The ATG1/13 kinase complex that regulates autophagy also includes ATG11 and ATG101, with ATG11 helping regulate the phosphorylation and turnover of ATG1 and ATG13, tether the complex to developing autophagic structures, and promote the delivery of autophagic vesicles to the vacuole. ATG11 and other autophagy factors also direct the elimination of mitochondria via a vacuolar mitophagic process.
Abstract
Autophagy-mediated turnover removes damaged organelles and unwanted cytoplasmic constituents and thus plays critical roles in cellular housekeeping and nutrient recycling. This “self eating” is tightly regulated by the AUTOPHAGY-RELATED1/13 (ATG1/13) kinase complex, which connects metabolic and environmental cues to the vacuolar delivery of autophagic vesicles. Here, we describe the Arabidopsis thaliana accessory proteins ATG11 and ATG101, which help link the ATG1/13 complex to autophagic membranes. ATG11 promotes vesicle delivery to the vacuole but is not essential for synthesizing the ATG12-ATG5 and ATG8-phosphatidylethanolamine adducts that are central to autophagic vesicle assembly. ATG11, ATG101, ATG1, and ATG13 colocalize with each other and with ATG8, with ATG1 tethered to ATG8 via a canonical ATG8-interacting motif. Also, the presence of ATG11 encourages starvation-induced phosphorylation of ATG1 and turnover of ATG1 and ATG13. Like other atg mutants, ATG11-deficient plants senesce prematurely and are hypersensitive to nitrogen and fixed-carbon limitations. Additionally, we discovered that the senescence-induced breakdown of mitochondria-resident proteins and mitochondrial vesicles occurs via an autophagic process requiring ATG11 and other ATG components. Together, our data indicate that ATG11 (and possibly ATG101) provides important scaffolds connecting the ATG1/13 complex to both general autophagy and selective mitophagy.
INTRODUCTION
Plant cells employ multiple catabolic mechanisms to remove dysfunctional and unneeded constituents and to recycle intracellular nutrients. A major route involves macroautophagy (hereafter referred to as autophagy), which encapsulates and delivers cytoplasmic material to the vacuole for breakdown (reviewed in Li and Vierstra, 2012; Liu and Bassham, 2012; Reggiori and Klionsky, 2013). Central to this process is the de novo formation of a cup-shaped vesicle called the phagophore (or isolation membrane) by a set of AUTOPHAGY-RELATED (ATG) proteins that localize within a common phagophore assembly site (PAS). The phagophore elongates, engulfs cytoplasmic material, and eventually seals to create a double membrane-enclosed compartment called the autophagosome, which subsequently fuses with the vacuole/lysosome to release the internal vesicle as an autophagic body. The autophagic body and its cargo are then degraded by vacuolar hydrolases, and the resulting metabolites are exported back to the cytoplasm for reuse.
Autophagy occurs at a low basal level under nutrient-rich conditions and is actively increased upon nutrient stress, consistent with its involvement in the nonselective turnover of bulk cytoplasm and in promoting cell survival under nutrient limitations through accelerated recycling (Li and Vierstra, 2012; Liu and Bassham, 2012; Reggiori and Klionsky, 2013). These roles are especially apparent in plants, where the accumulation of autophagic bodies is strongly upregulated during senescence or under nitrogen or fixed-carbon starvation (Doelling et al., 2002; Hanaoka et al., 2002; Xiong et al., 2005; Chung et al., 2009; Suttangkakul et al., 2011). This nonselective autophagy is also activated during the hypersensitive response following pathogen invasion, presumably to discourage pathogen spread by promoting programmed cell death at the infection site (Hayward and Dinesh-Kumar, 2011; Lenz et al., 2011). In addition, recent studies have provided evidence for several types of selective autophagy that specifically target organelles and macromolecular protein complexes when defective or in excess, cytotoxic protein aggregates (aggrephagy), and even invading pathogens (xenophagy) (Johansen and Lamark, 2011). Examples of this selectivity in plants include the clearance of damaged/excess chloroplasts (chlorophagy; Ishida et al., 2008; Wada et al., 2009) and peroxisomes (pexophagy; Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013), ribosomes (ribophagy; Hillwig et al., 2011), endoplasmic reticulum components during endoplasmic reticulum stress (Liu et al., 2012), insoluble ubiquitylated proteins generated by environmental stress (Zhou et al., 2013), and free phototoxic porphyrins (Vanhee et al., 2011).
From extensive studies with yeast (Saccharomyces cerevisiae) and mammalian cells and subsequent analyses with Arabidopsis thaliana, a complex ATG system has been revealed that stimulates autophagy, organizes the PAS, accumulates membrane to create the phagophore, recruits appropriate cargo, promotes phagophore enclosure, and delivers autophagosomes to the vacuole to release the autophagic bodies for eventual breakdown (Li and Vierstra, 2012; Reggiori and Klionsky, 2013). Central to this system is a pair of ubiquitin-like proteins, ATG8 and ATG12, that become attached via an ATP-dependent conjugation cascade to the lipid phosphatidylethanolamine (PE) and the ATG5 protein, respectively. ATG8 and ATG12 share a common activating enzyme, ATG7, but are then transferred to distinct conjugating enzymes, ATG3 and ATG10, respectively. Activated ATG12 is linked to ATG5 via an isopeptide bond between the C-terminal Gly of ATG12 and a unique Lys in ATG5. The resulting ATG12-ATG5 conjugate then docks with the phagophore via ATG16 and stimulates the lipidation of ATG8 at its C-terminal Gly. The ATG8-PE adduct decorates the emerging phagophore and provides a docking surface for factors that help expand and seal the vesicle and for receptors that recruit specific cargo. Often, an ATG8 interaction motif (AIM; also known as the LC3-interacting region) is present in these factors that specifically binds to the ATG8 moiety (Noda et al., 2010). Several of these AIM-containing receptors (e.g., neighbor of BRCA1 [NBR1] and sequestosome-1) also have affinity for ubiquitin, thus providing a mechanism to remove insoluble protein aggregates and organelles/protein complexes that are too large for the 26S proteasome following their ubiquitylation (Johansen and Lamark, 2011).
Regulating the formation of autophagic vesicles in both fungi and metazoans is an intricate web of sensor kinases that respond to nutrient availability, including Target of Rapamycin (TOR), Protein Kinase A, and the sugar nonfermenting–type kinases (likely AKIN10 and AKIN11 in Arabidopsis; Reggiori and Klionsky, 2013; Wong et al., 2013; Xiong et al., 2013). These kinases converge on a complex consisting of the Ser/Thr kinase ATG1 (Uncoordinated51-like kinase [ULK1] in mammals) and its accessory regulator ATG13, whereby collective changes in the phosphorylation status of ATG1 and ATG13 help control autophagic flux, presumably through the regulation of downstream effectors (Mizushima, 2010; Suttangkakul et al., 2011). For example, phosphorylation of ATG13 by TOR under nutrient-rich conditions dampens the autophosphorylation of ATG1 that is necessary for autophagy induction (Kraft et al., 2012). The kinase motif in ATG1 is located at the N-terminal end and appears important for releasing ATG components from the maturing autophagosome (Cheong et al., 2008). Its C-terminal end contains an early autophagy targeting/tethering domain that binds to the curved membrane surface of the vesicles that coalesce into the nascent phagophore, and to other factors (e.g., ATG13) that stimulate its kinase activity (Cheong et al., 2008; Ragusa et al., 2012). Ultimately, tight regulation of ATG1/13 signaling is critical, as too much or too little autophagy induced by modifying ATG1 and ATG13 is deleterious (Chen and Klionsky, 2011).
Besides ATG1 and ATG13, the ATG1/13 complex includes a number of accessory proteins, including ATG11, ATG17, ATG29, and ATG31 in yeast and the ATG11 ortholog Focal Adhesion Kinase Family–Interacting Protein of 200 kD (FIP200) and ATG101 in mammals (Mizushima, 2010). The functions of ATG29, ATG31, and ATG101 are unclear; mammalian ATG101 binds ATG13 and might stabilize the ATG1/13 complex and protect it from turnover (Hosokawa et al., 2009; Mercer et al., 2009). By contrast, data from yeast suggest that ATG11 and ATG17 act as scaffolds to help connect the ATG1/13 complex to the PAS. In particular, the extensive coiled-coil region in ATG17 assumes an elongated crescent shape that docks ATG29 and ATG31 on its concave surface. When dimerized, this complex could form a bridge that clusters membrane vesicles essential for phagophore assembly (Ragusa et al., 2012). Consistent with this action, ATG17 is one of the earliest components of the yeast PAS, with its elimination completely disrupting PAS assembly (Suzuki et al., 2007). ATG11 is also critical for PAS architecture and appears to play a direct role in scaffolding the ATG1/13 complex to the PAS (Yorimitsu and Klionsky, 2005). Moreover, yeast ATG11 interacts with a variety of receptors for selective autophagy, including ATG32, ATG36, and ATG19, that are required for mitochondrial turnover (mitophagy), pexophagy, and the cytoplasm-to-vacuole targeting of functional proteins, respectively (Reggiori and Klionsky, 2013).
In a previous study, we characterized the ATG1 and ATG13 subunits of the Arabidopsis ATG1/13 kinase complex and showed that it is essential for autophagic transport, the survival of plants when nutrient limited, and proper senescence (Suttangkakul et al., 2011). Whereas a family of four genes encodes ATG1, a pair of genes encodes ATG13. atg13a atg13b null plants fail to accumulate autophagic bodies inside the vacuole but still assemble the ATG12-ATG5 and ATG8-PE adducts, suggesting that the kinase complex works downstream of PAS/phagophore assembly but before the fusion of autophagosomes with the vacuole. Surprisingly, nutrient starvation induces the rapid degradation of both ATG1 and ATG13 via an autophagic route, thus providing a dynamic mechanism to feedback regulate autophagy depending on the plant’s nutritional status (Suttangkakul et al., 2011).
Here, we further describe the organization and regulation of the ATG1/13 complex through analysis of its interactions with, and possible regulation by, accessory proteins. Although Arabidopsis and other plants appear to be missing ATG17, they express orthologs of yeast ATG11 and mammalian ATG101 that interact with ATG1 and/or ATG13 in planta. Like other well-characterized atg mutants, Arabidopsis seedlings missing ATG11 have severely dampened tolerance to nitrogen and fixed-carbon deprivation, have much-reduced deposition of autophagic bodies, and are compromised in the stress-induced phosphorylation of ATG1 and turnover of ATG1 and ATG13, suggesting a critical role for ATG11 in autophagosome formation and proper ATG1/13 complex assembly/activity. Consistent with the involvement of yeast ATG11 in mitophagy (Kanki et al., 2009; Okamoto et al., 2009), Arabidopsis atg11 mutants are attenuated in the turnover of mitochondrial proteins and in the autophagic transport of mitochondria into vacuoles. Taken together, ATG11 appears to be a multiple-function scaffold in Arabidopsis that regulates signaling by the ATG1/13 kinase, is necessary for proper autophagic vesicle formation, and is involved in the selective clearance of mitochondria.
RESULTS
Identification of Arabidopsis ATG11 and Organization of the ATG11 Family
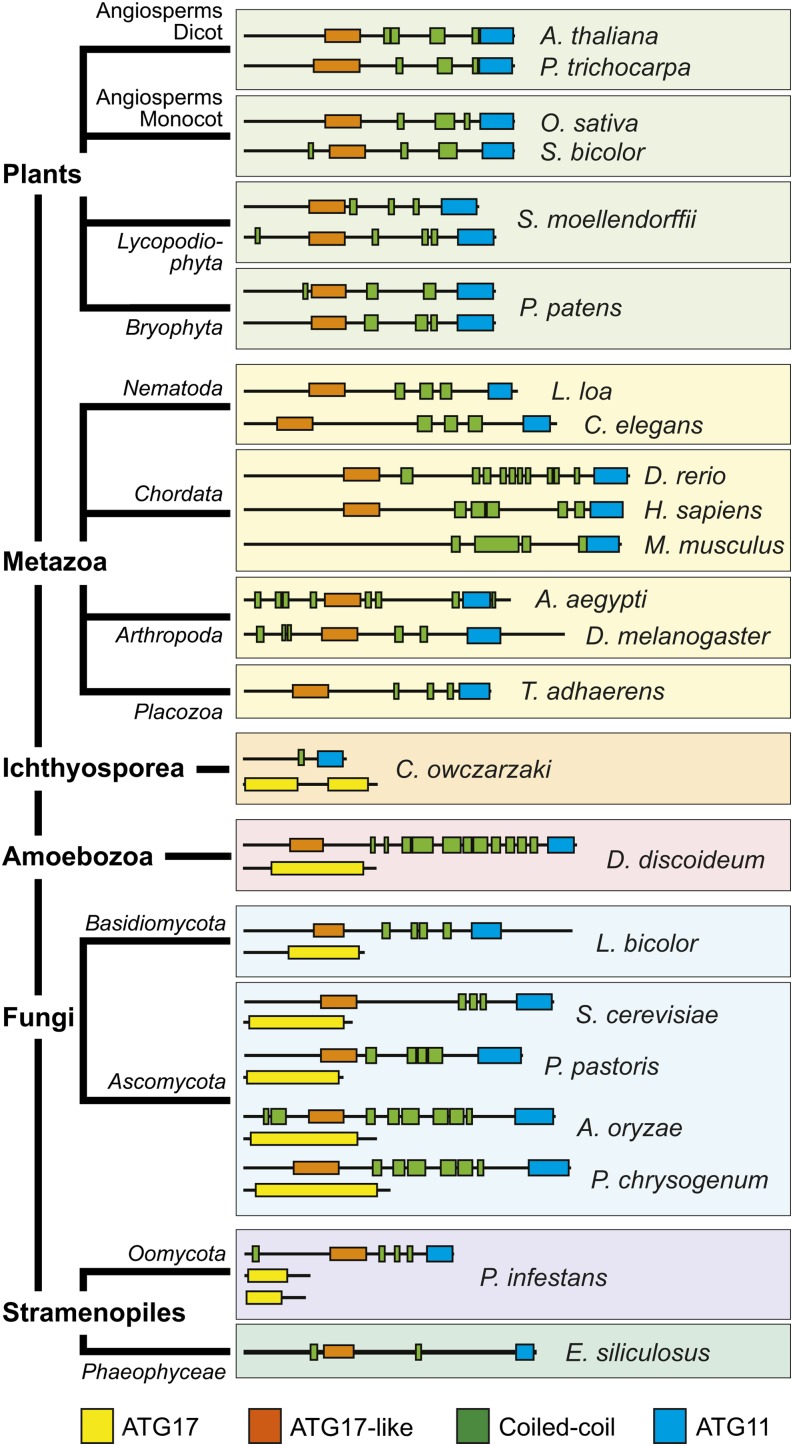
Besides ATG1 and ATG13, the ATG1/13 kinase complex in metazoans and fungi includes additional subunits ATG11/FIP200, ATG17, ATG101, and the yeast-specific ATG29 and ATG31 proteins (Mizushima, 2010; Reggiori and Klionsky, 2013). In agreement with Meijer et al. (2007), we failed to detect close relatives of ATG17, ATG29, and ATG31 by DELTA-BLAST searches of the Arabidopsis Columbia-0 (Col-0) proteome database. However, we did identify likely orthologs of ATG11/FIP200 and ATG101. Using the ∼160-amino acid ATG11 domain from yeast and metazoan representatives as queries, a single Arabidopsis protein (At4g30790) of similar overall length (1148 amino acids) was found, which contains a comparably positioned ATG11 domain with 18 and 15% identity to those in yeast ATG11 and human FIP200, respectively (Supplemental Figure 1). The Arabidopsis protein also includes proximal repeats of a predicted coiled-coil motif (residues 620 to 644, 649 to 676, 814 to 876, and 958 to 992), which are also characteristic of the ATG11 family (Figures 1 and 2A) and have been implicated in homodimerization and interactions with other ATG components (Yorimitsu and Klionsky, 2005).
Figure 1.
Phylogenetic Distribution of ATG11/FIP200 and ATG17 Proteins among Eukaryotes.
The representatives from plants, metazoa, Ichthyosporea, amoebozoa, fungi, and stramenopiles were clustered based on their amino acid sequences and the presence/absence and arrangement of the signature ATG17, ATG17-like, coiled-coil, and ATG11 domains. See Supplemental Table 4 for the species names and UniProt accession numbers.
Figure 2.
Interactions of Arabidopsis ATG11 with Itself, Subunits of the ATG1/13 Kinase Complex, and ATG8-Decorated Autophagic Structures.
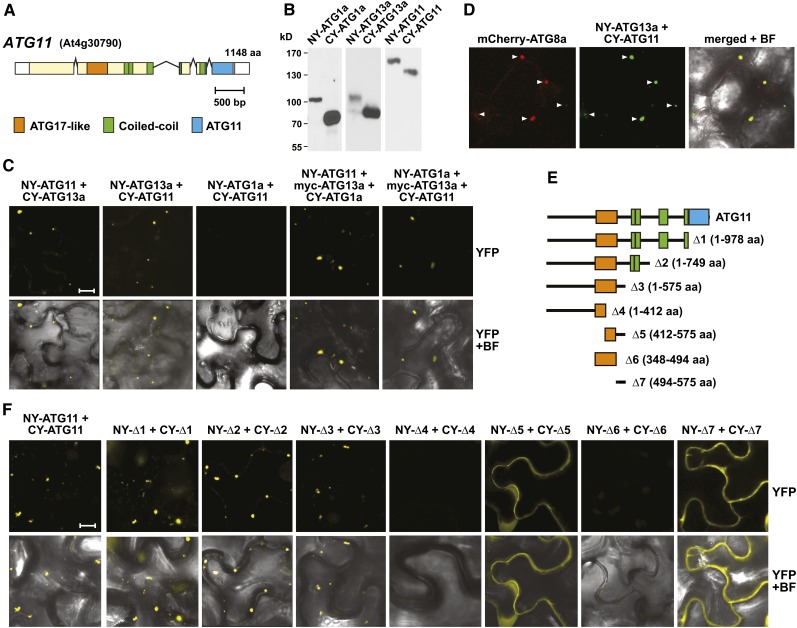
(A) Domain organization of ATG11. Lines represent introns, and colored and white boxes represent coding and untranslated regions, respectively. Positions of the signature ATG17-like, coiled-coil, and ATG11 domains are indicated. aa, amino acids.
(B) and (C) Interaction of ATG11 with ATG1 and ATG13 in planta by BiFC.
(B) Demonstration that each possible orientation of the NY and CY constructions for ATG1a, ATG11, and ATG13a could be expressed in N. benthamiana. Crude extracts prepared from leaves 36 h after infiltration were immunoblotted with anti-GFP antibodies.
(C) BiFC using N. benthamiana leaf epidermal cells. Leaves were coinfiltrated with plasmids expressing the N- and C-terminal fragments of YFP fused to ATG1a, ATG11, and ATG13a. To detect indirect interactions between ATG1a and ATG11, the cells were also infiltrated with a plasmid expressing ATG13a tagged with an N-terminal Myc epitope. Shown are reconstituted BiFC signals as detected by confocal fluorescence microscopy of leaf epidermal cells 36 h after coinfiltration along with a bright-field (BF) image of the cells. Bar = 10 μm.
(D) ATG11 and ATG13a colocalize with ATG8-decorated autophagic structures. The leaves of 4-d-old transgenic Arabidopsis seedlings stably expressing mCherry-ATG8a were coinfiltrated with plasmids expressing the NY-ATG13a and CY-ATG11 BiFC constructs and examined by confocal fluorescence microscopy as in (C). Arrowheads indicate colocalized structures.
(E) and (F) Localization of the homodimerization domain within ATG11 by BiFC.
(E) Diagram of the ATG11 truncations.
(F) BiFC signals in N. benthamiana leaf epidermal cells from paired ATG11 truncations fused to the N- and C-terminal fragments of YFP along with a bright-field image of the cells. Bar = 10 μm.
Surprisingly, when the At4g30790 protein sequence was used in a reciprocal BLAST search of yeast, the canonical ATG17 protein was detected (<1e-51) in addition to ATG11. This homology was generated by a short cryptic ATG17-like domain (residues 348 to 494) with weak identity (13%) toward the N terminus of At4g30790 (Supplemental Figure 2). Especially obvious was the presence of the conserved sequence Y-X-X-X-L/V/I-X-E-V/I-X-R-R-R/K, which is characteristic of the ATG17 family.
To further clarify this organization, we extensively searched other eukaryotic genomes using the ATG11 and ATG17 domains as separate queries. Likely relatives containing either or both domains were detected in various lineages (Figure 1; Supplemental Figures 3 and 4). For example, representative Ichthyosporea, amoebozoa, fungi, and stramenopiles typically encode separate polypeptides with ATG11/coiled-coil or the longer ATG17 domain signatures. At first glance, metazoans and plants appeared to encode only ATG11/coiled-coil–type proteins, but careful hand analyses of the ATG11 homologs revealed that they, like Arabidopsis At4g30790, also contain the short ATG17-like region (∼150 amino acids) with its core Y-X-X-X-L/V/I-X-E-V/I-X-R-R-R/K sequence upstream of a coiled-coil region (Figure 1; Supplemental Figure 2). Moreover, this shorter ATG17-like domain was also detected in the fungal ATG11 family, even though the host species also encode separate, canonical ATG17 proteins. Taken together, these different architectures implied that the ATG11 family is actually a hybrid of ATG11 and ATG17 containing sequential coiled-coil motifs bracketed on each side by ATG17-like and ATG11 domains. The plant ATG11/17 hybrids are expressed from a single gene in most angiosperms analyzed, but two representative seedless plants (Physcometrilla patens and Selaginella moellendorffii) have two highly related loci that appear, based on sequence similarity, to have emerged from a recent duplication. Thus, despite its low overall sequence identity to fungal ATG11 and its mammalian relative FIP200, it appears that the Arabidopsis At4g30790 and its land plant orthologs represent close relatives; thus, we designated them as ATG11 proteins hereafter.
Previous studies with yeast and mammalian cells demonstrated that ATG11 and FIP200 associate with various other ATG components. As examples, yeast ATG11 interacts with ATG1, ATG17, ATG20, ATG29, and various cargo receptors, whereas mammalian FIP200 interacts with ATG13 and possibly ULK1/2 (ATG1; Yorimitsu and Klionsky, 2005; Hara et al., 2008; Ganley et al., 2009; Jung et al., 2009; Reggiori and Klionsky, 2013). To confirm that Arabidopsis ATG11 assembles into the ATG1/13 kinase complex, we set up binding assays between it and ATG1a or ATG13a. Yeast two-hybrid (Y2H) assays failed, even though ATG1a, ATG13a, and ATG11 succeeded with other binding partners (see below). In contrast, binding was successfully demonstrated by bimolecular fluorescence complementation (BiFC), which is based upon the reconstitution of split yellow fluorescent protein (YFP) into a fluorophore via interactions between the appended protein pairs (Hu et al., 2002). In preliminary studies, we found that all three proteins N-terminally tagged with either the N- or C-terminal half of YFP (NY or CY, respectively) could be correctly synthesized in Nicotiana benthamiana leaves following transient expression (Figure 2B). When NY-ATG11 or CY-ATG11 was coexpressed with reciprocally tagged ATG13a, reconstituted YFP fluorescence was easily detected as punctate intracellular spots (1 to 5 μm) by confocal fluorescence microscopy of epidermal cells (Figure 2C; Supplemental Figure 5A). In most cases, the spots appeared within the cytoplasm and outside of the vacuole. Identical BiFC assays in N. benthamiana with ATG1a and ATG11 pairs failed, suggesting that these two proteins do not interact directly. However, indirect interaction could be detected using ATG13a as a bridge when NY-ATG1a and CY-ATG11 were coexpressed with Myc-tagged ATG13a (Figure 2C). When the fluorescent BiFC signal of the NY-ATG11 and CY-ATG13 pair was colocalized in Arabidopsis with the autophagic vesicle marker mCherry-ATG8 (Yoshimoto et al., 2004; Thompson et al., 2005), a strong coherence was observed, indicating that these puncta likely represent developing PAS, phagophores, and/or autophagosome structures (Figure 2D).
Studies with other eukaryotes also showed that ATG11 homodimerizes and that this self-interaction is possibly important for tethering the rest of the ATG1/13 complex to developing autophagic structures (Yorimitsu and Klionsky, 2005). Y2H assays with activation and binding domain pairs and BiFC studies also detected this self-association for Arabidopsis ATG11 (Figures 2E and 2F; Supplemental Figure 6). To pinpoint the region involved, we tested a series of ATG11 truncations with a focus on the ATG17-like, coiled-coil, and ATG11 domain–containing regions (Figure 2E). Both Y2H and BiFC showed that none of these signature regions were required; instead, the dimerization site was localized to a short 81-amino acid stretch (residues 494 to 575) just distal to the ATG17-like domain (Figure 2F; Supplemental Figure 6). Even this region in isolation [Δ7(494-575)] dimerized. Whereas full-length ATG11 or truncations containing the complete ATG17-like domain along with the homodimerization domain [i.e., Δ1(1-978), Δ2(1-749), or Δ3(1-575)] self-associated by BiFC in small puncta expected for autophagic structures, the Δ5(412-575) truncation, which is missing part of the ATG17-like domain but has the homodimerization domain, displayed diffuse cytoplasmic fluorescence like Δ7(494-575) (Figure 2F). This redistribution implied that binding of ATG11 to the PAS and/or autophagic membranes requires the complete ATG17-like domain and/or the N-terminal residues preceding it.
Using this same set of ATG11 truncations above, we also attempted to determine by BiFC where ATG13 binds to ATG11. Whereas C-terminal truncations of CY-ATG11 missing the ATG11 domain and the coiled-coil repeats still interacted with NY-ATG13a [e.g., Δ3(1-575)], a set of truncations removing the ATG11 dimerization site and the upstream ATG17-like domain or including just these segments did not [e.g., Δ4(1-412), Δ6(348-494), and Δ7(494-575); Supplemental Figure 7A]. Together, the BiFC data suggest that ATG11 associates with ATG13 through the combined action of multiple regions within its N-terminal half.
The Arabidopsis Gene Encoding ATG101
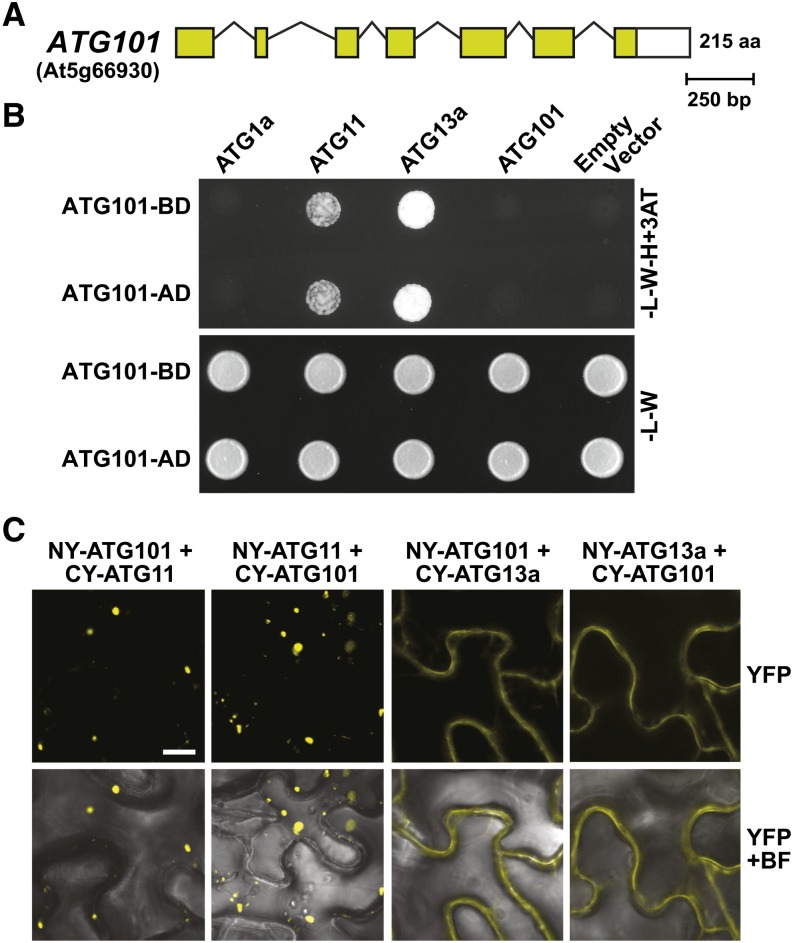
The likely Arabidopsis homolog of ATG101 is a 215-amino acid polypeptide encoded by a single locus (At5g66930), which has 26, 27, and 23% sequence identity to its Schizosaccharomyces pombe, Drosophila melanogaster, and human counterparts, respectively. (Figure 3A; Supplemental Figures 8 and 9). Searches using Arabidopsis ATG101 as the query identified orthologs in many other plant species (57 to 78% identity), including seedless plants (P. patens and S. moellendorffii) and numerous angiosperms (both eudicot and monocot) that clustered phylogenetically in a group separate from their relatives in Chlamydomonas reinhardtii, fungi, and metazoans (Supplemental Figure 8). In most plant species (the exception being S. moellendorffii), ATG101 is encoded by a single gene. Outside of a unique ∼13-residue insertion near the N terminus of the metazoan relatives, ATG101 polypeptides are mostly contiguous and of similar sizes (200 to 220 amino acids), with conserved regions scattered throughout the sequence (Supplemental Figure 9). Other than potentially acting as a proteolytic regulator (Hosokawa et al., 2009; Mercer et al., 2009), the amino acid sequence of ATG101 is devoid of recognizable domains to imply specific activit(ies).
Figure 3.
Interaction of ATG101 with Other Subunits of the ATG1/13 Kinase Complex.
(A) Structure of the ATG101 locus (At5g66930). Lines represent introns, and the colored and white boxes represent coding and untranslated regions, respectively. Amino acid (aa) sequence length is indicated on the right.
(B) Y2H interactions of ATG101 with other subunits of the ATG1/13 complex. Full-length ATG101 designed as N-terminal fusions to either the GAL4 activating (AD) or binding (BD) domain was coexpressed with complementary AD or BD fusions of ATG1a, ATG11, and ATG13a. Shown are cells grown on selection medium lacking Trp and Leu (−L−W) or lacking Trp, Leu, and His and containing 3-amino-1,2,4-triazole (−L−W−H+3AT).
(C) Interaction of ATG101 with ATG11 or ATG13a in planta by BiFC. N. benthamiana leaf epidermal cells were coinfiltrated with plasmids expressing the N- and C-terminal fragments of YFP fused to ATG101, ATG11, and ATG13a. Reconstituted BiFC signals, as detected by confocal fluorescence microscopy of leaf epidermal cells 36 h after infiltration, are shown along with a bright-field (BF) image of the cells. Bar = 10 μm.
Mammalian ATG101 interacts with ATG13 and FIP200 (ATG11) in the complete ATG1/13 complex (Hosokawa et al., 2009; Mercer et al., 2009; Behrends et al., 2010). Confirmation that Arabidopsis ATG101 is indeed part of the plant ATG1/13 particle was provided by Y2H and BiFC assays. As shown in Figure 3B, ATG101 in either Y2H orientation interacted with ATG11 and ATG13a but not with itself or ATG1a. Similarly, ATG101 associated with ATG11 and ATG13a in either orientation by BiFC following coexpression in N. benthamiana leaves (Figure 3C; Supplemental Figure 5B). Similar to the distribution of the ATG11/ATG13 pair, the reconstituted YFP fluorescent signals for the ATG11/ATG101 pair were in punctate cytoplasmic foci, consistent with their likely association with autophagic structures. Conversely, signals for the ATG13a/ATG101 pair appeared diffuse within the cytoplasm, suggesting that ATG11 but not ATG13 helps anchor ATG101 to these sites.
To help locate where ATG101 binds ATG11, we tested by BiFC how the set of ATG11 truncations defined above would interact with full-length ATG101 (Figure 2E). As can be seen in Supplemental Figure 7C, C-terminal truncations missing the ATG11 domain and the coiled-coil region retained binding [e.g., Δ3(1-575)], whereas mutants affecting the region upstream did not [e.g., Δ4(1-412), Δ6(348-494), and Δ7(494-575)], suggesting that the ATG101 interaction sites are within or upstream of the ATG11 dimerization motif.
Expression of ATG11 and ATG101
Based on transcriptome analyses within the eFP DNA microarray database (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), both ATG11 and ATG101, like ATG1a to ATG1d and ATG13a/b (Suttangkakul et al., 2011), are widely expressed in a number of Arabidopsis tissues at various developmental stages, consistent with the importance of the ATG1/13 complex in particular, and autophagy in general, in cellular housekeeping/recycling (Supplemental Figure 10). High transcript levels were evident in senescing leaves and in mature floral tissues, including petals and stamens. Interestingly, in comparison with ATG1c and ATG13a, high expression of ATG11 and ATG101 was detected in maturing and dry seeds, suggesting a role for these cofactors during seed development, such as nucellus absorption and/or storage protein accumulation (Herman and Larkins, 1999). Conversely, ATG1c transcripts were highest in mature pollen (Supplemental Figure 10).
Expression correlations based on the ATTED-II database (http://atted.jp) identified 15 other ATG genes among the top 300 genes that coexpress significantly with ATG11, including loci encoding other subunits of the ATG1/13 complex (ATG1b, ATG1c, and ATG101), ATG5, ATG6, ATG7, ATG9, ATG12a, three of the nine ATG18 paralogs, and four of the nine ATG8 paralogs (Supplemental Table 1). In addition, ATG11 displayed strong coexpression correlations with two genes (At3g43230 and At5g16680) encoding FYVE (for Fab1, YOTB/ZK632.12, Vac1, and EEA1)-type zinc finger proteins (ranks 1 and 18; Supplemental Table 2). Related FYVE proteins in other organisms have been proposed to aid in the curvature and sealing of autophagic vesicles by using the FYVE domain to bind the phosphatidylinositol 3-phosphate moiety that decorates the enveloping autophagic membrane (Pankiv et al., 2010).
Reverse Genetic Analysis of ATG11
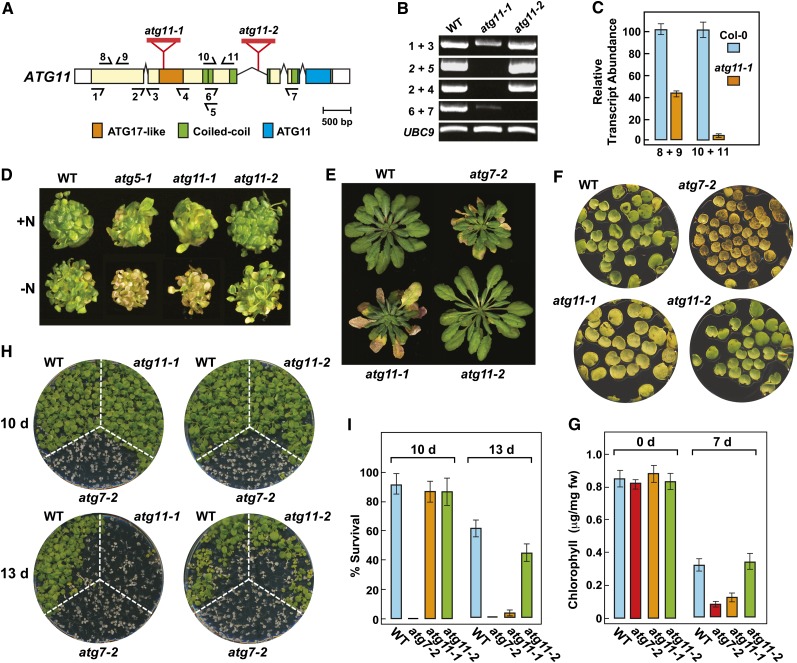
To better define the importance of ATG11, we searched the publicly available T-DNA libraries for insertions within the transcribed region that would likely compromise functional expression. The most promising were the atg11-1 and atg11-2 alleles, which interrupt either the exonic region encoding the ATG17 domain (insertion within codon 436) or the second intron intervening the region encoding the third coiled-coil motif, respectively (Figure 4A). RT-PCR revealed that both alleles blocked transcription across the respective insertion sites (primers 2+4/5 and 6+7, respectively) but that the mutant plants accumulated relatively high levels of transcript containing sequence upstream of the insertion sites (Figures 4B and 4C). Consequently, while atg11-1 plants might express at most only a short N-terminal portion of the ATG11 polypeptide upstream of the ATG17-like domain, atg11-2 plants could express a substantially larger fragment encompassing the N-terminal region of ATG11, including the ATG17-like domain, the sequence involved in homodimerization and binding to ATG13 and ATG101, and possibly three of the four coiled-coil motifs (Figure 4A).
Figure 4.
Genotypic and Phenotypic Analyses of Arabidopsis atg11 Mutants.
(A) Diagram of the ATG11 locus showing domain organization and the positions of relevant T-DNA insertion mutations. Lines represent introns, and the colored and white boxes represent coding and untranslated regions, respectively. Positions of the signature ATG17-like, coiled-coil, and ATG11 domains are indicated. Red triangles identify the T-DNA insertion sites for the atg11-1 and atg11-2 alleles. Half-arrows at bottom and top indicate positions of the primers used for RT-PCR in (B) and quantitative RT-PCR in (C), respectively.
(B) RT-PCR analysis of the ATG11 transcript in the atg11-1 and atg11-2 mutants. Total RNA isolated from wild-type or homozygous mutant plants was subjected to RT-PCR using the primer pairs indicated in (A). RT-PCR with primers specific for UBC9 was included to confirm the analysis of equal amounts of RNA.
(C) Quantitative real-time RT-PCR analysis of transcripts arising 5′ and 3′ to the T-DNA insertion site in atg11-1. Primer locations are indicated in (A).
(D) Enhanced sensitivity to nitrogen starvation. Plants were germinated and grown for 1 week on nitrogen-containing liquid medium and then transferred to either nitrogen-containing (+N) or nitrogen-deficient (−N) medium for an additional 2 weeks. Lines tested include the wild type, the atg11-1 and atg11-2 alleles shown in (A), and previously described autophagy-defective mutants atg5-1 and atg7-2 (Thompson et al., 2005; Chung et al., 2010).
(E) Premature senescence under an SD photoperiod. Plants were grown on soil at 21°C under SD conditions for 10 weeks.
(F) Accelerated senescence of detached leaves. The first pair of true leaves was cut from 2-week-old seedlings grown with 1% Suc and incubated for 7 d in the dark at 24°C.
(G) Relative chlorophyll content of detached leaves shown in (D). Each bar represents the mean ± sd from three independent experiments analyzing at least 30 leaves each.
(H) Enhanced sensitivity to fixed-carbon starvation. Seedlings were grown under an LD photoperiod without added Suc for 2 weeks and then placed in darkness for 10 or 13 d before returning to LD conditions for a 12-d recovery.
(I) Survival of fixed-carbon–deprived plants shown in (E). Each bar represents the mean percentage survival ± sd of three independent experiments examining at least 15 seedlings each.
atg11-1 plants also accumulated a sizable transcript downstream of the T-DNA insertion site, albeit at ∼32-fold lower levels than wild-type plants (Figure 4C). This transcript could be translated theoretically into a polypeptide containing part of the ATG17-like, coiled-coil, and ATG11 domains (Figure 4B). To assess whether this possible C-terminal fragment of ATG11 along with the shorter N-terminal fragment might be functional if expressed at sufficient levels in atg11-1 plants, we tested the self-interaction of these fragments and their interaction with full-length ATG13a in planta using BiFC. Neither fragment (encoding residues 1 to 435 or 436 to 1148) was able to interact with ATG13a by BiFC, and only the C-terminal fragment self-interacted, but displayed a diffuse cytoplasmic distribution instead of the punctate pattern normal for full-length ATG11 (Supplemental Figure 11). Consequently, it is likely that the atg11-1 mutant approximates a null allele, given the absence of known functional domains in the predicted N-terminal fragment of ATG11 and the expectation that substantially lower levels of a mislocalized C-terminal fragment would be expressed.
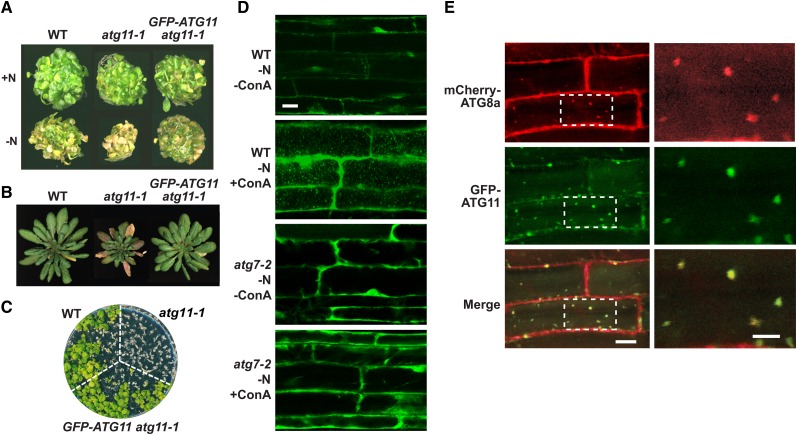
Homozygous atg11-1 and atg11-2 plants developed and grew normally under standard growth chamber conditions. However, like other atg mutants (Doelling et al., 2002; Hanaoka et al., 2002; Chung et al., 2010), the atg11-1 line, but not the atg11-2 line, developed smaller rosettes that senesced earlier following bolting under both long-day (LD) and short-day (SD) photoperiods, suggesting that only the atg11-1 plants have compromised autophagy. For example, the atg11-1 mutant, like the null atg7-2 allele, which is blocked in ATG8 and ATG12 conjugation (Chung et al., 2010), began leaf senescence about 2 weeks before wild-type and atg11-2 plants under SD conditions (Figure 4E).
To examine whether the atg11 mutation compromises autophagy, we challenged the two atg11 alleles to a set of diagnostic phenotypic assays, including sensitivity and survival of seedlings under nitrogen- and fixed carbon–limiting conditions and accelerated chlorosis of detached leaves (Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011). For nitrogen starvation, plants were first grown under continuous illumination on nitrogen-rich liquid medium containing Suc for 7 d and then transferred to either fresh nitrogen-rich medium or nitrogen-deficient medium for an additional 14 d (Figure 4D). For fixed-carbon starvation, plants were grown on solid Murashige and Skoog (MS) medium without added Suc under an LD photoperiod for 2 weeks and then placed in the dark for 10 or 13 d before return to LD conditions (Figures 4H and 4I). For accelerated chlorosis, the first pair of true leaves was excised from 2-week-old seedlings, floated on buffer in the dark, and then assayed for chlorophyll content after 7 d (Figures 4F and 4G). In all cases, atg11-1 plants showed enhanced sensitivity to these nutrient stresses, like other strong atg mutants such as atg7-2.
Previous studies showed that double atg13a atg13b mutants eliminating the core ATG13 subunit of the ATG1/13 kinase complex were less sensitive to fixed-carbon starvation than were mutants compromising central components involved in ATG8/12 conjugation (Suttangkakul et al., 2011). This appears also true for atg11-1 plants, which better tolerated 10 d in the dark as compared with atg7-2 plants (88% versus 0% survival; Figures 4H and 4I). Taken together, our data indicate that disrupting the ATG1/13 kinase complex only partially dampens autophagy as compared with inhibiting ATG8/12 conjugation. Surprisingly, despite our predictions that the atg11-2 protein would be missing much of the C-terminal end of ATG11 (including the signature ATG11 domain) even if expressed, homozygous atg11-2 plants responded nearly indistinguishably from wild-type plants under all growth conditions tested. Consequently, it is likely that the C-terminal half of ATG11, which is not needed for homodimerization or binding to ATG13 and ATG101, is also not essential for the phenotypic aspects of the ATG1/13 kinase.
ATG11 Associates with Autophagic Bodies
ATG1a colocalizes with ATG8 in vacuolar autophagic bodies, which accumulate in high numbers when plants are starved for nitrogen or fixed carbon and then treated with the inhibitor concanamycin A (ConA), which slows autophagic body turnover (Suttangkakul et al., 2011). To show that ATG11 also associates with autophagic bodies, we generated homozygous atg11-1 transgenic lines that replaced ATG11 with a GFP-ATG11 reporter. As shown in Figures 5A to 5C, plants expressing GFP-ATG11 under the control of the UBQ10 promoter rescued the nitrogen and fixed-carbon sensitivity and early-senescence phenotypes of the atg11-1 mutant, indicating not only that the GFP fusion is functional but also that the atg11-1 phenotypes were caused solely by the lack of ATG11. When UBQ10:GFP-ATG11 atg11-1 root cells exposed to nitrogen-deficient medium were examined by confocal fluorescence microscopy, substantial diffuse GFP fluorescence was observed in the cytoplasm (Figure 5D). However, upon treatment with ConA, numerous ∼1-μm fluorescent vesicles reminiscent of autophagic bodies accumulated in the vacuole. Support that these puncta represent autophagic bodies was generated by the analysis of UBQ10:GFP-ATG11 roots also homozygous for atg7-2; few vacuolar bodies were evident even after nitrogen starvation and ConA treatment (Figure 5D). Confirmation was then provided by colocalization of GFP-ATG11 with the mCherry-ATG8a reporter that decorates autophagic bodies (Yoshimoto et al., 2004; Thompson et al., 2005; Suttangkakul et al., 2011). Most vesicles from root cells exposed to nitrogen starvation and ConA displayed coincident GFP and mCherry fluorescence (Figure 5E; Supplemental Movie 1).
Figure 5.
ATG11 Associates with Autophagic Vesicles.
(A) to (C) Rescue of the atg11-1 phenotype with the UBQ10:GFP-ATG11 transgene.
(A) Growth of wild-type, atg11-1, and UBQ10:GFP-ATG11 atg11-1 plants under nitrogen starvation. Plants were germinated and grown for 1 week on nitrogen-containing (+N) liquid medium and then transferred to either nitrogen-containing or nitrogen-deficient (−N) liquid medium for an additional 2 weeks.
(B) Senescence under an SD photoperiod. Plants were grown on soil at 21°C under an 8-h-light/16-h-dark cycle for 10 weeks.
(C) Sensitivity to fixed-carbon starvation. Seedlings were grown under an LD photoperiod (16 h of light/8 h of dark) without added Suc for 2 weeks and then placed in darkness for 13 d before returning to LD conditions for a 12-d recovery.
(D) Deposition of GFP-ATG11–containing vesicles in the vacuole by a process that requires ATG7. Wild-type and atg7-2 plants expressing UBQ10:GFP-ATG11 were grown for 6 d on nitrogen-containing medium and then transferred to nitrogen-deficient medium without or with 1 μM ConA for an additional 24 h. Root cells were imaged by confocal fluorescence microscopy. Bar = 10 μm.
(E) GFP-ATG11 colocalizes with mCherry-ATG8a in autophagic bodies. Plants stably expressing both reporters were grown for 6 d on nitrogen-containing medium and then exposed for 8 h to nitrogen-deficient medium supplemented with 1 μM ConA before confocal microscopy. Boxes outlined in the left panels were magnified three times in the right panels. Bars = 10 and 5 μm for the left and right panels, respectively.
ATG11 Is Required for Autophagic Body Deposition but Not ATG8/12 Conjugation
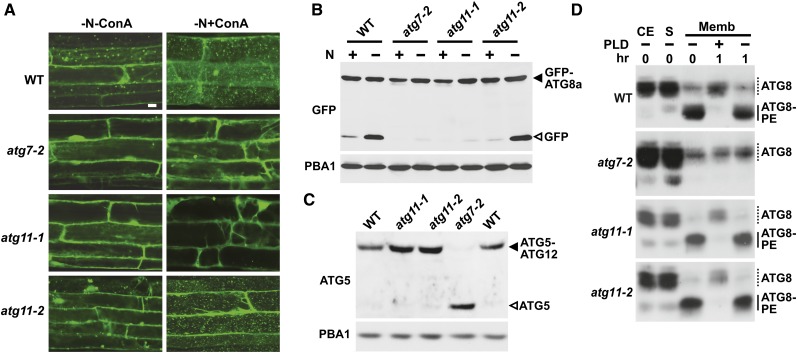
To determine which aspects of autophagy require ATG11, we examined homozygous atg11-1 and atg11-2 plants for their ability to synthesize the requisite ATG8-PE and ATG12-ATG5 conjugates and then generate ATG8-decorated autophagic bodies. Confocal fluorescence microscopy of plants expressing GFP-ATG8a showed that ATG11 is needed for autophagic body deposition. Whereas such vacuolar bodies were easily detected in wild-type root cells starved for nitrogen and pretreated with ConA, they were not evident in similarly treated atg11-1 roots, as seen for atg7-2 roots (Figure 6A). Chung et al. (2010) previously showed that the autophagic transport of ATG8 into the vacuole can also be monitored by the accumulation of free, and presumably stable, GFP during the autophagic turnover of the GFP-ATG8a reporter. We found that this accumulation of released GFP, which was readily detected by immunoblot analysis of crude extracts from wild-type plants subjected to nitrogen starvation (Chung et al., 2010; Suttangkakul et al., 2011), was substantially dampened in atg11-1 plants as well as in atg7-2 plants (Figure 6B).
Figure 6.
Lack of ATG11 Blocks Autophagic Body Deposition in the Vacuole but Not Modification of ATG8 or ATG12.
(A) Deposition of autophagic bodies inside the vacuole. Transgenic seedlings expressing GFP-ATG8a were grown for 6 d on nitrogen-containing solid medium with 1% Suc and then exposed for 24 h to nitrogen-deficient liquid medium without or with the addition of 1 μM ConA before confocal fluorescence microscopic analysis of root cells. Lines tested include the wild type and atg7-2, atg11-1, and atg11-2 mutants each expressing GFP-ATG8a. Bar = 10 μm.
(B) Detection of the free GFP released during the vacuolar degradation of GFP-ATG8a. Seven-day-old seedlings described in (A) were grown on nitrogen-containing liquid medium and then exposed to nitrogen-deficient medium for 16 h. Total protein was subjected to immunoblot analysis with anti-GFP antibodies. Closed and open arrowheads indicate GFP-ATG8a and free GFP, respectively. Immunoblotting with anti-PBA1 antibodies was used to confirm nearly equal loading in (B) and (C).
(C) Immunoblot detection of the ATG12-ATG5 conjugate. Total protein from 7-d-old seedlings grown on MS solid medium with 1% Suc was subjected to immunoblot analysis with anti-ATG5 antibodies. Closed and open arrowheads indicate the ATG12-ATG5 conjugate and free ATG5, respectively.
(D) Immunoblot detection of ATG8-PE adducts. Seedlings were grown on nitrogen-containing liquid medium for 7 d and then exposed to nitrogen-deficient medium for 2 d before extraction. Crude extracts (CE) were separated into the soluble (S) and membrane (Memb) fractions by centrifugation. The membrane fraction was solubilized in Triton X-100 and incubated with or without phospholipase D (PLD) for 1 h. Samples were then subjected to SDS-PAGE in the presence of 6 M urea and immunoblotted with antibodies against ATG8a. Dashed lines indicate free ATG8; solid lines indicate ATG8-PE adducts.
Our phenotypic analyses suggested that the atg11-2 allele does not substantially compromise ATG8-mediated autophagy (Figure 4). This lack of effect was also seen for the autophagy-dependent delivery of GFP-ATG8a to the vacuole. Both the accumulation of GFP-ATG8–decorated autophagic bodies and the accumulation of free GFP from the GFP-ATG8a reporter were indistinguishable from those seen in the wild type (Figures 6A and 6B).
Previous genetic studies with ATG13a/b implied that the ATG1/13 complex is not needed for conjugating ATG8 to PE and ATG12 to ATG5 (Suttangkakul et al., 2011). From the analysis of atg11-1 plants, it appears that ATG11 is also not required. The ATG12-ATG5 conjugate can be observed by immunoblot analysis of crude extracts with anti-ATG5 antibodies as a protein ∼10 kD larger than the free, 40-kD form of ATG5. This adduct, which is not evident in atg7-2 seedlings, was readily detected in wild-type, atg11-1, and atg11-2 seedlings (Figure 6C). To assess the lipidation status of ATG8, we first enriched for total membranes from nitrogen-starved seedlings by centrifugation, solubilized the ATG8-PE adduct with Triton X-100, and then assayed for the adduct following clarification based on its sensitivity to phospholipase D cleavage. The ATG8-PE conjugate was easily detected by immunoblot assays as a faster migrating ATG8 species during SDS-PAGE in the presence of 6 M urea (Chung et al., 2010; Suttangkakul et al., 2011). As shown in Figure 6D, atg11-1 seedlings, like wild-type and atg11-2 seedlings, accumulated the faster migrating, phospholipase D–sensitive ATG8-PE species, which was not evident in atg7-2 seedlings.
Proper Phosphorylation and Autophagic Turnover of ATG1a Require ATG11
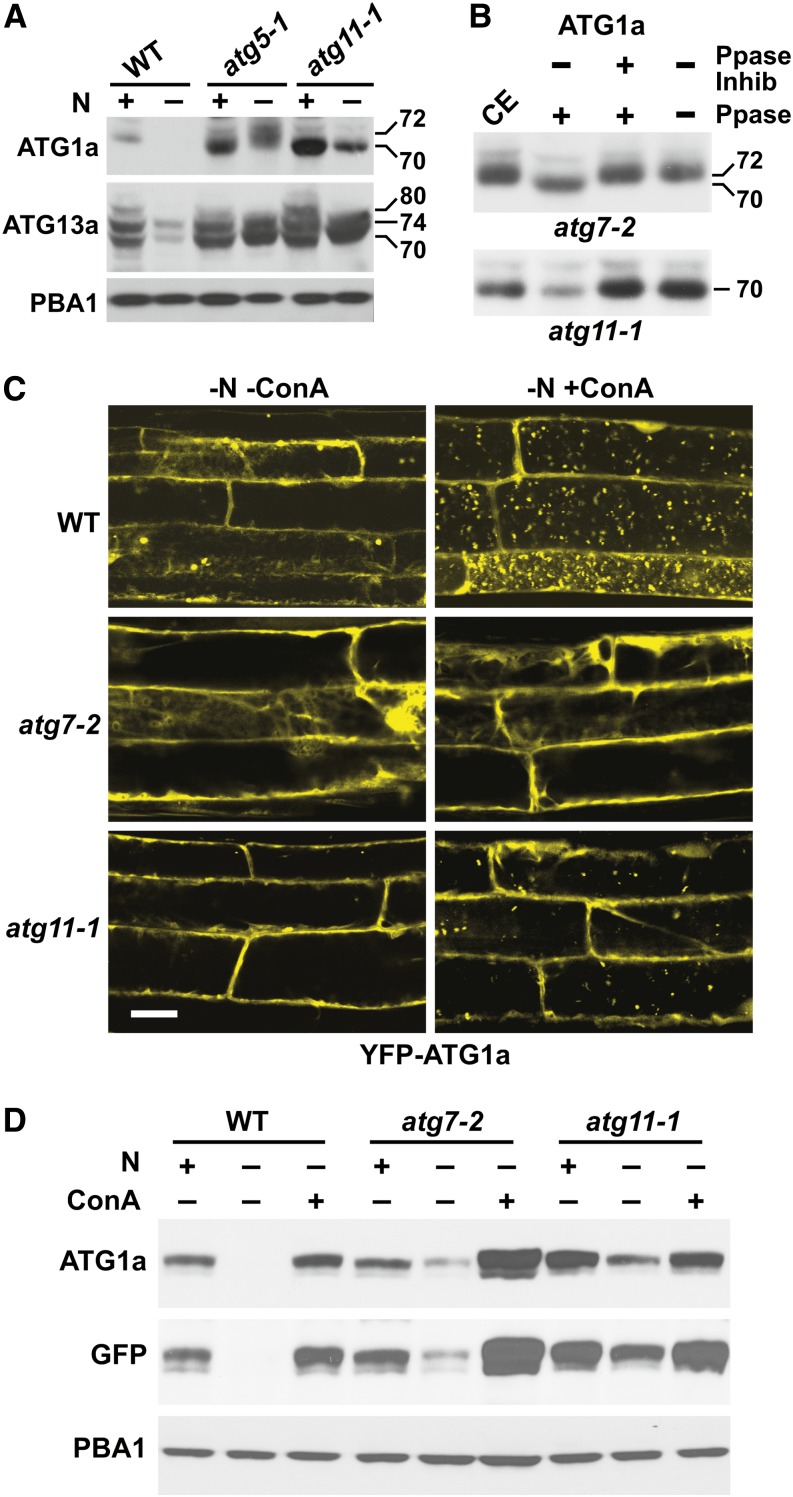
We demonstrated recently that both ATG1a and ATG13a are rapidly turned over during nitrogen or fixed-carbon starvation via an autophagic route that requires the ATG8/12 conjugation machinery (Suttangkakul et al., 2011). We show here that this turnover also involves ATG11. Under well-fertilized growth conditions, atg5-1 and atg11-1 plants both accumulated more ATG1a and ATG13a than the wild type, which is consistent with constitutive turnover of these proteins by autophagy. Whereas the levels of the ATG1a and ATG13a proteins dropped rapidly in wild-type plants exposed to nitrogen-deficient medium, their levels were substantially stabilized in the atg11-1 background, as seen for atg5-1 (Figure 7A).
Figure 7.
ATG11 Is Required for Proper Phosphorylation and Autophagic Turnover of ATG1a.
(A) and (B) ATG11 regulates the phosphorylation of ATG1a and ATG13a/b.
(A) Loss of ATG11 affects the phosphorylation status of ATG1a and ATG13a/b, as observed by changes in SDS-PAGE migration pattern and abundance of isoforms. Wild-type, atg5-1, and atg11-1 seedlings were grown for 7 d under continuous light on nitrogen-containing liquid medium and then transferred for 1 d to nitrogen-containing or nitrogen-deficient (+/−) medium. Crude extracts were subjected to immunoblot analysis with anti-ATG1a or anti-ATG13a antibodies. Immunoblot analysis with anti-PBA1 antibodies was used to confirm equal protein loading.
(B) ATG1a is dephosphorylated in the atg11-1 mutant. Seven-day-old atg7-2 and atg11-1 plants were incubated for 3 d in MS liquid medium without nitrogen and Suc. Crude extracts (CE) were treated with λ-phosphatase (Ppase) in the presence or absence of phosphatase inhibitor for 1 h and then subjected to immunoblot analysis with anti-ATG1a antibodies. Positions of the 72-kD phosphorylated and 70-kD unphosphorylated forms of ATG1a are indicated.
(C) Loss of ATG11 reduces the accumulation of ATG1a in autophagic bodies during nitrogen starvation. Wild-type, atg7-2, and atg11-1 seedlings expressing YFP-ATG1a were grown for 6 d on nitrogen-containing solid medium containing 1% Suc and then exposed to nitrogen-deficient liquid medium without or with 1 μM ConA for 1 d. Root cells were imaged by confocal fluorescence microscopy of root cells. Bar = 10 μm.
(D) Loss of ATG11 reduces the nutrient stress–induced turnover of ATG1a. Wild-type, atg7-2, and atg11-1 plants expressing YFP-ATG1a were grown for 6 d on nitrogen-deficient liquid medium and then incubated in nitrogen-containing or nitrogen-deficient medium without or with 1 μM ConA for 16 h. YFP-ATG1a protein levels were revealed by immunoblots with anti-ATG1a and anti-GFP antibodies. Immunoblot analysis with anti-PBA1 antibodies was used to confirm equal protein loading.
Suttangkakul et al. (2011) previously showed that nitrogen starvation decreases the electrophoretic mobility of ATG1a and reduces the SDS-PAGE banding complexity of ATG13 by loss of the 80-kD species (Figure 7A). Both changes have been attributed to altered phosphorylation, with starvation inducing the phosphorylation of ATG1a and the dephosphorylation of ATG13. Surprisingly, the 70- to 72-kD mobility shift for ATG1a, which represents phosphate addition, requires ATG11 but not the ATG8/12 conjugation machinery. Whereas the size shift for ATG1a was still seen in nitrogen-starved atg7-2 seedlings, as evidenced by the increased SDS-PAGE migration of ATG1a upon treatment of the sample with λ-phosphatase, it was not evident in starved atg11-1 seedlings treated similarly (Figures 7A and 7B). In contrast, the phosphorylation status of ATG13a appeared unaffected by the loss of ATG11, with the 74- and 70-kD forms becoming more prevalent in the absence of nitrogen in the atg11-1 seedlings, like wild-type and atg5-1 seedlings (Figure 7A).
The turnover of ATG1a coincides with its appearance in autophagic bodies (Suttangkakul et al., 2011). Confocal microscopy of the YFP-ATG1a reporter revealed that this transport also requires ATG11. In root cells from wild-type seedlings expressing YFP-ATG1a, numerous fluorescent vacuolar vesicles accumulated upon nitrogen starvation and ConA pretreatment (Figure 7C). Conversely, these puncta were not seen in vacuoles from YFP-ATG1a atg7-2 roots, and only a few accumulated in vacuoles from YFP-ATG1a atg11-1 roots. The reduced autophagic turnover of ATG1a in the atg11-1 background was also revealed by immunoblot analysis of the YFP-ATG1a reporter. More reporter was observed in nitrogen-starved atg7-2 or atg11-1 seedlings as compared with similarly starved wild-type seedlings, using both anti-ATG1a and anti-GFP antibodies for detection (Figure 7D). However, a modest drop in ATG1a was still evident (with or without nitrogen) in the atg mutants, suggesting that other mechanisms also control its levels during nutrient stress.
ATG1 and ATG11 Bind to ATG8 on Autophagic Vesicles
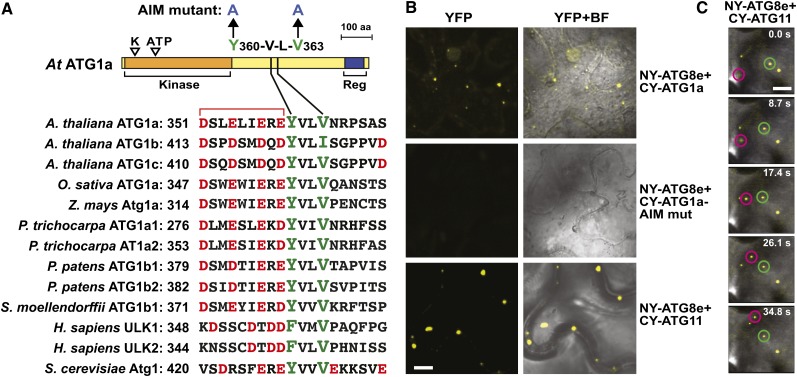
The interaction of ATG11 with ATG1a and ATG13a either directly or indirectly, and the simultaneous entry of ATG1a and ATG11 into vacuoles via autophagic bodies during nitrogen starvation (Figures 2C, 5D, and 7C; Suttangkakul et al., 2011), implied that the entire ATG1/13 complex (including associated factors) binds to autophagic vesicles and is cleared by the vacuole together, thus providing feedback control over the entire particle. Such coordinated turnover has also been reported for yeast ATG1 and mammalian ULK1, with their link to autophagic vesicles being mediated by an AIM in ATG1/ULK1 interacting with lipidated ATG8 lining the engulfing phagophore (Kraft et al., 2012; Nakatogawa et al., 2012). To explore whether a similar route exists in Arabidopsis, we examined by BiFC whether ATG1a binds ATG8e in planta. As shown in Figure 8B, coexpression of CY-ATG1a with NY-ATG8e in N. benthamiana leaf epidermal cells readily reconstituted YFP fluorescence indicative of binding, with the punctate pattern implicating autophagic structures. Alignments of plant, yeast, and metazoan ATG1 polypeptides identified a highly conserved YVLV sequence preceded by an acidic patch, which is related to the canonical AIM and thus might drive the association of ATG1a with ATG8 (Figure 8A). This motif was confirmed as important by BiFC of an Arabidopsis ATG1a mutant bearing a double Ala substitution (Y360A/V363A) of the AIM sequence; ATG1a containing this AVLA sequence failed to bind ATG8e (Figure 8B).
Figure 8.
ATG1a and ATG11 Both Interact with ATG8e.
(A) Schematic representation of Arabidopsis ATG1a and the sequence alignment of its putative AIM with those in other ATG1 family members from Arabidopsis and with orthologs from other eukaryotes. K and ATP indicate the catalytic site Lys and the ATP binding site in the kinase domain, respectively. Reg indicates the C-terminal early autophagy targeting/tethering regulatory domain. Residues targeted by mutagenesis are marked with arrows. The conserved aromatic/hydrophobic residues and the upstream and downstream acidic residues in the AIM are highlighted in green and red, respectively. The numbers on the right indicate the position of the first residue within the context of each full-length protein. aa, amino acids.
(B) Interaction of Arabidopsis ATG8e with ATG1a and ATG11 in planta by BiFC. N. benthamiana leaf epidermal cells were coinfiltrated with plasmids expressing the N-terminal fragment of YFP (NY) fused to ATG8e and the C-terminal YFP (CY) fragment fused to ATG11, ATG1a, or an ATG1a mutant affecting the AIM motif (Y360-A/L363-A). Shown are confocal fluorescence microscopic images of coinfiltrated cells along with a bright-field (BF) image of the cells. Bar = 5 μm
(C) Time-lapse images showing that ATG11 interacts with ATG8e in fast-moving particles. The NY-ATG8e and CY-ATG11 transgenes were coexpressed as in (B) and visualized over time by confocal fluorescence microscopy. The time duration in seconds is shown for each panel. Bar = 5 μm.
When ATG13a and ATG101 were similarly tested by BiFC for binding to ATG8e, we found that neither interacted. However, when CY-ATG11 was coexpressed with NY-ATG8e, reconstituted YFP fluorescence was clearly evident in epidermal cells as fast-moving fluorescent puncta (Figures 8B and 8C). BiFC mapping of the ATG8 interaction site(s), using the same set of ATG11 truncations described above (Figure 2E), identified potential binding regions within the ATG11 domain and the homodimerization region (Supplemental Figure 7C). Two possible AIM sequences were detected in these regions from sequence alignments of ATG11 orthologs (YFIV [1130 to 1133] and FDDI [567 to 570], respectively) that could provide these docking sites (Supplemental Figure 1). Interestingly, full-length ATG11 and ATG8e colocalized in small puncta akin to those seen when full-length ATG11 associated with itself or with ATG13a and ATG101 (Supplemental Figure 7C), thus providing further support that these spots represent assembly sites for the ATG1/13 complex on autophagic structures. However, when CY fusions constructed with just the ATG11 domain or the ATG11 dimerization site were paired with NY-ATG8e, we saw dispersed cytoplasmic BiFC signal. This diffuse distribution might reflect a need for ATG11, and possibly the entire ATG1/13 complex, to properly localize lipidated ATG8. More broadly, ATG11 appears to provide a second contact site between the ATG1/13 complex and ATG8-decorated autophagic vesicles.
ATG11 and ATG7 Are Required for Senescence-Induced Mitophagy
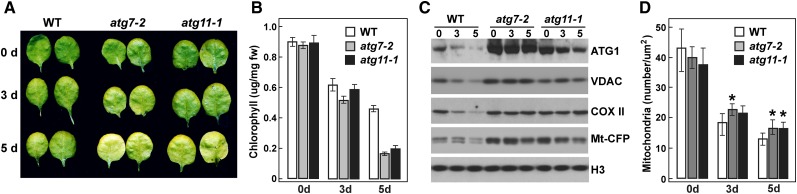
Previous studies in nonplant systems implicated ATG11 as an adaptor for selective autophagic routes such as mitophagy (Reggiori and Klionsky, 2013). As examples, yeast ATG11 interacts with the mitophagy receptor ATG32 to remove aberrant or superfluous mitochondria when nutrients become limiting (Kanki et al., 2009; Okamoto et al., 2009), whereas mice missing FIP200 hyperaccumulate mitochondria damaged by depolarization (Liang et al., 2010). A similar clearance of plant mitochondria was possible based on the observed drop in mitochondrion numbers in Arabidopsis under conditions that stimulate autophagy, including fixed-carbon starvation and senescence (Journet et al., 1986; Keech et al., 2007).
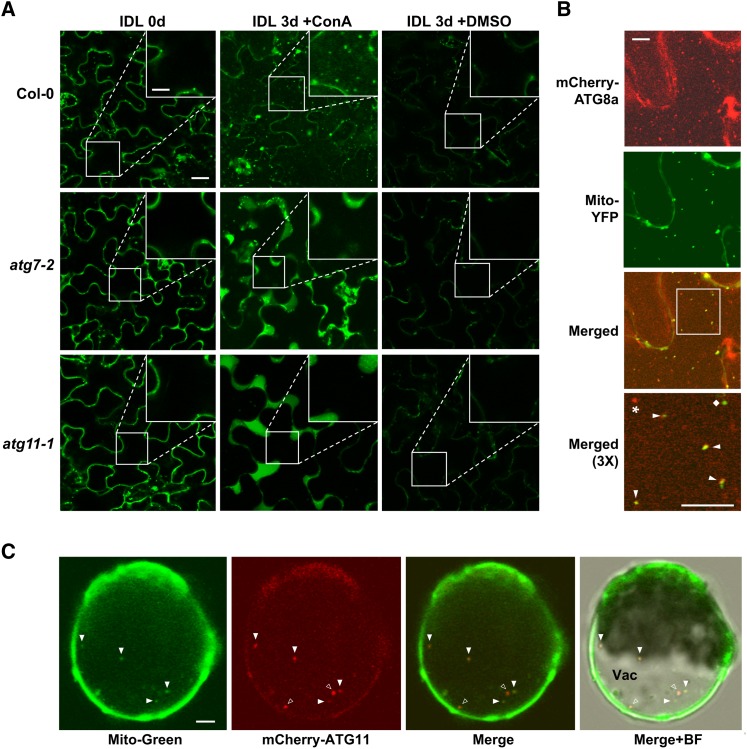
To test for mitophagy in Arabidopsis, we stimulated mitochondrial turnover by the previously described individually darkened leaf (IDL) protocol (Weaver and Amasino, 2001; Keech et al., 2007). As expected (Doelling et al., 2002; Wada et al., 2009), such treatments induced senescence in the darkened leaves, with the loss of chlorophyll noticeably faster in various atg mutant backgrounds, including atg7-2 and atg11-1, than in the wild type (Figures 9A and 9B). Autophagic activity was concomitantly upregulated in IDLs, which could be seen by the rapid loss of ATG1a in wild-type but not atg7-2 and atg11-1 leaves (Figure 9C). IDL treatment also induced the loss of mitochondria based on analysis of the fluorescent mitochondrial reporter Mito-CFP, which was generated by fusing the 29-residue mitochondrial targeting sequence from the cytochrome c oxidase (COX) IV subunit to the N terminus of CFP (Nelson et al., 2007). When cytoplasmic Mito-CFP foci were counted in the confocal microscopy images of epidermal cells from atg11-1 and atg7-2 leaves, a significant stabilization of mitochondrion numbers after IDL treatment was evident as compared with the wild type (Figure 9D). Importantly, when the Mito-CFP signal was tracked in wild-type IDLs pretreated with ConA, fluorescent vacuolar puncta indicative of mitophagy became obvious (Figure 10A), which was not seen in atg7-2 or atg11-1 leaves. This deposition was also observed in plants expressing two other mitochondrial reporters (Mito-YFP and CIB22-GFP; Nelson et al., 2007; Han et al., 2010) but not in plants expressing free GFP (Supplemental Figure 11 and Supplemental Movies 3 and 4).
Figure 9.
Dark-Induced Senescence Induces Mitochondrion Turnover via an Autophagic Process.
The third and fourth rosette leaves of 4-week-old plants were individually darkened for 3 to 5 d prior to examination.
(A) Representative IDLs from wild-type, atg7-2, and atg11-1 plants showing accelerated chlorosis.
(B) Chlorophyll contents of IDLs shown in (A). Each bar represents the mean ± sd from three independent experiments measuring 12 leaves each. fw, fresh weight.
(C) Loss of mitochondrial markers during IDL senescence. IDLs were harvested at the indicated times of dark exposure from wild-type, atg7-2, and atg11-1 plants expressing Mito-CFP, homogenized, and the crude extracts were subjected to immunoblot analyses with antibodies against ATG1a, VDAC, COX II, and GFP. Immunoblot analysis with anti-histone H3 antibodies was used to confirm nearly equal protein loading.
(D) Quantification of mitochondrion numbers during IDL senescence. Mitochondria within representative 2500-μm2 regions of leaf cells from (C) were counted using the Particle Analysis function of ImageJ. Each bar represents the mean ± sd from two independent experiments counting at least eight regions each. Asterisks indicate statistically significant differences as determined by Student’s t test (P < 0.05).
Figure 10.
Association of Mitochondria with Autophagic Bodies during Leaf Senescence.
(A) Accumulation of Mito-CFP in vacuolar puncta during dark-induced leaf senescence (IDL) by a process that requires the ATG system. Four-week-old wild-type, atg7-2, and atg11-1 plants expressing Mito-CFP were subjected to IDL for 3 d followed by a 20-h incubation with 1 μM ConA or DMSO before confocal fluorescence microscopy of epidermal cells from the third and fourth rosette leaves. Bars = 10 μm and 3.4 μm in the inset. Insets show 3× magnifications of the vacuole.
(B) Colocalization of Mito-YFP with the autophagic membrane marker mCherry-ATG8a in autophagic bodies. Wild-type plants expressing both reporters were subjected to IDL senescence as in (A). The vacuolar region of leaf epidermal cells was imaged by confocal fluorescence microscopy. Bar = 10 μm. A 3× magnification of the merged signals (outlined by the white box) is included to confirm colocalization of the two proteins in autophagic bodies (arrowheads). A free Mito-YFP–labeled mitochondrion and an autophagic body not containing mitochondria are indicated with the diamond and the star, respectively.
(C) Colocalization of the mitochondrial stain MitoTracker Green FM with mCherry-ATG11. Arabidopsis leaf protoplasts stably expressing mCherry-ATG11 were treated for 30 min with MitoTracker Green FM, washed twice, and then incubated for 24 h with ConA in the absence of Suc before confocal fluorescence microscopy. Closed arrowheads indicate vacuolar puncta containing both fluorescent markers; open arrowheads are puncta preferentially containing mCherry-ATG11. BF, bright field; Vac, vacuole. Bar = 5 μm.
Subsequent immunoblot assays for several mitochondrial proteins showed that the IDL treatment induced their rapid loss via a route requiring autophagy. Examples include the COX II subunit and the voltage-dependent anion channel (VDAC) on the outer membrane, in addition to the Mito-CFP reporter; the levels of all three proteins drop substantially in wild-type leaves subjected to IDL but not in similarly treated atg11-1 and atg7-2 leaves (Figure 9C). To directly demonstrate mitophagy, we used fluorescence confocal microscopy to examine a transgenic line coexpressing Mito-YFP and mCherry-ATG8a. Analyses of epidermal cells from IDL leaves exposed to ConA detected numerous vacuolar puncta containing both reporters (Figure 10B). For further confirmation, we examined the association of ATG11 with mitochondria by using MitoTracker Green FM to specifically label mitochondrial membranes in vivo (Colcombet et al., 2013). When protoplasts isolated from a transgenic line expressing mCherry-ATG11 were also stained for MitoTracker Green FM, punctate structures containing both fluorescent signals were readily detected in the vacuole of protoplasts after Suc starvation and ConA treatment (Figure 10C). Collectively, our results demonstrate that mitochondrion-resident proteins and mitochondrial vesicles are sequestered and delivered to the vacuole for breakdown during senescence via a mitophagic pathway requiring ATG11 and the ATG8/12 conjugation pathway.
DISCUSSION
Given its central position within the ATG system, its potential interaction with a host of autophagic components, and the phenotypic response of atg1Δ mutants in yeast and metazoans (Reggiori and Klionsky, 2013), the ATG1/13 kinase complex is proposed to be a central regulator that connects developmental cues and nutritional status to PAS/autophagic vesicle assembly and flux. Our cell biological and genetic studies here and elsewhere (Suttangkakul et al., 2011) on the Arabidopsis complex confirm this importance in plants, with the demonstrations that it promotes autophagic body delivery to the vacuole and is needed for survival under nutrient limitation. Besides ATG1 and ATG13, ATG11 and ATG101 are also components of the Arabidopsis complex, as shown here by Y2H interactions and by BiFC in planta. Other accessory proteins are possible based on studies outside of the plant kingdom, but they have thus far eluded our detection by sequence homology searches. ATG11 associates with ATG8-decorated autophagic vesicles and is eventually deposited inside the vacuole along with ATG1 and ATG13. It also assists in the starvation-induced turnover of ATG1a and ATG13a/b, presumably by helping tether the pair to autophagic structures via a connection that might involve the binding of ATG11 to ATG8-PE. Genetic elimination of ATG11, like ATG13a/b, accelerates leaf senescence, indicating that this accessory protein is also needed during developmentally induced nutrient recycling.
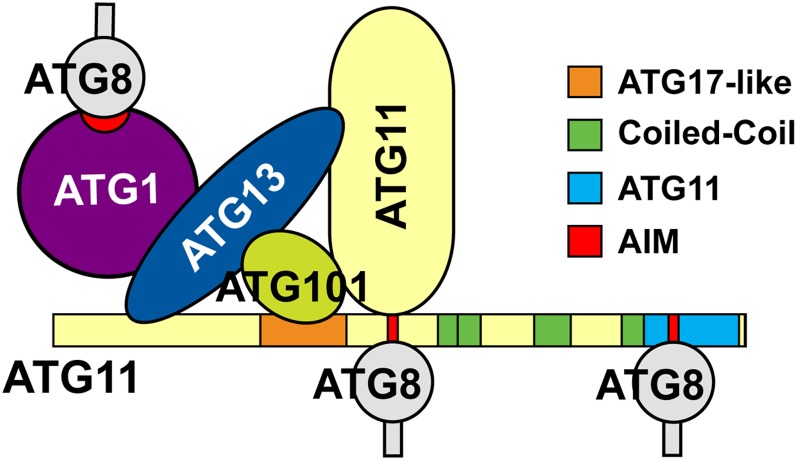
A striking feature of ATG11-type proteins is their ability to interact with a diverse assortment of other ATG components, implying that they mainly act as scaffolds to help assemble and integrate the ATG system (Yorimitsu and Klionsky, 2005). Our Arabidopsis studies demonstrate that the plant version directly binds to ATG13, ATG101, and ATG8 and indirectly to ATG1 through ATG13 (Figure 11). A similar use of ATG13 as a bridge was reported for connecting yeast ATG1 to ATG17 (Kabeya et al., 2005). Based on the organization of ATG11/FIP200-related proteins (especially the extended coiled-coil region), the unique elongated crescent structure of its dimeric relative ATG17, and their early association with the PAS (Ragusa et al., 2012), Arabidopsis ATG11 likely helps in the hierarchical assembly of other ATG proteins into the PAS, particularly the ATG1/13 kinase. Binding of ATG11 to ATG8 (presumably through an AIM sequence) might be particularly relevant as a way to tether components like the ATG1/13 kinase to developing phagophores as they become decorated with ATG8-PE. In this regard, it is interesting that the autophagic degradation of ATG1 and ATG13 is slower in the absence of ATG11, suggesting that one important role for ATG11 is to regulate turnover of the ATG1/13 kinase complex during nutrient starvation through its simultaneous interactions with ATG8-PE and ATG13. However, it should be mentioned that ATG1 also has an AIM sequence capable of binding ATG8, thus providing a second way to tether the ATG1/13 kinase to autophagic membranes.
Figure 11.
Schematic of the Interactions between ATG1, ATG8, ATG13, ATG11, and ATG101 as Determined by Y2H and/or BiFC Assays.
The AIM sequences potentially involved in ATG8 binding are shown in red.
Of functional interest is the ability of ATG11 to dimerize through a site just distal to the ATG17-like domain (Figure 2). Preliminary mapping of yeast ATG11 revealed that this region is also critical for its association with other ATG proteins, including ATG1, ATG17, and ATG20 (Yorimitsu and Klionsky, 2005), whereas our study revealed that it (or its surroundings) bind ATG13a, ATG101, and ATG8e (Figures 2 and 3; Supplemental Figure 7). Conversely, the function(s) of the ATG11 domain has not yet been defined. In yeast, this domain binds cargo receptors of the cytoplasm-to-vacuole targeting pathway and mitophagy, ATG19 and ATG32, respectively (Yorimitsu and Klionsky, 2005; Aoki et al., 2011). Moreover, a Caenorhabditis elegans ATG11 variant without the ATG11 domain failed to restore the defective degradation of sequestosome-1–containing aggregates, implying a direct role in aggrephagy (Lin et al., 2013). Collectively, these data suggest that the ATG11 domain participates in selective autophagy. Such a selective role might explain why the atg11-2 allele is phenotypically indistinguishable from the wild type in our various assays designed to emphasize bulk autophagy. Consequently, assays more relevant to certain types of selective autophagy might be necessary to uncover phenotypes specific for the atg11-2 allele.
A number of studies have implicated the ATG1/13 kinase in the integration of multiple signals to regulate autophagic flux (Mizushima, 2010; Wong et al., 2013). Possible upstream controls include affecting the interactions between ATG1 with ATG13, the autophosphorylation of ATG1, and the phosphotransferase activity of ATG1, all of which could be driven by the phosphorylation of ATG13 via TOR and other nutrient-sensing kinases. Ultimately, ATG1 is thought to pass on the phosphorylation signal to downstream partners within the ATG system, the nature of which are currently unknown. Our analyses of the atg11-1 mutant revealed that the ATG11 protein affects this cascade in Arabidopsis, with its absence blocking some or all ATG1a phosphorylation, as detected by SDS-PAGE mobility shifts of the protein. While several mechanisms are possible, one likely scenario is that ATG11 is required to scaffold both ATG1a and ATG13 within the PAS and/or autophagic vesicles where these phosphate additions/subtractions are mediated. In addition to promoting signaling, these changes in phosphorylation state might also be required for the starvation-induced turnover of both ATG1 and ATG13.
It should be noted that the functions of ATG11/FIP200 appear to differ among eukaryotes. As examples, although ATG1 phosphorylation requires ATG11/FIP200 in both Arabidopsis and mammals, its accelerated turnover during starvation appears to require ATG11/FIP200 in Arabidopsis only (Hara et al., 2008; this article). With respect to downstream autophagic events, the synthesis of ATG12-ATG5 and ATG8-PE conjugates is not affected in Arabidopsis or yeast atg11 mutants (Shintani and Klionsky, 2004; this article), whereas the lipidation of ATG8 is greatly reduced in mammalian FIP200-deficient cells (Hara et al., 2008). Also, while loss of yeast Atg11 does not affect nonselective bulk autophagy involving autophagic bodies (Reggiori and Klionsky, 2013), the vacuolar deposition of autophagic bodies is substantially blocked in the Arabidopsis atg11-1 mutant (this article), and autophagosome formation is mostly abolished in the absence of mammalian FIP200 (Hara et al., 2008). Consequently, in line with the dynamic evolution of ATG11/17 protein sequences among eukaryotes, there also appears to be a substantial divergence of their roles within the autophagy system.
Whereas deletion of ATG components within the ATG8/12 conjugation cascade appears to completely inhibit the accumulation of autophagic bodies in Arabidopsis vacuoles (Yoshimoto et al., 2004; Phillips et al., 2008; Chung et al., 2010), the atg11-1 and atg13a atg13b mutants still accumulate a modest number upon ConA treatment (Figure 7C; Suttangkakul et al., 2011), implying that some autophagic transport occurs without these subunits. This residual activity is likely responsible for the slightly higher tolerance of atg11-1 and atg13a atg13b plants to fixed-carbon starvation (Figure 4E). It could reflect a basal activity for ATG1 in the absence of ATG13 and ATG11; at present, testing this scenario will likely require the phenotypic analysis of quadruple mutants eliminating all four Arabidopsis ATG1 isoforms (Suttangkakul et al., 2011). Conversely, the residual activity could represent an ATG1/13 kinase–independent route for assembling autophagic vesicles. In support of this, glucose deprivation has been reported to stimulate autophagy in mice via a process that does not require ULK1 (ATG1; Wong et al., 2013). A plant-specific alternative route for vesicle delivery independent of ATG1/13 is also possible (Reyes et al., 2011). It has also been proposed that the C. elegans and mammalian forms of ATG11 (Unc51 and FIP200, respectively) function outside of autophagy (Mizushima, 2010; Wong et al., 2013). However, given the nearly coincident phenotypes of the Arabidopsis atg11-1 and atg13a atg13b mutants and mutants affecting other aspects of ATG8-mediated autophagy (e.g., atg7-2 and atg5-1), a nonautophagic activity for the plant ATG1/13 kinase complex either does not exist or is phenotypically irrelevant under the conditions tested here.
Our phylogenetic analysis of ATG11- and ATG17-related proteins implies a complex evolution of these ATG components among eukaryotes. Ichthyosporea, amoebozoa, fungi, and some stramenopiles encode separate proteins with signature ATG11 and ATG17 domains. In contrast, metazoans and plants appear to lack canonical ATG17-type proteins but instead encode a hybrid ATG11/17 protein with a similar architecture to those found in fungi. These hybrids contain the signature coiled-coil and ATG11 domains and are preceded by a short ATG17-like domain. Consequently, these metazoan/plant/fungal polypeptides likely share some of the functional properties of both ATG11/FIP200 and ATG17 and possibly represent the ancestral scaffolds for both bulk and selective autophagy. The separate ATG17 protein could have arisen via gene duplication of ATG11/FIP200 followed by loss of the C-terminal region containing the coiled-coil and ATG11 domains and elaboration of the ATG17 domain. This shorter ATG17 version then subfunctionalized to direct nonselective autophagy, as appears to be the case in yeast (Reggiori and Klionsky, 2013). Notably, the ATG17-interacting proteins ATG29 and ATG31 have been detected thus far only in yeast, Ashbya gossypii, and Kluyveromyces lactis, whereas most other fungi appear to lack either or both accessory proteins, suggesting that the ATG17/29/31 complex is a relatively new addition to the ATG system.
All our BiFC interaction studies are consistent with localization of the ATG1/13 kinase complex within the PAS. If true, dozens of PAS foci that are capable of nucleating phagophores/autophagosomes would be predicted in Arabidopsis, which is in agreement with the numbers reported for mammalian cells but is much higher than in yeast, where a single PAS adjacent to the vacuole is typically seen (Itakura et al., 2012). Tethering of the ATG1/13 complex to the PAS might be achieved by direct binding of either or both ATG1 and ATG11 to ATG8-PE lining the emerging phagophore via ATG8 interaction sites within each polypeptide. Notably, interactions between ATG11 and ATG8 have also been detected in C. elegans and mammalian cells (Behrends et al., 2010; Alemu et al., 2012; Lin et al., 2013), indicating that this association is a conserved feature within the ATG11 family. Studies with yeast and mammalian cells in addition to our study have suggested that these ATG1–ATG8/ATG11–ATG8 interactions serve two purposes: one is to trigger the vacuolar degradation of ATG1 (and possibly the whole ATG1/13 complex), thus providing a feedback control on autophagy, and the other is to target ATG1 to phagophores/autophagosomes to promote vesicle maturation/transport (Suttangkakul et al., 2011; Kraft et al., 2012; Nakatogawa et al., 2012).
In agreement with previous studies with yeast and mammalian counterparts (Kraft et al., 2012; Nakatogawa et al., 2012), we mapped the essential ATG8 interaction site in Arabidopsis ATG1a to a canonical AIM sequence, Tyr-Val-Leu-Val preceded by acidic residues, which sits between the kinase and regulatory domains. For ATG11, the ATG8 interaction sequence(s) appears more complex and might involve several sites somewhere downstream of the ATG17 domain as well as within the ATG11 domain. Two potential AIM sequences conserved within the ATG11 family were detected in these regions that could provide the docking sites. Surprisingly, whereas BiFC assays between ATG8e and truncations of ATG11 that include the coiled-coil region display a punctate distribution suggestive of PAS/phagophores/autophagosomes, assays using the regions downstream of the ATG17 domain or the full ATG11 domain by itself revealed a diffuse cytoplasmic distribution for the ATG8e/ATG11 pair. This anomalous distribution was also seen for ATG11 dimers when the ATG17 domain was compromised, suggesting that the N terminus of ATG11, possibly the ATG17 domain, is essential for its association with PAS but not for its dimerization. Whereas ATG101 interacted with ATG11 by BiFC in punctate structures, the interaction of ATG101 with ATG13 also displayed this diffuse cytoplasmic distribution, suggesting that ATG11 links ATG101 to autophagic structures as well.
Plant mitochondria are highly dynamic organelles, and their quantity, shape, and size are tightly controlled by a set of sophisticated mechanisms involving mitochondrial fusion/fission and turnover. While the mechanisms underpinning fusion/fission are relatively well understood (Scott and Logan, 2010), little is known about how plant cells remove mitochondria, especially when damaged or in excess. Here, we show that autophagy is involved by following the fate of mitochondrial reporters in an IDL system. Particularly conclusive was the colocalization of fluorescent mitochondrial markers and the mitochondrial membrane stain MitoTracker Green FM within ATG8-labeled vesicles inside the vacuole, via a process dependent on the autophagic machinery. The markers detected inside the autophagic bodies could represent whole mitochondria or fragments generated by fission. As in yeast and mammalian cells (Kanki et al., 2009; Okamoto et al., 2009; Itakura et al., 2012), ATG11 is involved, given the substantial stabilization of mitochondrial reporters upon IDL treatment and their failure to enter the vacuole in the atg11-1 as well as atg7-2 backgrounds.
At present, we do not understand how ATG11 participates in plant mitophagy. The yeast mitophagy pathway involves a direct interaction between ATG11 and the integral outer membrane protein ATG32. Yeast ATG11 also recruits by directly binding Dnm1, a component of the mitochondrial fission machinery, possibly as a way to promote the budding of mitochondrial fragments sufficiently small enough to fit into the engulfing phagophore (Mao et al., 2013). Although the mitochondrial fission machinery appears to be highly conserved among eukaryotes, our DELTA-BLAST searches of the Arabidopsis Col-0 proteome failed to detect close relatives of ATG32.
In mammals, FIP200 (ATG11) could participate in removing depolarized or damaged mitochondria by two routes. One involves the mitochondrial protein Nix that binds ATG8 via an AIM sequence (Novak et al., 2010). The other first requires ubiquitylation of the mitochondrial surface by the recruitment of the ubiquitin ligase Parkin in a process dependent on the mitochondria-associated kinase PTEN-INDUCED KINASE1 (PINK1; Itakura et al., 2012). Presumably, the ubiquitylated surface proteins are then recognized by autophagic receptors that bind both ubiquitin and ATG8. Whereas Arabidopsis homologs of Nix, Parkin, or PINK1 are not apparent, a functional ortholog of the ubiquitin binding autophagic receptor NBR1 has been described (Svenning et al., 2011), which might help recognize ubiquitylated mitochondria. Clearly, the identification of mitochondrial factors that bind ATG11 might help identify components critical to plant mitophagy.
Taken together, our studies show that ATG11 is an important modulator of the ATG1/13 kinase, possibly through its ability to scaffold the complex to the PAS and/or autophagic vesicles, to direct its autophagic turnover, and to regulate the phosphorylation state of ATG1. Its link to mitophagy adds another plant organelle/particle to the list selectively cleared from the cytoplasm via an autophagic route (Li and Vierstra, 2012). We now require an understanding of how the plant autophagic machinery recognizes mitochondria to fully appreciate the mechanisms by which plants remove damaged mitochondria and regulate their abundance during development and nutrient stress.
METHODS
Identification and Phylogenetic Analysis of ATG11/17 Genes
All nucleic acid sequences and plant materials were derived from the Arabidopsis thaliana Col-0 ecotype. Potential Arabidopsis ATG101, ATG11/FIP200, and ATG17 orthologs were identified by using full-length human ATG101, the ATG11 domain protein sequences from yeast (Saccharomyces cerevisiae) ATG11 and human FIP200, and the full-length yeast ATG17 protein sequence as queries to scan the Col-0 ecotype protein database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) by DELTA-BLAST. The full coding sequences of Arabidopsis ATG11 and ATG101 were obtained by RT-PCR using total leaf RNA from young seedlings as the template in combination with gene-specific primers. See Supplemental Table 3 for all primers used in this study. The ATG11 and ATG101 cDNAs were subcloned into the pCR8/GW/TOPO vector (Invitrogen) and verified as correct by DNA sequence analysis. The resulting plasmids were used as the entry vectors to build the Gateway constructions for BiFC and Y2H assays and to create the various ATG11-based transgenes. ATG11 truncations were generated by PCR with specific primers using the cDNA as template. The ATG1a AIM mutant (V420/423L) was created by the QuickChange protocol using the ATG1a cDNA as template.
For phylogenetic analyses, the ATG11/FIP200 and ATG17 domains were predicted by Pfam (http://pfam.sanger.ac.uk/) or UniProt (www.uniprot.org/; Supplemental Tables 4 and 5). The ATG17-like domain in ATG11 proteins was also predicted from plant, stramenopile, and Ascomycota representatives by Pfam, whereas the putative ATG17 domain in ATG11 proteins from Basidiomycota and in FIP200 proteins from metazoan species was identified by alignment with the ATG17-like domain from Arabidopsis and Aspergillus oryzae ATG11. Amino acid sequence alignments were generated by ClustalX (Chenna et al., 2003) under the default settings. Maximum likelihood trees were created by MEGA 5.1 (www.megasoftware.net), using the Jones/Taylor/Thornton model of protein evolution, partial deletion of gaps, uniform rates among sites, and 500 bootstrap replicates.
Y2H Assays
The Y2H assays were performed as described previously by Suttangkakul et al. (2011). Briefly, full-length cDNAs for Arabidopsis ATG1a, ATG13a, ATG11, and ATG101 and truncations of ATG11 were recombined into either the GAL4 activation domain plasmid (pDEST22) or the GAL4 binding domain plasmid (pDEST32) using the LR Clonase reaction (Invitrogen). Both plasmids were then cotransformed into yeast strain MaV203. Protein–protein interactions were determined by measuring the growth of transformants after 2 d on medium lacking His, Leu, and Trp and containing 25 mM 3-amino-1,2,4-triazole.
Plant Materials
The atg11-1 (SAIL_1166_G10) and atg11-2 (SAIL_1257_G01) T-DNA insertion mutants in the Col-0 background were obtained from the ABRC at Ohio State University. The positions of the T-DNA insertions were confirmed by genomic PCR using a 5′ gene-specific primer and a T-DNA left border–specific primer; the exact insertion sites were identified by sequencing the PCR products. Each mutant was backcrossed three times to wild-type Col-0 and then selfed to obtain homozygous lines before various phenotypic analyses. The autophagy mutants atg5-1 and atg7-2 (Col-0 background) were as described previously (Thompson et al., 2005; Chung et al., 2010). Transgenic lines stably expressing the autophagic markers GFP-ATG8a were from Thompson et al. (2005), and those expressing mCherry-ATG8a and YFP-ATG1a were from Suttangkakul et al. (2011). Germplasm expressing the mitochondrial markers Mito-CFP (CS16262) and Mito-YFP (CS16264; Nelson et al., 2007) was obtained from the ABRC. The CIB22-GFP transgenic line was a gift from Li-Jia Qu (Han et al., 2010).
RT-PCR Analyses of atg11 Mutants
Total RNA was extracted from 2-week-old seedlings using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s protocol. One microgram of total RNA was treated with DNase RQ1 (Promega) and then added to 20-μL reactions containing SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primers to synthesize the RNA/DNA hybrid. cDNAs were generated by 35 cycles of PCR using Taq polymerase and the indicated primer pairs (Figure 2B). RT-PCR of the UBC9 transcript was used as an internal control. Quantitative RT-PCR of ATG11 transcripts was performed as described previously (Suttangkakul et al., 2011). The relative abundance of target transcripts was determined by the comparative threshold cycle method, using the levels of Arabidopsis UBC9 transcript as the internal control (Chung et al., 2009).
Plant Growth Conditions and Phenotypic Assays
Unless noted otherwise, seedlings were germinated on 0.7% (w/v) agar plates containing full-strength MS salts and 1% (w/v) Suc and then grown at 21°C under an LD (16-h-light/8-h-dark) photoperiod. After ∼2 weeks, plants were transplanted to soil and grown at 21°C under LD conditions. To assay natural senescence, plants were grown at 21°C under SD (8-h-light/16-h-dark) conditions for 10 weeks.
The effects of fixed-carbon starvation were determined according to Chung et al. (2010). Seedlings were grown in LD conditions for 2 weeks on MS solid medium without Suc. Plates were then wrapped with aluminum foil and placed in the dark for varying lengths of time before returning to LD conditions for 2 weeks. Seedling recovery was scored by obvious regreening and the appearance of new leaves. The effects of nitrogen starvation on seedling growth were determined according to Phillips et al. (2008). Briefly, 1-week-old seedlings grown under continuous light in liquid MS medium containing 1% Suc were transferred for an additional 2 weeks to either MS medium or nitrogen-deficient MS liquid medium supplemented with 1% Suc. For detached leaf senescence assays, the third and fourth leaves were excised from 2-week-old seedlings grown on solid agar medium (MS salts plus 1% Suc) and floated on 10 mM MES, pH 5.7, in the dark at 24°C for 7 d. Residual chlorophyll content was measured spectrophotometrically following extraction in 80% acetone (Chung et al., 2010).
IDL assays were performed as described previously (Weaver and Amasino, 2001) using seedlings grown for 4 weeks on soil at 21°C under LD conditions. The expanding third and fourth rosette leaves were individually covered with aluminum foil, whereas the rest of the plant remained exposed to LD light conditions.
Confocal Laser Scanning Microscopy and Image Analysis
Plants expressing GFP-ATG11 and mCherry-ATG11 under the control of the UBQ10 promoter were generated with the full-length ATG11 cDNA subcloned into the pCR8/GW/TOPO vector (Invitrogen), and then the LR Clonase reaction (Invitrogen) was used to place them in the pMDC99-UBQ10:GFP and pMDC99-UBQ10:mCherry destination vectors, respectively (Suttangkakul et al., 2011). The sequence-verified construction was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into the homozygous atg11-1 line by the floral dip method. Homozygous UBQ10:GFP-ATG11 atg11-1 and UBQ10:mCherry-ATG11 atg11-1 plants were identified in the T3 generation by hygromycin resistance, genomic PCR, and confocal fluorescence microscopy. The background of reporter UBQ10:GFP-ATG11 atg11-1 was combined with the various atg mutants by introgression.
Confocal fluorescence microscopy of stable transgenic lines expressing GFP-ATG8a, mCherry-ATG8a, GFP-ATG11, or YFP-ATG1a in various genetic backgrounds was performed according to Suttangkakul et al. (2011). Briefly, 6-d-old seedlings grown on MS agar medium containing 1% Suc were transferred for 24 h to either MS medium or nitrogen-deficient MS medium with 1% Suc. ConA (1 μM) or an equivalent volume of DMSO was added to the medium at the indicated times prior to examination. Cell images were obtained with a Zeiss 510-Meta laser scanning confocal microscope, using 488-nm light combined with the BP 500-530 IR filters for the GFP and YFP images, 543-nm light combined with BP 565-615 IR filters for the mCherry images, and 453-nm light combined with the BP 480-520 IR filters for the CFP images. YFP and mCherry signals were detected simultaneously in the multiple-track mode. Microscopy images were processed using the LSM510 image browser (http://www.zeiss.com/lsm) and converted to TIFF (for images) or AVI (for movies) formats.
Protein–protein interactions were assessed in planta by BiFC (Martin et al., 2009). cDNAs encoding full-length ATG11, ATG1a, ATG1a-AIM-mut, ATG13a, ATG101, or ATG8e and various truncated forms of ATG11 were appended in-frame via the LR Clonase reaction to the N-terminal half of EYFP (NY) in the pSITE-nEYFP-C1 vector (ABRC CD3-1648) or to the C-terminal half of EYFP (CY) in the pSITE-cEYFP-C1 vector (ABRC CD3-1649). For the Myc-ATG13a construction, the cDNA encoding full-length ATG13a was placed into pEarleyGate 203 (Earley et al., 2006) via the LR Clonase reaction. The sequence-verified CY and NY transgenes expressed under the control of the cauliflower mosaic virus 35S promoter were introduced into the Agrobacterium strain GV3101 and then used to infiltrate Nicotiana benthamiana leaves as described (Martin et al., 2009) or to infiltrate transgenic mCherry-ATG8a Arabidopsis plants as described (Marion et al., 2008). Leaf regions were examined ∼36 to 48 h after infiltration by confocal fluorescence microscopy.
Mitophagy was observed by confocal fluorescence microscopic imaging of lines expressing the Mito-CFP, Mito-YFP, and CIB22-GFP reporters under the control of the 35S promoter (Nelson et al., 2007; Han et al., 2010). For analyses involving MitoTracker Green FM (Molecular Probes), isolated protoplasts were incubated with 1 μM of the stain for 30 min at 25°C. The protoplasts were then washed twice and resuspended with MS liquid medium supplemented with 0.4 M mannitol and 1 μM ConA for 24 h before confocal fluorescence microscopy. MitoTracker Green FM was excited at 488 nm and detected using BP 480-520 IR filters (Colcombet et al., 2013). Mitochondrial numbers and sizes in root cells expressing Mito-CFP were analyzed from the photographs using ImageJ (http://rsb.info.nih.gov/ij/) as described previously (Zhang and Hu, 2009). Images (50 μm × 50 μm) were converted to eight-bit grayscale and then counted for mitochondria either manually or by the Particle Analyzer function of ImageJ. At least eight images were used for each data point, with statistical significance provided by Student’s t test (P < 0.05).
Immunoblot Analyses and Lipidation and Phosphorylation Assays
The detection of free ATG8 and the ATG8-PE adduct was performed according to Chung et al. (2010). The total membrane fraction was collected from crude protein extracts by a 10-min centrifugation at 100,000g. The pellet was solubilized in Triton X-100 and clarified, and the resulting supernatant was incubated for 1 h at 37°C without or with the addition of Streptomyces chromofuscus phospholipase D (Enzo Life Sciences). Samples were subjected to SDS-PAGE in the presence of 6 M urea and immunoblotted with anti-ATG8a antibodies. Phosphate bound to ATG1a was assayed by incubating crude extracts with 4 units/μL λ protein phosphatase (New England Biolabs) for 1 h followed by SDS-PAGE as described previously (Suttangkakul et al., 2011). Immunoblot analyses employed antibodies against ATG5 and ATG8a (Thompson et al., 2005; Phillips et al., 2008), ATG1a and ATG13a (Suttangkakul et al., 2011), and PBA1 (Smalle et al., 2002). Anti-VDAC antibodies were provided by Tom Elthon. GFP and COX II were detected with antibodies obtained from Roche Applied Science (11814460001) and Agrisera (AS04 053A), respectively. Antibodies against histone H3 (ab1791) and CFP/YFP (ab290) were obtained from Abcam.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers listed in Supplemental Tables 4 and 5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of the ATG11 Domains within ATG11/FIP200-Related Proteins.
Supplemental Figure 2. Amino Acid Sequence Alignment of the ATG17 and ATG17-Like Domains within ATG11/ATG17-Related Proteins.
Supplemental Figure 3. Phylogenetic Analysis of ATG11 Domain–Containing Proteins.
Supplemental Figure 4. Phylogenetic Analysis of ATG17 Domain–Containing Proteins.
Supplemental Figure 5. BiFC Controls Used to Examine the in Planta Interactions of ATG11 or ATG101 with Other Components of the ATG1/13 Kinase Complex.
Supplemental Figure 6. Y2H and BiFC Controls Used to Map the ATG11 Dimerization Region.
Supplemental Figure 7. BiFC Mapping of the Interaction Site(s) between ATG11 and ATG13a, ATG101, or ATG8e.
Supplemental Figure 8. Phylogenetic Analysis of ATG101 Proteins.
Supplemental Figure 9. Amino Acid Sequence Alignment of ATG101 Orthologs.
Supplemental Figure 10. Expression of ATG11, ATG101, ATG1c, and ATG13a in Arabidopsis Tissues at Various Developmental Stages.
Supplemental Figure 11. Truncated Transcripts Possibly Generated 5′ and 3′ to the T-DNA Insertion Site in atg11-1 Do Not Generate Interaction-Competent ATG11 Proteins Even if Expressed.
Supplemental Figure 12. Association of the Mitochondrial Markers Mito-YFP and CIB22-GFP with Autophagic Bodies during IDL-Induced Senescence.
Supplemental Tables 1 and 2. Coexpression of ATG11 with Genes Encoding Other Components of the ATG8-Mediated Autophagic Pathway and with Other Arabidopsis Genes.
Supplemental Table 3. Oligonucleotide Primers Used in This Study.
Supplemental Table 4. Species Names, Data Sources, and Domain Positions for the Various ATG11, ATG17, and FIP200 Proteins Used for the Phylogenetic Analysis.
Supplemental Table 5. Species Names and Data Sources for ATG101 Proteins Used for the Phylogenetic Analysis.
Supplemental Movie 1. Colocalization of the Autophagy Marker mCherry-ATG8 with ATG11.
Supplemental Movie 2. Distribution of Mito-YFP–Labeled Mitochondria in Leaf Epidermal Cells.
Supplemental Movie 3. Distribution of Mito-YFP–Labeled Mitochondria in Epidermal Cells from IDLs following ConA Treatment.
Supplemental Movie 4. Mito-YFP–Labeled Mitochondria in Mesophyll Cells from IDLs following ConA Treatment.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 3.
Supplemental Data Set 2. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 4.
Supplemental Data Set 3. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 8.
Supplementary Material
Acknowledgments
We thank Wisconsin colleagues Joseph M. Walker, Yinzhou Zhu, Richard S. Marshall, Andrew F. Bent, Yangrong Cao, Junqi Song, and Colleen M. McMichael for technical help and reagents and Marisa S. Otegui and Sarah J. Swanson for advice on confocal microscopy imaging. We thank Li-Jia Qu (Peking University) for providing the CIB22-GFP seeds and Tom Elthon (University of Nebraska, Lincoln) for providing the anti-VDAC antibody. This work was supported by the USDA-National Institute of Food and Agriculture (Grant 2008-02545 to R.D.V.) and the National Science Foundation Arabidopsis 2010 (Grant MCB-0929100 to R.D.V.) and Plant Genome (Grant IOS-1339325 to R.D.V.) research programs.
AUTHOR CONTRIBUTIONS
F.L. designed and performed the bulk of the research, analyzed the data, and wrote the article. T.C. contributed genetic materials and cell biological reporters. R.D.V. contributed to experimental design and data analysis and wrote the article.
Glossary
- PAS
phagophore assembly site
- PE
phosphatidylethanolamine
- AIM
ATG8 interaction motif
- Col-0
Columbia-0
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- LD
long-day
- SD
short-day
- MS
Murashige and Skoog
- ConA
concanamycin A
- IDL
individually darkened leaf
- VDAC
voltage-dependent anion channel
Footnotes
Online version contains Web-only data.
References
- Alemu E.A., Lamark T., Torgersen K.M., Birgisdottir A.B., Larsen K.B., Jain A., Olsvik H., Øvervatn A., Kirkin V., Johansen T. (2012). ATG8 family proteins act as scaffolds for assembly of the ULK complex: Sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287: 39275–39290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. (2011). Phosphorylation of serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. (2010). Network organization of the human autophagy system. Nature 466: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Klionsky D.J. (2011). The regulation of autophagy: Unanswered questions. J. Cell Sci. 124: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. (2003). Multiple sequence alignment with the CLUSTAL series of programs. Nucleic Acids Res. 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. (2008). The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., Berthomé R., Lurin C. (2013). Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 10: 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., Lam H., Wang J., Ding X., Chen S., Jiang X. (2009). ULK1-ATG13-FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284: 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Qin G., Kang D., Chen Z., Gu H., Qu L.J. (2010). A nuclear-encoded mitochondrial gene AtCIB22 is essential for plant development in Arabidopsis. J. Genet. Genomics 37: 667–683. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576. [DOI] [PubMed] [Google Scholar]
- Herman E.M., Larkins B.A. (1999). Protein storage bodies and vacuoles. Plant Cell 11: 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig M.S., Contento A.L., Meyer A., Ebany D., Bassham D.C., Macintosh G.C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 108: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. (2009). Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979. [DOI] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798. [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Koyama-Honda I., Mizushima N. (2012). Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125: 1488–1499. [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.P., Bligny R., Douce R. (1986). Biochemical changes during sucrose deprivation in higher plant cells. J. Biol. Chem. 261: 3193–3199. [PubMed] [Google Scholar]
- Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20: 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. (2005). Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16: 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. (2009). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O., Pesquet E., Ahad A., Askne A., Nordvall D., Vodnala S.M., Tuominen H., Hurry V., Dizengremel P., Gardeström P. (2007). The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ. 30: 1523–1534. [DOI] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H.N., Kim S.H., Shin K.D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., et al. (2012). Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31: 3691–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830. [DOI] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Liang C.C., Wang C., Peng X., Gan B., Guan J.L. (2010). Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285: 3499–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Yang P., Huang X., Zhang H., Lu Q., Zhang H. (2013). The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J. Cell Biol. 201: 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Wang K., Liu X., Klionsky D.J. (2013). The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell 26: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179. [DOI] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- Meijer W.H., van der Klei I.J., Veenhuis M., Kiel J.A. (2007). ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3: 106–116. [DOI] [PubMed] [Google Scholar]
- Mercer C.A., Kaliappan A., Dennis P.B. (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5: 649–662. [DOI] [PubMed] [Google Scholar]
- Mizushima N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22: 132–139. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ohbayashi S., Sakoh-Nakatogawa M., Kakuta S., Suzuki S.W., Kirisako H., Kondo-Kakuta C., Noda N.N., Yamamoto H., Ohsumi Y. (2012). The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 287: 28503–28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8 family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385. [DOI] [PubMed] [Google Scholar]
- Novak I., et al. (2010). Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11: 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17: 87–97. [DOI] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., Johansen T. (2010). FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188: 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M.J., Stanley R.E., Hurley J.H. (2012). Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 151: 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. (2013). Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 194: 341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Chung T., Holding D., Jung R., Vierstra R., Otegui M.S. (2011). Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 23: 769–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, I., and Logan, D.C. (2010). Mitochondrial dynamics. In Plant Mitochondria, F. Kempken, ed (Berlin: Springer), pp. 31–63. [Google Scholar]
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., Hayashi M., Sakamoto W., Ohsumi Y., Nishimura M. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279: 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Babiychuk E., Kushnir S., Durski A., Vierstra R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218. [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L.M., Amasino R.M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127: 876–886. [PMC free article] [PubMed] [Google Scholar]
- Wong P.M., Puente C., Ganley I.G., Jiang X. (2013). The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy 9: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546. [DOI] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J. (2005). Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16: 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.