In the Brassicaceae, the pollen coat of compatible pollen grains contains signaling factors that induce Ca2+ export from the stigmatic papilla cells. This Ca2+ export is performed by at least one Ca2+ pump, ACA13, and is required for pollen germination and successful pollination.
Abstract
In the Brassicaceae, intraspecific non-self pollen (compatible pollen) can germinate and grow into stigmatic papilla cells, while self-pollen or interspecific pollen is rejected at this stage. However, the mechanisms underlying this selective acceptance of compatible pollen remain unclear. Here, using a cell-impermeant calcium indicator, we showed that the compatible pollen coat contains signaling molecules that stimulate Ca2+ export from the papilla cells. Transcriptome analyses of stigmas suggested that autoinhibited Ca2+-ATPase13 (ACA13) was induced after both compatible pollination and compatible pollen coat treatment. A complementation test using a yeast Saccharomyces cerevisiae strain lacking major Ca2+ transport systems suggested that ACA13 indeed functions as an autoinhibited Ca2+ transporter. ACA13 transcription increased in papilla cells and in transmitting tracts after pollination. ACA13 protein localized to the plasma membrane and to vesicles near the Golgi body and accumulated at the pollen tube penetration site after pollination. The stigma of a T-DNA insertion line of ACA13 exhibited reduced Ca2+ export, as well as defects in compatible pollen germination and seed production. These findings suggest that stigmatic ACA13 functions in the export of Ca2+ to the compatible pollen tube, which promotes successful fertilization.
INTRODUCTION
In flowering plants, successful pollination is the first step in reproduction, leading to fertilization, which requires coordination and communication between the pollen grain and the stigma. In the Brassicaceae, the stigma is dry-type and covered with a layer of epidermal cells called papilla cells. In self-incompatible Brassica species, when a self-pollen grain lands on a papilla cell (incompatible pollination), pollen hydration and pollen tube germination are prevented. In the self-recognition system, the interaction between a male determinant, S-locus protein 11 (SP11; S-locus cysteine-rich protein [SCR]) and a female determinant, S-locus receptor kinase (SRK), induces SRK autophosphorylation, leading to rejection of self-pollen (Takayama et al., 2001; Takayama and Isogai, 2005; Iwano and Takayama, 2012). By contrast, when a cross-pollen grain lands on a papilla cell and is recognized as compatible by the papilla cell (compatible pollination), the papilla cell supplies water to the dehydrated pollen grain, which hydrates and germinates a pollen tube. The pollen tube then invades the papilla cell wall and grows toward the ovule cells. However, the recognition mechanisms in compatible pollination remain unclear, and the signaling pathway in the papilla cell induced after compatible pollination is also unclear.
Since Brewbaker and Kwack (1963) revealed that Ca2+ is essential for in vitro pollen tube cultures, the relationship between Ca2+ concentration ([Ca2+]) and pollen tube growth has been extensively examined (Steinhorst and Kudla, 2013). In compatible pollination using pollen grains of transgenic Arabidopsis thaliana expressing a Ca2+ sensor protein, a cytoplasmic [Ca2+] ([Ca2+]cyt) increase was observed at the potential germination site and at the tip region of the pollen tube (Iwano et al., 2004, 2009). Ratiometric ion imaging using fluorescent dye has revealed that the apical domain of a pollen tube grown in vitro contains a tip-focused [Ca2+] gradient (Pierson et al., 1994, 1996; Cheung and Wu, 2008). Studies using a Ca2+-sensitive vibrating electrode showed that Ca2+ influx in the tip region of the pollen tube is essential for pollen tube growth (Pierson et al., 1994; Holdaway-Clarke et al., 1997; Franklin-Tong et al., 2002). Stretch-activated Ca2+ channels have been found in the plasma membrane using patch-clamp electrophysiology (Kühtreiber and Jaffe, 1990; Dutta and Robinson, 2004). A cyclic nucleotide-gated channel and a Glu receptor-like channel were identified as Ca2+-permeable channels in the plasma membrane that are essential for pollen tube growth (Frietsch et al., 2007; Michard et al., 2011). These reports suggest that an influx of extracellular Ca2+ occurs when a pollen grain lands on a papilla cell after compatible pollination, similar to the in vitro pollen tube germination and growth condition. In Brassica rapa, elemental analysis of papilla cells and pollen grains during pollination was done using x-ray microanalysis in combination with variable pressure scanning electron microscopy. In the pollen grains, K, Cl, and Ca levels rose with pollen germination only after cross-pollination (Iwano et al., 1999). The elevation of these elements in the pollen grains implies that K+, Cl−, and Ca2+ are transported from the papilla cells to the pollen grains after cross-pollination. From these results, we speculated that Ca2+ transport from a papilla cell to a pollen grain is required for pollen germination.
In papilla cells of the Brassicaceae, x-ray microanalysis showed Ca elevation near the surface of the papilla cells after cross-pollination (Elleman and Dickinson, 1999; Iwano et al., 1999). Inside the papilla cell, [Ca2+]cyt fluctuated simultaneously with pollen hydration, pollen tube germination, and pollen tube penetration at the pollen attachment site (Iwano et al., 2004). However, a continuous elevation of [Ca2+]cyt was not observed in the papilla cell after pollination and the maximum [Ca2+]cyt was estimated to be submicromolar. Therefore, we speculated that unknown molecules in the pollen grain elicit a Ca2+-dependent signaling pathway in the papilla cells and that preexisting molecules in the papilla cells keep intracellular [Ca2+] at low levels during compatible pollination, while extracellular [Ca2+] is high. Cell biological analysis of B. rapa showed that cross-pollination induced the concentration of actin bundles at the pollen attachment site, whereas self-pollination induced actin depolymerization; in addition, the actin-depolymerizing drug cytochalasin D significantly inhibited pollen germination during cross-pollination (Iwano et al., 2007). The pollen surface is covered with exine and a protein- and lipid-rich substance called the pollen coat. The pollen coat alone from cross-pollen grains induced actin polymerization, while self-pollen coat alone induced the disappearance of actin bundles (Iwano et al., 2007). Furthermore, electron microscopic observation in Brassica oleracea revealed that applying the isolated pollen coat to the stigma caused cell wall expansion (Elleman and Dickinson, 1996). Therefore, we hypothesized that the pollen coat contains molecules that initiate a signaling cascade, leading to successful compatible pollination.
Here, we established a biological assay that is used to show that Ca2+ is exported from the papilla cell during compatible pollination and showed that compatible pollen coat contains molecules eliciting the Ca2+ transport. To identify the Ca2+ transporter(s) functioning in the papilla cell during pollination, we first performed a microarray analysis of stigmas after pollination and after adhesion of the pollen coat in Arabidopsis and identified autoinhibited Ca2+-ATPase13 (ACA13), which was induced by pollination and pollen coat adhesion, as a candidate. Second, we examined the expression site of the ACA13 gene using real-time PCR, promoter β-glucuronidase (GUS) staining, and in situ hybridization. Third, we characterized the function of ACA13 using a T-DNA line and found that ACA13 is required for pollen germination and pollen tube growth. Furthermore, the localization of ACA13 on the plasma membrane of the papilla cell was examined using a green fluorescent protein (GFP) reporter assay and immune electron microscopy. We found that ACA13 accumulates around the pollen tube attachment site of the pollinated papilla cell. Moreover, a complementation test in the yeast strain K616 showed that ACA13 functions as a Ca2+ pump. From these results, we conclude that the stigmatic ACA13 functions as a Ca2+ pump, which is required for successful pollination.
RESULTS
Calcium Green Assay
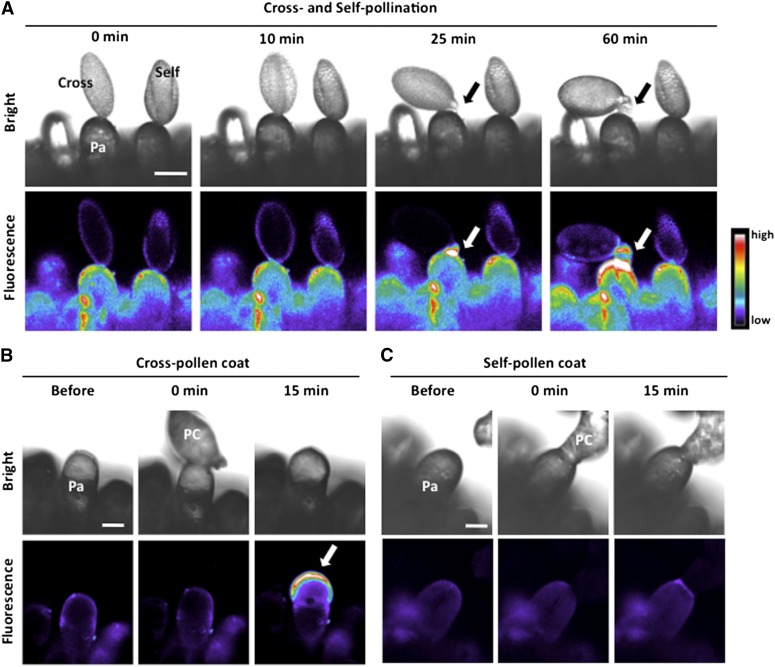
Previous microscopy studies suggested that Ca2+, K+, and Cl− are transported from a papilla cell to a pollen grain during pollination in the Brassicaceae (Iwano et al., 1999). To confirm the Ca2+ transport, we established a pollination assay using a water-soluble Ca2+-sensitive dye, Calcium Green 5N. A drop of Calcium Green solution was applied to the stigma surface in B. rapa and air-dried, and bright-field and fluorescence images of the papilla cell were captured under a confocal laser microscope after a cross- or a self-pollen grain was applied to the papilla cell, using a micromanipulator. The fluorescence intensity at the surface of the papilla cell increased markedly from the time of pollen germination after cross-pollination, but not after self-pollination (Figure 1A). While the timing of germination varied between 20 and 40 min, the cross-pollination-specific increase in fluorescence intensity was invariably observed for all stigmas when germination successfully occurred (n = 20). To examine whether the pollen coat alone induces the increase of fluorescence intensity, we isolated pollen coats from cross- and self-pollen grains, as previously reported (Stephenson et al., 1997; Takayama et al., 2000), and applied them to papilla cells coated with Calcium Green. As shown in Figures 1B and 1C, only the cross-pollen coat induced a marked increase of fluorescence intensity on the papilla cell (n = 25). Therefore, this increase suggests that the pollen coat alone induces Ca2+ export to the cell wall after cross-pollination. These data support the hypothesis that the pollen coat elicits a signaling cascade leading to successful pollination.
Figure 1.
Calcium Green Assay of Cross- and Self-Pollination in B. rapa.
Papilla cells coated with Calcium Green were used, and representative images are displayed. Fluorescence images of Calcium Green appear in pseudo color. Pa, papilla cell; PC, pollen coat. Bars = 20 µm.
(A) Bright-field and fluorescence images of papilla cells before pollination and at 0, 10, 25, and 60 min after cross- and self-pollination. Arrows indicate a germinated pollen tube (cross-pollen, n = 20; self-pollen, n = 20).
(B) Bright-field and fluorescence images of papilla cells before and at 0 and 15 min after adhesion of cross-pollen coat. Pollen coat spreading, and a remarkable increase of fluorescence, was observed (white arrow) (n = 25).
(C) Bright-field and fluorescence images of papilla cells before and at 0 and 15 min after adhesion of self-pollen coat (n = 25).
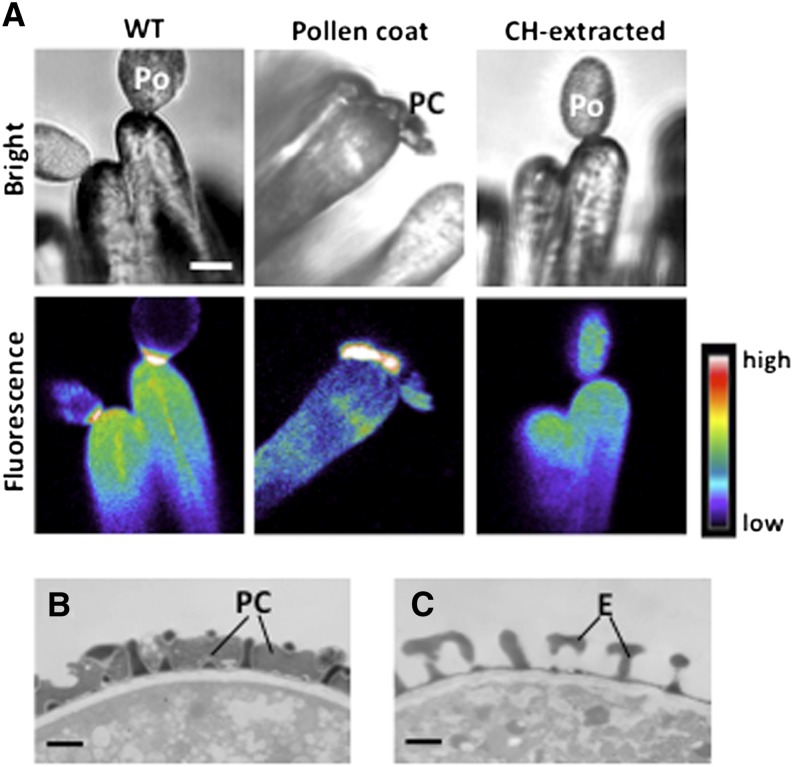
Next, to examine whether the increase of fluorescence intensity of Calcium Green during compatible pollination also occurs in Arabidopsis, we applied self-compatible pollen grains to papilla cells coated with Calcium Green. We collected pollen grains from flowers using a vacuum, isolated pollen coats using the same method as for B. rapa, and applied them to papilla cells coated with Calcium Green. The fluorescence intensity of Calcium Green increased markedly within 15 min after pollination or after adhesion of the pollen coat, which corresponded to the time of pollen germination (Figures 2A and 2B; Supplemental Movie 1). Increased fluorescence intensity was observed in all samples in which pollen grains successfully germinated (n = 8). In the Brassicaceae, pollen coat can be removed by extraction with cyclohexane (Doughty et al., 1993). The cyclohexane-extracted pollen grains are viable but do not hydrate on the papilla cell after pollination. When cyclohexane-extracted pollen grains were applied to papilla cells coated with Calcium Green, the fluorescence intensity did not increase at 15 min after pollination (Figure 2C). From these results in B. rapa and Arabidopsis, the pollen coat induced an increase in Calcium Green fluorescence, suggesting that Ca2+ is exported from the papilla cell. These results confirm the hypothesis that pollen coat elicits a signaling cascade leading to pollen germination and led us to speculate the involvement of a Ca2+ transporter(s) in the papilla cell in compatible pollination in the Brassicaceae.
Figure 2.
Calcium Green Assay during Pollination in Arabidopsis.
(A) Representative bright-field and fluorescence images of papilla cells coated with Calcium Green at 15 min after pollination with nontreated pollen (WT), after adhesion of pollen coat, and after pollination with cyclohexane (CH)–extracted pollen. Each Calcium Green fluorescence image is shown in pseudo color (wild-type pollen, n = 8; pollen coat, n = 5; cyclohexane-extracted pollen, n = 5). Bar = 10 µm.
(B) and (C) Transmission electron micrographs of wild-type pollen (B) and CH-extracted pollen (C). Bars = 1 µm.
Po, pollen grain; PC, pollen coat; E, exine.
Identification of Ca2+ Transporters Functioning during Pollination Using Microarray Analysis
To identify molecules that promote Ca2+ export in the stigma during compatible pollination, we performed microarray analysis with Arabidopsis, harvesting 50 stigmas from mature wild-type pistils at stage 13 (Smyth et al., 1990). First, to identify genes induced in the stigma after pollination, we pollinated with wild-type pollen grains and collected stigmas immediately (0 min) or at 15 min after pollination. As described above, at around 15 min after pollination, the fluorescence of Calcium Green increased markedly at the attachment site between the pollen grain and the papilla cell, which corresponds with the time at which pollen has hydrated on the papilla cell and begins to germinate (Iwano et al., 2004). The samples were labeled with cyanine-3 (Cy3)-CTP (0 min) and cyanine-5 (Cy5)-CTP (15 min), and a two-color oligonucleotide microarray analysis covering 28,500 genes was performed. For 27,205 valid probes, we calculated normalized ratios from the expression values at 0 min (control data) and 15 min (raw data) samples in three replicates and identified 398 genes whose mean normalized ratios were more than 2-fold higher at 15 min after pollination than that at 0 min after pollination (Supplemental Data Set 1).
However, the increased expression level in stigmas after pollination may also have included expression of genes in the attached pollen grains. Since pollen coat alone also induced a high level of Calcium Green fluorescence, we performed microarray analysis using stigmas after pollen coat adhesion to identify genes induced exclusively in the stigma during the compatible response. We used stigmas before (control data) and at 15 min after adhesion of pollen coat (raw data) in three replicates and identified 474 genes whose expression more than doubled at 15 min after pollen coat adhesion (Supplemental Data Set 2). Among these, 255 matched genes that were upregulated after pollination (Supplemental Data Set 3).
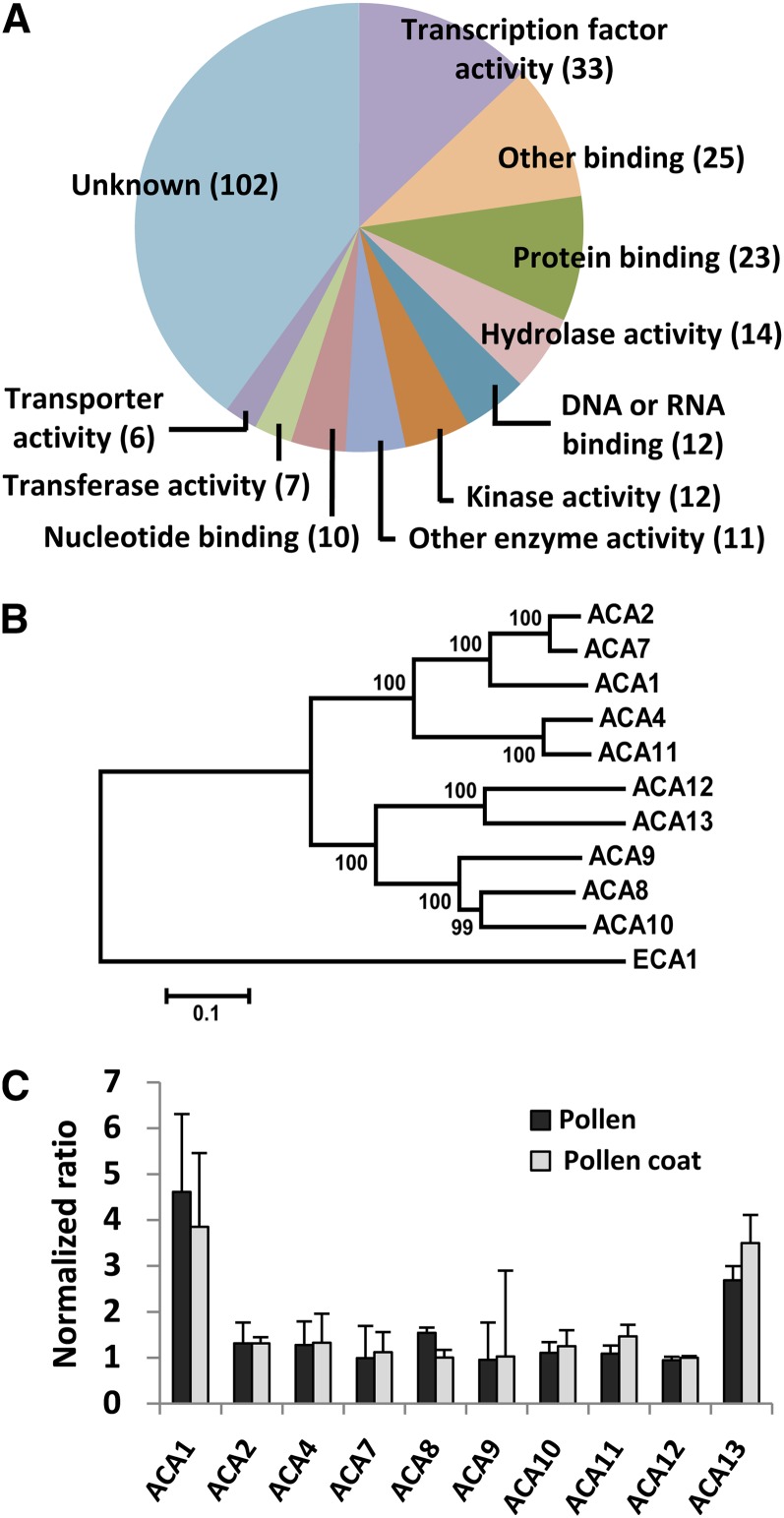
To identify genes involved with Ca2+ transport in the stigma after compatible pollination, we examined the function of the 255 genes which were upregulated both by pollination and by adhesion of pollen coat (Figure 3A; Supplemental Data Set 3). We found six genes related to transport, including two Ca2+ pumps, ACA1 and ACA13, which belong to the ACA family with 10 members (Figure 3B). The expression changes of these ACA genes after pollination and after adhesion of pollen coat are shown in Figure 3C. The expression of ACA1 15 min after pollination or pollen coat adhesion was 4.6 or 3.9 times, respectively, higher than that just after pollination or in no-treatment stigmas. Likewise, ACA13 expression after pollination or pollen coat adhesion increased ∼2.7 times or 3.5 times, respectively, relative to that just after pollination or in no-treatment stigmas. As it has been reported that ACA1 is localized to plastids (Huang et al., 1993) or endoplasmic reticulum (Dunkley et al., 2006), we focused in this study on the analysis of ACA13 function.
Figure 3.
Transcription after Pollination and Pollen Coat Adhesion.
(A) Functional categorization of 255 genes that were upregulated in stigma after both pollination and pollen coat adhesion, based on molecular functions in Gene Ontology annotations from TAIR.
(B) Phylogenetic analysis of ACAs with ECA1 as an outgroup. Bootstrap values from 2000 replicates are shown as percentage values at the nodes. The scale measures evolutionary distance based on the amino acid sequences. The tree corresponded with that of George et al. (2008). Bar = 0.1 amino acid substitutions per site.
(C) Transcription of ACAs in stigma. Ratios show the signal intensities at 15 min after either pollination (Pollen) or pollen coat adhesion (Pollen coat), relative to those either at 0 min after pollination or before adhesion with pollen coat. Data are means ± sd for three replicates.
Statistical Analysis of Microarray Data
To examine whether the increase of ACA13 (AT3G22910) expression in pollination experiments or pollen coat adhesion experiments is significant, microarray data were statistically analyzed. First, the log-transformed normalized ratios of pollination experiments and pollen coat experiments [=log(raw/control)] for each locus were plotted (Supplemental Figure 1). There are 27,205 loci, of which 255 loci had mean normalized ratios >2 in each of three experiments. As there is a monotonic trend (indicating data error) in the Experiment 3 pollination data, we removed these data from subsequent analyses.
We conducted five experiments using samples of pollination and pollen coat and regarded these five experiments as independent samples. We then analyzed the differences between raw and control data using a paired t test (n = 5). In our analysis, we used the natural logarithm of raw and control values. We were not interested in cases in which raw values were smaller than those of the control. Using a one-sided test, we evaluated the null hypothesis that raw values were smaller than or equal to control values: The t test statistic of ACA13 was 5.01, and the P value was 0.0037. When the 27,205 loci were ranked in order of ascending P value, ACA13 was ranked 1171st (i.e., within the top 4.3% [=1171/27,205] among all loci) (Supplemental Figure 2).
Our t test did not distinguish between pollination experiments and pollen coat experiments. If the normalized ratio had differed systematically between pollination experiments and pollen coat experiments, we would not be able to rule out the possibility that results obtained from the t test represent a spurious manifestation of the intrinsic structure. To eliminate this possibility, we examined the differences between pollination experiments and pollen coat experiments by adjusting the differences in average structures using ANOVA, yielding a P value of 0.00061 for ACA13. When the 27,205 loci were ranked in ascending order of P values, ACA13 was ranked 180th, within the top 0.66% (=180/27,205) (Supplemental Figure 3). This ANOVA still rejected the hypothesis “raw value is smaller than or equal to control value,” indicating that the result is robust against any systematic difference between experiments that might exist.
Although both tests gave P values considerably smaller than 5%, neither result was sufficiently significant to endure multiple-testing correction. However, applying a standard multiple-testing correction, such as the Bonferroni procedure or false discovery rate control, to such high-dimensional data with very small sample sizes makes the test excessively conservative and of no statistical power. Given the current lack of an optimal multiple correction in this situation, the above procedure would be a practical approach for prioritizing loci that are actually novel.
Expression of ACA13
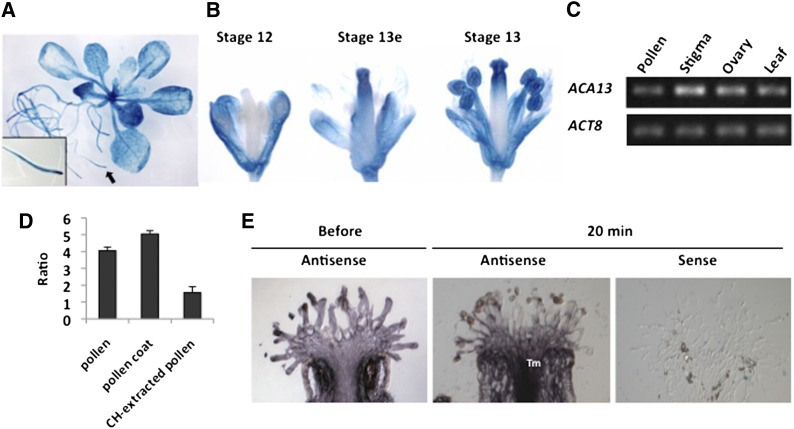
As the statistical analysis indicated that ACA13 was upregulated after pollination and after pollen coat adhesion, we examined the expression site of ACA13 protein in Arabidopsis. A DNA fragment corresponding to the 1940 bp upstream of the ACA13 start codon was fused to the GUS reporter gene and transformed into Arabidopsis. GUS expression was mainly observed in the stigma and anther of the flower and in the root tip region (Figures 4A and 4B). GUS expression in stigma was high at the early stage of flower stage 13.
Figure 4.
Transcription of ACA13.
(A) and (B) ACA13:GUS activity.
(A) GUS expression in 3-week-old plants. A root tip (arrow) is enlarged in the inset.
(B) Expression in flowers at stage 12, early stage 13 (stage 13e), and stage 13.
(C) RT-PCR of ACA13 in pollen, stigma, ovary, and leaf. ACT8 was amplified as control.
(D) Real-time PCR of ACA13 in stigma. Ratios of threshold cycle at 15 min either after pollination with wild-type pollen or cyclohexane (CH)–extracted pollen or after adhesion with pollen coat to threshold cycle either at 0 min after pollination or before pollen coat adhesion are shown. Data are means ± sd for three replicates.
(E) In situ hybridization to localize ACA13 in the stigma before and at 20 min after pollination. Tm, transmitting tract.
To confirm these results, the expression pattern of ACA13 mRNA was evaluated by real-time RT-PCR using ACA13-specific primers. Expression in the stigma was higher than that in pollen, ovary, and leaves (Figure 4C). Real-time PCR was used to confirm the upregulation of ACA13 gene expression. In the stigma at 15 min after pollination, the expression was 4 times higher than that immediately after pollination, and at 15 min after adhesion of pollen coat, it was ∼5 times higher than that before pollination (Figure 4D). By contrast, in the stigma pollinated with cyclohexane-extracted pollen grains whose pollen coat was removed, there was no marked increase in ACA13 expression. This result supported the above data that pollen coat induced ACA13 gene expression. Furthermore, in situ hybridization showed that ACA13 is expressed in the papilla cells and that ACA13 gene expression increased not only in the papilla cells but also in the transmitting tract after pollination (Figure 4E). These RT-PCR, real-time PCR, and in situ hybridization results suggest that ACA13 is a Ca2+ transporter functioning in the stigma during compatible pollination in Arabidopsis.
Gene Disruption of ACA13
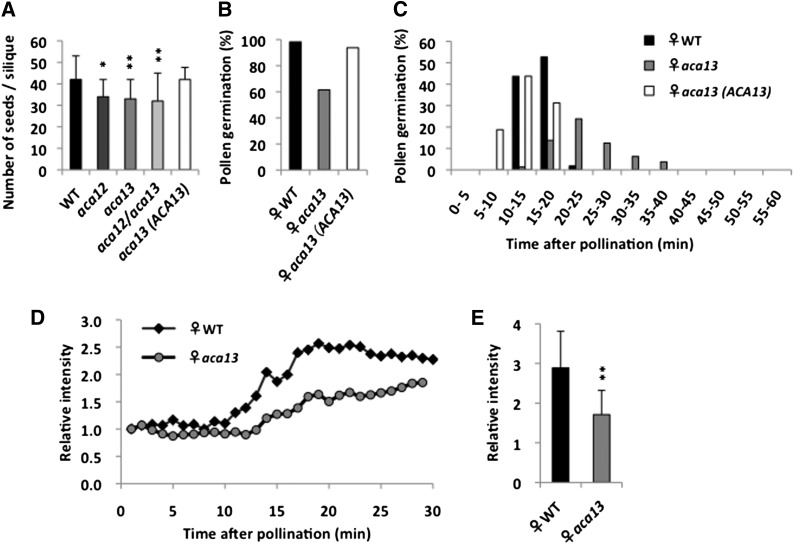
To examine ACA13 function, a homozygous line harboring a T-DNA insertion that inactivates ACA13 was identified by PCR (Supplemental Figure 4). In this line, aca13, the number of seeds per silique was significantly less than that in the wild type (P < 0.01) (Figure 5A). However, we thought that other ACA family members might complement the loss of ACA13 function in aca13. Next, we generated an aca12 aca13 double mutant by crossing aca13 with an ACA12 T-DNA insertion line (Supplemental Figure 4) because ACA12 has the highest sequence similarity with ACA13 among the ACAs. In the aca12 aca13 double mutant, however, the number of seeds per silique was almost the same as that in the aca13 line (Figure 5A; Supplemental Figure 4).
Figure 5.
T-DNA Insertion Mutant Phenotypes.
(A) Number of seeds per silique of the wild type, aca12, aca13, aca12 aca13, and aca13 expressing BrSLG:ACA13-Venus [aca13(ACA13)] after hand pollination. Data shown are means ± sd (n = 24). Asterisks indicate significantly lower number of seeds compared with the wild type (*P < 0.05; **P < 0.01).
(B) and (C) Pollen germination rate by pollination assay.
(B) Germination rate of wild-type pollen on papilla cells of the wild type, aca13, and aca13(ACA13) at 60 min after pollination [the wild type, n = 55; aca13, n = 59; aca13(ACA13), n = 16].
(C) Timing of wild-type pollen germination on papilla cells of the wild type, aca13, and aca13(ACA13) [the wild type, n = 55; aca13, n = 59 aca13(ACA13), n = 16].
(D) and (E) Calcium Green assay of the wild type and aca13 after pollination with wild-type pollen. Fluorescence intensity on papilla cells immediately under the applied pollen grain was measured.
(D) Typical changes in fluorescence intensity on wild-type and aca13 papilla cells after pollination. Relative intensity shows the ratio of fluorescence intensity at each time point to that at 0 min after pollination.
(E) Fluorescence intensities at the time of pollen germination. Relative intensity shows the ratio of fluorescence intensity at 15 min to that at 0 min after pollination for the wild type. Asterisks indicate significantly lower relative intensity from aca13 than from wild-type papilla cells (**P < 0.01). Data are means ± sd (the wild type, n = 8; aca13, n = 12).
To determine whether ACA13 contributes to pollen germination during pollination, we performed a pollination assay using a micromanipulator. When wild-type pollen grains were applied to wild-type papilla cells, the germination rate was more than 95% at 60 min after pollination, and the timing of germination was 10 to 20 min after pollination (Figures 5B and 5C). By contrast, when wild-type pollen grains were applied to aca13 papilla cells, the germination rate was reduced to 60% at 60 min after pollination, and germination was delayed (Figures 5B and 5C). Moreover, in the Calcium Green assay, the fluorescence increase on aca13 papilla cells was lower than on wild-type papilla cells (Figure 5D). We next generated stable transgenic Arabidopsis plants that express high levels of ACA13-Venus, a yellow fluorescent protein, under the SLG promoter in the papilla cell and crossed these transgenic plants with aca13. The reduced seed number per silique, the reduced pollen germination rate, and the delayed germination both recovered when this BrSLG:ACA13-Venus gene was introduced into aca13 (Figures 5A to 5C). These results demonstrate that ACA13 contributes to pollen germination during pollination.
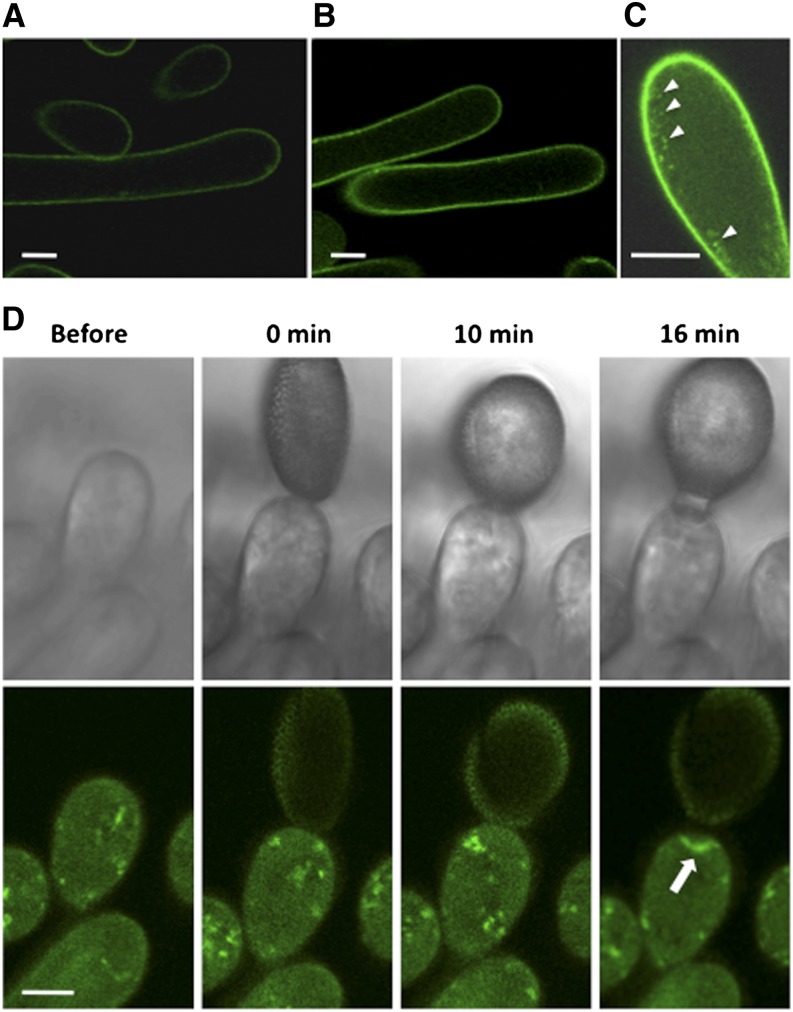
Subcellular Localization of ACA13
To determine the subcellular localization of ACA13, an ACA13-Venus fusion protein was expressed in stable transgenic Arabidopsis plants under the promoter of either the ACA13 or the SLG gene. When the pistil was mounted on a glass slide and immersed in water, ACA13-Venus fluorescence was observed on the papilla cell membrane in both transgenic plants using a confocal microscope whose pinhole diameter was 1.49 Airy units (Figures 6A and 6B). When the pinhole diameter was 3.04 Airy units and the optical section was thick, dot-like signals were also visible, in both transgenic plants, in the cytoplasm (Figure 6C). Next, to examine the localization of ACA13-Venus during pollination, a pollen grain was applied to a papilla cell expressing ACA13-Venus using a micromanipulator, and the ACA13-Venus signal was observed in an optically thick section before and after pollination. Before pollination, dot-like signals were observed throughout the papilla cell. However, when the pollen germinated and the pollen tube penetrated into the papilla cell, the dot-like signals were reduced and ACA13-Venus accumulated around the pollen tube (arrow in Figure 6D). This behavior of the ACA13-Venus signal was consistently observed in independent flowers (n = 15).
Figure 6.
Localization of Venus-Tagged ACA13 in Papilla Cells.
(A) Fluorescence image of papilla cells expressing ACA13:ACA13-Venus.
(B) Fluorescence image of papilla cells expressing BrSLG:ACA13-Venus.
(C) Fluorescence image of a papilla cell expressing BrSLG:ACA13-Venus. The pinhole diameter was 3.04 Airy units. Arrowheads show dot-like ACA13-Venus signals.
(D) Behavior of ACA13-Venus during pollination. ACA13-Venus accumulated at tube penetration site (arrow) (n = 15). The pinhole diameter was 3.04 Airy units.
Bars = 10 µm.
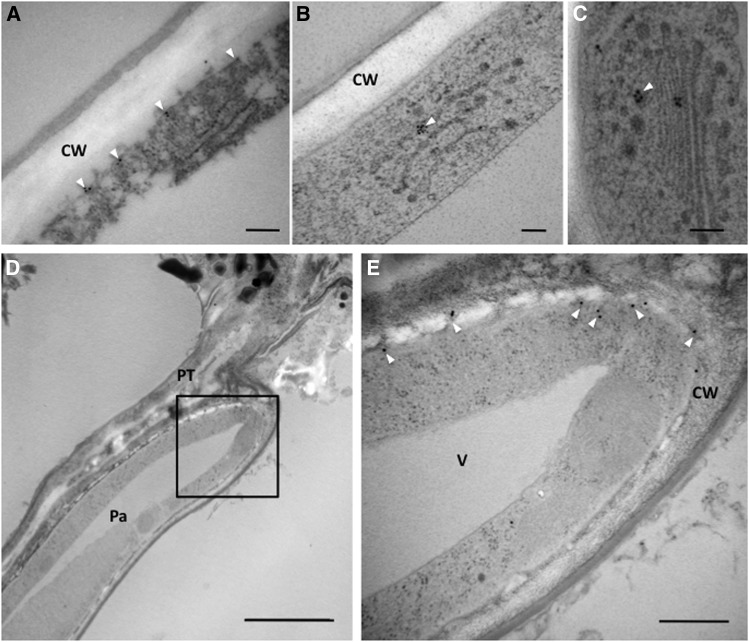
To examine the precise localization of ACA13, immune electron microscopy of transgenic plants expressing ACA13-Venus under the control of the ACA13 promoter was performed, using anti-GFP rabbit antibody as the primary antibody and 15-nm gold particle–conjugated anti-rabbit antibody as the secondary antibody. Before pollination, the ACA13-Venus fusion protein was mainly associated with the plasma membrane and on vesicles near the Golgi bodies (Figures 7A to 7C). However, when the pollen tube was growing through the cell wall of the papilla cell after pollination, many signals were observed at the plasma membrane beneath the pollen tube (Figures 7D and 7E). Furthermore, when independent papilla cell sections were observed (n = 20), the number of gold particles at the plasma membrane beneath the pollen tube after germination was 3.4 particles/µm, which was significantly higher than that before pollination (1.1 particles/µm) (P < 0.005). These observations were consistent with the accumulation of ACA13-Venus fluorescence observed in live-cell monitoring (Figure 6). Together, our imaging data indicate that ACA13 in the stigmatic papilla cells accumulates at the plasma membrane around the penetration site of the pollen tube during pollen tube germination and penetration.
Figure 7.
Localization of ACA13 in a Papilla Cell before and after Pollination.
ACA13 was detected using anti-GFP rabbit antibody as the primary antibody and 15-nm gold particle–conjugated anti-rabbit antibody as the secondary antibody. Gold particles are indicated by arrowheads.
(A) to (C) Papilla cell before pollination. Gold particles were detected at the plasma membrane (A) and on vesicles near the Golgi body ([B] and [C]) in the papilla cell. Bars = 100 nm.
(D) Papilla cell at 20 min after pollination. Bar = 2.5 µm.
(E) Enlargement of the square in (D). Bar = 500 nm.
CW, cell wall; PT, pollen tube; Pa, papilla cell; V, vacuole.
Function of ACA13 as a Ca2+ Pump
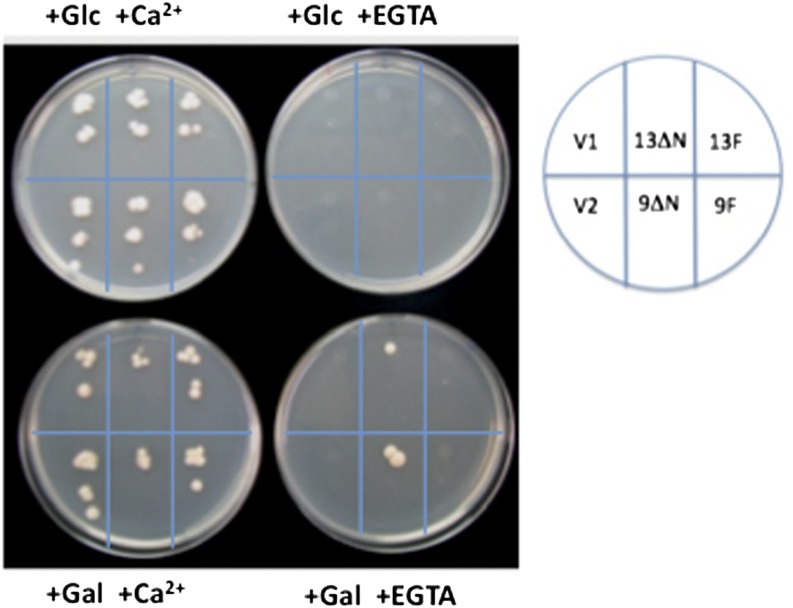
To confirm a Ca2+ pump function for ACA13, we performed a complementation test using the yeast strain K616, which lacks its own Ca2+ pumps, PMR1 and PMC (Cunningham and Fink, 1994). K616 cells normally fail to grow on Ca2+-depleted medium. If ACA13 functions as a Ca2+ pump, expression of ∆N-ACA13 (ACA13 lacking the 65 N-terminal amino acid residues, which comprise a predicted calmodulin-regulated autoinhibitor domain) should allow the K616 strain to grow on Ca2+-depleted medium, as has been demonstrated for the plasma membrane Ca2+ pumps ACA8 and ACA9 (Bonza et al., 2004; Schiøtt et al., 2004). As shown in Figure 8, the deregulated ∆N-ACA13 indeed rescued the growth of K616 on Ca2+-depleted medium. This result confirms that ACA13 is a calmodulin-activated calcium pump, similar to ACA8 and ACA9.
Figure 8.
Complementation of Yeast K616 by ACA13.
Growth of Saccharomyces cerevisiae mutant K616 (pmc1 pmr1 cnb1) on medium containing either Gal or Glc and either CaCl2 or EGTA was complemented with GAL1:ΔN-ACA13 and GAL1:ΔN-ACA9 in the pYES2 vector. V1 and V2, pYES2 empty vector; 13ΔN and 9ΔN, either GAL1:ΔN-ACA13 or GAL1:ΔN-ACA9 in pYES2; 13F and 9F, either GAL1:ACA13 or GAL1:ACA9 (full-length) in pYES2.
Furthermore, to confirm that ACA13-Venus is functional, constructs encoding either full-length ACA13 or ∆N-ACA13 and Venus were also tested. As shown in Supplemental Figure 5, ∆N-ACA13 rescued the growth of K616 on Ca2+-depleted medium. Therefore, the complementation test showed that ACA13-Venus is indeed functional.
DISCUSSION
During pollination, pollen grains from various species may come into contact with papilla cells. However, successful pollination occurs only when the pollen grain is recognized as compatible by interaction between the stigmatic papilla cell and the pollen grain (Dickinson, 1995). While compatible interactions are likely to involve compatibility factors on both the pollen side and the stigma side, the molecular mechanisms underlying compatible interactions are not thoroughly understood. In this study, we established a bioassay using Calcium Green in B. rapa and Arabidopsis and found that the fluorescence of Calcium Green on the papilla cell surface increased at the pollen attachment site after cross-pollination and compatible pollination. In addition, we found that this increase could be induced by the pollen coat of the cross and compatible pollen. By contrast, neither a self-pollen grain nor the self-pollen coat in B. rapa induced this increase in fluorescence. This result led us to speculate that Ca2+ could be transported from the papilla cell to the cell wall, and then transferred from the cell wall to the pollen grain during compatible pollination, and that compatibility factors on the pollen coat could induce this transport. On the other hand, the pollen S-determinant SP11/SCR prevented the Ca2+ transport to the cell wall, leading to the rejection of self-pollen.
This hypothesis was supported by transcriptional analyses of stigmas after pollination and after pollen coat adhesion in Arabidopsis. We found that almost 65% of the genes that were upregulated at 15 min after pollination were also upregulated after pollen coat adhesion. Since pollen germination begins 15 min after compatible pollination, many of the upregulated genes are likely to be involved with pollen hydration and germination and with the disorganization of the papilla cell wall for pollen tube penetration. Actually, the expansin and xyloglucan:xyloglucosyl transferase genes, which are directly related to loosening or degradation of the cell wall, were upregulated. Furthermore, the gene encoding formin-like protein 7 (FH7) was upregulated. Formin-like protein, an actin-nucleating protein, binds to the barbed ends of actin filaments and activates de novo actin nucleation (Cvrcková et al., 2004). Arabidopsis possesses 21 members of the formin-like protein family, which can be divided into two clades (Class I and Class II). It has been reported that the Class I FH5 stimulates actin assembly most prevalently around the subapical membrane and in apical vesicular trafficking, supporting pollen tube growth (Cheung et al., 2010). FH7 also belongs to Class I. We showed previously that actin bundle formation in the papilla cell is promoted by both compatible pollination and adhesion of compatible pollen coat (Iwano et al., 2007). Therefore, it is possible that FH7 stimulates both actin assembly and apical vesicular trafficking in the papilla cell and thus contributes to successful pollination.
During compatible pollination, [Ca2+]cyt in the papilla cell fluctuated with pollen hydration, germination, and pollen tube penetration. In the transcriptional analysis, 25 calcium-related genes, including those encoding C2 domain-containing proteins, Ca2+ binding EF hand family proteins, and calmodulin-related proteins, were upregulated by both compatible pollination and the compatible pollen coat (Supplemental Data Set 4). Furthermore, harpin-induced proteins, WRKY DNA binding proteins, a calcium-dependent protein kinase, protein phosphatase 2C, and a mitogen-activated protein kinase kinase, all of which are associated with changes in [Ca2+]cyt in plant–microbe interactions (Tena et al., 2011), were also upregulated. These results suggest that analogous Ca2+-related signaling pathways exist in both compatible pollination and plant–microbe interactions.
The Calcium Green assay and the transcriptional analysis data suggested that compatibility factors, molecules inducing Ca2+ export from the papilla cell, exist in the compatible pollen coat and that they induce a signaling pathway that contributes to successful pollination. In the Brassicaceae, the pollen coat comprises mainly a complex mixture of lipid and protein with a sporophytic, rather than a gametophytic, origin. The lipid mixture is mostly derived from lipid droplets or plastoglobules, which accumulate in special plastids called elaioplasts inside the cells of the anther tapetum. The lipid mixture is enriched in acylated compounds, such as sterol esters and phospholipids (Murphy and Vance, 1999). The main protein component of pollen coat is oleosin-like protein, which may facilitate the creation of water channels through the lipidic pollen coat (Murphy and Ross, 1998; Murphy, 2006). In Arabidopsis eceriferum mutants, which are defective in long-chain lipid metabolism, the absence of long-chain lipids results in an absent pollen coat or a reduction in the lipid content of the pollen coat, and the pollen fails to hydrate on the stigma (Preuss et al., 1993; Hülskamp et al., 1995; Wolters-Arts et al., 1998; Fiebig et al., 2000). In the glycine-rich protein17 (grp17) mutant, which encodes a defective GRP17 oleosin-domain protein, the initiation of pollen hydration is delayed (Mayfield and Preuss, 2000), and in the extracellular lipase4 mutant, whose esterase activity is reduced, the time for pollen hydration is significantly longer than that of wild-type pollen (Updegraff et al., 2009). In this study, we could not determine whether the compatibility factor in the pollen coat is a lipid or a protein, but further screening using the Calcium Green assay in male sterile mutants will be useful in identifying this factor.
We found that pollen coat induces a Ca2+ signaling pathway, which is crucial to successful pollination. Ca2+ has an important role as a secondary messenger in various signal transduction processes. However, a high [Ca2+]cyt is toxic for living cells, and [Ca2+]cyt is maintained at the submicromolar level. Therefore, fine-tuning of [Ca2+]cyt using Ca2+ pumps, antiporters, and uniporters is essential for maintaining Ca2+ homeostasis in animals and plants (Petersen et al., 2005; Clapham, 2007). In this study, we found that a Ca2+ pump gene, ACA13, is elicited by compatible pollen coat treatment. We confirmed that ACA13 is localized on the plasma membrane of stigmatic papilla cells and has Ca2+ transport activity. In the T-DNA line of ACA13, the germination rate of pollen grains was reduced and the initiation of pollen germination was delayed, probably because of the lack of Ca2+ supply from the papilla cell to the pollen grain. Alternatively, the loss of Ca2+-transporting activity on the plasma membrane of the papilla cell might cause a high [Ca2+]cyt in the papilla cell, generating conditions unsuitable for pollen germination. On the other hand, fertility was not completely abolished in either aca13 or aca12 aca13 double mutants. This result suggests that other ACA family members complement the loss of ACA13 function in aca13 and aca12 aca13.
ACA13 was upregulated at 15 min after both pollination and adhesion of pollen coat. On the other hand, live-cell imaging using plants expressing ACA13:ACA13-Venus and immune electron microscopy showed that ACA13 was already present on the plasma membrane of papilla cells before pollination. From these data, together with the observation that Ca2+ was exported continuously from the papilla cell under the growing pollen tube after pollination, in a Calcium Green assay of Arabidopsis (Supplemental Movie 1), we propose that the number of ACA13 molecules in the papilla cell before pollination is insufficient for successful pollination and that rapid upregulation of ACA13 expression provides a requisite reinforcement of the ACA13 protein level.
Furthermore, we found that ACA13 localized not only to the plasma membrane but also to vesicles near Golgi bodies. When the pollen grain germinated, ACA13 signals concentrated around the pollen tube in both live-cell imaging and immune electron microscopy experiments. These observations show the involvement of the secretory pathway in ACA13 accumulation on the plasma membrane around the growing pollen tube. In Brassica species, ARM repeat containing 1 (ARC1) has been identified as one of the direct downstream effectors of the SRK signaling pathway (Gu et al., 1998). ARC1 is a U-box protein with E3 ubiquitin ligase activity (Stone et al., 2003) and interacts with Exo70A1, a putative component of the exocyst complex (Hsu et al., 2004; Samuel et al., 2009). It has therefore been hypothesized that Exo70A1 functions in polarized secretion during compatible pollination, while SRK phosphorylates ARC1 to induce ubiquitination and degradation of Exo70A1 and to inhibit the secretory pathway, leading to rejection of self-pollen (Samuel et al., 2009). In this study, the expression of SYP121 and SYP122 at 15 min after pollination or pollen coat adhesion, respectively, was upregulated. SYP121 and SYP122 encode SNARE proteins localized to the plasma membrane and are associated with the secretory pathway (Rehman et al., 2008). Collectively, the above findings and our data suggest that the secretory pathway functions during compatible pollination to direct the accumulation of the papilla cell’s Ca2+ pump, ACA13, to regions of the plasma membrane near the pollen attachment site and around the germinated pollen tube and that the ACA13 accumulated at these sites effectively exports Ca2+ from the papilla cell to support successful pollination.
METHODS
Plant Materials and Growing Conditions
S8- and S9-allele homozygotes of Brassica rapa and Arabidopsis thaliana plants with ecotype Columbia-0 (Col-0) background were used in this study. Arabidopsis T-DNA lines ACA12 (SALK_098383) and ACA13 (SAIL_878_B06) were obtained from the Nottingham Arabidopsis Stock Centre. By crossing aca12 and aca13, homozygous aca12 aca13 plants were produced in the F2 population. Homozygous insertions were confirmed by PCR amplification with EX Taq (TaKaRa) polymerase and the following primers. For aca12: LBa1, a left border primer for SALK lines, left primer (LP) (5′-CTGTGCATATCACTGTCGCTG-3′) and right primer (RP) (5′-GAAGTCAGTAGACGCCGTGAC-3′); for aca13: SAIL 2, a left border for SAIL lines, LP (5′-GACACTAACGAGCAAACTCCG-3′), and RP (5′-TCTCGCCGTGAAAATGTTATC-3′).
B. rapa was cultivated in a greenhouse at temperatures ranging from 18 to 25°C, and Arabidopsis wild-type plants, T-DNA insertion lines, and transgenic plants were grown at 23°C under 14 h light and 10 h dark in a growth chamber.
Phylogenetic Analysis
Amino acid sequences of ACAs and ER-type Ca2+-ATPase1 (ECA1), obtained from The Arabidopsis Information Resource (TAIR), were multiply aligned using ClustalW 2.1 (Larkin et al., 2007) with the Gonnet matrix (Gonnet et al., 1992) for evaluating evolutionary divergence (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Supplemental Data Set 5). The Type IIA Ca2+-ATPase ECA1 was used as an outgroup of ACAs, which belong to the other group, Type IIB (Geisler et al., 2000). A phylogenetic tree was generated by the neighbor-joining method (Saitou and Nei, 1987) using MEGA 6 software (Tamura et al., 2013). Evolutionary distances were computed by the Poisson correction method (Zuckerkandl and Pauling, 1965) and are displayed as branch length. A bootstrap test (Felsenstein, 1985) with 2000 replicates was performed, and the bootstrap values were calculated as percentages.
Calcium Green Assay
For B. rapa, newly opened flowers of S9-homozygotes, whose anthers were not dehisced, were selected and stuck to an agar plate. A drop of 10 µM Calcium Green solution (Calcium Green 5N, hexapotassium salt; Molecular Probes) containing 0.01% Tween 20 was put on the stigma, which was then air-dried for 1 h. The pistil was mounted on a cover slip, and a pollen grain or pollen coat from freshly dehisced anthers of S9- or S8-homozygotes was applied using a micromanipulator (MO-189; Narishige). The sample was observed with a confocal microscope (LSM710; Zeiss) excited with 488-nm light using a Plan Apo ×20/0.8 objective lens. Fluorescence emission of 500 to 530 nm was captured. Images were collected every 1 min. For Arabidopsis, the method was the same except that 10 µM Calcium Green solution containing 0.005% Tween 20 was used.
Plasmid Constructs and Transformation
DNA fragments encompassing the ACA13 promoter (ProACA13) and encoding ACA13 were obtained by PCR from genomic DNA of Arabidopsis Col-0 with the primers LP (5′-AAGCTTGAACGACGGAAGTCTCAATTATAA-3′) and RP (5′-TCTAGATGTTTCTGGTATTTGCTGAAC-3′) for ProACA13 and LP (5′-ATTCTAGATGCGGAGAAACGTATCTGA-3′) and RP (5′-TTGGATCCCCTCCAGATCCTCCTGATCTCTTCTTCCATTTCAGGTA-3′) for ACA13 and ligated separately to pGEM-T Easy (Promega). For the ACA13:GUS construct, the cauliflower mosaic virus 35S promoter of pBI121 (Promega) was replaced with cloned ProACA13 by digestion with HindIII and XbaI and ligation. For BrSLG:ACA13-Venus, the cloned ACA13 fragment excised by digestion with NotI and BamHI and the previously cloned BrSLG9 promoter (Iwano et al., 2004), excised by digestion with HindIII and Eco52I, were cloned into pBI121-Venus. For ACA13:ACA13-Venus, the cloned ProACA13 was subcloned into HindIII and XbaI sites of pCambia-1300 (Hajdukiewicz et al., 1994). From BrSLG:ACA13-Venus, a DNA fragment containing the ACA13-Venus fusion protein sequence and the nopaline synthase terminator was excised by digestion with XbaI and EcoRI and ligated into the XbaI and EcoRI sites of pCambia-1300 containing ProACA13. Arabidopsis plants were transformed with plasmid constructs using Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) according to the flower dip method described by Clough and Bent (1998).
Pollen Coat Isolation and Preparation of Cyclohexane-Extracted Pollen
Pollen coat was isolated from wild-type pollen of B. rapa and Arabidopsis according to a previous method (Stephenson et al., 1997; Takayama et al., 2000). Pollen coat was removed from wild-type pollen by treatment with cyclohexane (cyclohexane-extracted pollen) as described by Doughty et al. (1993).
Microarray Analysis
Freshly opened Arabidopsis Col-0 flowers at stage 13 (Smyth et al., 1990) were collected from 4- to 5-week-old plants, selected for nondehisced anthers, and stuck to an agar plate. We confirmed that no pollen grains were adhered to the stigmas and pollinated with intact wild-type pollen grains. In each experiment, 50 stigmas were collected at 0 and 15 min after pollination. To identify genes upregulated by pollen coat, we applied pollen coat to the stigmas and collected stigmas both at 15 min after treatment and without treatment. Each experiment was replicated twice. Total RNA was extracted with an RNeasy plant mini kit (Qiagen) according to the manufacturer’s protocol. For each sample, 500 ng of total RNA was used, and a microarray experiment was performed as described by Fujiwara et al. (2004). After cDNA was synthesized, cRNA was amplified and labeled with Cy3-CTP (Agilent Technologies), for samples at 0 min after pollination and nontreatment pollen coat samples, or with Cy5-CTP (Agilent Technologies), for samples at 15 min after pollination or pollen coat adhesion. The labeled cRNA was purified with an RNeasy mini kit, and, for each array, 1 µg Cy3- and Cy5-labeled cRNA was hybridized on a 1x44k Arabidopsis (version3) Gene Expression Microarray (Agilent Technologies), which contains 37,494 Arabidopsis oligonucleotide probes. After washing, the slides were scanned with an FLA-8000 scanner (Fujifilm), and signal intensity was quantified with Array Vision RLS 8.0 software (Genicon Sciences and Imaging Research).
With GeneSpring version 6.0 (Agilent Technologies), Lowess normalization was performed after eliminating error spots. From 37,494 total probes, 27,205 valid data were filtered with a present flag for each treatment. The expression ratios between either (1) 0 min (raw data) and 15 min (control data) after pollination with wild-type pollen or (2) before (raw data) and at 15 min (control data) after adhesion with pollen coat were calculated from the signal intensities of Cy3 (raw data) and Cy5 (control data).
For statistical analysis of microarray data, the log-transformed normalized ratio [=log(raw/control)] for each locus was calculated and mean normalized ratios >2 in each of three experiments were plotted. Next, we treated the experiments using pollen and pollen coat as independent samples and analyzed the differences between raw and control data using a paired t test. We then examined the differences between pollen and pollen coat experiments by evaluating the differences in average structures using ANOVA.
Functional categorization of genes was performed based on molecular functions in Gene Ontology annotations from TAIR and followed by manual correction.
RT-PCR and Real-Time PCR
Total RNA was extracted from stigmas and other tissues of Col-0 wild type and T-DNA mutants with an RNeasy plant mini kit according to the manufacturer’s protocol. For RT-PCR, DNase-treated total RNA (∼50 ng for expression analysis of ACA13 in tissues and 500 ng for T-DNA mutants) was used for cDNA synthesis using oligo(dT)12-18 primer and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. PCR was performed for 35 cycles using either KAPA Taq (Kapa Biosystems) or Ex Taq (TaKaRa) polymerase and the following primers: for aca12, LP (5′-AGCTGCAGTAACCATTGTCGT-3′) and RP (5′-ATGGAGTAAGCCAGCGTCAG-3′); for aca13, LP (5′-CATTTTTGTTCTCCGGGACT-3′) and RP (5′-CGGAAGTAACCGCCATTTTA-3′). For real-time PCR, ∼100 ng DNase-treated total RNA was used for cDNA synthesis, as described above. The real-time PCR reaction was performed with a QuantiTect SYBR Green PCR kit (Qiagen) in an ABI PRISM 7000 (Applied Biosystems) according to the manufacturers’ protocols. The following primers were used: LP (5′-GTGTTCAACGTTCTGTGATG-3′) and RP (5′-GCAACGGAGAGATTTAGCTG-3′). The relative expression level was calculated by 2[threshold cycle (Ct) of 0 min/nontreatment sample – Ct of 15 min after treatment sample] of three biological replicates. For each biological replicate, three technical replicates were performed.
As an expression control, ACT8 was used for both RT-PCR and real-time PCR with the primers LP (5′-AGCACTTTCCAGCAGATGTG-3′) and RP (5′-GAAAGAAATGTGATCCCGTCATG-3′).
Live-Cell Imaging
To determine the localization of ACA13-Venus, pistils were mounted on a cover slip and immersed in water. The samples were observed with a confocal microscope (LSM710; Zeiss) excited with 514-nm light, using a Zeiss C Apo ×63/1.20 water immersion objective lens. Fluorescence emission of 519 to 590 nm was captured. The pinhole diameter was 1.49 to 3.04 Airy units.
To observe the behavior of ACA13-Venus, a stigma excised before anther dehiscence was mounted on a cover glass and fixed with double-sided tape and then covered with a piece of 1% agar. After a wild-type or an aca13 pollen grain was placed on an ACA13-Venus–expressing papilla cell using a micromanipulator, fluorescence was monitored and images were collected every 1 min under dry conditions using a Plan Apo ×20/0.8 objective lens. The pinhole diameter was 3.04 Airy units.
Immune Electron Microscopy
Stigmas before and after pollination were rapidly frozen (KF80; Leica) and subsequently treated with an acetone solution containing 0.01% OsO4 at −80°C for 3 d. The samples were brought slowly to room temperature and then embedded in Spurr’s resin. Ultrathin sections were cut and mounted on Formvar-covered nickel grids.
Sections were incubated with 3% hydrogen peroxide solution for 30 min and washed with distilled water. After blocking with 0.1% gelatin in PBST (PBS containing 0.05% Tween 20), the sections were incubated with anti-GFP antibody (1/100) (Secant Chemicals) and then with 15-nm gold-conjugated goat anti-rabbit IgG (1/75) (Biocell Research Laboratories). After immunolabeling, the sections were stained with uranyl acetate and lead citrate. As a cytochemical control, specimens were incubated without primary antibody or with nonimmune rabbit IgG. Samples were observed with a transmission electron microscope (H-7100; Hitachi).
In Situ Hybridization
In situ hybridization was performed according to the methods described by Doughty et al. (1998) and Tarutani et al. (2010) with minor modifications. Stigmas were collected before and at 20 min after pollination from Arabidopsis flowers at early stage 13 (Smyth et al., 1990), fixed, and embedded in paraffin. The samples were cut in 8-µm sections and hybridized with ACA13 oligo-DNA probes labeled with digoxigenin. The sense (5′-GGTAAAGGAACAAACAACTATGTATGTGATGTATATCACCAGGTGATTACTTTTGACAAG-3′) and antisense (5′-CTTGTCAAAAGTAATCACCTGGTGATATACATCACATACATAGTTGTTTGTTCCTTTACC-3′) probes (Nihon Gene Research Laboratories) were detected and visualized using 5-bromo-4-chloro-3′-indolyphosphate and nitro-blue tetrazolium (Vector Laboratories).
Complementation of Yeast Strain K616
Saccharomyces cerevisiae strain K616 (MATa pmr1:HIS3 pmc1:TRP1 cnb1:LEU2 and ura3), described by Cunningham and Fink (1994), was transformed with pYES2 vector (Invitrogen), which contains URA3 for selection of transformants and the Gal-inducible GAL1 promoter. Downstream of the GAL1 promoter, full-length ACA13, full-length ACA13-Venus, ΔN-ACA13 (lacking sequence encoding the first 65 amino acids), ΔN-ACA13-Venus, or Venus was fused, and the Ca2+ transport ability of ACA13 and these derivatives was tested. The methods were as described by Schiøtt et al. (2004). Constructs encoding either full-length ACA9 or ΔN-ACA9 (lacking the first 93 amino acids) and pYES2 empty vector were also used as controls.
Transformed K616 cells were grown on medium lacking uracil and containing either 2% Glc or 2% Gal, and either 10 mM CaCl2 or 10 mM EGTA, at 30°C for 3 d.
Statistical Analysis
Statistical analyses, other than those of microarray data, were performed using Student's t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ACA1 (At1g27770; NM_179385), ACA2 (At4g37640; NM_119927), ACA4 (At2g41560; NM_129719), ACA7 (At2g22950; NM_127860), ACA8 (At5g57110; NM_180869), ACA9 (At3g21180; NM_113013), ACA10 (At4g29900; NM_119136), ACA11 (At3g57330; NM_115593), ACA12 (At3g63380; NM_116203), ACA13 (At3g22910; NM_113191), ECA1 (At1g07810; NM_100655), and ACT8 (AT1G49240; NM_103814). The microarray data sets were deposited into the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE54637.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Plot of Normalized Ratios.
Supplemental Figure 2. Histogram of P Values for 27,205 Loci According to a Paired t Test.
Supplemental Figure 3. Histogram of P Values for 27,205 Loci According to ANOVA.
Supplemental Figure 4. T-DNA Mutants of ACA12 and ACA13.
Supplemental Figure 5. Complementation of Yeast K616 by ACA13-Venus.
Supplemental Data Set 1. Genes Upregulated in Stigmas after Pollination.
Supplemental Data Set 2. Genes Upregulated in Stigmas after Pollen Coat Adhesion.
Supplemental Data Set 3. Genes Upregulated in Stigmas Both after Pollination and after Pollen Coat Adhesion.
Supplemental Data Set 4. Ca2+-Related Genes Upregulated in Stigmas Both after Pollination and after Pollen Coat Adhesion.
Supplemental Data Set 5. Multiple Sequence Alignment of ACAs and ECA1 Using ClustalW 2.1.
Supplemental Movie 1. Calcium Green Assay during Compatible Pollination in Arabidopsis thaliana.
Supplementary Material
Acknowledgments
We thank Kyle W. Cunningham for the gift of yeast K616 and Yoshie Ryokume, Momoko Okamura, Fumiyo Yamamoto, and Maki Matsumura-Kawashima for their technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (21112003 to M.I.; 23113002 to S.T.) and Grants-in-Aid for Scientific Research (23570056 to M.I.; 21248014 and 25252021 to S.T.).
AUTHOR CONTRIBUTIONS
M. Iwano and S.T. initiated the project. M. Iwano, P.K.-N., H.N., H.S.-A., and S.T. designed experiments. M. Iwano, M. Igarashi, Y.T., P.K.-N., H.N., R.Y., T.E., and H.M. performed the experiments. M. Igarashi, M.U., and G.T. analyzed the microarray data. M. Iwano, Y.T., P.K.-N., and M. Igarashi wrote the article.
Glossary
- Cy3
cyanine-3
- Cy5
cyanine-5
- Col-0
Columbia-0
- RP
right primer
- LP
left primer
- TAIR
The Arabidopsis Information Resource
Footnotes
Online version contains Web-only data.
References
- Bonza M.C., Luoni L., De Michelis M.I. (2004). Functional expression in yeast of an N-deleted form of At-ACA8, a plasma membrane Ca(2+)-ATPase of Arabidopsis thaliana, and characterization of a hyperactive mutant. Planta 218: 814–823. [DOI] [PubMed] [Google Scholar]
- Brewbaker J.L., Kwack B.H. (1963). The essential role of calcium ion in pollen germination and pollen tube growth. Am. J. Bot. 50: 859–865. [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Niroomand S., Zou Y., Wu H.M. (2010). A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. USA 107: 16390–16395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham D.E. (2007). Calcium signaling. Cell 131: 1047–1058. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cunningham K.W., Fink G.R. (1994). Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F., Novotný M., Pícková D., Zárský V. (2004). Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson H. (1995). Dry stigmas, water and self-incompatibility in Brassica. Sex. Plant Reprod. 8: 1–10. [Google Scholar]
- Doughty J., Dixon S., Hiscock S.J., Willis A.C., Parkin I.A., Dickinson H.G. (1998). PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10: 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty J., Hedderson F., McCubbin A., Dickinson H. (1993). Interaction between a coating-borne peptide of the Brassica pollen grain and stigmatic S (self-incompatibility)-locus-specific glycoproteins. Proc. Natl. Acad. Sci. USA 90: 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley T.P.J., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103: 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R., Robinson K.R. (2004). Identification and characterization of stretch-activated ion channels in pollen protoplasts. Plant Physiol. 135: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman C.J., Dickinson H.G. (1996). Identification of pollen components regulating pollination-specific responses in the stigmatic papillae of Brassica oleracea. New Phytol. 133: 197–205. [DOI] [PubMed] [Google Scholar]
- Elleman C.J., Dickinson H.G. (1999). Commonalities between pollen/stigma and host/pathogen interactions: Calcium accumulation during stigmatic penetration by Brassica oleracea pollen tubes. Sex. Plant Reprod. 12: 194–202. [Google Scholar]
- Fiebig A., Mayfield J.A., Miley N.L., Chau S., Fischer R.L., Preuss D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong V.E., Holdaway-Clarke T.L., Straatman K.R., Kunkel J.G., Hepler P.K. (2002). Involvement of extracellular calcium influx in the self-incompatibility response of Papaver rhoeas. Plant J. 29: 333–345. [DOI] [PubMed] [Google Scholar]
- Frietsch S., Wang Y.-F., Sladek C., Poulsen L.R., Romanowsky S.M., Schroeder J.I., Harper J.F. (2007). A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 104: 14531–14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Tanaka N., Kaneda T., Takayama S., Isogai A., Che F.-S. (2004). Rice cDNA microarray-based gene expression profiling of the response to flagellin perception in cultured rice cells. Mol. Plant Microbe Interact. 17: 986–998. [DOI] [PubMed] [Google Scholar]
- Geisler M., Axelsen K.B., Harper J.F., Palmgren M.G. (2000). Molecular aspects of higher plant P-type Ca(2+)-ATPases. Biochim. Biophys. Acta 1465: 52–78. [DOI] [PubMed] [Google Scholar]
- George L., Romanowsky S.M., Harper J.F., Sharrock R.A. (2008). The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 146: 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet G.H., Cohen M.A., Benner S.A. (1992). Exhaustive matching of the entire protein sequence database. Science 256: 1443–1445. [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke T.L., Feijó J.A., Hackett G.R., Kunkel J.G., Hepler P.K. (1997). Pollen tube growth and the intracellular cytostolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood E.E., Gelvin S.B., Melchers L.S., Hoekema A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2: 208–218. [Google Scholar]
- Hsu S.C., TerBush D., Abraham M., Guo W. (2004). The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 233: 243–265. [DOI] [PubMed] [Google Scholar]
- Huang L., Berkelman T., Franklin A.E., Hoffman N.E. (1993). Characterization of a gene encoding a Ca(2+)-ATPase-like protein in the plastid envelope. Proc. Natl. Acad. Sci. USA 90: 10066–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M., Kopczak S.D., Horejsi T.F., Kihl B.K., Pruitt R.E. (1995). Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 8: 703–714. [DOI] [PubMed] [Google Scholar]
- Iwano M., Takayama S. (2012). Self/non-self discrimination in angiosperm self-incompatibility. Curr. Opin. Plant Biol. 15: 78–83. [DOI] [PubMed] [Google Scholar]
- Iwano M., Entani T., Shiba H., Kakita M., Nagai T., Mizuno H., Miyawaki A., Shoji T., Kubo K., Isogai A., Takayama S. (2009). Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 150: 1322–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Shiba H., Matoba K., Miwa T., Funato M., Entani T., Nakayama P., Shimosato H., Takaoka A., Isogai A., Takayama S. (2007). Actin dynamics in papilla cells of Brassica rapa during self- and cross-pollination. Plant Physiol. 144: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Shiba H., Miwa T., Che F.-S., Takayama S., Nagai T., Miyawaki A., Isogai A. (2004). Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 136: 3562–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Wada M., Morita Y., Shiba H., Takayama S., Isogai A. (1999). X-ray microanalysis of papillar cells and pollen grains in the pollination process in Brassica using variable-pressure scanning electron microscope. J. Electron Microsc. (Tokyo) 48: 909–917. [Google Scholar]
- Kühtreiber W.M., Jaffe L.F. (1990). Detection of extracellular calcium gradients with a calcium-specific vibrating electrode. J. Cell Biol. 110: 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Mayfield J.A., Preuss D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2: 128–130. [DOI] [PubMed] [Google Scholar]
- Michard E., Lima P.T., Borges F., Silva A.C., Portes M.T., Carvalho J.E., Gilliham M., Liu L.H., Obermeyer G., Feijó J.A. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437. [DOI] [PubMed] [Google Scholar]
- Murphy D.J. (2006). The extracellular pollen coat in members of the Brassicaceae: Composition, biosynthesis, and functions in pollination. Protoplasma 228: 31–39. [DOI] [PubMed] [Google Scholar]
- Murphy D.J., Vance J. (1999). Mechanisms of lipid-body formation. Trends Biochem. Sci. 24: 109–115. [DOI] [PubMed] [Google Scholar]
- Murphy D.J., Ross J.H.E. (1998). Biosynthesis, targeting and processing of oleosin-like proteins, which are major pollen coat components in Brassica napus. Plant J. 13: 1–16. [PubMed] [Google Scholar]
- Petersen O.H., Michalak M., Verkhratsky A. (2005). Calcium signalling: Past, present and future. Cell Calcium 38: 161–169. [DOI] [PubMed] [Google Scholar]
- Pierson E.S., Miller D.D., Callaham D.A., Shipley A.M., Rivers B.A., Cresti M., Hepler P.K. (1994). Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: Effect of BAPTA-type buffers and hypertonic media. Plant Cell 6: 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E.S., Miller D.D., Callaham D.A., van Aken J., Hackett G., Hepler P.K. (1996). Tip-localized calcium entry fluctuates during pollen tube growth. Dev. Biol. 174: 160–173. [DOI] [PubMed] [Google Scholar]
- Preuss D., Lemieux B., Yen G., Davis R.W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Rehman R.U., Stigliano E., Lycett G.W., Sticher L., Sbano F., Faraco M., Dalessandro G., Di Sansebastiano G.P. (2008). Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant Cell Physiol. 49: 751–766. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiøtt M., Romanowsky S.M., Baekgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. (2004). A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA 101: 9502–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorst L., Kudla J. (2013). Calcium - A central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 1833: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Stephenson A.G., Doughty J., Dixon S., Elleman C., Hiscock S., Dickinson H.G. (1997). The male determinant of self-incompatibility in Brassica oleracea is located in the pollen coating. Plant J. 12: 1351–1359. [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Isogai A. (2005). Self-incompatibility in plants. Annu. Rev. Plant Biol. 56: 467–489. [DOI] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F.-S., Kai N., Watanabe M., Suzuki G., Hinata K., Isogai A. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97: 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.-S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani Y., Shiba H., Iwano M., Kakizaki T., Suzuki G., Watanabe M., Isogai A., Takayama S. (2010). Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466: 983–986. [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff E.P., Zhao F., Preuss D. (2009). The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 22: 197–204. [DOI] [PubMed] [Google Scholar]
- Wolters-Arts M., Lush W.M., Mariani C. (1998). Lipids are required for directional pollen-tube growth. Nature 392: 818–821. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Molecules as documents of evolutionary history. J. Theor. Biol. 8: 357–366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.