.0 (MAS 5.0). The nine CEL files of data are available from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/geoprofiles) as the Gene Expression Omnibus (GEO) series GSM1060242, GSM1060243, and GSM1060244 for bud stage, GSM1060245, GSM1060246, and GSM1060247 for in vivo–pollinated pollen tube, and GSM1060248, GSM1060249, and GSM1060250 for in vitro–cultured pollen grain/tube translatomes (polysomal mRNA transcript data)
Microarray Data Analyses Using GeneSpring
Eleven microarray data sets were collected from NCBI (http://www.ncbi.nlm.nih.gov/geoprofiles) for comparison to our data set. These included GEO files for total cellular mRNA of ovaries (GSM67078, GSM67079, GSM67080, and GSM67081), stigmata (GSM67082, GSM67083, GSM67110, and GSM67112), and seedlings (GSM67084, GSM67086, and GSM67087) (Swanson et al., 2005). These were coanalyzed with the pollen translatome data generated in this study using GeneSpring (GX 9) software. First, the RMA algorithm was used to normalize the data (Irizarry et al., 2003) and then all genes with a normalized expression value >100 were selected for further analysis. The gene list of pollen–pistil interaction was from Boavida et al. (2011) and the semi–in vivo gene list was also acquired for comparison (Qin et al., 2009). Venn diagram analysis was used for comparison with the different microarray data sets. To identify the significantly differentially expressed genes enriched in the in vivo sample, the microarray data from the bud, in vivo, and in vitro samples were analyzed by one way ANOVA, with the criteria for significance set as a P value < 0.005 and signal fold-change > 5 in both the in vivo/in vitro and in vivo/bud sample comparisons.
Microarray Analyses Using R Statistical Software in the Bioconductor Package
Eighteen microarray data sets were analyzed with the R program. These included the nine CEL files generated in this study and the 10 CEL files generated for 7-d-old control-grown seedlings (nonstress) from immunopurified polysomal mRNA from the Arabidopsis 35S:HF-RPL18 transgenic line (Branco-Price et al., 2008) (GEO accession number GSE9719). All CEL file data were normalized, and MAS5.0 was used to determine the “present,” “marginal,” or “absent” (P, M, A) calls for each probe pair set. RMA was used to calculate SLR values for the different sample comparisons. FDRs for identification of significant differences between probe pair sets in the samples were generated using P-value distributions (Smyth, 2004). Probe pair sets that met the MAS5.0 criteria for selection and had an FDR < 0.05 and |SLR| ≥ 1 in at least one comparison (n = 4158 probe pair sets) used as the data set for fuzzy k-means cluster analyses (Mustroph et al., 2009). The genes in each cluster were used for GO analysis to define the gene functions.
Quantitative RT-PCR Analysis of RNA
Total RNA was extracted by use of the RNeasy mini kit (Qiagen). For RT-PCR, 92 ng of total RNA was treated with DNaseI (Invitrogen Life Technologies) for 15 min, and first-strand cDNA was synthesized with 10 μM oligo(dT) primer and MMLV reverse transcriptase (Invitrogen Life Technologies). For quantitative RT-PCR, gene-specific primers pairs (Supplemental Table 3) were designed using a primer design program (Applied Biosystems). For each gene, six reactions were performed, including three technical replicates and two biological replicates. The genes encoding cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPDH; AT3G04120) and ACTIN2 (ACT2; AT3G18780) were used as controls for normalization of mRNA content between samples in pilot RT-PCRs. Quantitative RT-PCR was performed using the FastStart Universal SYBR Green Master (Roche) and an amplification procedure according to the ABI 7500 real-time PCR system (Applied Biosystems) (Yang et al., 2008). The amount of target (2–ΔCT) was obtained by normalizing to GAPDH or ACT2 as the endogenous reference.
Aniline Blue Staining of Pollen Tubes and GUS Staining of the Pollen Grains
Aniline blue detection of the callose plugs produced in pollen tubes was performed according to the procedure described previously (Szumlanski and Nielsen, 2009). To detect pollen tube behavior of iv4 and iv6 mutants, the T-DNA insertion mutants SALK_047724C and SAIL_62_A10 were used for aniline blue staining. SAIL line mutants carried the qrt1 mutation and a T-DNA insert which encodes the GUS reporter gene driven by the ProLAT52 (pCSA110). Heterozygous quartets of pollen (2 GUS+: 2 GUS-) were treated with X-Gluc for 8 h (Johnson et al., 2004), and GUS staining was visualized with a LSM 510 Meta confocal laser scanning microscope (Zeiss). To analyze the pollen tube behavior of iv2 mutant, the T-DNA insertion mutant SAIL_270_A07 was used for GUS staining.
Methods for Cryo-Scanning Electron Microscopy
Ovaries harvested after hand-pollination for 12 to 16 h and wild-type and iv mutant styles were isolated and loaded on stubs. The samples were frozen by liquid nitrogen slush and then transferred to a sample preparation chamber at −160°C. After 5 min, the temperature was raised to −85°C and the samples were sublimated for 15 min. After coating with Pt at −130°C, the samples were transferred to cryo-stages in a scanning electron microscopy chamber and observed at −160°C using a cryo-scanning electron microscope (FEI Quanta 200 SEM/Quorum Cryo System PP2000TR; FEI) at 20 kV.
Reciprocal Cross for Studying in Vivo Pollen Tube Growth
Flowers were emasculated at stage 12 (Smyth et al., 1990) for use as a female, and the pollen grains from stage 15 (Smyth et al., 1990) flowers were gently applied to the stigma for pollination.
Accession Numbers
Sequence data for the genes used in this study can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: AT2G23590 (methyl esterase 8), AT1G17190 (glutathione S-transferase), and AT4G30290 (xyloglucan endotransglucosylase/hydrolase). The nine CEL files of data are available from the NCBI (http://www.ncbi.nlm.nih.gov/geoprofiles) as the GEO series GSM1060242, GSM1060243, and GSM1060244 for bud stage, GSM1060245, GSM1060246, and GSM1060247 for in vivo–pollinated pollen tube, and GSM1060248, GSM1060249, and GSM1060250 for in vitro–cultured pollen grain/tube translatomes (polysomal mRNA transcript data)
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scheme for Extraction of Polysome-mRNA from in Vitro–Germinated Pollen (in Vitro), Pollinated (in Vivo Stage, ST14 and ST15a), and Nonpollinated (Bud Stage, ST10-12) Floral Buds.
Supplemental Figure 2. Three Venn Diagram Comparisons.
Supplemental Figure 3. Comparison of Fold Change Data Generated with GeneSpring and SLR Data Generated with R Program.
Supplemental Figure 4. The Silique Phenotypes and Seed Number per Silique of Several Self-Pollinated iv Mutants.
Supplemental Figure 5. T-DNA Insertion Patterns and Investigation of iv2-, iv4-, and iv6-Specific mRNAs Detected in Flowers of Corresponding Homozygous Mutants.
Supplemental Table 1. Number of the Flowers Used for the Polysome-mRNA Extraction of the Bud Stage, in Vivo Stage, and in Vitro–Germinated Pollen in Each of Three Biological Replicate Samples.
Supplemental Table 2. Investigation of Pollen Viability and Seed Abortion Pattern of Selected iv Mutants.
Supplemental Table 3. Primer Pairs Used in This Study.
Supplemental Methods. Analysis for T-DNA Mutants, Genomic DNA Extraction and DNA Gel Blot Analysis, and RT-PCR Analysis of RNA
Supplemental Data Set 1. The Average SLR Values of Comparisons in 20 Clusters and GO Analysis of Genes Used on Figure 3.
Supplemental Data Set 2. Characterization of the 519 Genes Abundant in the in Vivo Pollen Tube Polysomal mRNA Population and the 41 Genes Chosen for Further Study.
Supplemental Data Set 3. GO Analysis of the in Vivo 519 Genes Used in Figure 4A.
Supplementary Material
Acknowledgments
We thank Mei-Jane Fang (DNA Analysis Core Laboratory, Academia Sinica) for assistance with quantitative RT-PCR analysis, Shu-Jen Chou (DNA Microarray Core Laboratory, Academia Sinica) for assistance with microarray analysis, and Wann-Neng Jane (Plant Cell Biology Core Laboratory, Academia Sinica) for assistance with scanning electron microscopy. This work was supported by research grants from Academia Sinica (Taiwan), the National Science and Technology Program for Agricultural Biotechnology (098S0030055-AA; Taiwan), the National Science Council (99-2321-B-001-036-MY3 and 102-2321-B-001-040-MY3) to G.-Y.J., and the U.S. National Science Foundation (IOS-075081 and MCB-1021969) to J.B.-S.
AUTHOR CONTRIBUTIONS
S.-Y.L., P.J., J.B.-S., and G.-Y.J. conceived and designed the experiments. S.-Y.L., P.-W.C, and M.-H.C. performed the experiments. S.-Y.L., J.B.-S., and G.-Y.J. wrote the article.
Glossary
- CSF
crude extract-Suc gradient fraction-FLAG fraction
- CF
crude extract-FLAG fraction
- C
crude cell extract
- IP
immunopurification
- SLR
signal log2 ratio
- FDR
false discovery rate
- GO
Gene Ontology
- RMA
robust multichip average
- PEB
polysome extraction buffer
- aRNA
antisense RNA
- NCBI
National Center for Biotechnology Information
- GEO
Gene Expression Omnibus
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alandete-Saez M., Ron M., Leiboff S., McCormick S. (2011). Arabidopsis thaliana GEX1 has dual functions in gametophyte development and early embryogenesis. Plant J. 68: 620–632. [DOI] [PubMed] [Google Scholar]
- Alvarez J.P., Goldshmidt A., Efroni I., Bowman J.L., Eshed Y. (2009). The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21: 1373–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud D., Déjardin A., Leplé J.-C., Lesage-Descauses M.-C., Pilate G. (2007). Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J. (2013). Microgenomics: Genome-scale, cell-specific monitoring of multiple gene regulation tiers. Annu. Rev. Plant Biol. 64: 293–325. [DOI] [PubMed] [Google Scholar]
- Beale K.M., Leydon A.R., Johnson M.A. (2012). Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr. Biol. 22: 1090–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Hamamura Y., Ingouff M., Higashiyama T. (2008). Double fertilization - Caught in the act. Trends Plant Sci. 13: 437–443. [DOI] [PubMed] [Google Scholar]
- Blasiak J., Mulcahy D.L., Musgrave M.E. (2001). Oxytropism: A new twist in pollen tube orientation. Planta 213: 318–322. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., Borges F., Becker J.D., Feijó J.A. (2011). Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 155: 2066–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M., Brownfield L., Twell D. (2009). Male gametophyte development: A molecular perspective. J. Exp. Bot. 60: 1465–1478. [DOI] [PubMed] [Google Scholar]
- Bosch M., Hepler P.K. (2006). Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta 223: 736–745. [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K.A., Jang C.J.H., Larive C.K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56: 743–755. [DOI] [PubMed] [Google Scholar]
- Cai S., Lashbrook C.C. (2008). Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: Enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146: 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Falasca G., Costantino P., Cardarelli M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K., Lord E.M. (2011). Pollen tube growth and guidance: roles of small, secreted proteins. Ann. Bot. (Lond.) 108: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Li H.-J., Shi D.-Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.-C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572. [DOI] [PubMed] [Google Scholar]
- Crawford B.C.W., Yanofsky M.F. (2011). HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138: 2999–3009. [DOI] [PubMed] [Google Scholar]
- Ding Z., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3: 941. [DOI] [PubMed] [Google Scholar]
- Dixon D.P., Lapthorn A., Edwards R. (2002). Plant glutathione transferases. Genome Biol. 3: S3004–, 3010.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M.F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68: 321–354. [DOI] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A.R., Holdsworth M.J., Baker A., Theodoulou F.L. (2007). The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144: 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler J.E., Quatrano R.S. (1997). Plant cell morphogenesis: Plasma membrane interactions with the cytoskeleton and cell wall. Annu. Rev. Cell Dev. Biol. 13: 697–743. [DOI] [PubMed] [Google Scholar]
- Fujitani Y., Nakajima N., Ishihara K., Oikawa T., Ito K., Sugimoto M. (2006). Molecular and biochemical characterization of a serine racemase from Arabidopsis thaliana. Phytochemistry 67: 668–674. [DOI] [PubMed] [Google Scholar]
- Gerola P.D., Mol C.A., Newbigin E., Lush W.M. (2000). Regulation of LAT52 promoter activity during pollen tube growth through the pistil of Nicotiana alata. Sex. Plant Reprod. 12: 347–352. [Google Scholar]
- Goubet F., Misrahi A., Park S.K., Zhang Z., Twell D., Dupree P. (2003). AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 131: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon V.N., Astwood J.D., Garner E.C., Dunker A.K., Taylor L.P. (2000). Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in petunia. Plant Physiol. 123: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S.P., Rochon Y., Franza B.R., Aebersold R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe N.J., Olsen L.J. (2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4: 339–348. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Hamamura Y. (2008). Gametophytic pollen tube guidance. Sex. Plant Reprod. 21: 17–26. [Google Scholar]
- Higashiyama T., Kuroiwa H., Kuroiwa T. (2003). Pollen-tube guidance: Beacons from the female gametophyte. Curr. Opin. Plant Biol. 6: 36–41. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R., Tanaka C.K., Vensel W.H., Hurkman W.J., McCormick S. (2005). Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5: 4864–4884. [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-C., Chang L.-C., Wang M.-L., Guo C.-L., Chung M.-C., Jauh G.-Y. (2011). Identification and exploration of pollen tube small proteins encoded by pollination-induced transcripts. Plant Cell Physiol. 52: 1546–1559. [DOI] [PubMed] [Google Scholar]
- Irish V.F., Sussex I.M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Sorenson R., Bailey-Serres J. (2013). Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 74: 1016–1028. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Maruyama D., Hamamura Y., Sakakibara T., Twell D., Higashiyama T. (2012). Fertilization recovery after defective sperm cell release in Arabidopsis. Curr. Biol. 22: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Berger F. (2011). Green love talks; cell-cell communication during double fertilization in flowering plants. AoB Plants 2011: plr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Mollet J.-C., Dong J., Zhang K., Park S.-Y., Lord E.M. (2003). Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 100: 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa K., Matsui A., Yokoyama R., Kuriyama T., Yoshizumi T., Matsui M., Suwabe K., Watanabe M., Nishitani K. (2009). The AtXTH28 gene, a xyloglucan endotransglucosylase/hydrolase, is involved in automatic self-pollination in Arabidopsis thaliana. Plant Cell Physiol. 50: 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens H.F., Schrauwen J. (1966). Measurement of oxygen tension changes in the style during pollen tube growth. Planta 71: 98–106. [DOI] [PubMed] [Google Scholar]
- Lu Y., Chanroj S., Zulkifli L., Johnson M.A., Uozumi N., Cheung A., Sze H. (2011). Pollen tubes lacking a pair of K+ transporters fail to target ovules in Arabidopsis. Plant Cell 23: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett G. (2008). The role of Rab GTPases in cell wall metabolism. J. Exp. Bot. 59: 4061–4074. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J.P. (1993). Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5: 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E., Lima P.T., Borges F., Silva A.C., Portes M.T., Carvalho J.E., Gilliham M., Liu L.-H., Obermeyer G., Feijó J.A. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437. [DOI] [PubMed] [Google Scholar]
- Muschietti J., Dircks L., Vancanneyt G., McCormick S. (1994). LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6: 321–338. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S., Bräutigam A., Colby T., Schmidt J., Panstruga R. (2005). A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem. Biophys. Res. Commun. 337: 1257–1266. [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67. [DOI] [PubMed] [Google Scholar]
- Okuda S., Suzuki T., Kanaoka M.M., Mori H., Sasaki N., Higashiyama T. (2013). Acquisition of LURE-binding activity at the pollen tube tip of Torenia fournieri. Mol. Plant 6: 1074–1090. [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Brass L., Edlund A.F., Preuss D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59. [DOI] [PubMed] [Google Scholar]
- Peng J., Ilarslan H., Wurtele E.S., Bassham D.C. (2011). AtRabD2b and AtRabD2c have overlapping functions in pollen development and pollen tube growth. BMC Plant Biol. 11: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C., Pinto F., Feijó J.A., Becker J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138: 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro D.M., Araújo W.L., Fernie A.R., Schippers J.H.M., Mueller-Roeber B. (2012). Translatome and metabolome effects triggered by gibberellins during rosette growth in Arabidopsis. J. Exp. Bot. 63: 2769–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T., Yokoyama R., Nishitani K., Voesenek L.A., Pierik R. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.K., Ito T., Ishiguro S., Okada K. (2008). MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana. Plant Cell Physiol. 49: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.K., Okada K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518. [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: e3. [DOI] [PubMed] [Google Scholar]
- Song X.F., Yang C.Y., Liu J., Yang W.C. (2006). RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol. 141: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D.F., Huang A.H.C. (2007). Maize pollen coat xylanase facilitates pollen tube penetration into silk during sexual reproduction. J. Biol. Chem. 282: 625–636. [DOI] [PubMed] [Google Scholar]
- Swanson R., Clark T., Preuss D. (2005). Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 18: 163–171. [Google Scholar]
- Szumlanski A.L., Nielsen E. (2009). The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21: 526–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2011). Attraction of tip-growing pollen tubes by the female gametophyte. Curr. Opin. Plant Biol. 14: 614–621. [DOI] [PubMed] [Google Scholar]
- Tang W., Ezcurra I., Muschietti J., McCormick S. (2002). A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14: 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.P., Hepler P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 461–491. [DOI] [PubMed] [Google Scholar]
- Twell D., Wing R., Yamaguchi J., McCormick S. (1989). Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217: 240–245. [DOI] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713. [DOI] [PubMed] [Google Scholar]
- Wang H.J., Huang J.C., Jauh G.Y. (2010). Pollen germination and tube growth. Adv. Bot. Res. 54: 1–52. [Google Scholar]
- Wang H.J., Wan A.R., Jauh G.Y. (2008). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.L., Hsu C.M., Chang L.C., Wang C.S., Su T.H., Huang Y.J., Jiang L., Jauh G.Y. (2004). Gene expression profiles of cold-stored and fresh pollen to investigate pollen germination and growth. Plant Cell Physiol. 45: 1519–1528. [DOI] [PubMed] [Google Scholar]
- Wu H.M., Wang H., Cheung A.Y. (1995). A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82: 395–403. [DOI] [PubMed] [Google Scholar]
- Wu J.-Y., Qu H.-Y., Shang Z.-L., Tao S.-T., Xu G.-H., Wu J., Wu H.-Q., Zhang S.-L. (2011). Reciprocal regulation of Ca2+-activated outward K+ channels of Pyrus pyrifolia pollen by heme and carbon monoxide. New Phytol. 189: 1060–1068. [DOI] [PubMed] [Google Scholar]
- Xiao C.M., Mascarenhas J.P. (1985). High temperature-induced thermotolerance in pollen tubes of tradescantia and heat-shock proteins. Plant Physiol. 78: 887–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.J.W., Perry P.J., Ciani S., Pandian S., Schmidt W. (2008). Manganese deficiency alters the patterning and development of root hairs in Arabidopsis. J. Exp. Bot. 59: 3453–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.W., Wilen R.W., Bonham-Smith P.C. (2004). High temperature stress of Brassica napus during flowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J. Exp. Bot. 55: 485–495. [DOI] [PubMed] [Google Scholar]
- Zanetti M.E., Chang I.-F., Gong F., Galbraith D.W., Bailey-Serres J. (2005). Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li S., Zhou L.Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. (2011). Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 68: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Zhou S., et al. (2011). Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23: 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.